
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 12 September 2022
Sec. Genomic Assay Technology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.986322
This article is part of the Research Topic Multi-Omics Analysis During the Animal Development View all 4 articles
Hepatocellular carcinoma (HCC) is one of the most fatal malignant tumors worldwide. Circular RNAs (circRNAs) are a special type of RNA that lacks the 5′ and 3’ ends. The functional roles of circRNAs in HCC remain largely unknown. Using high-throughput sequencing, we found several differentially expressed circRNAs in HCC tissues compared with nearby normal tissues. Among them, circRNA derived from the LIFR gene, named circLIFR, was significantly downregulated in HCC. Intriguingly, circLIFR overexpression in SK-Hep-1 cells promoted cell growth and invasion. RNA pull-down and mass spectrometry detection revealed circLIFR interacting with TANK binding kinase 1 (TBK1). Anti-TBK1 RIP confirmed the interaction between circLIFR and TBK1. TBK1 is a serine/threonine kinase that regulates several signaling pathways, including the NF-κB pathway. TBK1 inhibitors inhibit NF-κB activation. Overexpression of circLIFR overcame the in-hibitory function of TBK1, resulting in the upregulation of several genes, including MMP13, MMP3, VEGF, and MAPK. This study shows that the downregulation of circLIFR in HCC has a can-cer-promoting effect by interacting with TBK1 to promote the activation of downstream NF-κB pathway genes related to cell proliferation, migration, and invasion. This novel finding reveals the diversity of circRNA functions in HCC and provides novel insights into the role of circRNAs.
The patocellular carcinoma (HCC) is the fifth most common malignancy world-wide, accounting for 70%–90% of all primary liver cancers (Li et al., 2020). The average survival times of patients with early liver cancer and advanced liver cancer receiving less spe-cialized treatment are 6–9 months and 1–2 months, respectively. More than 60% of pa-tients are diagnosed withadvanced liver cancer that has metastasized, resulting in an overall 5-year survival rate of <16% (Liu et al., 2020; Vibert et al., 2020). Few HCC biomarkers have sufficient diag-nostic performance for early HCC(Liu et al., 2020). Surgical resection of HCC has a 10-year recur-rence-free rate of approximately 22–25% compared with 50–70% following liver transplantation (Vibert et al., 2020).
Circular RNAs (circRNAs) are special closed RNAs that lack the 5′ and 3’ ends (Zhou and Yu, 2017). CircRNAs show higher stability than linear RNAs because of their single-stranded closed circular structures (Guarnerio et al., 2016; Sun et al., 2020). CircRNAs are usually generated by the back-splicing of mRNA gene exons. The function of circRNAs mainly includes but is not limited to: 1) acting as a sponge of miRNAs, 2) interacting with proteins, 3) regulating translation of proteins, and 4) regulating the transcription of linear RNAs. Some circRNAs play several roles (Li et al., 2019).
CircRNAs are closely linked to liver cancer and can influence the proliferation, invasion, and death of liver cells, being potential biomarkers and therapeutic targets. Previous research has shown that circRNAs in liver cancer are closely linked to the development of HCC(Cheng et al., 2020a; Cheng et al., 2020b; Zhang et al., 2020). CircRNAs are differentially expressed in this disease, such as circ-ITCH, circ-MALAT1, etc. (Chen et al., 2020; Wu et al., 2020). The specific functional mechanism of circRNAs in HCC is not yet clear.
A circRNA (circBase ID: hsa_circ_0072309) derived from the LIFR gene, named circLIFR, has been described to, when overexpressed, promote cell proliferation, inva-sion, and migration. CircLIFRs have been reported in several studies. Yan et al. re-ported that circLIFR inhibits the proliferation and invasion of breast cancer cells by targeting miR-492 (Yan et al., 2019). Chen et al. found that circLIFR inhibits cancer progression by sponging miR-100 in renal carcinoma cell lines (Chen et al., 2019). Yuan et al. reported that circLIFR inhibits proliferation and invasion of glioblastoma (Yuan et al., 2021). Zhao et al. revealed that cir-cLIFR promotes apoptosis in ischemic stroke by sponging miR-100 (Zhao et al., 2020). Zhang reported that circLIFRis downregulated in bladder cancer (Zhang et al., 2021).
In this study, we investigated the effects of circLIFRon HCC cells. We constructed plasmid vectors to investigate the function and effect ofcircLIFR on HCC at the cellular level. The interaction between circLIFR and TBK1 was also verified to further study the mechanism of circLIFR on HCC cells. Our results pro-vide novel insights into the role of circRNAs in this disease.
