
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 06 October 2022
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.999695
Background: This study analyzed the phenotypes and genotypes of 41 Chinese families with inherited retinal dystrophy (IRD) and RPGR gene mutations.
Methods: This retrospective analysis evaluated a cohort of 41 patients who were subjected to a specific Hereditary Eye Disease Enrichment Panel (HEDEP) analysis. All (likely) pathogenic variants were determined by Sanger sequencing, and co-segregation analyses were performed on the available family members. All cases were subjected to Sanger sequencing for RPGR open reading frame 15 (ORF15) mutations.
Results: A total of 41 probands from different families with a clinical diagnosis of retinitis pigmentosa (RP; 34 cases) and cone-rod dystrophy (CORD; 7 cases) were included in this cohort. According to clinical information, 2, 18, and 21 cases were first assigned as autosomal dominant (AD), sporadic, and X-linked (XL) inheritance, respectively. Several cases of affected females who presented with a male phenotype have been described, posing challenges at diagnosis related to the apparent family history of AD. Mutations were located in RPGR exons or introns 1–14 and in ORF15 of 12 of 41 (29.3%) and 29 of 41 (70.7%) subjects, respectively. Thirty-four (likely) pathogenic mutations were identified. Frameshifts were the most frequently observed variants, followed by nonsense, splice, and missense mutations. Herein, a detailed description of four RP patients carrying RPGR intronic mutations is reported, and in vitro splice assays were performed to confirm the pathogenicity of these intronic mutations.
Conclusion: Our findings provide useful insights for the genetic and clinical counseling of patients with XL IRD, which will be useful for ongoing and future gene therapy trials.
X-linked retinitis pigmentosa (XL RP) is the most severe subtype of RP (OMIM 268000) and accounts for 10%–20% of all RP cases (Breuer et al., 2002; Bader et al., 2003). Male patients with XL RP generally develop a more severe phenotype, whereas female carriers of XL RP show a wide spectrum of clinical features that can vary from asymptomatic to severe symptoms similar to those observed in male patients (Wu et al., 2010; Nanda et al., 2018; Talib et al., 2018). At present, mutations in three genes, RP2 (OMIM #312600), OFD1 (OMIM #312610), and RPGR (OMIM #312610), have been reported to result in XL RP (Breuer et al., 2002; Bader et al., 2003). Among them, RPGR variations remain the most common genetic cause, and are found in approximately 30%–80% of XL RP cases (Vervoort et al., 2000; Pelletier et al., 2007). Previous studies have indicated that RPGR variants are also responsible for other XL inherited retinal dystrophies (IRDs), including cone dystrophy 3(COD3, OMIM #602093), cone-rod dystrophy 2 (CORD2, OMIM #120970) (Mears et al., 2000; Ebenezer et al., 2005), and macular degeneration (Ayyagari et al., 2002).
RPGR was previously identified in the RP3 region of Xp11.4 (Meindl et al., 1996; Roepman et al., 1996). The two main isoforms are the constitutively expressed RPGR1-19 isoform, which consists of 19 exons, and the retina-specific RPGRORF15 isoform, which shares exons 1 to 14 with RPGR1-19, and includes a terminal exon (open reading frame 15, ORF15). ORF15 consists of exon 15 and extends into intron 15, encoding a highly repetitive and purine-rich 567-aa protein. Approximately 60% of disease-causing mutations in RPGR are found in ORF15 (Vervoort et al., 2000; Breuer et al., 2002).
This study reports the clinical and genetic findings in a Chinese cohort of 40 male probands and a female proband with RP (34 cases) or CORD (7 cases) caused by variations in RPGR. Among them, we report two families with a provisional diagnosis of autosomal dominant RP (ADRP) associated with affected female carriers and characterized by a phenotypic manifestation in these patients. We describe four patients with RP who carried RPGR intronic mutations in detail and confirmed the pathogenicity of these intronic mutations using an in vitro splice assay.
This study recruited 40 male Chinese probands and a female proband of Han ethnicity affected with IRD, including RP and CORD, whose genetic causes were RPGR mutations, from the Department of Ophthalmology, Peking University Third Hospital. This study conforms to the tenets of the Declaration of Helsinki. All experiments involving DNA from a patient and their relatives were approved by the Peking University Third Hospital Medical Ethics Committee (No. 2012093). Written informed consent was obtained from all participants or guardians on behalf of minors. The ethics committee approved the consent procedure.
Detailed medical and family histories were obtained from probands and/or their family members. All patients underwent standard ophthalmic examinations, including best-corrected visual acuity (BCVA), slit-lamp biomicroscopy, intraocular pressure measurement, dilated indirect ophthalmoscopy and fundus photography, electroretinography (ERG), and visual field tests, if possible. The patients underwent a systematic physical examination before genetic testing. Inheritance patterns were classified based on criteria described by Stone et al. with minimal modification (Stone et al., 2017): (i) autosomal dominant (AD; a minimum of three generations with at least one instance of male-to-male transmission); (ii) autosomal recessive (AR; several affected patients in a single sibship with healthy parents); (iii) X-linked (affected males in multiple sibships connected to each other through unaffected or mildly affected females and no male-to-male transmission); and (iv) uncertain inheritance (sporadic patients and other multiplex kindreds).
Blood samples were obtained from all probands and their family members. Genomic DNA (gDNA) was extracted using standard protocols ((D2492, Omega Bio-Tek). The patients were subjected to a specific hereditary eye disease enrichment panel (HEDEP) for targeted exon enrichment analysis, which captured 483 IRD genes (Liu et al., 2021). Next-generation sequencing (NGS) was performed using an Illumina HiSeq X platform (Illumina, San Diego, CA, USA). HEDEP sequencing data were analyzed as previously described (Zhang et al., 2016; Liu et al., 2021).
All variants considered (likely) pathogenic by NGS in this study were validated further by PCR-based Sanger sequencing. The primers used in this study are listed in Supplementary Table S1. Touchdown PCR amplification consisted of a denaturizing step at 95°C for 5 min, followed by 35 cycles of amplification (at 95°C for 30 s, at 64∼57°C for 30 s starting from 64°C, decreasing by 0.5°C every cycle for 14 cycles, which was maintained at 57°C for 21 cycles, followed by 72°C for 40 s), with a final extension at 72°C for 5 min. Purified PCR products were sequenced using the ABI BigDye Terminator cycle sequencing kit on an ABI 3130XL genetic analyzer (ABI Applied Biosystem, Foster City, CA, USA). Sanger sequencing was also used to determine whether the variant co-segregated with the disease phenotype in available family members.
