- 1Key Laboratory of Tropical and Subtropical Fishery Resources Application and Cultivation, Ministry of Agriculture and Rural Affairs, Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, Guangdong, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, Jiangsu, China
- 3College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
- 4Zhejiang Fisheries Technical Extension Center, Hangzhou, China
Introduction: Bone morphogenetic proteins (BMPs) play a crucial role in bone formation and differentiation. Recent RNA-Seq results suggest that BMPs may be involved in the sex differentiation of P. sinensis, yet more relevant studies about BMPs in P. sinensis are lacking.
Methods: Herein, we identified BMP gene family members, analyzed the phylogeny, collinear relationship, scaffold localization, gene structures, protein structures, transcription factors and dimorphic expression by using bioinformatic methods based on genomic and transcriptomic data of P. sinensis. Meanwhile, qRT-PCR was used to verify the RNA-Seq results and initially explore the function of the BMPs in the sex differentiation of P. sinensis.
Results: A total of 11 BMP genes were identified, 10 of which were localized to their respective genomic scaffolds. Phylogenetic analysis revealed that BMP genes were divided into eight subfamilies and shared similar motifs (“WII”, “FPL”, “TNHA”, “CCVP”, and “CGC”) and domain (TGF-β superfamily). The results of the sexually dimorphic expression profile and qRT-PCR showed that Bmp2, Bmp3, Bmp15l, Bmp5, Bmp6 and Bmp8a were significantly upregulated in ovaries, while Bmp2lb, Bmp7, Bmp2bl and Bmp10 were remarkable upregulated in testes, suggesting that these genes may play a role in sex differentiation of P. sinensis.
Discussion: Collectively, our comprehensive results enrich the basic date for studying the evolution and functions of BMP genes in P. sinensis.
1 Introduction
The transforming growth factor-beta (TGF-β) superfamily is a large group of extracellular growth factors controlling many aspects of development (Chang, Brown et al., 2002). Researches have shown that a total of 33 members of the TGF-β superfamily are identified in Humans, including 10 members of bone morphogenetic proteins (BMPs), 10 members of growth differentiation factors (GDFs), and 13 additional members (Morikawa et al., 2016). As a dominant subfamily of the TGF-β superfamily, the activity of BMPs was initially investigated in the 1960s (Urist, 1965). BMPs play a major role in osteoblast differentiation and bone formation (Lademann et al., 2020; Yang et al., 2021), and are associated with cancer progression, activating the proliferation and increasing the invasiveness of cancer cells (Ehata and Miyazono, 2022). Moreover, BMPs modulate vascular calcification by regulating the phenotypic plasticity of multipotent progenitor lineages (Yang et al., 2021). Simultaneously, BMPs are also involved in inflammation, glucose homeostasis, and energy metabolism (Grgurevic et al., 2016).
Recently, studies have shown that BMP family members have a role in sex differentiation. Bmp2 and retinoic acid (RA) act collectively to control female germ cell fate in mice (Miyauchi et al., 2017). Bmp2, in cooperation with Estradiol-17β, promotes the transformation of somatic cells into primordial granulosa cells and the formation of primordial follicle in mouse ovaries (Chakraborty et al., 2022). In Gallus gallus domestics, Bmp7 exhibits female-biased expression during sex differentiation and was reduced upon aromatase inhibitor-induced female-to-male reversal (Hoshino et al., 2005). Knocking out Bmp15 in female zebrafish leads to normal development, but these animals’ sexual revert to fertile males during the juvenile period (Dranow et al., 2016). In Paralichthys olivaceus, overexpression of Gdf9 individually or co-expression with Bmp15 lead to upregulation of most steroid genes, whereas overexpression of single Bmp15 decreases the expression of steroid genes (Yu et al., 2020). Meanwhile, BMP signaling mediates the differentiation of primordial germ cells of Gryllus bimaculatus (Donoughe et al., 2014). Significantly, Amh, Gsdf, and Gdf9 share the same domain with BMPs, belonging to the TGF-β superfamily (Yu et al., 2020; Pan et al., 2021). In Epinephelus coioides, overexpressing Amh could lead to a decrease in female-associated genes and estradiol levels and an increase in male-associated genes and testosterone levels, thereby causing female-to-male reversal in vivo (Han et al., 2018). Also, Gsdf and Gdf9 are key members of vertebrates’ sex differentiation pathways (Nagahama et al., 2021). Taken together, BMPs are connected to the process of sex differentiation and seem to be conserved in multiple species.
Likewise, sex differentiation in the Chinese soft-shell turtle (Pelodiscus sinensis) is the consequence of multi-gene regulation. P. sinensis is an economically vital aquatic animal, with an annual production of about 330,000 tons (Yearbook, 2019). The growth pattern of P. sinensis presents sexual dimorphism, with males growing 1.5 times faster than females (Zhou and Zhu, 2011). Consequently, it is necessary to explore the master genes that regulate sex differentiation in P. sinensis, thereby producing all-male populations. Dmrt1 is the first major gene identified as involved in male sex differentiation in P. sinensis, and knockdown of Dmrt1 causes male-to-female reversal (Sun et al., 2017). Loss of function of Amh resulted in complete sex reversal of ZZ embryos, downregulation of Cyp19a1, and upregulation of Sox9 (Zhou et al., 2019). Rspo1 is essential for female sex differentiation in P. sinensis, and knocking down Rspo1 led to a partial female-to-male reversal (Zhang et al., 2021). Particularly, a novel study indicates that Bmp2 was significantly more expressed in the gonads of female P. sinensis than in males by RNA-Seq, suggesting that Bmp2 may be a potential gene for sex differentiation in P. sinensis (Zhu et al., 2022). Nevertheless, further information about BMP family genes in P. sinensis remains necessary.
In the present study, we performed genome-wide identification of the members of BMP gene family in P. sinensis using bioinformatics methods and analyzed the phylogenetic relationship, collinear relationship, scaffold locations, gene structures, protein structures, transcription factors, and gonad expression profiles of BMP genes. Moreover, qRT-PCR was used to examine the relative expression of BMP gene family members in male and female gonads at different developmental stages of P. sinensis. This study enriches our knowledge of BMP genes and contributes to further studies on the regulation mechanisms of sex differentiation in P. sinensis.
2 Materials and methods
2.1 Genome-wide identification of BMP genes
All genome sequences, protein sequences, and annotation files of P. sinensis (https://www.ncbi.nlm.nih.gov/genome/?term=Pelodiscus+sinensis) and Homo sapiens (https://www.ncbi.nlm.nih.gov/genome/?term=human) were obtained from the National Center for Biotechnology Information (NCBI) database. The Hidden Markov Model file of BMP genes (PF00019) was downloaded from the Pfam protein family database (http://pfam.xfam.org/family/PF00019) and was used to screen the members of BMP gene family in P. sinensis and human genome with HMMER 3.0 software (Finn et al., 2011). Subsequently, the candidate sequences were further analyzed using the NCBI Conserved Domain tool (Marchler-Bauer et al., 2017) and SMART database (Letunic et al., 2021) to proofread the process of the gene family member screening and remove redundant sequences, and the confirmed members of the BMP gene family contained the complete TGF-β superfamily domain.
2.2 Phylogenetic analysis and collinear analysis of BMP gene family
Amino acid sequences of typical vertebrates were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov), including Homo sapiens, Mus musculus, Gallus gallus, Anas platyrhynchos, Danio rerio, Oryzias latipes, Xenopus tropicalis, Bufo gargarizans, Terrapene carolina triunguis, Chelydra serpentina, Dermochelys coriacea, Mauremys reevesii, Caretta caretta, Chelonia mydas, Mauremys mutica, and Chrysemys picta bellii (All protein accession numbers and sequences used for the analysis were shown in Annex 1). Multiple sequence alignment of full-length proteins among species was assessed using MUSCLE (Edgar, 2004), and an unrooted phylogenetic tree was constructed using the Maximum Likelihood method with a Jones-Taylor-Thornton (JTT) + Gamma Distributed (G) model and 1,000 bootstrap replications using MEGA 7 software (Kumar et al., 2016). The phylogenetic tree was visually improved using the online website ChiPlot (https://www.chiplot.online/). Collinear analysis was performed among human, chicken, P. sinensis, and zebrafish using NCBI genome browser (https://www.ncbi.nlm.nih.gov) and Ensembl (https://useast.ensembl.org/index.html).
2.3 Analysis of scaffold location, gene structure, conserved motif, and domain
First, BMP genes were mapped to the scaffold locations, and a gene density file of each scaffold was generated according to the genome annotation files. TBtools as a visualization tool to display BMP genes structures (Chen et al., 2020). The protein sequences of BMP genes were submitted to MEME Suite database for predicting conserved motifs, and the motif numbers were searched until exceeding the default thresholds (Bailey et al., 2015). Meanwhile, the NCBI Conserved Domain tool was used to screen the conserved domains of BMP genes with the default parameters (Marchler-Bauer et al., 2017). The visualization of the conserved motifs and domains of BMP genes were applied by TBtools software.
2.4 Multiple sequence alignment and prediction of protein structure
The amino acid sequences of conserved domains, from the TGF-β superfamily, of BMP genes were obtained by SAMRT (Letunic et al., 2021), and BioEdit software was utilized for multiple sequence alignment. Meanwhile, WebLogo (Crooks et al., 2004) was used to show the characteristics of amino acid sequences. The prediction of protein secondary and tertiary structures were performed using SOPMA (Geourjon and Deléage, 1995) and SWISS MODLE (Waterhouse et al., 2018), respectively.
2.5 Analysis of transcription factors
TBtools software was applied to extract a 2-kilobase (kb) promoter region upstream of the transcription start site of the BMP genes. Transcription factors were predicted by JASPAR (Castro-Mondragon et al., 2022). In short, all promoter sequences were used to search BMP genes sites with all available matrices in the taxon of vertebrates via the Scan tool and a relative score cutoff of 0.8 was used to identify putative sites. TBtools was used to map the transcription factors to the 2-kb promoter region of BMP genes.
