
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 30 September 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.996203
 Simon Dontoro Dekomah1,2†
Simon Dontoro Dekomah1,2† Zhenzhen Bi1,2†
Zhenzhen Bi1,2† Richard Dormatey1,2
Richard Dormatey1,2 Yihao Wang1,2
Yihao Wang1,2 Fasih Ullah Haider2,3
Fasih Ullah Haider2,3 Chao Sun1,2
Chao Sun1,2 Panfeng Yao1
Panfeng Yao1 Jiangping Bai1,2*
Jiangping Bai1,2*The second messenger calcium (Ca2+) is a ubiquitous intracellular signaling molecule found in eukaryotic cells. In plants, the multigene family of calcium-dependent protein kinases (CDPKs) plays an important role in regulating plant growth, development, and stress tolerance. CDPKs sense changes in intracellular Ca2+ concentration and translate them into phosphorylation events that initiate downstream signaling processes. Several functional and expression studies on different CDPKs and their encoding genes have confirmed their multifunctional role in stress. Here, we provide an overview of the signal transduction mechanisms and functional roles of CDPKs. This review includes details on the regulation of secondary metabolites, nutrient uptake, regulation of flower development, hormonal regulation, and biotic and abiotic stress responses.
Calcium is an important messenger in plants that coordinates a wide range of physiological processes in response to external and endogenous factors. A transient increase in cytosolic calcium often occurs during plant stress responses (Mahajan et al., 2008) and produces specific calcium signatures that can be detected by various calcium sensors to trigger downstream effects. These effects are manifested in changes in protein phosphorylation and gene expression patterns (Kudla et al., 2010). Ca2+-dependent protein kinases (CDPKs), calmodulin (CaM), calmodulin-like proteins (CMLs), calcineurin B-like proteins (CBLs), and their interacting kinases (CIPKs) are four major classes of calcium-sensing proteins in plants (Hashimoto and Kudla 2011; Batistic and Kudla 2012). CDPKs are Ca2+-regulated serine/threonine protein kinases encoded by a large family of multigene found in plants, green algae, protists, and oomycetes (Valmonte et al., 2014). Among calcium sensors, CDPKs are the only ones that combine sensing activity via EF-hand calcium-binding motifs with response activity via the protein kinase domain in a single protein (Yip Delormel and Boudsocq, 2019).
CDPKs consists of four characteristic domains including a variable N-terminal domain containing myristoylation and palmitoylation sites, a catalytic ser/thr protein kinase domain that phosphorylates the serine and threonine residues of its substrates, an autoinhibitory domain acting as a pseudosubstrate blocking enzyme activity in the absence of Ca2+ stimulation, and a C-terminal regulatory calmodulin-like domain with one to four conserved EF-hand motifs for calcium-binding (Cheng et al., 2002; Liese and Romeis, 2013; Valmonte et al., 2014; Xu et al., 2015). CDPKs recognize Ca2+ signals via the EF-hand motif, a distinctive and conserved helix-loop-helix structure composed of 12 amino acids that confer the Ca2+-binding activity. Because EF-hands typically occur in pairs as a discrete domain, the majority of CDPK family members have two, four, or six EF-hands (Shi et al., 2018; Yip Delormel and Boudsocq, 2019). In certain conditions, the pairing exhibits positive interaction, reducing the Ca2+ signal required to achieve protein saturation. Ca2+ binding to the CDPK cause a structural change that promotes the CDPK’s interaction with its target proteins or alters the CDPKs enzyme activity. CDPKs are one general class of EF-hand-containing proteins found in plants that translate chemical signals into diverse biochemical responses (Shi et al., 2018; Yip Delormel and Boudsocq, 2019). Many CDPK genes have been identified in plants, 34 in Arabidopsis thaliana (Cheng et al., 2002), 31 in rice (Oryza sativa) (Ray et al., 2007), 29 in tomato (Solanum lycopersicum) (Wang et al., 2016), 18 in melon (Cucumis melo) (Zhang et al., 2017), 31 and 32 in Cucurbita maxima and C. pepo, respectively (Wei et al., 2019), and 19 in cucumber (Cucumis sativus) (Xu et al., 2015).
CDPKs have been found in various subcellular locations, such as the plasma membrane, cytosol, nucleus, endoplasmic reticulum, peroxisomes, outer mitochondrial membrane, and oil bodies, implying that they play roles in a variety of physiological processes (Dammann et al., 2003; Harper et al., 2004). They are involved in starch and protein accumulation in immature rice seeds, tolerance to cold, salt, drought stress, and abscisic acid (ABA) response in Arabidopsis, rice, and watermelon (Citrullus lanatus) (Mehlmer et al., 2010; Asano et al., 2011; Asano et al., 2012b; Wei et al., 2019), defense response in tobacco (Nicotiana tabacum) (Romeis et al., 2000) and tomato (Chico et al., 2002), nodulation in Medicago truncatula (Gargantini et al., 2006), and pollen tube growth in petunia and abscisic acid (ABA) response in Arabidopsis (Nakata et al., 2009). CDPKs regulate seed germination (Jiang et al., 2013), pigment accumulation, and fruit development (Pawełek et al., 2015), and promote grain filling in rice which shortens the crop duration (Manimaran et al., 2015).
Plants are constantly faced with environmental stresses, and adequate stress management requires a high energy expenditure and often results in slowed or completely impaired growth. Therefore, it is in the best interest of plants to develop mechanisms to prioritize simultaneous external challenges to balance growth and stress responses (Seybold et al., 2019) so that plants can escape, avoid, or tolerate environmental changes (Blum, 1996; Lerner, 2018). Several mechanisms come into play, depending largely on the type, severity, and duration of stress, and may also vary among plant species and developmental stages (Pinheiro and Chaves, 2011). Among the known mechanisms, pathways involving CDPKs are one of the best-studied cases. CDPKs are promising candidates for a role in coordinating the trade-off between stress tolerance and growth (Claeys and Inzé, 2013). Therefore, this review will focus on the progress of CDPK research, particularly on how CDPKs regulate plant stress tolerance and promote growth and development.
The CDPK gene family has a long evolutionary history that can be traced back to the earliest land plants, such as pteridophytes (Selaginella moellendorffii) and bryophytes (Physcomitrella patens), both of which have at least 35 CDPKs in their genomes (Hamel et al., 2014). CDPK homologs were previously thought to be plant-specific, but they have now been discovered in ciliates and apicomplexan parasites (Zhang and Choi. 2001; Billker et al., 2009). The first CDPK gene appeared prior to the basal split between green plants and alveolate protists (Zhang and Choi. 2001). The cucumber’s genome encodes 19 CsCDPKs and 18 CmCDPKs in melon, compared to 34 AtCPKs in Arabidopsis (Harper and Harmon 2005; Xu et al., 2015; Zhang et al., 2017), 25 in potato (Bi et al., 2021), 30 PtCDPKs in Poplar (Zuo et al., 2013), and 31 OsCDPKs in rice (Ray et al., 2007). The lower number of CDPK genes in some plants e.g. cucumber and melon may be attributed to a lack of whole genome duplication (WGD) events following the gamma WGD, as seen in Arabidopsis, or the WGD after the split between eudicots and monocots, as seen in rice (Jiao et al., 2011). Another possible explanation could be the cucumber or melon genome’s low number of segmental duplications and tandem gene duplications (Asano et al., 2012a). However, recent duplication events in Arabidopsis, Populus, and rice may have resulted in an increase in the number of CDPK family members in their genomes (Remington et al., 2004).
The evolutionary history of CDPKs phylogenetic groupings and gene structure can be used to predict the functional specificity between paralogs. The Arabidopsis genome encodes 34 CDPKs, grouped into four distinct clusters (Boudsocq and Sheen, 2013) which is similar to several angiosperms CDPK family’s basal architecture such as tomato (Hu et al., 2016), Chinese cabbage (Wu et al., 2017), potato (Dekomah et al., 2022), maize (Li et al., 2018). However, recent studies in sweet potato (Ipomoea spp.) revealed more than four clusters, with three CDPKs forming a fifth cluster (V) (IbCDPK35, ItfCDPK35, ItbCDPK35) that was speculated to have resulted from chromosomal hybridization (Li et al., 2022). With gene structure, it has been proven that when CDPK sequences within a genome belonged to the same evolutionary cluster they showed similar intron-exon patterns. Although this is not always the case, but highly homologous CDPKs have very similar intron-exon patterns. One example is the comparison of AtCPK4 and AtCPK11 to AtCPK12. In terms of protein sequence, these three sequences share a lot of similarities. AtCPK4 and AtCPK11 have very similar intron-exon patterns, but the AtCPK12 gene structure differs from these two (Valmonte et al., 2014). In terms of function, AtCPK4 and AtCPK11 display functional similarity, but AtCPK12 was different (Valmonte et al., 2014). CDPKs, CMLs, CaMs, and CBLs account for more than one-third of all EF-hand containing domain found in plant genomes (Mohanta et al., 2017). The overall trend in the evolutionary process of green plants is a significant and sustained increase in the number of EF-hand domains, whereas the number of EF-hands was initially very low in early algae, indicating that Ca2+ sensing appeared to experience differential expansion and functional specialization in CDPKs, CMLs, CaMs, and CBLs. Abiotic stresses and plant morphological complexity may also be linked to changes in the number of Ca2+-sensing genes and EF-hand motifs (Tong et al., 2021). Considering the evolutionary history of CDPKs it is established that, this multigene family is conserved among angiosperms based on their basal architecture and gene structure which has contributed to expanding their signaling roles.
Plant phenotypes have been significantly altered during evolution to adapt to environmental changes by transforming the form and function of genes. Most land plants have undergone polyploidization, which has resulted in Whole-Genome Duplication (WGD) and allowed duplicated genes to diverge in function. Each of these genes went through one of three transformations: neofunctionalization, subfunctionalization, or non-functionalization (pseudogenization). These outcomes allowed duplicated genes to acquire functional diversification, culminating in more complex organisms (Wu et al., 2017; Rensing, 2014). Neofunctionalization is an adaptive process in which one copy of a duplicated gene mutates and ultimately performs a novel function (that the ancestral sequence cannot perform. This is one of the mechanisms that can result in the long-term retention of both copies of a gene after duplication. A classic example was found in the functional diversification of cucumber CsCDPKs paralogs gene pair (CsCDPK1 and CsCDPK3) in cluster A. Both genes lost motif 9 whiles sharing the same intron phase, with one of the duplicates changing its expression pattern to be more tissue (fruit) specific indicating neofunctionalization and may play a significant role in fruit development (Xu et al., 2015). The duplicate gene pair in grapevine (VvCPK5 and VvCPK11) in cluster C underwent neofunctionalization where VvCPK5 evolved to become more specific in pollen development while VvCPK11 retained its housekeeping role (Chen et al., 2013). Subfunctionalization is a related process in which duplicated gene copies diverge to perform specific functions than the ancestral gene, which can help to explain the duplicates’ short-term fate and can be considered as a transition state towards neofunctionalization (Rastogi and Liberles, 2005). In the case of CsCDPK8 and CsCDPK17 in cluster B, CsCDPK17 is primarily expressed in tendrils, whereas CsCDPK8 is expressed at a relatively similar level in all tissues maintaining their housekeeping role, suggesting subfunctionalization (Xu et al., 2015). For VvCPK8 and VvCPK9 in cluster A, both genes were found to express in fewer tissues such as pollens compared to VvCPK13 in the same cluster A which retained its ancestral function in stress response (Chen et al., 2013).