RPGR ORF15 (NG 009553.1; NM001034853) was not effectively covered by NGS and was therefore amplified and sequenced using primers located outside the repetitive stretch as described previously (Bader et al., 2003). The forward primer was 5′-CAGAGATCCTATCAGATGACC-3′, and the reverse primer was 5′-TGTCTGACTGGCCATAATCG-3′, with a PCR product of 1630 bp. PCR products were sequenced using three reverse primers (5′-GTTTGCCATATTTCACAGATCC-3′, 5′-TCCTTCCTCCTCTTCCCCCTCCCA-3′, and 5′-CCTTCCTCCTCTTCCCCCTCA-3′). Sequencing results were analyzed by Sequencher (Gene Codes Corporation, Ann Arbor, MI, USA).
HEDEP sequencing data were analyzed as described previously (Liu et al., 2021). Sequence changes were classified according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology variant interpretation guidelines. In this study, only variants identified as pathogenic or likely pathogenic were reported.
The pathogenicity of RPGR intronic mutations was assessed using a population-based genome dataset (the Single Nucleotide Polymorphism Database). The potential effect of the variants on RNA splicing was assessed using the Human Splice Finder (HSF) tool (Desmet et al., 2009).
For use in the RNA splicing assay, a shortened RPGR intron DNA sequence around the splice acceptor site (intron A, 200 bp) was generated and combined with another shortened intron DNA sequence around the splice donor site (intron B, 200 bp). Introns A and B were combined to generate a shortened RGPR intron. The plasmid pmCherryN1 was linearized using the SbfI restriction enzyme (R3733S, New England Biolabs), followed by purification with a gel extraction kit (D2500, Omega). The shortened RGPR intron was cloned into the SbfI site using the pEASY-Basic Seamless Cloning and Assembly Kit (CU201, TransGen Biotech) and then transformed into Top10-competent bacteria. Clones were amplified in liquid Luria-Bertani (LB) broth, followed by the extraction of plasmid DNA (D6943; OMEGA). Site-directed mutagenesis was performed on the plasmid using the StarMut Site-directed Mutagenesis Kit (T111, Genstar, Beijing, China). The resulting DNA was sequenced to confirm nucleotide changes. HEK293T cells were cultured in regular Dulbecco’s modified Eagle’s medium (DMEM; C11995500BT, Gibco) and seeded on coated glass slides in 6-well plates. The cells were transfected with recombinant vectors using PEI (B600070, ProteinTech Group, Chicago, IL, USA), according to the manufacturer’s protocol. After 24 or 72 h of transfection, mCherry expression was observed using a fluorescence microscope (Nikon, Japan).
The results were evaluated using a two-tailed unpaired Student’s t-test and are presented as the mean ±SEM. GraphPad Prism 9 (GraphPad Software, La Jolla, CA, USA) was used for the statistical analyses. Statistical significance was set at p < 0.05.
In this cohort, 40 male Chinese probands and one female proband from 41 families with a clinical diagnosis of RP (34 cases) or CORD (7 cases) were found to harbor a disease-causing mutation in the RPGR gene. Table 1 provides an overview of the cohort’s primary clinical information. According to their clinical information, the inheritance pattern of 2 cases was assigned as AD without male-to-male transmission, with 18 cases assigned as sporadic, and 21 cases assigned as XL. Apart from the proband of family P25 (proband P25), whose onset age was 20 years, the average age at the onset of the first symptom was 4.85 years (range = 1∼11 years). Apart from proband P33, all probands complained that night blindness was present at disease onset, and three patients suffering from CORD, simultaneously complained of poor visual acuity. Twenty-seven patients had progressive visual acuity. Eighteen patients at study entry experienced a sudden decline in central vision in their 20 and 30s, with a BCVA for light perception (LP) of 0.25. Three patients suffered from unilateral or bilateral blindness; among them, the left eye of proband P11 was blind owing to untreated retinal detachment when he was 14, the right eye of proband P31 was blind when he was 50, and proband P31 suffered from bilateral blindness when he was 60. Fundus examination and imaging results of all probands are listed in Table 1. Four probands (P09, P11, P27, and P31) complained of high myopia; the mothers of probands P09 and P11, as carriers, also suffered from high myopia and/or poor visual acuity. The mother of proband P18, who was diagnosed as an RPGR carrier, also complained of high myopia.

TABLE 1. Clinical information of 41 Chinese X-linked inherited retinal dystrophy families caused by RPGR mutations.
We identified RPGR (likely) causative variants in 41 Chinese probands affected by RP or CORD (Table 2). All samples were subjected to HEDEP analysis, and (likely) pathogenic variants in the RPGR exons or introns 1–14 were identified in 12 patients. All patients in this cohort were subjected to direct PCR-based Sanger sequencing of the entire RPGR ORF15. We identified (likely) pathogenic variants of RPGR ORF15 in 29 unsolved cases. All identified RPGR variants were segregated with IRD phenotypes among the available family members. Proband P27 had RPGR variants that might have been inherited from her affected father; however, her father was not subjected to HEDEP or other genetic testing methods. The other 40 male probands had RPGR variants inherited from their mothers. Thirty-four mutations were identified in this cohort (Table 2), of which one was novel and 21 were first reported in our previous study (Liu et al., 2021). In this study, 29 patients carried variants in RPGR ORF15, 4 had intronic variants, and 8 harbored variants in exons 1–14. In terms of the type of mutation, frameshifts were the most frequently observed variants in this cohort (n = 15), followed by nonsense mutations (n = 13), splice variants (n = 4), and missense mutations (n = 2).
The presence of affected carriers in XL RP families can misleadingly suggest AD inheritance (De Silva et al., 2021). As illustrated in Figure 1, the pedigrees of patients P19 and P20 showed that males and females were affected, with individuals affected in each generation, suggesting AD inheritance. However, there was no male-to-male transmission; therefore, XL inheritance was not excluded. Genetic testing confirmed that RPGR ORF15 variants resulted in RP phenotypes in these two families; therefore, their inheritance pattern was revised to XL.