CircRNAs are expressed in atissue/disease-specific manner. To identify differen-tially expressed circRNAs in HCC, high-throughput RNA sequencing (RNA-seq) was per-formed on six tissue samples (three cancerous tissues and three paired normal tis-sues from adjacent sites). The results revealed that 56 circRNAs were upregulated and 70 circRNAs downregulated in cancerous tissues compared to normal tissues (Figures 1A,B). The top 10 significantly different circRNAs are shown (Table 1). Among these differen-tially expressed circRNAs, we selected three downregulated circRNAs and verified their expression in 10 other HCC and paired normal liver tissues. The results show that all these threecircRNAs were significantly downregulated in HCC tissues compared to their paired adjacent normal tissues (Figure 1C). In particular, circRNA from the LIFR gene (hsa_circ_0072309,circLIFR) was significantly downregulated in HCC samples and SK-hep-1 cells. Sanger sequencing revealed the interface position se-quence of cir-cLIFR (Figure 1D).
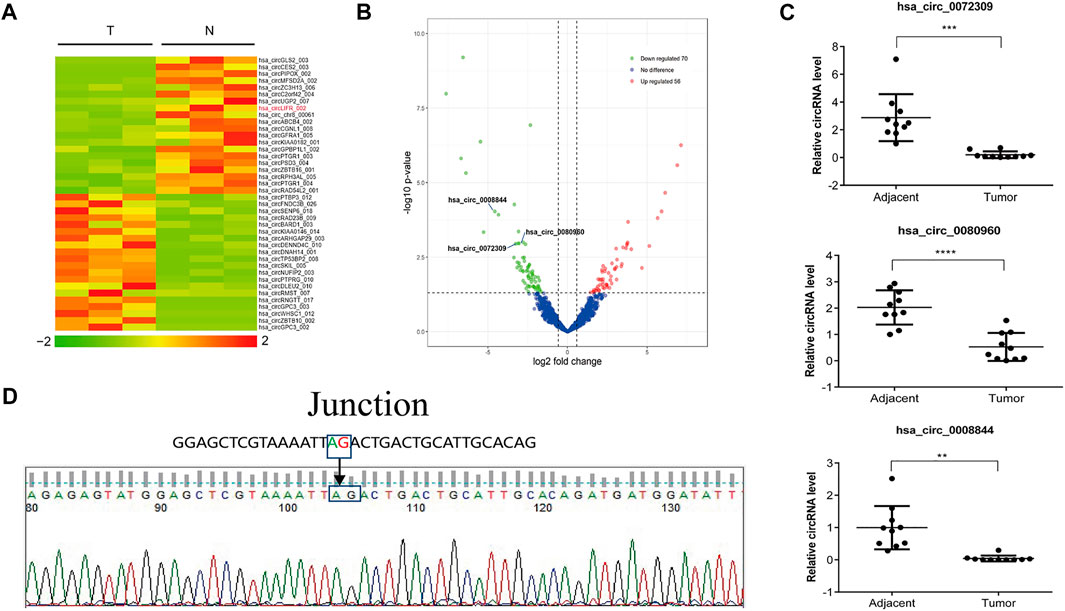
FIGURE 1. Identification of circLIFR by RNA-seq in liver cancer tissues.(A) Heat map of circRNA expression fold-change. Red indicates a higher fold-change; (B)Volcano plots of differentially expressed circRNAs in the three pairs of matched liver cancer tissues and adjacent normal tissues; (C) Expression levels of circRNA in paired HCC and adjacent tissues collected from 10 HCC patients were measured by qRT-PCR assay; (D) Sanger sequencing of circLIFR.
LIFR acts as a tumor suppressor in several human cancers. To investigate the function of circLIFR in HCC, we constructed an overexpression vector for circLIFR and transfected Sk-Hep-1 HCC cells to evaluate cell growth and migration.
Intriguingly, overexpression of circLIFR promoted cell growth in Sk-Hep-1 (Figure 2A) and HepG2 cells (Supplementary Figure S1A). Wound-healing and transwell assays show that overexpression of circLIFR promoted migration and invasion of Sk-Hep-1 cells (Figures 2B,C) and HepG2 cells (Supplementary Figures S1B,C). Colony formation assays also show that circLIFRpromoted colony formation in Sk-Hep-1 cells (Figure 2D) and HepG2 cells (Supplementary Figure S1D). Cell cycle and apoptosis detec-tion also showsthat overexpression of circLIFR promoted cell progression through the cell cycle and reduced the apoptosis rate (Figures 2E,F) and HepG2 (Supplementary Figures S1E,F). The cell cycle and apoptosis analysis results are shown in Supplementary Figure S2.