2.6 Analysis of sexually dimorphic expression profiles of BMP genes based on transcriptomic data
The transcriptomic data of the gonads in P. sinensis was downloaded from NCBI database of Zhu et al. (2022); SRA accession: PRJNA838782). Trimmomatic (Bolger et al., 2014) was used to remove low-quality reads and adapters, and Hisat2 v2.1.0 (Kim et al., 2015) was used to for mapping to the reference genome of P. sinensis. The expression levels of BMP genes were calculated and normalized with the fragments per kilobase million (FPKM) values, using StringTie (v2.2.0) with default parameters (Pertea et al., 2016). Then, the differentially expressed levels of mRNAs were evaluated using edgeR (Robinson, McCarthy, and Smyth 2010). And the screening criteria were a fold change ≥2 and p <0.05. Finally, a heatmap was generated using TBtools.
2.7 Quantitative real-time PCR (qRT-PCR) of BMP gene family
2.7.1 Sample collection
According to previous studies (Lei et al., 2022) (Zhou et al., 2019), the gonads of P. sinensis were undifferentiated at 9-day and the 16-day was the critical period for gonad differentiation, under incubation conditions at a temperature of 31°C and 75% humidity. To verify the reliability of RNA-Seq results and explore the functions of Bmp gene family members initially, the relative expression of the Bmp gene family members was detected in the gonads of 9-day, 16-day male and female embryos and in 6-month-old male and female gonads by qRT-PCR.
All experimental procedures were performed in accordance with the regulations for animal care of the Pearl River Fisheries Research Institute (Guangzhou, China). All experimental P. sinensis and embryos were obtained from Caixing Industrial Co. (Huizhou, China). According to the sample collection method of Lei et al. (2022), embryos were collected on 9 and 16-day and gonads (including kidneys and gonads) were rapidly isolated under a light microscope, and stored in liquid nitrogen for RNA extraction. According to the Tissue DNA Extraction CZ Kit (Mabio, China) instructions, the rest of the tissue was used as DNA extraction to identify the sex of the embryos by PCR (Li et al., 2020). Three 6-month-old male and female P. sinensis were anesthetized using 0.05% MS-222 (20 mg/kg, Sigma, MO, United States) by intraperitoneal injection and sacrificed. Subsequently, testes and ovaries were quickly collected, and stored in liquid nitrogen for RNA extraction.
2.7.2 RNA extraction, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Total RNA extraction of all samples was performed according to the RNAiso Plus (Takara, Beijing, China) instructions. After RNA quality testing by RNA electrophoresis (Bio-Rad, PowerPacTM, CA, United States) and NanoDrop 2000 Spectrophotometer (Thermo Fisher, NanoDropOne, MA, United States), cDNA synthesis was performed following the protocol of the reverse transcription kit (Takara, Beijing, China).
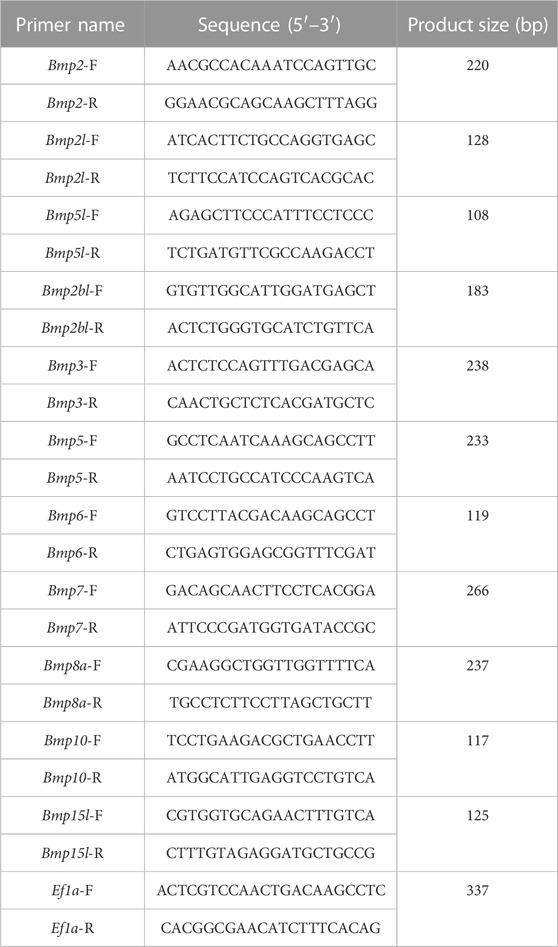
Follow the instructions of iTaq Universal SYBR Supermix (BIO-RAD, CA, United States) for qRT-PCR. All the primers were designed based on the nucleotides of P. sinensis BMPs family genes on NCBI, the Ef1a gene was chosen as a reference gene to calculate the relative expression of the target genes since it showed a stable expression pattern in P. sinensis. The qRT-PCR volume was 20 μL, including 10 μL of SYBR Supermix, 1 μL of cDNA, 1 μL of each primer (2 μM) and 7 μL of nuclease-free water. The PCR cycling conditions for all target genes and Ef1a were as follows: 95°C for 10°min; 40 cycles of 95°C for 15 s, 55°C–60°C for 15 s, and 72°C for 15 s; and melting curve analysis at 95°C 15 s, 60°C for 60 s, and 95°C 15 s. Each sample was analyzed in triplicate. The sequences of the primers for the target genes and reference genes are given in Table 1. Transcript expression levels were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Differential expression analysis was performed by ANOVA (Momen et al., 2001). The results are presented as the means ± SEM of three replicates, and the statistical significance is represented by a p-value <0.05.
3 Results
3.1 Identification of the members of BMP gene family in the P. sinensis genome
A total of 11 BMP gene family members named Bmp2 (XM_006136012.3), Bmp2l (XM_006123882.3), Bmp2lb (XM_014571013.2), Bmp2bl (XM_006117827.3), Bmp3 (XM_006110911.3), Bmp5 (XM_006110971.2), Bmp6 (XM_025179361.1), Bmp7 (XM_006125345.3), Bmp8a (XM_025179496.1), Bmp10 (XM_006110793.3), and Bmp15l (XM_025185441.1) were initially identified within the genome of P. sinensis. Bmp6 was discarded due to the absence of a complete protein sequence, and 10 members of the BMP gene family were finally selected for subsequent analysis.
3.2 Phylogenetic analysis of identified BMP genes
To investigate the phylogenetic relationships of BMP genes in P. sinensis, an unrooted phylogenetic tree was constructed (Figure 1). The BMP genes of P. sinensis were clustered into eight subfamilies including Bmp2, Bmp2l, Bmp3, Bmp5, Bmp7, Bmp8a, Bmp10, and Bmp15. The majorities of BMP genes in P. sinensis were first clustered with turtles, followed by frogs and birds. Significantly, Bmp2l, Bmp2l,b and Bmp2bl of P. sinensis were clustered into one subfamily, suggesting that the function of these genes may have changed during the evolution and development of P. sinensis compared to the members of Bmp2 subfamilies.
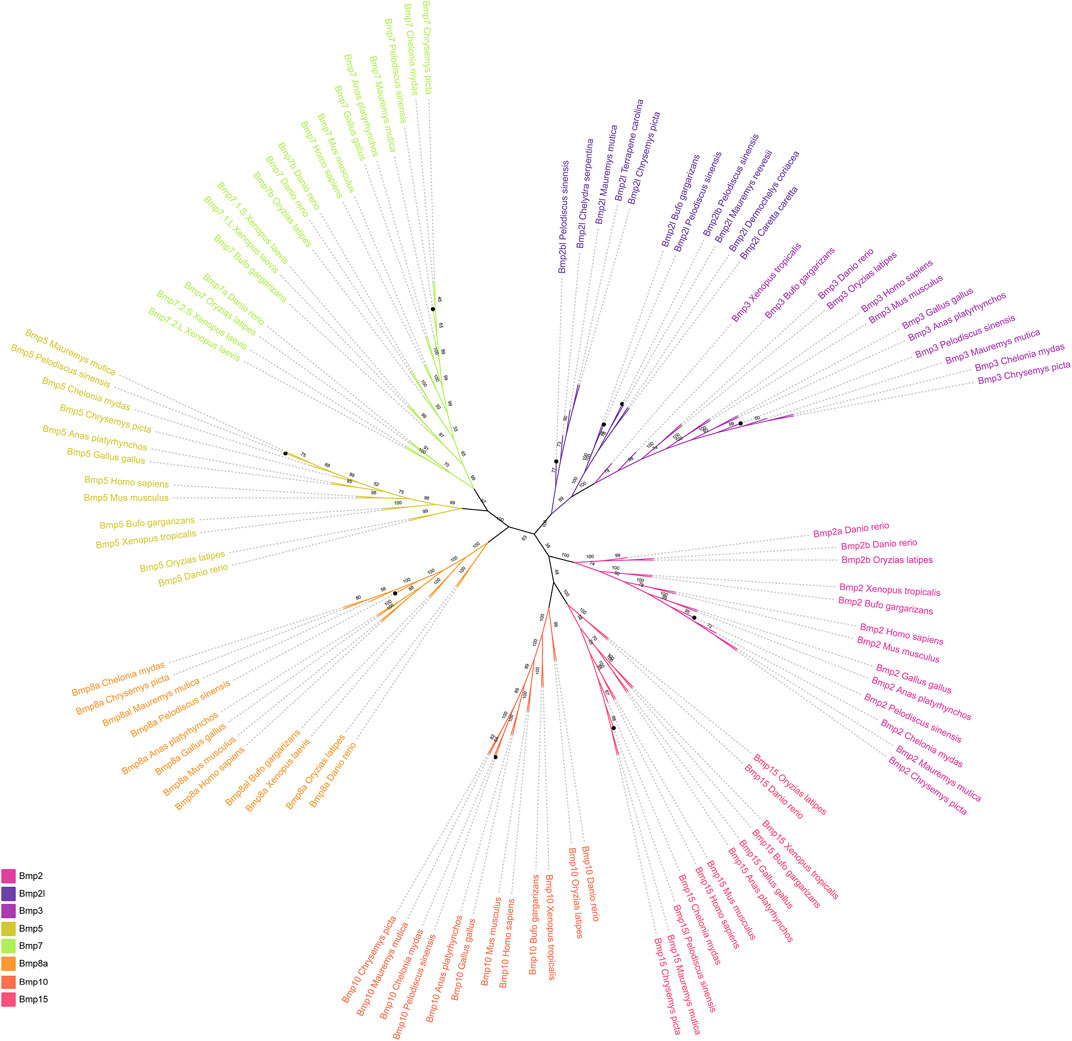
FIGURE 1. Phylogenetic tree of the BMP gene family among different vertebrates. Different colors represent different BMP gene subfamilies. The black circles indicate the BMP genes in P. sinensis.
