
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 30 January 2023
Sec. Computational Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1124638
This article is part of the Research TopicGenetic and non-genetic Determinants of InflammationView all 5 articles
Background: The purpose of this Mendelian randomization (MR) study was to assess the causal relationship between circulating cytokines and periodontitis.
Materials and methods: Based on the aggregated statistics of the largest publicly available genome-wide association study (GWAS), we applied a bidirectional two-sample MR. MR analyses were conducted using Inverse variance weighted (IVW), Robust Adjusted Profile Score (RAPS), Maximum likelihood (ML), Weighted median and MR-Egger, and results obtained from IVW served as the primary outcome. Cochran Q test was used to test the heterogeneity. MR-Egger intercept test and MR polymorphism residual and outlier test (MR-PRESSO) were used for polymorphism analysis. Leave-one-out sensitivity and funnel plots were used for sensitivity analysis.
Results: The IVW method indicated that interleukin 9 (IL9) had a positive causal relationship with periodontitis [odds ratio (OR) = 1.199, 95% confidence interval (CI) = 1.049–1.372, p = 0.008], and interleukin 17 (IL17) had a negative causal relationship with periodontitis (OR = 0.847, 95% CI = 0.735–0.976, p = 0.022). In bidirectional MR, periodontitis was not causally related to any of the cytokines in our study.
Conclusion: Our findings provided evidence in support of potential causal associations between circulating IL9/IL17 and periodontitis.
Periodontitis is the sixth most prevalent disease, and severe periodontitis affecting 10.8% of the world’s population (Peres et al., 2019). Major characteristics of periodontitis include destruction of periodontal soft tissues and resorption of alveolar bone, which eventually lead to tooth loss (Hienz et al., 2015). Disruption of oral microbial homeostasis is a major cause for periodontitis (Valm, 2019). In addition, genetic variance, dietary habits, and other diseases also contribute to the development of periodontitis. Identifications of risk factors at the early stage of periodontitis and taking effective intervention strategies accordingly may help to reduce periodontitis-associated tooth loss.
Cytokines play a key role in the development of periodontitis and influence the progression of periodontitis, and are risk factors. Under pathological stimuli, the periodontium stages immune responses that are characterized by accumulation of immune cells and production of cytokines (Cekici et al., 2014). In barrier tissues such as intestine, oral cavity, and skin, cytokines in response to pathological stimuli can become a bridge between parenchymal and non-parenchymal cells, producing an immediate immune response (Kwiecien et al., 2019). At the same time, tissue-resident immune cells and migrating immune cells secrete different types of cytokines in a large amount to participate in the immune response. The cytokines, in turn, exert regulatory effects on each other and are critical for the progression of inflammation. Due to continuous stimulation and damage to the periodontal tissue caused by the pathogenic oral flora and mastication, immune cells such as macrophages and T cells are recruited, and cytokines such as interleukin (IL) 1, IL6 and tumor necrosis factor (TNF) are released in large quantities (Pan et al., 2019). These cytokines can activate the corresponding signaling pathways and further aggravate inflammatory response in periodontal tissue. For example, IL1 activates the nuclear factor kappa-B ligand (RANKL) pathway to promote osteoclast formation, resulting in resorption and destruction of alveolar bone (Huynh et al., 2017).
Previous studies have demonstrated important roles of cytokines and related pathways in the progression of periodontitis. For example, TNF-α inhibits osteoblast differentiation through inhibition of insulin-like growth factor-1 (IGF1), osterix and the runt-related transcription factor-2 (Runx2) (Osta et al., 2014). In vitro experiments have shown that IL17 can regulate autophagy and osteoclastogenesis through Beclin1/TAB3/p-ERK pathway (Zhong et al., 2022). However, the findings of these studies were conducted in observational studies that assumed no confounding effects, and therefore could be limited by selection bias, other related diseases, and heterogeneity of comparison groups (Andrews et al., 2020).
Several studies have investigated the relationship between cytokines and periodontitis using meta-analysis. For example, a meta-analysis has included 21 case-control studies to explore the association between IL6 polymorphism and periodontitis susceptibility (Zhu et al., 2016). Another meta-analysis has included 53 studies to investigate the role of multiple cytokine gene polymorphisms in the pathogenesis of periodontitis (Nikolopoulos et al., 2008). However, significant heterogeneity and potential publication bias may occur when analyzing genetic polymorphism of various cytokines from different databases (Liu and Li, 2021). Therefore, evidence for a causal link between cytokines and periodontitis is still lacking.
To assess the causal relationship between exposure and outcome, Mendelian randomization (MR) uses genetic variation (single nucleotide polymorphisms, SNPs) as instrumental variables (IV). At conception, genetic variants are randomly assigned, similar to randomized controlled trials, reducing the effect of confounding factors. Genetic variation was only related to exposure, not outcome, and genetic variation affected outcome only through exposure, not through other pathways (Lawlor et al., 2008). Recently, MR methods have been applied to investigate the genetic relationship between cytokines and the danger of cancer (Bouras et al., 2022), and the causal association between cytokines and risk of multiple sclerosis has also identified by the MR approach (Lu et al., 2021).
In this study, we investigated the genetic causal association between 41 cytokines and periodontitis by applying two sample bidirectional MR, aimed to provide a new perspective on the prevention, diagnosis and treatment of periodontitis.
This bidirectional MR study was undertaken following a framework as delineated in Figure 1. The MR approach was based on three assumptions: 1) genetic variation used for IV was associated with exposure; 2) genetic variation was independent of confounding factors; 3) genetic variation affected the outcome only through the exposure and not through other pathways (Lawlor et al., 2008).
