- 1Henan Provincial People’s Hospital, Henan Eye Hospital, Henan Eye Institute, People’s Hospital of Zhengzhou University, Henan University People’s Hospital, Zhengzhou, China
- 2Zhengzhou University People’s Hospital, Henan Provincial People’s Hospital, Henan Eye Hospital, Henan Eye Institution, Zhengzhou, China
- 3Henan University People’s Hospital, Henan Provincial People’s Hospital, Henan Eye Hospital, Henan Eye Institute, Zhengzhou, China
Background: Keratoconus (KC) is a complex corneal disorder with a strong genetic component. The present study aimed to identify candidate genes related to KC in Chinese families.
Methods: Family-based exome sequencing was performed in ten patients suffering from KC who belong to five families with two affected members in each. The candidate rare variants were identified with multi-step bioinformatics analysis. The STRING website was used to perform the protein interaction of the identified genes.
Results: Our analyses identified 32 candidate rare variants in 13 genes by family-based exome sequencing. The molecular analyses of identified genes showed that EPCAM directly interacted with CTNNB1 of the Hippo signaling pathway and focal adhesion pathway, and directly interacted with CTNNB1, CDH1 of the WNT signaling pathway. SHROOM3 directly interacted with ROCK2, ROCK1 of the focal adhesion pathway. SYNE1 directly interacted with MUSK of the extracellular matrix organization pathway. TEK directly interacted with VEGFA, SHC1, PIK3R1, GRB2 of the focal adhesion pathway. TTN directly interacted with CAPN3 of the extracellular matrix organization pathway.
Conclusion: The EPCAM, SHROOM3, SYNE1, TEK, and TTN genes were potential high-risk candidate pathogenic genes of familial KC. The findings might significantly improve our understanding of the genetic etiology of the disease, providing novel insights on KC pathogenesis.
Introduction
Keratoconus (KC) is characterized by progressive corneal protrusion and thinning, leading to irregular astigmatism and impairment of visual function (Rabinowitz, 1998). The estimated prevalence of KC in the whole population is 1.38 per 1,000 people (Hashemi et al., 2020). The disease usually begins at puberty and progresses into the third or fourth decades (Santodomingo-Rubido et al., 2022). Currently, no curative treatments are available for KC (Mohammadpour et al., 2018). The progressive corneal thinning can be stabilized with corneal cross-linking when it is recognized at an early stage (Gomes et al., 2015). However, corneal transplantation is necessary for advanced cases (Sarezky et al., 2017). Thus, an early diagnosis of KC is crucial for improving its prognosis. Notably, understanding the pathogenesis of KC could help in the achievement of an early diagnosis and timely treatment of the disease.
KC is considered as a complex corneal disorder determined by a combination of environmental and genetic factors (Lucas and Burdon, 2020). Environmental factors included eye rubbing, allergies, diabetes, and sleeping position, as highlighted in previous studies (Ahuja et al., 2020; Mazharian et al., 2020; Ates et al., 2021). The higher concordance rate in monozygotic twins (Tuft et al., 2012), and a positive family history of 5%–23% in KC cases (Rabinowitz et al., 2021) suggested a strong genetic component in the development of KC. Indeed, many researchers have identified KC susceptibility genes by genome-wide association studies (Hosoda et al., 2020; Hardcastle et al., 2021), linkage studies (Hughes et al., 2011; Karolak et al., 2015) and candidate gene sequencing analyses (Abdelghany et al., 2021; Lopes et al., 2021). The genetic studies on KC significantly contributed to the biological basis of its pathogenesis. However, the genetic basis of KC susceptibility has not been fully understood due to the genetic heterogeneity and population differences, and the pathogenesis underlying the genetic variants remains unclear.
Currently, several genetic studies have been performed on KC in Chinese populations, and identified some genetic variants accounted for the disease (Hao et al., 2017; Xu et al., 2020; Zhang et al., 2020; Lin et al., 2022; Yuan et al., 2022). Nevertheless, the majority of the studies were performed in sporadic cases or one pedigree. The complex etiology of KC with a strong genetic heterogeneity still needs to be fully elucidated. Thus, the aim of this study was to identify candidate genes potentially related to KC predisposition in families with KC. Consequently, family-based exome sequencing of ten patients with KC from five Chinese families were performed in the present study, and bioinformatics approaches were used to determine the genetic factors contributing to the onset of the disease.
Materials and methods
Family recruitment
A total of ten patients with KC from five families with two affected members in each were selected for the current study. The diagnosis of KC was based on clinical manifestations such as localized stromal thinning, conical protrusion, Vogt’s striae, Fleischer’s ring, or anterior stromal scar, as well as signs of corneal topography revealing an asymmetric bowtie pattern with or without skewed axes (Mas Tur et al., 2017). Patients whose KC was caused by trauma, other disease, or surgery were excluded from the study. The study was approved by the Institutional Review Board of Henan Eye Hospital [ethical approval number: HNEECKY-2019(5) and performed in accordance with the guidelines of the Declaration of Helsinki. Written informed consent was obtained from each subject.