3.3 Collinear analysis
Comparative gene collinear analysis was performed between P. sinensis, Homo sapiens, Gallus and Danio rerio (Figures 2A–G). The results showed that the Hao1-Bmp2-Fermt1-Lrrn4-Crls1-Mcm8-Trmt6 block was highly conserved between Homo and P. sinensis, and four adjacent genes of P. sinensis Bmp2 (Hao1, Tmx4, Plcb1and Pclb4) were identical in Danio rerio and Gallus gallus (Figure 2A). The Bmp3-Cfap299-Fgf5-Prdm8b-Antxr2 block was highly conserved in P. sinensis, Homo sapiens and Danio rerio. Only one adjacent gene (Rasgef1b) was identical in Gallus gallus (Figure 2B). The Fam83b- Hcrtr2- Gfral- Hmgcll1-Bmp5-Col21a1-Dst-Bend6 block was highly conserved in Homo, P. sinensis and Danio rerio. However, no conserved adjacent gene was found in Gallus gallus (Figure 2C). The adjacent genes of Bmp7 (Spo11, Rae1, Rbm38) (Figure 2D) and Bmp8a (Macf1, Ppie, Pabpc4, Heyl, Nt5c1a, Hpcal4) (Figure 2E) were highly conserved in Gallus gallus, P. sinensis and Homo. Two adjacent genes of P. sinensis Bmp10 (Arhgap25, Cds2) were identical in Danio rerio, and three adjacent genes (Cds2, Pcna, Tmem230) were identical in Gallus gallus, and P. sinensis Bmp10 shared only one conserved adjacent gene (Arhgap25) in Homo sapiens (Figure 2F). P. sinensis Bmp15l shared only one conserved adjacent gene in Homo sapiens (Shroom4) and Danio rerio (Hdac8). Compared with Gallus gallus, P. sinensis Bmp15l shared three conserved adjacent genes (Phka1, Hdac8, Shroom4) (Figure 2G).

FIGURE 2. Collinear analysis of Bmp2(A), Bmp3(B), Bmp5(C), Bmp7(D), Bmp8a(E), Bmp10(F), Bmp15(G) and their adjacent genes in Homo sapiens, Gallus gallus, P. sinensis and Danio rerio. Direction of the arrows indicates gene orientation.
3.4 Scaffold localization of BMP genes
To determine the scaffold locations of P. sinensis BMP genes, 10 members were mapped to 10 different genomic scaffolds (Figure 3). Bmp2bl, Bmp2, Bmp7, Bmp3, Bmp10, Bmp8a, Bmp2lb, Bmp5, Bmp15l, and Bmp21 were located on scaffold_1, scaffold_130, scaffold_287, scaffold_360, scaffold_489, scaffold_493, scaffold_801, scaffold_876, scaffold_1809, and scaffold_2314, respectively.
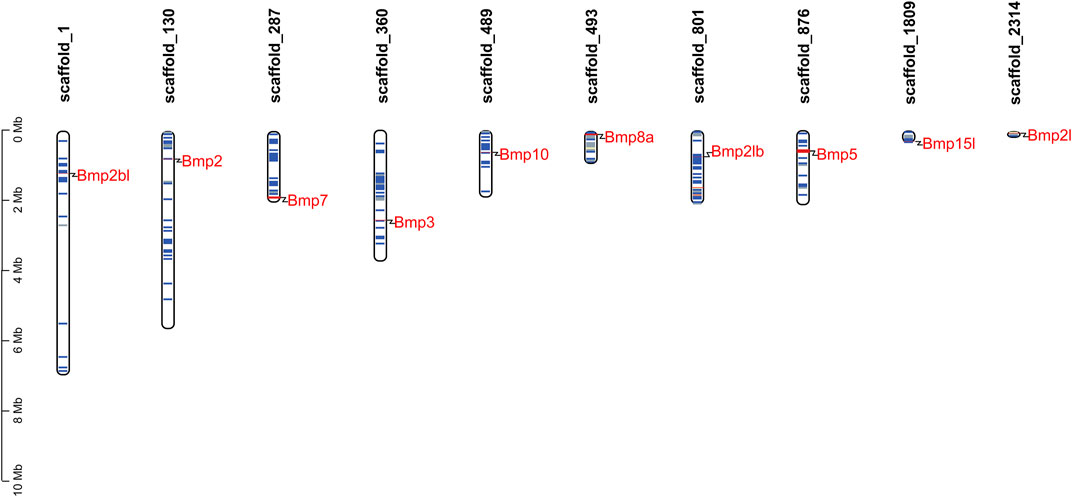
FIGURE 3. Scaffold distribution of BMP genes in P. sinensis. The blue lines represent gene density of scaffolds, and the red lines indicate higher gene density. The gene density is defined as the number of genes within a 50 kb genome.
3.5 Gene structures and conserved motifs
The results of the gene structure analysis showed that the gene length, coding sequence (CDS) number, and introns of BMP genes demonstrated significant differences in P. sinensis. Bmp2, Bmp3, and Bmp10 had the lowest numbers of CDS (2), with Bmp5, Bmp7, and Bmp8a having the highest CDS numbers (7). Furthermore, Bmp8a and Bmp15l lack of the 3′untranslated region (3′UTR) (Figure 4A). Similarly, in Homo, Bmp2 and Bmp10 had the lowest numbers of CDS (2), with Bmp5, Bmp7, Bmp8a and Bmp8b having the highest CDS numbers (7) (Figure 4B). Analysis of the conserved motifs of BMP proteins showed that 13 putative motifs Fwere unevenly distributed in P. sinensis BMP genes. The number of motifs ranged from 1 (Bmp15l) to 11 (Bmp5). Motif 1 was shared by 9 BMP genes, and motif 2 was shared by all (Figure 5A). In Homo, the number of motifs ranged from 3 (Bmp15) to 11 (Bmp5, Bmp6, Bmp7, Bmp8a, Bmp8b), and there are three motifs (Motif 1, Motif 2, Motif 5) shared in all BMP proteins (Figure 5B) (The sequences of all motifs are provided in Annex 2).
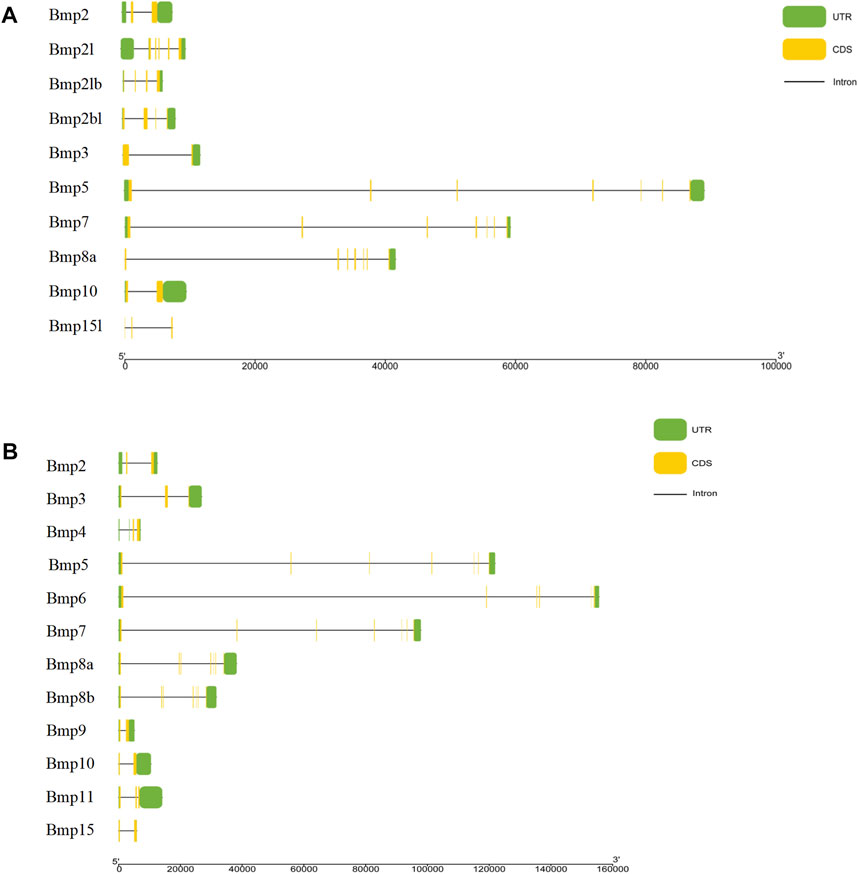
FIGURE 4. Gene structures of BMP genes. (A). Gene structures of BMP genes in P. sinensis. (B). Gene structures of BMP genes in Homo. The green boxes show untranslated regions, the yellow boxes show coding sequences, and the black lines show introns.