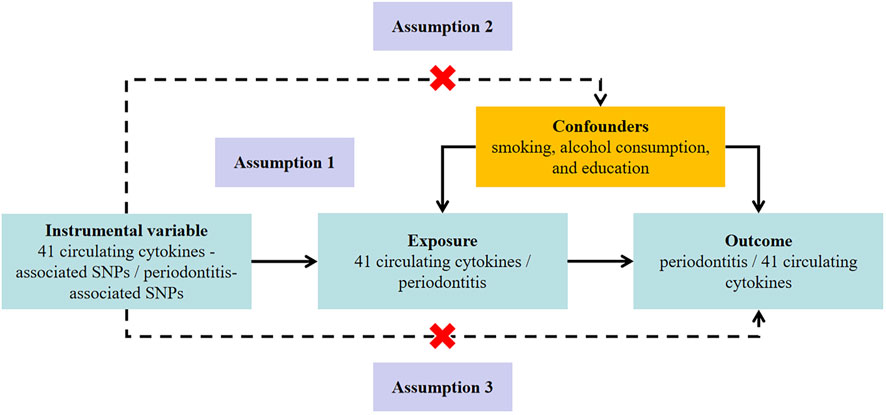
FIGURE 1. Study design to investigate the association between circulating cytokines and risk of periodontitis basing on bidirectional Mendelian randomization assumptions. The MR study must satisfy three assumptions. Assumption 1: genetic variation used for instrumental variable is associated with exposure; Assumption 2: genetic variation is independent of confounding factors; Assumption 3: genetic variation affects the outcome only through the exposure and not through other pathways. MR, Mendelian randomization; SNP, single-nucleotide polymorphism.
The summary level datasets of cytokines were obtained from the most recent GWAS, which included 8,293 Finnish individuals from the Cardiovascular Risk in Young Finns Study (YFS) and the FINRISK studies (including “FINRISK 2002” and “FINRISK 1997”) (Ahola-Olli et al., 2017). The cytokine quantification was performed from EDTA treated plasma in FINRISK 1997, heparinized plasma in FINRISK 2002, and sera in YFS. The genetic associations were adjusted for age, sex, body mass index and the first 10 genetic principal components with genomic-control correction. In YFS and FINRISK 2002, a total of 48 cytokines were measured using Bio-Rad’s premixed Bio-Plex Pro Human Cytokine 27-plex Assay and 21-plex Assay, and Bio-Plex 200 reader with Bio-Plex 6.0 software. Only measures within the cytokine-specific detection range were included in the analyses. Cytokines with >90% of values missing were excluded (7 out of 48). In FINRISK 1997, a total of 17 cytokines overlapped with those measured in FINRISK2002 and YFS and were thus included in the GWAS (Ahola-Olli et al., 2017).
Summary statistics for periodontitis were obtained from the latest meta-analysis of the largest sample size to date for the GWAS (GLIDE) consortium of dental endpoints involved 17,353 clinically diagnosed cases and 28,210 controls (Shungin et al., 2019). After excluding the Hispanic Community Health Study/Latino Study (HCHS/SOL) in the GLIDE consortium, there were 12,289 cases and 22,326 controls of European ancestry. Clinical diagnostic criteria from the Centers for Disease Control and Prevention/American Academy of Periodontology and self-reported diagnoses from the Brigham and Women’s Hospital Women’s Health Study were primarily used, while additional inclusion criteria were defined as one of the following: periodontal probing depth ≥5 mm on two or more tooth surfaces, or ≥4 mm on four or more tooth surfaces, ≥5.5 mm on two or more tooth surfaces or “gum surgery” from dental records (Shungin et al., 2019).
To ensure the veracity and accuracy of the conclusions regarding the causal relationship between cytokines and periodontitis risk, the following quality control steps were used to select the best genetic tool. First, a set of SNPs below the genome-wide threshold of statistical significance (5 × 10−8) was used as IV, since only 26 SNPs for cytokines reached genome-wide significance. To obtain more comprehensive results, we also selected another set of SNPs below the genome-wide significance level (5 × 10−6) as IVs. Second, the PLINK clustering method (R2 < 0.001, clustering distance = 10,000 kb) was used to assess the linkage disequilibrium (LD) among the included SNPs. Third, SNPs for cervical cancer will not be included in the instrumental variables. Fourth, the F-statistic = R2 × (N-2)/(1-R2) was calculated for each IV used for cytokines as a measure of instrumental strength, where R2 is the phenotypic variation explained by each genetic variant in the exposure and N is the sample size (Palmer et al., 2012). Generally, F-statistics >10 was set as the threshold of strong IV (Burgess et al., 2017). Finally, when exposure-related SNPs were not present in the resultant GWAS, proxy SNPs significantly associated with the variant of interest were selected (r2 > 0.8). Ultimately in our analysis, 8 SNPs (p < 5 × 10−6) were used as IVs in periodontitis. 1 to 6 SNPs (p < 5 × 10−8) and 4 to 16 SNPs (p < 5 × 10−6) were used as IVs in cytokines. The detailed information on the IVs was displayed in Supplementary Tables S1–S3.
