- Key Laboratory of Ruminant Molecular and Cellular Breeding, School of Agriculture, Ningxia University, Yinchuan, China
The quality of colostrum is a key factor contributing to healthy calf growth, and pasteurization of colostrum can effectively reduce the counts of pathogenic microorganisms present in the colostrum. Physiological changes in calves fed with pasteurized colostrum have been well characterized, but little is known about the underlying molecular mechanisms. In this study, key genes and functional pathways through which pasteurized colostrum affects calf growth were identified through whole blood RNA sequencing. Our results showed that calves in the pasteurized group (n = 16) had higher body height and daily weight gain than those in the unpasteurized group (n = 16) in all months tested. Importantly, significant differences in body height were observed at 3 and 4 months of age (p < 0.05), and in daily weight gain at 2, 3, and 6 months of age (p < 0.05) between the two groups. Based on whole blood transcriptome data from 6-months old calves, 630 differentially expressed genes (DEGs), of which 235 were upregulated and 395 downregulated, were identified in the pasteurized compared to the unpasteurized colostrum groups. Most of the DEGs have functions in the immune response (e.g., CCL3, CXCL3, and IL1A) and metabolism (e.g., PTX3 and EXTL1). Protein-protein interaction analyses of DEGs revealed three key subnetworks and fifteen core genes, including UBA52 and RPS28, that have roles in protein synthesis, oxidative phosphorylation, and inflammatory responses. Twelve co-expression modules were identified through weighted gene co-expression network analysis. Among them, 17 genes in the two modules that significantly associated with pasteurization were mainly involved in the tricarboxylic acid cycle, NF-kappa B signaling, and NOD-like receptor signaling pathways. Finally, DEGs that underwent alternative splicing in calves fed pasteurized colostrum have roles in the immune response (SLCO4A1, AKR1C4, and MED13L), indicative of potential roles in immune regulation. Results from multiple analytical methods used suggest that differences in calf growth between the pasteurized and unpasteurized groups may be due to differential immune activity. Our data provide new insights into the impact of pasteurization on calf immune and metabolic-related pathways through its effects on gene expression.
1 Introduction
The calf rearing stage is crucial and healthy calf growth has a positive impact on improving mating performance of breeding cows, as well as on overall production performance (Cummins et al., 2016). Colostrum is the milk secreted by cows for up to 2–3 days after parturition. It contains high amounts of immunoglobulins, antimicrobial peptides and growth factors. As the first meal for newborn calves, colostrum has a vital role in calf nutrition and immune defense, growth and development (Playford and Weiser, 2021). The immune status of calves after birth directly depends on the amount and quality of colostrum they consume (Silva et al., 2021). Colostrum quality significantly affects the formation of intestinal microbiota and the daily weight gain of calves, which is the highest when the colostrum density is > 1.070 g/cm3 (Puppel et al., 2020). In calves, adequate passive immunity increases daily gain and weaning weight, reduces the age at first mating and first calving, and increases milk production in the first lactation (Godden et al., 2019). Calves with serum IgG concentrations <10 g/L in the first 24–48 h after birth are considered to have failed to recieve passive immunization, and this increases the incidence of disease and mortality in the first week of life (Godden et al., 2019). Therefore, high quality colostrum can enhance calf immunity, health and growth performance, which is essential to maintain the economic efficiency of dairy farms.
However, colostrum is susceptible to contamination by milking equipment, storage containers, and feeding equipment, resulting in high levels of pathogenic microorganisms in colostrum such as Mycobacterium avium ssp. paratuberculosis (Sweeney, 1996), Mycoplasma ssp. (Stabel et al., 2004), Escherichia coli, and Salmonella (Streeter et al., 1995; Pithua et al., 2011; Slosarkova et al., 2021). These pathogens may bind immunoglobulins in the gut of the calves and affect the uptake and transport of immunoglobulins by intestinal epithelial cells, leading to passive immunization failure (Elizondo-Salazar and Heinrichs, 2009). Numerous studies have demonstrated that pasteurization can inactivate crucial pathogens such as E. coli, Salmonella, and Listeria monocytogenes while maintaining the quality of colostrum (Stabel et al., 2004; McMartin et al., 2006; Elizondo-Salazar and Heinrichs, 2009; Heinrichs and Elizondo-Salazar, 2009). In addition, pasteurization is effective in reducing the concentration of certain heat-stable pathogens such as M. avium ssp. paratuberculosis (Fechner et al., 2019). Pasteurized colostrum has a positive impact on healthy calf growth, and feeding pasteurized colostrum is effective in reducing the risk of upper respiratory disease and diarrhea, and mortality in calves (Armengol and Fraile, 2016; Kargar et al., 2020). Pasteurization treatment may disrupt the function of heat-unstable bioactive compounds such as immunoglobulins, proteases, and cholesterol (Sousa et al., 2014; Parron et al., 2016). However, previous study showed no significant effect of heat treatment on the composition of the colostrum metabolome and no detectable changes in the serum metabolome profile of raw colostrum compared to heat-treated colostrum fed calves (Xu W. et al., 2021).
Pasteurized colostrum is helpful for establishing passive immunity in calves. Pathogenic microorganisms are greatly reduced in pasteurized colostrum compared to unpasteurized colostrum, but changes related to gene expression and immune pathways are not known. We therefore hypothesized that feeding pasteurized colostrum to calves at birth would influence the expression of genes associated with immune responses and metabolism. In this study, RNA-seq technology and bioinformatic analysis were used to study the blood gene expression profiles of calves fed with pasteurized or unpasteurized colostrum as well as examined the effect of pasteurization treatment on calf growth traits. The study results provide better understanding of the effects of pasteurized colostrum on health status and growth performance, which may facilitate optimization of current calf feeding regimes.
2 Materials and methods
2.1 Animal and feeding management
Animal experiments were approved by the Animal Ethical and Welfare Committee of Ningxia University (approval number: NXUC20210718). The experiment was conducted on a cattle farm in Ningxia, China. Holstein female calves were selected as experimental subjects based on the following criteria: Birth weight greater than 30 kg and less than 40 kg, and calves with serum total protein less than 5.5 g/dL at 24 h were excluded (Windeyer et al., 2014). Calves with fever, diarrhea, disability, and other diseases were also excluded. A total of 32 healthy female calves (average weight 39.8 ± 1.22 kg) were randomly assigned to two groups (pasteurized and unpasteurized colostrum, 16 calves per group), and fed in individual pens where manure was removed daily to keep the pens clean and dry. Colostrum and milk were provided in buckets, which were cleaned and disinfected after each use. The trial period was 180 days in total. All tested calves were fed 6–8 L colostrum for 12 h after birth (colostrum processing for the pasteurized group is explained in Section 2.2). Subsequently, from day 2 to day 60, an average of 3 L (2.5–3.5 L) milk was fed every 12 h. From day 61 to day 180, pelleted feed and oat hay were offered ad libitum to all the calves (Supplementary Table S1). To reduce the stress of the change in diet, milk feeding was reduced daily for both groups (by 1.5 L per day, see Supplementary Table S2 for detailed feeding schedule) until the end of the 180-day trial. Starting on day 60, all calves were fed ad libitum on pellet feed and water.
2.2 Colostrum and milk management
The milk used was produced on the farm itself. Colostrum was from cows free of paratuberculosis and having somatic cell count less than 400,000 cells/ml, and no visible signs of inflammation or disease. Serum samples were used to test for the presence of Mycobacterium avium spp. paratuberculosis with the PARACHEK two Bovine Mycobacterium paratuberculosis Test Kit (Prionics AG, Schlieren-Zurich, Switzerland). Colostrum with a specific gravity of more than 1.065 was used (Fleenor and Stott, 1980; Armengol and Fraile, 2016). Colostrum was pasteurized by heating continuously at 60°C for 1 h, followed by storage at −20°C. Frozen colostrum was heated continuously at 40°C for 30 min and then fed to calves. Details of the treatment process are shown in Supplementary Figure S1. The temperature and time of pasteurization during colostrum and milk handling were strictly observed.
2.3 Sampling and phenotyping
For all calves in this study, traits related to growth, such as height, weight and daily weight gain were measured. The mean of each parameter for all calves in each group was taken as the phenotypic value. The data were analyzed for significance using the general linear model in SPSS 22.0, and the results were represented as “mean ± SE”, with p < 0.05 indicating significant differences. At the end of the test period, 5 ml whole blood was drawn from the caudal vein of each calf using disposable blood collection needles into EDTA anticoagulant tubes, and stored at −80°C until used.
2.4 RNA isolation and transcriptome sequencing
Total RNA was extracted from blood samples using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), in accordance with the manufacturer’s instructions. RNA concentration and integrity were assessed using an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, United States). RNA samples with RIN ≥7 and 28S/18S values in the range 1.8–2.0 were used for sequencing. Four samples were randomly selected from each of the pasteurized and unpasteurized groups for sequencing. RNA sequencing libraries were constructed and sequenced by CapitalBio Technology Co., Ltd. (Beijing China) using the Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, United States).
2.5 Quality control, alignment, and identification of differentially expressed genes
The sequencing quality of the raw data was evaluated using FastQc v0.11.7 (Schmieder and Edwards, 2011), and reads were trimmed using Trimmomatic v0.39 (Bolger et al., 2014). The clean reads with average base quality greater than 20 were selected for subsequent analysis. The trimmed clean reads were aligned to the bovine reference genome (Bos taurus UMD3.1) using Hisat2 v2.2.1 (Kim et al., 2019) to obtain the sam files. The sam files were converted to the bam files using samtools v1.9 (Li et al., 2009). Meanwhile, the bam file index was generated, and expressed genes were counted using StringTie v2.1.2 (Pertea et al., 2015) to obtain the count matrix. Gene expression was quantified based on transcript per million values. Differentially expressed genes (DEGs) were detected using the R package DESeq2 v1.20 (Anders and Huber, 2010), with a differential gene screening threshold of |log2FC|≥0.585 and p-value corrected using the Benjamini–Hochberg false discovery rate of <0.05.