Since it diverged from Carica papaya, the genome of A. thaliana has undergone a paleohexaploidy (β) duplication, as well as two subsequent genome duplications (α and γ), in addition to rapid DNA sequence divergence and extensive gene loss (Jiao et al., 2011). The evolutionary history of grapevine CDPKs occurred in the absence of the alpha (α) and beta (β) whole-genome duplications experienced by A. thaliana (Chen et al., 2013). The CDPK family in angiosperms is characterized by the presence of numerous paralogs with high levels of homology. These closely related genes most likely arose as a result of recent duplication events and, as a result, have not yet expanded significantly (Hamel et al., 2014). In some instances, duplication appears to predate the monocot-eudicot split (e.g., OsCPK17/27, AtCPK1/2, and PtCPK1/2), whereas, in others, duplication appears to be specific to a particular lineage (OsCPK12 and AtCPK29 versus PtCPK29-1/29-2) (Hamel et al., 2014). Some duplicated regions in CDPK-SnRK protein kinases found in A. thaliana indicated that CDPK-SnRK protein kinases are paralogs that emerged by divergence after genome duplication events (Hrabak et al., 2003). Arabidopsis and maize CPK genes have undergone both segmental and tandem duplication, which has contributed to the CPK family’s expansion. In Populus, segmental duplication was the major factor in the expansion of CPK genes (Zuo et al., 2013). A study of gene chromosomal distribution reveals that few CDPK genes occur as tandem duplicates within angiosperms. CPK20-1/20-3 and CPK20-2/20-4 in poplar (Zuo et al., 2013) are notable exceptions, as well as a cluster of five Group II CDPKs in Arabidopsis (CPK21/22/23/27/31) (Cheng et al., 2002; Hrabak et al., 2003). As identified for mitogen-activated protein kinases (MAPKs) (Hamel et al., 2014), the expansion of the CDPK family from angiosperms depended largely on large-scale DNA rearrangements, namely whole-genome or large segmental duplications. No tandem duplication of CPK genes has occurred in the rice genome (Asano et al., 2012a). Two duplicated gene pairs (TkCPK26/TkCPK27; TkCPK27/TkCPK) were found in Taraxacum koksaghyz indicating accelerated evolution rates (Zhu et al., 2010). Most ancestral CDPKs of plants had diverse functions in the development of reproductive structures and maintenance of other cellular processes; however, several duplication events over the years have contributed to expanding, gaining, or retention of CDPK functions in response to changing environmental conditions.
Plants have a large number of calcium-binding proteins that act as cellular Ca2+ sensors as well as the first point of information translation (McCormack et al., 2005; Kim et al., 2007; Kudla et al., 2010). As one of the Ca2+ sensors, CDPKs have four domains including; a calmodulin-like domain (CaM-LD) or calcium-binding domain, a serine/threonine protein kinase domain, a variable N-terminal domain (VNTD), and an autoinhibitory junction domain (AI-JD) (Batistic and Kudla 2012) (Figure 1A). At its C-terminus, the auto-inhibitory-junction domain contains a pseudosubstrate auto-inhibitor (20∼30 amino acids) and a junction region, whereas the CaM-LD domain contains four EF-hand Ca2+-binding motifs classified into two lobes, with the N-lobe having a lower calcium affinity than the C-lobe (Christodoulou et al., 2004; Liese and Romeis 2013; Yip Delormel and Boudsocq, 2019). The covalent tethering of the CaM-LD to its regulatory-junction region in CDPKs is a unique feature of the CaM superfamily (Chandran et al., 2006). In an inactive state, the auto-inhibitor binds to the adjacent kinase domain in the active site and prevents it from performing its activity (Figure 1B). When activated by increased Ca2+, Ca2+ ions loads into the EF-lobes, causing a structural change of CaM-LD. Meanwhile, the EF-lobes cling to the junction region, allowing the auto-inhibitor to be released. The kinase domain is then exposed and phosphorylated by an uncharacterized cellular kinase (Liese and Romeis 2013; Christodoulou et al., 2004; Klimecka and Muszy’nska, 2007) (Figure 1C). The activated CDPK can then recognize and phosphorylate its targets (Figure 1C).
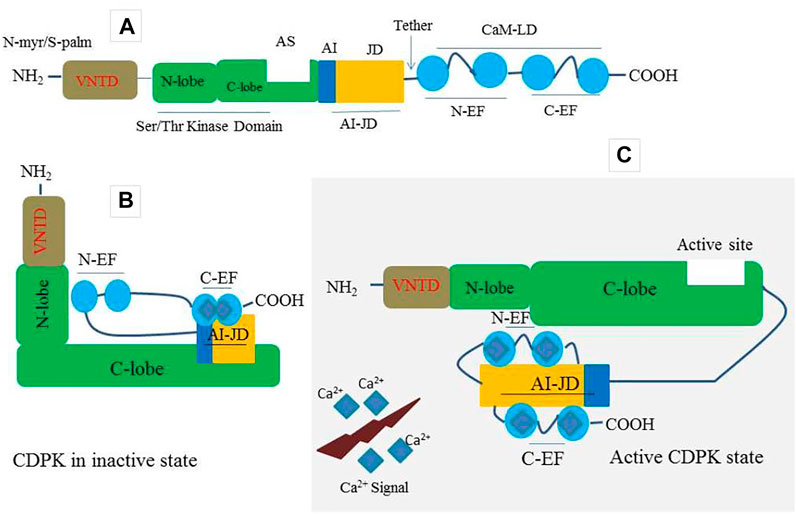
FIGURE 1. A prototype of CDPK protein structure and its decoding mechanism. (A) A typical CDPK comprises a variable N-terminal domain (VNTD) which contains (Myristoylation site and Palmitoylation site), serine/threonine kinase domain, active site (AS), auto-inhibitory junction domain (AI-JD) made up of [auto-inhibitor (AJ) and junction domain (JD)], and a C-terminal CaM-like domain (CaMLD) which harbor (four EF-hands) for Ca2+ binding. (B) CDPK in an inactive state with AI acting as a restraining molecule. In a resting position, the AI-JD interacts with the C-lobe and C-EF by binding in the AS whiles the N-lobe interacts with N-EF. (C) CDPK in an active state. Upon increase in cytosolic Ca2+ concentration, the AI-JD detaches from the AS as a result of large conformation changes that occurred in the kinases domain. Ca2+ ions bind with the C-terminal lobe (C-EF) and N-terminal lobe (N-EF) and then interact with AI-JD on both sides.
CDPK proteins contain a variable N-terminal domain (VNTD) at the N- terminus (Hamel et al., 2014) (Figure 1A), which varies in amino acid sequence and length (Cheng et al., 2002). Although there is little information about the function of the VNTD, recent studies suggest that this region is involved in substrate specificity (Ito et al., 2010; Asai et al., 2013). Most flowering plants CDPK VNTDs also have predicted N-myristoylation and S-palmitoylation sites, which promote protein targeting to lipid membranes (Boudsocq and Sheen 2013; Zuo et al., 2013; Hamel et al., 2014; Ma et al., 2015) (Figure 1A). The membrane-anchoring of a CDPK was first demonstrated experimentally in rice (Mart’n and Busconi 2000); however, this phenomenon has since been demonstrated for many Arabidopsis candidates, as well as CDPKs from other flowering plant species (Boudsocq and Sheen 2013; Liese and Romeis 2013). Importantly, CDPKs that are membrane-targeted can also move away from membranes in response to stress signals (Gao et al., 2013). This feature most likely allows CDPKs to shuttle between various subcellular compartments in order to perform a broader range of cellular functions.
CDPKs have been linked to the regulation of secondary metabolite production through phosphorylation of the enzyme PAL (phenylalanine ammoniumlyase) (Zhang and Liu, 2015). AtCPK1 could phosphorylate PAL in vitro, the first-step enzyme in Phe-derived phytoalexin biosynthesis. Overexpression of AtCPK1 in soybean cells causes the accumulation of phytoalexins, the antimicrobial compounds, indicating the involvement of AtCPK1 in defense (Cheng et al., 2001; Veremeichik et al., 2019). In addition, heterologous expression of AtCPK1 in Rubia cordifolia cells was found to have a strong stimulatory effect on anthraquinone (AQ) production (Shkryl et al., 2011). Furthermore, by phosphorylating WRKY33, the pathogen-responsive CPK5 and CPK6 in Arabidopsis regulate camalexin biosynthesis (Zhou et al., 2020). It was shown that phosphorylation of WRKY33 by MPK3/6 promotes its transactivation, whereas CPK5/CPK6 phosphorylation increases its DNA binding ability (Zhou et al., 2020), implying that CPKs and MAPKs may function synergistically (Yang et al., 2020; Zhou et al., 2020). Aleynova-Shumakova et al. (2014) found that overexpression of VaCPK20 or VaCPK29 increased resveratrol production in Vitis amurensis cells whiles some CDPKs such as HbCPK9/15 are associated with latex biosynthesis (Zhu et al., 2010; Xiao et al., 2017). On the other hand, FaCDPK4 and FaCDPK11 have been linked to drought stress tolerance where ABA is used as a signaling molecule to increase phenolics, anthocyanins, ascorbic acid, and sugar compounds (Crizel et al., 2020).
It is well understood that nitrate (NO3−) availability causes extensive transcriptional changes, which are mediated in part by changes in [Ca2+] in various cellular compartments. NO3− induces distinct Ca2+ signatures, culminating in the activation of CDPKs and CIPKs, which phosphorylate channels, metabolic enzymes, and transcription factors (Liu et al., 2020). AtCPK10/AtCPK30/AtCPK32, for example, phosphorylates NIN-LIKE PROTEIN (NLP) transcription factors to modulate transcriptional responses to nitrate availability. An Arabidopsis triple-mutant of cpk10 cpk30 cpk32 3-day-old seedlings placed on a 3MBiP medium in the presence of 5 mM KNO3 for 5-8 days exhibits down-regulation of nitrate-responsive genes as well as impaired cotyledon greening and expansion (Liu et al., 2017). The triple-mutant-specific deficiency of cotyledon and leaf expansion in response to nitrate may be partly correlated with decreased expression of CYP735A2 for trans-zeatin synthesis (Kiba et al., 2013; Liu et al., 2017).
One of the most important macronutrients in crop production is nitrogen. A. thaliana roots gain access to ammonium (NH4+) by promoting the expression of ammonium transporters (AMTs) of the Ammonium Transporter/methylammonium permease/rhesus (AMT/MEP/Rh) protein superfamily (Loqué and von Wirén, 2004). AMTs are potent transporters with Km values in the micromolar (μM) range, exhibiting high substrate specificity for NH4+ and its methylated analog, methylammonium (MeA) (Loqué et al., 2006; Pantoja, 2012). Six AMTs have been identified in Arabidopsis. Of these AMT1; 1, AMT1; 2, and AMT1; 3, are the major transporters for high-affinity ammonium uptake into roots, with ammonium substrate affinities of 50, 234, and 61 μM respectively (Loqué et al., 2006; Yuan et al., 2007). AMT1; 1 and AMT1; 3 localized predominantly in the rhizodermal and cortical cells, form homo- and hetero-oligomers at the plasma membrane, and contribute 30–35% of the high-affinity ammonium uptake capacity (Yuan et al., 2007; Yuan et al., 2013). The function of AMT1:1 depends on CPK32-mediated phosphorylation at the non-conserved serine residue S450 in its C-terminal domain (Qin et al., 2020). The phosphorylated variant AMT1; 1S450E but not the non-phosphorylatable variant AMT1; 1S450A was found to wholly complement MeA insensitivity and restore increased 15NH4+ uptake in both amt1;1 and cpk32 mutants in transgenic plants (Qin et al., 2020). This suggests that CDPK positively regulates the uptake of both nitrate and ammonium in plant roots.