To evaluate the genetic causal effects, several different methods were applied, which provided valid evidence under different conditions, with Inverse variance weighted (IVW) serving as the primary outcome (Burgess et al., 2017). In order to aggregate the Wald values for individual SNP and generate an estimated total value of the impact, a meta-analysis methodology was used in the IVW model. In the absence of horizontal pleiotropy and heterogeneity, IVW linear regression could provide an unbiased causal estimate (Burgess et al., 2017). For traits with only one SNP, the Wald ratio test was applied. By assuming that exposure and result have a linear relationship and that the estimates of the genetic association have a normal bivariate distribution, the Maximum likelihood method (ML) generates estimated causal associations (Milligan, 2003). MR-Egger can offer a reliable, unbiased estimate even if all of the SNPs in the selection are invalid (Burgess and Thompson, 2017). Although as much as 50% of the data used in the study comes from variations of interest that are invalid IVs, the weighted median can still produce reliable estimates of the causal effects (Bowden et al., 2016a). When the causal estimates of most instrumental variables are similar, the weighted model approach remains valid even if other instrumental variables do not qualify for the causal inference of the MR technique (Hartwig et al., 2017). The Robust Adjusted Profile Score (MR-RAPS) is statistically robust to weak instruments and robust in the presence of systematic pleiotropy. MR-RAPS can lessen horizontal pleiotropy by taking into account the measurement error in the association of SNPs and exposure (Zhao et al., 2019).
The Cochran Q statistic and I2 statistic were used to test the heterogeneity. Random-effect IVW is employed if there is heterogeneity among the SNPs included in each analysis (Bowden et al., 2016b). To determine if a specific genetic locus had an impact on random estimates, the leave-one-out sensitivity method was used. Additionally, scatterplots and funnel plots were created for further analysis. The requirement that exposure-related SNPs exclusively affect the outcome through themselves is crucial for the prerequisite of the MR method. To determine whether IV increases the degree of polymorphic impact of the results through more than one biological pathway, we tested for polymorphism using MR-Egger regression and Mendelian randomized polymorphism RESidual Sum and Outlier (MR-PRESSO) (Verbanck et al., 2018). Furthermore, we searched all SNPs in the PhenoScanner database (Kamat et al., 2019) to evaluate whether those SNPs were significantly associated (p < 5 × 10−8) with periodontal disease risk factors, including smoking, alcohol consumption, and education. The causal associations were re-estimated after removing pleiotropic SNPs in sensitivity analyses to avoid potential pleiotropic effects.
The threshold for statistical significance for the association between cytokines and periodontitis was set to 1.2 × 10−3 (p = 0.05/41) using a Bonferroni-adjusted p-value. If p < 0.05 but above the Bonferroni-adjusted significance threshold, this was considered suggestive of evidence for a potential association. All the analyses and relevant figures were made by R 4.1.2 (R Foundation for Statistical Computing, Vienna, Austria), using the “TwoSampleMR (0.5.6)”, “MR-PRESSO (1.0)”, “mr.raps (0.2)”, and “ggforestplot (0.1.0)” packages. Reporting follows the STROBE-MR statement (Skrivankova et al., 2021). The study protocol was not pre-registered. Power calculations were performed in mRnd, a Web-based application (https://shiny.cnsgenomics.com/mRnd/) (Brion et al., 2013) assuming a 5% type I error rate. Statistical significance was set to a two-sided α of 0.05.
Only 26 among the 41 cytokines have independent genome-wide significant SNP when the cut-off is set to p < 5 × 10−8, whereas all 41 have independent SNPs when using a higher cut-off (p < 5 × 10−6) (Supplementary Figure S1, Figure ). At genome-wide significance, 1 to 6 independent genome-wide significant SNPs are used for cytokines explained 0.36%–19.48% of the variance in cytokines, corresponding to F-statistic of 29.98–1731.64 (Supplementary Table S1). In the higher cut-off value (p < 5 × 10−6), 4 to 16 SNPs are used as IVs in cytokines, and 8 SNPs are used as IVs in periodontitis. The genetic variance they explained are 0.86%–22.99% and 0.53%, corresponding to F-statistic of 71.44–2023.73 and 44.23 respectively (Supplementary Table S2, Supplementary Table S3). In the PhenoScanner search, we find that both rs3748034 and rs7088799 are associated with alcohol consumption and education, so we re-analyze after eliminating these SNPs (Supplementary Table S4). In the periodontitis GWAS the targeted SNPs are not found, rs138987090 and rs78422482 were replaced by proxy SNP rs149524246 and rs115349882 with R2 > 0.8 respectively. Details about identified SNPs are presented in Supplementary Tables S1–S3.