2.6 Functional annotation of DEGs and protein-protein interaction network analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway functional annotation of DEGs was performed with DAVID v6.8 (Sherman et al., 2022). GO annotations were grouped into three broad categories, namely, cellular components, molecular function, and biological processes. Significance levels for GO terms and KEGG pathways were tested with a threshold of corrected p set at <0.05. Protein–protein interaction (PPI) networks were constructed using STRING v11.5 with a confidence level of >0.9 and visualized using Cytoscape v3.8.0 (Shannon et al., 2003). The Molecular Complex Detection (MCODE) program in Cytoscape was used to filter key subnetworks and nodes in the PPI network (degree cutoff = 2, node score cutoff = 0.2, k-core = 2, maximum depth = 100). The CytoHubba plugin in Cytoscape was used to detect hub genes through four methods, network topology analysis-Degree, edge percolated component (EPC), maximal clique centrality (MCC), and maximum neighborhood component (MNC). Overlapping genes screened using the four methods were selected as core genes.
2.7 Weighted gene Co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) was used to explore the association between gene networks and sample phenotypes, as well as the core genes in the network. Gene co-expression networks were constructed using the WGCNA v1.69 package in R (Zhang and Horvath, 2005; Langfelder and Horvath, 2008). The soft threshold was determined to be seven when the correlation coefficient was 0.85. The minimum number of genes in the module was set to 300. To merge potentially parallel modules, the threshold for the height of cut was set to 0.25. To further understand the role of expressed genes in the modules most closely associated with pasteurization treatment, GO and KEGG analyses were performed using DAVID v6.8, with corrected p < 0.05 set as the significance threshold. The results were visualized using the ggplot2 v3.3.2 package in R (Hamilton and Ferry, 2018).
2.8 Gene set enrichment analysis
All expressed genes were used for gene set enrichment analysis (GSEA). Genes were sorted according to the degree of differential expression, and then a predetermined set of gene enrichments were examined at the top or bottom of this sorted table to determine whether the pathway in question was activated or repressed under experimental conditions. Analysis was conducted using the clusterProfiler v4.0.0 package in R (Yu et al., 2012), and FDR <0.05 was chosen as the selection threshold.
2.9 Alternative splicing analysis
Alternative splicing (AS) events in each sample were classified and counted using Asprofile (v1.0.4) software (Florea et al., 2013). rMATs v4.1.0 (Shen et al., 2014) was used to identify AS events. The frequency of AS events was quantified as the percent spliced in (PSI) based on the sorted bam files. rMATs utilizes proportions of exon-exon junction reads to calculate PSI (IncLevel) values. Significantly different AS events were assessed by ΔPSI (|IncLevel1-IncLevel2|) > 0.05 and FDR <0.01, and IncLevel1-IncLevel2 represented the difference in PSI values between the pasteurized and unpasteurized groups. By referring to the KOBAS v3.0 (Bu et al., 2021), KEGG enrichment analysis (p-value < 0.05) of differentially splicing genes was performed to identify significantly enriched pathways and key genes.
3 Results
3.1 Effect of pasteurized colostrum on growth traits of calves
The differences in body weight between calves in the pasteurized and unpasteurized groups in all months tested was significant (p < 0.05; Table 1). Calves in the pasteurized group had significantly higher body heights than calves in the unpasteurized group at 3 and 4 months of age (p < 0.05), but not at 1, 2, 5, and 6 months of age. With respect to daily weight gain, the pasteurized group calves had generally higher daily weight gains than the unpasteurized group; with significant differences detected at 2, 3, and 6 months of age (p < 0.05), but not significant at other months (p >0.05).
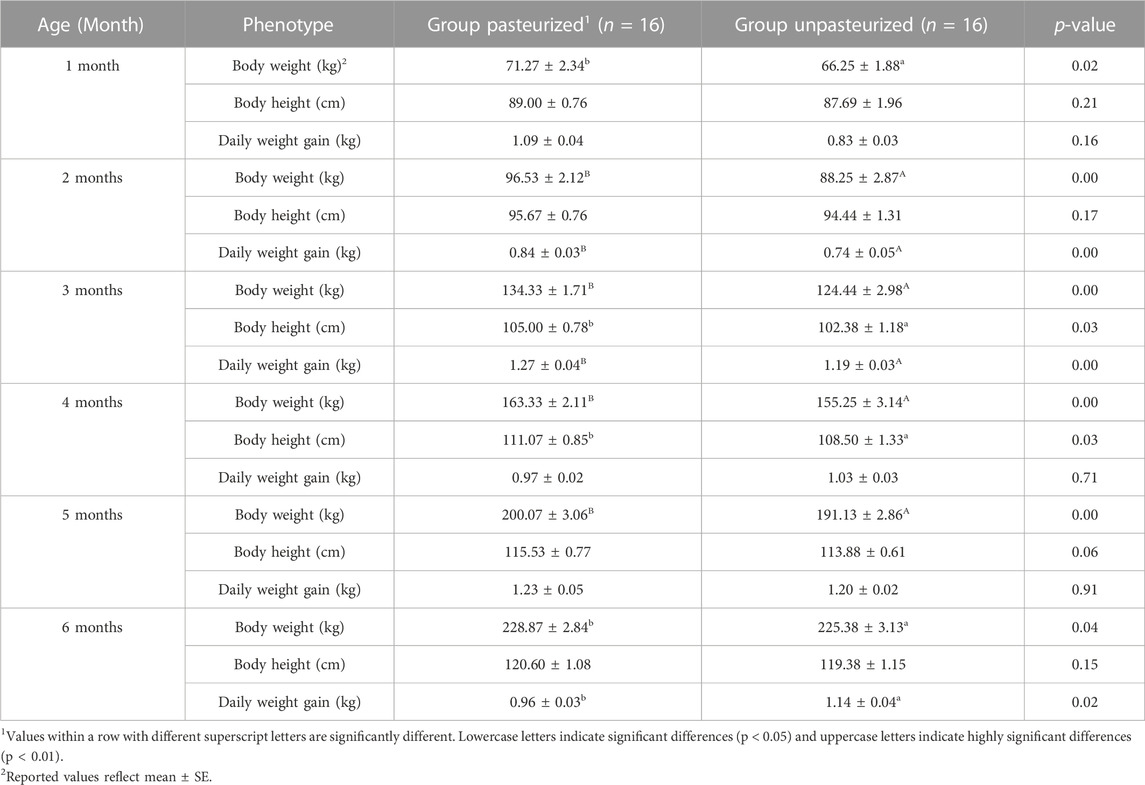
TABLE 1. Effect of pasteurized colostrum on body height, body weight and daily weight gain of calves.
3.2 Sequencing data mapping statistics
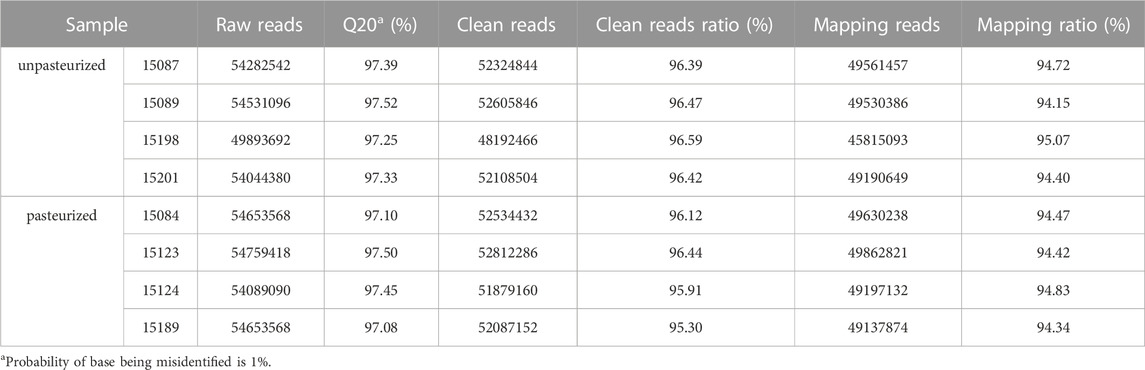
Transcriptome sequencing was performed on eight whole blood samples. An average of 53.86 million raw reads were generated per sample. After filtration, the clean reads ranged from 48,192,466 to 52,812,286 (>96.21%). The Q20 values ranged from 97.08% to 97.52%. On average, 94.55% of the reads were mapped to the reference genome (Table 2).