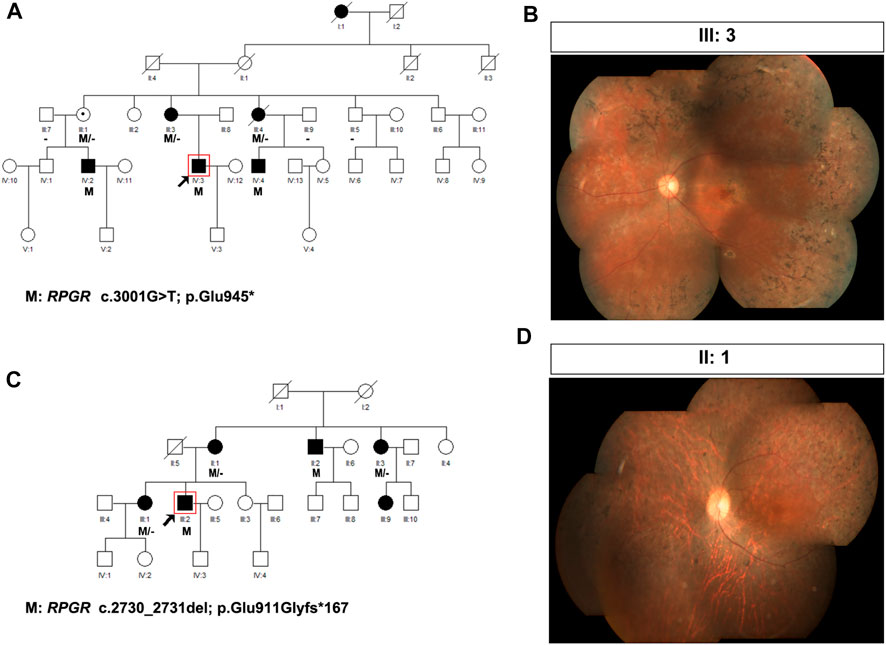
FIGURE 1. Pedigree and images of families P19 and P20. (A) Pedigree of family P19; (B) fundus photograph of the left eye of individual III:3 from family P19; (C) pedigree of family P20; and (D) fundus photograph of the right eye of individual II:1 from family P20. Black squares (males) and circles (females) represent individuals affected with RP. Dotted circles (females) represent individuals who are RPGR mutation carriers but who are not affected by RP. Unaffected individuals are not shaded. Black lines indicate deceased individuals The arrow marks the proband.
In family P19, the proband was a 27-year-old man with night blindness since childhood, progressive decline in visual acuity, tunnel vision, and visual field defects. Sequencing analysis identified c.3001G>T (p. Glu945*), an OFR15 mutation, which was the same pathogenic mutation found in his mother (individual III:3), two aunts (individuals III:1 and III:4), and two affected male cousins (Table 2; Figure 1A). In this family, III:3, an RPGR variant carrier, complained of night blindness since childhood, and her BCVA was 0.05 bilaterally. Fundus examination revealed classic symptoms of RP, consisting of retinal arteriolar attenuation, waxy pallor of the optic discs, and scattered bone spicule pigmentation around the mid-peripheral retina or the whole retina (Figure 1B). Individual III:4, a female carrier, also suffered from RP, whereas individual III:1 was unaffected. In family P20, the proband, an RP patient, had an RPGR frameshift mutation, c.2730_2731del (p. Glu911Glyfs*167), inherited from his mother (individual II:1). The same pathogenic mutation was also identified in his uncle (individual II:2), aunt (individual II:3), and older sister (individual III:1, Figure 1C). In this family, individual II:1, as an RPGR variant carrier, complained of night blindness since childhood and suffered from a sudden decline in vision in her 40s. Fundus examination revealed classic symptoms of RP (Figure 1D). Individuals II:3 and III:1 were female carriers both affected by RP. Another individual, III:9, a 24-year-old woman, also complained of night blindness, although she had not yet undergone a genetic screening test. Clinical diagnosis and genetic testing confirmed that individual II:2 was affected by RPGR-related XL RP.
In family P27, the proband was a 68-year-old woman with end-stage disease. Her vision was hand movement (HM) in the left eye and LP in the right eye (Table 1). This proband began suffering from night blindness at the age of 4 years, and complained of high myopia (-7D) bilaterally. She had undergone cataract surgery bilaterally at the age of 54 years. She had a 40-year-old son (individual IV:1) with the same mutation, c.607G>C (p. Ala203Pro) in exon 6 (Figure 2A). Proband P27 had an XL RP male phenotype fundus appearance similar to that of the affected son (Figure 2B). The proband’s ERG responses were completely extinguished (Figure 2C) and SD-OCT imaging demonstrated thinning of the outer retinal layer (Figure 2D). In this family, several individuals, including her father (individual II:1), the father’s younger twin brother (individual II:2), her son (individual IV:1), and her nephew (individual IV:3) were diagnosed with RP. Sequencing analysis indicated that IV:3 carried the RPGR hemizygous variant, c.607G>C (p. Ala203Pro), whereas his mother, as well as the proband’s younger sister (individual III:3), was unaffected, although the younger sister had the same heterozygous variant as the proband.
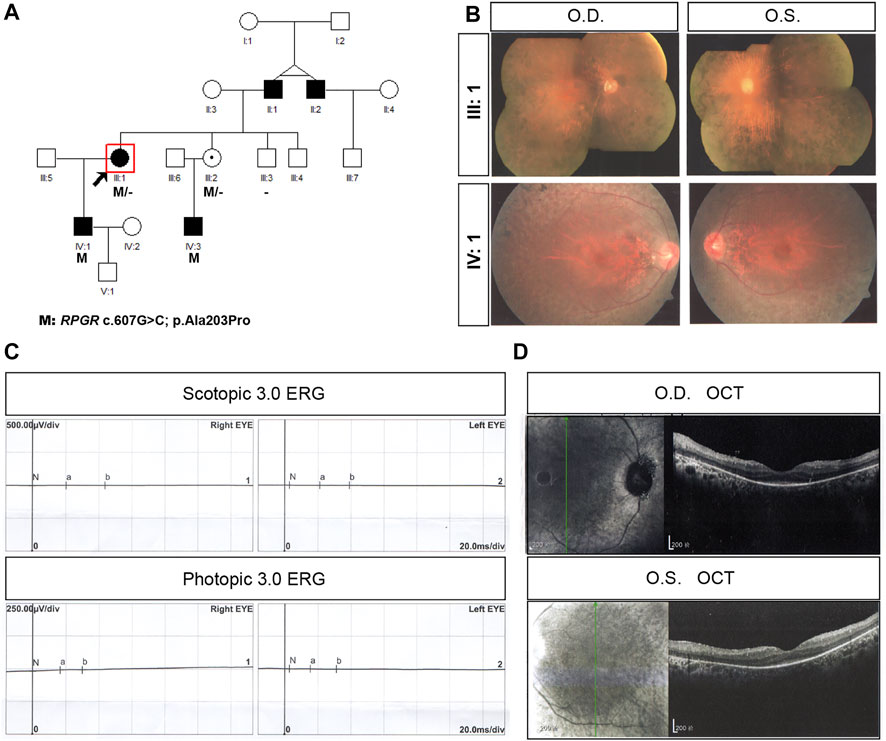
FIGURE 2. Pedigree and images of family P27. (A) Pedigree of family P27; (B) fundus photograph of both eyes of the proband (individual III:1) and IV:1 from the family; (C) scotopic and photopic electroretinogram results of the proband; and (D) F optical coherence tomography image of the both eyes from the proband. Black squares (males) and circles (females) represent individuals affected with RP. Dotted circles (females) represent individuals who are RPGR mutation carriers but who are not affected by RP. Unaffected individuals are not shaded. Black lines indicate deceased individuals. The arrow marks the proband.