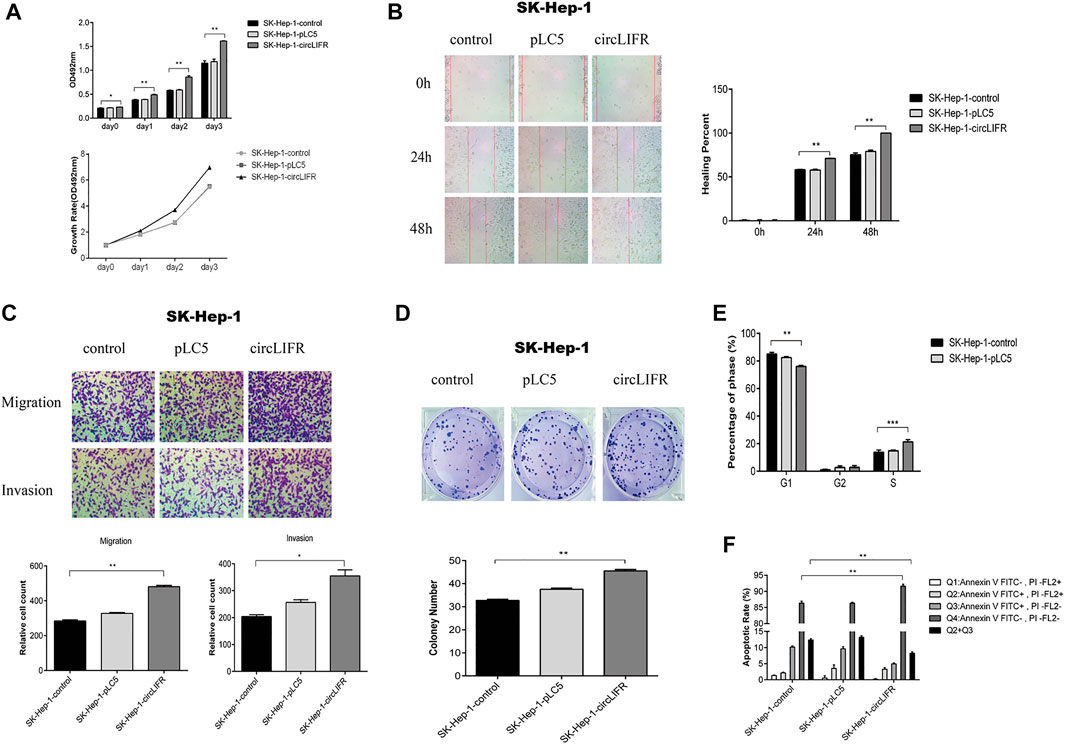
FIGURE 2. CircLIFR promotes proliferation, migration, and invasion in SK-Hep-1 cells.(A) The proliferation of SK-Hep-1 cells transfected with control overexpression circLIFR detected by CCK-8 assays; (B) Wound-healing assay for migration ability of SK-Hep-1 cells after overexpression of circLIFR; Original magnification: ×10. **p < 0.01 (C) Transwell assay shows that the overexpression circLIFR promoted metastasis and invasion of SK-Hep-1 cells. Original magnification: ×10. *p < 0.05; **p < 0.01; (D) Colony forming ability of SK-Hep-1 hepatoma cells transfected with circLIFR in the control group. *p < 0.05,**p < 0.01 (Student’s t-test); (E) Cell cycle assay for SK-Hep-1 cells transfected with control overexpression of circLIFR; (F) Flow cytometry to detect the effect of overexpression of circLIFR in the control group on apoptosis and renewal of SK-Hep-1 cells.
All Functional analysis of the endogenous low-expression circLIFR shows that it had a tumor-promoting effect. CircRNAs are non-coding RNAs that often function by interacting with proteins. To explore the cancer-promoting mechanism of circLIFR, capture analysis of interacting proteins, including TBK1, was performed by RNA pull-down (Figure 3A). We identified 10 interacting proteins, including TBK1 (Table 2). The protein interaction network for these captured proteins was predicted by PPI, and the results show that TBK1 was the center of the protein interaction network (Figure 3B). TBK1 is often highly expressed in tumors, and TCGA data analysis shows that the ex-pression of TBK1 in most tumor types was higher than in paracancerous controls (Figure 3C). Therefore, we further verified and analyzed the interaction between circLIFR and TBK1 and determined whether this interaction mediates the tumor-promoting effect of circLIFR.
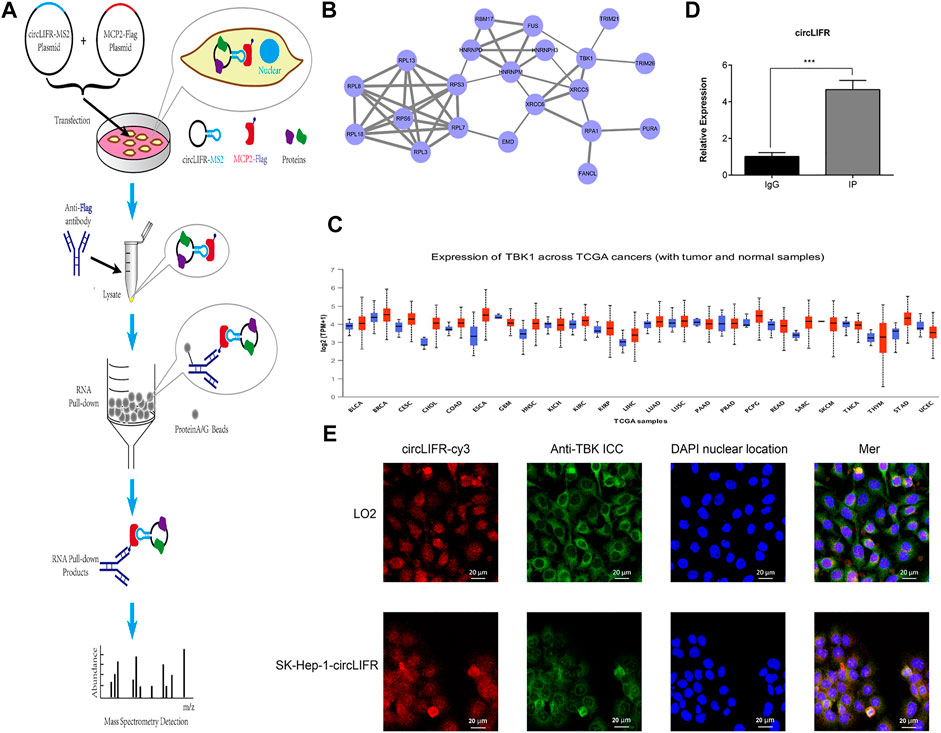
FIGURE 3. RNA pulldown for CircLIFR. (A) Schematic diagram for pulldown detection of the circLIFR gene; (B) PPI analysis of the detected proteins by mass spectrometry; (C) pan-cancer expression level of TBK1 in TCGA cohort, (obtained from the UALCAN website); (D) RIP test verified that overexpression of circLIFR significantly promoted specific binding between TBK1 proteins.*p < 0.05; **p < 0.01; (Student’s t-test); (E) FISH and IF detection for circLIFR and TBK1 in LO2 cells that enhanced expressing circLIFR.