Pollen tube growth, which involves ions (Ca2+, H+, K+, and Cl−) and water fluxes regulated by a calcium gradient at the tube tip, is essential for successful fertilization. Some Arabidopsis CPKs are only expressed in pollen tubes, revealing major roles for CDPKs in this process (Gutermuth et al., 2013; Zhao et al., 2013). The CPK11-CPK24-SPIK pathway has also been identified in pollen tube cells (Zhao et al., 2013; Shi et al., 2018). Using patch-clamp analysis, it was discovered that an increased cytoplasmic Ca2+ concentration inhibited K+ influx in pollen tube protoplasts. Among 16 examined pollen-tube-expressed CDPKs, six AtCPK mutants differed significantly from wild-type plants in pollen germination or pollen tube growth (Zhao et al., 2013). The measurement of the K+ influx currents in cpk11 and cpk24 mutants revealed that both CPK11 and CPK24 are required for Ca2+-dependent inhibition of K+ influx channels and showed that these two CPKs function in the same pathway (Zhao et al., 2013). CPK11 can bind to and phosphorylate CPK24 in vivo (Shi et al., 2018). Further electrophysiological experiments revealed that the shaker pollen K+ influx channel (SPIK) may act as the substrate protein in this pathway, implying that CDPK can regulate pollen tube elongation through K+ influx regulation (Zhao et al., 2013).
CDPKs have been implicated in mediating Ca2+-regulated pollen tube growth as an important node in Ca2+ signaling pathways (Estruch et al., 1994; Moutinho et al., 1998). The influence of CPK in promoting pollen tube germination and growth was first found in maize (Zea mays), where inhibition of a pollen-specific CPK hindered both germination and polar growth (Estruch et al., 1994). Moreover, PiCDPK1 and PiCDPK2 have been shown to regulate pollen tube growth polarity and extension, respectively (Yoon et al., 2006). Genetic evidence has proven that CPKs are involved in pollen tube growth, AtCPK17 and AtCPK34 essentially help in maintaining pollen tube tip growth rate and facilitating response to tropism signals (Myers et al., 2009). Further research discovered two pollen-specific water and nonionic channels, NIP4; 1 and NIP4; 2, as substrates of AtCPK34 that regulate pollen germination and tube growth (DiGiorgio et al., 2016). A negative gradient of anions at the pollen tube tip, which has an inverse correlation with Ca2+ concentration, is also required for pollen tube growth and is maintained by the anion efflux transporter S-type anion channel SLOW ANION CHANNEL-ASSOCIATED 3 (SLAH3). CPK2 and CPK20 were reported to enhance pollen tube growth at the pollen tip by activating SLAH3 (Gutermuth et al., 2013). In addition, the cpk11/24 double mutant showed improved pollen tube growth but impaired Ca2+-dependent inhibition of inward K+ channels, indicating that these CPKs negatively regulate pollen tube elongation (Zhao et al., 2013). The maize gene ZmCPK32 is expressed in pollen tube growth; ZmCPK32 has CPK activity and is localized to the plasma membrane and punctate internal membrane compartments. qPCR and in situ hybridization revealed that ZmCPK32 is expressed at high levels in mature pollen grains, suggesting that it may be functionally linked with pollen tube development (Li et al., 2018). Transient expression of ZmCPK32 in tobacco repressed pollen tube germination and growth, which depended on its kinase activity. Again, the constitutively active form of ZmCPK32 reduces seed germination rate, whereas cytosol-localized ZmCPK32 had a minor effect on pollen tube elongation (Li et al., 2018). These findings support the theory that different CPK members may be linked to different aspects of pollen tube growth.
CDPKs have been found in floral transition, which is initiated by the florigen complex. Florigen is made up of two transcription factors, flowering locus T and D (FT/FD), whose interaction is known to require Ca2+-dependent phosphorylation of FD on T282 by 14-3-3 proteins (Kawamoto et al., 2015b). In a screen of ten nuclear CPKs, AtCPK33 and AtCPK6 were identified as the major protein kinases interacting with and phosphorylating FD on T282 in vitro. However, single and double cpk6, 33 mutants showed only a weak delayed in flowering time when compared to the fd mutant (Kawamoto et al., 2015b), showing that additional CPKs are likely involved in this process. Transgenic lines expressing a dominant negative variant of AtCPK33 that can interact with FD without phosphorylating it displayed a distinct late-flowering phenotype (Kawamoto et al., 2015a).
CDPKs are expressed in plant roots, stems, leaves, fruits, and seeds. IbCDPK28 expression levels increased significantly in sweet potato tuberous roots during tuberization, and may play a functional role in regulating tuberous root growth (Li et al., 2022). The expression of potato CDPK isoforms has been linked to the stolon-to-tuber transition, with StCDPK1 being involved in gibberellic acid (GA) signaling, indicating a potential role in tuberization (Gargantini et al., 2009). An integrated network centered on the small GTPase ROP (rho of plants) revealed a role for CDPKs in root hair development. The GDP dissociation inhibitor (RhoGDI) and the guanine nucleotide exchange factor (RopGEF) regulate the transition of ROP from GDP-bound inactive to GTP-bound active states (Yip Delormel and Boudsocq, 2019). ROP11 and RopGEF1 impede root hair development, whereas RhoGDI promotes it (Yip Delormel and Boudsocq, 2019). AtCPK3 has been shown to phosphorylate RhoGDI at three residues: S45/S48/T52 (Wu et al., 2013). Unlike a phosphodead variant, a phosphomimic variant, which had a higher affinity for ROP, could rescue the gdi mutant’s defective root hair morphology, demonstrating a positive role of AtCPK3 by stimulating RhoGDI. Table 1 provides an overview of some identified CDPK genes in plants and their known biological functions.
The major biological function of CDPKs is associated with plant response to abiotic and biotic stresses. Differential expression of CDPKs has been found in response to a variety of stimuli, including ABA, drought, salinity, cold, pathogens, and wounding (Ray et al., 2007; Romeis et al., 2001; Wan et al., 2007). In rice, stress-responsive cis-elements have been identified as the promoters of its CDPK genes (Wan et al., 2007). Table 2 shows some known CDPKs involved in signal transduction pathways leading to various stress responses.
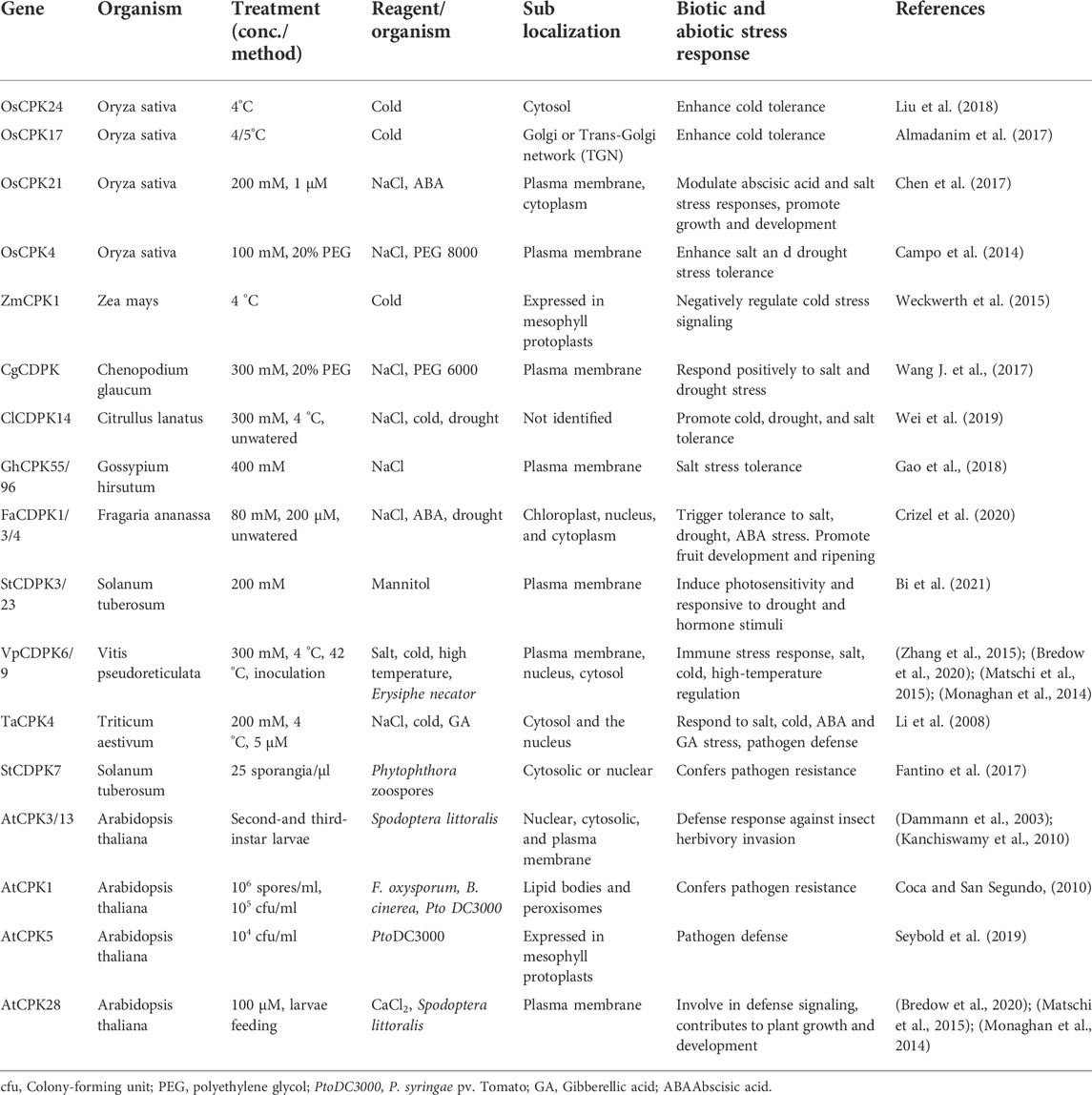
TABLE 2. Physiological role of CDPKs in plants and their subcellular location under biotic and abiotic stresses.
ABA acts as a chemical signal in response to environmental stresses, triggering the activation of numerous physiological and developmental processes in plants adaptation to stress conditions (Finkelstein et al., 2002; Ton et al., 2009). Several studies have recently described a group of CDPKs as parts of the ABA signaling pathway controlling plant responses to abiotic stresses. For example, AtCPK32 binds to and phosphorylates the ABA-responsive transcription factor ABF4, and AtCPK32-overexpressing plants showed a hypersensitive phenotype to ABA (Choi et al., 2005). As mentioned above, AtCPK23, together with AtCPK21, regulates the anion channel SLAC1 and SLAH3 to control ABA-mediated stomatal movement (Geiger et al., 2010), where the ABA- receptor RCAR1-ABI1 pathway regulates AtCPK21 expression (Geiger et al., 2011). Moreover, AtCPK3 and AtCPK6 are both positive regulators of ABA signaling in stomatal movement (Mehlmer et al., 2010; Xu et al., 2010), while AtCPK6 plays a positive role in regulating methyl jasmonate signaling in guard cells resulting in stomatal closure (Ye et al., 2013). The Arabidopsis mutants lacking AtCPK10 expression were impaired in their ability to inhibit ABA-induced stomatal opening (Zou et al., 2010). Additionally, AtCPK12 interacts with, phosphorylates, and activates the type 2C protein phosphatase ABI2 and two ABA-dependent transcription factors ABF1 and ABF4 (Zhao et al., 2011). ABF1 and ABF4 are ABA-dependent, simple leucine zipper transcription factors that play a positive role in ABA signal transduction (Choi et al., 2000). However, ABI2 plays a negative role in ABA signal transduction (Cutler et al., 2010). When AtCPK12 is downregulated, AtCPK12-induced stimulation of ABI2 phosphatase activity can be abolished, whereas when AtCPK12 is upregulated, AtCPK12-induced repression of transcription factors ABF1 and ABF4 can be relieved. As a result, ABA signaling can be enhanced in both cases, which may explain why both AtCPK12 overexpression and RNAi lines exhibit ABA hypersensitive phenotypes (Zhao et al., 2011). Exogenous ABA treatment increased the expression of BrrCDPK38/42 and FaCDPK4/11 in Brassica and Fragaria, respectively (Wang et al., 2017; Crizel et al., 2020). In rice, OsCPK14 and OsCPK21 interact with and phosphorylate OsDi19–4 transcription factor (Wang et al., 2016) and 14–3-3 protein (OsGF14e) (Chen et al., 2017), respectively, and positively regulate ABA signaling and other abiotic factors.