3.3 DEGs and functional enrichment analysis
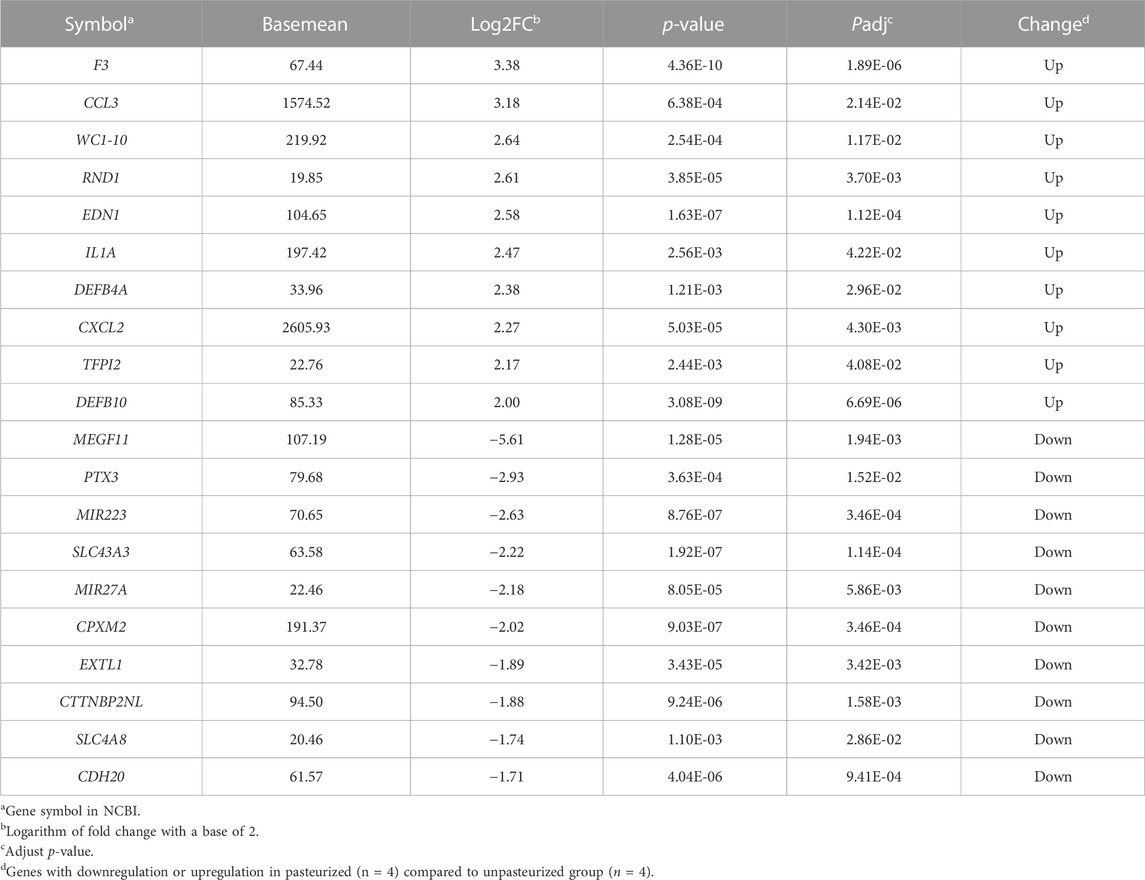
In our study, a total of 20,679 expressed genes were detected and 630 DEGs were obtained. Of these, 235 DEGs were up-regulated and 395 were down-regulated in the pasteurized group (compared to the unpasteurized group) (Figure 1A). The top 10 upregulated and downregulated genes in the pasteurized groups are listed in Table 3. Among these genes, chemokine ligand 3 (CCL3), CXC motif chemokine ligand 2 (CXCL2), interleukin one alpha (IL1A), multiple EGF like domains 11 (MEGF11), and pentraxin 3 (PTX3) are associated with growth as well as the immune response in calves.
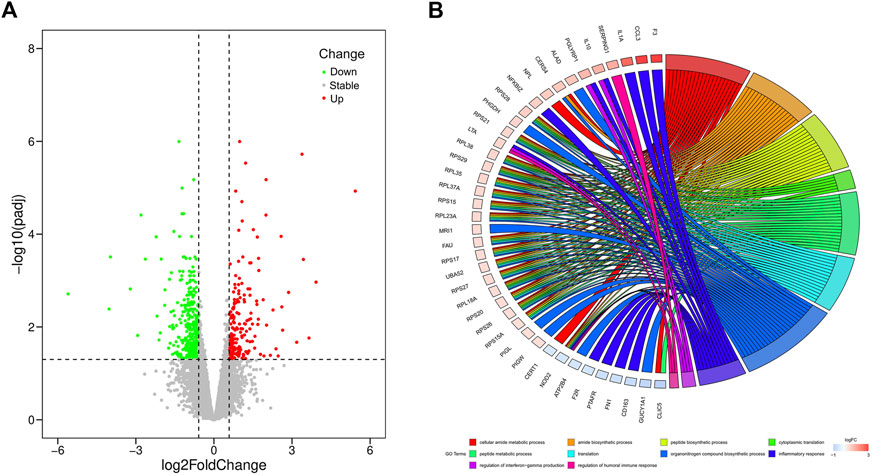
FIGURE 1. Volcano and circos plots of differentially expressed genes. (A) Volcano plot for expressed genes, it represents the -log10 (p-value) (y-axis) of genome-wide genes in relation to their respective log2 (fold change) (x-axis). Red dots represent up-regulated genes, green dots represent down-regulated genes, and gray dots represent genes with no significant difference in expression (|log2FC|≥0.585 and FDR<0.05). (B) Circos plot for functional enrichment of DEGs. The color gradient from blue to red on the left side of the graph represents the change of genes from down-regulation to up-regulation, and the terms on the right side are represented by different colors.
To investigate the relationship between DEGs and pasteurization, we performed GO enrichment analysis (Figure 1B). These DEGs were mainly enriched in protein anabolic pathways such as cytoplasmic translation, peptide biosynthesis, and peptide metabolism. They were also involved in pathways related to the immune response, such as the humoral immune response, negative regulation of the immune effector process, and the regulation of IL8 production. Some identified DEGs are among genes of signaling pathways involved in cellular substance metabolism, such as cellular amide metabolic processes, amide biosynthetic processes, and calcium ion transmembrane import into the cytosol. KEGG analysis revealed that 13 pathways were significantly enriched between the pasteurized and unpasteurized groups (Supplementary Figure S2) (corrected p < 0.05). Most enriched pathways were related to immune responses, including the cytokine–cytokine receptor interactions, tumor necrosis factor (TNF) signaling pathway, and chemokine signaling pathway. Some pathways were related to metabolism, including ribosome, thermogenic, and gap junction pathways. From these results we inferred that growth and developmental differences between pasteurized and unpasteurized groups may be attributed to protein function, immune response, and metabolic efficiency.
3.4 Protein-protein interaction network analysis
PPI is mainly used to study the interaction network between proteins, which helps to identify core regulatory genes. PPI analysis led to the construction of a PPI network containing 158 nodes and 443 edges. The top 15 DEGs (Supplementary Table S3) were identified through four methods (Degree, EPC, MCC, and MNC) in the CytoHubba plugin and overlapping DEGs were considered core genes (UBA52, RPS28, RPS14, FAU, RPS15, RPS17, RPS29, RPS20, RPL18A, RPS21, RPL38, RPL37A, RPL35, RPL36, and RPS15A). Most genes encoded ribosomal proteins, which play key roles in ribosome assembly and function. They also regulate protein synthesis and metabolic activities in response to cell growth and proliferation, and therefore play key roles in calf growth. In addition, three key subnetworks (Figures 2A–C) were identified through MCODE analysis: subnetwork 1 (Score = 18.889) had 19 nodes and 170 edges, subnetwork 2 (Score = 6.000) had six nodes and 15 edges, and subnetwork 3 (Score = 5.667) had seven nodes and 17 edges. Our analysis revealed that the core genes were mainly present in subnetwork one.
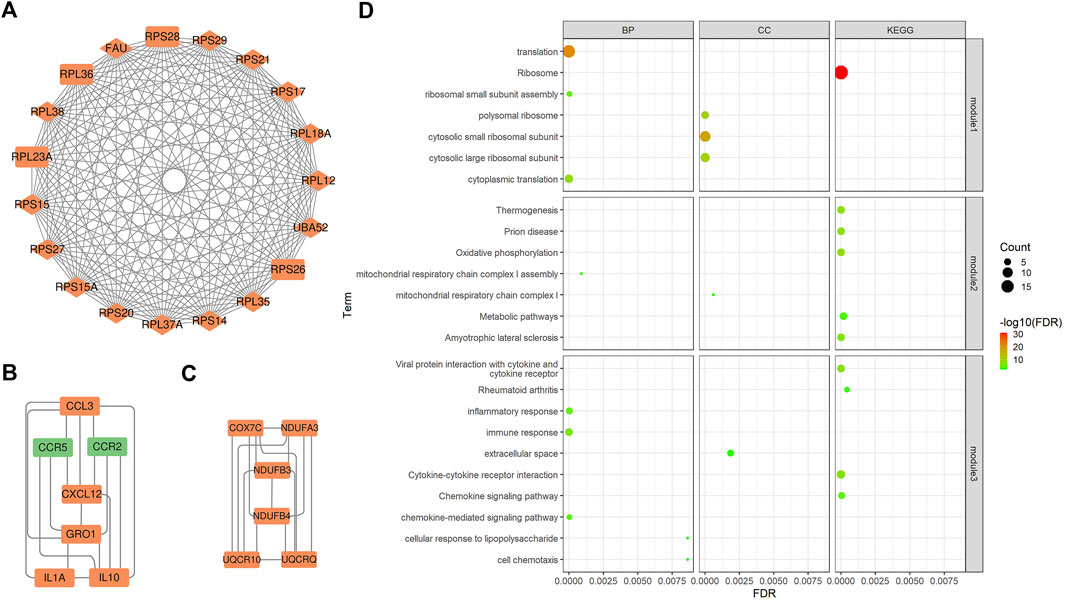
FIGURE 2. Key subnetworks from PPI networks and their enrichment analysis. (A), (B), and (C) Three key subnetworks. The red and green nodes represent the up- and down-regulated genes in the shapes of “diamond” and “rectangle”, represent core genes and DEGs. (D)The bubble plots of functional enrichment analysis of the three key subnetworks. The size of the dots represents the number of genes in the pathway and the color represents the pathway significance.