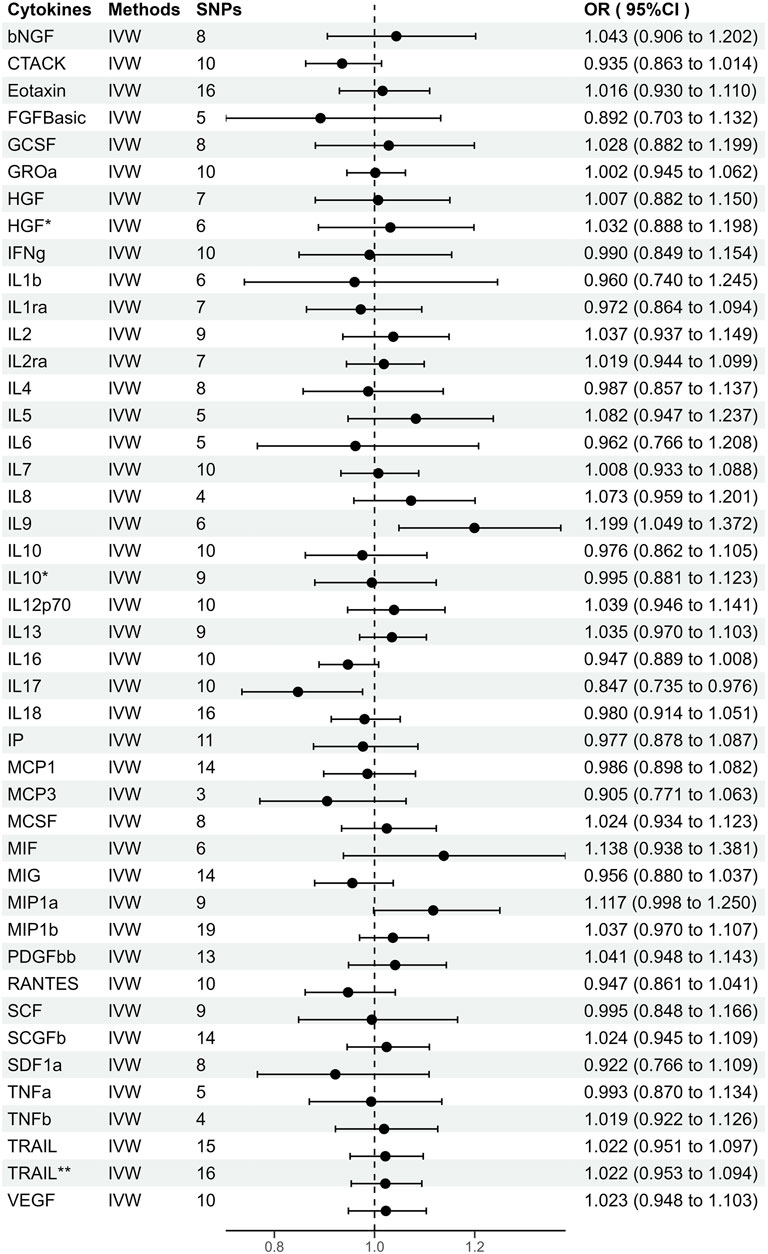
FIGURE 2. Odds ratios (95% Confidence interval) from Mendelian randomization analysis showing associations between genetically predicted cytokine levels (cut-off = p < 5 × 10−6) and risks for periodontitis. The causal association between circulating cytokine levels and periodontitis was mainly calculated through two-sample MR analysis using IVW method. Estimates with 95% CI are expressed as odds ratios for periodontitis per 1-SD increase of genetically predicted circulating cytokine levels. *, Results of MR analysis after exclusion of SNPs associated with confounding factors. **, Results of MR analysis after using a proxy SNP.
A priori statistical power is calculated (Brion et al., 2013). Given α = 5%, we had ≥80% power for the primary analysis when the expected odds ratios (ORs) for periodontitis are ≥1.38 or ≤0.71 for 41 circulating cytokines. Detailed information on the power calculation is included in Supplementary Table S5.
As shown in Supplementary Figure S1, genetically predicted circulating cytokines are not significantly associated with periodontitis. When using SNPs obtained with the higher cut-off (p < 5 × 10−6), the MR study reveals that genetically predicted circulating IL9 is potentially associated with periodontitis (OR = 1.199, 95%CI: 1.049–1.372 p = 0.008) based on the IVW method (Table 1). The Maximum likelihood, MR-RAPS, and MR-PRESSO method also provide similar results. Furthermore, based on the IVW method, the MR analysis show that there is a potential negative correlation between genetically predicted circulating IL17 and Periodontitis (OR = 0.847, 95%CI: 0.735–0.976, p = 0.022) (Table 1). The similar results are also obtained in the Maximum likelihood, the weighted median, and MR-PRESSO methods. The results are shown in Table 1 and Figure 2. Cochran Q statistic and I2 statistic indicate low heterogeneity and more reliability of these SNPs (all p > 0.05) (Table 1). The MR-Egger intercept suggests that stem cell factor (SCF) predicted by SNPs (p < 5 × 10−6) might be pleiotropic SNPs, but MR-PRESSO global test does not identify any pleiotropic SNPs. Beyond that, there is no evidence of horizontal pleiotropy for SNPs. Detailed information about the results is included in Supplementary Tables S6–S7. The estimated effect sizes for SNPs of cytokines on periodontitis are displayed in scatter plot (Figure 3). The funnel plots show general symmetry, suggesting little evidence of heterogeneity (Supplementary Figure S2). Leave-one-out sensitivity analyses suggest that there is no individual SNP with a strong influence on the overall effect (Supplementary Figure S3).
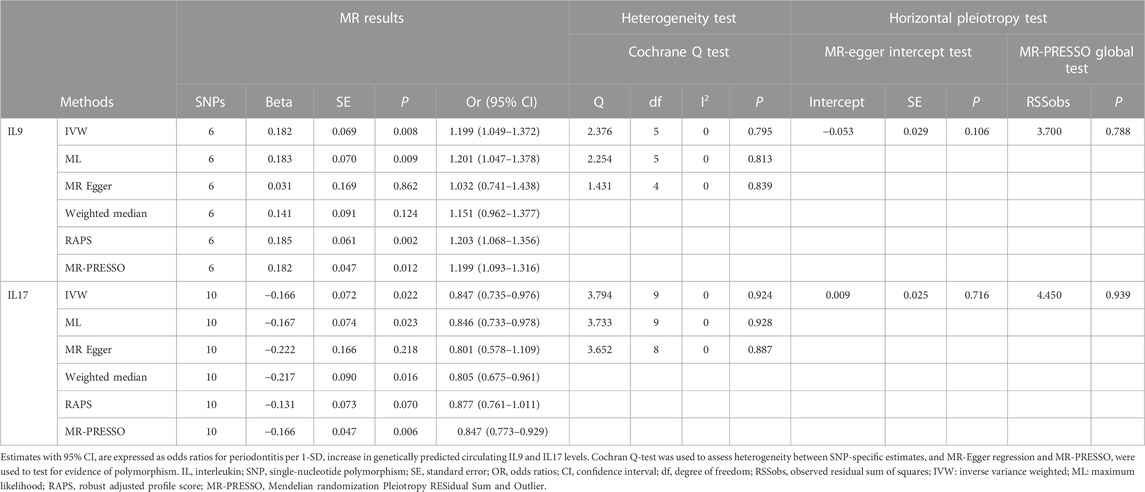
TABLE 1. Estimation of associations between circulating IL9, IL17 and risk of periodontitis using Mendelian randomization analysis.