The interaction between endogenous TBK1 and circLIFR was verified by An-ti-TBK1RIP assays (Figure 3D). FISH and IF colorization analyses also confirmed the colo-calization of circLIFR and TBK1 (Figure 3E). Through immunofluorescence fluorescence in situ hybridization (IF-FISH), we can perform co-localization and qualitative analysis of nucleic acid molecules and proteins in tissues and cells, which can be used to indi-cate the interaction between nucleic acids and proteins from the point of view ofco-localization. Consequently, we detected the interaction between circLIFR and TBK1 protein by FISHwith a red fluorescent cy3 probe, IF antigen localization with green fluorescent substance-labeled anti-TBK1 protein antibody, and nuclear locali-za-tion by DAPI staining. The results show that in LO2 cells and SK-Hep-1 cells over-ex-pressing circLIFR, the localization of circLIFR and TBK1 protein was basically the same, suggesting that there is an interaction between circLIFR and TBK1 protein.
The nuclear factor-κB (NF-κB) transcription factor family has been considered the central mediator of the inflammatory process and linked to the cancer development (DiDonato et al., 2012). Moreover, TBK1 also plays an importantrole in the NF-κB signaling pathway (Kim et al., 2010).
NF-κB is an important downstream target of TBK1. To verify the function of cir-cLIFR binding to TBK1, a promoter-reporter gene that binds to NF-κB was investigated. circLIFRbinds to TBK1 and regulates the NF-κB pathway. The results show that over-expression of circLIFR promoted the expression of the NF-κB reporter gene, indicating that circLIFR promotes NF-κB activity (Figure 4A). The addition of TBK1 inhibitors re-duced the expression of the NF-κB reporter gene. When circLIFR was overexpressed and TBK1 was suppressed simultaneously, the effect of the NF-κB reporter gene was compensated (Figure 4A).
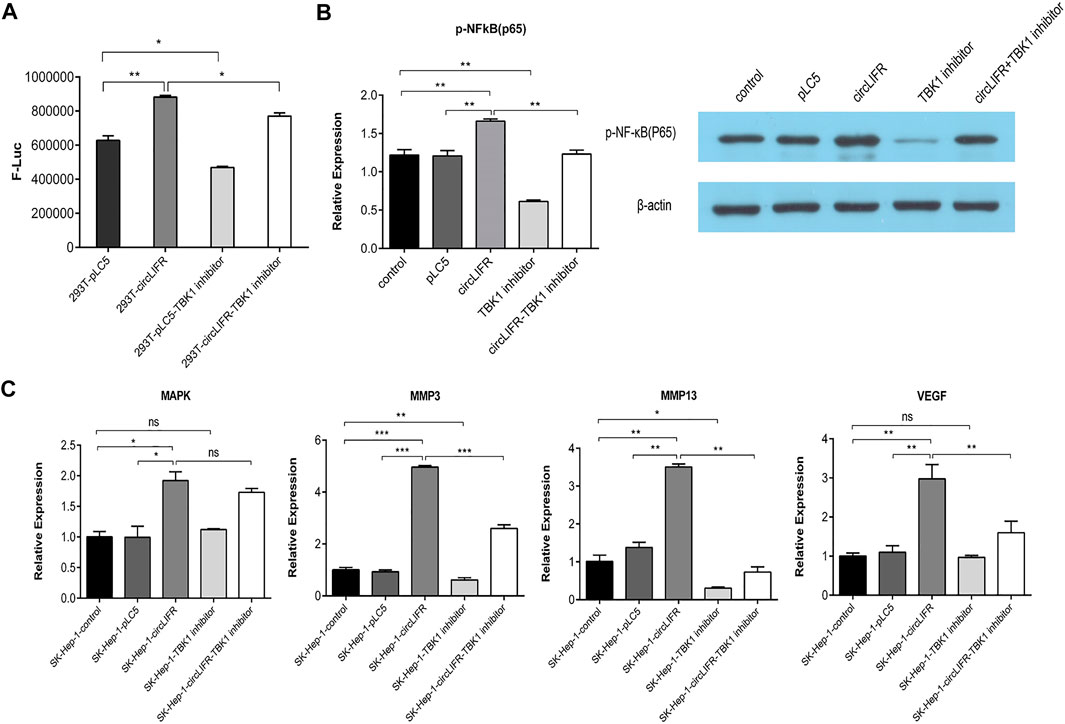
FIGURE 4. Interaction between circLIFR and TBK1 protein promotes NFκB signal pathway activation and regulates the expression of genes related to proliferation and migration. (A) NF-κB luciferase reporter system shows that overexpression of circLIFR counteract the inhibitory effect of TBK1 inhibitor and activated the NF-κB pathway to affect the activity of NF-κB by binding to TBK1; (B) Western blotting analysis suggests that circLIFR overexpression activated the NF-κB pathway; (C) qRT-PCR analysis shows that the overexpression of circLIFR interacted with TBK1 to affect the levels of oncogenes downstream of NF-κB.