To understand the biological functions involved in the key subnetworks, a functional enrichment analysis (corrected p < 0.05) was performed (Figure 2D). According to the results, genes in subnetwork one were significantly enriched in ribosome-related pathways, such as polysomes ribosomes, ribosomes, and small ribosomal subunits, as well as in translation-related pathways including, translation, cytoplasmic translation, and RNA binding. Genes in subnetwork two were significantly enriched in electron transport-related pathways, such as mitochondrial respiratory chain complex I and oxidative phosphorylation pathways. Genes in subnetwork three were significantly enriched in pathways related to immune responses, such as the chemokine signaling, the MAPK cascade and the NF-kappa B signaling pathways.
3.5 Weighted gene Co-expression network analysis
WGCNA is a powerful tool for finding clusters (modules) of highly correlated genes (Xing et al., 2021; Yang et al., 2021; Yang et al., 2022). The soft threshold power was chosen to be seven when 0.85 was used as the correlation coefficient threshold (Figure 3A). Twelve co-expression modules were constructed through WGCNA analysis (Figure 3B). The major modules were the turquoise module (7287 genes), blue module (1431 genes), brown module (817 genes), yellow module (663 genes), and green module (573 genes). These modules were independent of each other (Figure 3C).
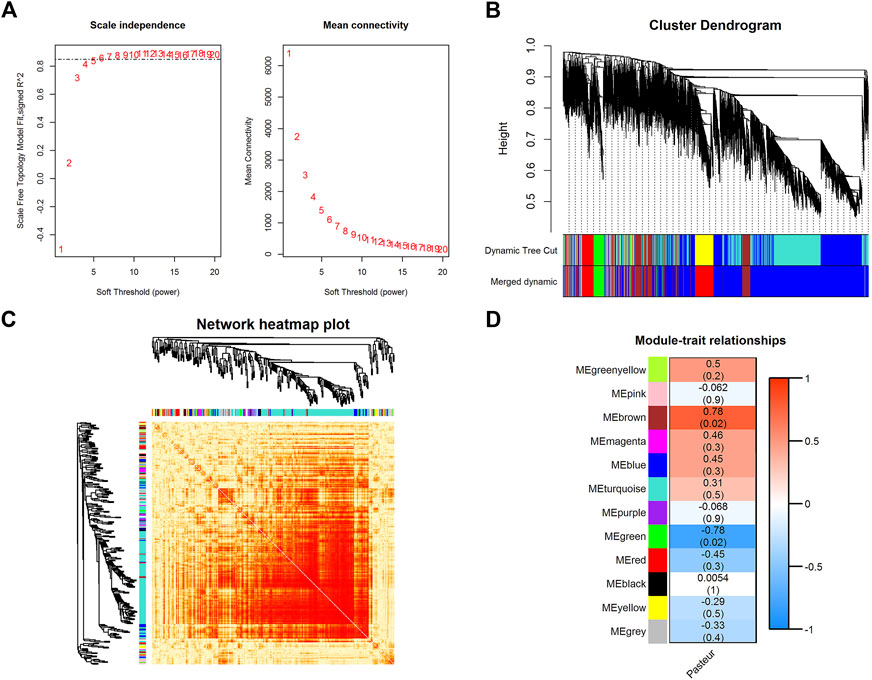
FIGURE 3. WGCNA of all expressed genes. (A) The analysis of the scale-free fit index and mean connectivity for diverse soft-thresholding powers. (B)Hierarchical clustering dendrograms showing 12 modules of co-expressed genes. Each branch in the clustering tree represents a gene, while co-expression modules were constructed in different colors. (C) Network heatmap in the co-expression modules (the yellow color scale indicates the degree of overlap between functional modules). (D) Module-trait association heatmap where each row corresponds to a module eigengene and the columns correspond to a trait. Each cell contains the correlation and p-value for the corresponding differentiation stages. The scale bar indicates the color coding for the correlations, with blue to red indicating low to high correlations, respectively.
After the gene co-expression modules were obtained, the modules were correlated with phenotype. The results showed that the brown module (r = 0.78, p = 0.02) and the green module (r = −0.78, p = 0.02) were significantly correlated with the phenotype (Figure 3D; Supplementary Figure S3), containing 817 and 573 genes, respectively. Among these two modules, PFKFB3, CALHM2, FBXO8, UTP15, CRK, METTL9, TMEM80, FBXW7, CLEC4A, NUDT4, TCN2, ROPN1L, GRAMD1A, AIMP2, CDKN1A, TDRD6, and MST1 had a high gene significance with phenotype (Supplementary Table S4) (|Gene significance| > 0.8), and were considered as key genes (Supplementary Figures S4, S5). KEGG enrichment analysis of key genes (corrected p < 0.05) (Supplementary Figures S6, S7) in both modules revealed that both modules were significantly enriched in pathways related to mitochondrial function, such as mitochondrial matrix, oxidative phosphorylation, and reactive oxygen species production pathways. They were also closely related to substance and energy metabolism, such as carbon metabolism, fatty acid metabolism, the biosynthesis of amino acids, and the tricarboxylic acid cycle. In addition, they were involved in immune-related pathways such as the IL17 signaling pathway, the TNF signaling pathway, the NF-kappa B signaling pathway, and the NOD-like receptor (NLR) signaling pathway. These results suggest that key genes in both modules were involved in substance and energy metabolism, and immune-related pathways, which may be closely related to the effects of colostrum pasteurization treatment on calf vital activity.
3.6 Gene set enrichment analysis
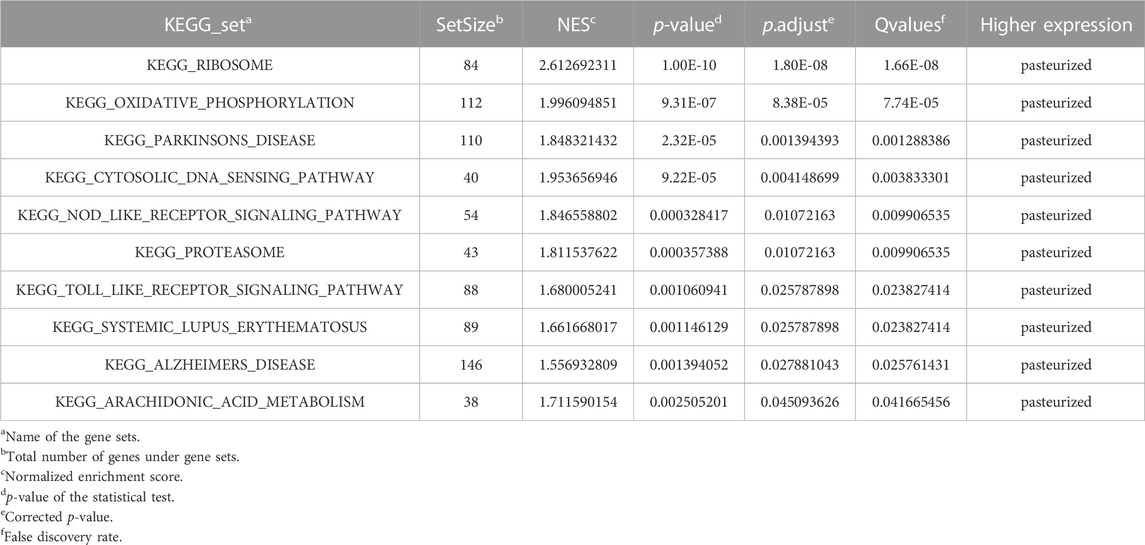
All 10 gene sets were significantly enriched in the pasteurized group (Table 4; Supplementary Figure S8). These gene sets are mainly involved in cellular substance and energy metabolism pathways, such as oxidative phosphorylation, proteasome metabolism, and arachidonic acid metabolism pathways. They are also involved in immune-related pathways, such as the cytosolic DNA sensing pathway, the NLR signaling pathway, and the TOLL-like receptor signaling pathway. These results have shown that colostrum pasteurization affected late growth in calves, probably mainly through modulation of immune and metabolic pathways.
3.7 Alternative splicing analysis
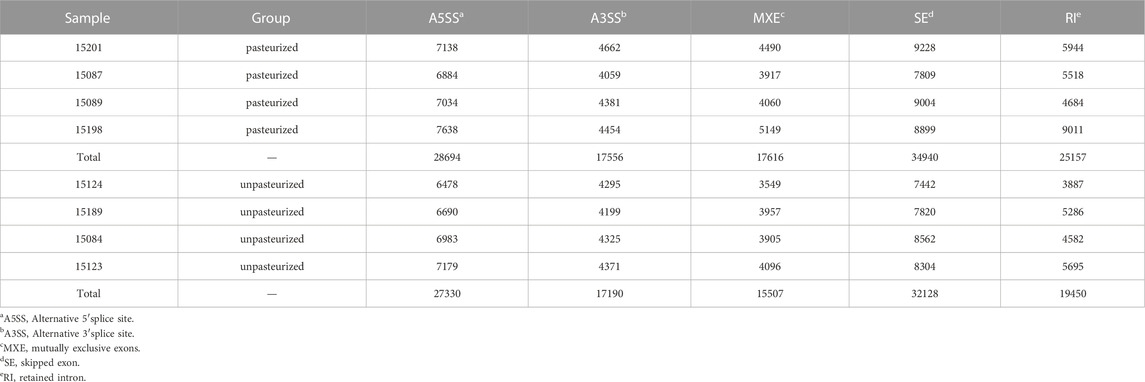
In an mRNA precursor, different exons usually have different AS patterns. There are five common types of AS patterns, namely skipped exon (SE), alternative 5′splice site (A5SS), alternative 3′splice site (A3SS), mutually exclusive exons (MXE), and retained intron (RI) patterns. In the blood of calves fed pasteurized colostrum, the incidence of A5SS and SE patterns were the highest, followed by RI, A3SS, and MXE patterns (Table 5). rMATS detected differential AS events, including 136 genes of the SE type, 10 genes of the MXE type, and one gene of the SE type (Supplementary Table S5).