Figure 3 shows the clinical information of the four RP patients and their genetic diagnoses. These four probands were male, and their ages ranged from 12 to 45 years old. All probands complained of night blindness since childhood and all had poor visual acuity, with BCVA ranging from 0.01 to 0.6. Probands P01, P03, and P04 suffered from a progressive loss of visual acuity, whereas proband P01 also suffered from a sudden decline in central vision 15 years prior. Probands, except for P04, had no family history of a similar disorder, and the grandfather (maternal) of proband P04 was also affected by night blindness. Fundus examination and imaging revealed the classic symptoms of RP, and full-field ERG showed markedly reduced rod responses in all patients. The observed phenotypes were consistent with a diagnosis of RP in all patients. Genetic analysis showed that all probands had intronic variants in the RPGR gene, which were inherited from their mother (Figure 4), although she did not manifest any symptoms despite being an RPGR carrier.
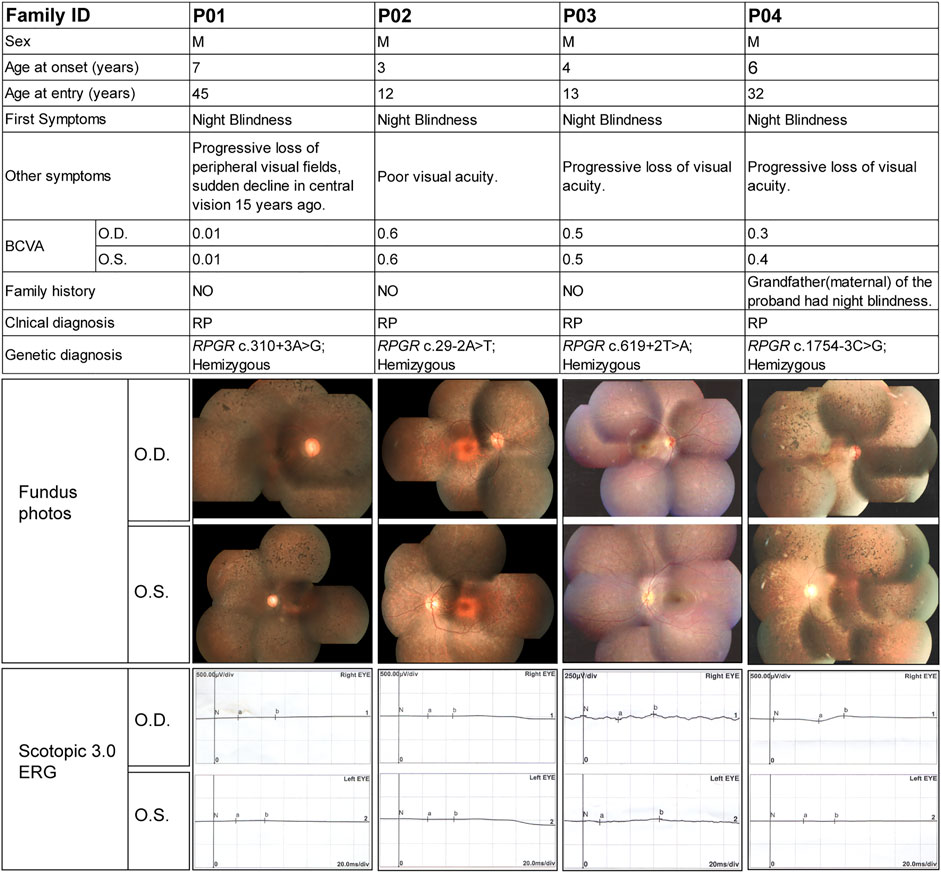
FIGURE 3. Summary of the clinical and genetic findings of four patients with RP and RPGR intronic variants. The composite image shows the most important available clinical findings. Each column represents a single patient with the following information in each row: family ID; sex; age of onset (as reported subjectively by the patient) and at entry; first symptom (first manifestation of the disease as reported by the patient); other symptoms; best corrected visual acuity (BCVA); family history; clinical diagnosis; genotype; fundus images; and scotopic and photopic electroretinogram results. O.D., right eye; O.S., left eye; F, female; M, male.
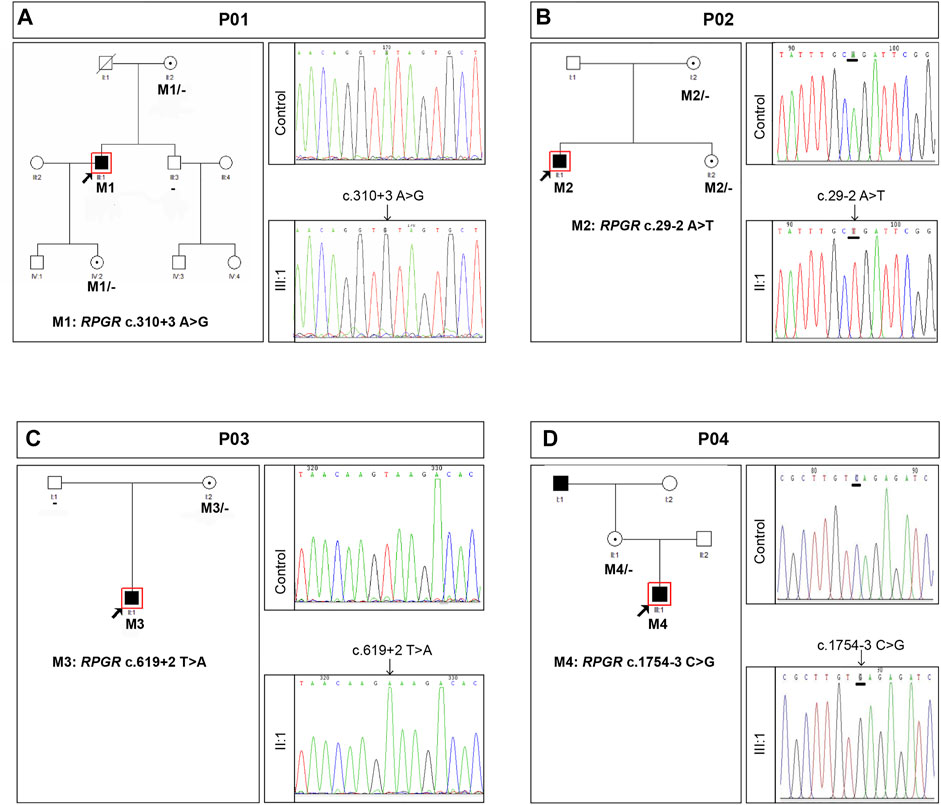
FIGURE 4. Summary of the genetic findings of four patients with RP and RPGR intronic variants. Pedigree and Sanger sequencing results of families P01 (A), P02 (B), P03 (C), and P04 (D). Black squares (males) and circles (females) represent individuals affected with RP. Dotted circles (females) represent individuals who are RPGR mutation carriers but who are not affected by RP. Unaffected individuals are not shaded. Black lines indicate deceased individuals. The arrow marks the proband.