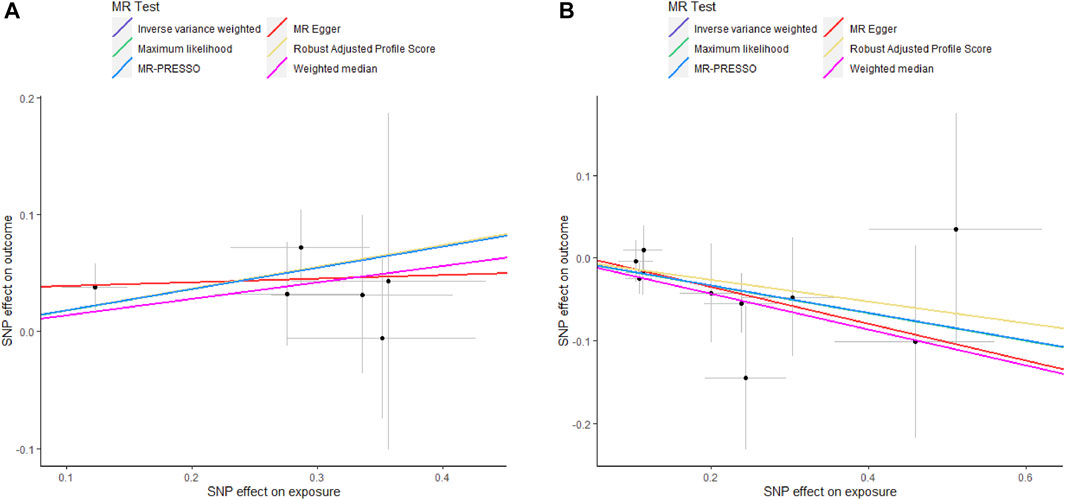
FIGURE 3. Scatter plots of potential effects of SNPs on IL9 and IL17 versus periodontitis. A, Scatter plot of potential effects of SNPs on IL9 versus periodontitis. B, Scatter plot of potential effects of SNPs on IL17 versus periodontitis. Scatter plots presented per-allele association with outcome risk plotted against the per-allele association with one standard deviation of exposure (with vertical and horizontal gray lines showing the 95% CI for each SNP). Analyses were conducted using IVW, Weighted median, Wald ratio, RAPS, MR Egger, MR-PRESSO, and ML. The slope of each line corresponding to the estimated MR effect per method. SNP, single-nucleotide polymorphism; IVW, Inverse variance weighted; ML, Maximum likelihood; MR-PRESSO, Mendelian Randomization Pleiotropy RESidual Sum and Outlier; RAPS, Robust Adjusted Profile Score.
As shown in Figure 4, genetically predicted periodontitis is not significantly associated with any of the 41 circulating cytokines based on any method. MR-PRESSO and the MR-Egger intercept do not identify any pleiotropic SNP. Except for IL1b as outcome, Cochran Q statistic and I2 statistic indicate low heterogeneity and more reliability of these SNPs (all p > 0.05). Since the presence of heterogeneity, we adopt the results of the random-effect IVW effects model, which does not identify causal relationship between genetically predicted periodontitis and IL1b. Detailed information about the results is included in Supplementary Table S1.

FIGURE 4. Beta (±1.96*Standard error) from Mendelian randomization analysis showing associations between genetically predicted periodontitis (Cut-off = p < 5 × 10−6) and risks for cytokine levels. The causal association between periodontitis and circulating cytokine levels was mainly calculated through two-sample MR analysis using IVW method. *, Results of MR analysis after exclusion of SNPs associated with confounding factors. **, Results of MR analysis after using a proxy SNP.
In this study, we use MR analysis to investigate the potential causal relationship between the genetic basis of circulating levels of 41 cytokines and the risk of periodontitis. We find that genetic proxies of circulating concentration of IL9 are positively associated with the risk of periodontitis, and those of IL17 are negatively with the risk of periodontitis.