NF-κB subunit p65 is a known target of TBK1. To analyze the regulatory role of circLIFR-TBK1 in NF-κB, western blot analysis was used to detect the phosphorylation state of p65. The results show that circLIFR overexpression promotes p65phosphorylation, whereas the TBK1 inhibitor inhibits the phosphorylation of p65. Overexpression of circLIFR also reversed the effects of the TBK1 inhibitor on p65 phosphorylation. These results suggest that circLIFR-TBK1 regulates the activation of the NF-κB pathway, whereas circLIFR reverses the effect of the TBK1 inhibitor (Figure 4B).
NF-κB is an important transcription factor that mediates the expression of many genes involved in cell proliferation, migration, and invasion. MMP13, MMP3, VEGF, and MAPK were used as target molecules in qPCR detection to show the effect of cir-cLIFR-TBK1 on downstream genes. The results show that Overexpression of circLIFR enhanced the expression of several downstream genes, whereas TBK1 inhibitors downregulated these genes. Overexpression of circLIFR reversed the effect of the TBK1 inhibitor (Figure 4C).
CircRNAs play an important role in the metastasis and invasion of several cancer cells (Kristensen et al., 2018). A study reported that circFNDC3B is significantly downregulated in BCa tissues and is associated with pathological T staging, grading, lymphatic invasion, and overall patient survival (Liu et al., 2018). Other studies have reported that circ_0072309 inhibits the progression of NSCLC by blocking miR-580-3p (Yuan et al., 2021). Tao C. et al. reported that the circular RNA hsa-circ-0072309 plays an anti-tumor role by sponging miR-100 through deactivation of the PI3K/AKT and mTOR pathways in renal carcinoma cell lines (Chen et al., 2019). These findings indicate that circRNAs play an important regulatory role in tumor-igenesis. The present study found that circLIFR was downregulated in HCC. Our find-ings reveal that circLIFR functions as a tumor promoter in HCC by promoting cell pro-liferation, migration, and invasion, contrary to expectations from the sequences ob-tained. Our study findings are also contrary to an earlier study by Qin et al., which re-ported that LIFR functions as a metastasis suppressor in HCC by negatively regulating the phosphoinositide 3-kinase/AKT pathway (Luo et al., 2015).
The expression of cirLIFR in vivo in paracancerous tissues was higher than that in cancerous tissues. The regulatory mechanism of circLIFR in cell proliferation in para-cancerous tissues remains unclear. The LIFR gene is significantly underexpressed in liver cancer and has a significant tumor suppressor effect in liver cancer. The expres-sion of circLIFR decreases with a decrease in transcription factors.
TBK1 is often highly expressed in tumors, and our TCGA data analysis shows that the expression of TBK1 in most tumor types was higher than in paracancerous controls (Figure 3C). Anti-TBK1RIP assays were used to determine the interaction between endog-enous TBK1 and circLIFR. TBK1 regulates gene transcription and immune signaling pathways by regulating downstream target molecules such as NF-κB and IRF3. These results suggest that there is an interaction between circLIFR and TBK1, which may further regulate the NF-κB pathway. Abnormal activation of the NF-κBsignaling pathway is related to the occurrence and development of cancer and affects genes re-lated to cell proliferation, migration, and invasion (Sims et al., 2013; Bennett et al., 2017). This mechanism may ex-plain the increased cell proliferation and migration following circLIFR overexpression observed.
This study also found that downregulation of circLIFR in HCC promoted tumor proliferation, migration, and invasion. Further research in this area is required.
