- 1Zhejiang Provincial Key Laboratory of Plant Evolutionary Ecology and Conservation, Taizhou University, Taizhou, Zhejiang, China
- 2School of Advanced Study, Taizhou University, Taizhou, Zhejiang, China
Dodders (Cuscuta australis R. Br.) are holo-parasitic stem angiosperms with an extensive host range that have significant ecological and economic potential impact on the ecosystem and the agricultural system. However, how the host plant responds to this biotic stress remains mostly unexplored. To identify the defense-related genes and the pathways in white clover (Trifolium repens L.) induced by dodder parasitism, we performed a comparative transcriptome analysis of the leaf and root tissues from white clover with and without dodder infection by high throughput sequencing. We identified 1,329 and 3,271 differentially expressed genes (DEGs) in the leaf and root tissues, respectively. Functional enrichment analysis revealed that plant-pathogen interaction, plant hormone signal transduction, and phenylpropanoid biosynthesis pathways were significantly enriched. Eight WRKY, six AP2/ERF, four bHLH, three bZIP, three MYB, and three NAC transcription factors showed a close relationship with lignin synthesis-related genes, which defended white clover against dodder parasitism. Real-time quantitative PCR (RT-qPCR) for nine DEGs, further validated the data obtained from transcriptome sequencing. Our results provide new insights into understanding the complex regulatory network behind these parasite-host plant interactions.
Introduction
Cuscuta spp. (family Convolvulaceae), commonly known as dodders, are obligate holoparasites with limited photosynthesis (Revill et al., 2005), that acquire all their nutrients and water from host plants via haustoria (Dawson et al., 1994; KC, 2002; Runyon et al., 2006). With a broad host range of crops, Cuscuta spp. have been among the most devastating agricultural pests, damaging the worldwide annual output of agricultural crops by millions of dollars (Press & Phoenix, 2005; Bardgett et al., 2006; Zagorchev et al., 2021a). White clover (Trifolium repens L.) is a preferred host of C. australis in the field (Yu et al., 2019). Understanding the molecular mechanisms underlying the interaction between Cuscuta spp. and their host plants would provide the technical foundation for the effective management of these crop pests.
Since Cuscuta spp. could adversely affect host plants like herbivores and pathogens, some reports have shown that the host plants defend against parasitic plants very similarly to those induced by herbivores and pathogens (Pennings & Callaway, 2002; Runyon et al., 2010a; Runyon et al., 2010b). Runyon et al. found that parasitism by C. pentagona sequentially induced the jasmonic acid (JA) and salicylic acid (SA) defense pathways in tomato. JA peaked 12 h before SA, suggesting that the defense response is coordinated by the sequential induction of both phytohormones (Runyon et al., 2010b). During caterpillar feeding on dodder, the host JA pathway regulates the transcriptomic changes both in dodder and the host plant (Qin et al., 2019). The contact and penetration of the haustoria, removal of the resources from the host, and the subsequent hormonal changes trigger the host defense responses (Saric-Krsmanovic et al., 2020; Zagorchev et al., 2021b). The host plants’ response to the Cuscuta spp. infection includes a hypersensitive-like response (HLR) and the expression of pathogenesis-related (PR) genes (Bringmann et al., 1999; Borsics & Lados, 2002). Most of the plant disease resistance genes (R genes) encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins, which can be further subdivided into TIR-NB-LRR proteins (TNLs) and CC-NB-LRR proteins (CNLs) (McHale, 2006; Kourelis & van der Hoorn, 2018). They are involved in the plant immune responses to attacks by invaders, including viruses, bacteria, oomycetes, fungi, nematodes, herbivores, and parasitic plants. The first receptor (CUSCUTA RECEPTOR 1, CuRe 1), an LRR-receptor-like serine/threonine-protein kinase (RLP) was identified in tomato, which initiates the pathogen-associated molecular pattern (PAMP)-triggered immunity of tomato against C. reflexa (Hegenauer andKaiser, 2016; Hegenauer et al., 2020). Based on the comparative transcriptomics of dodder-resistant and -susceptible tomato cultivars, it was found that the Cuscuta R-gene for Lignin-based Resistance 1 (CuRLR1, a CC-NBS-LRR) functioned as a receptor for recognizing C. campestris signals or effectors, resulting in lignification-based resistance (Jhu et al., 2022).
Secondary metabolites are vital in the plant’s environmental adaption and act as defense compounds against abiotic and biotic stress (Ramakrishna & Ravishankar, 2011; Dhakshinamoorthy et al., 2014). Phenolic secondary metabolites like phenols and lignin are important in the banana’s defense mechanisms against the nematode Radopholus similis. The phenol content in nematode-infected plants was twice the amount in the uninfected plants at 3 weeks post infection, due to the biosynthesis and accumulation of secondary metabolites at the infection sites (Dhakshinamoorthy et al., 2014). Lignin, a heterogeneous polymer of aromatic subunits derived from phenylalanine, primarily comprised three major monomers: p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monolignols that are synthesized via the phenylpropanoid pathway (Raes et al., 2003). Lignification has long been believed as a host defense mechanism against pathogens (Vance CP et al., 1980; Bhuiyan et al., 2009; Bellincampi et al., 2014). In cotton, the GhLAC15 expression enhanced Verticillium wilt resistance by increasing the defense-induced cell wall lignification (Zhang et al., 2019). The lignin accumulation in plant roots and stems reportedly confer host resistance against parasitic plants (Pérez-de-Luque et al., 2007; Jhu et al., 2022). For example, the host lignin composition affects haustorium induction in the root parasitic plants Striga hermonthica and Phtheirospermum japonicum (Cui et al., 2018). A recent report showed that the resistant tomato cultivar Heinz presented a lignin-based resistance against the stem parasite field dodder (C. campestris) infection (Jhu et al., 2022). Additionally, the three transcription factors, MYB55, WRKY16, and Lignin Induction Factor 1 (LIF1, an AP2-like transcription factor) are important in the lignin biosynthesis (Jhu et al., 2022).
Herbivores-attacking induce defense responses not only in the attacked (local) tissues but also in the other non-attacked distal (systemic) plant parts, indicating that a systemic signal is induced in the herbivores-damaged tissues and the signal can be translocated to the other parts of the whole plant to activate defense (Wu & Baldwin, 2010; Zhuang et al., 2018). For example, above-ground chewing herbivores (caterpillar and weevil) and sucking herbivore (aphid) feeding on leaves not only induced above-ground defense response metabolites (quercetin, quercitrin, isoquercetin, and kaempferitrin) in leaves but also induced below-ground defense chemicals in roots (Xiao et al., 2019). Stronger metabolic changes were found in the apex and root tissues than in undamaged leaflets, indicating rapid and significant whole-plant responses to leaves damage caused by caterpillars (Adam D. Steinbrenner et al., 2011). Cuscuta campestris has been proven to transfer N-systemic signals between hosts grown in N-heterogeneous soil, resulting in large transcriptome and DNA methylome changes in the recipient hosts (Zhang et al., 2021). Whether the stem parasitic dodder parasitism of the host stem elicits a defense response from the host roots is largely unknown. In the current study, we analyzed the DEGs involved in the plant hormone signal transduction, plant-pathogen interaction, and phenylpropanoid biosynthesis pathways, and also including the R genes and transcription factors of both the roots and leaves of white clover after dodder parasitism, based on transcriptome sequencing. The results will provide important information for our understanding of how host plants respond to parasitic plants.
Materials and methods
Plant materials and treatment
The seeds of white clover (T. repens L.) and dodder (Cuscuta australis) were collected from Luohe Park, Linhai, Taizhou City, Zhejiang Province, China. White clover seeds were sown in pots filled with peat soil, sand, and vermiculite (2:2:1) and grown in a walk-in climate chamber kept at 25°C/20°C in day (16 h)/night (8 h) conditions. Dodder seeds were sown into peat soil. A 10–15-cm-long dodder seedling was wound counterclockwise around the white clover stems to allow the parasite to infect the host plants. Three pots were infected with dodder and the other three pots were set as controls that without dodder infection. Leaf and root tissues of the white clover were sampled after 2 weeks. Leaves or roots from nine white clover plants were pooled as one biological replicate. Three biological replicates were collected for each: (a) parasitized leaves (designated Pleaf1, Pleaf2, and Pleaf3), (b) control leaves (designated Cleaf1, Cleaf2, and Cleaf3), (c) parasitized roots (designated Proot1, Proot2, and Proot3) and (d) control roots (designated Croot1, Croot2, and Croot3). All collected samples were immediately frozen in liquid nitrogen and then stored at −80°C for further analyses.