Exome sequencing
Total genomic DNA was extracted from peripheral blood samples according to the manufacturer’s recommendations. DNA quality was examined by Qubit 3.0 and confirmed by electrophoresis prior to library construction. The DNA was fragmented to an average size of 180–280 bp and subjected to DNA library creation with established Illumina paired-end protocols. The library was then subjected to exome sequence capture by Agilent SureSelect Human All ExonV6 Kit (Agilent Technologies, Santa Clara, CA, United States) according to the manufacturer’s instructions. The Illumina Novaseq 6000 platform (Illumina Inc., San Diego, CA, United States) was used for genomic DNA sequencing in Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
Bioinformatics analysis
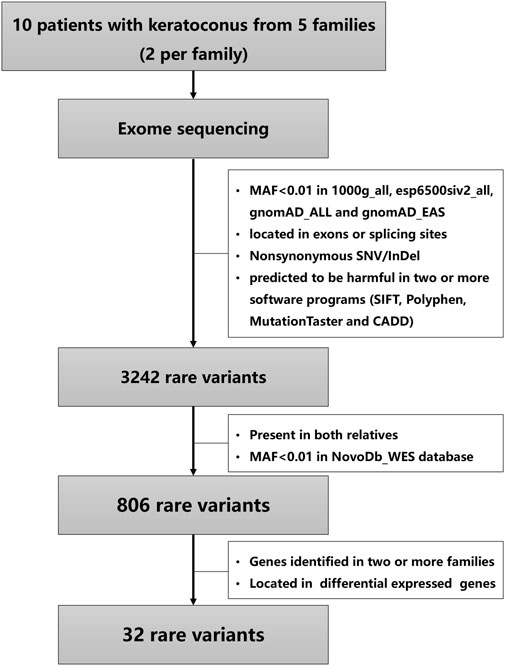
After quality control, the sequencing reads were mapped to hg19 (GRCh37) using Burrows-Wheeler Aligner (Li and Durbin, 2009), and duplicate reads were marked using Sambamba tools (Tarasov et al., 2015). SAMtools (Li et al., 2009) were used to perform variant calling to identify single nucleotide variants (SNVs) and small insertions or deletions (InDels). The raw calls of SNVs and InDels were further filtered with the following inclusion thresholds: (1) read depth >4; (2) Root-Mean-Square mapping quality of covering reads >30; (3) variant quality score >20. Then annotation of the variants was performed using ANNOVAR (Wang et al., 2010). As is shown in Figure 1, the variants were firstly filtered using the following criteria: (1) variants with a minor allele frequency less than 0.01 in 1,000 genomic data (1000 g_all), esp6500siv2_all, and gnomAD data (gnomAD_ALL and gnomAD_EAS); (2) variants located in exons or splicing sites; (3) variants predicted to influence splicing or amino acid alternation; (4) variants predicted to be harmful in more than half of the software programs (SIFT, Polyphen, MutationTaster and CADD) according to the scores. Secondly, the variants presented in both relatives and variants with a minor allele frequency less than 0.01 in NovoDb_WES database (2,573 healthy Chinese control individuals) were selected. Finally, the variants located in genes presented in two or more families and differentially expressed in KC were considered as candidate rare variants (Lee et al., 2009; Mace et al., 2011; Bykhovskaya et al., 2016; Kabza et al., 2017; Khaled et al., 2018; You et al., 2018; Sharif et al., 2019; Shinde et al., 2020; Sun et al., 2022). The STRING website was used to predict the relationships of proteins with previously reported genes in KC and known KC-associated pathways, including extracellular matrix organization, WNT signaling, Hippo signaling, focal adhesion and TGF-β pathways (Cai et al., 2020; Hao et al., 2021).
Results
Clinical characteristics of patients with Keratoconus
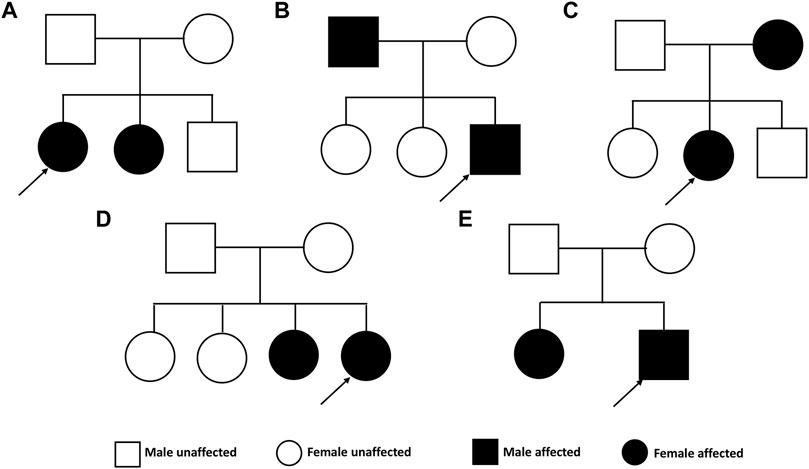
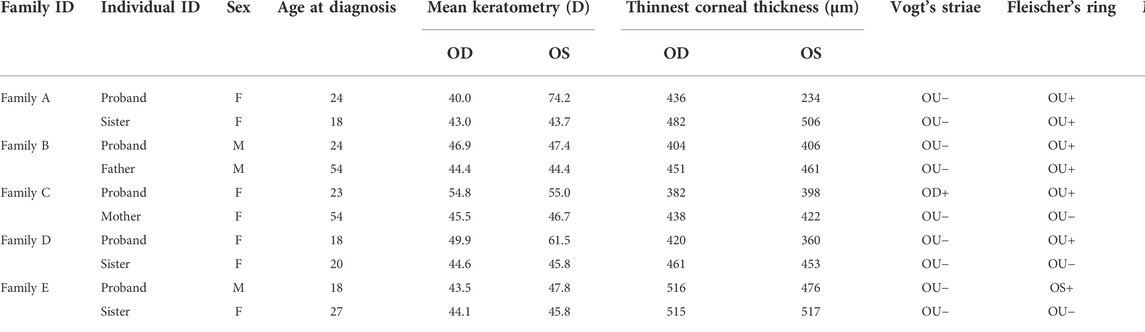
The pedigrees of the five investigated families are presented in Figure 2. The clinical characteristics of the ten patients with KC are listed in Table 1. The mean age at diagnosis was 28 years (range from 18 to 54). In this study, three patients were male and seven patients were female. The clinical investigation revealed the presence of Vogt’s striae in one patient, Munson’s sign in four patients, and Fleischer’s ring in seven patients.