Drought is one of several factors affecting crop production and productivity in developing countries (Aravind et al., 2017). The effects of drought stress on plants include reduction in water potential, closure of stomata due to increased accumulation of abscisic acid (ABA), and reduced photosynthesis, biomass, and grain yield (Shikha et al., 2017; Van Gioi et al., 2017). To minimize the adverse effects of drought, the role of CDPKs in regulating plant stress adaptation was investigated to gain more insight into their signaling processes. CPK33 interferes with stomatal closure and slow anion currents (Li et al., 2016). Drought stress and ABA treatment resulted in two unique cpk33 mutants with significantly smaller stomatal apertures than wild-type plants, as well as drought tolerance and increased slow anion channel activity. CPK33 over-expression lines displayed the reverse phenotypes as mutants, including impaired stomatal closure, increased water loss, and reduced drought tolerance. It was discovered that thiamine thiazole synthase 1 (THI1) physically interacts with CPK33 (Li et al., 2016). THI1 inhibited CPK33 auto-phosphorylation activity in an in vitro kinase assay, which is consistent with the phenotypes of THI1 over-expression lines. Similarly, AtCPK8 has been shown to have positive functions in ABA- and H2O2-mediated stomatal movement in response to water deficit, leading to stomatal closure (Zou et al., 2015). Under drought stress, Arabidopsis CPK23 and CPK21 phosphorylate and activate SLAC1 which leads to membrane depolarization and activation of the K+ release channel GORK (Geiger et al., 2011). These changes eventually result in the closure of stomata and reduction or even cessation of transpiration for the adaptation to drought (Geiger et al., 2010) (Figure 2).
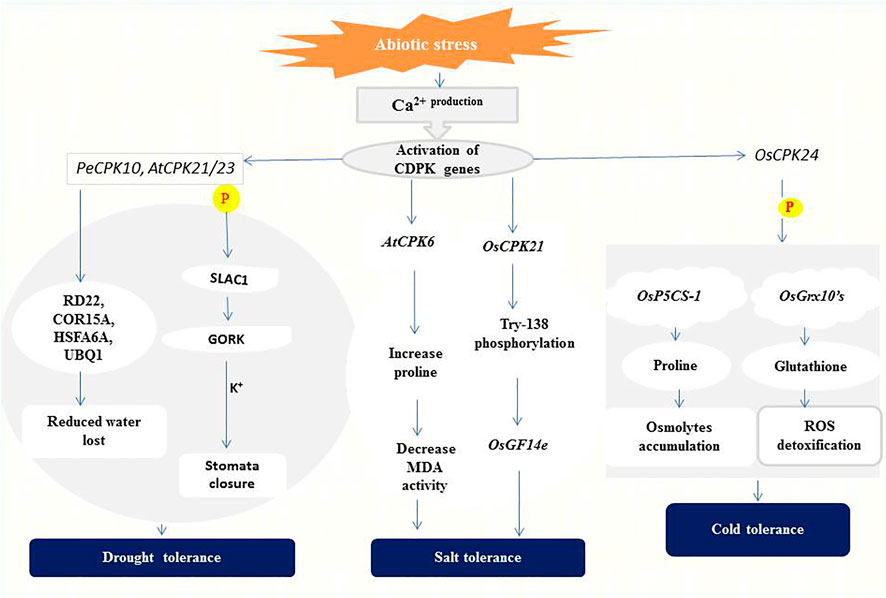
FIGURE 2. A representation of CDPK functions in abiotic stress signaling. CPK21/23 phosphorylates (P) SLAC1 to activate K+ ions efflux generation leading to stomatal closure (Geiger et al., 2010). PeCPK10 on the other hand induces drought tolerance through the expression of drought-responsive genes as stated above (Chen et al., 2013). Overexpression of AtCPK6 elevates proline level thereby repressing Malondialdehyde (MDA) generation (Xu et al., 2010). Whiles OsCPK21 interacts with OsGF14e by partially phosphorylating Try-138 to initiate the salt tolerance response (Chen et al., 2017). OsCPK24 phosphorylates (P) OsP5CS-1 to trigger proline production and also interacts with OsGrx10’s leading to glutathione generation which detoxifies reactive oxygen species (ROS) (Liu et al., 2018).
The expression of CDPKs in enhancing drought tolerance has been confirmed in several plant species. In ginger (Zingiber officinale), its ZoCDPK1 positively regulates signaling pathways in the response to drought and salt stress in a DRE/CRT-independent manner (Vivek et al., 2013). Overexpression of ZoCDPK1 in tobacco plants increases the expression level of the stress-related genes ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1) and RD21A (RESPONSIVE TO DEHYDRATION 21A), conferring drought stress tolerance. The rice OsCPK10 promotes drought tolerance by protecting cell membranes through enhanced ability to remove toxic reactive oxygen species (ROS) (Fang et al., 2015; Yin et al., 2015). ZmCPK12 is strongly induced by drought in maize seedlings, and overexpression of ZmCPK12 in Arabidopsis improved plant survival during drought (Wang and Song, 2013). Also, Populus euphratica PeCPK10 improves drought tolerance by activating drought-dependent genes such as RD22, COR15A, UBQ1, etc., and stimulates the expression of numerous ABA-dependent genes (AB11, AB15, NF-YB7, etc.) when overexpressed in Arabidopsis (Chen et al., 2013) (Figure 2).
In terms of transcriptional control of CDPKs under drought conditions, cis-elements in their promoters play an essential role. Many Glycine max GmCDPKs carry multiple stress-responsible cis-elements; moreover, most paralogs show little concordance in the distribution of cis-elements, suggesting that the promoters of paralogous GmCDPKs diverged after genome duplication events (Hettenhausen et al., 2016). Further investigation of the drought-inducible GmCDPK genes revealed the presence of ABREs (ABA response elements) and/or MBS (MYB binding sites) in their promoters, and nearly 80% of all GmCDPKs responded to either drought, ABA, or both (Hettenhausen et al., 2016). Functional analysis of turnip (Brassica rapa) BrrCDPKs revealed the up-regulation of many turnip CDPKs under various stress conditions, including drought stress (Wang et al., 2017). In cereals such as foxtail millet (Setaria italica), studies of the CDPK gene family revealed 13 and 11 SiCDPKs were up-and down-regulated, respectively, under drought, and most SiCDPKs were up-regulated by ABA, except SiCDPK4 and SiCDPK20 (Yu et al., 2018).
Soil salinity has emerged as a major threat to global crop production, affecting more than 800 million hectares of cropland, representing over 6% of the world’s land area (Munns and Tester, 2008). Salinity causes osmotic stress and decreases water uptake by plants (Liu et al., 2015). In addition, salinity also causes over-accumulation of Na+ and Cl− ions which creates an ionic imbalance in the plant. Plants normally mitigate the lethal effects of elevated Na+ in cells through two mechanisms: Na+ exclusion and Na+ sequestration (Nongpiur et al., 2016). Calcium acts as a second messenger in activating CDPKs gene expression, leading to the synthesis of compatible solutes to stop the deleterious effects of elevated Na+ in the cytoplasm (Mahajan et al., 2008). CPK13 prevents stomata opening through its inhibition of guard cell-expressed KAT2 (K+ transporter 2) and KAT1 (K+ transporter 1) channels (Ronzier et al., 2014). CPK12 on the other hand confers salt tolerance by regulating ion homeostasis and H2O2 production in roots, whilst downregulation of CPK12 led to high salt sensitivity in seedling growth and accumulation of high levels of Na+ and H2O2 (Zhang et al., 2018). Moreover, Negrão et al. (2011) discovered that OsCPK17 co-localized with four QTLs that correlate with ion homeostasis and salinity response. In addition, a potential overlap has been found between the signaling networks of salt stress and ethylene in cotton (Gossypium hirsutum). GhCPKs can be upregulated within an hour by ethephon (ETH) treatment under salt stress (Gao et al., 2018).
Rice has two positive regulators of salt tolerance: OsCPK7 and OsCPK12. OsCPK7 is mostly expressed in vascular bundles, where water stress is most severe when rice plants are stressed by salt and drought. Transgenic rice overexpressing OsCPK7 displayed increased salt resistance as well as increased induction of some stress-responsive genes, such as rab16A (Saijo et al., 2001). OsCPK12 promotes salt stress responses by upregulating ROS-scavenging enzymes (OsAPx2 and OsAPx8) and suppressing stress-induced ROS overproduction. OsCPK12 expression is linked to salt tolerance in rice, with mutations and RNA interference silencing resulting in a decreased tolerance to salt stress (Asano et al., 2012b). In the meantime, overexpression of OsCPK4 in rice induces the expression of genes involved in lipid metabolism and protection against oxidative stress under salt stress (Campo et al., 2014). According to Xu et al. (2010), high salt concentration is lethal to wild-type Arabidopsis plants, but transgenic lines overexpressing AtCPK6 accumulate more proline and less Malondialdehyde (MDA) which depicts improved stress tolerance (Xu et al., 2010). OsCPK21 interacts with Os14-3-3 (OsGF14e) to positively take part in ABA signaling and salt stress, partly by phosphorylating Try-138 (Chen et al., 2017) as indicated in (Figure 2). Other CDPKs identified in salt stress regulation in plants species include; ClCDPK6 in watermelon and its ortholog CsCDPK14 in cucumber (Xu et al., 2015; Wei et al., 2019) and VpCDPK9 in grape (Zhang et al., 2015).
Low temperatures are a major obstacle to the expansion of cultivated areas of some subtropical and tropical crops such as rubber (Hevea brasiliensis). Understanding plant response and adaptation would contribute to the development of cold-tolerant crops. The rubber, HbCPK (Ca2+ sensors) is downregulated after exposure to low temperatures (Xiao et al., 2017). Similarly, low temperatures control the expression of several CDPKs in A. thaliana, O. sativa, and P. trichocarpa (Xiao et al., 2017). In Brachypodium distachyon, the expression patterns of BdCDPKs vary with different gene members in response to cold and heat stress where several are down-regulated shortly after treatment and four (BdCDPK16/23/25/30) being strongly induced, suggesting that some BdCDPKs function early, while others respond late (Wen et al., 2020). Cold stress increased or decreased the expression of the ZmCPK1 and ZmCPK25 genes in maize. ZmCPK1 has an inverse relation with the regulation of the cold stress signaling mechanism. Studies on transgenic Arabidopsis also revealed that ZmCPK1 negatively regulates the expression of ethylene response factor (ZmERF3) genes and impairs cold stress tolerance (Weckwerth et al., 2015).