KEGG enrichment analysis was performed on genes associated with differential AS events (Figure 4). The enriched pathways, namely, the RIG-I-like receptor signaling pathway, the cytosolic DNA-sensing pathway, the NLR signaling pathway, endocytosis, and the Th17 cell differentiation pathway, which influence the immune response activity and are associated with substance and energy metabolism-related pathways, such as ribosome biogenesis in eukaryotes, peroxisome, thiamine metabolism, and fatty acid biosynthesis.
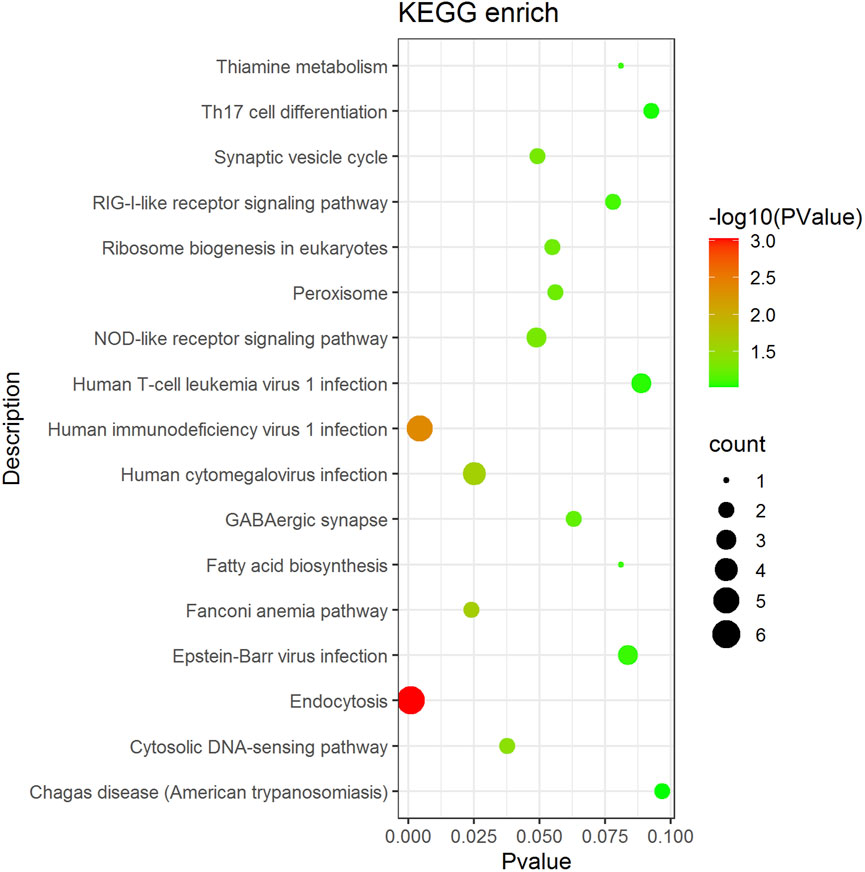
FIGURE 4. Main pathway enrichment of AS genes. The dot size represents the number of genes in the pathway, the color represents the pathway significance and the vertical coordinate represents the KEGG pathway.
4 Discussion
Colostrum quality can directly affect the growth and health of calves (Armengol and Fraile, 2020), and consequently farm sustainability. In the last few years, studies have used different histological methods to identify the effects of different milk qualities on animal growth. However, the results obtained from studies based on different animal species and tissue types were highly variable (Crisa et al., 2016; Lu et al., 2016; Xu W. et al., 2021; Keel et al., 2021). Colostrum absorption has an impact on the serum composition of newborn calves; first, colostrum contains a high amount of immunoglobulins and newborn calves must acquire passive immunity from colostrum (Blum, 2006; Mason et al., 2022). Second, colostrum is enriched with nutrient-active substances, including lactic acid, amino acids, and glycerol. Study has shown that following the intake of colostrum, calves have higher serum concentrations of glutamate, histidine, asparagine, thymidine, palmitic, and linolenic acids; these are metabolites associated with the biosynthesis of unsaturated fatty acids as well as nitrogen and galactose metabolism (Qi et al., 2018). In addition, colostrum contains other potentially bioactive substances, such as hormones and growth factors. These bioactive substances stimulate and promote intestinal epithelium growth in newborns and improve the function and absorption capacity of the gastrointestinal tract (Hammon et al., 2013; Fischer et al., 2018). Therefore, early intake of pasteurized colostrum is essential for the health, as well as the growth of newborn calves. Considering the above reasons, blood was selected as the sample for RNA-seq in this study to research the effect of gene expression changes caused by pasteurized colostrum on the growth traits in calves.
As expected, no significant difference was observed between the two groups of calves in terms of newborn weight, however the test calves fed pasteurized colostrum had significantly higher body weights, in all months tested, than the control group (unpasteurized colostrum). The pasteurized group had significantly higher body heights at 3 and 4 months of age than the unpasteurized group. Significant differences in daily weight gain were observed between the pasteurized and unpasteurized groups at 2, 3 and 6 months of age, indicating that pasteurized colostrum is crucial for the growth and development of calves. Study has proven that pasteurized colostrum not only reduces pre-weaning calf morbidity and diarrhea but also increases body weight and daily weight gain (Kargar et al., 2020). The results of the present experiment are generally consistent with those of the previous study. These results highlight that pasteurized colostrum has positive effects on the growth and development of calves.
In this study, transcriptome sequencing was used to screen the effect of pasteurized versus unpasteurized colostrum on blood-expressed genes in calves. In total, 630 DEGs (235 up-regulated and 395 down-regulated genes) were detected, and GO and KEGG enrichment analyses showed that DEGs were mainly enriched in biological processes such as the immune response, substance and energy metabolism. The exposure of calves to pathogenic bacteria through colostrum may lead to disease (Godden et al., 2019), so the associated immune response may be related to pathogenic bacteria in colostrum. Furthermore, by PPI analysis, we identified some key genes, such as IL10, NOD2, CCL3, and COX7C. Although these DEGs were not validated by qRT-PCR in this study, they have been reported to be associated with organismal immunity. IL10 may be an important immune response regulator in the central nervous system, mainly involved in chronic infections and neurodegenerative processes, and thus may affect internal environment homeostasis and behavior (Saraiva and O'Garra, 2010; Kwilasz et al., 2015). In humans and mice, IL10 was found to have a crucial role in the prevention of inflammatory bowel disease (IBD) (Kuhn et al., 1993). IL10 is also involved in immune regulation in newborns (Bullens et al., 2012; Raedler et al., 2013), suggesting that colostrum containing high amounts of immunoglobulins may be involved in the IL10-related cytokine–cytokine receptor interaction pathway in newborn calves. NOD2 and IBD are closely related, NOD2 has a host defense function in the gut mucosa and inhibits IBD by down-regulating RIP2 expression (Watanabe et al., 2019). NOD2 regulates NF-κB and MAPK pathways through RIP2, which in turn induces gene expression of pro-inflammatory cytokines and mediators (McDaniel et al., 2016; Zaidi and Wine, 2018). CCL3 is a chemokine involved in various inflammatory responses through chemokine receptors (namely CCR1 and CCR5), and has a key role in several human diseases such as cancer and HIV infection (Jones et al., 2011; Kanzler et al., 2012; Kanegasaki et al., 2014). COX7C is a subunit of cytochrome c oxidase and is responsible for electron transfer from cytochrome c to oxygen (Hofmann et al., 1998). In addition, studies have shown that COX7C is a key regulator in the physiological processes of numerous diseases, where upregulation of this gene is associated with the development of many cancer tumors (Wang et al., 2017; Wang et al., 2022) and its downregulation is a feature of many chronic kidney diseases (Zaza et al., 2013; Wu et al., 2020).
WGCNA and GSEA analyses revealed that most pathways enriched were related to the immune response and metabolism, in which the NLR signaling, fatty acid metabolism and cytosolic DNA sensing pathways were mostly observed. The NLR signaling pathway has an important role in the regulation of innate immune antiviral effects. Study has shown that NLRs are closely linked to IBD and the role of NOD2 recognition of intestinal microbiota, which is vital for maintaining intestinal immune cell homeostasis (Jiang et al., 2013). NOD2 signaling promotes hyper-reactive macrophages and colitis in IL10-deficient mice, and NOD2 deficiency in IL10−/− mice leads to a significant improvement in chronic colitis (Jamontt et al., 2013). Fatty acids, major energy substances in mammals, act as signaling molecules involved in energy metabolism in humans and animals. The cytosolic DNA sensing pathway is an evolutionarily conserved mechanism that initiates a rapid innate immune response to microbial pathogens (Kwon and Bakhoum, 2020). This pathway may be related to the immune response induced by pathogenic microorganisms in colostrum.