Candidate rare variants identified in Keratoconus families
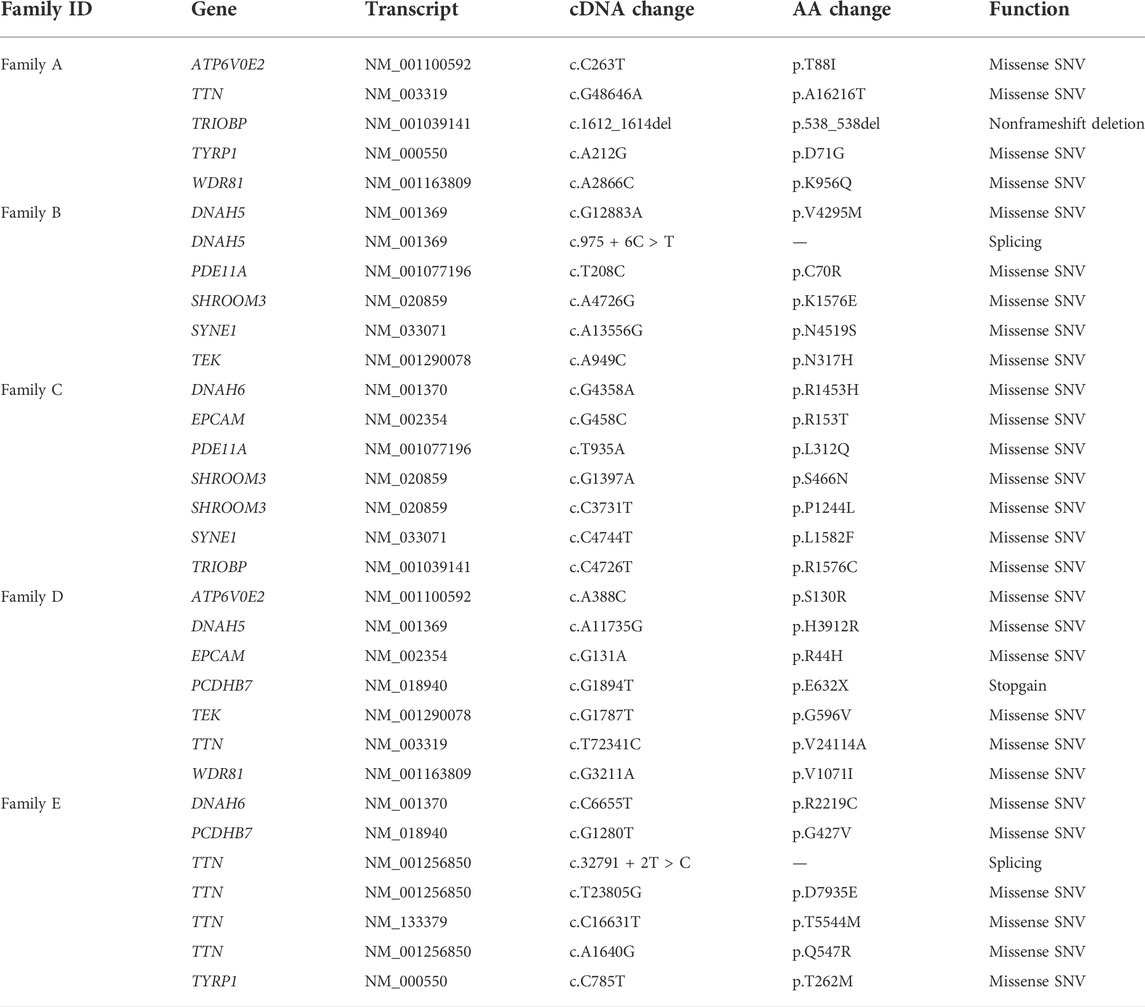
The variants were filtered to screen the candidate rare variants in KC families according to the analysis workflow in Figure 1. Firstly, the variants focusing on frequency, location, and effects of the variants were filtered. Consequently, a total of 3,242 rare variants were identified. The screening of the variants that occurred in both the affected relatives with a minor allele frequency less than 0.01 in NovoDb_WES database identified 806 rare variants. Then the variants located in genes presented in two or more families and differentially expressed in KC were selected as candidate rare variants. Finally, 32 variants in 13 genes were identified, including 28 missense variants, one nonframeshift deletion variants, one stop gained variant, and two splicing variants (Table 2). Among the identified genes, the dynein axonemal heavy chain 6 (DNAH6), epithelial cell adhesion molecule (EPCAM), and titin (TTN) were reported to be upregulated in KC (Kabza et al., 2017). The ATPase H+ transporting V0 subunit e2 (ATP6V0E2), dynein axonemal heavy chain 5 (DNAH5), phosphodiesterase 11A (PDE11A), spectrin repeat containing nuclear envelope protein 1 (SYNE1), TEK receptor tyrosine kinase (TEK), TRIO and F-actin binding protein (TRIOP), and tyrosinase related protein 1 (TYRP1) genes were reported to be downregulated (Kabza et al., 2017; You et al., 2018; Shinde et al., 2020). However, the expressions of protocadherin beta 7 (PCDHB7), shroom family member 3 (SHROOM3), and WD Repeat Domain 81 (WDR81) genes exhibited inconsistent results in different studies (Kabza et al., 2017; You et al., 2018; Sharif et al., 2019; Shinde et al., 2020; Sun et al., 2022). In addition, the candidate variants in TTN were identified in three families (60%), while variants in other genes were identified in two families (40%).
