- 1Shenzhen Key Laboratory of Marine Bioresources and Ecology, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2College of Physics and Optoelectronic Engineering, Shenzhen University, Shenzhen, China
- 3College of Animal Science and Technology, Nanjing Agricultural University, Nanjing, China
- 4The Fifth Affiliate Hospital of Xinjiang Medical University, Urumqi, China
- 5Animal Diseases Control and Prevention Centre of Xinjiang Uygur Autonomous Region, Urumqi, China
- 6Ili Kazak Autonomous Prefecture Institute of Animal Science, Ili, China
- 7College of Animal Science and Technology, Xinjiang Agricultural University, Urumqi, China
- 8Institute of Animal Husbandry, Xinjiang Academy of Animal Science, Urumqi, China
Bone morphogenetic protein receptor type-1B (BMPR1B) is one of the major gene for sheep prolificacy. However, few studies investigated its regulatory region. In this study, we reported that miR-1306 is a direct inhibitor of BMPR1B gene in the ovine granulosa cells (ovine GCs). We detected a miRNA response element of miR-1306 in the 3’ untranslated region of the ovine BMPR1B gene. Luciferase assay showed that the ovine BMPR1B gene is a direct target of miR-1306. qPCR and western blotting revealed that miR-1306 reduces the expression of BMPR1B mRNA and protein in the ovine granulosa cells. Furthermore, miR-1306 promoted cell apoptosis by suppressing BMPR1B expression in the ovine granulosa cells. Overall, our results suggest that miR-1306 is an epigenetic regulator of BMPR1B, and may serve as a potential target to improve the fecundity of sheep.
Introduction
Bone morphogenetic protein (BMP)/Smad is one of the key signaling pathways for the regulation of mammalian fertility and plays important roles in gonadal development, steroid hormone production, follicular development, ovulation, luteal formation, and female reproductive diseases (Wang et al., 2010; Otsuka, 2018; Hoffmann et al., 2020; Wu et al., 2020). This signaling pathway is closely related to fecundity in sheep. Genes involved in this signaling pathway are widely expressed in ovarian tissue (Bahire et al., 2019), among which BMP15, and BMP receptor type-1B (BMPR1B) are the major genes of reproductive traits and their expression in ovarian tissues is related to fecundity (Xu et al., 2010; Foroughinia et al., 2017; Bahire et al., 2019).
In high and low fecundity Hu sheep ovarian antral follicles, the mRNA expression of BMPR1B, BMP4, BMPR2, GDF9, and Smad4 in high fecundity Hu sheep are significantly higher than that in low fecundity Hu sheep (Xu et al., 2010). Whole-genome sequencing and genome-wide association analysis in the pig show that a couple of members of BMP signaling pathways, like BMP5, BMP6, BMP7, BMP15, and BMPR1B are candidate genes related to reproductive traits (Schneider et al., 2014; Li et al., 2017). Among the major genes (BMP15, BMPR1B, GDF9, and B4GALT2) certified to affect sheep reproductive traits, only BMP15 and BMPR1B are core members of BMP signaling pathways (Abdoli et al., 2016).
BMPR1B was one of the most important major gene to be associated with prolificity in sheep, the A746G mutation (FecB) in the coding region is the only causal mutation site in BMPR1B (Mulsant et al., 2001). Ewes carrying FecB mutations have a higher ovulation rate and litter size (the set of offspring which mammals produce at one birth), fewer apoptotic granulocytes, and higher BMPR1B levels in the ovaries (Regan et al., 2015). The FecB gene has been detected in multiple breeds of sheep. It was found in the famous multiple-litter sheep breeds such as Hu sheep and Small-tailed Han sheep, among which the genotype frequencies of the three genotypes of Hu sheep FecB locus BB, B+ and ++ were 0.80, 0.16, and 0.04, respectively; The genotype frequencies of the three genotypes in Small Tail Han sheep were 0.26, 0.64 and 0.10, respectively (Montgomery et al., 2001; Tang et al., 2018). The FecB gene was also detected in Marwari, Bharat sheep (Kumar et al., 2006), Assaf sheep (Gootwine, 2011), and GMM sheep (Bahire et al., 2019). According to a recent study, BMPR1B is an important antiapoptotic factor in ovine GCs. BMPR1B can promote follicular development and increase fecundity via the inhibition of the apoptosis of Granulosa cells (Yao et al., 2019). Although the correlation between the FecB mutation polymorphism in the BMPR1B coding region and litter size has been shown, however few studies have investigated its regulatory region. In this study, we aimed to analyze the regulatory function of miRNAs on BMPR1B in the ovine granulosa cells to provide a basis for an investigation into the regulatory mechanism of BMPR1B transcription in sheep.
Materials and methods
Ethics statement
All experiments in this study were approved by the Animal Protection and Utilization Committee of Nanjing Agricultural University and were carried out in strict accordance with the policies of the National Laboratory Animal Administration (Order No. 2 of the China Science and Technology Commission, 14 November 1988, institution certification number: SYXK 2017-0027).