Several studies have shown that cold-induced expression of CDPKs could lead to a slowdown of cellular metabolism or mounting protection against cold stress. In rice, Ma et al. (2015) discovered that the perception of low temperature is associated with changes in Ca2+ influx and that this transition is regulated by COLD1, a G-protein signaling regulator. The rice OsCDPK13 and OsCPK17 are considered essential signaling components in cold response (Komatsu et al., 2007; Almadanim et al., 2017), and OsCPK17 relays cold stress signals and negatively regulates overall metabolism by reducing the activity of enzymes involved in sugar and nitrogen metabolism (Almadanim et al., 2017). On the other hand, overexpression of OsCPK24 in rice significantly increased proline and glutathione contents during cold treatment likely through up-regulating proline synthetase gene OsP5CS-1 and promoting Ca2+-responsive thioltransferase activity of OsGrx10 (Liu et al., 2018), as shown in (Figure 2). Both proline and glutathione are well-known protectants against stress because proline contributes to cellular osmotic changes while glutathione helps detoxify ROS (Verbruggen and Hermans, 2008; Hayat et al., 2012). PeCPK10 in Populus euphratica (Chen et al., 2013) and VaCPK20 in Vitis amurensis (Dubrovina et al., 2015) have also been described as positive regulators of cold stress tolerance.
Plants have evolved a multilayered inducible immune system that ensures their survival when attacked by microbial pathogens, including recognition and response to the threat or danger. Intracellular immune responses are rapidly mediated by receptor-mediated perceptions, manifested by adjustments in ion fluxes across membranes, transcriptional reprogramming, and activation of phosphorylation cascades (Romeis and Herde, 2014), where CDPKs and one of their substrates Rboh are involved (Kobayashi et al., 2012; Yoshioka et al., 2016). Wang et al. (2017) discovered that Pseudomonas syringae treatment in tomato (pstDC3000) increased the expression of most CDPKs. Phytohormones such as ET, JA, and SA with functional roles in biotic stress response can trigger the expression of some BrrCDPKs, and the expression of BrrCDPK4/10/17 was two times higher by pstDC3000 infection (Dempsey et al., 1999; van Loon et al., 2006). Both hormones and pstDC3000 infection also upregulate BrrRbohD1/2 expression (Figure 3), indicating the involvement and the interaction between BrrCDPK4/10/17 and BrrRbohD1/D2 in plant resistance against pstDC3000 (Wang et al., 2017).
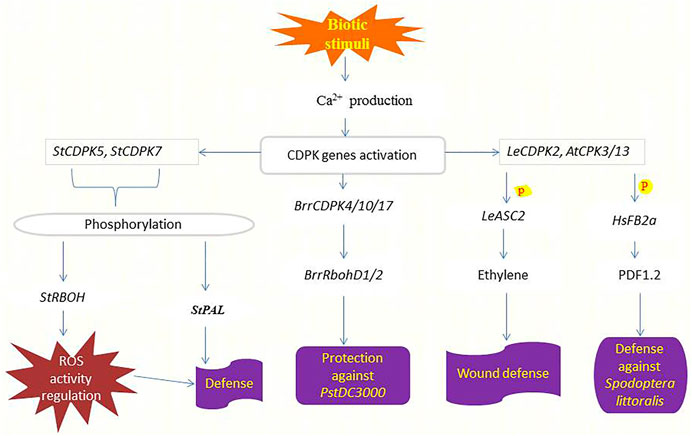
FIGURE 3. Functions of CDPKs in defending crops against pathogens and herbivore attacks. Upon sensing biotic stimuli or danger, the production of Ca2+ increases leading to the activation of various CDPKs responsible for inducing pathogen or herbivory resistance. StCDPK5 phosphorylates StRBOH to regulate ROS activity hence conferring defense response (Kobayashi et al., 2007), likewise, StCDPK7 phosphorylates StPAL to induce defense against Phytophthora infestans (Fantino et al., 2017). BrrCDPKs interact with BrrRbohD1/2 to repress the reproduction of pstDC3000 (Wang et al., 2017). Elevated cytosolic Ca2+ triggers the production of LeCDPK2 upon wounding which phosphorylates (P) LeASC2 leading to the generation and accumulation of ethylene and finally the induction of defense-related genes for protection (Kamiyoshihara et al., 2010). AtCPK3/13 also responds to wounding by phosphorylating (P) HsFB2a thereby regulating the transcript level of the PDF1.2 to induce resistance against Spodoptera littoralis attacks (Kanchiswamy et al., 2010).
The potato and tobacco orthologs of StCDPK7 and NtCDPK2 play functional roles in plant defense responses triggered by fungal elicitors (Romeis et al., 2001; Coca and San Segundo, 2010). Other CDPKs involved in the plant response to Phytophthora infestans in potato, probably target different proteins depending on their subcellular localization. StCDPK5 is localized in the plasma membrane and phosphorylates StRBOH regulating ROS activity (Kobayashi et al., 2007). Overexpression of StCDPK5 in potato results in tolerance to the hemibiotrophic pathogen P. infestans but enhances susceptibility to the necrotrophic pathogen Alternaria solani (Kobayashi et al., 2012). Besides that, StCDPK7 phosphorylates StPAL1 in vitro. Both proteins are cytosolically localized and up-regulated in response to P. infestans infection, and phosphorylation of StPAL1 mediated by StCDPK7 may affect PAL activity and localization associated with defense response (Fantino et al., 2017). Figure 3 summarizes the function of CDPKs in plant defense against pathogenic diseases. Future research will be required to determine whether the different CDPKs communicate with each other to control the projection of defense signals and plant resistance.
After herbivore feeding, there is a significant Ca2+ influx restricted to some cell layers in the damaged zone (Maffei et al., 2007). Phosphorylation cascade mediated by mitogen-activated protein kinases (MAP) and JA pathway is used to mediate these initial signals from insect attack within the plant (Howe and Jander, 2008). Kanchiswamy et al. (2010) identified the involvement of two Arabidopsis CPKs (CPK3 and CPK13) in the herbivory-induced signaling network through heat shock factor (HsfB2a)-mediated regulation of the defense-related transcriptional pathway. Mutant plants of cpk3 and cpk13 exposed to insect attack (Spodoptera littoralis) had lower transcript levels of PDF1.2 compared with wild-type plants, as shown in (Figure 3). It is shown that AtCPK3 can be activated by flg22 in protoplasts (Mehlmer et al., 2010) and induces the expression of the flg22-responsive gene NHL10 (Boudsocq et al., 2010). The activation of defense responses mediated by AtCPK3 and AtCPK13 is independent of phytohormone signaling pathways (Kanchiswamy et al., 2010). In comparison, LeCDPK2 in tomato phosphorylates the ethylene biosynthetic enzyme LeACS2 at the same site, that is, phosphorylated in vivo after wounding, indicating that LeCDPK2 plays a role in ethylene production in response to wounding (Kamiyoshihara et al., 2010) (Figure 3). Wounding also activates extracellular alkalinization by interfering with plasma membrane H+ -ATPase, which in tomato is mediated by membrane-anchored LeCPK1 (Rutschmann et al., 2002; Boudsocq and Sheen, 2013).
However, the first confirmation that CDPKs affect herbivore performance in plants was recently obtained in Nicotiana attenuate, where knockdown of NaCDPK4 and NaCDPK5 by virus-induced gene silencing (VIGS) increased JA and JA-Ile levels and distorted formation of secondary protective metabolites, leading to improved immunity to feeding by the specialized insect Manduca sexta (Yang et al., 2012; Hettenhausen et al., 2013). Interestingly, NaCDPK4/5 silenced plants also showed shortened stem elongation (Heinrich et al., 2013), which is identical to the cpk28 Arabidopsis single mutant (the closest homologs of AtCPK28 in N. attenuate are NaCDPK4 and NaCDPK5). Reduced stem elongation in cpk28 mutants is characterized by increased secondary growth, altered vascular architecture, and lignification (Matschi et al., 2013), suggesting that AtCPK28 activity may strongly influence developmental processes rather than specific defense-related phytohormones signaling pathways (Schulz et al., 2013). AtCPK28 explicitly phosphorylate BOTRYTIS-INDUCED KINASE1 (BIK1), a kinase required for PAMP-induced ignition of defense signaling (Matschi et al., 2015).
Several CDPKs have been shown to play deleterious roles in plant protection. HvCDPK3 promotes powdery mildew fungus invasion of barley (Hordeum vulgare) host cells in both compatible and incompatible interactions, while transient expression of constitutively active HvCDPK4 triggered a cell death response in N. benthamiana and H. vulgare (Freymark et al., 2007). Overexpression of OsCPK12 renders rice susceptible to both virulent and avirulent tuber leaf fungi, possibly due to ABA hypersensitivity and a decrease in ROS production (Asano et al., 2012b). When OsCPK13 is ectopically expressed in sorghum (Sorghum bicolor), it results in cell death, PR protein aggregation, and upregulation of certain protective genes (Mall et al., 2011).
Calcium-dependent protein kinases (CDPKs) in plants have received considerable attention in recent years because of their role in signal transduction in response to adverse conditions. Under stress conditions, various Ca2+ signals are generated, which are identified and decoded by specific Ca2+ transducers. This triggers various physiological and biochemical responses in the plant. However, the functional specificity of the different CDPK isoforms derives from their ability to recognize Ca2+ signals and bind Ca2+ with different affinities, interact with different proteins, and bind precise and exclusive substrates at different subcellular sites. This versatility is critical because plants in the field are very often exposed to a range of different biotic and abiotic stresses. Recent research has shown that plant responses to a mixture of different biotic and abiotic stressors are unique and cannot be directly inferred from the study of individual stressors. Such considerations are critical if we are to develop plants that are more resilient to more than one stress in order to reduce the adverse effects of expected global climate change on agricultural productivity worldwide.
Over the years, the focus has been on the study of biotic and abiotic stress tolerance in plants, including pathogens and herbivores defenses, hormonal signaling, drought, salt, and cold resistance, and other plant developmental processes. Despite extensive studies on the expression profiles of CDPK genes; their precise role in regulating adverse stress transduction pathways remains unclear. Considering the importance of CDPK genes in crop production, the developed transgenic plants should be tested in commercial fields to explore their interaction with an open environment. The positive functional role of CDPK genes in higher plants such as trees (rubber tree), legumes (soybean), tubers (potato), cereals (rice, maize), ginger, tomato, and others provide a solid foundation for future research to explore and exploit their full utility in food crops to mitigate the adverse effects of biotic and abiotic stresses. Progress in this area will help minimize the high cost of control measures associated with biotic stress in agriculture, resulting in high economic returns for industry players.
SDD and ZB developed the concept and prepared the original draft, RD, YW, FUH, CS, and PY reviewed and edited the manuscript, JB secured funding. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the National Natural Science Foundation of China (Grant No. 31960442, 32060502), the Special Fund for Discipline Construction of Gansu Agricultural University, Lanzhou China (GAU-XKJS-2018-084), China Agriculture Potato Research System of MOF and MARA (CARS-09-P14), the Gansu Provincial Science and Technology Department (120YF8WA137). Provided funds for drafting the manuscript and publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aleynova-Shumakova, O., Dubrovina, A., Manyakhin, A., Karetin, Y., and Kiselev, K. (2014). VaCPK20 gene overexpression significantly increased resveratrol content and expression of stilbene synthase genes in cell cultures of Vitis amurensis Rupr. Appl. Microbiol. Biotechnol. 98, 5541–5549. doi:10.1007/s00253-014-5625-7
Almadanim, M. C., Alexandre, B. M., Rosa, M. T., Sapeta, H., Leitão, A. E., Ramalho, J. C., et al. (2017). Rice calcium‐dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose‐phosphate synthase and is required for a proper cold stress response. Plant Cell. Environ. 40, 1197–1213. doi:10.1111/pce.12916
Aravind, J., Rinku, S., Pooja, B., Shikha, M., Kaliyugam, S., Mallikarjuna, M. G., et al. (2017). Identification, characterization, and functional validation of drought-responsive MicroRNAs in subtropical maize inbreds. Front. Plant Sci. 8, 941. doi:10.3389/fpls.2017.00941
Asai, S., Ichikawa, T., Nomura, H., Kobayashi, M., Kamiyoshihara, Y., Mori, H., et al. (2013). The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 288, 14332–14340. doi:10.1074/jbc.M112.448910
Asano, T., Hayashi, N., Kikuchi, S., and Ohsugi, R. (2012a). CDPK-mediated abiotic stress signaling. Plant Signal. Behav. 7, 817–821. doi:10.4161/psb.20351
Asano, T., Hakata, M., Nakamura, H., Aoki, N., Komatsu, S., Ichikawa, H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. doi:10.1007/s11103-010-9717-1
Asano, T., Hayashi, N., Kobayashi, M., Aoki, N., Miyao, A., Mitsuhara, I., et al. (2012b). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69, 26–36. doi:10.1111/j.1365-313X.2011.04766.x
Batistič, O., and Kudla, J. (2012). Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 1820, 1283–1293. doi:10.1016/j.bbagen.2011.10.012
Bi, Z., Wang, Y., Li, P., Sun, C., Qin, T., and Bai, J. (2021). Evolution and expression analysis of CDPK genes under drought stress in two varieties of potato. Biotechnol. Lett. 43, 511–521. doi:10.1007/s10529-020-03037-2
Billker, O., Lourido, S., and Sibley, L. D. (2009). Calcium-dependent signaling and kinases in apicomplexan parasites. Cell. Host Microbe 5, 612–622. doi:10.1016/j.chom.2009.05.017
Blum, A. (1996). “Crop responses to drought and the interpretation of adaptation,” in Drought tolerance in higher plants: Genetical, physiological and molecular biological analysis (Berlin, Germany: Springer), 57–70.