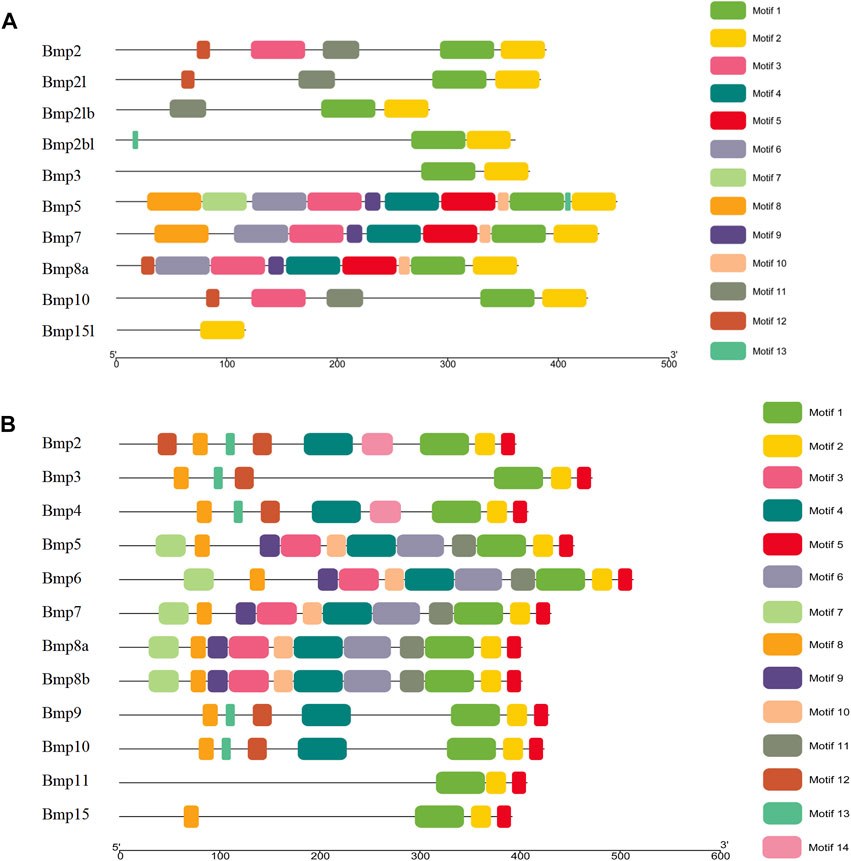
FIGURE 5. Conserved motifs of BMP genes in P. sinensis (A) and Homo (B). Different colored boxes indicate different motifs; the direction of the proteins is from the N-terminal to the C-terminal.
3.6 Conserved domains and multiple sequence alignment
BMP genes of P. sinensis all possessed a conserved protein domain of the TGF-β superfamily, and 7 proteins (70%) had two common domains, the TGF-β superfamily and TGF-β propeptide superfamily. However, Bmp2bl, Bmp3, and Bmp15l only had one domain (Figure 6A). Likewise, all BMPs in Homo shared the conserved protein structural domain of the TGF-β superfamily, and there was only one domain in Bmp3 and Bmp15 (Figure 6B). The results of multiple sequence alignment of the conserved protein domain (TGF-β superfamily) showed that conservative motifs of “WII”, “FPL”, “TNHA”, “CCVP”, and “CGC” were shared by almost all BMP genes in P. sinensis. However, a few conserved motifs of BMP genes have undergone mutation and evolution. For instance, the motifs of Bmp2bl and Bmp15l changed from “TNHA” to “ESRE” and “PNHA”, respectively. Further, the motif of “FPL” was missing in Bmp2bl and Bmp15l (Figure 7A). In Home, “WIIAP”, “FPL”, “HAIVQ”, “ISVLY”, and “CGC” were shared by almost all BMP genes (Figure 7B). In P. sinensis and Homo, the conservative motif of “CGC” was changed to “CAC” in Bmp3 and “CTC” in Bmp15, respectively.
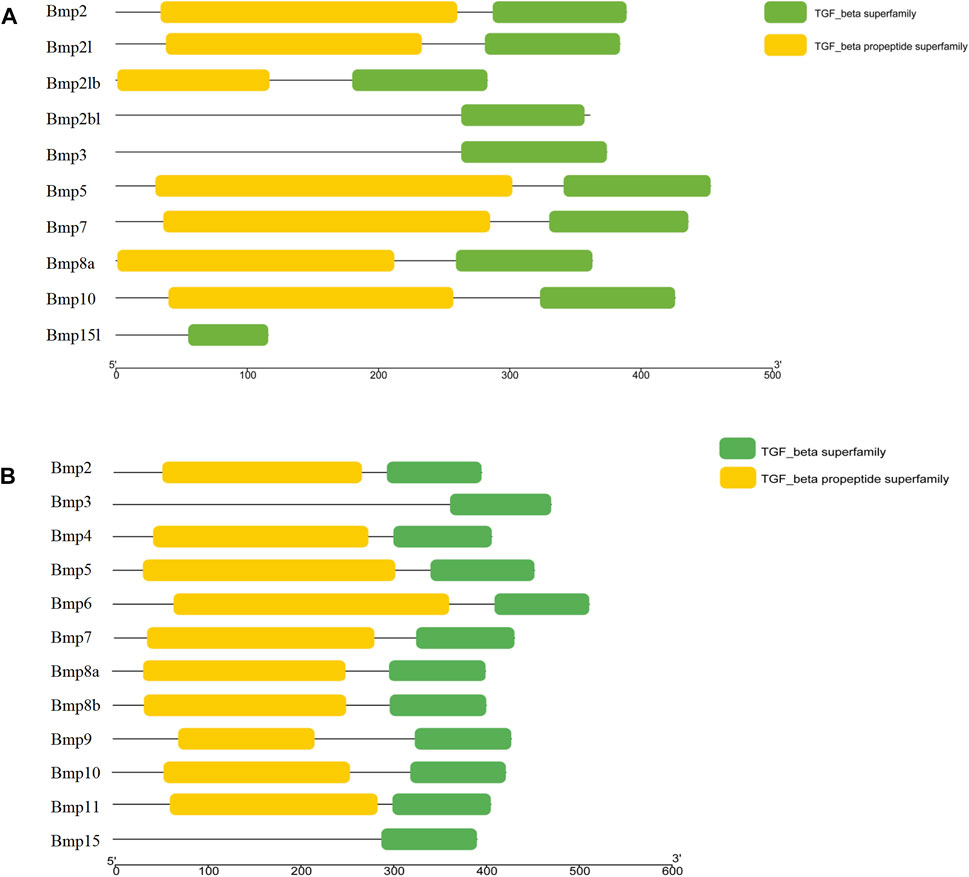
FIGURE 6. Conserved domains of BMP genes in P. sinensis(A) and Homo (B). The green boxes and yellow boxes indicate the protein domains of the TGF-β superfamily and the TGF-β propeptide superfamily, respectively.
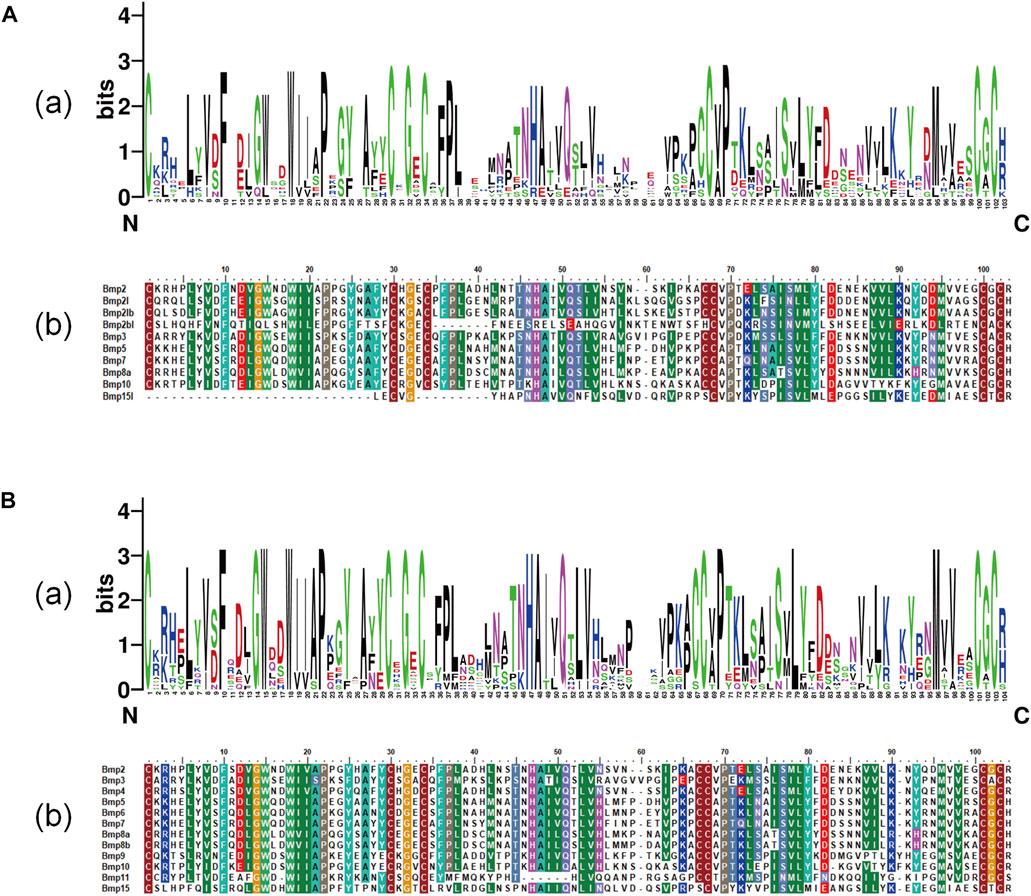
FIGURE 7. Multiple sequence alignment of TGF-β superfamily domains of BMP genesin P. sinensis (A) and Homo (B). (A) and (B) show the sequence logo and amino acid sequence alignment of the TGF-β superfamily domain of BMP genes, respectively. The direction of amino acid sequences is from the N-terminal to the C-terminal.