RNA extraction and transcriptome sequencing
Three biological replicates each were taken for leaf and root samples. Total RNA was extracted using a TRIzol Kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The concentration, fragment size, and the total RNA integrity were measured using an Agilent 2,100 platform (Agilent Technologies Inc., Santa Clara, CA, United States). The degradation and contamination of total RNA were assessed by 1.2% agarose gel electrophoresis. The leaf and root cDNA libraries were generated using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, United States) and the resulting cDNAs libraries were sequenced on an Illumina Hiseq platform and the paired-end reads were generated (Novogene, China).
Transcriptome data analysis
Firstly, adapters, poly-N, and low-quality reads were removed from raw reads to generate clean reads. Simultaneously, Q20, Q30, and the GC-content of the clean reads were calculated. All the downstream analyses were based on clean reads with high quality. Then, clean reads were used for transcriptome de novo assemble using Trinity (v2.4.0) with min_kmer_cov set to 2 as default along with all the other parameters being set as default, which provided a reference sequence for the downstream analysis (Grabherr et al., 2011). The longest transcripts in the cluster units were regarded as unigenes to eliminate redundant sequences. The function of all the unigenes were annotated based on seven databases: NCBI non-redundant protein sequences (Nr), Clusters of orthologous groups of proteins (KOG), A manually annotated and reviewed protein sequence database (Swiss-Prot), NCBI non-redundant nucleotide sequences (Nt), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Protein family (Pfam). Clean reads for every leaf and root sample were mapped to the assembled reference sequence by RSEM (v1.2.15) with bowtie2 set to mismatch 0 as default (Li & Dewey, 2011), resulting in the read count of each unigene in every leaf and root samples, respectively. The expression levels of each unigene were normalized based on the fragments per kilobase of transcript per million mapped reads (FPKM). DEGs in the Pleaf vs. Cleaf and Proot vs. Croot were analyzed using the DESeq R package (1.12.0) (Love et al., 2014) with the thresholds of padj <0.05 and the absolute value of log2 (parasitized/unparasitized) > 1. Gene Ontology (GO) enrichment analysis of the DEGs was performed using the GOseq R packages based Wallenius non-central hypergeometric distribution (Young et al., 2010). GO terms with p-value <0.05 were considered as significantly enriched. Pathway enrichment of DEGs was performed using the KEGG database (Kanehisa et al., 2008). Then, the KOBAS software (v2.0.12) was used to test the statistical enrichment of the DEGs in the KEGG pathways (Mao et al., 2005). The pathways with p-value <0.05 were considered as significantly enriched.
Transcription factors (TFs) identification and analysis
Transcription factors were identified based on the DEGs using the iTAK software (1.2) (Zheng et al., 2016) with the plant TFDB (Jin et al., 2017). The Pearson’s correlation coefficients between TFs and genes were calculated and the correlation network was visualized using the Cytoscape software (version 3.8.2) (Shannon et al., 2003).
RT-qPCR validation of the transcriptome data
To validate the reliability of the transcriptome data, nine DEGs that enriched to the main pathways like “plant hormone signal transduction,” “plant–pathogen interaction,” and “starch and sucrose metabolism” were selected for expression analysis by RT-qPCR. The RNA samples used for RT-qPCR were consistent with those for RNA-seq. Total RNA was reversed transcribed into cDNA using a Fastking RT kit (with gDNase; Tiangen, Beijing, China). Then, qPCR was carried out on a CFX Connect Real-time system (Bio-Rad, United States) using a FastFire qPCR premix (SYBR Green) kit (Tiangen, Beijing, China). The qPCR procedure was set as follows: 95°C for 1 min, and 40 cycles of 95°C for 5 s and 60°C for 15 s. EF1 was used as the reference gene (Wang et al., 2020). Three biological and technical replicates each were performed for the leaf and root samples. The relative expression levels of DEGs were calculated using the 2−ΔΔCT method (Livak & Schmittgen, 2001). The primers used for RT-qPCR were designed by primer premier 6 and the primers are listed in Supplementary Table S1.
Results
General analysis of white clover transcriptome data
To further improve our understanding of the molecular response of white clover to dodder parasitism, we performed a comparative transcriptome analysis of the dodder-infected and uninfected root and leaf samples. Three biological replicates from each treatment resulted in 12 cDNA libraries. The transcriptome sequencing generated 2,096 and 2,235 million raw reads in the leaf and root samples, respectively (Supplementary Table S2). After removing the adapter sequences and the low-quality sequences, we retained 27.14 and 28.92 Gb clean data for subsequent analyses in the leaf and root samples, respectively. Quality assessment of the transcriptome data showed that the error rate was very low (0.01%), while it was very high (>97%) for the Q30, thereby indicating a high-quality transcriptome data being generated in this study. Then, all the clean reads were assembled using Trinity, thus generating 288,139 transcripts (N50: 1,420) with a mean length of 1,047 bp (Supplementary Figure S1A). Clustering resulted in 101,498 unigenes (N50: 1,253) with a mean length of 910 bp; while the length of the unigenes ranged from 301 to 106,769 bp (Supplementary Figure S1B). To functionally annotate the assembled unigenes, we compared using a sequence similarity against seven protein databases using BLASTX. As a result, we annotated 77,231 unigenes (76.09%) in at least one database, and 7,981 unigenes (7.86%) in all the other databases (Supplementary Figure S1C). According to NR annotation, we compared 24.2% of the annotated unigenes to Trifolium pretense, 21.7% to T. subterraneum, 16.9% to Quercus suber, 12.2% to Mesicago truncatula, 4.4% to Cicer arietinum, and 20.5% to other species (Suppementary Figure S1D). All the RNA sequencing raw data can be found in the NCBI BioProject database under accession number PRJNA700502 and PRJNA828029.
DEGs in response to dodder parasitism
The DEGs were determined with the criteria of |log 2 (FoldChange)| > 1 and padj <0.05. We identified 1,329 DEGs in the leaves from white clover with dodder parasitism, of which 796 and 533 DEGs were up- and downregulated, respectively (Figure 1A). We identified a higher number of DEGs (3,271) in the roots of white clover with dodder parasitism, of which 1,089 and 2,182 were up- and downregulated, respectively (Figure 1B). A venny diagram showed that 31 and 36 DEGs were common in up- and downregulated DEGs in leaves and roots, respectively (Figures 1C, D).
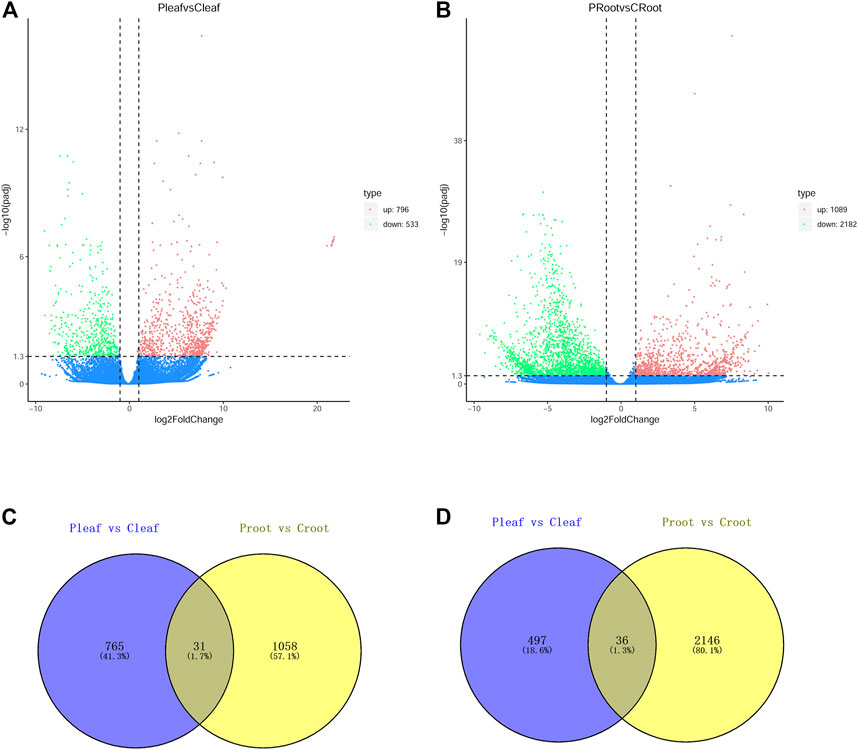
FIGURE 1. DEGs in leaves and roots of white clover with dodder parasitism. (A) Volcano plots displaying the up- and downregulated DEGs in Pleaf vs. Cleaf; (B) Volcano plots displaying the up- and downregulated DEGs in Proot vs. Croot; (C) Venn diagram showing the shared and unique upregulated DEGs between Pleaf vs. Cleaf and Proot vs. Croot; (D) Venn diagram showing the shared and unique downregulated DEGs between Pleaf vs. Cleaf and Proot vs. Croot.