Thirteen pairs of human primary HCC and paired adjacent non-tumorous liver tissues were collected from the Wen Zhou Traditional Chinese Medicine Hospital. Human materials were obtained with informed consent, and the study was approved by the Wenzhou Hospital of Traditional Chinese Medicine Ethics Committee. Three paired HCC and non-tumorous liver tissues were used for high-throughput RNA se-quencing, and the other 10 paired HCC and non-tumorous liver tissues were used for QPCR detection of selected circRNAs.
Total RNA from three paired HCC and adjacent noncancerous liver tissues was extracted using TRIZOL (Life technologies). Ribosome-depleted RNA samples were fragmented and used for first- and second-strand complementary DNA (cDNA) syn-thesis with random hexamer primers. PCR was used for amplification. The purification and screening of a suitably sized library were carried out. Finally, the library was se-quenced using X. A standard of fold-change ≥ 1.5 or ≤ −1.5 was set as the cut-off value. Significant differential expression of circRNAs was observed.
Total RNA was extracted from tissues and cultured cells using TRIZOL reagent (Life Technologies) following the manufacturer’s instructions. Total RNA was reverse transcribed to cDNA using specific primers in line with the Geneseed® II First Strand cDNA Synthesis Kit (Geneseed). qRT-PCR was performed using the Geneseed® qPCR SYBR® Green Master Mix (Geneseed). The primers used were as follows:
GAPDH forward: AGAAGGCTGGGGCTCATTTG;
Reverse: GCAGGAGGCATTGCTGATGAT;
circLIFR forward: 5′-GGAGCTCGTAAAATTAGACTG-3'.
Reverse: 5′-AATGTTGATAACAGCCACTGGA-3'.
HCC cell lines (SK-Hep-1, HepG2) and the human normal liver cell line (LO2) were purchased from ATCC and cultured in Roswell Park Memorial Institute-1640 medium (RPMI; Invitrogen, CA, United States).
To recapitulate circRNA, the 3276bp sequence of LIFR was amplified using Pri-merSTAR Max DNA Polymerase Mix (Takara). The PCR products were inserted into the pCDNA3.1 (+)-IRES-EGFP vector.
Human HCC cell lines SK-hep-1 and HepG2 were inoculated into two six-well plates. Plasmids and Lipofectamine 2000 (3 µg plasmid in 4 μL) were mixed at a cell confluence of 70–90% and added to serum-free medium into cell samples. Cell trans-fection was verified using PCR. The extracted RNA was detected using NanoDropND2000 and 1% agarose gel electrophoresis.
Cells were seeded onto a six-well plate after 48 h of transfection. Pipette tips (10 μl) were used to make cell scratches perpendicular to plates. The old culture medium was removed, the cells were washed thrice with sterile PBS, damaged cells were re-moved, and a new culture medium was added. The cell scratch mobility was observed and calculated at 0, 24, and 48 h.
Cells (1 × 104) were seeded 48 h after transfection. The CCK-8 solution (10 μL) was added to each well at (24, 48, and 72 h) and incubated for 2 h. The absorbance was read at 450 nm using a microplate reader (BioTek). The following formulas were used for the calculation of:
Cell proliferation rate: Proliferation rate (same treatment sample) = Day NOD value/Day0 average OD value.
Cell inhibition rate formula: Inhibition rate (at the same time point) = (1-experimental group proliferation rate)/control group average proliferation rate.
After 48 h, the transfected cells were digested with trypsin-EDTA solution and resuspended in serum-free DMEM after centrifugation. Next, 100 µl of the cell suspen-sion was added to the upper part of the transwell chamber. The lower chamber was filled with 600 μm complete DMEM medium. After 24 h, the filters were fixed with 4% paraformaldehyde and stained with crystal violet. The cells that passed through the lower chamber were counted.
The apoptotic rate of the modulated cells was determined. The Annexin V-fluorescein isothiocyanate Apoptosis Detection Kit (KeyGen, Nanjing, China) was used to perform annexin V/propidium iodide staining. Flow cytometry was used to de-tect red fluorescence at 488 nm excitation wavelength. The results were analyzed using the cell cycle simulation software ModFit.
Approximately 200 transfected cells were seeded per well in six-well plates, incu-bated for 10–15 days. Formed colonies were stained with 0.1% crystal violet solution and counted.
MS2 was then inserted into circLIFR (circLIFR-MS2). circLIFR-MS2 was con-structed into the pCDNA3.1 (+)-IRES-EGFP vector. MS2 binding protein, mCP2 was cloned into the TetOn3G vector and fused with Flag and mCherry (Flag-mCP2-mCherry). Both circLIFR-MS2 and Flag-mCP2-mCherry were transfected into the Sk-Hep1 cells. The expression of circLIFR-MS2 and Flag-mCP2-mCherry was examined using a fluorescence microscope. An anti-FLAG antibody was used to purify the Flag-mCP2-mCherry/circLIFR-MS2 complex. Proteins interacting with cir-cLIFR-MS2 were extracted and used for mass spectrometry.