Animals and samples collection
A total of 256 fresh sheep ovaries were collected from the Hauling slaughter house (Urumqi, Xinjiang). The ovaries were immediately soaked in 37°C saline that contained penicillin and streptomycin (Invitrogen, China) and taken to the laboratory for subsequent operations within 2 h.
Cell culture
The isolation and culture of ovine Granulosa cells were carried out according to an experiment manual (Yao et al., 2019). After resuscitation, HEK293T cells were inoculated in Dulbecco’s modified Eagle’s medium that contained 1% double antibody (Invitrogen, China) and 10% fetal bovine serum (Invitrogen). Ovine follicular Granulosa cells and HEK293T cells were inoculated in a T25 cell culture flask (Corning) and cultured in a 37°C incubator that contained 5% CO2.
Dual-luciferase assay
The cells were recaptured and placed in 6- and 12-well cell culture plates (Invitrogen, China) at the appropriate cell density. After 12 h, the cells were completely attached and transfected with Lipofectamine 3000 Transfection Reagent (Invitrogen, China), according to the kit instructions. The miR-1306-5p mimic sequence was CCACCUCCCCUGCAAACGUCCA: mimics NC was UUCUCCGAACGUGUCACGUTT. Control or mimics were transfected one per well at a concentration of 20uM. Expression vector pcDNA3.1-BMPR1B was prepared previously by our group (Yao et al., 2019). Cells were transfected with 2 ug of pcDNA3.1 and pcDNA3.1-BMPR1B one per well. After 24 h of transfection, the cells were collected, and the fluorescence intensity of firefly and Renilla in each sample was measured by a dual-luciferase assay system kit (Promega company, US) and enzyme labeling instrument. The operation steps are detailed in the instructions.
qRT-PCR
Total RNA was extracted from ovine Granulosa cells according to the instructions of a High Purity RNA Extraction Kit (Beijing Bioteke, China). The RNA thus obtained was reverse transcribed using PrimeScript RT Master Mix Reverse Transcriptase Kit (TaKaRa, Japan), and cDNA was generated using a SYBR Green Master Mix Kit (Vazyme, China) for quantitative analysis. The relative standard curve method (2- △△t method) was used to calculate the relative expression of the target gene, based on the expression of the GAPDH internal reference gene.
Western blotting
Antibodies for BMPR1B (ab155058) and GAPDH (ab9482) were obtained from Abcam (UK). Western blotting was performed according to a previously reported method (Liu et al., 2016).
Apoptosis assay
The Annexin V-FITC/PI apoptosis detection kit (Vazyme, China) was used for granulosa cell apoptosis analysis. The experimental processes were operated following the kit instructions and the experimental process is detailed as follows. Cells were digested using trypsin and collected. Gently blow the cells with 500 μl PBS until they are resuspended and repeated once; after centrifugation of 1,000 g for 5 min, suck up the supernatant, resuspend the cells with 100 μl binding buffer, add 5 μl annexin V-FITC and 5 μl PI in turn in dark; After 10 min of incubation, add 400 μL binding buffer on the machine to measure the apoptosis rate of ovine Granulosa cells.
Statistical analysis
Experimental data were analyzed for statistical significance with t-tests using SPSS v20.0 statistical software (SPSS Inc., Chicago, IL, United States). All experiments were performed in three biological replicates. Differences were considered significant and very significant at p < 0.05 and p < 0.01, respectively. Graphs were drawn using Prism 5.0 software.
Results
miR-1306 is a candidate miRNA that targets ovine BMPR1B
Through hybridization to incompletely complementary sequences in the 3' untranslated region (UTR) of their target messenger RNAs (mRNAs), miRNAs can negatively control gene transcription (Bartel, 2009; Djuranovic et al., 2012). Three types of online software (TargetScan, miRDB, and miR-walk) were used to predict the miRNAs in the 3'-UTR of BMPR1B, 98 common potential miRNAs were detected (Figure 1A). Among these, similar to the BMPR1B gene (Yao et al., 2019), miR-1306 has been identified as a key regulator of ovarian GC apoptosis in mammals (Yang et al., 2019). Results obtained using the online program RNAhybrid revealed that the binding site for miR-1306 is located at nucleotide 963–969 of the 3'-UTR of BMPR1B and that the minimum free energy for binding is -23.7 kcal/mol; this indicates that miR-1306 can stably bind with the BMPR1B 3′-UTR (Figures 1B,C). Based on these findings, miR-1306 was identified as a candidate miRNA targeting the ovine BMPR1B gene and was selected for further analysis.