Identification of the R genes in response to dodder parasitism
Among the upregulated DEGs in the Pleaf vs. Cleaf, we identified 2 R genes, 6 protein kinase genes, 7 PR genes, 17 ubiquitin pathway genes, and 11 calcium signaling genes (Supplementary Table S3). While we identified 49 R genes, 108 protein kinase genes, 6 PR genes, 9 ubiquitin pathway genes, and 11 calcium signaling genes in the downregulated DEGs (Supplementary Table S4). In Proot vs. Croot, we identified 64 R genes, 45 protein kinase genes, 14 PR genes, 14 ubiquitin pathway genes, and 18 calcium signaling genes among the upregulated DEGs (Supplementary Table S5), while we found 22 R proteins, 39 protein kinase genes, 61 PR genes, 55 ubiquitin pathway genes, and 44 calcium signaling genes among the downregulated DEGs (Supplementary Table S6).
GO and KEGG enrichment analysis of DEGs
To know the putative function classification and pathways of the DEGs, we performed GO enrichment analysis and KEGG pathway enrichment analysis, respectively. A total of 1836 GO terms were enriched based on the upregulated DEGs in Pleaf vs. Cleaf, of which 188 terms were significantly enriched (p-value <0.05; Supplementary Figure S2). In the biological process (BP) category, the DEGs were mainly enriched in metabolic process and biosynthetic process, including metabolic process (GO:0008152), cellular process (GO:0009987), organic substance metabolic process (GO:0071704), primary metabolic process (GO:0044238), and cellular metabolic process (GO:0044237). In the cellular component (CC) category, the DEGs were significantly enriched in cell (GO:0005623), cell part (GO:0044464), intracellular (GO:0005622) and intracellular part (GO:0044424), macromolecular complex (GO:0032991), cytoplasm (GO:0005737), and cytoplasmic part (GO:0044444). In the molecular function (MF) category, the DEGs were significantly enriched in structural molecule activity (GO:0005198), structural constituent of ribosome (GO:0003735) and oxidoreductase activity (GO:0016491; Supplementary Figure S2A). A total of 1,297 terms were enriched based on the downregulated DEGs in leaves, of which 88 terms were significantly enriched, including 48 MF, 35 BP and 5 CC (Supplementary Figure S2B). A total of 1792 GO terms were enriched based on the upregulated DEGs in Proot vs. Croot, of which 101 terms were significantly enriched. The upregulated genes were mostly enriched in BP, including response to stimulus (GO:0050896), cell communication (GO:0007154), signal transduction (GO:0007165), single organism signaling (GO:0044700), signalng (GO:0023052), DNA integration (GO:0015074) and DNA metabolic process (GO:0006259; Supplementary Figure S2C). Eighty-five terms were significantly enriched on the downregulated DEGs in roots, including 39 BP, 39 MF and 7 CC. In BP, 457genes were enriched in nodulation (GO:0009877) and nodulation morphogenesis (GO:0009878; Supplementary Figure S2).
We also subjected the DEGs to KEGG pathway enrichment analysis. In Pleaf vs. Cleaf, the upregulated DEGs were mainly enriched in ribosome (ko03010), cyanoamino acid metabolism (ko00460), galactose metabolism (ko00052) and phenylpropanoid biosynthesis (ko00940; Figure 2A), while the downregulated DEGs were significantly enriched in plant-pathogen interaction (ko04626), inositol phosphate metabolism (ko00562), DNA replication (ko03030) and phenylpropanoid biosynthesis (ko00940; Figure 2B). In Proot vs. Croot, the upregulated DEGs were significantly enriched in plant-pathogen interaction (ko04626), ABC transporters (ko02010), plant hormone signal transduction (ko04075) and endocytosis (ko04144; Figure 2C), while the downregulated DEGs were significantly enriched in linoleic acid metabolism “(ko00591), phenylpropanoid biosynthesis (ko00940), starch and sucrose metabolism (ko00500), isoquinoline alkaloid biosynthesis (ko00950), tropane, piperidine and pyridine alkaloid biosynthesis (ko00960), amino sugar and nucleotide sugar metabolism (ko00520), alpha-Linolenic acid metabolism (ko00592), tyrosine metabolism (ko00350), glucosinolate biosynthesis (ko00966), glycine, serine and threonine metabolism (ko00260), sulfur metabolism (ko00920), taurine and hypotaurine metabolism (koko00430), nitrogen metabolism (ko00910), diterpenoid biosynthesis (ko00904), inositol phosphate metabolism (ko00562) and DNA replication (ko03030; Figure 2D).
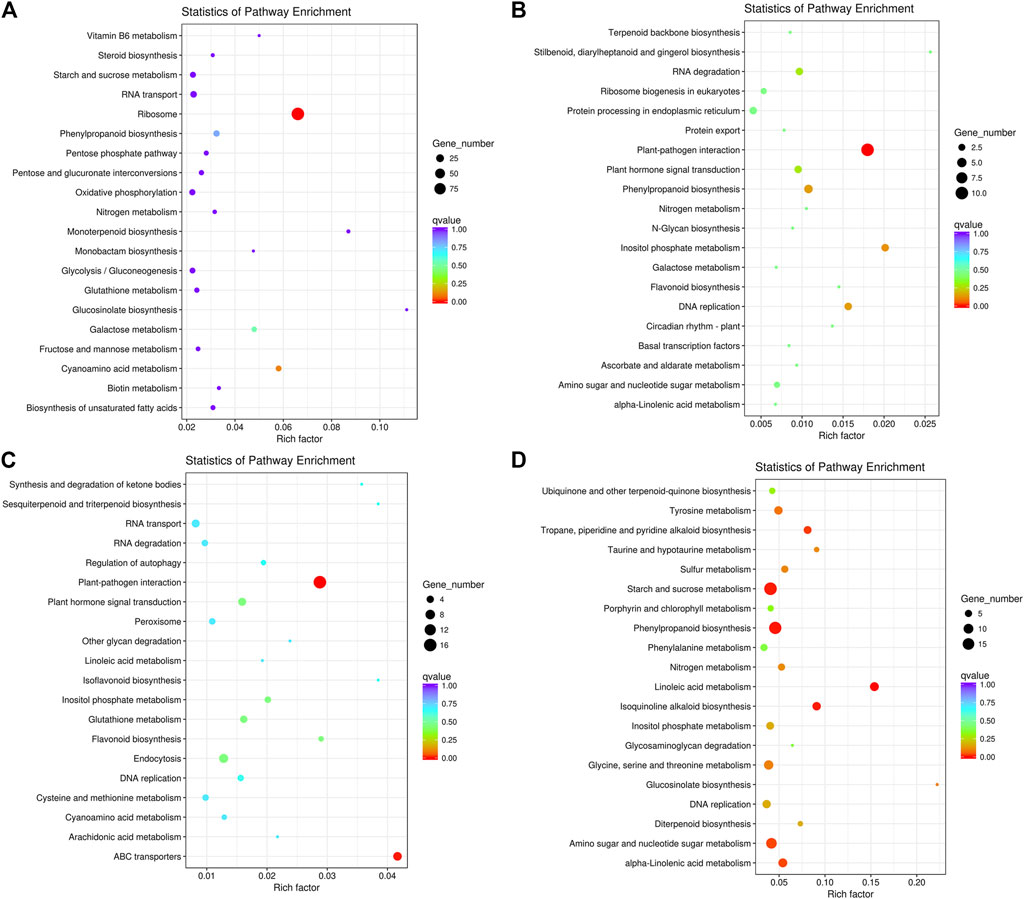
FIGURE 2. The top 20 enriched KEGG pathways of the DEGs in white clover with dodder parasitism. (A) upregulated DEGs in leaves; (B) downregulated DEGs in leaves; (C) upregulated DEGs in roots; (D) downregulated DEGs in roots. The Rich factor is the ratio of the number of differentially expressed genes annotated in this pathway to the number of all genes annotated in this pathway. The higher the Rich factor, the greater the degree of pathway enrichment. A q-value is an adjusted p-value; taking into account the false discovery rate and range from 0 to 1 and a lower value indicates greater pathway enrichment.
DEGs related to plant-pathogen interaction pathway
We identified 14 and 24 DEGs related to plant-pathogen interaction in the leaves and roots, respectively (Figure 3 and Supplementary Table S7). In Pleaf vs. Cleaf, most of the DEGs (10 DEGs) were downregulated, including one PR1, two CNGF, two RPM1/RPS3, one CML, and four FLS2, while only four DEGs were upregulated, including CALM, HtpG/HSP90A, CML, and CNGF (Figure 3A). Contrarily, more DEGs (16 DEGs) were upregulated in the Proot vs. Croot, including 13 RPM1/RPS3, two MEKK1P, and one PBS1, while only eight DEGs were downregulated, including three RPM1/RPS3, two CNGF, two RBOH, and one CALM (Figure 3B).