Peptides were dissolved in (0.1% formic acid and 2% acetonitrile) solution, cen-trifuged at 4°C, 13,200 rpm, for 20 min, and the supernatant was identified by mass spectrometry. The separated peptides were directly uploaded using a ThermoScientificQExactive mass spectrometer for online detection. The MMFileConversion software was used to convert the results to the MGF format file and then subjected to MASCOT (http://www.matrixscience.com/) to search the UniProt database. The comparison da-tabase for this experiment was Homosapien (https://www.uniprot.org/taxonomy/9606).
The cells were co-transfected with control MS2bs-Rluc, MS2bs-circLIFRmt, or MS2bs-circLIFR using Lipofectamine 2000. RIP was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Germany) 48 h later. For the RIP assay on TBK1, RIP was performed with an anti-TBK1 antibody (Millipore, Billerica United States, MA) 48 h after transfection.
The 293T cells were cultured and inoculated in a 24-well plate. The cells were al-lowed to grow to 70% Mel 80% before being transfected into the hsa _circ_0072309 plasmid. The cells were treated with a TBK1 inhibitor for 1 and 24 h after transfection. The luciferase reporter gene was detected after complete lysis of the cells 48 h after transfection.
Proteins were separated using 10% SDS-PAGE and transferred to a PVDF mem-brane (Millipore). The film was then sealed in Tris-buffered saline at room temperature. Incubation was performed using primary antibodies. The membrane was then washed with TBST 3 times and incubated with the secondary antibody at room temperature for 2 h. Finally, chemiluminescence detection was performed.
At harvest, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Then, the cells were incubated with denatured probe (hsa_circ_0072309) at 37°C overnight (12–18 h). The primary antibody was diluted with 3% bovine serum albumin (BSA) and incubated overnight with a fluorescent sec-ondary antibody (1:200) in the dark at RT for 1h. DAPI was used for nuclear staining. A TCS SP2 AOBS laser scanning confocal microscope was used for confocal microsco-py.
All experimental data were analyzed by spss21.0 software. The results are ex-pressed as mean ± standard deviation (x ± s). The means of two independent samples were compared using t-tests. Data were compared between multiple groups using one-way ANOVA and the LSD method. Results were considered statistically signifi-cant at p < 0.05.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Wenzhou Hospital of Traditional Chinese Medicine Ethics Committee (protocol code WZY2022-LW-003-01 and on 10 May, 2022) for studies involving humans. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Methodology, JJ and QW; software, JT and XY; validation, JT and PR; formal analysis, PR; investigation, WC; resources, WC; data curation, MS and QW; writing—original draft prep-aration, JJ; writing—review and editing, JJ; supervision, CW; project administration, CW; funding acquisition, JJ and CW. All authors have read and agreed to the published version of the manuscript.
The present study was supported by the National Natural Science Foundation of China grant number 81772735, the National College Students Innovation and Entrepreneurship Training Pro-gram grant numbers 202010343021 202110343004, the Zhejiang Medical and Health Science and Technology Plan Project grant number KYB451, the Zhejiang University Students Science and Technology Innovation Activity Plan and new Seedling Talent Plan grant number 2020R413023, the Zhejiang Natural Science Foundation Youth Project grant number LQ19H200002, and the Zhejiang Natural Science Foundation Public Welfare Technology Project grant number LGF18H200003 and the Wenzhou Science and Technology Plan Project grant number Y20170009.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.986322/full#supplementary-material
Bennett, L., Mallon, E. A., Horgan, P. G., Paul, A., McMillan, D. C., and Edwards, J. (2017). The relationship between members of the canonical NF-κB pathway, components of tumour microenvironment and survival in patients with invasive ductal breast cancer. Oncotarget 8, 33002–33013. doi:10.18632/oncotarget.16031
Chen, L., Kong, R., Wu, C., Wang, S., Liu, Z., Liu, S., et al. (2020). Circ-MALAT1 functions as both an mRNA translation brake and a microRNA sponge to promote self-renewal of hepatocellular cancer stem cells. Adv. Sci. 7, 1900949. doi:10.1002/advs.201900949
Chen, T., Shao, S., Li, W., Liu, Y., and Cao, Y. (2019). The circular RNA hsa-circ-0072309 plays anti-tumour roles by sponging miR-100 through the deactivation of PI3K/AKT and mTOR pathways in the renal carcinoma cell lines. Artif. Cells Nanomed. Biotechnol. 47, 3638–3648. doi:10.1080/21691401.2019.1657873