Alternative splicing (AS) is widely present in eukaryotic organisms and specific splicing isoforms are produced in different tissues or developmental stages. Thus, gene expression changes in AS events are associated with organismal life activities and diseases. In this study, feeding calves colostrum with or without pasteurization revealed that some genes involved in signaling pathways in calves underwent AS, such as genes involved in endocytosis (BOLA-NC1, AP2M1, SPG21, GIT2, and SMAD2) and the NLR signaling pathway (RIPK1, TMEM173, and MEFV). Major histocompatibility complex class I (MHC-I) molecules are cell surface glycoproteins that play a key role in triggering immune responses (Shi et al., 2018). Of them, BOLA-NC1 is one of the non-classical (MHC-Ib) isoforms. BOLA-NC1 is a critical immunomodulatory molecule that interacts with inhibitory receptors expressed by natural killer cells, T lymphocytes, and antigen-presenting cells, thereby inhibiting these immune cells (Parasar et al., 2016; Geng and Raghavan, 2021). RIPK1 is an upstream regulator that controls cell survival and inflammatory signaling as well as multiple cell death pathways, including apoptosis and necroptosis (Xu D. C. et al., 2021). RIPK1 deficiency leads to the accumulation of aspartate in starved cells and tissues, which in turn inhibits AMPK signaling pathway and cellular autophagy, and increases tricarboxylic acid (TCA) activity and production of the energy metabolite ATP, thereby resulting in impaired regulation of starvation stress under starvation conditions (Mei et al., 2021). Pathogen DNA from infection or DNA from damaged cells stimulates STING (stimulator of interferon genes)-dependent type I interferon production and promotes inflammation (Ishikawa and Barber, 2008). Furthermore, the TMEM173 gene encodes the STING protein, a key player in host defense against pathogens (Patel and Jin, 2019; Zhang et al., 2020).
A comparison of differentially spliced genes and DEGs revealed five overlapping genes, namely SLCO4A1, AKR1C4, MED13L, SENP5, and C16orf72. Among them, AKR1C4 play roles in immune and metabolic processes (Onsurathum et al., 2018). AKR1C4 is a liver-specific gene that regulates vital metabolism pathways, such as steroid hormone metabolism, bile acid synthesis and xenobiotic metabolism (Rižner and Penning, 2014). SLCO4A1, a potential biomarker for various cancers, is involved in neutrophil-mediated immune responses (Ban et al., 2017; Wang et al., 2021). MED13L play key roles in gene transcription and in the inflammatory response through participation in the Wnt signaling pathway. MED13L can also participate in the inflammatory response and positively regulate the secretion of pro-inflammatory factors by regulating and activating transcription factor 4 expression (Iwasaki et al., 2014; Asadollahi et al., 2017). In addition, MED13L was reported to be involved in FGF and Rb/E2F-related pathways, thus playing a crucial role in the regulation of immune responses (Angus and Nevins, 2012; Utami et al., 2014). Genes that undergo AS are mainly involved in intercellular inflammatory responses, signaling, and metabolic activities. However, the mechanisms underlying the role of AS genes in the immune response to pathogenic bacteria in colostrum are not well studied and need to be explored further.
Our results exemplify the intricate relationship between metabolism and immune, especially in DEGs functional enrichment. The immune system requires large amounts of energy to function, and immune cells can reprogram their metabolism to meet these energy demands. A common example of metabolic reprogramming is the coupling of the mitochondrial TCA cycle to oxidative phosphorylation (OXPHOS), which is an efficient way of energy production (Chou et al., 2022). During Brucella infection, TMEM173 activation polarizes M2 macrophages to M1 macrophages, inhibits prolyl hydroxylase (PHD) activity, and increases mitochondrial ROS, thereby stabilizing hypoxia-inducible factor-1α (HIF-1α). This leads to a decrease in OXPHOS and an increase in glycolysis, thereby altering cellular immunometabolism (Downey et al., 2014; Gomes et al., 2021). The NLR signaling pathway recognizes specific pathogen-associated molecules in the cytoplasm or host-derived injury signals and activates inflammatory responses, and the NLRP3 is a member of the NLR family, NLPR3 activation could result in the production of pro-inflammatory cytokines (Saxena and Yeretssian, 2014; Kim et al., 2016). Salmonella infection causes disruption of glycolysis, leading to a decrease in NAD+/NADH, which activates NLRP3 inflammasome (Palsson-McDermott et al., 2015; Sanman et al., 2016). NLRP3 inflammasome promotes glycolysis while reducing OXPHOS, resulting in the accumulation of the TCA cycle intermediate succinate, succinate inhibits PHD and promotes HIF-1α stabilization, leading to the upregulation of glycolytic enzymes (Selak et al., 2005; Tannahill et al., 2013; Parsamanesh et al., 2019). Thus, the activation of the NLRP3 inflammasome creates a metabolic cycle. During the metabolic activity of calves, pathogenic microorganisms interfere with relevant metabolic circuits, leading to the accumulation of specific endogenous substances that induce an inflammatory response.
Although we identified DEGs and pathways underlying the effects of pasteurized colostrum on calf growth, our study has some limitations. First, the sample size used for RNA-seq was too small and needs to be further expanded to improve our study reproducibility. Second, we used blood as the study sample, which is a non-invasive and cost-saving method; however, the results of the assay may be compromised compared to more invasive tissue samples. Our future work will focus on the validation of the DEGs and pathways in additional animals.
5 Conclusion
The present study identified DEGs in the blood of calves fed with pasteurized and unpasteurized colostrum. The DEGs were mainly enriched in immune responses and energy metabolism pathways, which have key roles in regulating calf growth. Our study identifies the molecular mechanisms involved in the beneficial effects of pasteurized colostrum on calf growth, which will help optimize calf colostrum management practices.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA893578.
Ethics statement
The animal study was reviewed and approved by the Animal Ethical and Welfare Committee of Ningxia University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
XK and YS designed this study. CL, YD, YZ, XW, XZ, and ZS collected samples and recorded their phenotypes. WM and LZ extracted RNA. CL and SL analyzed the RNA-seq data. CL and CY enabled data visualization. CL wrote the manuscript. YM helped for revising the reviewed manuscript. XK supervised the study and revised the manuscript. All authors read and approved the manuscript.
Funding
This research was financially supported by the National Key Research and Development Program of China (2018YFD0501603), Key Research and Development Program of Ningxia Province (2018BBF33006, 2017BY078), National Modern Agriculture Industry Technology System (CARS-36), The National Natural Science Foundation of China (32160776) and Natural Science Foundation of Ningxia Province (2021AAC03027).
Acknowledgments
We thank all the breeding staff who helped with our experiment, and we would like to thank the reviewers for their time and valuable comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1075950/full#supplementary-material.
References
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Nat. Preced 11, R106. doi:10.1038/npre.2010.4282.2
Angus, S. P., and Nevins, J. R. (2012). A role for Mediator complex subunit MED13L in Rb/E2F-induced growth arrest. Oncogene 31, 4709–4717. doi:10.1038/onc.2011.622
Armengol, R., and Fraile, L. (2016). Colostrum and milk pasteurization improve health status and decrease mortality in neonatal calves receiving appropriate colostrum ingestion. J. Dairy Sci. 99, 4718–4725. doi:10.3168/jds.2015-10728
Armengol, R., and Fraile, L. (2020). Feeding calves with pasteurized colostrum and milk has a positive long-term effect on their productive performance. Animals 10, 1494. doi:10.3390/ani10091494
Asadollahi, R., Zweier, M., Gogoll, L., Schiffmann, R., Sticht, H., Steindl, K., et al. (2017). Genotype-phenotype evaluation of MED13L defects in the light of a novel truncating and a recurrent missense mutation. Eur. J. Med. Genet. 60, 451–464. doi:10.1016/j.ejmg.2017.06.004
Ban, M. J., Ji, S. H., Lee, C. K., Bae, S. B., Kim, H. J., Ahn, T. S., et al. (2017). Solute carrier organic anion transporter family member 4A1 (SLCO4A1) as a prognosis marker of colorectal cancer. J. Cancer Res. Clin. Oncol. 143, 1437–1447. doi:10.1007/s00432-017-2393-7
Blum, J. W. (2006). Nutritional physiology of neonatal calves. J. Animal Physiology Animal Nutr. 90, 1–11. doi:10.1111/j.1439-0396.2005.00614.x
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-I: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi:10.1093/nar/gkab447