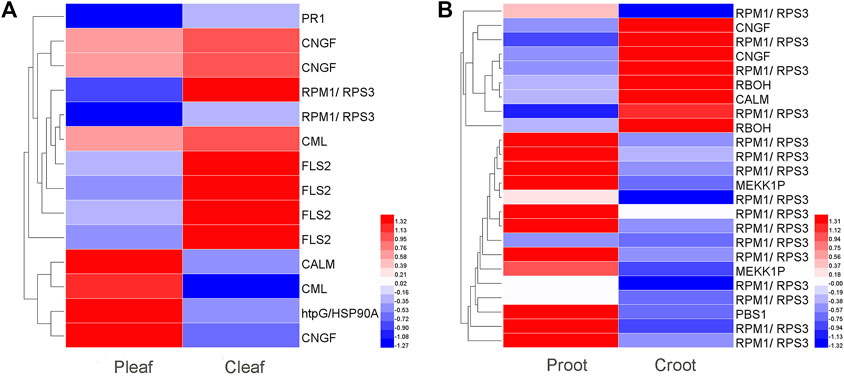
FIGURE 3. DEGs enriched in the plant–pathogen interaction pathway in white clover after dodder parasitism. (A) Leaves; (B) Roots.
DEGs related to plant hormone signal transduction pathway
We detected 6 and 13 DEGs to be involved in plant hormone signal transduction in the leaves and roots, respectively. These DEGs were related to various phytohormones like abscisic acid (ABA), auxin, cytokinin (CTK), ethylene (ET), salicylic acid (SA), and jasmonic acid (JA) (Figure 4 and Supplementary Table S8). ABA, auxin, CTK, and SA related genes were differentially expressed in both Pleaf vs. Cleaf and Proot vs. Croot, while ET and JA related genes were specific to the Proot vs. Croot. Specifically, three DEGs were involved in the ABA metabolism, including one upregulated SNRK2 (Cluster-28897.34756) gene in the leaves and two downregulated genes, ABF (Cluster-28897.5726) and SNRK2 (Cluster-57825.0) in the roots. In the leaf samples, we found that two DEGs involved in auxin metabolism or signaling were significantly affected by the dodder infection, with the GH3 (Cluster-28897.5110) and SAUR (Cluster-40615.0) genes being significantly upregulated and downregulated, respectively. In the root samples, three DEGs were related to auxin signaling, including one upregulated ARF (Cluster-28897.37861) gene, and two downregulated genes, AUX1 (Cluster-45618.0) and SAUR (Cluster-28897.6014). Three SA related DEGs (one PR1 and two TGA) were both downregulated in leaves and roots. The specific JA related DEG JAR1 was also downregulated in roots.
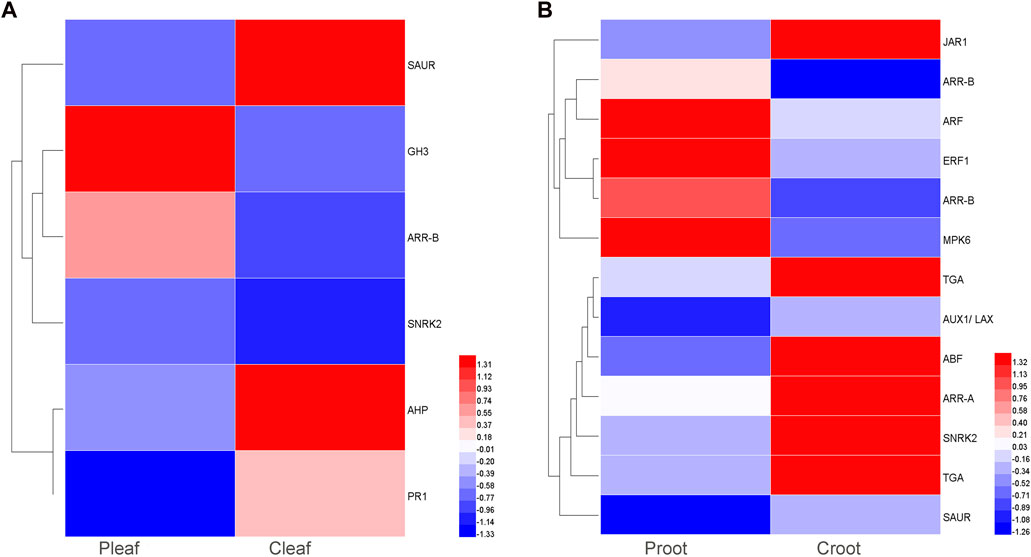
FIGURE 4. DEGs enriched in the plant hormone signal transduction pathway in white clover after dodder parasitism. (A) Leaves; (B) Roots.
DEGs related to phenylpropanoid biosynthesis pathway
We detected 16 and 19 DEGs to be involved in phenylpropanoid biosynthesis in the leaves and roots, respectively (Figures 5A, B and Supplementary Table S9). Among these DEGs, 10 were structural enzymes in the coumarine biosynthesis pathway. The other 25 DEGs were structural enzymes closely involved in the lignin biosynthesis, including one phenylalanine ammonia-lyase (PAL), three caffeic acid 3-O-methyltransferase (COMT) genes, one caffeoyl-CoA O-methyltransferase (CCoAOMT) gene, one coniferyl-aldehyde dehydrogenase (REF1) gene, six cinnamyl-alcohol dehydrogenase (CAD) genes, eight peroxidase (PRDX6) genes, and three coniferyl-alcohol glucosyltransferase (UGT72E) genes (Figure 5C).
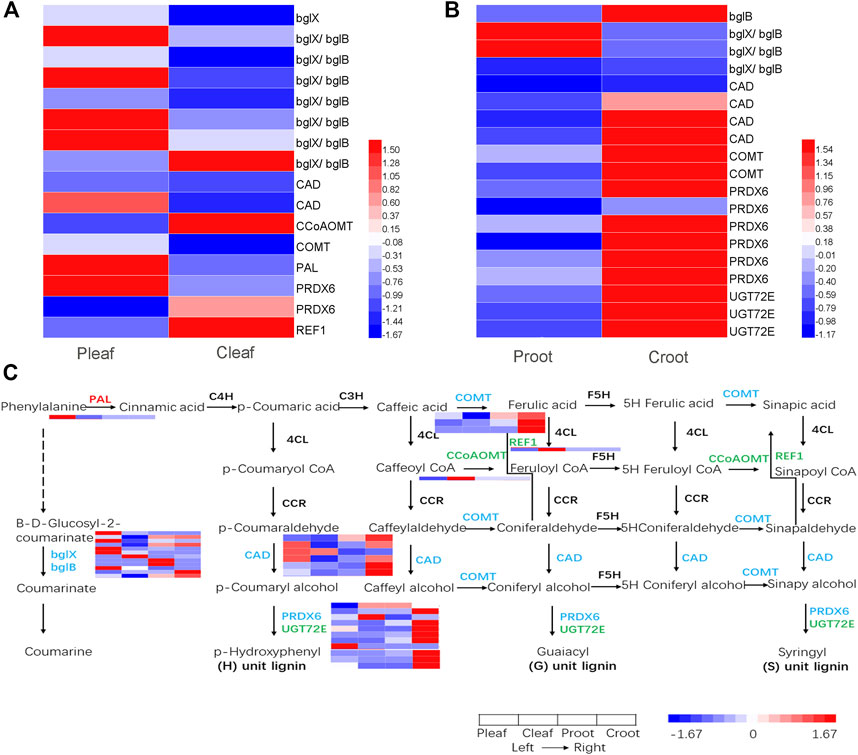
FIGURE 5. DEGs enriched in the phenylpropanoid biosynthesis pathway in the (A) leaves and (B) roots from white clover after dodder parasitism. (C) Pathway of phenylpropanoid biosynthesis. The red, green, and blue colorations of the gene names indicate upregulated, downregulated, and both upregulated and downregulated, respectively.
Differentially expressed transcription factors (TFs) in response to dodder parasitism
We searched the dodder parasitism-induced transcriptome for TFs among the DEGs. Consequently, we annotated 109 of the DEGs into 35 transcription factor families, including 9 WRKYs, 7 AP2/ERFs, 5 bHLHs, 5 bZIPs, 4 MYBs and 5 NACs. We analyzed the correlation between the expression levels of these transcription factors and genes involved in phenylpropanoid synthesis. Accordingly, 9 WRKYs, 7 AP2/ERFs, 5 bHLHs, 4 bZIPs, 4 MYBs, and 4 NACs showed a close relationship with the phenylpropanoid synthesis, with the correlation coefficients >0.9, and the expression levels of majority TFs showed a positive correlation with those of the phenylpropanoid synthesis genes (Figure 6, Supplementary Table S10). Additionally, 25 TFs were highly related to the lignin synthesis, including 8 WRKYs, 6 AP2/ERFs, 4 bHLHs, 3 bZIPs, 3 MYBs, and 3 NACs, thereby suggesting that these TFs are important in the white clover defense against dodder parasitism.