Molecular analysis of the identified genes
Prediction of protein-protein interactions of the thirteen genes (ATP6V0E2, DNAH5, DNAH6, EPCAM, PCDHB7, PDE11A, SHROOM3, SYNE1, TEK, TRIOBP, TTN, TYRP1, and WDR81) was conducted using the online STRING software. The protein-protein interactions of the thirteen genes with previously reported 88 genes in KC were listed in Figure 3. The results showed that the genes directly interact with previously reported genes such as SOD1, CAST. Besides, the protein-protein interactions of identified genes with five KC-associated pathways (extracellular matrix organization, WNT signaling, Hippo signaling, focal adhesion and TGF-β pathways) were also analyzed. According to the interaction network shown in Figure 4, EPCAM directly interacted with CTNNB1 of the Hippo signaling and focal adhesion pathways, and directly interacted with CTNNB1, CDH1 of the WNT signaling pathway. SHROOM3 directly interacted with ROCK2, ROCK1 of the focal adhesion pathway. SYNE1 directly interacted with MUSK of the extracellular matrix organization pathway. TEK directly interacted with VEGFA, SHC1, PIK3R1, GRB2 of the focal adhesion pathway. TTN directly interacted with CAPN3 of the extracellular matrix organization pathway. However, other eight genes did not show any interaction with the investigated pathways (Figure 5). Our results highlighted that EPCAM, SHROOM3, SYNE1, TEK, and TTN were potential high-risk candidate pathogenic genes of KC that exert their effects by the disruption of the extracellular matrix organization, WNT signaling, Hippo signaling, focal adhesion and TGF-β pathways.
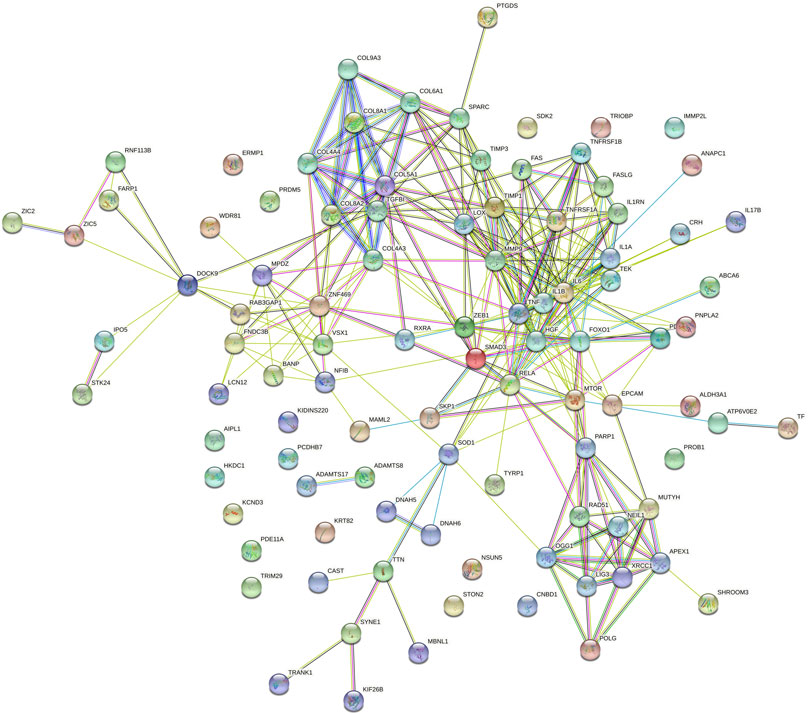
FIGURE 3. Protein-to-protein interactions of identified thirteen candidate genes with previously reported genes in keratoconus.
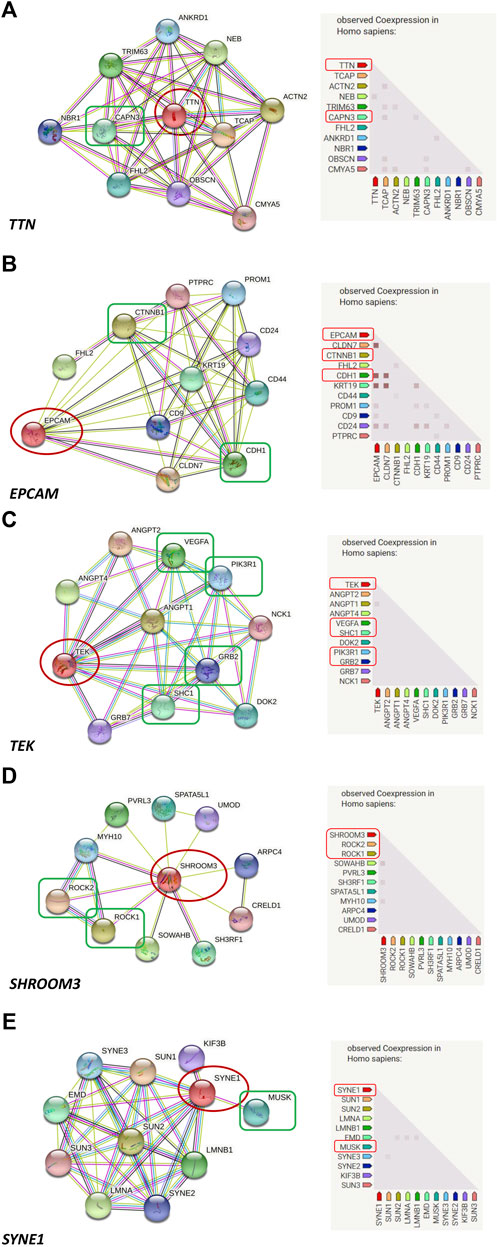
FIGURE 4. Protein-to-protein interactions of candidate genes with known keratoconus associated pathways. ((A): EPCAM directly interacts with CTNNB1 of the Hippo signaling and Focal adhesion pathways, and directly interacts with CTNNB1, CDH1 of the WNT signaling pathway; (B): SHROOM3 directly interacts with ROCK2, ROCK1 of the Focal adhesion pathway; (C): SYNE1 directly interacts with MUSK of the extracellular matrix organization pathway; (D): TEK directly interacts with VEGFA, SHC1, PIK3R1, GRB2 of the Focal adhesion pathway; (E): TTN directly interacts with CAPN3 of the extracellular matrix organization pathway).