Intronic variants, including c.310 + 3A>G, c.29-2A>T, c.619 + 2T>A, and c.1754-3C>G, have been previously reported to be disease-causing (Buraczynska et al., 1997; Wang et al., 2018; Liu et al., 2021). Splice prediction analysis using HSF indicated that the pathogenic variants c.310 + 3A>G and c.619 + 2T>A may influence the donor splice site, whereas c.29-2A>T and c.1754-3C>G may influence the acceptor splice site (Supplementary Data). Additionally, these intronic pathogenic variants were investigated using in vitro splicing assays. Shortened versions of introns 4, 1, 6, and 14 were created for the splice assay (Figure 5). A wild-type (WT) or mutant variant of the shortened intron was created and inserted into the mCherry coding sequence of a reporter construct (Figure 5A). Sanger sequencing results of reporter plasmids with the four intronic variants are shown in Figures 5B–E. If not successfully spliced, the presence of the intron would result in a premature stop codon and therefore would not lead to mCherry expression. Successful splicing would enable the complete removal of the intron and would therefore lead to mCherry expression. As illustrated in Figure 6A, mCherry expression was observed in HEK293T cells transfected with WT-intron-pmCherryN1, whereas mCherry expression was not observed in HEK293T cells transfected with mutant-intron-pmCherryN1. After 72 h of transfection, the number of mCherry+ cells/5,000 μm2 in HEK293T cells transfected with mutant-intron-pmCherryN1 plasmids was statistically (p < 0.001) lower than that in 293T cells transfected with WT-intron-pmCherryN1 plasmids (Figure 6B), indicating that these four intronic variants disrupted RPGR splicing.
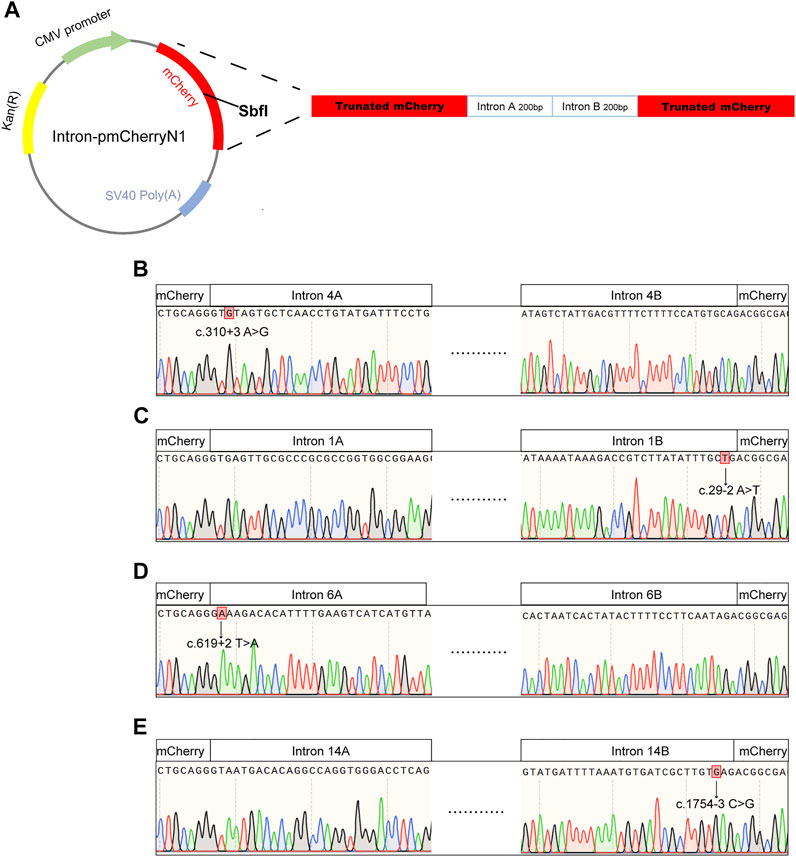
FIGURE 5. Sequences used for in vitro RNA splicing assays. (A) Schematic view of the pmCherryN1 plasmid used in the splicing assays; a shortened RPGR intron was generated for use in the assay by selecting the DNA sequence around the splice acceptor site (intron A), which was combined with the DNA sequence around the splice donor site (intron B). Sanger sequencing indicated that intron 4A contained the c. c.310 + 3A>G variant (B), intron 1B contained the c.29-2A>T variant (C), intron 6A contained the c.619 + 2T>A variant (D), and intron 14B contained the c.1754-3C>G variant (E).