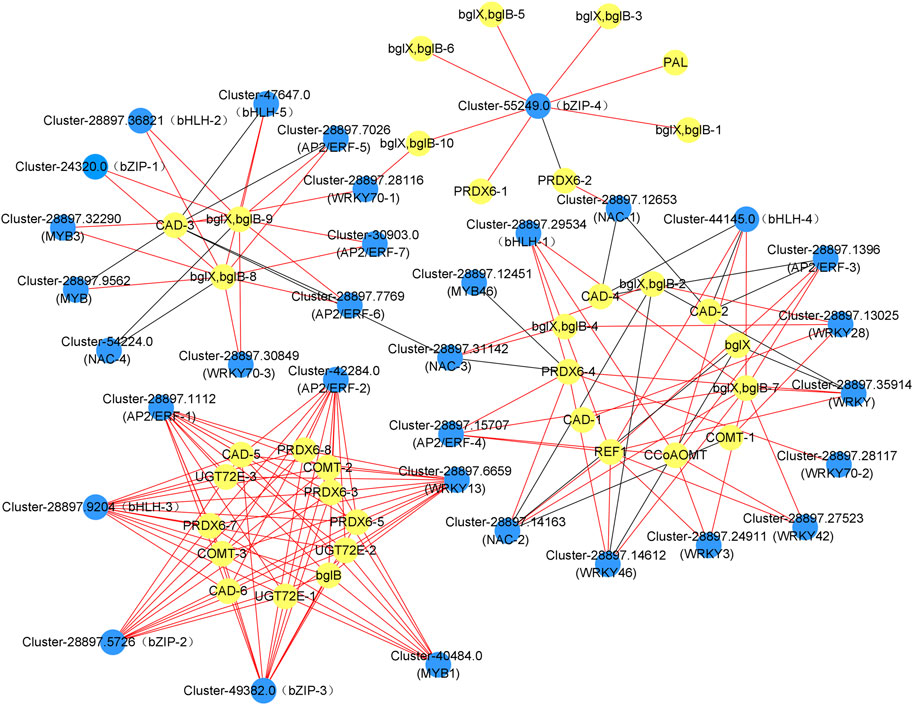
FIGURE 6. Co-expression analysis of the genes involved in the phenylpropanoid biosynthesis pathway and transcription factors (TFs) in the leaves and roots from white clover after dodder parasitism. Blue nodes indicate TFs. Yellow nodes indicate genes related to phenylpropanoid biosynthesis. Red and dark grey lines indicate positive corrections and negative corrections, respectively.
Validation of DEGs via RT-qPCR
To validate the DEGs obtained by RNA-seq, we selected nine DEGs for real-time reverse transcription polymerase chain reaction (RT-qPCR) analysis, including four genes (Cluster-28897.18901-FLS2, Cluster-28897.29775-RPM1, Cluster-28897.145389-RPM1, and Cluster-28897.25841-MEKK1P) involved in plant-pathogen interaction, three genes (Cluster-28897.22619-PR1, Cluster-32110.0-ARR-B, and Cluster-30903.0-ERF1) involved in plant hormone signal transduction, and two genes (Cluster-24876.0-PG and Cluster-28897.24433-FRK) involved in starch and sucrose metabolism (Figure 7). The results showed that the expression patterns of these DEGs were consistent with those obtained from RNA-seq.
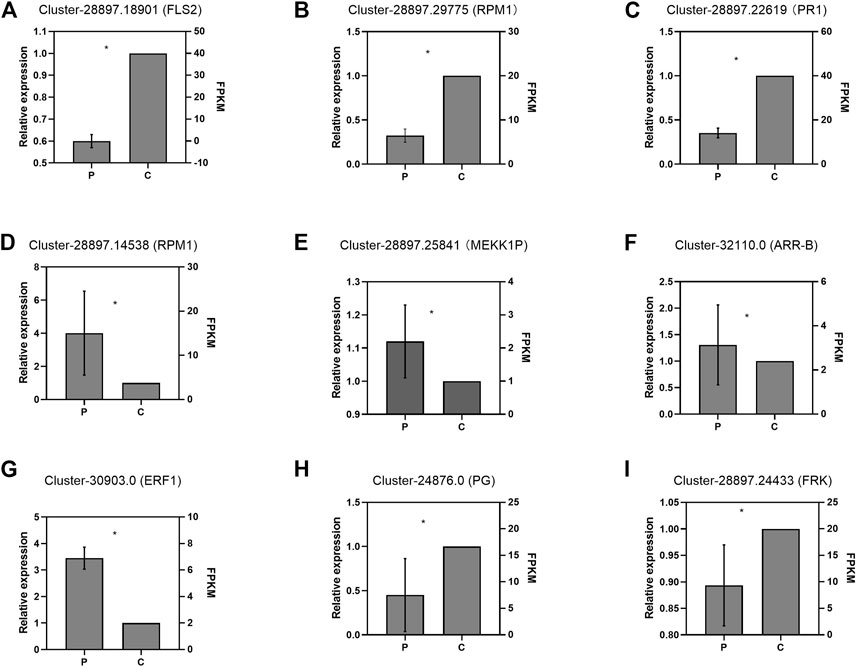
FIGURE 7. Validation of the expression profiles of partial DEGs by RT-qPCR: Panels (A)–(C) and (D)–(I) indicate the DEGs in the leaves and roots, respectively. P and C indicate the parasitized and non-parasitized hosts, respectively. Each value is calculated from three biological and technical replicates. The error bars represent the standard deviations (SD). Values with the different letters indicate significant differences according to the Independent-Samples t-Test (p < 0.05). Cluster-28897.18901 (FLS2): Plant-pathogen interaction/LRR receptor-like serine/threonine-protein kinase ERL1/LRR receptor-like serine/threonine-protein kinase FLS2; Cluster-28897.29775(RPM1): Plant-pathogen interaction/disease resistance protein RPM1; Cluster-28897.22619(PR1): Plant hormone signal transduction/pathogenesis-related protein 1; Cluster-28897.14538(RPM1): Plant-pathogen interaction/disease resistance protein RPM1; Cluster-28897.25841(MEKK1P): Plant-pathogen interaction/mitogen-activated protein kinase kinase 1; Cluster-32110.0(ARR-B): Plant hormone signal transduction/two-component response regulator ARR12-like protein; Cluster-30903.0 (ERF1): Plant hormone signal transduction/ethylene-responsive transcription factor 1b-like protein; Cluster-24876.0 (PG):Starch and sucrose metabolism/probable polygalacturonase; Cluster-28897.24433(FRK):Starch and sucrose metabolism/pfkB family carbohydrate kinase.
Discussion
A previous report suggested that the host plants have a similar response mechanism against parasite plants and pathogen infection (Runyon et al., 2010a). Many genes are vital for recognizing PAMPs and subsequent activating anti-pathogen defense mechanisms, like R genes, protein kinase genes, and PR genes (Tariq et al., 2018). The physiological and biochemical impact of parasitism on hosts have been continually characterized and it has been established in quantitative terms that Cuscuta parasitism reduces the lushness of growth of hosts (Shen et al., 2007; Li et al., 2015). Cuscuta parasitism results in significant root biomass reduction and changes the biomass allocation patterns, resulting in drastic increase of shoot: root dry weight ratio in parasitized hosts (Shen et al., 2005). There is a trade-off between plant growth and defense (Ballhorn et al., 2014). Competition or competitive behaviors can also affect the plant at various organizational levels resulting in morphological responses (plant growth), biochemical responses (plant defense) and resource allocation (Fernandez et al., 2016). In this study, more R genes were downregulated in parasitized leaves while more R genes were upregulated in parasitized roots, suggesting a trade-off between growth and defense in different tissues of host.
Plant hormones are signaling compounds that regulate crucial aspects of growth, development and different biotic and abiotic stress responses (Smith et al., 2009; Wani et al., 2016; Chen LM. et al., 2021). In this study, the expression of 19 genes related to six hormones (Auxin, CTK, ABA, ET, JA and SA) metabolism or signaling were significantly affected by dodder parasitism (Figure 4). The induction of plant defense is regulated by complex signaling networks, in which the phytohormones JA and SA play vital roles. Generally, JA-mediated pathway is involved in the defense response against herbivores, while the SA-mediated pathway helps regulate defense against microbial pathogens (Smith et al., 2009). However, studies on plant response to parasitic plants suggested that the crucial interaction between JA and SA is necessary to mediate effective defense. In tomato, JA and SA were sequentially induced to mediate effective defense against the C. pentagona infection. In the current study, one gene related to SA was differentially expressed in dodder affected leaves, while two genes related to SA and one gene related to JA were differentially expressed in dodder affected roots. Furthermore, the ABA content also increased after dodder attack (Runyon et al., 2010b). In this study, one and two genes related to ABA were differentially expressed in dodder affected leaves and roots, respectively, thus suggesting the complex signaling pathways regulated by plant hormones against parasitic plant infection.