Cheng, F., Wang, L., and Zhang, J. (2020). Circular RNA 0016788 displays as a biomarker for tumor progression and poor prognosis in surgical hepatocellular carcinoma patients. J. Clin. Lab. Anal. 34, 23300. doi:10.1002/jcla.23300
Cheng, X., Tian, P., Zheng, W., and Yan, X. (2020). Piplartine attenuates the proliferation of hepatocellular carcinoma cells via regulating hsa_circ_100338 expression. Cancer Med. 9, 4265–4273. doi:10.1002/cam4.3043
DiDonato, J. A., Mercurio, F., and Karin, M. (2012). NF-κB and the link between inflammation and cancer. Immunol. Rev. 246, 379–400. doi:10.1111/j.1600-065X.2012.01099.x
Guarnerio, J., Bezzi, M., Jeong, J. C., Paffenholz, S. V., Berry, K., Naldini, M. M., et al. (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165, 289–302. doi:10.1016/j.cell.2016.03.020
Kim, H. R., Lee, S. H., and Jung, G. (2010). The hepatitis B viral X protein activates NF-kappaB signaling pathway through the up-regulation of TBK1. FEBS Lett. 584, 525–530. doi:10.1016/j.febslet.2009.11.091
Kristensen, L. S., Hansen, T. B., Veno, M. T., and Kjems, J. (2018). Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 37, 555–565. doi:10.1038/onc.2017.361
Li, F., Bai, L., Li, S., Chen, Y., Xue, X., and Yu, Z. (2020). Construction and evaluation of a prognosis lncRNA model for hepatocellular carcinoma. J. Cell. Biochem. 122, 983–992. doi:10.1002/jcb.29608
Li, Z., Ruan, Y., Zhang, H., Shen, Y., Li, T., and Xiao, B. (2019). Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 110, 3630–3638. doi:10.1111/cas.14211
Liu, H., Bi, J., Dong, W., Yang, M., Shi, J., Jiang, N., et al. (2018). Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol. Cancer 17, 161. doi:10.1186/s12943-018-0908-8
Liu, J., Li, K., Cai, J., Zhang, M., Zhang, X., Xiong, X., et al. (2020). Landscape and regulation of m6A and m6Am methylome across human and mouse tissues. Mol. Cell 77, 426–440.e6. doi:10.1016/j.molcel.2019.09.032
Luo, Q., Wang, C., Jin, G., Gu, D., Wang, N., Song, J., et al. (2015). LIFR functions as a metastasis suppressor in hepatocellular carcinoma by negatively regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis 36, 1201–1212. doi:10.1093/carcin/bgv108
Sims, J. T., Ganguly, S. S., Bennett, H., Friend, J. W., Tepe, J., and Plattner, R. (2013). Imatinib reverses doxorubicin resistance by affecting activation of STAT3-dependent NF-κB and HSP27/p38/AKT pathways and by inhibiting ABCB1. PLoS One 8, 55509. doi:10.1371/journal.pone.0055509
Sun, J. Y., Zhang, X. Y., Cao, Y. Z., Zhou, X., Gu, J., and Mu, X. X. (2020). Diagnostic and prognostic value of circular RNAs in hepatocellular carcinoma. J. Cell. Mol. Med. 24, 5438–5445. doi:10.1111/jcmm.15258
Vibert, E., Schwartz, M., and Olthoff, K. M. (2020). Advances in resection and transplantation for hepatocellular carcinoma. J. Hepatol. 72, 262–276. doi:10.1016/j.jhep.2019.11.017
Wu, M., Deng, X., Zhong, Y., Hu, L., Zhang, X., Liang, Y., et al. (2020). MafF is regulated via the circ-ITCH/miR-224-5p Axis and acts as a tumor suppressor in hepatocellular carcinoma. Oncol. Res. 28, 299–309. doi:10.3727/096504020X15796890809840
Yan, L., Zheng, M., and Wang, H. (2019). Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag. Res. 11, 1033–1041. doi:10.2147/CMAR.S186857
Yuan, F., Sun, Q., Xu, Y., Zhang, H., Deng, G., Yang, J., et al. (2021). Hsa_circ_0072309 inhibits proliferation and invasion of glioblastoma. Pathol. Res. Pract. 222, 153433. doi:10.1016/j.prp.2021.153433
Zhang, H., Xiao, X., Wei, W., Huang, C., Wang, M., Wang, L., et al. (2021). CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM-p73 axis in bladder cancer. Mol. Cancer 20, 70. doi:10.1186/s12943-021-01360-4
Zhang, X. J., Cao, G., Fu, J., Zhuang, H. J., and Shi, J. (2020). The role of hsa_circ_0000285 in metastasis of hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 24, 3579–3585. doi:10.26355/eurrev_202004_20819
Zhao, Y., Li, J., Li, J., Xu, L., and Lian, W. (2020). The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100. Eur. Rev. Med. Pharmacol. Sci. 24, 4420–4429. doi:10.26355/eurrev_202004_21024
Keywords: hepatocellular carcinoma, circLIFR, metastasis, TBK1, CircRNAs
Citation: Ji J, Tang J, Ren P, Cai W, Shen M, Wang Q, Yang X and Chen W (2022) Downregulation of circLIFR exerts cancer-promoting effects on hepatocellular carcinoma in vitro. Front. Genet. 13:986322. doi: 10.3389/fgene.2022.986322
Received: 05 July 2022; Accepted: 15 August 2022;
Published: 12 September 2022.
Edited by:
Jiaqiang Wang, Northeast Agricultural University, ChinaReviewed by:
Yuxue Wang, Hubei University of Chinese Medicine, ChinaCopyright © 2022 Ji, Tang, Ren, Cai, Shen, Wang, Yang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Chen, Y2hlbndlaTgwOEBtYWlsLnhqdHUuZWRkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.