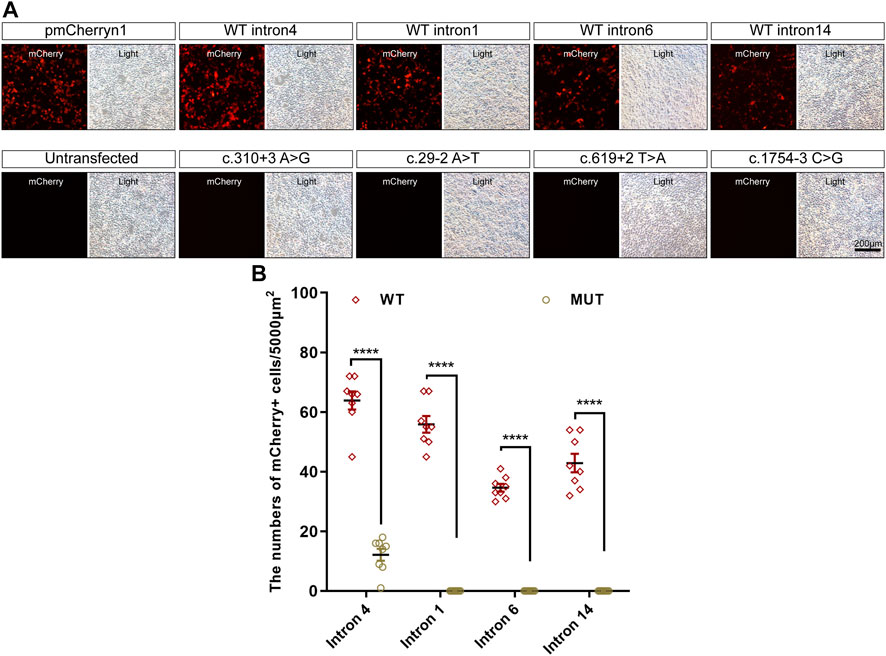
FIGURE 6. Expression of mCherry in in vitro splicing assays. (A) mCherry expression in HEK293T cells transfected with the pmCherryN1 vector containing WT or mutant introns, 24 h after transfection; scale bar = 200 μm. (B) The number of mCherry+ cells per 5,000 μm2 in HEK293T cells transfected with mutant-intron-pmCherryN1 plasmids was statistically lower than that in 293T cells with WT-intron-pmCherryN1 plasmids. Error bars indicate the SEM, and the significance was calculated by a two-tailed unpaired t-test. ****p < 0.001.
This study describes the natural history of 34 RP cases and 7 CORD cases associated with RPGR mutations in a large Chinese cohort (40 male probands and 1 female proband from 41 different families), thereby confirming that the inheritance mode of these families was XL. We found that 70.73% (29/41) of the cases carried RPGR ORF15 mutations. All RPGR mutations causing a CORD phenotype were found in exon ORF15. In this study, we identified 34 disease-causing mutations, of which one was novel and 21 were first reported in our previous study (Liu et al., 2021). These results expand the IRD mutation spectrum and provide new targets for the diagnosis and treatment of IRD.
Previously, when patients were diagnosed with XL IRDs, PCR-based Sanger sequencing was used directly to screen the disease-causing mutations in RPGR and RP2 for these patients (Yang et al., 2014), because the majority of XLRP cases are attributed to mutations in RPGR (30%–80% of XLRP patients (Vervoort et al., 2000; Pelletier et al., 2007)) and RP2 (7%–18% of XLRP patients (Sahel et al., 2014)). However, as a result of family planning over the past 30 years in China, some patients are the only child in a family, making it impossible to determine the exact inheritance pattern and recurrence risk in offspring. In this cohort, we found that 18 families had sporadic inheritance. A previous study (Martin-Merida et al., 2019) of a large cohort of 877 cases of sporadic retinitis pigmentosa (sRP) showed that 84.5% (279/330) of the solved sRP cases carried homozygous or compound heterozygous variants in the AR gene. This result indicates that the majority of sRP cases have an AR inheritance. In this cohort, genetic testing showed that 18 sporadic cases were revised as XL inheritance, which may result from the following facts: 1) the parents of these probands were unaffected and did not carry any disease-causing variant whereas the probands were affected by a new pathogenic mutation; and 2) the mother and the other female elders, but no males, carried a disease-causing variant, resulting in the affected probands inheriting this variant from their mother.
Despite the XL inheritance of RPGR, typical RP characteristics were observed in female carriers (Wu et al., 2010; Nanda et al., 2018; Salvetti et al., 2021). Therefore, the XL RP cases might be mistaken as having an AD inheritance mode, resulting in an incorrect prediction of recurrence risks and errors in patient prognosis. Previous studies reported that mutations in exons 1–14 and ORF15 can result in severe RP phenotypes in female carriers (Nanda et al., 2018). In this cohort, several female carriers in two families with a clinical diagnosis of AD RP carried c.3001G>T (p. Glu945*) and c.2730_2731del (p. Glu911Glyfs*167) in the RPGR ORF15. The p. Glu945* mutation was first reported in our previous study of family P19. Different from her two affected sisters, individual III:3, who had this heterozygous mutation, was unaffected, indicating that, in a given family, the same RPGR mutation can result in different phenotypes, even in female carriers. In family P27, c.607G>C (p. Ala203Pro) caused typical RP phenotypes in the proband, but not in individual III:3. Previous studies (Aguirre et al., 2002; Koenekoop et al., 2003; Nanda et al., 2018; Kurata et al., 2019; Salvetti et al., 2021) showed that the intrafamilial spectrum of severity in female carriers of XL retinal disorders partially resulted from X-chromosome inactivation. Based on the Drosophila model, Dobyns (2006) proposed a penetrance and severity index for X-linked diseases, as opposed to the terminology of X-linked dominant, semidominant, or recessive inheritance. Random X-inactivation can result in a mosaic pattern of cells expressing WT and mutant genes (Lyon, 2002). Although a previous study (Talib et al., 2018) indicated that random X-inactivation cannot fully explain the clustering of nearly complete penetrance in several families based on their investigation of the phenotypes of 125 female carriers of phenotypic variants in RPGR, the intrafamilial variability of severity is likely to be due to genetic and/or environmental modifiers as well as potentially skewed X inactivation. Previously, a large number of RPGR variants were reported to result in a Severe “Male Pattern” in female carriers (Nanda et al., 2018; Salvetti et al., 2021) and variants c.3001G>T (p. Glu945*), c.2730_2731del (p. Glu911Glyfs*167), and c.607G>C (p. Ala203Pro) were reported to be associated with RP phenotypes in female carriers. A previous study (Talib et al., 2018) also indicated that 23% of female RPGR carriers displayed complete RP or COD/CORD phenotypes, and our study also found several female carriers with RP, indicating that affected female RPGR carriers are not uncommon. Even in a single family, patients with the same variants may have different phenotypes, for example, in family P19, individual III:4, a female carrier, also suffered from RP, whereas III:1 was unaffected, illustrating the clinical heterozygosity of IRD and emphasizing the significance of genetic testing. Currently, several gene therapy trials for RPGR-XLRP male patients are ongoing or have been completed (https://clinicaltrials.gov), and affected female carriers should be recruited for, and may benefit from, gene therapy.