The phenylpropanoid pathway is crucial for land plants to survive severe challenges, including desiccation stress, temperature stress, UV radiation, pests and pathogen infections. The phenylpropanoid pathway produces lignin, flavonoids, and other metabolites (Yadav et al., 2020). It has been reported that plants defend root and stem parasitic plants through the accumulation of lignin at the infection sites on roots and stems of host plants, respectively. For example, lignification of endodermal and pericycle host cells prevent parasite intrusion into the root vascular cylinder at early infection stages (Perez et al., 2005). An increased deposition of lignin was accumulated in the resistant rice cultivar upon root parasitic S. hermonthica infection. And the increased deposition of lignin was accompanied by induction of the expression of corresponding enzyme-encoding genes in the phenylpropanoid pathway (Mutuku et al., 2019). The resistant tomato cultivar Heinz exhibited a lignification-based resistance against the stem parasitic field dodder (C. campestris) infection (Jhu et al., 2022). PAL is important for providing precursors for various downstream metabolites. Downregulation of PAL can significantly decrease the lignin content in both Arabidopsis and Populus (Rohde et al., 2004; Wang CT et al., 2018; Zhang et al., 2020). In this study, the PAL expression (Cluster-28897.11757) was significantly upregulated in Pleaf vs. Cleaf, thereby suggesting that sufficient precursors were available for the downstream lignin biosynthesis. Additionally, two CAD genes (Cluster-28897.12905 and Cluster-28897.21230), one COMT (Cluster-28897.5861) and one PRDX6 (Cluster-33784.0) were upregulated in Pleaf vs. Cleaf (Figure 5A), which are important in the lignin monolignol synthesis in Arabidopsis (Rohde et al., 2004) and Populus (Zhang et al., 2020). Various reports have shown that a mobile signal was produced in the herbivores damaged local leaves and traveled to systemic leaves to activate defenses (Grenn & Ryan, 1972). For example, above-ground herbivores attacked Triadica sebifera leaves induced above- and below-ground defensive responses (Xiao et al., 2019). In this study, dodder parasitism on host stem also resulting in many transcriptomic changes in host roots, suggested that systemic signal produced at the dodder infection sites and transmitted to the distal roots. In terms of lignin biosynthesis, four CAD genes (Cluster-28897.16469, Cluster-28897.21230, Cluster-51220.0 and Cluster-28897.21232), two COMT genes (Cluster-57653.0 and Cluster-51096.0), six PRDX6 genes (Cluster-28897.5019, Cluster-28897.34522, Cluster-28897.21461, Cluster-28897.22464, Cluster-28897.10573, and Cluster-28897.5566) and three UGT72E genes (Cluster-28897.10004, Cluster-28897.3248 and Cluster-19366.0) showed differential expression patterns in Proot vs. Croot. Interestingly, more DEGs involved in the lignin biosynthesis displayed downregulation in Proot vs. Croot (Figure 5B), which indicated that the parasitized host might allocate more resources to leaves than roots to meet the requirement of lignin biosynthesis in leaves.
In plants, transcription factors of the WRKY, AP2/ERF, bZIP, bHLH, MYB, and NAC families are important in gene regulatory networks against various biotic and abiotic stresses in different plants (Shen et al., 2012; Wang et al., 2018; Li et al., 2020; Yan et al., 2020; Gao et al., 2021; Herath & Verchot, 2021). Our study found that 9 WRKYs, 7 AP2/ERFs, 5 bHLHs, 4 bZIPs, 4 MYBs, and 4 NACs were closely correlated with genes involved in the phenylpropanoid synthesis and the lignin synthesis. Accumulation studies have shown that WRKYs are responsible for plant defense response to abiotic and biotic stresses (Wani et al., 2021). For example, in cotton, GhWRKY41 directly activates the expression of GhC4H and Gh4CL, thereby contributing to the accumulation of flavonoids and lignin and enhancing the defense against Verticillium dahlia (Xiao et al., 2023). In a recent report, the resistant tomato cultivar Heinz showed a lignin-based resistance against field dodder parasitism (Jhu et al., 2022), which was regulated by a CC-NBS-LRR gene and three transcription factors, SIWRKY16, SIMYB55, and an AP2 transcription factor LIF1. Specifically, SIWRKY16 was upregulated in the Heinz tomato after the C. campestris parasitism (Jhu et al., 2022). AP2/ERF transcription factors have been reported to play an important role in lignin biosynthesis (Ma et al., 2005). For example, in Eirobotrya japonica, EjAP2-1 was identified as a novel regulator of fruit lignification via interaction with EjMYB (Zeng et al., 2015); in rice, the overexpression of Arabidopsis SHINE (an AP2/ERF gene) resulted in a 45% reduction in lignin content compared with the wild type production (Ambavaram et al., 2011), suggesting that AP2/ERF had potential functions in lignin regulation. It is known that bZIP transcription factors can regulate plant defense against pathogen infection. For example, in potato, StbZIP61 function together with StNPR3L to regulate the temporal activation of SA biosynthesis, resulting in SA-mediated immunity against Phytophthora infestans infection (Zhou et al., 2018). bHLH gene family has been reported to play vital roles in response to abiotic and biotic stresses. For example, the expression of HabHLH024 was upregulated in both roots and leaves of a resistant sunflower cultivar under root parasite Orobanche cumana infection (Li et al., 2020). MYB transcription factors have been reported to play a key role in response to the infection of the root parasitic weed O. cumana on the sunflower (Li et al., 2020). Additionally, transcriptomic analyses of the parasitic plant C. japonica on both host and non-host plants have shown that the expression of MYB genes is upregulated in host plants compared to non-host plants. This upregulation of MYB genes is positively related to the genes involved in the phenylpropanoid biosynthesis pathway, which ultimately leads to stress responses in host plants (Guo et al., 2022). NAC transcription factors have been demonstrated to participate in plant-pathogen interaction as both positive or negative regulators of the downstream defense-related genes (Bian et al., 2020). NAC and MYB transcription factors are involved in the regulation of lignin biosynthesis (Chen L. et al., 2021). Recently, the co-expression network among MYB, NAC, and lignin biosynthesis genes was conducted in two rattan species, Calamus simplicifolius, and D. jenkinsiana. In Daemonorops jenkinsiana, four NACs were co-expressed with ten lignin biosynthesis genes, C3H, CAD, CCoAOMT, CSE, F5H, C4H, CCR, PAL, LACCASES, and PEROXIDASES, while 59 MYBs were co-expressed with 76 lignin biosynthesis genes. Among the co-expression relationships, 4CL, C4H, C3H, CCR, CAD, HCT, LACCASES, and PEROXIDASES had binding sites for MYB transcription factors. This result suggests that these NACs and MYBs were profoundly involved in regulating the lignin biosynthesis pathway (Wang JP et al., 2018). Our result demonstrated that eight WRKYs, six AP2/ERFs, four bHLHs, three bZIPs, three MYBs, and three NACs were co-expressed with lignin biosynthesis genes. For example, WRKY28 (Cluster-28897.13025) was co-expressed with CAD-1 (Cluster-28897.12905) and COMT-1 (Cluster-28897.5861); MYB1 was co-expressed with PRDX6-3/7 (Cluster-288978.5019/Cluster-28897.10573), UGT72E-1/2 (Cluster-288978.10004/Cluster-28897.3248), CAD-5 (Cluster-51220.0), and COMT-2 (Cluster-57653.0); NAC-1 (Cluster-28897.12653) was co-expressed with PRDX6-2 (Cluster-28897.20416) and CAD-2 (Cluster-28897.21230), suggesting that these TFs play a regulatory role in the lignin biosynthetic pathway and thus response against dodder parasitism (Figures 6, 8).
Conclusion
A comparative transcriptomics of the leaves and roots from white clover both with and without dodder parasitism were conducted to explore the defense-related genes and pathways. We identified 1,329 and 3,271 DEGs in the leaf and roots samples, respectively. The KEGG pathway enrichment analysis revealed that the DEGs were mainly involved in ribosome, plant-pathogen interaction, plant hormone signal transduction, and phenylpropanoid biosynthesis. The results showed that 8 WRKYs, 6 AP2/ERFs, 4 bHLHs, 3 bZIPs, 3 MYBs and 3 NACs were closely correlated with lignin biosynthesis, which may play vital roles in the white clover response against dodder parasitism. Thus, our results provide new insights into understanding the complex regulatory network behind the parasite-host plant interactions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary Material.