Bullens, D. M., Kasran, A., Dilissen, E., and Ceuppens, J. L. (2012). Neonatal IL-10 production and risk of allergy development. Clin. Exp. Allergy 42, 483–484. doi:10.1111/j.1365-2222.2011.03952.x
Chou, W. C., Rampanelli, E., Li, X., and Ting, J. P. Y. (2022). Impact of intracellular innate immune receptors on immunometabolism. Cell. Mol. Immunol. 19, 337–351. doi:10.1038/s41423-021-00780-y
Crisa, A., Ferre, F., Chillemi, G., and Moioli, B. (2016). RNA-Sequencing for profiling goat milk transcriptome in colostrum and mature milk. Bmc Veterinary Res. 12, 264. doi:10.1186/s12917-016-0881-7
Cummins, C., Berry, D. P., Sayers, R., Lorenz, I., and Kennedy, E. (2016). Questionnaire identifying management practices surrounding calving on spring-calving dairy farms and their associations with herd size and herd expansion. Animal 10, 868–877. doi:10.1017/S1751731116000124
Downey, C. M., Aghaei, M., Schwendener, R. A., and Jirik, F. R. (2014). DMXAA causes tumor site-specific vascular disruption in murine non-small cell lung cancer, and like the endogenous non-canonical cyclic dinucleotide STING agonist, 2 ' 3 '-cGAMP, induces M2 macrophage repolarization. Plos One 9, e99988. doi:10.1371/journal.pone.0099988
Elizondo-Salazar, J. A., and Heinrichs, A. J. (2009). Feeding heat-treated colostrum or unheated colostrum with two different bacterial concentrations to neonatal dairy calves. J. Dairy Sci. 92, 4565–4571. doi:10.3168/jds.2009-2188
Fechner, K., Dreymann, N., Schimkowiak, S., Czerny, C. P., and Teitzel, J. (2019). Efficacy of dairy on-farm high-temperature, short-time pasteurization of milk on the viability of Mycobacterium avium ssp. paratuberculosis. J. Dairy Sci. 102, 11280–11290. doi:10.3168/jds.2019-16590
Fischer, A. J., Malmuthuge, N., Guan, L. L., and Steele, M. A. (2018). Short communication: The effect of heat treatment of bovine colostrum on the concentration of oligosaccharides in colostrum and in the intestine of neonatal male Holstein calves. J. Dairy Sci. 101, 401–407. doi:10.3168/jds.2017-13533
Fleenor, W. A., and Stott, G. H. (1980). Hydrometer test for estimation of immunoglobulin concentration in bovine colostrum. J. Dairy Sci. 63, 973–977. doi:10.3168/jds.S0022-0302(80)83034-7
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2, 188. doi:10.12688/f1000research.2-188.v1
Geng, J., and Raghavan, M. (2021). Conformational sensing of major histocompatibility complex (MHC) class I molecules by immune receptors and intracellular assembly factors. Curr. Opin. Immunol. 70, 67–74. doi:10.1016/j.coi.2021.03.014
Godden, S. M., Lombard, J. E., and Woolums, A. R. (2019). Colostrum management for dairy calves. Veterinary Clin. North America-Food Animal Pract. 35, 535–556. doi:10.1016/j.cvfa.2019.07.005
Gomes, M. T. R., Guimarães, E. S., Marinho, F. V., Macedo, I., Aguiar, E. R., Barber, G. N., et al. (2021). STING regulates metabolic reprogramming in macrophages via HIF-1α during Brucella infection. PLoS Pathog. 17, e1009597. doi:10.1371/journal.ppat.1009597
Hamilton, N. E., and Ferry, M. (2018). ggtern: Ternary diagrams using ggplot2. J. Stat. Softw. 87, 1–17. doi:10.18637/jss.v087.c03
Hammon, H. M., Steinhoff-Wagner, J., Flor, J., Schonhusen, U., and Metges, C. C. (2013). Lactation biology symposium: Role of colostrum and colostrum components on glucose metabolism in neonatal calves. J. Animal Sci. 91, 685–695. doi:10.2527/jas.2012-5758
Heinrichs, A. J., and Elizondo-Salazar, J. A. (2009). Reducing failure of passive immunoglobulin transfer in dairy calves. Rev. De. Med. Veterinaire 160, 436–440. doi:10.1017/s0022029921000686
Hofmann, S., Lichtner, P., Schuffenhauer, S., Gerbitz, K. D., and Meitinger, T. (1998). Assignment of the human genes coding for cytochrome c oxidase subunits Va (COX5A), VIc (COX6C) and VIIc (COX7C) to chromosome bands 15q25, 8q22-->q23 and 5q14 and of three pseudogenes (COX5AP1, COX6CP1, COX7CP1) to 14q22, 16p12 and 13q14-->q21 by FISH and radiation hybrid mapping. Cytogenet Cell Genet. 83, 226–227. doi:10.1159/000015185
Ishikawa, H., and Barber, G. N. (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. doi:10.1038/nature07317
Iwasaki, Y., Suganami, T., Hachiya, R., Shirakawa, I., Kim-Saijo, M., Tanaka, M., et al. (2014). Activating transcription factor 4 links metabolic stress to interleukin-6 expression in macrophages. Diabetes 63, 152–161. doi:10.2337/db13-0757
Jamontt, J., Petit, S., Clark, N., Parkinson, S. J., and Smith, P. (2013). Nucleotide-binding oligomerization domain 2 signaling promotes hyperresponsive macrophages and colitis in IL-10-deficient mice. J. Immunol. 190, 2948–2958. doi:10.4049/jimmunol.1201332
Jiang, W., Wang, X. Q., Zeng, B. H., Liu, L., Tardivel, A., Wei, H., et al. (2013). Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J. Exp. Med. 210, 2465–2476. doi:10.1084/jem.20122490
Jones, K. L., Maguire, J. J., and Davenport, A. P. (2011). Chemokine receptor CCR5: From AIDS to atherosclerosis. Br. J. Pharmacol. 162, 1453–1469. doi:10.1111/j.1476-5381.2010.01147.x
Kanegasaki, S., Matsushima, K., Shiraishi, K., Nakagawa, K., and Tsuchiya, T. (2014). Macrophage inflammatory protein derivative ECI301 enhances the alarmin-associated abscopal benefits of tumor radiotherapy. Cancer Res. 74, 5070–5078. doi:10.1158/0008-5472.CAN-14-0551
Kanzler, I., Liehn, E. A., Koenen, R. R., and Weber, C. (2012). Anti-inflammatory therapeutic approaches to reduce acute atherosclerotic complications. Curr. Pharm. Biotechnol. 13, 37–45. doi:10.2174/138920112798868557
Kargar, S., Roshan, M., Ghoreishi, S. M., Akhlaghi, A., Kanani, M., Shams-Abadi, A. R. A., et al. (2020). Extended colostrum feeding for 2 weeks improves growth performance and reduces the susceptibility to diarrhea and pneumonia in neonatal Holstein dairy calves. J. Dairy Sci. 103, 8130–8142. doi:10.3168/jds.2020-18355
Keel, B. N., Lindholm-Perry, A. K., Oliver, W. T., Wells, J. E., Jones, S. A., and Rempel, L. A. (2021). Characterization and comparative analysis of transcriptional profiles of porcine colostrum and mature milk at different parities. Bmc Genomic Data 22, 25. doi:10.1186/s12863-021-00980-5
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Kim, Y. K., Shin, J. S., and Nahm, M. H. (2016). NOD-like receptors in infection, immunity, and diseases. Yonsei Med. J. 57, 5–14. doi:10.3349/ymj.2016.57.1.5
Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K., and Muller, W. (1993). Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274. doi:10.1016/0092-8674(93)80068-p
Kwilasz, A. J., Grace, P. M., Serbedzija, P., Maier, S. F., and Watkins, L. R. (2015). The therapeutic potential of interleukin-10 in neuroimmune diseases. Neuropharmacology 96, 55–69. doi:10.1016/j.neuropharm.2014.10.020
Kwon, J., and Bakhoum, S. F. (2020). The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 10, 26–39. doi:10.1158/2159-8290.CD-19-0761
Langfelder, P., and Horvath, S. (2008). Wgcna: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559. doi:10.1186/1471-2105-9-559
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Lu, J., Liu, L., Pang, X. Y., Zhang, S. W., Jia, Z. H., Ma, C. L., et al. (2016). Comparative proteomics of milk fat globule membrane in goat colostrum and mature milk. Food Chem. 209, 10–16. doi:10.1016/j.foodchem.2016.04.020
Mason, W., Cuttance, E., and Laven, R. (2022). The transfer of passive immunity in calves born at pasture. J. Dairy Sci. 105, 6271–6289. doi:10.3168/jds.2021-21460
Mcdaniel, D. K., Eden, K., Ringel, V. M., and Allen, I. C. (2016). Emerging roles for noncanonical NF-κB signaling in the modulation of inflammatory bowel disease pathobiology. Inflamm. Bowel Dis. 22, 2265–2279. doi:10.1097/MIB.0000000000000858
Mcmartin, S., Godden, S., Metzger, L., Feirtag, J., Bey, R., Stabel, J., et al. (2006). Heat treatment of bovine colostrum. I: Effects of temperature on viscosity and immunoglobulin G level. J. Dairy Sci. 89, 2110–2118. doi:10.3168/jds.S0022-0302(06)72281-0
Mei, X., Guo, Y., Xie, Z., Zhong, Y., Wu, X., Xu, D., et al. (2021). RIPK1 regulates starvation resistance by modulating aspartate catabolism. Nat. Commun. 12, 6144. doi:10.1038/s41467-021-26423-4
Onsurathum, S., Haonon, O., Pinlaor, P., Pairojkul, C., Khuntikeo, N., Thanan, R., et al. (2018). Proteomics detection of S100A6 in tumor tissue interstitial fluid and evaluation of its potential as a biomarker of cholangiocarcinoma. Tumor Biol. 40, 1010428318767195. doi:10.1177/1010428318767195
Palsson-Mcdermott, E. M., Curtis, A. M., Goel, G., Lauterbach, M. A. R., Sheedy, F. J., Gleeson, L. E., et al. (2015). Pyruvate kinase M2 regulates hif-1 alpha activity and IL-1 beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 21, 347–380. doi:10.1016/j.cmet.2015.01.017
Parasar, P., Wilhelm, A., Rutigliano, H. M., Thomas, A. J., Teng, L. H., Shi, B., et al. (2016). Expression of bovine non-classical major histocompatibility complex class I proteins in mouse P815 and human K562 cells. Res. Veterinary Sci. 107, 161–170. doi:10.1016/j.rvsc.2016.06.004
Parron, J. A., Ripolles, D., Perez, M. D., Calvo, M., Rasmussen, J. T., and Sanchez, L. (2016). Effect of heat treatment on antirotaviral activity of bovine and ovine whey. Int. Dairy J. 60, 78–85. doi:10.1016/j.idairyj.2016.02.030