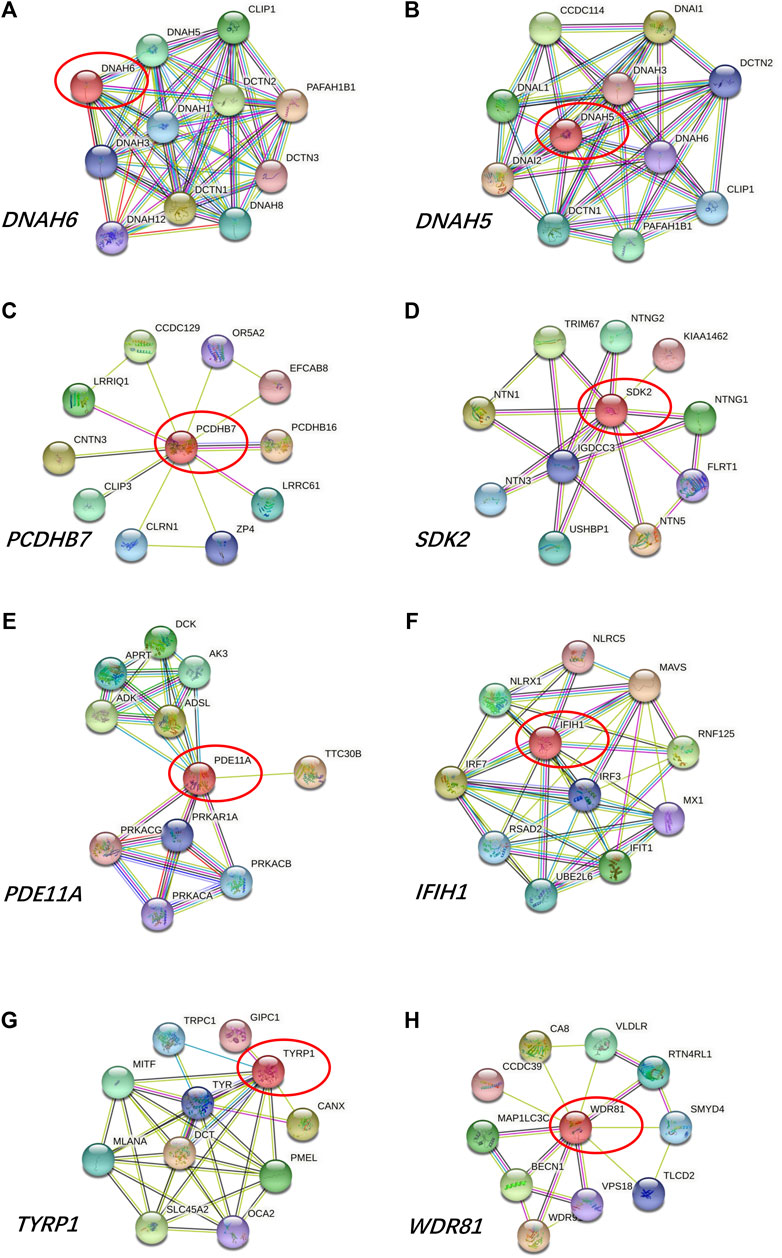
FIGURE 5. Protein-to-protein interactions of (A) ATP6V0E2, (B) DNAH5, (C) DNAH6, (D) PCDHB7, (E) PDE11A, (F) TRIOBP, (G) TYRP1, and (H) WDR81 genes.
Discussion
KC is a complex disease, with numerous genetic and environmental factors potentially involved in its pathogenesis (Lucas and Burdon, 2020). Although multiple studies on the etiology of KC have been performed, no consensus has been reached to date. In the present study, a family-based exome sequencing in five Chinese KC families was performed, with the aim to identify potential candidate genes contributing to KC susceptibility. By applying several filtering strategies, we identified 32 variants located in ATP6V0E2, DNAH5, DNAH6, EPCAM, PCDHB7, PDE11A, SHROOM3, SYNE1, TEK, TRIOBP, TTN, TYRP1, and WDR81 genes as candidate rare variants. Bioinformatics analysis revealed that the EPCAM, SHROOM3, SYNE1, TEK, and TTN genes were potential high-risk candidate pathogenic genes of KC because of their relationships with known KC-associated pathways.
Genetic factors are implicated in the pathogenesis of KC, and multiple studies have identified numerous loci that might contribute to KC by high-throughput sequencing (Hosoda et al., 2020; Xu et al., 2020; Hardcastle et al., 2021). Although some genetic studies on KC families have been performed, and identified several candidate genes for KC (Froukh et al., 2020; Shinde et al., 2021), the family-based exome sequencing in Chinese KC families is limited. Our present study performed a family-based exome sequencing in five Chinese KC families. A total of 32 candidate rare variants located in 13 genes were finally identified by a series of filtering steps. Protein-protein interactions of the thirteen genes with previously reported genes in KC showed direct or indirect interaction with previously reported genes, indicating potential associations with KC. In addition, the EPCAM, SHROOM3, SYNE1, TEK, and TTN genes were considered as potential high-risk candidate pathogenic genes in KC after analyzing the protein-protein interactions of the thirteen genes with known KC-associated pathways.