3.7 Prediction of secondary and tertiary structures
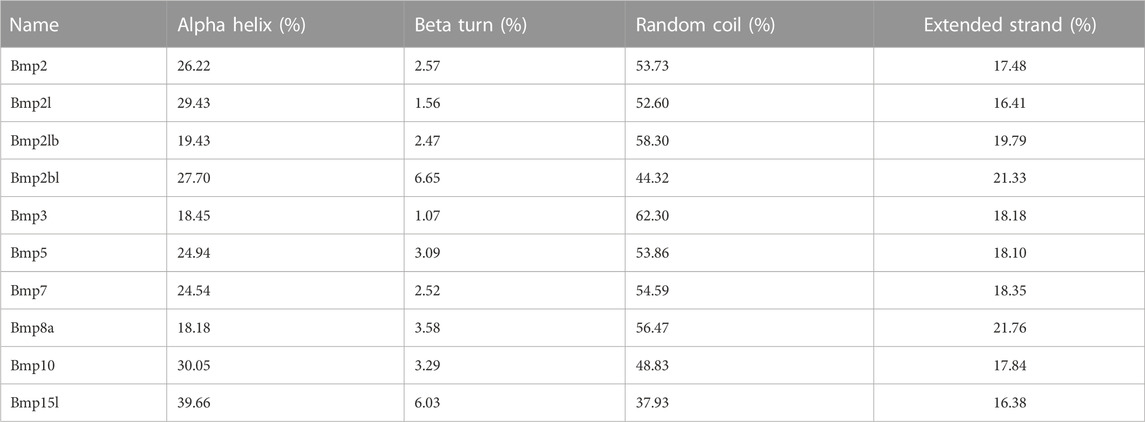
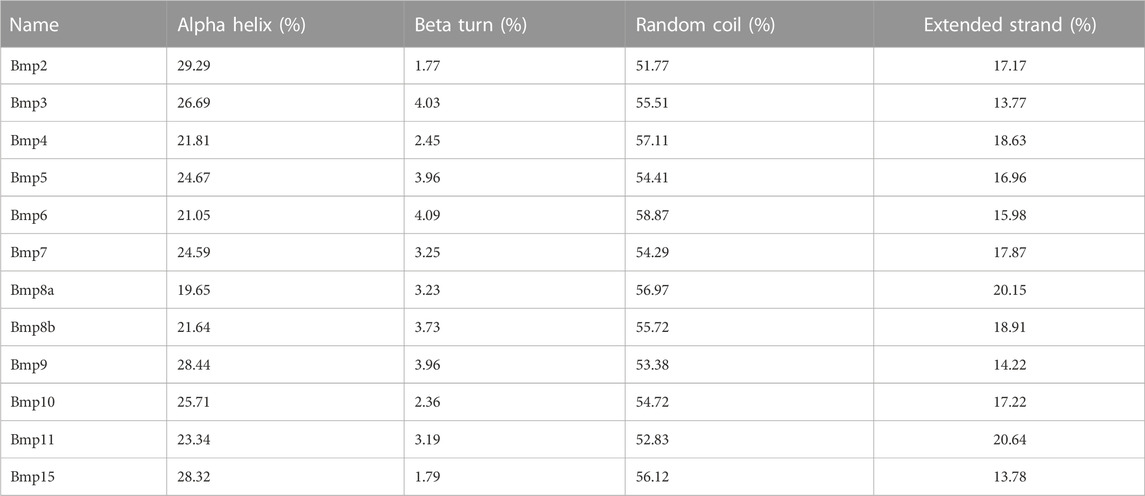
P. sinensis and Homo sapiens BMP proteins are mainly composed of helixes, beta turns, random coils and extended strands (Figures 8A, B). In P. sinensis, alpha helixes ranged from 18.18 to 39.66%, beta turns from 1.07 to 6.65%, random coils from 37.93 to 62.30%, and extended strands from 16.38 to 21.76% (Table 2). In Homo sapiens, alpha helixes ranged from 19.65 to 29.29%, beta turns from 1.77 to 4.09%, random coils from 51.77 to 58.87%, and extended strands from 13.77 to 20.64% (Table 3).
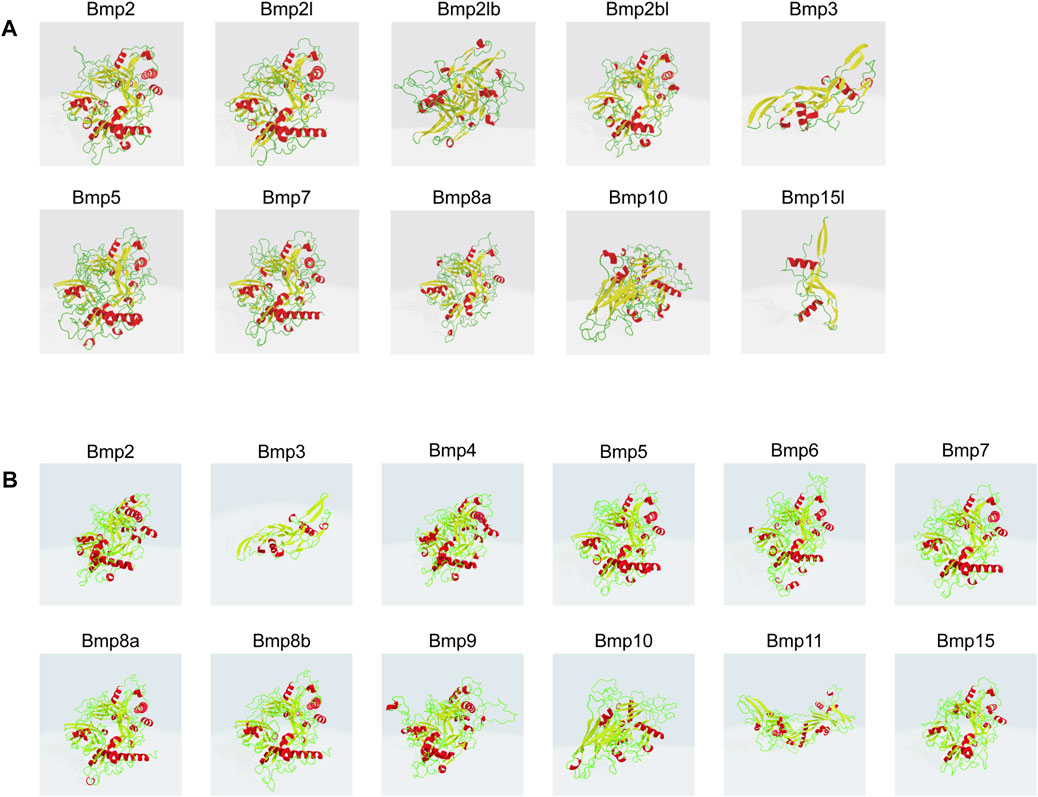
FIGURE 8. Three-dimensional modeling of BMP proteins in P. sinensis (A) and Homo (B). Red indicates alpha helix, yellow represents beta turn, green shows random coil.
3.8 Prediction of transcription factors
The 2-kb 5’ upstream region of P. sinensis and Homo sapiens BMP genes was selected to analyze transcription factors. There are 640 and 682 types unique transcription factors predicted to interact with the 2-kb promoter region of the upstream transcription start site of the BMP genes in P. sinensis (Supplementary Table S1) and Homo sapiens (Supplementary Table S2), respectively. Additionally, the top 20 transcription factors were chosen for visualization (Figures 9A, B). The transcription factor PRDM9 and ZNF148 were the most frequently transcription factor in P. sinensis and Homo, respectively. The zinc finger protein (ZFP) was associated with the broadest varieties of transcription factors of the top 20, both in P. sinensis and Homo. Also, the transcription factors involved in classic BMP signaling were obtained, containing SMAD2 and SMAD4 in these two species. Intriguingly, the transcription factors associated with sex differentiation including RA receptor alpha (RARA), FOXL2, and DMRT1 were also predicted in both P. sinensis and Homo.
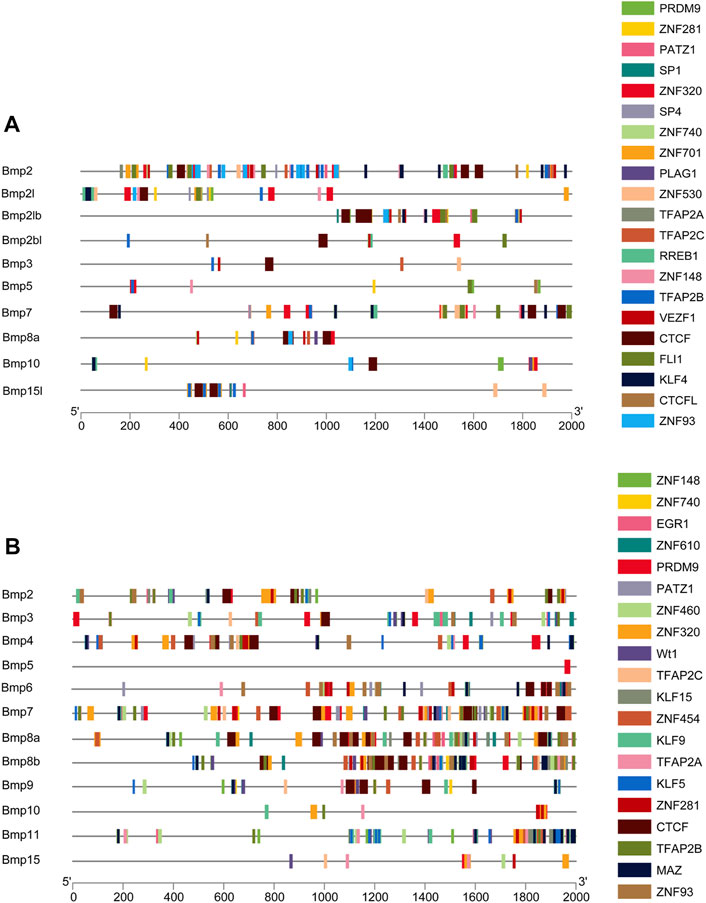
FIGURE 9. Promoter analysis of BMP genes in P. sinensis (A) and Homo (B). Different colored rectangles indicate different transcription factors.
3.9 Expression analysis
To study sexual dimorphisms in the expression profiles of BMP genes in P. sinensis male and female gonads, we analyzed transcriptome data of the early developmental gonads. As shown in Figure 8, Bmp2, Bmp3, and Bmp15l were strongly expressed in the ovaries. The expression of Bmp5, Bmp6 and Bmp8a in ovaries was higher than in testes (8 times, 6 times and 7 times, respectively), and Bmp2l, Bmp2lb, and Bmp7 in testes were expressed more than in ovaries (8 times, 11 times, and 37 times, respectively). Nevertheless, Bmp2bl and Bmp10 were barely expressed in ovaries (Figure 10).
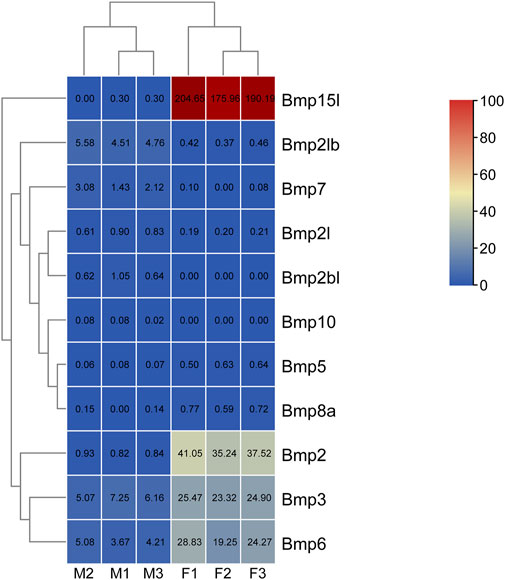
FIGURE 10. Expression analysis of BMP genes in P. sinensis. The expression levels of BMP genes are presented with FPKM values. The color scale ranges from 0 to 100, red indicates high expression, and blue indicates low expression. F1, ovary1; F2, ovary2; F3, ovary3; M1, testis1; M2, testis2; M3, testis3.