Parsamanesh, N., Moossavi, M., Bahrami, A., Fereidouni, M., Barreto, G., and Sahebkar, A. (2019). NLRP3 inflammasome as a treatment target in atherosclerosis: A focus on statin therapy. Int. Immunopharmacol. 73, 146–155. doi:10.1016/j.intimp.2019.05.006
Patel, S., and Jin, L. (2019). TMEM173 variants and potential importance to human biology and disease. Genes Immun. 20, 82–89. doi:10.1038/s41435-018-0029-9
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi:10.1038/nbt.3122
Pithua, P., Wells, S. J., Godden, S. M., and Stabel, J. R. (2011). Evaluation of the association between fecal excretion of Mycobacterium avium subsp paratuberculosis and detection in colostrum and on teat skin surfaces of dairy cows. Javma-Journal Am. Veterinary Med. Assoc. 238, 94–100. doi:10.2460/javma.238.1.94
Playford, R. J., and Weiser, M. J. (2021). Bovine colostrum: Its constituents and uses. Nutrients 13, 265. doi:10.3390/nu13010265
Puppel, K., Golebiewski, M., Konopka, K., Kunowska-Slosarz, M., Slosarz, J., Grodkowski, G., et al. (2020). Relationship between the quality of colostrum and the formation of microflora in the digestive tract of calves. Animals 10, 1293. doi:10.3390/ani10081293
Qi, Y. X., Zhao, X. W., Huang, D. W., Pan, X. C., Yang, Y. X., Zhao, H. L., et al. (2018). Exploration of the relationship between intestinal colostrum or milk, and serum metabolites in neonatal calves by metabolomics analysis. J. Agric. Food Chem. 66, 7200–7208. doi:10.1021/acs.jafc.8b01621
Raedler, D., Illi, S., Pinto, L. A., Von Mutius, E., Illig, T., Kabesch, M., et al. (2013). IL10 polymorphisms influence neonatal immune responses, atopic dermatitis, and wheeze at age 3 years. J. Allergy Clin. Immunol. 131, 789–796. doi:10.1016/j.jaci.2012.08.008
Rižner, T. L., and Penning, T. M. (2014). Role of aldo–keto reductase family 1 (AKR1) enzymes in human steroid metabolism. Steroids 79, 49–63. doi:10.1016/j.steroids.2013.10.012
Sanman, L. E., Qian, Y., Eisle, N. A., Ng, T. M., Van Der Linden, W. A., Monack, D. M., et al. (2016). Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. Elife 5, e13663. doi:10.7554/eLife.13663
Saraiva, M., and O'garra, A. (2010). The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181. doi:10.1038/nri2711
Saxena, M., and Yeretssian, G. (2014). NOD-like receptors: Master regulators of inflammation and cancer. Front. Immunol. 5, 327. doi:10.3389/fimmu.2014.00327
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi:10.1093/bioinformatics/btr026
Selak, M. A., Armour, S. M., Mackenzie, E. D., Boulahbel, H., Watson, D. G., Mansfield, K. D., et al. (2005). Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 7, 77–85. doi:10.1016/j.ccr.2004.11.022
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shen, S. H., Park, J. W., Lu, Z. X., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 111, E5593–E5601. doi:10.1073/pnas.1419161111
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). David: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi:10.1093/nar/gkac194
Shi, B., Thomas, A. J., Benninghoff, A. D., Sessions, B. R., Meng, Q. G., Parasar, P., et al. (2018). Genetic and epigenetic regulation of major histocompatibility complex class I gene expression in bovine trophoblast cells. Am. J. Reproductive Immunol. 79, e12779. doi:10.1111/aji.12779
Silva, F. L. M., Miqueo, E., Da Silva, M. D., Torrezan, T. M., Rocha, N. B., Salles, M. S. V., et al. (2021). Thermoregulatory responses and performance of dairy calves fed different amounts of colostrum. Animals 11, 703. doi:10.3390/ani11030703
Slosarkova, S., Pechova, A., Stanek, S., Fleischer, P., Zouharova, M., and Nejedla, E. (2021). Microbial contamination of harvested colostrum on Czech dairy farms. J. Dairy Sci. 104, 11047–11058. doi:10.3168/jds.2020-19949
Sousa, S. G., Delgadillo, I., and Saraiva, J. A. (2014). Effect of thermal pasteurisation and high-pressure processing on immunoglobulin content and lysozyme and lactoperoxidase activity in human colostrum. Food Chem. 151, 79–85. doi:10.1016/j.foodchem.2013.11.024
Stabel, J. R., Hurd, S., Calvente, L., and Rosenbusch, R. F. (2004). Destruction of Mycobacterium paratuberculosis, Salmonella spp., and Mycoplasma spp. in raw milk by a commercial on-farm high-temperature, short-time pasteurizer. J. Dairy Sci. 87, 2177–2183. doi:10.3168/jds.S0022-0302(04)70038-7
Streeter, R. N., Hoffsis, G. F., Bech-Nielsen, S., Shulaw, W. P., and Rings, D. M. (1995). Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 56, 1322–1324.
Sweeney, R. W. (1996). Transmission of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 12, 305–312. doi:10.1016/s0749-0720(15)30408-4
Tannahill, G., Curtis, A., Adamik, J., Palsson-Mcdermott, E., Mcgettrick, A., Goel, G., et al. (2013). Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242. doi:10.1038/nature11986
Utami, K. H., Winata, C. L., Hillmer, A. M., Aksoy, I., Long, H. T., Liany, H., et al. (2014). Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum. Mutat. 35, 1311–1320. doi:10.1002/humu.22636
Wang, L., Collings, C. K., Zhao, Z. B., Cozzolino, K. A., Ma, Q. H., Liang, K. W., et al. (2017). A cytoplasmic COMPASS is necessary for cell survival and triple-negative breast cancer pathogenesis by regulating metabolism. Genes & Dev. 31, 2056–2066. doi:10.1101/gad.306092.117
Wang, X. S., Wu, S. L., Peng, Z., and Zhu, H. H. (2021). SLCO4A1 is a prognosis-associated biomarker involved in neutrophil-mediated immunity in thyroid cancer. Int. J. General Med. 14, 9615–9628. doi:10.2147/IJGM.S339921
Wang, X., Wang, L. T., and Yu, B. (2022). UBE2D1 and COX7C as potential biomarkers of diabetes-related sepsis. Biomed Res. Int. 2022, 9463717. doi:10.1155/2022/9463717
Watanabe, T., Minaga, K., Kamata, K., Sakurai, T., Komeda, Y., Nagai, T., et al. (2019). RICK/RIP2 is a NOD2-independent nodal point of gut inflammation. Int. Immunol. 31, 669–683. doi:10.1093/intimm/dxz045
Windeyer, M. C., Leslie, K. E., Godden, S. M., Hodgins, D. C., Lissemore, K. D., and Leblanc, S. J. (2014). Factors associated with morbidity, mortality, and growth of dairy heifer calves up to 3 months of age. Prev. Vet. Med. 113, 231–240. doi:10.1016/j.prevetmed.2013.10.019
Wu, B. Y., Chen, S. R., Zhuang, L. H., and Zeng, J. Y. (2020). The expression level of COX7C associates with venous thromboembolism in colon cancer patients. Clin. Exp. Med. 20, 527–533. doi:10.1007/s10238-020-00644-1
Xing, K., Liu, H., Zhang, F., Liu, Y., Shi, Y., Ding, X., et al. (2021). Identification of key genes affecting porcine fat deposition based on co-expression network analysis of weighted genes. J. Anim. Sci. Biotechnol. 12, 100. doi:10.1186/s40104-021-00616-9
Xu, D. C., Zou, C. Y., and Yuan, J. Y. (2021a). Genetic regulation of RIPK1 and necroptosis. Annu. Rev. Genet. 55, 235–263. doi:10.1146/annurev-genet-071719-022748 55
Xu, W., Mann, S., Curone, G., and Kenez, A. (2021b). Heat treatment of bovine colostrum: Effects on colostrum metabolome and serum metabolome of calves. Animal 15, 100180. doi:10.1016/j.animal.2021.100180
Yang, C. Y., Han, L. Y., Li, P., Ding, Y. L., Zhu, Y., Huang, Z. W., et al. (2021). Characterization and duodenal transcriptome analysis of Chinese beef cattle with divergent feed efficiency using RNA-seq. Front. Genet. 12, 741878. doi:10.3389/fgene.2021.741878
Yang, C. Y., Zhu, Y., Ding, Y. L., Huang, Z. W., Dan, X. G., Shi, Y. G., et al. (2022). Identifying the key genes and functional enrichment pathways associated with feed efficiency in cattle. Gene 807, 145934. doi:10.1016/j.gene.2021.145934
Yu, G. C., Wang, L. G., Han, Y. Y., and He, Q. Y. (2012). clusterProfiler: an R Package for comparing biological themes among gene clusters. Omics-a J. Integr. Biol. 16, 284–287. doi:10.1089/omi.2011.0118
Zaidi, D., and Wine, E. (2018). Regulation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κβ) in inflammatory bowel diseases. Front. Pediatr. 6, 317. doi:10.3389/fped.2018.00317
Zaza, G., Granata, S., Masola, V., Rugiu, C., Fantin, F., Gesualdo, L., et al. (2013). Downregulation of nuclear-encoded genes of oxidative metabolism in dialyzed chronic kidney disease patients. Plos One 8, e77847. doi:10.1371/journal.pone.0077847
Zhang, B., and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, 17. doi:10.2202/1544-6115.1128
Keywords: pasteurization, Holstein, growth traits, immunity, colostrum
Citation: Li C, Li S, Yang C, Ding Y, Zhang Y, Wang X, Zhou X, Su Z, Ming W, Zeng L, Ma Y, Shi Y and Kang X (2023) Blood transcriptome reveals immune and metabolic-related genes involved in growth of pasteurized colostrum-fed calves. Front. Genet. 14:1075950. doi: 10.3389/fgene.2023.1075950
Received: 07 November 2022; Accepted: 24 January 2023;
Published: 06 February 2023.
Edited by:
Eveline M. Ibeagha-Awemu, Agriculture and Agri-Food Canada (AAFC), CanadaCopyright © 2023 Li, Li, Yang, Ding, Zhang, Wang, Zhou, Su, Ming, Zeng, Ma, Shi and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Kang, a2FuZ3hsOTUyN0AxMjYuY29t
 Chenglong Li
Chenglong Li Shuzhen Li
Shuzhen Li Chaoyun Yang
Chaoyun Yang Xiaowei Wang
Xiaowei Wang Xiaonan Zhou
Xiaonan Zhou Yuangang Shi
Yuangang Shi Xiaolong Kang
Xiaolong Kang