Author contributions
JL conceived and designed the research. AZ and LZ analyzed data and wrote the manuscript. JL and LZ revised the manuscript. BY and QL assist during the experiments. All authors read andapproved the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32271590), and the Ten Thousand Talent Program of Zhejiang Province (2019R52043).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1106936/full#supplementary-material
References
Ambavaram, M. M., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931. doi:10.1104/pp.110.168641
Ballhorn, D. J., Godschalx, A. L., Smart, S. M., Kautz, S., and Schadler, M. (2014). Chemical defense lowers plant competitiveness. Oecologia 176 (3), 811–824. doi:10.1007/s00442-014-3036-1
Bardgett, R. D., Smith, R. S., Shiel, R. S., Peacock, S., Simkin, J. M., Quirk, H., et al. (2006). Parasitic plants indirectly regulate below-ground properties in grassland ecosystems. Nature 439 (7079), 969–972. doi:10.1038/nature04197
Bellincampi, D., Cervone, F., and Lionetti, V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5, 228. doi:10.3389/fpls.2014.00228
Bhuiyan, N. H., Selvaraj, G., Wei, Y., and King, J. (2009). Role of lignification in plant defense. Plant Signal Behav. 4 (2), 158–159. doi:10.4161/psb.4.2.7688
Borsics, T., and Lados, M. (2002). Dodder infection induces the expression of a pathogenesis-related gene of the family PR-10 in alfalfa. J. Exp. Bot. 53 (375), 1831–1832. doi:10.1093/jxb/erf039
Bringmann, G. S. J., Ruckert, M., Wiesen, B., Ehrenfeld, K., Ppc, F. C., Proksch, P., et al. (1999). Host-derived acetogenins involved in the incompatible parasitic relationship between Cuscuta reflexa (Convolvulaceae) and Ancistrocladus heyneanus (Ancistrocladaceae). Plant Biol. 1, 581–584. doi:10.1111/j.1438-8677.1999.tb00787.x
Chen, L., Wu, F., and Zhang, J. (2021b). NAC and MYB families and lignin biosynthesis-related members identification and expression analysis in melilotus albus. Plants (Basel) 10 (2), 303. doi:10.3390/plants10020303
Chen, L. M., Li, X. W., He, T. J., Li, P. J., Liu, Y., Zhou, S. X., et al. (2021a). Comparative biochemical and transcriptome analyses in tomato and eggplant reveal their differential responses to Tuta absoluta infestation. Genomics 113 (4), 2108–2121. doi:10.1016/j.ygeno.2021.05.002
Cui, S., Wada, S., Tobimatsu, Y., Takeda, Y., Saucet, S. B., Takano, T., et al. (2018). Host lignin composition affects haustorium induction in the parasitic plants Phtheirospermum japonicum and Striga hermonthica. New Phytol. 218 (2), 710–723. doi:10.1111/nph.15033
Davidson, Nadia M., and Oshlack, A. (2014). Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. GeneBiology 15, 410. doi:10.1186/s13059-014-0410-6
Dawson, J., Musselman, L., Wolswinkel, P., and Doerr, I. (1994). Biology and control of Cuscuta. Rev. Weed Sci. 6, 265–317.
Dhakshinamoorthy, S., Mariama, K., Elsen, A., and De Waele, D. (2014). Phenols and lignin are involved in the defence response of banana (Musa) plants to Radopholus similis infection. Nematology 16 (5), 565–576. doi:10.1163/15685411-00002788
Fernandez, C., Monnier, Y., Santonja, M., Gallet, C., Weston, L. A., Prevosto, B., et al. (2016). The impact of competition and allelopathy on the trade-off between plant defense and growth in two contrasting tree species. Front. Plant Sci. 7, 594. doi:10.3389/fpls.2016.00594
Gao, S., Wang, F., Niran, J., Li, N., Yin, Y., Yu, C., et al. (2021). Transcriptome analysis reveals defense-related genes and pathways against Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum L.). PLoS One 16 (3), e0240279. doi:10.1371/journal.pone.0240279
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi:10.1038/nbt.1883
Grenn, T. R., and Ryan, C. A. (1972). Wound-induced proteinase inhibitor in plant leaves: A possible defense mechanism against insects. Science 175 (4023), 776–777. doi:10.1126/science.175.4023.776
Guo, C., Qin, L., Ma, Y., and Qin, J. (2022). Integrated metabolomic and transcriptomic analyses of the parasitic plant Cuscuta japonica choisy on host and non-host plants. BMC Plant Biol. 8 (1), 393. doi:10.1186/s12870-022-03773-9
Hegenauer, V., Slaby, P., Korner, M., Bruckmuller, J. A., Burggraf, R., Albert, I., et al. (2020). The tomato receptor CuRe1 senses a cell wall protein to identify Cuscuta as a pathogen. Nat. Commun. 11 (1), 5299. doi:10.1038/s41467-020-19147-4
Hegenauer, V., Furst, U., Kaiser, B., Smoker, M., Zipfel, C., Felix, G., et al. (2016). Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 353 (6298), 478–481. doi:10.1126/science.aaf3919
Herath, V., and Verchot, J. (2021). Transcriptional regulatory networks associate with early stages of Potato Virus X infection of Solanum tuberosum. Int. J. Mol. Sci. 22 (6), 2837. doi:10.3390/ijms22062837
Jhu, M. Y., Farhi, M., Wang, L., Philbrook, R. N., Belcher, M. S., Nakayama, H., et al. (2022). Heinz resistant tomato cultivars exhibit a lignin-based resistance to field dodder (Cuscuta campestris) parasitism. Plant Physiol. 189, 129–151. doi:10.1093/plphys/kiac024
Jin, J. P. T. F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J. C., Gao, G., et al. (2017). PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45 (D1), D1040–D1045. doi:10.1093/nar/gkw982
Kaiser, B., Vogg, G., Furst, U. B., and Albert, M. (2015). Parasitic plants of the genus Cuscuta and their interaction with susceptible and resistant host plants. Front. Plant Sci. 6, 45. doi:10.3389/fpls.2015.00045
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi:10.1093/nar/gkm882
Kc, V. (2002). Attachment of the parasitic weed dodder to the host. Protoplasma 219, 227–237. doi:10.1007/s007090200024
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 Years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30 (2), 285–299. doi:10.1105/tpc.17.00579
Li, B., and Dewey, C. N. (2011). Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinforma. 12 (323), 323. doi:10.1186/1471-2105-12-323
Li, J., Yang, B., Yan, Q., Zhang, J., Yan, M., and Li, M. (2015). Effects of a native parasitic plant on an exotic invader decrease with increasing host age. AoB Plants 7, plv031. doi:10.1093/aobpla/plv031
Li, W., Pang, S., Lu, Z., and Jin, B. (2020). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants (Basel) 9 (11), 1515. doi:10.3390/plants9111515
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21 (19), 3787–3793. doi:10.1093/bioinformatics/bti430
McHale, L., Tan, X., Koehl, P., and Michelmore, R. W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7 (4), 212. doi:10.1186/gb-2006-7-4-212
Mutuku, J. M., Cui, S., Hori, C., Takeda, Y., Tobimatsu, Y., Nakabayashi, R., et al. (2019). The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiol. 179 (4), 1796–1809. doi:10.1104/pp.18.01133
Pennings, S. C., and Callaway, R. M. (2002). Parasitic plants: Parallels and contrasts with herbivores. Oecologia 131 (4), 479–489. doi:10.1007/s00442-002-0923-7
Perez, D. E. L. A., Rubiales, D., Cubero, J. I., Press, M. C., Scholes, J., Yoneyama, K., et al. (2005). Interaction between Orobanche crenata and its host legumes: Unsuccessful haustorial penetration and necrosis of the developing parasite. Ann. Bot. 95 (6), 935–942. doi:10.1093/aob/mci105
Pérez-de-Luque, A., Lozano, M. D., Moreno, M. T., Testillano, P. S., and Rubiales, D. (2007). Resistance to broomrape (Orobanche crenata) in faba bean (Vicia faba): Cell wall changes associated with prehaustorial defensive mechanisms. Ann. Appl. Biol. 151 (1), 89–98. doi:10.1111/j.1744-7348.2007.00164.x
Press, M. C., and Phoenix, G. K. (2005). Impacts of parasitic plants on natural communities. New Phytol. 166 (3), 737–751. doi:10.1111/j.1469-8137.2005.01358.x
Qin, Y., Zhang, J., Hettenhausen, C., Liu, H., Li, S., Shen, G., et al. (2019). The host jasmonic acid pathway regulates the transcriptomic changes of dodder and host plant under the scenario of caterpillar feeding on dodder. BMC Plant Biol. 19 (1), 540. doi:10.1186/s12870-019-2161-8