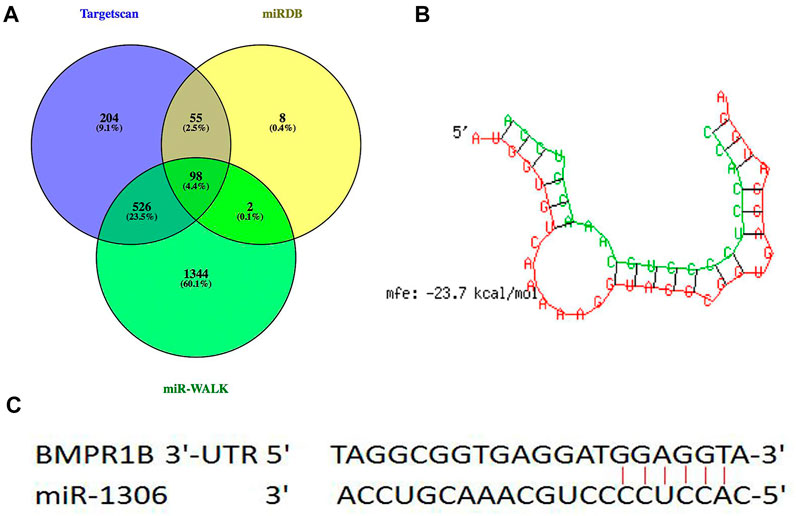
FIGURE 1. Candidate miRNAs that target BMPR1B. (A) miRNAs directly targeting BMPR1B were predicted using TargetScan, miRDB, and miR-WALK software. (B) RNAhybrid analysis. (C) Schematic diagram indicating the binding site sequences.
BMPR1B was a direct target of miR-1306 in ovine granulosa cells
To investigate whether the BMPR1B gene of Hu sheep is targeted by miR-1306, we performed luciferase assays using wild-type and mutant-type BMPR1B gene 3'-UTR luciferase reporter vectors (Figure 2A). These vectors co-transfected with miR-1306 mimics. The assay results indicated that the luciferase activity in cells containing the wild-type vector was markedly reduced on co-transfection with miR-1306 mimics (Figure 2B), thereby indicating that miR-1306 can inhibit the transcriptional activity of the 3′-UTR region of BMPR1B. In contrast, we observed no significant changes when the miR-1306 mimics were co-transfected with the mutant-type vector (Figure 2C), thereby indicating that miR-1306 inhibits luciferase activity of Hu sheep GCs by binding directly to the 3'-UTR region of BMPR1B. Thus, our findings provide convincing evidence that the ovine BMPR1B gene is a direct target of miR-1306.
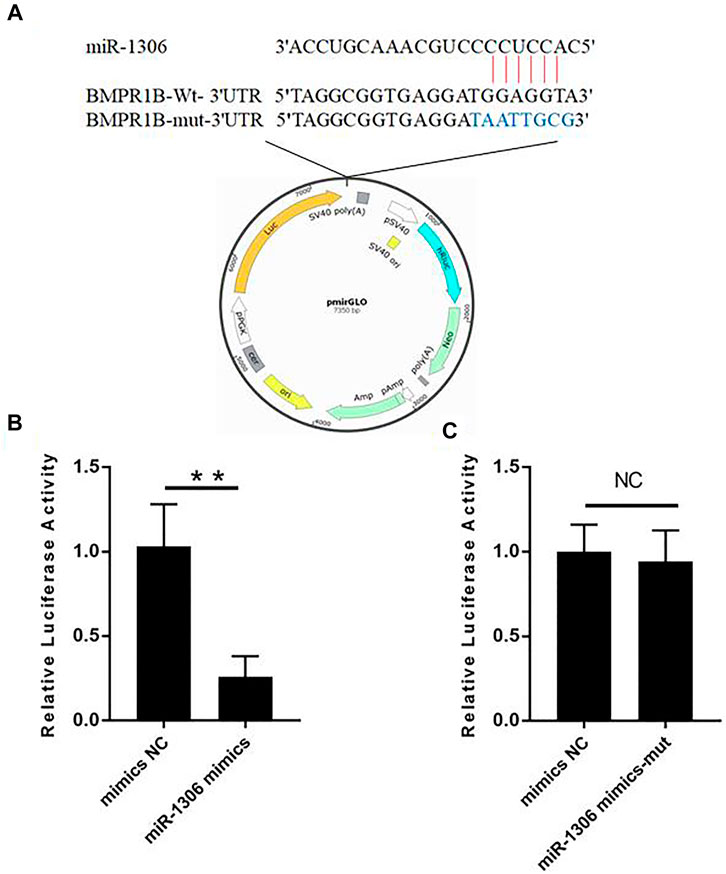
FIGURE 2. Hu sheep BMPR1B is a direct target of miR-1306. (A) BMPR1B 3'-UTR reporter vector. (B, C) Results of luciferase activity analyses. miR-1306 mimics and reporter vectors with wild-type (B) or mutant-type (C) miR-1306 binding sites were used to co-transfect HEK293T cells and determine luciferase activity. Bars represent the mean ± SEM of three repilicates.**p < 0.01. NS, no significant.
miR-1306 regulated endogenous BMPR1B expression in ovine granulosa cells
To further investigate the mechanisms whereby miR-1306 affects the expression levels of endogenous BMPR1B, ovine GCs were treated with miR-1306 mimics. As shown in Figure 3A, the overexpression of miR-1306 significantly down-regulated BMPR1B mRNA levels. Interestingly, in response to transfection with miR-1306 mimics, we observed that the protein levels of BMPR1B were markedly reduced in ovine GCs (Figure 3B), thereby indicating that miR-1306 can regulate BMPR1B expression in ovine GCs.