RPGR ORF15 is a hotspot for pathogenic mutations (Vervoort et al., 2000; Breuer et al., 2002) and splice site pathogenic variants are uncommon within any part of the RPGR (Cehajic-Kapetanovic et al., 2020). The pathogenicity of c.310 + 3A>G, c.29-2A>T c.619 + 2T>A, and c.1754-3C>G has been validated in previous studies (Buraczynska et al., 1997; Wang et al., 2018; Liu et al., 2021). However, few functional studies have validated the pathogenicity of the intronic variants of RPGR. The splicing process is a complex event that is important for proper protein synthesis, and splicing mutations may occur in introns and exons, which disrupt existing splice sites, create new sites, or activate cryptic sites (Anna and Monika, 2018). The most common mutations affect +1 and +2 residues at the 5′ donor splice site and −1 and −2 residues at the 3′ acceptor splice site (Anna and Monika, 2018). There are a large number of online tools that can be used to predict the pathogenicity of splicing variants (Anna and Monika, 2018); however, the exact splicing effect should be tested at different levels: DNA, RNA, RNA-protein interactions, or at the protein level (Thery et al., 2011; Fredericks et al., 2015). Analysis of RNA extracted from a patient’s cells or tissues is the simplest and most effective method to determine whether the selected variant affects splicing (Anna and Monika, 2018). In this study, RNA from the patients’ retinal tissues was not available. Therefore, the minigene assay may be an alternative method. In the minigene assay, an amplified fragment of an analyzed gene, such as a specific exon with surrounding intronic sequences with and without mutations, is cloned into a special expression plasmid, enabling the analysis of pre-mRNA splicing. Because of the large size of RPGR introns, we developed a simplified molecular assay in which we designed reporter constructs to investigate the effect of variants on RNA splicing in vitro, according to a previous study (Cehajic-Kapetanovic et al., 2020). By comparing the mutant and WT constructs, we found that a single nucleotide variation influenced splicing, also indicating the practicability of our approach, which can be used to validate splicing effects and validate which exons would be skipped.
In summary, the findings in this cohort provide useful insights for the genetic and clinical counseling of XL RP and CORD patients, which would be useful for ongoing and future gene therapy trials. Herein, we also describe the genotype and phenotype correlations of several female RPGR carriers who showed a male phenotype, reminding us of the significance of genetic testing in this scenario. Our study highlights the potential value of a molecular splice assay to confirm a new disease-causing intronic variant of RPGR.
The original contributions presented in the study are included in the article/Supplementary Material. further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Peking University Third Hospital Medical Ethics Committee. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
LXZ: design of study, data collection, data analysis, preparation and review of the manuscript. YLP and WLK: design of study, data analysis, and manuscript preparation and review. JRX and MX: data collection and analysis.
This work was supported by the National Natural Science Foundation of China (grant no: 81770966 to LY), the Beijing Natural Science Foundation of China (grant no: 19JCZDJC64000(Z)), and the Capital’s Funds for Health Improvement and Research (grant no: 2022-2G-4254 to LW). The sponsors and/or funding organizations had no role in the design or conduct of this research.
The authors thank Dr. He Huang for his helpful insight when developing the project and revising this manuscript and we thank all patients and their family members participating in this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.999695/full#supplementary-material
Aguirre, G. D., Yashar, B. M., John, S. K., Smith, J. E., Breuer, D. K., Hiriyanna, S., et al. (2002). Retinal histopathology of an XLRP carrier with a mutation in the RPGR exon ORF15. Exp. Eye Res. 75 (4), 431–443. doi:10.1006/exer.2002.2037
Anna, A., and Monika, G. (2018). Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 59 (3), 253–268. doi:10.1007/s13353-018-0444-7
Ayyagari, R., Demirci, F. Y., Liu, J., Bingham, E. L., Stringham, H., Kakuk, L. E., et al. (2002). X-linked recessive atrophic macular degeneration from RPGR mutation. Genomics 80 (2), 166–171. doi:10.1006/geno.2002.6815
Bader, I., Brandau, O., Achatz, H., Apfelstedt-Sylla, E., Hergersberg, M., Lorenz, B., et al. (2003). X-Linked retinitis pigmentosa: RPGR mutations in most families with definite X linkage and clustering of mutations in a short sequence stretch of exon ORF15. Invest. Ophthalmol. Vis. Sci. 44 (4), 1458–1463. doi:10.1167/iovs.02-0605
Breuer, D. K., Yashar, B. M., Filippova, E., Hiriyanna, S., Lyons, R. H., Mears, A. J., et al. (2002). A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am. J. Hum. Genet. 70 (6), 1545–1554. doi:10.1086/340848
Buraczynska, M., Wu, W., Fujita, R., Buraczynska, K., Phelps, E., Andreasson, S., et al. (1997). Spectrum of mutations in the RPGR gene that are identified in 20% of families with X-linked retinitis pigmentosa. Am. J. Hum. Genet. 61 (6), 1287–1292. doi:10.1086/301646
Cehajic-Kapetanovic, J., McClements, M. E., Whitfield, J., Shanks, M., Clouston, P., and MacLaren, R. E. (2020). Association of a novel intronic variant in RPGR with hypomorphic phenotype of X-linked retinitis pigmentosa. JAMA Ophthalmol. 138 (11), 1151–1158. doi:10.1001/jamaophthalmol.2020.3634
De Silva, S. R., Arno, G., Robson, A. G., Fakin, A., Pontikos, N., Mohamed, M. D., et al. (2021). The X-linked retinopathies: Physiological insights, pathogenic mechanisms, phenotypic features and novel therapies. Prog. Retin. Eye Res. 82, 100898. doi:10.1016/j.preteyeres.2020.100898
Desmet, F. O., Hamroun, D., Lalande, M., Collod-Beroud, G., Claustres, M., and Beroud, C. (2009). Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37 (9), e67. doi:10.1093/nar/gkp215