EPCAM is a cell surface molecule involved in cell-to-cell adhesion, and plays significant roles in the modulation of proliferation, differentiation, and migration of epithelial cells (Huang et al., 2018). The molecular analysis of EPCAM indicated its interactions with CTNNB1 and CDH1 which are involved in WNT signaling, Hippo signaling, and focal adhesion pathways, suggesting a potential role of EPCAM in the pathogenesis of KC. SHROOM3 directly interacted with proteins involved in the focal adhesion pathway, and is a central regulator of morphogenetic cell shape changes in certain tissues (Tariq et al., 2011). As far as we know, KC is a bilateral and usually asymmetrical disease in which the ectatic cornea becomes conical in shape. Therefore, we speculated that the SHROOM3 gene might play roles in the pathogenesis of KC because of its interaction with proteins of the focal adhesion pathway and its potential functions in regulating cell shape. The SYNE1 gene encodes nesprin-1, a scaffold protein associated with anchoring the plasma membrane to the actin cytoskeleton and involved in the binding between the cytoskeleton, nuclear envelope and other subcellular compartments (Swan et al., 2018). Moreover, SYNE1 interacted with MUSK which is a gene involved in the extracellular matrix organization pathway, indicating a relationship between SYNE1 and KC. The TEK gene encodes a tyrosine kinase receptor and plays a central role in vascular stability (Gal et al., 2020). The gene directly interacted with VEGFA, SHC1, PIK3R1, GRB2 of the focal adhesion pathway, as revealed by the molecular analysis, which was associated with the pathogenesis of KC. Mutations in TEK might result in pathogenic effects by disrupting the focal adhesion pathway, leading to KC. Titin (TTN) is the largest protein in the human body, which is encoded by 364 exons of the TTN gene. It is reported that the TTN protein plays important roles in the regulation of the cytoskeleton organization in cardiomyocyte (Loescher et al., 2021). Among its interaction proteins, CAPN3 is involved in the extracellular matrix organization which is identified as a related pathway with KC. Thus, we speculated that TTN might be considered as a candidate gene for KC due to its indirect interaction with the extracellular matrix organization. In addition, TTN is considered as a major determinant of cardiomyocyte stiffness, and mutations in TTN might result in dilated cardiomyopathy in which myocardial stiffness has an important role in its pathogenesis (Begay et al., 2015). KC is a corneal disorder with its corneal stiffness changed. Mutations in TTN were both existed and predicted to be causative in dilated cardiomyopathy and KC, indicating that there might some similar molecular mechanism between them. Although no studies reported mutations in the identified six genes in KC, the genes might be involved in the pathogenesis of KC through their indirect interactions with the known KC-associated pathways.
Additionally, variants in ATP6V0E2, DNAH5, DNAH6, PCDHB7, PDE11A, TRIOBP, TYRP1, and WDR81 genes were detected in the present study. However, these eight genes showed no correlations with the investigated pathways related to KC in our study. And there were no studies reported their associations with KC.
Our study has several limitations that should be taken into consideration. Firstly, we only analyzed five KC families in the study due to the limited KC families recruited in our hospital. Secondly, the molecular mechanism of the candidate genes was not explored, and further studies should be performed to explore the mechanism of KC caused by those genes. Thirdly, the putative predisposition variants in noncoding or uncaptured regions of the genome (promoter or intronic variants) were not detectable by exome sequencing.
In conclusion, our family-based exome sequencing studies combined with bioinformatics analysis identified the EPCAM, SHROOM3, SYNE1, TEK, and TTN genes as potential high-risk candidate pathogenic genes of familial KC. The results obtained significantly improved our understanding of the genetic etiology of the disease, providing novel insights on KC pathogenesis.
Data availability statement
All relevant data were presented in the article. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Henan Eye Hospital (HNEECKY-2019(5)). Written informed consent has been obtained from the participants to publish this paper.
Author contributions
SR conceived and designed this experiment. LX, KY, and SY collected the DNA samples and analyzed the data. YG, QF, YW, and DZ helped collect the patients’ information and analyzed the data. LX wrote the manuscript. KY and SR revised the manuscript. All the authors read and approved the final manuscript.
Funding
This research was funded by Henan Natural Science Foundation (No. 222300420536), Special Program for Basic Research of Henan Eye Hospital (No. 20JCZD003), Henan Young Health Science and Technology Innovation Outstanding Program (No. YXKC2020023), Henan Provincial Building Key Program (No. SBGJ202002028, SBGJ202102051), Henan Provincial Medical Science and Technology Joint Program (No. LHGJ20200066, LHGJ20210080), Henan Provincial Scientific and Technological Research Project (No. 222102310307, 222102310599), Youth Special Program for Basic Research of Henan Eye Hospital (No. 21JCQN006, 21JCQN008).