To further validate the RNA-Seq results and initially explore the expression of the Bmp gene family in male and female gonads at different developmental stages, the expression patterns of the BMPs in male and female gonads at 9-day, 16-day and 6-month-old was examined by qRT-PCR. Consistent with the RNA-Seq results, the relative expression of Bmp2, Bmp3, Bmp15l, Bmp5, Bmp6 and Bmp8a were extremely significantly higher in the ovary than in the testis of six-month-old P. sinensis (p <0.05). In contrast, Bmp2l, Bmp2lb, Bmp7, Bmp2bl and Bmp10 were male-biased genes in 6-month-old P. sinensis by qRT-PCR, which was matched with the RNA-Seq results. Moreover, the expression of these male-biased genes was very weak compared with female-biased genes (Figure 11A). As the gonads develop, the expression of Bmp2, Bmp3 and Bmp5 in the ovary gradually increases. Interestingly, the expression of Bmp2 and Bmp5 showed a significant difference at 9-day (p <0.05). The relative expression of Bmp2l, Bmp2lb and Bmp2bl was highest at 16-day in testes from different developmental periods. Another interesting finding was that on 9-day, the relative expression of Bmp7 was extremely significantly higher in female gonads than in male (p <0.01), which is the exact opposite of the expression pattern of Bmp7 in six-month-old gonads (Figure 11B).
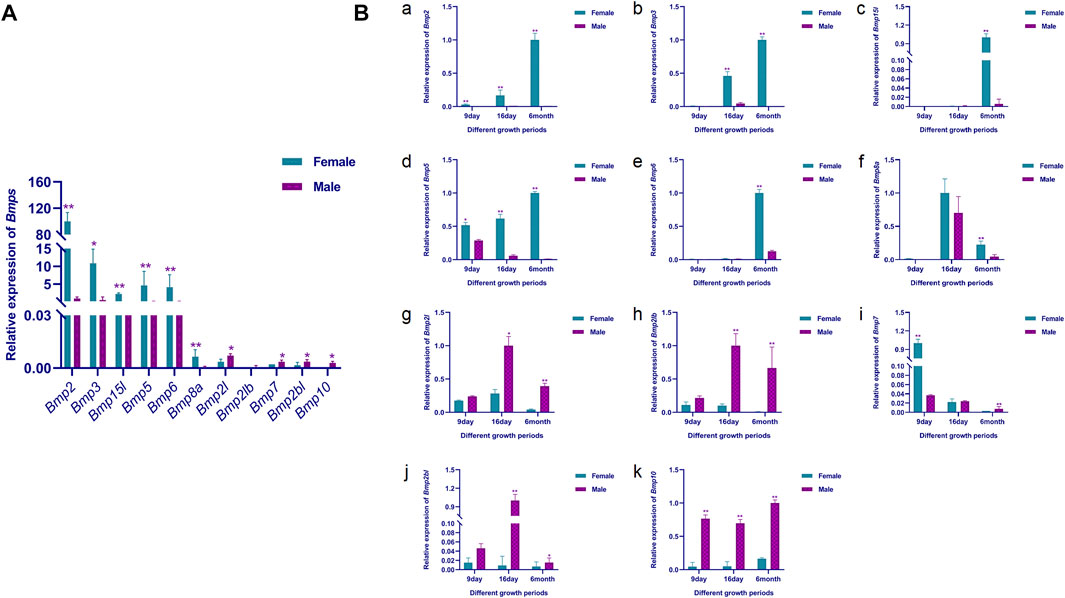
FIGURE 11. qRT-PCR validation results. (A). Relative expression of BMPs in the male and female gonads of the 6-month-old P. sinensis. (B). Relative expression of BMPs in male and female gonads at different developmental periods (A–K) Represent the relative expression of Bmp2, Bmp3, Bmp15l, Bmp5, Bmp6, Bmp8a, Bmp2l, Bmp2lb, Bmp7, Bmp2bl and Bmp10 in gonads at different developmental stages, respectively.11 Tables.
4 Discussion
BMPs are key factors in bone formation and differentiation (Lademann et al., 2020), but their roles in sex differentiation are less reported, especially in turtles. In the present study, we successfully identified 11 members of the BMP gene family in the P. sinensis genome. In contrast to other species, P. sinensis is missing some BMP genes, such as Bmp8b, and Bmp9 (Ehata and Miyazono, 2022). It remains unclear whether this is a loss of evolution or due to a lack of genomic data in P. sinensis. The subfamily of Bmp2l seems to be unique to turtles and frogs. They are distinct from the subfamily of Bmp2 and clustered individually as a branch. Conserved regions of genes can be identified through gene distribution and permutation (Xu et al., 2022). Collinear analysis demonstrated that adjacent genes block of Bmp5 were the most conserved in P. sinensis, Homo and Gallus gallus, implying that Bmp5 may be regulated by similar mechanisms and play similar functions in these species.
Gene duplication occurs in three main forms including tandem, fragment, and genome duplication, which are the primary drivers of gene family expansion (Lee et al., 2015) and provide the raw materials for the evolution of gene function (Jin S. et al., 2022). The presence of two or more genes within 200 kb on a chromosome means that there are tandem duplicate gene pairs in the region (Holub, 2001). Nevertheless, we found 10 BMP genes that were unevenly distributed on 10 different genomic scaffolds, showing that the expansion of BMP gene family was not through tandem duplication in P. sinensis. Additionally, they shared distinct gene structures, amino acid sequences, protein structures, conserved motifs, and domains, suggesting that they might have functional differences. Previous study showed that BMPs had a predomain (Mulloy and Rider, 2015), of the TGF-β propeptide superfamily, which was also found in Bmp2, Bmp2l, Bmp2lb, Bmp5, Bmp7, Bmp8a, and Bmp10.
Concurrently, transcription factors can bind to cis-acting elements to regulate gene expression and participate in developmental processes (Spitz and Furlong, 2012). Among the 640 predicted transcription factors in P. sinensis, the classical BMP signaling pathway members were included. Classical BMP signaling is a highly conserved cascade reaction involving BMP ligands, two types of receptors (type I and type II), and the signal transduction molecules Smads, and this signaling regulates multiple biological events (Massagué, 2000; Shi and Massagué, 2003). More importantly, the transcription factors related to sex differentiation were identified in P. sinensis and Homo. RA functions through its receptor RARA (Huebner et al., 2018). RA is critical for the production of oocytes and sperm in mammals (Endo et al., 2019). In Mauremys mutica, the expression levels of some meiosis genes in ovarian cells were significantly increased by RA treatment (Liu et al., 2022). Exogenous RA treatment activates Dmrt1a and Amh, thereby inhibiting germ cell differentiation, and genetic knockout of the RA degrading enzyme Cyp26a promoted meiosis and oogenesis in medaka (Adolfi et al., 2020). FOXL2 and DMRT1 are the critical members of the feminization and masculinization pathways, respectively (Li and Gui, 2018). In mammals, deletion of DMRT1 led to male to female sex reversal (Huang et al., 2017), and knockout of FOXL2 induced the transformation of ovarian cells into testis-like cells (Chen et al., 2022). In P. sinensis, Dmrt1 (Sun et al., 2017) and Foxl2 (Jin L. et al., 2022) have been shown to be essential genes for male and female differentiation, respectively. In fish, functional deficiency of DMRT1 resulted in testicular degeneration and proliferation of steroidogenic cells, and loss of FOXL2 caused complete sex reversal in females (Li et al., 2013). Likewise, similar experimental results were also presented in birds (Smith et al., 2009; Luo et al., 2020) and other reptiles (Ge et al., 2018).
Furthermore, it is well-known that the expression pattern of genes is closely related to their function (Zhang et al., 2022b). BMP genes presented a sexually dimorphic expression pattern in the male and female gonads of P. sinensis. As female-biased genes, the expression of Bmp2, Bmp3 and Bmp5 increased with ovarian development, and showed differential expression between males and females at 16-day, implying that these genes may be closely associated with ovarian differentiation and development in P. sinensis. In mouse, Bmp2 plays an important role in ovarian development, Foxl2 and Bmp2 act cooperatively to regulate Follistatin gene expression during ovarian development (Kashimada et al., 2011). Bmp2 interacts with the downstream effector Zglp1 to determine the oogenic fate of mice (Nagaoka et al., 2020). Loss of Sox9 enhanced the expression of Bmp2 and follistatin (Chaboissier et al., 2004). In the chicken embryo, Bmp3 is preferentially expressed in the developing ovary (Carré et al., 2011), and in Muscovy ducks, Bmp3 induced differentiated gonads to develop as females (Bai et al., 2020). Research has shown that Bmp5 might play a fully paracrine role in rodent ovarian folliculogenesis (Magro-Lopez and Muñoz-Fernández 2021). Simultaneously, Bmp6, Bmp8a and Bmp15l were significantly more expressed in the ovaries than in the testes of six-month-old P. sinensis, and it is speculated that these three genes may play an important role in the physiological activities of the ovaries. Bmp6 promotes 17β-estradiol and progesterone secretion in goat ovarian granulosa cells (Song et al., 2022). In rat, that Bmp8 may promote female fertility by inducing cumulus cell expansion through the Smad1/5/8 pathway (Wu and Luo 2014). Mutation analysis demonstrated that deletion of Bmp15 caused a failure of oocytes to enter early oogenesis (Kossack and Draper, 2019). In Cynoglossus semilaevis, Bmp15 was significantly expressed in female gonads, and knockdown of Gdf9 induced upregulation of Bmp15 (Shi et al., 2022). The 16-day of embryonic development is a critical period for the sex differentiation of P. sinensis (Zhou et al., 2019). Interestingly, the expression of Bmp2l, Bmp2lb and Bmp2bl was highest in testes at 16-day, suggesting that these genes may be involved in the sex differentiation of P. sinensis. The expression pattern of Bmp7 in gonads at different developmental stages indicates that Bmp7 may play different roles in gonads at different developmental periods. Bmp7 expression was significantly higher in females than in males at 9-day, suggesting that Bmp7 is involved in female-related physiological activities before gonadal differentiation. In addition, the expression of Bmp7 in the testis was significantly higher than that in the ovary in 6-month-old gonads, suggesting that Bmp7 may be involved in physiological activities related to testicular development or spermatogenesis in P. sinensis. In chickens, Bmp7 is specifically expressed in the ovary (Hoshino et al., 2005). In mouse, Bmp7 regulates germ cell proliferation in fetal gonads (Ross et al., 2007). Bmp7 was a downstream effector in the androgen signaling pathway of medaka and participated in the development of sex characteristics (Ogino et al., 2014). Mutation of Bmp7 accentuated the spermatogenic defects caused by the mutation of Bmp8a, implying that Bmp7 plays a role in spermatogenesis via a signaling pathway similar to that of Bmp8a (Zhao et al., 2001). Accordingly, the differential expression patterns of BMP genes in male and female gonads suggest a potential role in sex differentiation in P. sinensis and might function through the above transcription factors.