Dobyns, W. B. (2006). The pattern of inheritance of X-linked traits is not dominant or recessive, just X-linked. Acta Paediatr. Suppl. 95 (451), 11–15. doi:10.1080/08035320600618759
Ebenezer, N. D., Michaelides, M., Jenkins, S. A., Audo, I., Webster, A. R., Cheetham, M. E., et al. (2005). Identification of novel RPGR ORF15 mutations in X-linked progressive cone-rod dystrophy (XLCORD) families. Invest. Ophthalmol. Vis. Sci. 46 (6), 1891–1898. doi:10.1167/iovs.04-1482
Fredericks, A. M., Cygan, K. J., Brown, B. A., and Fairbrother, W. G. (2015). RNA-binding proteins: Splicing factors and disease. Biomolecules 5 (2), 893–909. doi:10.3390/biom5020893
Koenekoop, R. K., Loyer, M., Hand, C. K., Al Mahdi, H., Dembinska, O., Beneish, R., et al. (2003). Novel RPGR mutations with distinct retinitis pigmentosa phenotypes in French-Canadian families. Am. J. Ophthalmol. 136 (4), 678–687. doi:10.1016/s0002-9394(03)00331-3
Kurata, K., Hosono, K., Hayashi, T., Mizobuchi, K., Katagiri, S., Miyamichi, D., et al. (2019). X-Linked retinitis pigmentosa in Japan: Clinical and genetic findings in male patients and female carriers. Int. J. Mol. Sci. 20 (6), E1518. doi:10.3390/ijms20061518
Liu, X., Tao, T., Zhao, L., Li, G., and Yang, L. (2021). Molecular diagnosis based on comprehensive genetic testing in 800 Chinese families with non-syndromic inherited retinal dystrophies. Clin. Exp. Ophthalmol. 49 (1), 46–59. doi:10.1111/ceo.13875
Lyon, M. F. (2002). X-chromosome inactivation and human genetic disease. Acta Paediatr. Suppl. 91 (439), 107–112. doi:10.1111/j.1651-2227.2002.tb03120.x
Martin-Merida, I., Avila-Fernandez, A., Del Pozo-Valero, M., Blanco-Kelly, F., Zurita, O., Perez-Carro, R., et al. (2019). Genomic landscape of sporadic retinitis pigmentosa: Findings from 877 Spanish cases. Ophthalmology 126 (8), 1181–1188. doi:10.1016/j.ophtha.2019.03.018
Mears, A. J., Hiriyanna, S., Vervoort, R., Yashar, B., Gieser, L., Fahrner, S., et al. (2000). Remapping of the RP15 locus for X-linked cone-rod degeneration to Xp11.4-p21.1, and identification of a de novo insertion in the RPGR exon ORF15. Am. J. Hum. Genet. 67 (4), 1000–1003. doi:10.1086/303091
Meindl, A., Dry, K., Herrmann, K., Manson, F., Ciccodicola, A., Edgar, A., et al. (1996). A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat. Genet. 13 (1), 35–42. doi:10.1038/ng0596-35
Nanda, A., Salvetti, A. P., Clouston, P., Downes, S. M., and MacLaren, R. E. (2018). Exploring the variable phenotypes of RPGR carrier females in assessing their potential for retinal gene therapy. Genes (Basel) 9 (12), E643. doi:10.3390/genes9120643
Pelletier, V., Jambou, M., Delphin, N., Zinovieva, E., Stum, M., Gigarel, N., et al. (2007). Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: Genotype-phenotype correlations and impact on genetic counseling. Hum. Mutat. 28 (1), 81–91. doi:10.1002/humu.20417
Roepman, R., van Duijnhoven, G., Rosenberg, T., Pinckers, A. J., Bleeker-Wagemakers, L. M., Bergen, A. A., et al. (1996). Positional cloning of the gene for X-linked retinitis pigmentosa 3: Homology with the guanine-nucleotide-exchange factor RCC1. Hum. Mol. Genet. 5 (7), 1035–1041. doi:10.1093/hmg/5.7.1035
Sahel, J. A., Marazova, K., and Audo, I. (2014). Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb. Perspect. Med. 5 (2), a017111. doi:10.1101/cshperspect.a017111
Salvetti, A. P., Nanda, A., and MacLaren, R. E. (2021). RPGR-related X-linked retinitis pigmentosa carriers with a severe "male pattern. Ophthalmologica. 244 (1), 60–67. doi:10.1159/000503687
Stone, E. M., Andorf, J. L., Whitmore, S. S., DeLuca, A. P., Giacalone, J. C., Streb, L. M., et al. (2017). Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology 124 (9), 1314–1331. doi:10.1016/j.ophtha.2017.04.008
Talib, M., van Schooneveld, M. J., Van Cauwenbergh, C., Wijnholds, J., Ten Brink, J. B., Florijn, R. J., et al. (2018). The spectrum of structural and functional abnormalities in female carriers of pathogenic variants in the RPGR gene. Invest. Ophthalmol. Vis. Sci. 59 (10), 4123–4133. doi:10.1167/iovs.17-23453
Thery, J. C., Krieger, S., Gaildrat, P., Revillion, F., Buisine, M. P., Killian, A., et al. (2011). Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur. J. Hum. Genet. 19 (10), 1052–1058. doi:10.1038/ejhg.2011.100
Vervoort, R., Lennon, A., Bird, A. C., Tulloch, B., Axton, R., Miano, M. G., et al. (2000). Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 25 (4), 462–466. doi:10.1038/78182
Wang, L., Zhang, J., Chen, N., Wang, L., Zhang, F., Ma, Z., et al. (2018). Application of whole exome and targeted panel sequencing in the clinical molecular diagnosis of 319 Chinese families with inherited retinal dystrophy and comparison study. Genes (Basel) 9 (7), E360. doi:10.3390/genes9070360
Wu, D. M., Khanna, H., Atmaca-Sonmez, P., Sieving, P. A., Branham, K., Othman, M., et al. (2010). Long-term follow-up of a family with dominant X-linked retinitis pigmentosa. Eye (Lond) 24 (5), 764–774. doi:10.1038/eye.2009.270
Yang, L., Yin, X., Feng, L., You, D., Wu, L., Chen, N., et al. (2014). Novel mutations of RPGR in Chinese retinitis pigmentosa patients and the genotype-phenotype correlation. PLoS One 9 (1), e85752. doi:10.1371/journal.pone.0085752
Keywords: inherited retinal dystrophies, female carriers, hereditary eye disease enrichment panel, RPGR gene, in vitro splice assay
Citation: Liu X, Jia R, Meng X, Wang L and Yang L (2022) Analysis of RPGR gene mutations in 41 Chinese families affected by X-linked inherited retinal dystrophy. Front. Genet. 13:999695. doi: 10.3389/fgene.2022.999695
Received: 26 July 2022; Accepted: 12 September 2022;
Published: 06 October 2022.
Edited by:
Gabrielle Wheway, University of Southampton, United KingdomReviewed by:
Xunlun Sheng, Ningxia Hui Autonomous Region People’s Hospital, ChinaCopyright © 2022 Liu, Jia, Meng, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Likun Wang, d2FuZ2xrQGJqbXUuZWR1LmNu; Liping Yang, YWxleGxpcGluZ3lhbmdAYmptdS5lZHUuY24=
†ORCID: Xiaozhen Liu, orcid.org/0000-0001-9893-1986; Ruixuan Jia, orcid.org/0000-0002-8231-6898
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.