IL9 exhibits pro-inflammatory activities in a range of diseases, but its role in periodontitis is unclear. Numerous studies have indicated the involvement of IL9 in the pathogenesis of allergic diseases. In addition, IL9 promotes tumorigenesis through autocrine and anti-apoptotic activity (Feng et al., 2011). A recent study has demonstrated that IL9 plays a central role in osteoclast genesis by enhancing M-CSF/sRANKL-mediated osteoclast formation and function (Kar et al., 2021). Our study reveals a genetic link between IL9 and periodontitis. Using MR approach, we show that elevated circulating concentrations of IL9 predispose individuals to the development of periodontitis. A previous meta-analysis assessing genetic polymorphisms of IL9 in Chinese non-smokers has suggested that IL9 does not have a causal relationship with periodontitis (Zheng et al., 2020). We use MR to analyze the latest GWAS meta-analysis of the GLIDE Consortium, the largest sample size to date involving 17,353 clinically diagnosed cases and 28,210 controls (Shungin et al., 2019). Consistent with our results, a positive association between IL9 and the severity of periodontal disease has been established by assessing IL9 levels in gingival sulcus fluid samples and biopsies (Díaz-Zúñiga et al., 2017). Although IL9 has been understudied in the field of periodontitis, our study provides a genetic basis for a causal relationship between IL9 and periodontitis. Future studies are warranted to understand the regulation of IL9 in periodontitis in more details, to identify relevant target cells of IL9, and to delineate the mechanisms that mediate the effects of IL9 in periodontitis.
Another interesting finding of our study is an inverse association between genetically proxied circulating IL17 concentrations and periodontitis using the MR approach. IL17 is a widely reported pro-inflammatory cytokine involved in the pathogenesis inflammatory diseases and autoimmune diseases (McGeachy et al., 2019). IL17 also induces an increase in osteoclasts not only by stimulating the secretion of IL6 and RANKL, but also by altering the microbial composition of the oral cavity, increasing levels of Enterobacteriaceae, Enterococcus and Staphylococcus, which can aggravate periodontitis (Xiao et al., 2017). Our findings were in the opposite direction of the expected association. Supporting our results, deficiency of IL17 receptor A has been demonstrated to accelerate bone loss decreased neutrophil accumulation to the periodontal lesions in an experimental periodontitis model, indicating a protective effect of IL17 (Yu et al., 2007). Moreover, IL17 receptor A signaling plays a protective role in periapical bone destruction stimulated by infection (AlShwaimi et al., 2013). Using meta-analysis, polymorphisms of IL17F gene rs763780 and IL17A gene rs2275913 have recently been detected in patients with periodontitis, but there was no significant association between IL17 and periodontitis (Mazurek-Mochol et al., 2021). It is clear that IL17 has pleiotropic functions in the immune system (McGeachy et al., 2019), likely contributing to its different roles in different diseases. More studies are needed to precisely understand the mechanism of IL17 in periodontitis.
The main strengths of this study included the following. First, we used several MR techniques, including bidirectional MR to detect direction of association, PhenoScanner to exclude confounders, and sensitivity analyses to enhance the robustness of the results. Second, our study included a total of 41 cytokines from multiple classes, which permitted an examination of the wider inflammatory network in periodontitis. Third, our GWASs of exposure and outcome were the largest sample size to date, which added statistical power, and to minimize the risk of bias due to sample overlap, the GWAS of exposure and result came from entirely different consortia.
However, this study has potential limitations. First, we used a higher p < 5 × 10−6 cut-off for obtaining relatively abundant SNPs predicting the circulating cytokines. This could increase the risk of weak instrument bias for individual genetic variants, although our F-statistics suggested that these instruments had appropriate strength. Second, the potential synergistic effects between the studied cytokine networks were not taken into account. Third, our findings did not meet the strict multiple test threshold. It is necessary to test these findings with larger GWAS. Future MR studies using single- or multi-trait GWAS with larger number of cytokines may be able to address some of these issues.
As inflammatory regulators, cytokines are important risk factors for inflammatory diseases, and they can be exploited in GWAS to elucidate biochemical pathways underlying the link from locus to disease susceptibility. In our study, IL9 and IL17 were found to be genetically associated with periodontitis. These genetic associations may contribute to the understanding of pathological mechanisms, and provide innovative approaches for risk assessment, outcome prediction, and treatment for periodontitis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Conceptualization, S-JH and RL; formal analysis, S-JH, RL, SX, YL; methodology, S-JH, RL; writing—original draft preparation, S-JH, RL; writing—review and editing, S-JH, RL; supervision, S-JH, RL, S-ZD, and S-HL. All authors have read and agreed to the published version of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (81991503, 81991500, 81921002), the Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZDCX20212,500), and Clinical Research Plan of SHDC (SHDC2020CR5015).