5 Conclusion
We performed an initial characterization of BMP genes to explore their evolution and functions, especially in sex differentiation, in P. sinensis. Our study not only provides complete data of BMP genes in the P. sinensis genome but also offers a novel idea to study the regulatory mechanisms of sex differentiation in P. sinensis, even in other turtles.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XZ and WL designed the experiments. CC, YW, XL, HC, XH, LY, CW, and MQ collected samples. JZ, LL, YW, CW, and YL performed the experiments and analyzed the data; LL wrote the main manuscript. XZ and WL revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Key R and D Program of China (2018YFD0900201), the National Natural Science Foundation of China (32102792), Funding by Science and Technology Projects in Guangzhou (202102021220), the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2020TD35), the National Freshwater Genetic Resource Center (NFGR-2022), the Guangdong Agricultural Research System (2019KJ150), the China-ASEAN Maritime Cooperation Fund (CAMC-2018F).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1109478/full#supplementary-material
References
Adolfi, M. C., Herpin, A., Martinez-Bengochea, A., Kneitz, S., Regensburger, M., Grunwald, D. J., et al. (2020). Crosstalk between retinoic acid and sex-related genes controls germ cell fate and Gametogenesis in medaka. Front. Cell Dev. Biol. 8, 613497. doi:10.3389/fcell.2020.613497
Ampuja, M., and Kallioniemi, A. (2018). Transcription factors-Intricate players of the bone morphogenetic protein signaling pathway. Genes Chromosom. Cancer 57 (1), 3–11. doi:10.1002/gcc.22502
Bai, D. P., Chen, Y., Hu, Y. Q., He, W. F., Shi, Y. Z., Fan, Q. M., et al. (2020). Transcriptome analysis of genes related to gonad differentiation and development in Muscovy ducks. BMC genomics 21 (1), 1–17. doi:10.1186/s12864-020-06852-z
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi:10.1093/nar/gkv416
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi:10.1093/bioinformatics/btu170
Carré, G. A., Couty, I., Hennequet-Antier, C., and Govoroun, M. S. (2011). Gene expression profiling reveals new potential players of gonad differentiation in the chicken embryo. PLoS One 6 (9), e23959. doi:10.1371/journal.pone.0023959
Castro-Mondragon, J. A., Riudavets-Puig, R., Rauluseviciute, I., Lemma, R. B., Turchi, L., Blanc-Mathieu, R., et al. (2022). Jaspar 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50 (D1), D165–d173. doi:10.1093/nar/gkab1113
Chaboissier, M. C., Kobayashi, A., Vidal, V. I., Lützkendorf, S., van de Kant, H. J., Wegner, M., et al. (2004). Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131 (9), 1891–1901. doi:10.1242/dev.01087
Chakraborty, P., Anderson, R. L., and Roy, S. K. (2022). Bone Morphogenetic protein 2 and estradiol-17β -induced changes in ovarian transcriptome during primordial follicle formation. Biol. Reprod. 107, 800–812. doi:10.1093/biolre/ioac111
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit developed for interactive Analyses of Big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, H., Chen, Q., Zhu, Y., Yuan, K., Li, H., Zhang, B., et al. (2022). MAP3K1 variant causes Hyperactivation of Wnt4/β-Catenin/FOXL2 signaling contributing to 46,XY Disorders/differences of sex development. Front. Genet. 13, 736988. doi:10.3389/fgene.2022.736988
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14 (6), 1188–1190. doi:10.1101/gr.849004
Donoughe, S., Nakamura, T., Ewen-Campen, B., Green, D. A., Henderson, L., and Extavour, C. G. (2014). BMP signaling is required for the generation of primordial germ cells in an insect. Proc. Natl. Acad. Sci. U. S. A. 111 (11), 4133–4138. doi:10.1073/pnas.1400525111
Dranow, D. B., Hu, K., Bird, A. M., Lawry, S. T., Adams, M. T., Sanchez, A., et al. (2016). Bmp15 is an Oocyte-Produced signal required for maintenance of the Adult female sexual phenotype in zebrafish. PLoS Genet. 12 (9), e1006323. doi:10.1371/journal.pgen.1006323
Edgar, R. C. (2004). Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 5, 113. doi:10.1186/1471-2105-5-113
Ehata, S., and Miyazono, K. (2022). Bone morphogenetic protein signaling in cancer; some Topics in the recent 10 Years. Front. Cell Dev. Biol. 10, 883523. doi:10.3389/fcell.2022.883523
Endo, T., Mikedis, M. M., Nicholls, P. K., Page, D. C., and de Rooij, D. G. (2019). Retinoic acid and germ cell development in the ovary and testis. Biomolecules 9 (12), 775. doi:10.3390/biom9120775
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Ge, C., Ye, J., Weber, C., Sun, W., Zhang, H., Zhou, Y., et al. (2018). The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 360 (6389), 645–648. doi:10.1126/science.aap8328
Geourjon, C., and Deléage, G. (1995). Sopma: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11 (6), 681–684. doi:10.1093/bioinformatics/11.6.681
Grgurevic, L., Christensen, G. L., Schulz, T. J., and Vukicevic, S. (2016). Bone morphogenetic proteins in inflammation, glucose homeostasis and adipose tissue energy metabolism. Cytokine Growth Factor Rev. 27, 105–118. doi:10.1016/j.cytogfr.2015.12.009
Han, Y., Peng, C., Wang, L., Guo, J., Lu, M., Chen, J., et al. (2018). Female-to-male sex reversal in orange-spotted grouper (Epinephelus coioides) caused by overexpressing of Amh in vivo. Biol. Reprod. 99 (6), 1205–1215. doi:10.1093/biolre/ioy157
Holub, E. B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2 (7), 516–527. doi:10.1038/35080508
Hoshino, A., Koide, M., Ono, T., and Yasugi, S. (2005). Sex-specific and left-right asymmetric expression pattern of Bmp7 in the gonad of normal and sex-reversed chicken embryos. Dev. Growth Differ. 47 (2), 65–74. doi:10.1111/j.1440-169x.2004.00783.x
Hua, C., Brown, C. W., and Matzuk, M. M. (2002). Genetic analysis of the mammalian transforming growth factor-β superfamily. Endocr. Rev. 23 (6), 787–823. doi:10.1210/er.2002-0003
Huang, S., Ye, L., and Chen, H. (2017). Sex determination and maintenance: The role of DMRT1 and FOXL2. Asian J. Androl. 19 (6), 619–624. doi:10.4103/1008-682x.194420
Huebner, H., Hartner, A., Rascher, W., Strick, R. R., Kehl, S., Heindl, F., et al. (2018). Expression and regulation of retinoic acid receptor Responders in the human Placenta. Reprod. Sci. 25 (9), 1357–1370. doi:10.1177/1933719117746761
Jin, L., Sun, W., Bao, H., Liang, X., Li, P., Shi, S., et al. (2022b). The forkhead factor Foxl2 participates in the ovarian differentiation of Chinese soft-shelled turtle Pelodiscus sinensis. Dev. Biol. 492, 101–110. doi:10.1016/j.ydbio.2022.10.001
Jin, S., Deng, Z., Xu, S., Zhang, H., and Han, Z. (2022a). Genome-wide identification and low-salinity stress analysis of the Hsp70 gene family in swimming crab (Portunus trituberculatus). Int. J. Biol. Macromol. 208, 126–135. doi:10.1016/j.ijbiomac.2022.03.055
Kashimada, K., Pelosi, E., Chen, H., Schlessinger, D., Wilhelm, D., and Koopman, P. (2011). FOXL2 and BMP2 act cooperatively to regulate follistatin gene expression during ovarian development. Endocrinology 152 (1), 272–280. doi:10.1210/en.2010-0636
Kim, D., Langmead, B., and Salzberg, S. L. (2015). Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Kossack, M. E., and Draper, B. W. (2019). Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr. Top. Dev. Biol. 134, 119–149. doi:10.1016/bs.ctdb.2019.02.004
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis Version 7.0 for Bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lademann, F., Hofbauer, L. C., and Rauner, M. (2020). The bone morphogenetic protein pathway: The Osteoclastic Perspective. Front. Cell Dev. Biol. 8, 586031. doi:10.3389/fcell.2020.586031
Lee, T., Yang, S., Kim, E., Ko, Y., Hwang, S., Shin, J., et al. (2015). AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 43, D996–D1002. doi:10.1093/nar/gku1053
Lei, L., Chen, C., Zhu, J., Wang, Y., Liu, X., Liu, H., et al. (2022). Transcriptome analysis reveals key genes and pathways related to sex differentiation in the Chinese soft-shelled turtle (Pelodiscus sinensis). Comp. Biochem. Physiology Part D Genomics Proteomics 42, 100986. doi:10.1016/j.cbd.2022.100986
Letunic, I., Khedkar, S., and Bork, P. (2021). Smart: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49 (D1), D458–D460. doi:10.1093/nar/gkaa937
Li, M. H., Yang, H. H., Li, M. R., Sun, Y. L., Jiang, X. L., Xie, Q. P., et al. (2013). Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 154 (12), 4814–4825. doi:10.1210/en.2013-1451
Li, W., Zhu, X. P., Xu, H. Y., Zhao, J., and Chen, C. (2020). “A PCR Amplification primer, method and kit for Rapid identification of genetic Traits in the Chinese soft-shelled turtle,”. CN108841945B, China.