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi:10.1016/j.tplants.2012.08.008
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464, 418–422. doi:10.1038/nature08794
Bredow, M., Bender, K. W., Dingee, A. J., Holmes, D. R., Thomson, A., Ciren, D., et al. (2020). Phosphorylation-dependent sub-functionalization of the calcium-dependent protein kinase CPK28. J. bioRxiv 118, e2024272118. doi:10.1073/pnas.2024272118
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., and San Segundo, B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi:10.1104/pp.113.230268
Chandran, V., Stollar, E. J., Lindorff-Larsen, K., Harper, J. F., Chazin, W. J., Dobson, C. M., et al. (2006). Structure of the regulatory apparatus of a calcium-dependent protein kinase (CDPK): A novel mode of calmodulin-target recognition. J. Mol. Biol. 357, 400–410. [CrossRef] [PubMed]. doi:10.1016/j.jmb.2005.11.093
Chen, F., Fasoli, M., Tornielli, G. B., Dal Santo, S., Pezzotti, M., Zhang, L., et al. (2013). The evolutionary history and diverse physiological roles of the grapevine calcium dependent protein kinase gene family. PLoS ONE 8 (12), e80818. doi:10.1371/journal.pone.0080818
Chen, J., Xue, B., Xia, X., and Yin, W. J. (2013). A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem. Biophys. Res. Commun. 441, 630–636. doi:10.1016/j.bbrc.2013.10.103
Chen, Y., Zhou, X., Chang, S., Chu, Z., Wang, H., Han, S., et al. (2017). Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice. Biochem. Biophys. Res. Commun. 493, 1450–1456. doi:10.1016/j.bbrc.2017.09.166
Cheng, S.-H., Sheen, J., Gerrish, C., and Bolwell, G. P. (2001). Molecular identification of phenylalanine ammonia‐lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS Lett. 503, 185–188. doi:10.1016/s0014-5793(01)02732-6
Cheng, S.-H., Willmann, M. R., Chen, H.-C., and Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi:10.1104/pp.005645
Chico, J. M., Raıces, M., Téllez-Iñón, M. a. T., and Ulloa, R. M. (2002). A calcium-dependent protein kinase is systemically induced upon wounding in tomato plants. Plant Physiol. 128, 256–270. doi:10.1104/pp.128.1.256
Choi, H.-i., Hong, J.-h., Ha, J.-o., Kang, J.-y., and Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi:10.1074/jbc.275.3.1723
Choi, H. I., Park, H. J., Park, J. H., Kim, S., Im, M. Y., Seo, H. H., et al. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139, 1750–1761. doi:10.1104/pp.105.069757
Christodoulou, J., Malmendal, A., Harper, J. F., and Chazin, W. J. (2004). Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J. Biol. Chem. 279, 29092–29100. [CrossRef] [PubMed]. doi:10.1074/jbc.M401297200
Claeys, H., and Inzé, D. (2013). The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 162, 1768–1779. doi:10.1104/pp.113.220921
Coca, M., and San Segundo, B. (2010). AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 63, 526–540. doi:10.1111/j.1365-313X.2010.04255.x
Crizel, R. L., Perin, E. C., Vighi, I. L., Woloski, R., Seixas, A., da Silva Pinto, L., et al. (2020). Genome-wide identification and characterization of the CDPK gene family reveal their involvement in abiotic stress response in Fragaria x ananassa. Sci. Rep. 10, 11040–11117. doi:10.1038/s41598-020-67957-9
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi:10.1146/annurev-arplant-042809-112122
Dammann, C., Ichida, A., Hong, B., Romanowsky, S. M., Hrabak, E. M., Harmon, A. C., et al. (2003). Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132, 1840–1848. doi:10.1104/pp.103.020008
Dekomah, S. D., Wang, Y., Qin, T., Xu, D., Sun, C., Yao, P., et al. (2022). Identification and expression analysis of calcium-dependent protein kinases gene family in potato under drought stress. Front. Genet. 13, 874397. doi:10.3389/fgene.2022.874397
Dempsey, D. M. A., Shah, J., and Klessig, D. F. (1999). Salicylic acid and disease resistance in plants. CRC. Crit. Rev. Plant Sci. 18, 547–575. doi:10.1016/s0735-2689(99)00391-3
DiGiorgio, J. A., Bienert, G. P., Ayub, N. D., Yaneff, A., Barberini, M. L., Mecchia, M. A., et al. (2016). Pollen-specific aquaporins NIP4;1 and NIP4;2 are required for pollen development and pollination in Arabidopsis thaliana. Plant Cell. 28 (5), 1053–1077. doi:10.1105/tpc.15.00776
Dubrovina, A. S., Kiselev, K. V., Khristenko, V. S., and Aleynova, O. A. (2015). VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 185, 1–12. doi:10.1016/j.jplph.2015.05.020
Estruch, J. J., Kadwell, S., Merlin, E., and Crossland, L. (1994). Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 91 (19), 8837–8841. PMID:8090732. doi:10.1073/pnas.91.19.8837
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., et al. (2015). A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 66, 6803–6817. doi:10.1093/jxb/erv386
Fantino, E., Segretin, M. E., Santin, F., Mirkin, F. G., and Ulloa, R. M. (2017). Analysis of the potato calcium-dependent protein kinase family and characterization of StCDPK7, a member induced upon infection with Phytophthora infestans. Plant Cell. Rep. 36, 1137–1157. doi:10.1007/s00299-017-2144-x
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell. 14, S15–S45. doi:10.1105/tpc.010441
Freymark, G., Diehl, T., Miklis, M., Romeis, T., and Panstruga, R. (2007). Antagonistic control of powdery mildew host cell entry by barley calcium-dependent protein kinases (CDPKs). Mol. Plant. Microbe. Interact. 20, 1213–1221. doi:10.1094/MPMI-20-10-1213
Gao, W., Xu, F.-C., Guo, D.-D., Zhao, J.-R., Liu, J., Guo, Y.-W., et al. (2018). Calcium-dependent protein kinases in cotton: Insights into early plant responses to salt stress. BMC Plant Biol. 18, 15. doi:10.1186/s12870-018-1230-8
Gao, X., Chen, X., Lin, W., Chen, S., Lu, D., Niu, Y., et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9, e1003127. doi:10.1371/journal.ppat.1003127
Gargantini, P. R., Giammaria, V., Grandellis, C., Feingold, S. E., Maldonado, S., and Ulloa, R. M. (2009). Genomic and functional characterization of StCDPK1. Plant Mol. Biol. 70, 153–172. doi:10.1007/s11103-009-9462-5
Gargantini, P. R., Gonzalez-Rizzo, S., Chinchilla, D., Raices, M., Giammaria, V., Ulloa, R. M., et al. (2006). A CDPK isoform participates in the regulation of nodule number in Medicago truncatula. Plant J. 48, 843–856. doi:10.1111/j.1365-313X.2006.02910.x
Geiger, D., Maierhofer, T., Al-Rasheid, K. A., Scherzer, S., Mumm, P., Liese, A., et al. (2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4, ra32. doi:10.1126/scisignal.2001346
Geiger, D., Scherzer, S., Mumm, P., Marten, I., Ache, P., Matschi, S., et al. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U. S. A. 107, 8023–8028. doi:10.1073/pnas.0912030107
Gutermuth, T., Lassig, R., Portes, M. T., Maierhofer, T., Romeis, T., Borst, J. W., et al. (2013). Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell. 25, 4525–4543. doi:10.1105/tpc.113.118463
Hamel, L-P., Jen, S., and Armand, S. (2014). Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant Sci. 19, 79–89. doi:10.1016/j.tplants.2013.10.009
Harper, J. F., Breton, G., and Harmon, A. (2004). Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 55, 263–288. doi:10.1146/annurev.arplant.55.031903.141627
Harper, J. F., and Harmon, A. (2005). Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell. Biol. 6, 555–566. doi:10.1038/nrm1679
Hashimoto, K., and Kudla, J. (2011). Calcium decoding mechanisms in plants. Biochimie 93, 2054–2059. doi:10.1016/j.biochi.2011.05.019
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., and Ahmad, A. (2012). Role of proline under changing environments: A review. Plant Signal. Behav. 7, 1456–1466. doi:10.4161/psb.21949
Heinrich, M., Hettenhausen, C., Lange, T., Wünsche, H., Fang, J., Baldwin, I. T., et al. (2013). High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J. 73, 591–606. doi:10.1111/tpj.12058
Hettenhausen, C., Sun, G., He, Y., Zhuang, H., Sun, T., Qi, J., et al. (2016). Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 6, 18973–19014. doi:10.1038/srep18973
Hettenhausen, C., Yang, D. H., Baldwin, I. T., and Wu, J. (2013). Calcium-dependent protein kinases, CDPK4 and CDPK5, affect early steps of jasmonic acid biosynthesis in Nicotiana attenuata. Plant Signal. Behav. 8, e22784. doi:10.4161/psb.22784
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi:10.1146/annurev.arplant.59.032607.092825
Hrabak, E. M., Chan, C. W., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi:10.1104/pp.102.011999
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 7, 469. doi:10.3389/fpls.2016.00469
Ito, T., Nakata, M., Fukazawa, J., Ishida, S., and Takahashi, Y. (2010). Alteration of substrate specificity: The variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell. 22, 1592–1604. doi:10.1105/tpc.109.073577
Jaworski, K., Pawełek, A., Kopcewicz, J., and Szmidt-Jaworska, A. (2012). The calcium-dependent protein kinase (PnCDPK1) is involved in Pharbitis nil flowering. J. Plant Physiol. 169, 1578–1585. doi:10.1016/j.jplph.2012.05.025
Jiang, S., Zhang, D., Wang, L., Pan, J., Liu, Y., Kong, X., et al. (2013). A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. biochem. 71, 112–120. doi:10.1016/j.plaphy.2013.07.004
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi:10.1038/nature09916
Kamiyoshihara, Y., Iwata, M., Fukaya, T., Tatsuki, M., and Mori, H. (2010). Turnover of LeACS2, a wound‐inducible 1‐aminocyclopropane‐1‐carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J. 64, 140–150. doi:10.1111/j.1365-313X.2010.04316.x
Kanchiswamy, C. N., Takahashi, H., Quadro, S., Maffei, M. E., Bossi, S., Bertea, C., et al. (2010). Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 10, 97. doi:10.1186/1471-2229-10-97
Kawamoto, N., Endo, M., and Araki, T. (2015a). Expression of a kinase-dead form of CPK33 involved in florigen complex formation causes delayed flowering. Plant Signal. Behav. 10, e1086856. doi:10.1080/15592324.2015.1086856
Kawamoto, N., Sasabe, M., Endo, M., Machida, Y., and Araki, T. (2015b). Calcium dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci. Rep. 5, 8341. doi:10.1038/srep08341
Kiba, T., Takei, K., Kojima, M., and Sakakibara, H. (2013). Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell. 27, 452–461. doi:10.1016/j.devcel.2013.10.004
Kim, B. G., Waadt, R., Cheong, Y. H., Pandey, G. K., DominguezSolis, J. R., Schultke, S., et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52, 473–484. doi:10.1111/j.1365-313X.2007.03249.x
Klimecka, M., and Muszy ´nska, G. (2007). Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. 54, 219–233. [PubMed]. doi:10.18388/abp.2007_3242
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell. 19, 1065–1080. doi:10.1105/tpc.106.048884
Kobayashi, M., Yoshioka, M., Asai, S., Nomura, H., Kuchimura, K., Mori, H., et al. (2012). StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 196, 223–237. doi:10.1111/j.1469-8137.2012.04226.x
Komatsu, S., Yang, G., Khan, M., Onodera, H., Toki, S., and Yamaguchi, M. (2007). Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277, 713–723. doi:10.1007/s00438-007-0220-6
Kudla, J., Batistič, O., and Hashimoto, K. (2010). Calcium signals: The lead currency of plant information processing. Plant Cell. 22, 541–563. doi:10.1105/tpc.109.072686
Lerner, H. (2018). “Introduction to the response of plants to environmental stresses,” in Plant responses to environmental stresses (xfordshire, England, UK: Routledge), 1–26.