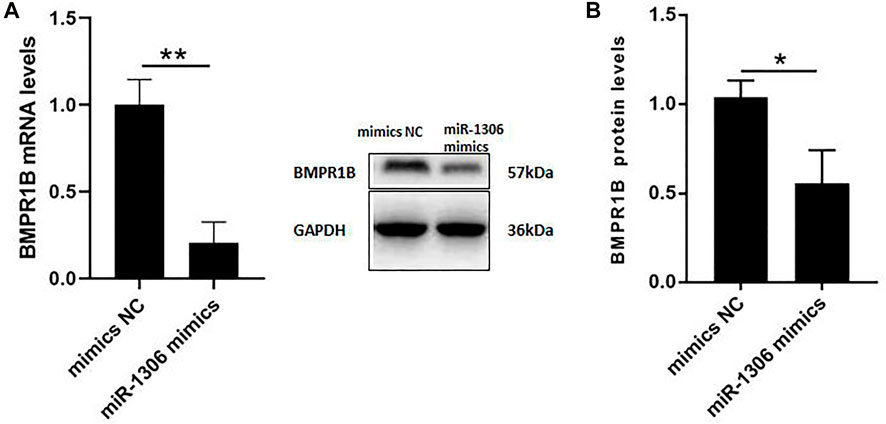
FIGURE 3. miR-1306 inhibits the expression of BMPR1B in sheep ovine Granulosa cells (A) BMPR1B mRNA levels. (B) BMPR1B protein levels. Bars represent the mean ± SEM of three repilicates. p < 0.05; **p < 0.01.
miR-1306 promotes apoptosis in ovine granulosa cells
To analyze the function of miR-1306 in ovine GCs, we treated these cells with miR-1306 mimics and mimics NC, respectively. FACS assays revealed that compared with the mimic NC group, overexpression of the miR-1306 mimic group markedly enhanced the apoptosis of ovine GCs (Figure 4), indicating that miR-1306 can function as an apoptotic factor in ovine GCs.
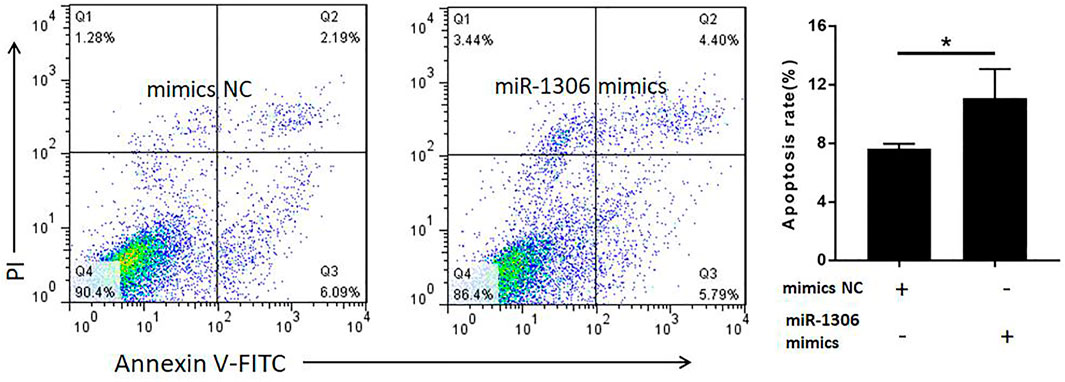
FIGURE 4. miR-1306 enhances apoptosis in ovine Granulosa cells miR-1306 promotes cell apoptosis. Bars represent the mean ± SEM of three repilicates. *p < 0.05.
miR-1306 promotes granulosa cells apoptosis by targeting BMPR1B
To determine whether miR-1306 regulates ovine GC apoptosis via its interaction with BMPR1B, we co-transfected ovine GCs with miR-1306 mimics and the BMPR1B overexpression vector pcDNA3.1-BMPR1B in vitro. As shown in Figure 5, overexpression of BMPR1B can rescue the cell apoptosis caused by miR-1306 in the ovine GCs.
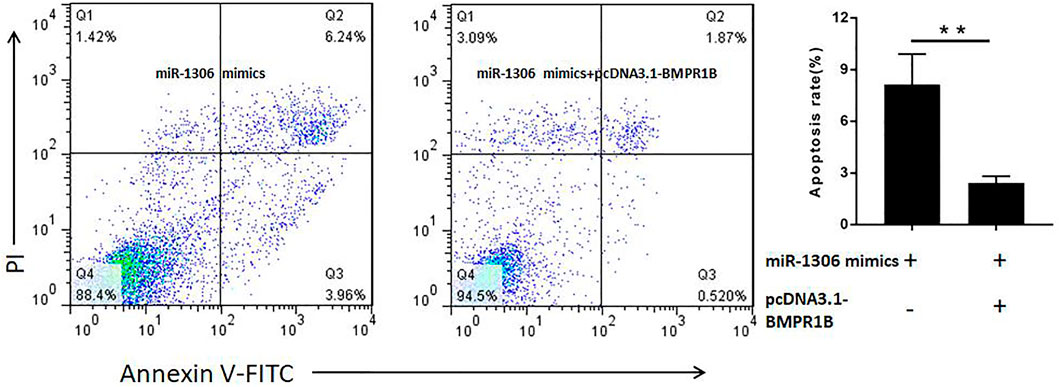
FIGURE 5. BMPR1B prevents the miR-1306-induced apoptosis of ovine Granulosa cells BMPR1B inhibits cell apoptosis caused by miR-1306. Bars represent the mean ± SEM of three repilicates.**p < 0.01.