Li, X. Y., and Gui, J. F. (2018). Diverse and variable sex determination mechanisms in vertebrates. Sci. China Life Sci. 61 (12), 1503–1514. doi:10.1007/s11427-018-9415-7
Liu, X., Liu, F., Xu, H., Yang, Y., Wang, Y., Hong, X., et al. (2022). Characterization of the in Vitro Cultured ovarian cells in the Asian yellow Pond turtle (Mauremys mutica). Biology 11 (10), 1404. doi:10.3390/biology11101404
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCT method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Luo, W., Gu, L., Li, J., and Gong, Y. (2020). Transcriptome sequencing revealed that knocking down FOXL2 affected cell proliferation, the cell cycle, and DNA replication in chicken pre-ovulatory follicle cells. PLoS One 15 (7), e0234795. doi:10.1371/journal.pone.0234795
Magro-Lopez, E., and Muñoz-Fernández, M. Á. (2021). The role of BMP signaling in female reproductive system development and function. Int. J. Mol. Sci. 22 (21), 11927. doi:10.3390/ijms222111927
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 (D1), D200–d203. doi:10.1093/nar/gkw1129
Massagué, J. (2000). How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1 (3), 169–178. doi:10.1038/35043051
Miyauchi, H., Ohta, H., Nagaoka, S., Nakaki, F., Sasaki, K., Hayashi, K., et al. (2017). Bone morphogenetic protein and retinoic acid synergistically specify female germ-cell fate in mice. Embo J. 36 (21), 3100–3119. doi:10.15252/embj.201796875
Momen, B., Anderson, P. D., and Helms, J. A. (2001). Application of response surface methodology and ANOVA to detect pollution effects on photosynthetic response under varying temperature and light regimes. For. Ecol. Manag. 152, 331–337. doi:10.1016/S0378-1127(00)00617-4
Morikawa, M., Derynck, R., and Miyazono, K. (2016). TGF-Β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 8 (5), a021873. doi:10.1101/cshperspect.a021873
Mulloy, B., and Rider, C. C. (2015). The bone morphogenetic proteins and their Antagonists. Vitam. Horm. 99, 63–90. doi:10.1016/bs.vh.2015.06.004
Nagahama, Y., Chakraborty, T., Paul-Prasanth, B., Ohta, K., and Nakamura, M. (2021). Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 101 (3), 1237–1308. doi:10.1152/physrev.00044.2019
Nagaoka, S. I., Nakaki, F., Miyauchi, H., Nosaka, Y., Ohta, H., Yabuta, Y., et al. (2020). ZGLP1 is a determinant for the oogenic fate in mice. Science 367 (6482), eaaw4115. doi:10.1126/science.aaw4115
Ogino, Y., Hirakawa, I., Inohaya, K., Sumiya, E., Miyagawa, S., Denslow, N., et al. (2014). Bmp7 and Lef1 are the downstream effectors of androgen signaling in androgen-induced sex characteristics development in medaka. Endocrinology 155 (2), 449–462. doi:10.1210/en.2013-1507
Pan, Q., Kay, T., Depincé, A., Adolfi, M., Schartl, M., Guiguen, Y., et al. (2021). Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Philos. Trans. R. Soc. Lond B Biol. Sci. 376 (1832), 20200091. doi:10.1098/rstb.2020.0091
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11 (9), 1650–1667. doi:10.1038/nprot.2016.095
Robinson, M. D., Davis, J. M., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Ross, A., Munger, S., and Capel, B. (2007). Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex. Dev. 1 (2), 127–137. doi:10.1159/000100034
Shi, R., Li, X., Cheng, P., Yang, Q., Chen, Z., Chen, S., et al. (2022). Characterization of growth differentiation factor 9 and bone morphogenetic factor 15 in Chinese tongue sole (Cynoglossus semilaevis): Sex-biased expression pattern and promoter regulation. Theriogenology 182, 119–128. doi:10.1016/j.theriogenology.2022.02.003
Shi, Y., and Massagué, J. (2003). Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113 (6), 685–700. doi:10.1016/s0092-8674(03)00432-x
Smith, C. A., Roeszler, K. N., Ohnesorg, T., Cummins, D. M., Farlie, P. G., Doran, T. J., et al. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461 (7261), 267–271. doi:10.1038/nature08298
Song, S., Ding, W., Yao, H., Wang, L., Li, B., Wang, Y., et al. (2022). BMP6 promotes the secretion of 17 beta-estradiol and progesterone in goat ovarian granulosa cells. Animals 16, 2132. doi:10.3390/ani12162132
Spitz, F., and Furlong, E. E. (2012). Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13 (9), 613–626. doi:10.1038/nrg3207
Sun, W., Cai, H., Zhang, G., Zhang, H., Bao, H., Wang, L., et al. (2017). Dmrt1is required for primary male sexual differentiation in Chinese soft-shelled turtle Pelodiscus sinensis. Sci. Rep. 7 (1), 4433. doi:10.1038/s41598-017-04938-5
Urist, M. R. (1965). Bone: Formation by autoinduction. Science 150 (3698), 893–899. doi:10.1126/science.150.3698.893
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–w303. doi:10.1093/nar/gky427
Wu, F.-J., and Luo, C.-W. (2014). Characterization of the signaling and ovarian functions of bone morphogenetic protein 8. Endocr. Abstr. Biosci. 35, 665. doi:10.1530/endoabs.35.P665
Xu, S., Zhang, S., Zhang, W., Liu, H., Wang, M., Zhong, L., et al. (2022). 'Genome-Wide identification, phylogeny, and expression profile of the Dmrt (Doublesex and Mab-3 related transcription factor) gene family in channel Catfish (Ictalurus punctatus). Front. Genet. 13, 891204. doi:10.3389/fgene.2022.891204
Yang, D., Yang, X., Dai, F., Wang, Y., Yang, Y., Hu, M., et al. (2021). The role of bone morphogenetic protein 4 in ovarian function and Diseases. Reprod. Sci. 28 (12), 3316–3330. doi:10.1007/s43032-021-00600-8
Yu, H., Wang, Y., Wang, M., Liu, Y., Cheng, J., and Zhang, Q. (2020). Growth differentiation factor 9 (gdf9) and bone morphogenetic protein 15 (bmp15) are potential intraovarian regulators of steroidogenesis in Japanese flounder (Paralichthys olivaceus). Gen. Comp. Endocrinol. 297, 113547. doi:10.1016/j.ygcen.2020.113547
Zhang, C., Wang, W., Wang, D., Hu, S., Zhang, Q., Wang, Z., et al. (2022a). Genome-wide identification and characterization of the WRKY gene family in Scutellaria baicalensis Georgi under Diverse Abiotic stress. Int. J. Mol. Sci. 23 (8), 4225. doi:10.3390/ijms23084225
Zhang, C., Xu, X., Xu, X., Li, Y., Zhao, P., Chen, X., et al. (2022b). Genome-wide identification, evolution analysis of cytochrome P450 monooxygenase multigene family and their expression patterns during the early somatic embryogenesis in Dimocarpus longan Lour. Gene 826, 146453. doi:10.1016/j.gene.2022.146453
Zhang, Y., Xiao, L., Sun, W., Li, P., Zhou, Y., Qian, G., et al. (2021). Knockdown of R-spondin1 leads to partial sex reversal in genetic female Chinese soft-shelled turtle Pelodiscus sinensis. Gen. Comp. Endocrinol. 309, 113788. doi:10.1016/j.ygcen.2021.113788
Zhao, G. Q., Chen, Y. X., Liu, X. M., Xu, Z., and Qi, X. (2001). Mutation in Bmp7 exacerbates the phenotype of Bmp8a mutants in spermatogenesis and epididymis. Dev. Biol. 240 (1), 212–222. doi:10.1006/dbio.2001.0448
Zhou, X., and Zhu, Y. (2011). Sex-specific growth characteristics of Chinese soft-shelled turtle. Pelodiscus Sin. Chin. Aquac. 32 (05), 11–13.
Zhou, Y., Sun, W., Cai, H., Bao, H., Zhang, Y., Qian, G., et al. (2019). The role of Anti-Müllerian Hormone in testis differentiation reveals the significance of the TGF-β pathway in Reptilian sex determination. Genetics 213 (4), 1317–1327. doi:10.1534/genetics.119.302527
Keywords: Pelodiscus sinensis, bone morphogenetic protein, gene family, genome-wide, transcriptome, sex differentiation
Citation: Lei L, Zhu J, Chen C, Wang Y, Wu C, Qi M, Wang Y, Liu X, Hong X, Yu L, Chen H, Wei C, Liu Y, Li W and Zhu X (2023) Genome-wide identification, evolution and expression analysis of bone morphogenetic protein (BMP) gene family in chinese soft-shell turtle (Pelodiscus sinensis). Front. Genet. 14:1109478. doi: 10.3389/fgene.2023.1109478
Received: 27 November 2022; Accepted: 19 January 2023;
Published: 01 February 2023.
Edited by:
Zheng-Yong Wen, Neijiang Normal University, ChinaReviewed by:
Yunyun Lv, Neijiang Normal University, ChinaShan He, Huazhong Agricultural University, China
Bingjian Liu, Zhejiang Ocean University, China
Zhong-duo Wang, Guangdong Ocean University, China
Wanbo Li, Jimei University, China
Copyright © 2023 Lei, Zhu, Chen, Wang, Wu, Qi, Wang, Liu, Hong, Yu, Chen, Wei, Liu, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinping Zhu, emh1eGlucGluZ0BwcmZyaS5hYy5jbg==; Wei Li, bGl3ZWlAcHJmcmkuYWMuY24=
†These authors contributed equally to this work
 Luo Lei1,2†
Luo Lei1,2† Ming Qi
Ming Qi Xinping Zhu
Xinping Zhu