Li, A. L., Zhu, Y. F., Tan, X. M., Wang, X., Wei, B., Guo, H. Z., et al. (2008). Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 66, 429–443. doi:10.1007/s11103-007-9281-5
Li, C. L., Wang, M., Wu, X. M., Chen, D. H., Lv, H. J., Shen, J. L., et al. (2016). THI1, a thiamine thiazole synthase, interacts with Ca2+-dependent protein kinase CPK33 and modulates the S-Type anion channels and stomatal closure in Arabidopsis. Plant Physiol. 170, 1090–1104. [CrossRef] [PubMed]. doi:10.1104/pp.15.01649
Li, J., Li, Y., Deng, Y., Chen, P., Feng, F., Chen, W., et al. (2018). A calcium-dependent protein kinase, ZmCPK32, specifically expressed in maize pollen to regulate pollen tube growth. PLoS One 13, e0195787. doi:10.1371/journal.pone.0195787
Li, X., Zhao, L., Zhang, H., Liu, Q., Zhai, H., Zhao, N., et al. (2022). Genome-wide identification and characterization of CDPK family reveal their involvements in growth and development and abiotic stress in sweet potato and its two diploid relatives. Int. J. Mol. Sci. 23, 3088. doi:10.3390/ijms23063088
Liese, A., and Romeis, T. (2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 1833, 1582–1589. doi:10.1016/j.bbamcr.2012.10.024
Liu, K. H., Diener, A., Lin, Z., Liu, C., and Sheen, J. (2020). Primary nitrate responses mediated by calcium signalling and diverse protein phosphorylation. J. Exp. Bot. 71, 4428–4441. doi:10.1093/jxb/eraa047
Liu, K. H., Niu, Y., Konishi, M., Wu, Y., Du, H., Sun, C. H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 18, 311–316. [CrossRef] [PubMed]. doi:10.1038/nature22077
Liu, P., Yin, L., Wang, S., Zhang, M., Deng, X., Zhang, S., et al. (2015). Enhanced root hydraulic conductance by aquaporin regulation accounts for silicon alleviated salt-induced osmotic stress in Sorghum bicolor L. Environ. Exp. Bot. 111, 42–51. doi:10.1016/j.envexpbot.2014.10.006
Liu, Y., Xu, C., Zhu, Y., Zhang, L., Chen, T., Zhou, F., et al. (2018). The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 60, 173–188. doi:10.1111/jipb.12614
Loqué, D., and von Wirén, N. J. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi:10.1093/jxb/erh147
Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., et al. (2006). Additive contribution of AMT1; 1 and AMT1; 3 to high‐affinity ammonium uptake across the plasma membrane of nitrogen‐deficient Arabidopsis roots. Plant J. 48, 522–534. doi:10.1111/j.1365-313X.2006.02887.x
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). COLD1 confers chilling tolerance in rice. Cell. 160, 1209–1221. doi:10.1016/j.cell.2015.01.046
Maffei, M. E., Mithöfer, A., and Boland, W. (2007). Before gene expression: Early events in plant–insect interaction. Trends Plant Sci. 12, 310–316. doi:10.1016/j.tplants.2007.06.001
Mahajan, S., Pandey, G. K., and Tuteja, N. (2008). Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 471, 146–158. doi:10.1016/j.abb.2008.01.010
Mall, T. K., Dweikat, I., Sato, S. J., Neresian, N., Xu, K., Ge, Z., et al. (2011). Expression of the rice CDPK-7 in sorghum: Molecular and phenotypic analyses. Plant Mol. Biol. 75, 467–479. doi:10.1007/s11103-011-9741-9
Manimaran, P., Mangrauthia, S. K., Sundaram, R., and Balachandran, S. (2015). Constitutive expression and silencing of a novel seed specific calcium dependent protein kinase gene in rice reveals its role in grain filling. J. Plant Physiol. 174, 41–48. doi:10.1016/j.jplph.2014.09.005
Martı´n, M. L., and Busconi, L. (2000). Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 24, 429–435. doi:10.1046/j.1365-313x.2000.00889.x
Matschi, S., Hake, K., Herde, M., Hause, B., and Romeis, T. (2015). The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell. 27, 591–606. doi:10.1105/tpc.15.00024
Matschi, S., Werner, S., Schulze, W. X., Legen, J., Hilger, H. H., and Romeis, T. (2013). Function of calcium‐dependent protein kinase CPK 28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896. doi:10.1111/tpj.12090
McCormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi:10.1016/j.tplants.2005.07.001
Mehlmer, N., Wurzinger, B., Stael, S., Hofmann‐Rodrigues, D., Csaszar, E., Pfister, B., et al. (2010). The Ca2+‐dependent protein kinase CPK3 is required for MAPK‐independent salt‐stress acclimation in. Plant J. 63, 484–498. doi:10.1111/j.1365-313X.2010.04257.x
Mohanta, T. K., Kumar, P., and Bae, H. (2017). Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin-like proteins in plants. BMC Plant Biol. 17, 38. [CrossRef]. doi:10.1186/s12870-017-0989-3
Monaghan, J., Matschi, S., Shorinola, O., Rovenich, H., Matei, A., Segonzac, C., et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell. Host Microbe 16, 605–615. doi:10.1016/j.chom.2014.10.007
Moutinho, A., Trewavas, A. J., and Malho, R. (1998). Relocation of a Ca2+-dependent protein kinase activity during pollen tube reorientation. Plant Cell. 10 (9), 1499–1510. PMID:9724696. doi:10.1105/tpc.10.9.1499
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi:10.1146/annurev.arplant.59.032607.092911
Myers, C., Romanowsky, S. M., Barron, Y. D., Garg, S., Azuse, C. L., Curran, A., et al. (2009). Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 59 (4), 528–539. doi:10.1111/j.1365-313X.2009.03894.x
Nakata, M., Yuasa, T., Takahashi, Y., and Ishida, S. (2009). CDPK1, a calcium-dependent protein kinase, regulates transcriptional activator RSG in response to gibberellins. Plant Signal. Behav. 4, 372–374. doi:10.4161/psb.4.5.8229
Negrão, S., Courtois, B., Ahmadi, N., Abreu, I., Saibo, N., and Oliveira, M. (2011). Recent updates on salinity stress in rice: From physiological to molecular responses. Crit. Rev. Plant Sci. 30, 329–377. doi:10.1080/07352689.2011.587725
Nongpiur, R., Lata Singla-Pareek, S., and Pareek, A. (2016). Genomics approaches for improving salinity stress tolerance in crop plants. Curr. Genomics 17, 343–357. doi:10.2174/1389202917666160331202517
Pantoja, O. (2012). High affinity ammonium transporters: Molecular mechanism of action. Front. Plant Sci. 3, 34. doi:10.3389/fpls.2012.00034
Pawełek, A., Szmidt-Jaworska, A., Świeżawska, B., and Jaworski, K. (2015). Genomic structure and promoter characterization of the CDPK kinase gene expressed during seed formation in Pharbitis nil. J. Plant Physiol. 189, 87–96. doi:10.1016/j.jplph.2015.08.008
Pinheiro, C., and Chaves, M. (2011). Photosynthesis and drought: Can we make metabolic connections from available J. Exp. Bot. 62, 869–882. doi:10.1093/jxb/erq340
Qin, D.-B., Liu, M.-Y., Yuan, L., Zhu, Y., Li, X.-D., Chen, L.-M., et al. (2020). CALCIUM-DEPENDENT PROTEIN KINASE 32-mediated phosphorylation is essential for the ammonium transport activity of AMT1; 1 in Arabidopsis roots. J. Exp. Bot. 71, 5087–5097. doi:10.1093/jxb/eraa249
Rastogi, S., and Liberles, D. A. (2005). Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 5, 1–7.