Acknowledgments
We acknowledge all the subjects for providing blood samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelghany, A. A., Toraih, E. A., Abdelaziz, E. Z., El-Sherbeeny, N. A., and Fawzy, M. S. (2021). Association of collagen gene (COL4A3) rs55703767 variant with response to riboflavin/ultraviolet A-induced collagen cross-linking in female patients with keratoconus. Cornea 40 (1), 88–98. doi:10.1097/ICO.0000000000002489
Ahuja, P., Dadachanji, Z., Shetty, R., Nagarajan, S. A., Khamar, P., Sethu, S., et al. (2020). Relevance of IgE, allergy and eye rubbing in the pathogenesis and management of Keratoconus. Indian J. Ophthalmol. 68 (10), 2067–2074. doi:10.4103/ijo.IJO_1191_19
Ates, K. M., Estes, A. J., and Liu, Y. (2021). Potential underlying genetic associations between keratoconus and diabetes mellitus. Adv. Ophthalmol. Pract. Res. 1, 100005. doi:10.1016/j.aopr.2021.100005
Begay, R. L., Graw, S., Sinagra, G., Merlo, M., Slavov, D., Gowan, K., et al. (2015). Role of titin missense variants in dilated cardiomyopathy. J. Am. Heart Assoc. 4 (11), e002645. doi:10.1161/JAHA.115.002645
Bykhovskaya, Y., Gromova, A., Makarenkova, H. P., and Rabinowitz, Y. S. (2016). Abnormal regulation of extracellular matrix and adhesion molecules in corneas of patients with keratoconus. Int. J. Keratoconus Ectatic Corneal Dis. 5 (2), 63–70. doi:10.5005/jp-journals-10025-1123
Cai, J., Estes, A., and Liu, Y. (2020). Omics analyses in keratoconus: From transcriptomics to proteomics. Curr. Ophthalmol. Rep. 8 (4), 216–225. doi:10.1007/s40135-020-00253-x
Froukh, T., Hawwari, A., and Al Zubi, K. (2020). Whole exome sequencing highlights variants in association with Keratoconus in Jordanian families. BMC Med. Genet. 21 (1), 177. doi:10.1186/s12881-020-01112-z
Gal, Z., Gezsi, A., Molnar, V., Nagy, A., Kiss, A., Sultesz, M., et al. (2020). Corrigendum: Investigation of the possible role of Tie2 pathway and TEK gene in asthma and allergic conjunctivitis. Front. Genet. 11, 702. doi:10.3389/fgene.2020.00702
Gomes, J. A., Tan, D., Rapuano, C. J., Belin, M. W., Ambrosio, R., Guell, J. L., et al. (2015). Global consensus on keratoconus and ectatic diseases. Cornea 34 (4), 359–369. doi:10.1097/ICO.0000000000000408
Hao, X. D., Chen, P., Zhang, Y. Y., Li, S. X., Shi, W. Y., and Gao, H. (2017). De novo mutations of TUBA3D are associated with keratoconus. Sci. Rep. 7 (1), 13570. doi:10.1038/s41598-017-13162-0
Hao, X. D., Gao, H., Xu, W. H., Shan, C., Liu, Y., Zhou, Z. X., et al. (2021). Systematically displaying the pathogenesis of keratoconus via multi-level related gene enrichment-based review. Front. Med. 8, 770138. doi:10.3389/fmed.2021.770138
Hardcastle, A. J., Liskova, P., Bykhovskaya, Y., McComish, B. J., Davidson, A. E., Inglehearn, C. F., et al. (2021). A multi-ethnic genome-wide association study implicates collagen matrix integrity and cell differentiation pathways in keratoconus. Commun. Biol. 4 (1), 266. doi:10.1038/s42003-021-01784-0
Hashemi, H., Heydarian, S., Hooshmand, E., Saatchi, M., Yekta, A., Aghamirsalim, M., et al. (2020). The prevalence and risk factors for keratoconus: A systematic review and meta-analysis. Cornea 39 (2), 263–270. doi:10.1097/ICO.0000000000002150
Hosoda, Y., Miyake, M., Meguro, A., Tabara, Y., Iwai, S., Ueda-Arakawa, N., et al. (2020). Keratoconus-susceptibility gene identification by corneal thickness genome-wide association study and artificial intelligence IBM Watson. Commun. Biol. 3 (1), 410. doi:10.1038/s42003-020-01137-3
Huang, L., Yang, Y., Yang, F., Liu, S., Zhu, Z., Lei, Z., et al. (2018). Functions of EpCAM in physiological processes and diseases (Review). Int. J. Mol. Med. 42 (4), 1771–1785. doi:10.3892/ijmm.2018.3764
Hughes, A. E., Bradley, D. T., Campbell, M., Lechner, J., Dash, D. P., Simpson, D. A., et al. (2011). Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet. 89 (5), 628–633. doi:10.1016/j.ajhg.2011.09.014
Kabza, M., Karolak, J. A., Rydzanicz, M., Szczesniak, M. W., Nowak, D. M., Ginter-Matuszewska, B., et al. (2017). Collagen synthesis disruption and downregulation of core elements of TGF-beta, Hippo, and Wnt pathways in keratoconus corneas. Eur. J. Hum. Genet. 25 (5), 582–590. doi:10.1038/ejhg.2017.4
Karolak, J. A., Rydzanicz, M., Ginter-Matuszewska, B., Pitarque, J. A., Molinari, A., Bejjani, B. A., et al. (2015). Variant c.2262A>C in DOCK9 leads to exon skipping in keratoconus family. Invest. Ophthalmol. Vis. Sci. 56 (13), 7687–7690. doi:10.1167/iovs.15-17538
Khaled, M. L., Bykhovskaya, Y., Yablonski, S. E. R., Li, H., Drewry, M. D., Aboobakar, I. F., et al. (2018). Differential expression of coding and long noncoding RNAs in keratoconus-affected corneas. Invest. Ophthalmol. Vis. Sci. 59 (7), 2717–2728. doi:10.1167/iovs.18-24267
Lee, J. E., Oum, B. S., Choi, H. Y., Lee, S. U., and Lee, J. S. (2009). Evaluation of differentially expressed genes identified in keratoconus. Mol. Vis. 15, 2480–2487.