Raes, J., Rohde, A., Christensen, J. H., Van de Peer, Y., and Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133 (3), 1051–1071. doi:10.1104/pp.103.026484
Ramakrishna, A., and Ravishankar, G. A. (2011). Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav. 6 (11), 1720–1731. doi:10.4161/psb.6.11.17613
Revill, M. J., Stanley, S., and Hibberd, J. M. (2005). Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta. J. Exp. Bot. 56 (419), 2477–2486. doi:10.1093/jxb/eri240
Rohde, A., Morreel, K., Ralph, J., Goeminne, G., Hostyn, V., De Rycke, R., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16 (10), 2749–2771. doi:10.1105/tpc.104.023705
Runyon, J. B., Mescher, M. C., and De Moraes, C. M. (2010a). Plant defenses against parasitic plants show similarities to those induced by herbivores and pathogens. Plant Signal Behav. 5 (8), 929–931. doi:10.4161/psb.5.8.11772
Runyon, J. B., Mescher, M. C., and De Moraes, C. M. (2006). Volatile chemical cues guide host location and host selection by parasitic plants. Science 313 (5795), 1964–1967. doi:10.1126/science.1131371
Runyon, J. B., Mescher, M. C., Felton, G. W., and De Moraes, C. M. (2010b). Parasitism by Cuscuta pentagona sequentially induces JA and SA defence pathways in tomato. Plant Cell Environ. 33 (2), 290–303. doi:10.1111/j.1365-3040.2009.02082.x
Saric-Krsmanovic, M., Dragumilo, A., Gajic Umiljendic, J., Radivojevic, L., Santric, L., and Durovic-Pejcev, R. (2020). Infestation of field dodder (Cuscuta campestris yunck.) promotes changes in host dry weight and essential oil production in two aromatic plants, peppermint and chamomile. Plants (Basel) 9 (10), 1286. doi:10.3390/plants9101286
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shen, H., Hong, L., Ye, W., Cao, H., and Wang, Z. (2007). The influence of the holoparasitic plant Cuscuta campestris on the growth and photosynthesis of its host Mikania micrantha. J. Exp. Bot. 58 (11), 2929–2937. doi:10.1093/jxb/erm168
Shen, H. L. C., Zhang, Y., Meng, X., Zhou, X., Chu, C., Wang, X., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80 (3), 241–253. doi:10.1007/s11103-012-9941-y
Shen, H., Ye, W., Hong, L., Cao, H., and Wang, Z. (2005). Influence of the obligate parasite Cuscuta campestris on growth and biomass allocation of its host Mikania micrantha. J. Exp. Bot. 56 (415), 1277–1284. doi:10.1093/jxb/eri128
Smith, J. L., De Moraes, C. M., and Mescher, M. C. (2009). Jasmonate- and salicylate-mediated plant defense responses to insect herbivores, pathogens and parasitic plants. Pest Manag. Sci. 65 (5), 497–503. doi:10.1002/ps.1714
Steinbrenner, Adam D., Gómez, Sara, Osorio, Sonia, Fernie, Alisdair R., and Orians, C. M. (2011). Herbivore-induced changes in tomato (Solanum lycopersicum) primary metabo-lism: A whole plant perspective. J. Chem. Ecol. 37, 1294–1303. doi:10.1007/s10886-011-0042-1
Tariq, R., Wang, C., Qin, T., Xu, F., Tang, Y., Gao, Y., et al. (2018). Comparative transcriptome profiling of rice near-isogenic line carrying Xa23 under infection of Xanthomonas oryzae pv. oryzae. Int. J. Mol. Sci. 19 (3), 717. doi:10.3390/ijms19030717
Vance, C. P., Tk, K., and Rt, S. (1980). Lignification as a mechanism of disease resistance. Ann. Rev. Phytopath 18, 259–288. doi:10.1146/annurev.py.18.090180.001355
Wang, B., Duan, H., Chong, P., Su, S., Shan, L., Yi, D., et al. (2020). Systematic selection and validation of suitable reference genes for quantitative real-time PCR normalization studies of gene expression in Nitraria tangutorum. Sci. Rep. 10 (1), 15891. doi:10.1038/s41598-020-73059-3
Wang, C. T., Liu, Y. W., Li, M., Zhao, D., Yang, J. F., Fu, J. D., et al. (2018). Maize WRKY Transcription Factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 19 (10), 3046. doi:10.3390/ijms19103046
Wang, J. P., Matthews, M. L., Williams, C. M., Shi, R., Yang, C., Tunlaya-Anukit, S., et al. (2018). Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 9 (1), 1579. doi:10.1038/s41467-018-03863-z
Wani, A. B., Chadar, H., Wani, A. H., Singh, S., and Upadhyay, N. (2016). Salicylic acid to decrease plant stress. Environ. Chem. Lett. 15 (1), 101–123. doi:10.1007/s10311-016-0584-0
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 40 (7), 1071–1085. doi:10.1007/s00299-021-02691-8
Wu, J., and Baldwin, I. T. (2010). New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24. doi:10.1146/annurev-genet-102209-163500
Xiao, L., Carrillo, J., Siemann, E., and Ding, J. (2019). Herbivore-specific induction of indirect and direct defensive responses in leaves and roots. AoB Plants 11 (1), plz003. doi:10.1093/aobpla/plz003
Xiao, S., Ming, Y., Hu, Q., Ye, Z., Si, H., Liu, S., et al. (2023). GhWRKY41 forms a positive feedback regulation loop and increases cotton defence response against Verticillium dahliae by regulating phenylpropanoid metabolism. Plant Biotechnol. J. doi:10.1111/pbi.14008
Yan, F., Zhu, Y., Zhao, Y., Wang, Y., Li, J., Wang, Q., et al. (2020). De novo transcriptome sequencing and analysis of salt-alkali-and drought-responsive genes in Sophora alopecuroides. BMC Genomics 21 (1), 423. doi:10.1186/s12864-020-06823-4
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11 (2), R14. doi:10.1186/gb-2010-11-2-r14
Yu, B. B., Brunel, C., Yang, B. F., Li, J., and Lu, H. (2019). Parasitism by Cuscuta australis affects the rhizhospher-ic soil bacterial communities of Trifolium repens L. Acta Agric. Scand. sec. B--Soil Plant Sci. 69, 649–656. doi:10.1080/09064710.2019.1637016
Zagorchev, L., Atanasova, A., Pachedjieva, K., Tosheva, A., Li, J., and Teofanova, D. (2021a). Salinity effect on germination and further development of parasitic Cuscuta spp. and related non-parasitic vines. Plants (Basel) 10 (3), 438. doi:10.3390/plants10030438
Zagorchev, L., Stoggl, W., Teofanova, D., Li, J., and Kranner, I. (2021b). Plant parasites under pressure: Effects of abiotic stress on the interactions between parasitic plants and their hosts. Int. J. Mol. Sci. 22 (14), 7418. doi:10.3390/ijms22147418
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi:10.1111/pbi.12351
Zhang, J., Tuskan, G. A., Tschaplinski, T. J., Muchero, W., and Chen, J. G. (2020). Transcriptional and post-transcriptional regulation of lignin biosynthesis pathway genes in Populus. Front. Plant Sci. 11, 652. doi:10.3389/fpls.2020.00652
Zhang, J., Xu, Y., Xie, J., Zhuang, H., Liu, H., Shen, G., et al. (2021). Parasite dodder enables transfer of bidirectional systemic nitrogen signals between host plants. Plant Physiol. 23 (4), 1395–1410. doi:10.1093/plphys/kiaa004
Zhang, Y., Wu, L., Wang, X., Chen, B., Zhao, J., Cui, J., et al. (2019). The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 20 (3), 309–322. doi:10.1111/mpp.12755
Zheng, Y. J. C., Sun, H., Rosli, H. G., Pombo, M. A., Zhang, P., Banf, M., et al. (2016). iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670. doi:10.1016/j.molp.2016.09.014
Zhou, X. T., Jia, L. J., Wang, H. Y., Zhao, P., Wang, W. Y., Liu, N., et al. (2018). The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant J. 95 (6), 1055–1068. doi:10.1111/tpj.14010
Keywords: parasitic plant dodder, transcriptome, plant hormone, transcription factors, lignin
Citation: Zhou L, Zawaira A, Lu Q, Yang B and Li J (2023) Transcriptome analysis reveals defense-related genes and pathways during dodder (Cuscuta australis) parasitism on white clover (Trifolium repens). Front. Genet. 14:1106936. doi: 10.3389/fgene.2023.1106936
Received: 24 November 2022; Accepted: 08 March 2023;
Published: 16 March 2023.
Edited by:
Milind B. Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaReviewed by:
Harleen Kaur, University of Alberta, CanadaMuthusamy Ramakrishnan, Nanjing Forestry University, China
Copyright © 2023 Zhou, Zawaira, Lu, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junmin Li, bGlqbUB0emMuZWR1LmNu
 Li Zhou1,2
Li Zhou1,2 Alexander Zawaira
Alexander Zawaira Qiuwei Lu
Qiuwei Lu Junmin Li
Junmin Li