Ray, S., Agarwal, P., Arora, R., Kapoor, S., and Tyagi, A. K. (2007). Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol. Genet. Genomics 278, 493–505. doi:10.1007/s00438-007-0267-4
Remington, D. L., Vision, T. J., Guilfoyle, T. J., and Reed, J. W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752. doi:10.1104/pp.104.039669
Rensing, S. A. (2014). Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 17, 43–48. doi:10.1016/j.pbi.2013.11.002
Romeis, T., and Herde, M. (2014). From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 20, 1–10. doi:10.1016/j.pbi.2014.03.002
Romeis, T., Ludwig, A. A., Martin, R., and Jones, J. D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20, 5556–5567. doi:10.1093/emboj/20.20.5556
Romeis, T., Piedras, P., and Jones, J. D. (2000). Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell. 12, 803–816. doi:10.1105/tpc.12.5.803
Ronzier, E., Corratge-Faillie, C., Sanchez, F., Prado, K., Briere, C., Leonhardt, N., et al. (2014). CPK13 a noncanonical Ca2+-dependent protein kinase specifically inhibits KAT2 and KAT1 shaker K+ channels and reduces stomatal opening. Plant Physiol. 166, 314–326. [CrossRef] [PubMed]. doi:10.1104/pp.114.240226
Rutschmann, F., Stalder, U., Piotrowski, M., Oecking, C., and Schaller, A. (2002). LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 129, 156–168. doi:10.1104/pp.000869
Saijo, Y., Kinoshita, N., Ishiyama, K., Hata, S., Kyozuka, J., Hayakawa, T., et al. (2001). A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell. Physiol. 42 (11), 1228–1233. doi:10.1093/pcp/pce158
Schulz, P., Herde, M., and Romeis, T. (2013). Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 163, 523–530. doi:10.1104/pp.113.222539
Seybold, H., Bortlik, J., Conrads, B., Hoehenwarter, W., and Romeis, T. (2019). Prioritization of abiotic and biotic stress responses by direct linkage of ABI1 phosphatase and CPK5 calcium-dependent protein kinase. J. bioRxiv 2019, 839662 doi:10.1101/839662
Shi, S., Li, S., Asim, M., Mao, J., Xu, D., Ullah, Z., et al. (2018). The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 19, 1900. [CrossRef] [PubMed]. doi:10.3390/ijms19071900
Shikha, M., Kanika, A., Rao, A. R., Mallikarjuna, M. G., Gupta, H. S., and Nepolean, T. (2017). Genomic selection for drought tolerance using genome-wide SNPs in maize. Front. Plant Sci. 8, 550. doi:10.3389/fpls.2017.00550
Shkryl, Y., Veremeichik, G., Bulgakov, V., and Zhuravlev, Y. N. (2011). Induction of anthraquinone biosynthesis in Rubia cordifolia cells by heterologous expression of a calcium‐dependent protein kinase gene. Biotechnol. Bioeng. 108, 1734–1738. doi:10.1002/bit.23077
Ton, J., Flors, V., and Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi:10.1016/j.tplants.2009.03.006
Tong, T., Li, Q., Jiang, W., Chen, G., Xue, D., Deng, F., et al. (2021). Molecular evolution of calcium signaling and transport in plant adaptation to abiotic stress. Int. J. Mol. Sci. 22, 12308. doi:10.3390/ijms222212308
Valmonte, G. R., Arthur, K., Higgins, Colleen M., and MacDiarmid, Robin M. (2014). Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell. Physiol. 55 (3), 551–569. doi:10.1093/pcp/pct200
Van Gioi, H., Mallikarjuna, M. G., Shikha, M., Pooja, B., Jha, S. K., Dash, P. K., et al. (2017). Variable level of dominance of candidate genes controlling drought functional traits in maize hybrids. Front. Plant Sci. 8, 940. doi:10.3389/fpls.2017.00940
van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi:10.1146/annurev.phyto.44.070505.143425
Verbruggen, N., and Hermans, C. (2008). Proline accumulation in plants: A review. Amino Acids 35, 753–759. doi:10.1007/s00726-008-0061-6
Veremeichik, G., Grigorchuk, V., Silanteva, S., Shkryl, Y., Bulgakov, D., Brodovskaya, E., et al. (2019). Increase in isoflavonoid content in Glycine max cells transformed by the constitutively active Ca2+ independent form of the AtCPK1 gene. Phytochemistry 157, 111–120. doi:10.1016/j.phytochem.2018.10.023
Vivek, P. J., Tuteja, N., and Soniya, E. V. (2013). CDPK1 from ginger promotes salinity and drought stress tolerance without yield penalty by improving growth and photosynthesis in Nicotiana tabacum. PLoS One 8, e76392. doi:10.1371/journal.pone.0076392
Wan, B., Lin, Y., and Mou, T. (2007). Expression of rice Ca2+-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS letters 581, 1179–1189.
Wang, C.-T., and Song, W. (2013). Calcium-dependent protein kinase gene ZmCPK12 from maize confers tolerance to drought and salt stresses in transgenic plants. Acta Physiol. Plant. 35, 1659–1666. doi:10.1007/s11738-012-1208-3
Wang, J., Cheng, G., Wang, C., He, Z., Lan, X., Zhang, S., et al. (2017). The bHLH transcription factor CgbHLH001 is a potential interaction partner of CDPK in halophyte Chenopodium glaucum. Sci. Rep. 7, 1–16. doi:10.1038/s41598-017-06706-x
Wang, J. P., Xu, Y. P., Munyampundu, J. P., Liu, T. Y., and Cai, X. Z. (2016). Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: Genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genomics 291, 661–676. doi:10.1007/s00438-015-1137-0
Wang, L., Yu, C., Xu, S., Zhu, Y., and Huang, W. (2016). OsDi19‐4 acts downstream of OsCDPK14 to positively regulate ABA response in rice. Plant Cell. Environ. 39, 2740–2753. doi:10.1111/pce.12829
Wang, Q., Yin, X., Chen, Q., Xiang, N., Sun, X., Yang, Y., et al. (2017). Genome-wide survey indicates diverse physiological roles of the turnip (Brassica rapa var. rapa) calcium-dependent protein kinase genes. Sci. Rep. 7, 1–14. doi:10.1038/s41598-017-16102-0
Wang, X., Lv, S., Han, X., Guan, X., Shi, X., Kang, J., et al. (2019). The calcium-dependent protein kinase CPK33 mediates strigolactone-induced stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 10, 1630. doi:10.3389/fpls.2019.01630
Weckwerth, P., Ehlert, B., and Romeis, T. (2015). ZmCPK1, a calcium‐independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signaling. Plant Cell. Environ. 38, 544–558. doi:10.1111/pce.12414
Wei, C., Zhang, R., Yang, X., Zhu, C., Li, H., Zhang, Y., et al. (2019). Comparative analysis of calcium-dependent protein kinase in cucurbitaceae and pt
Wen, F., Ye, F., Xiao, Z., Liao, L., Li, T., Jia, M., et al. (2020). Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in grass Brachypodium distachyon. BMC Genomics 21, 53. doi:10.1186/s12864-020-6475-6
Wu, P., Wang, W., Duan, W., Li, Y., and Hou, X. (2017). Comprehensive analysis of the CDPK-SnRK superfamily genes in Chinese cabbage and its evolutionary implications in plants. Front. Plant Sci. 8, 162. doi:10.3389/fpls.2017.00162
Wu, Y., Zhao, S., Tian, H., He, Y., Xiong, W., Guo, L., et al. (2013). CPK3-phosphorylated RhoGDI1 is essential in the development of Arabidopsis seedlings and leaf epidermal cells. J. Exp. Bot. 64, 3327–3338. doi:10.1093/jxb/ert171
Xiao, X. H., Yang, M., Sui, J. L., Qi, J. Y., Fang, Y. J., Hu, S. N., et al. (2017). The calcium‐dependent protein kinase (CDPK) and CDPK‐related kinase gene families in Hevea brasiliensis—Comparison with five other plant species in structure, evolution, and expression. FEBS open bio 7, 4–24. doi:10.1002/2211-5463.12163
Xu, J., Tian, Y. S., Peng, R. H., Xiong, A. S., Zhu, B., Jin, X. F., et al. (2010). AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 231, 1251–1260. doi:10.1007/s00425-010-1122-0
Xu, X., Liu, M., Lu, L., He, M., Qu, W., Xu, Q., et al. (2015). Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genomics 290, 1403–1414. doi:10.1007/s00438-015-1002-1
Yang, D.-H., Hettenhausen, C., Baldwin, I. T., and Wu, J. (2012). Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound-and herbivory-induced jasmonic acid accumulations. Plant Physiol. 159, 1591–1607. doi:10.1104/pp.112.199018
Yang, L., Zhang, Y., Guan, R., Li, S., Xu, X., Zhang, S., et al. (2020). Co‐regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J. Integr. Plant Biol. 62, 1780–1796. doi:10.1111/jipb.12973
Ye, W., Muroyama, D., Munemasa, S., Nakamura, Y., Mori, I. C., and Murata, Y. (2013). Calcium-dependent protein kinase CPK6 positively functions in induction by yeast elicitor of stomatal closure and inhibition by yeast elicitor of light-induced stomatal opening in Arabidopsis. Plant Physiol. 163, 591–599. doi:10.1104/pp.113.224055
Yin, X. M., Huang, L. F., Zhang, X., Wang, M. L., Xu, G. Y., and Xia, X. J. (2015). OsCML4 improves drought tolerance through scavenging of reactive oxygen species in rice. J. Plant Biol. 58, 68–73. doi:10.1007/s12374-014-0349-x
Yip Delormel, T., and Boudsocq, M. (2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224, 585–604. doi:10.1111/nph.16088
Yoon, G. M., Dowd, P. E., Gilroy, S., and McCubbin, A. G. (2006). Calcium dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell. 18 (4), 867–878. doi:10.1105/tpc.105.037135
Yoshioka, H., Adachi, H., Nakano, T., Miyagawa, N., Asai, S., Ishihama, N., et al. (2016). Hierarchical regulation of NADPH oxidase by protein kinases in plant immunity. Physiol. Mol. Plant Pathol. 95, 20–26. doi:10.1016/j.pmpp.2016.03.004
Yu, T. F., Zhao, W. Y., Fu, J. D., Liu, Y. W., Chen, M., Zhou, Y. B., et al. (2018). Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 9, 651. doi:10.3389/fpls.2018.00651
Yuan, L., Gu, R., Xuan, Y., Smith-Valle, E., Loqué, D., Frommer, W. B., et al. (2013). Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell. 25, 974–984. doi:10.1105/tpc.112.108027
Yuan, L., Loque, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell. 19, 2636–2652. doi:10.1105/tpc.107.052134
Zhang, H., Wei, C., Yang, X., Chen, H., Yang, Y., Mo, Y., et al. (2017). Genome-wide identification and expression analysis of calcium dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PLoS One 12, e0176352. doi:10.1371/journal.pone.0176352
Zhang, H., Zhang, Y., Deng, C., Deng, S., Li, N., Zhao, C., et al. (2018). The Arabidopsis Ca2+-dependent protein kinase CPK12 is involved in plant response to salt stress. Int. J. Mol. Sci. 19, 4062. doi:10.3390/ijms19124062
Zhang, K., Han, Y.-T., Zhao, F.-L., Hu, Y., Gao, Y.-R., Ma, Y.-F., et al. (2015). Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 15, 1–19. doi:10.1186/s12870-015-0552-z
Zhang, X., and Liu, C. J. (2015). Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 8, 17–27. doi:10.1016/j.molp.2014.11.001
Zhang, X. S., and Choi, J. H. (2001). Molecular evolution of calmodulin-like domain protein kinases (CDPKs) in plants and protists. J. Mol. Evol. 53, 214–224. doi:10.1007/s002390010211
Zhao, L. N., Shen, L. K., Zhang, W. Z., Zhang, W., Wang, Y., and Wu, W. H. (2013). Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell. 25, 649–661. [CrossRef] [PubMed]. doi:10.1105/tpc.112.103184
Zhao, R., Sun, H. L., Mei, C., Wang, X. J., Yan, L., Liu, R., et al. (2011). The Arabidopsis Ca2+‐dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post‐germination growth. New Phytol. 192, 61–73. doi:10.1111/j.1469-8137.2011.03793.x
Zhou, J., Wang, X., He, Y., Sang, T., Wang, P., Dai, S., et al. (2020). Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell. 32, 2621–2638. doi:10.1105/tpc.19.00971
Zhu, J. H., Chen, X., Chang, W. J., Tian, W. M., and Zhang, Z. L. (2010). Molecular characterization of HbCDPK1, an ethephon-induced calcium-dependent protein kinase gene of. Biosci. Biotechnol. Biochem. 74, 2183–2188. doi:10.1271/bbb.100293
Zou, J. J., Li, X. D., Ratnasekera, D., Wang, C., Liu, W. X., Song, L. F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell. 27, 1445–1460. doi:10.1105/tpc.15.00144
Zou, J. J., Wei, F. J., Wang, C., Wu, J. J., Ratnasekera, D., Liu, W. X., et al. (2010). Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 154, 1232–1243. doi:10.1104/pp.110.157545
Keywords: calcium, calcium-dependent protein kinases, stress tolerance, plants, biotic and abiotic stress
Citation: Dekomah SD, Bi Z, Dormatey R, Wang Y, Haider FU, Sun C, Yao P and Bai J (2022) The role of CDPKs in plant development, nutrient and stress signaling. Front. Genet. 13:996203. doi: 10.3389/fgene.2022.996203
Received: 17 July 2022; Accepted: 14 September 2022;
Published: 30 September 2022.
Edited by:
Vijay Gahlaut, Institute of Himalayan Bioresource Technology (CSIR), IndiaReviewed by:
Cleverson Carlos Matiolli, Universidade Nova de Lisboa, PortugalCopyright © 2022 Dekomah, Bi, Dormatey, Wang, Haider, Sun, Yao and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangping Bai, YmFpanBAZ3NhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.