Discussion
In the present study, we found that miR-1306 restrains the transcriptional activity of the BMPR1B by directly binding its 3′-UTR regions. Furthermore, the overexpression of miR-1306 significantly down-regulated BMPR1B mRNA and protein levels in ovine GCs. Finally, the apoptotic assay indicated that miR-1306 is an apoptotic factor in ovine granulosa cells, while the overexpression of BMPR1B prevents miR-1306-induced apoptosis.
3'-UTR is an important regulatory hub of mRNAs that can participate in the regulation of mRNAs level, translation, localization, stability, and polyadenylation status (Christine, 2018; Li F. et al., 2020). miRNA is a short non-coding RNA, that is, approximately 22 nt long and targets specific mRNAs 3'-untranslated regions (UTRs) and regulates gene expression (Bin Goh et al., 2012). miRNA can regulate various cellular processes, such as proliferation (Lozano-Velasco et al., 2015), differentiation (Wang et al., 2015), apoptosis (Cheng et al., 2005), and immune response (Okoye et al., 2014). Mechanistically, miRNAs mainly suppress the expression of target mRNAs via the reduction of translation or promotion of mRNA degradation (Du et al., 2020). In mammalian ovarian tissues, BMP/Smad is an important signaling pathway that affects reproductive performance and members of this pathway are direct targets for mRNAs. In ovine ovaries, BMPR2, SMAD1, SMAD4, and SMAD5 are potential target genes of miR-10b, miR-181a, miR-26a, miR-143, let-7a, miR-127, let-7f, and let-7c (Hu et al., 2016). The miR-17-92 cluster can regulate the cell proliferation and differentiation of bovine Granulosa cells by targeting BMPR2 (Andreas et al., 2016). miR-23a and miR-27a contribute to human Granulosa cell apoptosis by targeting SMAD5 (Nie et al., 2015). miR-130b targets SMAD5 to regulate the proliferation of Granulosa cells and cumulus cells, production of lactic acid in cumulus cells, biosynthesis of cholesterol, and ovulation in bovine ovaries (Sinha et al., 2017). SMAD4 is the most studied miRNA target gene among those in the ovarian BMP/Smad signaling pathway. miR-224 (Yao et al., 2010), miR-144 (Zhou et al., 2017), and miR-26b (Li et al., 2019) can regulate ovarian function by targeting SMAD4. In this study, we showed that miR-1306 direct targets BMPR1B gene in the ovine granulosa cells. However, few studies have investigated the regulation of BMPR1B by miRNAs. Currently, only studies that research how miR-125b targeting BMPR1B regulates apoptosis have been conducted in Granulosa cells (Yao et al., 2018; Yao et al., 2019). Together, our findings provide important insights into the epigenetic mechanisms underlying the regulation of BMPR1B expression in granulosa cells. miR-1306 is involved in various biological processes, for example, apoptosis, differentiation, proliferation, and pathological changes (Augustin et al., 2012; Du et al., 2018). The miR-1306 is differentially expressed in humans among those with Alzheimer’s disease, mild cognitive impairment, and vascular dementia, which suggests that miR-1306 plays an important role in the occurrence of these diseases (Li F. et al., 2020). Down-regulation of miR-1306 could aggravate the injury of OGD/rsh-sy5y in vitro; thus, miR-1306 plays a key role in cerebral I/R injury. Recently, a study found that Up-regulation of miR-1306 to lower cerebral ischemia/reperfusion injury in vitro by targeting BIK (Chen et al., 2019). Additionally, the expression of miR-1306 and congenital heart defects are closely related, which is significantly upregulated in twins (Pulakat et al., 2019). Chicken miR-1306, which generally targets Toll-interacting protein, has a key role in the host response against Salmonella enterica infection (Sun et al., 2019). Importantly, we predicted the binding site of miR-1306 on the 3'UTR of the BMPR1B gene. Further research found that the dual luciferase activity was significantly reduced after the binding site mutation of miR-1306 compared with the control group. These results suggest that miR-1306 directly binds to the 3'UTR of BMPR1B gene and reduces transcriptional activity. We observed that the mRNA and protein expression level of BMPR1B gene was strongly decreased after miR-1306 mimics was transfected into sheep ovarian granulosa cells. These data further demonstrated that miR-1306 binds to the 3'UTR of BMPR1B gene to regulate its expression.