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 (14), 1754–1760. doi:10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi:10.1093/bioinformatics/btp352
Lin, Q., Zheng, L., and Shen, Z. (2022). A novel variant in TGFBI causes keratoconus in a two-generation Chinese family. Ophthalmic Genet. 43 (2), 159–163. doi:10.1080/13816810.2021.2015788
Loescher, C. M., Hobbach, A. J., and Linke, W. A. (2021). Titin (TTN): From molecule to modifications, mechanics and medical significance. Cardiovasc. Res., cvab328 Advance online publication. doi:10.1093/cvr/cvab328
Lopes, A. G., de Almeida, G. C., Miola, M. P., Teixeira, R. M., Pires, F., Miani, R. A., et al. (2021). Absence of significant genetic alterations in the VSX1, SOD1, TIMP3, and LOX genes in Brazilian patients with Keratoconus. Ophthalmic Genet. 43, 73–79. doi:10.1080/13816810.2021.1992785
Lucas, S. E. M., and Burdon, K. P. (2020). Genetic and environmental risk factors for keratoconus. Annu. Rev. Vis. Sci. 6, 25–46. doi:10.1146/annurev-vision-121219-081723
Mace, M., Galiacy, S. D., Erraud, A., Mejia, J. E., Etchevers, H., Allouche, M., et al. (2011). Comparative transcriptome and network biology analyses demonstrate antiproliferative and hyperapoptotic phenotypes in human keratoconus corneas. Invest. Ophthalmol. Vis. Sci. 52 (9), 6181–6191. doi:10.1167/iovs.10-70981
Mas Tur, V., MacGregor, C., Jayaswal, R., O'Brart, D., and Maycock, N. (2017). A review of keratoconus: Diagnosis, pathophysiology, and genetics. Surv. Ophthalmol. 62 (6), 770–783. doi:10.1016/j.survophthal.2017.06.009
Mazharian, A., Panthier, C., Courtin, R., Jung, C., Rampat, R., Saad, A., et al. (2020). Incorrect sleeping position and eye rubbing in patients with unilateral or highly asymmetric keratoconus: A case-control study. Graefe's archive Clin. Exp. Ophthalmol. = Albrecht von Graefes Archiv fur klinische und Exp. Ophthalmol. 258 (11), 2431–2439. doi:10.1007/s00417-020-04771-z
Mohammadpour, M., Heidari, Z., and Hashemi, H. (2018). Updates on managements for keratoconus. J. Curr. Ophthalmol. 30 (2), 110–124. doi:10.1016/j.joco.2017.11.002
Rabinowitz, Y. S., Galvis, V., Tello, A., Rueda, D., and Garcia, J. D. (2021). Genetics vs chronic corneal mechanical trauma in the etiology of keratoconus. Exp. Eye Res. 202, 108328. doi:10.1016/j.exer.2020.108328
Santodomingo-Rubido, J., Carracedo, G., Suzaki, A., Villa-Collar, C., Vincent, S. J., and Wolffsohn, J. S. (2022). Keratoconus: An updated review. Cont. Lens Anterior Eye. 45 (3), 101559. doi:10.1016/j.clae.2021.101559
Sarezky, D., Orlin, S. E., Pan, W., and VanderBeek, B. L. (2017). Trends in corneal transplantation in keratoconus. Cornea 36 (2), 131–137. doi:10.1097/ICO.0000000000001083
Sharif, R., Khaled, M. L., McKay, T. B., Liu, Y., and Karamichos, D. (2019). Transcriptional profiling of corneal stromal cells derived from patients with keratoconus. Sci. Rep. 9 (1), 12567. doi:10.1038/s41598-019-48983-8
Shinde, V., Hu, N., Mahale, A., Maiti, G., Daoud, Y., Eberhart, C. G., et al. (2020). RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses. Sci. Rep. 10 (1), 9907. doi:10.1038/s41598-020-66735-x
Shinde, V., Sobreira, N., Wohler, E. S., Maiti, G., Hu, N., Silvestri, G., et al. (2021). Pathogenic alleles in microtubule, secretory granule and extracellular matrix-related genes in familial keratoconus. Hum. Mol. Genet. 30 (8), 658–671. doi:10.1093/hmg/ddab075
Sun, X., Zhang, H., Shan, M., Dong, Y., Zhang, L., Chen, L., et al. (2022). Comprehensive transcriptome analysis of patients with keratoconus highlights the regulation of immune responses and inflammatory processes. Front. Genet. 13, 782709. doi:10.3389/fgene.2022.782709
Swan, L., Cardinal, J., and Coman, D. (2018). SYNE1-related autosomal recessive cerebellar ataxia, congenital cerebellar hypoplasia, and cognitive impairment. Clin. Pract. 8 (3), 1071. doi:10.4081/cp.2018.1071
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J., and Prins, P. (2015). Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31 (12), 2032–2034. doi:10.1093/bioinformatics/btv098
Tariq, M., Belmont, J. W., Lalani, S., Smolarek, T., and Ware, S. M. (2011). SHROOM3 is a novel candidate for heterotaxy identified by whole exome sequencing. Genome Biol. 12 (9), R91. doi:10.1186/gb-2011-12-9-r91
Tuft, S. J., Hassan, H., George, S., Frazer, D. G., Willoughby, C. E., and Liskova, P. (2012). Keratoconus in 18 pairs of twins. Acta Ophthalmol. 90 (6), e482–6. doi:10.1111/j.1755-3768.2012.02448.x
Wang, K., Li, M., and Hakonarson, H. (2010). Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38 (16), e164. doi:10.1093/nar/gkq603
Xu, L., Yang, K., Fan, Q., Gu, Y., Zhang, B., Pang, C., et al. (2020). Exome sequencing identification of susceptibility genes in Chinese patients with keratoconus. Ophthalmic Genet. 41 (6), 518–525. doi:10.1080/13816810.2020.1799415
You, J., Corley, S. M., Wen, L., Hodge, C., Hollhumer, R., Madigan, M. C., et al. (2018). RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci. Rep. 8 (1), 389. doi:10.1038/s41598-017-18480-x
Yuan, S., Li, D., Ma, M., Zhou, L., Ma, Z., Shi, B., et al. (2022). Evaluating the association between MPDZ-NF1B rs1324183 and keratoconus in an independent northwestern Chinese population. BMC Ophthalmol. 22 (1), 129. doi:10.1186/s12886-022-02359-1
Keywords: keratoconus, family-based exome sequencing, candidate genes, genetic etiology, bioinformatics analysis
Citation: Xu L, Yang K, Yin S, Gu Y, Fan Q, Wang Y, Zhao D and Ren S (2022) Family-based exome sequencing identifies candidate genes related to keratoconus in Chinese families. Front. Genet. 13:988620. doi: 10.3389/fgene.2022.988620
Received: 07 July 2022; Accepted: 08 August 2022;
Published: 02 September 2022.
Edited by:
Silviu-Alin Bacanu, Virginia Commonwealth University, United StatesReviewed by:
Ziye Rong, Anhui Medical University, ChinaMukharram M. Bikbov, Ufa Eye Research Institute, Russia
Copyright © 2022 Xu, Yang, Yin, Gu, Fan, Wang, Zhao and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengwei Ren, eXNnenoyMDE4QDE2My5jb20=
 Liyan Xu1
Liyan Xu1 Kaili Yang
Kaili Yang Shengwei Ren
Shengwei Ren