We are grateful to the participants in the YFS, FINRISK, GLIDE, and other consortium or study and to all the researchers who worked on the data collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1124638/full#supplementary-material
Ahola-Olli, A. V., Würtz, P., Havulinna, A. S., Aalto, K., Pitkänen, N., Lehtimäki, T., et al. (2017). Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am. J. Hum. Genet. 100 (1), 40–50. eng. Epub 2016/12/19Cited in: Pubmed; PMID 27989323. doi:10.1016/j.ajhg.2016.11.007
AlShwaimi, E., Berggreen, E., Furusho, H., Rossall, J. C., Dobeck, J., Yoganathan, S., et al. (2013). IL-17 receptor A signaling is protective in infection-stimulated periapical bone destruction. J. Immunol. 191 (4), 1785–1791. eng. Epub 2013/07/19Cited in: Pubmed; PMID 23863904. doi:10.4049/jimmunol.1202194
Andrews, S. J., Goate, A., and Anstey, K. J. (2020). Association between alcohol consumption and alzheimer's disease: A mendelian randomization study. Alzheimers Dement. 16 (2), 345–353. eng. Epub 2019/12/02Cited in: Pubmed; PMID 31786126. doi:10.1016/j.jalz.2019.09.086
Bouras, E., Karhunen, V., Gill, D., Huang, J., Haycock, P. C., Gunter, M. J., et al. (2022) Circulating inflammatory cytokines and risk of five cancers: A mendelian randomization analysis. BMC Med. 20;3. Cited in: Pubmed; PMID 35012533. doi:10.1186/s12916-021-02193-0
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40 (4), 304–314. eng. Epub 2016/04/12Cited in: Pubmed; PMID 27061298. doi:10.1002/gepi.21965
Bowden, J., Del Greco, M. F., Minelli, C., Davey Smith, G., Sheehan, N. A., and Thompson, J. R. (2016). Assessing the suitability of summary data for two-sample mendelian randomization analyses using MR-egger regression: The role of the I2 statistic. Int. J. Epidemiol. 45 (6), 1961–1974. eng. Epub 2016/09/13Cited in: Pubmed; PMID 27616674. doi:10.1093/ije/dyw220
Brion, M. J., Shakhbazov, K., and Visscher, P. M. (2013). Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 42 (5), 1497–1501. eng. Epub 2013/10/26Cited in: Pubmed; PMID 24159078. doi:10.1093/ije/dyt179
Burgess, S., Small, D. S., and Thompson, S. G. (2017). A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 26 (5), 2333–2355. eng. Epub 2015/08/19Cited in: Pubmed; PMID 26282889. doi:10.1177/0962280215597579
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32 (5), 377–389. eng. Epub 2017/05/21Cited in: Pubmed; PMID 28527048. doi:10.1007/s10654-017-0255-x
Cekici, A., Kantarci, A., Hasturk, H., and Van Dyke, T. E. (2014). Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol. 2000 64 (1), 57–80. eng. Epub 2013/12/11. doi:10.1111/prd.12002
Díaz-Zúñiga, J., Melgar-Rodríguez, S., Rojas, L., Alvarez, C., Monasterio, G., Carvajal, P., et al. (2017). Increased levels of the T-helper 22-associated cytokine (interleukin-22) and transcription factor (aryl hydrocarbon receptor) in patients with periodontitis are associated with osteoclast resorptive activity and severity of the disease. J. Periodontal Res. 52 (5), 893–902. eng. Epub 2017/04/11Cited in: Pubmed; PMID 28393368. doi:10.1111/jre.12461
Feng, L. L., Gao, J. M., Li, P. P., and Wang, X. (2011). IL-9 contributes to immunosuppression mediated by regulatory T cells and mast cells in B-cell non-hodgkin's lymphoma. J. Clin. Immunol. 31 (6), 1084–1094. eng. Epub 2011/09/08Cited in: Pubmed; PMID 21898141. doi:10.1007/s10875-011-9584-9
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46 (6), 1985–1998. eng. Epub 2017/10/19Cited in: Pubmed; PMID 29040600. doi:10.1093/ije/dyx102
Hienz, S. A., Paliwal, S., and Ivanovski, S. (2015). Mechanisms of bone resorption in periodontitis. J. Immunol. Res. 2015, 615486. eng. Epub 2015/06/13. doi:10.1155/2015/615486
Huynh, N. C., Everts, V., Pavasant, P., and Ampornaramveth, R. S. (2017). Interleukin-1β induces human cementoblasts to support osteoclastogenesis. Int. J. Oral Sci. 9 (12), e5. Epub 2017/12/14. doi:10.1038/ijos.2017.45
Kamat, M. A., Blackshaw, J. A., Young, R., Surendran, P., Burgess, S., Danesh, J., et al. (2019). PhenoScanner V2: An expanded tool for searching human genotype-phenotype associations. Bioinformatics 35 (22), 4851–4853. eng. Epub 2019/06/25Cited in: Pubmed; PMID 31233103. doi:10.1093/bioinformatics/btz469
Kar, S., Gupta, R., Malhotra, R., Sharma, V., Farooque, K., Kumar, V., et al. (2021). Interleukin-9 facilitates osteoclastogenesis in rheumatoid arthritis. Int. J. Mol. Sci. 22, 10397. Cited in: Pubmed; PMID 34638736. doi:10.3390/ijms221910397
Kwiecien, K., Zegar, A., Jung, J., Brzoza, P., Kwitniewski, M., Godlewska, U., et al. (2019). Architecture of antimicrobial skin defense. Cytokine Growth Factor Rev. 49, 70–84. eng. Epub 2019/09/02Cited in: Pubmed; PMID 31473081. doi:10.1016/j.cytogfr.2019.08.001
Lawlor, D. A., Harbord, R. M., Sterne, J. A., Timpson, N., and Davey Smith, G. (2008). Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27 (8), 1133–1163. eng. Epub 2007/09/22. doi:10.1002/sim.3034
Liu, X., and Li, H. (2021). A systematic review and meta-analysis on multiple cytokine gene polymorphisms in the pathogenesis of periodontitis. Front. Immunol. 12, 713198. eng. Epub 2022/01/21. doi:10.3389/fimmu.2021.713198