Functionally, we showed that miR-1306 could promote granulosa cell apoptosis via directly binding to 3′UTR of BMPR1B gene. As is well known, the members of the BMP/Smad signaling pathway are associated with fecundity in domestic animals and are essential for steroidogenesis, follicular development, and ovulation and apoptosis (Abdoli et al., 2016; Liu et al., 2017; Li X. J. et al., 2020). Studies show that BMP2 (Liu et al., 2013), BMP4, BMP7 (Shimizu et al., 2012), and BMP15 (Shimizu et al., 2012) regulate granulosa cell apoptosis in mouse, cow, and human ovaries, respectively. BMPR2 and BMPR1B are two important receptors in the BMP/Smad signaling pathway. Previous studies have found that these two receptors have been found to regulate granulosa cell apoptosis in human (Cui et al., 2017) and sheep (Yao et al., 2019) ovaries, respectively.
In conclusion, our study provided evidence that miR-1306 regulates granulosa cell apoptosis through direct binding to the 3'UTR of the BMPR1B gene, and provides evidence of BMPR1B in regulating granulosa cell apoptosis in sheep.
Conclusion
In summary, we showed that miR-1306 regulates the expression of BMPR1B in the ovine granulosa cells by directly binding to the 3'-UTR region. Moreover, we found that the BMPR1B gene is a functional target of miR-1306 and that miR-1306 induces ovine granulosa cell apoptosis. Our findings provide important insights into the molecular mechanisms underlying the regulation of BMPR1B expression and will provide a theoretical basis for improving the reproductive performance of sheep.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by the Animal Protection and Utilization Committee of Nanjing Agricultural University and were carried out in strict accordance with the policies of the National Laboratory Animal Administration (Order No. 2 of the China Science and Technology Commission, 14 November 1988, institution certification number: SYXK 2017-0027).
Author contributions
Conceptualization; Data curation, AA; Methodology, AA, FZ, WA, and RO; Project administration, AS; Supervision, AS; Writing—original draft, AA; Writing—review and editing, AS and JA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdoli, R., Zamani, P., Mirhoseini, S. Z., Hossein-Zadeh, N. G., and Nadri, S. (2016). A review on prolificacy genes in sheep. Reprod. Domest. Anim. 51, 631–637. doi:10.1111/rda.12733
Andreas, E., Hoelker, M., Neuhoff, C., Tholen, E., Schellander, K., Tesfaye, D., et al. (2016). MicroRNA 17-92 cluster regulates proliferation and differentiation of bovine granulosa cells by targeting PTEN and BMPR2 genes. Cell Tissue Res. 366, 219–230. doi:10.1007/s00441-016-2425-7
Augustin, R., Endres, K., Reinhardt, S., Kuhn, P.-H., Lichtenthaler, S. F., Hansen, J., et al. (2012). Computational identification and experimental validation of microRNAs binding to the Alzheimer-related gene ADAM10. BMC Med. Genet. 13, 35. doi:10.1186/1471-2350-13-35
Bahire, S. V., Rajput, P. K., Kumar, V., Kumar, D., Kataria, M., and Kumar, S. (2019). Quantitative expression of mRNA encoding BMP/SMAD signalling genes in the ovaries of Booroola carrier and non-carrier GMM sheep. Reproduction Domest. Animals 54, 1375–1383. doi:10.1111/rda.13535
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233. doi:10.1016/j.cell.2009.01.002
Bin Goh, W. W., Oikawa, H., Sng, J. C. G., Sergot, M., and Wong, L. (2012). The role of miRNAs in complex formation and control. Bioinformatics 28, 453–456. doi:10.1093/bioinformatics/btr693
Chen, X., Li, C., Li, J., Sheng, L., and Liu, X. (2019). Upregulation of miR-1306-5p decreases cerebral ischemia/reperfusion injury in vitro by targeting BIK. Biosci. Biotechnol. Biochem. 83, 2230–2237. doi:10.1080/09168451.2019.1654846
Cheng, A. M., Byrom, M. W., Shelton, J., and Ford, L. P. (2005). Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33, 1290–1297. doi:10.1093/nar/gki200
Christine, M. (2018). What are 3′ UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728. doi:10.1101/cshperspect.a034728
Cui, X. R., Jing, X., Wu, X. Q., Bi, X. Y., Liu, J. F., Long, Z. J., et al. (2017). Abnormal expression levels of BMP15/Smad1 are associated with granulosa cell apoptosis in patients with polycystic ovary syndrome. Mol. Med. Rep. 16, 8231–8236. doi:10.3892/mmr.2017.7658
Djuranovic, S., Nahvi, A., and Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240. doi:10.1126/science.1215691
Du, X., Liu, L., Li, Q., Zhang, L., Pan, Z., and Li, Q. (2020). NORFA, long intergenic noncoding RNA, maintains sow fertility by inhibiting granulosa cell death. Commun. Biol. 3, 131. doi:10.1038/s42003-020-0864-x
Du, X., Pan, Z., Li, Q., Liu, H., and Li, Q. (2018). SMAD4 feedback regulates the canonical TGF-beta signaling pathway to control granulosa cell apoptosis. Cell Death Dis. 9, 151. doi:10.1038/s41419-017-0205-2
Foroughinia, G., Fazileh, A., and Eghbalsaied, S. (2017). Expression of genes involved in BMP and estrogen signaling and AMPK production can be important factors affecting total number of antral follicles in ewes. Theriogenology 91, 36–43. doi:10.1016/j.theriogenology.2016.12.023
Gootwine, E. (2011). Mini review: Breeding awassi and Assaf sheep for diverse management conditions. Trop. Anim. Health Prod. 43, 1289–1296. doi:10.1007/s11250-011-9852-y
Hoffmann, K., Berger, H., Kulbe, H., Thillainadarasan, S., Mollenkopf, H.-J., Zemojtel, T., et al. (2020). Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. Embo J. 39, e104013. doi:10.15252/embj.2019104013
Hu, X., Pokharel, K., Peippo, J., Ghanem, N., Zhaboyev, I., Kantanen, J., et al. (2016). Identification and characterization of miRNAs in the ovaries of a highly prolific sheep breed. Anim. Genet. 47, 234–239. doi:10.1111/age.12385
Kumar, S., Kolte, A. P., and Singh, V. K. (2006). Twinning in Marwari and Bharat merino ewes and its relationship with booroola FecB mutation. Indian J. Biotechnol. 5, 482–485.
Li, F., Xie, X.-Y., Sui, X.-F., Wang, P., Chen, Z., and Zhang, J.-B. (2020a). Profile of pathogenic proteins and MicroRNAs in plasma-derived extracellular vesicles in Alzheimer’s disease: A pilot study. Neuroscience 432, 240–246. doi:10.1016/j.neuroscience.2020.02.044
Li, W. Q., Li, W. N., Zou, L. H., Ji, S. M., Li, C. Y., Liu, K. H., et al. (2017). Membrane targeting of inhibitory Smads through palmitoylation controls TGF-β/BMP signaling. Proc. Natl. Acad. Sci. U. S. A. 114, 13206–13211. doi:10.1073/pnas.1710540114
Li, X., Du, X., Yao, W., Pan, Z., and Li, Q. (2019). TGF-beta/SMAD4 signaling pathway activates the HAS2-HA system to regulate granulosa cell state. J. Cell. Physiology 235 (59), 2260–2272. doi:10.1002/jcp.29134
Li, X. J., Ye, J. W., Han, X. L., Qiao, R. M., Li, X. L., Lv, G., et al. (2020b). Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 112, 199–206. doi:10.1016/j.ygeno.2019.01.014
Liu, J. Y., Tu, F., Yao, W., Li, X. Y., Xie, Z., Liu, H. L., et al. (2016). Conserved miR-26b enhances ovarian granulosa cell apoptosis through HAS2-HA-CD44-Caspase-3 pathway by targeting HAS2. Sci. Rep. 6, 21197. doi:10.1038/srep21197
Liu, Y., Du, S. Y., Ding, M., Dou, X., Zhang, F. F., Wu, Z. Y., et al. (2017). The BMP4-Smad signaling pathway regulates hyperandrogenism development in a female mouse model. J. Biol. Chem. 292, 11740–11750. doi:10.1074/jbc.M117.781369
Liu, Z. L., Castrillon, D. H., Zhou, W., and Richards, J. S. (2013). FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol. Endocrinol. 27, 238–252. doi:10.1210/me.2012-1296
Lozano-Velasco, E., Vallejo, D., Esteban, F. J., Doherty, C., Hernandez-Torres, F., Franco, D., et al. (2015). A pitx2-MicroRNA pathway modulates cell proliferation in myoblasts and skeletal-muscle satellite cells and promotes their commitment to a myogenic cell fate. Mol. Cell. Biol. 35, 2892–2909. doi:10.1128/MCB.00536-15
Montgomery, G. W., Galloway, S. M., Davis, G. H., and McNatty, K. P. (2001). Genes controlling ovulation rate in sheep. Reproduction 121, 843–852. doi:10.1530/rep.0.1210843
Mulsant, P., Lecerf, F., Fabre, S., Schibler, L., Monget, P., Lanneluc, I., et al. (2001). Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. U. S. A. 98, 5104–5109. doi:10.1073/pnas.091577598
Nie, M., Yu, S., Peng, S., Fang, Y., Wang, H., and Yang, X. (2015). miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol. Reprod. 93, 98. doi:10.1095/biolreprod.115.130690
Okoye, I. S., Coomes, S. M., Pelly, V. S., Czieso, S., Papayannopoulos, V., Tolmachova, T., et al. (2014). MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41, 503. doi:10.1016/j.immuni.2014.08.008
Otsuka, F. (2018). Modulation of bone morphogenetic protein activity by melatonin in ovarian steroidogenesis. Reprod. Med. Biol. 17, 228–233. doi:10.1002/rmb2.12089
Pulakat, L., Abu-Halima, M., Weidinger, J., Poryo, M., Henn, D., Keller, A., et al. (2019). Micro-RNA signatures in monozygotic twins discordant for congenital heart defects. Plos One 14, e0226164. doi:10.1371/journal.pone.0226164
Regan, S. L. P., McFarlane, J. R., O'Shea, T., Andronicos, N., Arfuso, F., Dharmarajan, A., et al. (2015). Flow cytometric analysis of FSHR, BMRR1B, LHR and apoptosis in granulosa cells and ovulation rate in merino sheep. Reproduction 150, 151–163. doi:10.1530/REP-14-0581
Schneider, H., Sedaghati, B., Naumann, A., Hacker, M. C., and Schulz-Siegmund, M. (2014). Gene silencing of chordin improves BMP-2 effects on osteogenic differentiation of human adipose tissue-derived stromal cells. Tissue Eng. Part A 20, 335–345. doi:10.1089/ten.TEA.2012.0563