Lu, H., Wu, P. F., Zhang, W., and Liao, X. (2021). Circulating interleukins and risk of multiple sclerosis: A mendelian randomization study. Front. Immunol. 12, 647588. eng. Epub 2021/05/04. doi:10.3389/fimmu.2021.647588
Mazurek-Mochol, M., Kozak, M., Malinowski, D., Safranow, K., and Pawlik, A. (2021). IL-17F gene rs763780 and IL-17a rs2275913 polymorphisms in patients with periodontitis. Int. J. Environ. Res. Public Health 18 (3), 1081. Epub 2021/02/04Cited in: Pubmed; PMID 33530501. doi:10.3390/ijerph18031081
McGeachy, M. J., Cua, D. J., and Gaffen, S. L. (2019). The IL-17 family of cytokines in health and disease. Immunity 50 (4), 892–906. eng. Epub 2019/04/18Cited in: Pubmed; PMID 30995505. doi:10.1016/j.immuni.2019.03.021
Milligan, B. G. (2003). Maximum-likelihood estimation of relatedness. Genetics 163 (3), 1153–1167. eng. Epub 2003/03/29. doi:10.1093/genetics/163.3.1153
Nikolopoulos, G. K., Dimou, N. L., Hamodrakas, S. J., and Bagos, P. G. (2008). Cytokine gene polymorphisms in periodontal disease: A meta-analysis of 53 studies including 4178 cases and 4590 controls. J. Clin. Periodontol. 35 (9), 754–767. eng. Epub 2008/08/05. doi:10.1111/j.1600-051X.2008.01298.x
Osta, B., Benedetti, G., and Miossec, P. (2014). Classical and paradoxical effects of TNF-α on bone homeostasis. Front. Immunol. 5, 48. eng. Epub 2014/03/05. doi:10.3389/fimmu.2014.00048
Palmer, T. M., Lawlor, D. A., Harbord, R. M., Sheehan, N. A., Tobias, J. H., Timpson, N. J., et al. (2012). Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 21 (3), 223–242. eng. Epub 2011/01/11Cited in: Pubmed; PMID 21216802. doi:10.1177/0962280210394459
Pan, W., Wang, Q., and Chen, Q. (2019). The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 11 (3), 30. Cited in: Pubmed; PMID 31685798. doi:10.1038/s41368-019-0064-z
Peres, M. A., Macpherson, L. M. D., Weyant, R. J., Daly, B., Venturelli, R., Mathur, M. R., et al. (2019). Oral diseases: A global public health challenge. Lancet 394 (10194), 249–260. Epub 2019/07/23. doi:10.1016/S0140-6736(19)31146-8
Shungin, D., Haworth, S., Divaris, K., Agler, C. S., Kamatani, Y., Keun Lee, M., et al. (2019). Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat. Commun. 10(1), 2773. Cited in: Pubmed; PMID 31235808. doi:10.1038/s41467-019-10630-1
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Davies, N. M., Swanson, S. A., VanderWeele, T. J., et al. (2021). Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. Bmj 375, n2233. Cited in: Pubmed; PMID 34702754. doi:10.1136/bmj.n2233
Valm, A. M. (2019). The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J. Mol. Biol. 431 (16), 2957–2969. eng. Epub 2019/05/20Cited in: Pubmed; PMID 31103772. doi:10.1016/j.jmb.2019.05.016
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50 (5), 693–698. eng. Epub 2018/04/25Cited in: Pubmed; PMID 29686387. doi:10.1038/s41588-018-0099-7
Xiao, E., Mattos, M., Vieira, G. H. A., Chen, S., Corrêa, J. D., Wu, Y., et al. (2017). Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe 22 (1), 120–128. e4. eng. Epub 2017/07/14Cited in: Pubmed; PMID 28704648. doi:10.1016/j.chom.2017.06.014
Yu, J. J., Ruddy, M. J., Wong, G. C., Sfintescu, C., Baker, P. J., Smith, J. B., et al. (2007). An essential role for IL-17 in preventing pathogen-initiated bone destruction: Recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood 109 (9), 3794–3802. eng. Epub 2007/01/05Cited in: Pubmed; PMID 17202320. doi:10.1182/blood-2005-09-010116
Zhao, Q., Chen, Y., Wang, J., and Small, D. S. (2019). Powerful three-sample genome-wide design and robust statistical inference in summary-data Mendelian randomization. Int. J. Epidemiol. 48 (5), 1478–1492. eng. Epub 2019/07/13Cited in: Pubmed; PMID 31298269. doi:10.1093/ije/dyz142
Zheng, Y., Chai, L., Fan, Y., Song, Y. Q., Zee, K. Y., Tu, W. W., et al. (2020). Th2 cell regulatory and effector molecules single nucleotide polymorphisms and periodontitis. J. Leukoc. Biol. 108 (5), 1641–1654. eng. Epub 2020/08/04Cited in: Pubmed; PMID 32745291. doi:10.1002/jlb.4ma0720-698rr
Zhong, J., Wang, Z., Yuan, W., Shen, Y., and Chen, L. (2022). Interleukin-17 promotes osteoclastogenesis and periodontal damage via autophagy in vitro and in vivo. Int. Immunopharmacol. 107, 108631. eng. Epub 2022/02/27Cited in: Pubmed; PMID 35219162. doi:10.1016/j.intimp.2022.108631
Keywords: periodontitis, cytokines, IL9, IL17, mendelian randomization
Citation: Huang S-J, Li R, Xu S, Liu Y, Li S-H and Duan S-Z (2023) Assessment of bidirectional relationships between circulating cytokines and periodontitis: Insights from a mendelian randomization analysis. Front. Genet. 14:1124638. doi: 10.3389/fgene.2023.1124638
Received: 15 December 2022; Accepted: 20 January 2023;
Published: 30 January 2023.
Edited by:
Rajiv Pathak, Albert Einstein College of Medicine, United StatesReviewed by:
Ritu Shekhar, University of Florida, United StatesCopyright © 2023 Huang, Li, Xu, Liu, Li and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng-Hui Li, c3VibWlzc2lvbjk5MDdAMTYzLmNvbQ==; Sheng-Zhong Duan, ZHVhbnN6QHNoc211LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.