Shimizu, T., Kayamori, T., Murayama, C., and Miyamoto, A. (2012). Bone morphogenetic protein (BMP)-4 and BMP-7 suppress granulosa cell apoptosis via different pathways: BMP-4 via PI3K/PDK-1/Akt and BMP-7 via PI3K/PDK-1/PKC. Biochem. Biophys. Res. Commun. 417, 869–873. doi:10.1016/j.bbrc.2011.12.064
Sinha, P. B., Tesfaye, D., Rings, F., Hossien, M., Hoelker, M., Held, E., et al. (2017). MicroRNA-130b is involved in bovine granulosa and cumulus cells function, oocyte maturation and blastocyst formation. J. Ovarian Res. 10, 37. doi:10.1186/s13048-017-0336-1
Sun, W., Liu, R., Li, P., Li, Q., Cui, H., Zheng, M., et al. (2019). Chicken gga-miR-1306-5p targets Tollip and plays an important role in host response against Salmonella enteritidis infection. J. Anim. Sci. Biotechnol. 10, 59. doi:10.1186/s40104-019-0365-2
Tang, J., Hu, W., Di, R., Liu, Q., Wang, X., Zhang, X., et al. (2018). Expression analysis of the prolific candidate genes, BMPR1B, BMP15, and GDF9 in Small Tail han ewes with three fecundity (FecB gene) genotype. Anim. (Basel) 8, E166. doi:10.3390/ani8100166
Wang, L., Zhang, X., Guo, Y., Chen, X., Li, R., Liu, L., et al. (2010). Involvement of BMPs/smad signaling pathway in mechanical response in osteoblasts. Cell. Physiol. biochem. 26, 1093–1102. doi:10.1159/000323987
Wang, Y. M., Ding, X. B., Dai, Y., Liu, X. F., Guo, H., and Zhang, Y. (2015). Identification and bioinformatics analysis of miRNAs involved in bovine skeletal muscle satellite cell myogenic differentiation. Mol. Cell. Biochem. 404, 113–122. doi:10.1007/s11010-015-2371-9
Wu, F.-J., Wang, Y.-W., and Luo, C.-W. (2020). Human BMP8A suppresses luteinization of rat granulosa cells via the SMAD1/5/8 pathway. Reproduction 159, 315–324. doi:10.1530/REP-19-0305
Xu, Y., Li, E., Han, Y., Chen, L., and Xie, Z. (2010). Differential expression of mRNAs encoding BMP/Smad pathway molecules in antral follicles of high- and low-fecundity Hu sheep. Anim. Reprod. Sci. 120, 47–55. doi:10.1016/j.anireprosci.2010.02.009
Yang, L., Du, X., Liu, L., Cao, Q., Pan, Z., and Li, Q. (2019). miR-1306 mediates the feedback regulation of the TGF-/SMAD signaling pathway in granulosa cells. Cells 8 (4), 298. doi:10.3390/cells8040298
Yao, G., Yin, M., Lian, J., Tian, H., Liu, L., Li, X., et al. (2010). MicroRNA-224 is involved in transforming growth factor-beta-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol. Endocrinol. 24, 540–551. doi:10.1210/me.2009-0432
Yao, Y., Niu, J., Sizhu, S., Li, B., Chen, Y., Li, R., et al. (2018). microRNA-125b regulates apoptosis by targeting bone morphogenetic protein receptor 1B in yak granulosa cells. DNA Cell Biol. 37, 878–887. doi:10.1089/dna.2018.4354
Yao, Y., Reheman, A., Xu, Y., and Li, Q. (2019). miR-125b contributes to ovarian granulosa cell apoptosis through targeting BMPR1B, a major gene for sheep prolificacy. Reprod. Sci. 26, 295–305. doi:10.1177/1933719118770544
Keywords: BMPR1B, granulosa cell, miR-1306, apoptosis, sheep
Citation: Abdurahman A, Aierken W, Zhang F, Obulkasim R, Aniwashi J and Sulayman A (2022) miR-1306 induces cell apoptosis by targeting BMPR1B gene in the ovine granulosa cells. Front. Genet. 13:989912. doi: 10.3389/fgene.2022.989912
Received: 09 July 2022; Accepted: 22 August 2022;
Published: 23 September 2022.
Edited by:
Kai Jiao, Augusta University, United StatesReviewed by:
Shun Yan, Augusta University, United StatesRaziye Işıkk, Sao Paulo State University, Brazil
Qian Wang, Yellow Sea Fisheries Research Institute (CAFS), China
Copyright © 2022 Abdurahman, Aierken, Zhang, Obulkasim, Aniwashi and Sulayman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ablat Sulayman, aWJsYXQyMDA5QHNpbmEuY24=
 Anwar Abdurahman
Anwar Abdurahman Wusimanjiang Aierken4
Wusimanjiang Aierken4