
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 21 November 2022
Sec. Livestock Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.985228
This article is part of the Research Topic Leveraging GxG and GxE Studies for a Climate-Resilient Agricultural System View all 7 articles
 Huie Wang1,2
Huie Wang1,2 Junhui Wen3
Junhui Wen3 Haiying Li4
Haiying Li4 Tao Zhu3
Tao Zhu3 Xiurong Zhao3
Xiurong Zhao3 Jinxin Zhang3
Jinxin Zhang3 Xinye Zhang3
Xinye Zhang3 Chi Tang1
Chi Tang1 Lujiang Qu1,3*
Lujiang Qu1,3* M. Gemingguli1,2*
M. Gemingguli1,2*Chicken plumage color is an inheritable phenotype that was naturally and artificially selected for during domestication. The Baicheng You chicken is an indigenous Chinese chicken breed presenting three main feather colors, lavender, black, and yellow plumages. To explore the genetic mechanisms underlying the pigmentation in Baicheng You chickens, we re-sequenced the whole genome of Baicheng You chicken with the three plumage colors. By analyzing the divergent regions of the genome among the chickens with different feather colors, we identified some candidate genomic regions associated with the feather colors in Baicheng You chickens. We found that EGR1, MLPH, RAB17, SOX5, and GRM5 genes were the potential genes for black, lavender, and yellow feathers. MLPH, GRM5, and SOX5 genes have been found to be related to plumage colors in birds. Our results showed that EGR1 is a most plausible candidate gene for black plumage, RAB17, MLPH, and SOX5 for lavender plumage, and GRM5 for yellow plumage in Baicheng You chicken.
Chicken plumage color is driven by natural, sexual, and artificial selection (Roulin and Ducrest, 2013). The diversity of the plumage colors is regulated by multiple genes, and a number of genetic mutations have been identified for certain feather color traits (Cieslak et al., 2011). Multiple genes have been found to be associated with animal pigmentation (http://www.ifpcs.org/colorgenes/) (Baxter et al., 2019). Given the complexity of pigmentation traits, the genetic foundation of some of them still remains unraveled.
Feather colors in birds are mainly determined by the distribution and quantity of eumelanin and phaeomelanin and the size, number, and type of compartments containing melanin in keratinocytes. Two types of melanin are produced by neural crest-derived melanocytes (Moreiras et al., 2021). The distribution of melanosomes within melanocytes is the result of competition between microtubules and actin-dependent transport (Gross et al., 2002). Melanosomes must be transported from the perinuclear region to melanocyte dendrites and then to keratinocytes (Alzahofi et al., 2020; Jiang et al., 2020). Some studies have shown that melanin-based color phenotypes are associated with mutations in melanin-producing genes. For example, melanocortin 1-receptor (MC1R), a seven-transmembrane helix-bearing G protein-coupled receptor on melanocytes, plays a crucial role in determining the melanin type and underlies the pigmentation traits in a wide range of animals. In chickens (Kerje et al., 2003; Takeuchi et al., 1996), Japanese quail (Nadeau et al., 2006), pigs (Fang et al., 2014), and horse (Ludwig et al., 2009), the extended black (E) locus controls coat color and pattern, and the molecular basis of its actions is rooted in the MC1R gene (Schiöth et al., 2003).
The Baicheng You chicken is an indigenous Chinese breed, and its main feather color types are black, yellow, and lavender. To identify the genetic foundation of the plumage colors, we employed a genome-wide selective sweep (Pavlidis et al., 2013) analysis by comparing chicken genomes with respect to the three colors to identify potential regions that are differentially selected for feather color (Chen et al., 2010; Pavlidis et al., 2013). Our results identified candidate genes related to the three types of feather colors in Baicheng You chicken.
We used a local Chinese Baicheng You chicken population to screen candidate genes for feather colors. First, 30 adult chickens were selected from differentiated sub-populations selected for the three target feather colors (Figure 1), lavender, black, and yellow, with 10 samples for each sub-population (ADMIXTURE and PCA; Supplementary Figure S1). Blood samples were obtained from the brachial veins by standard venipuncture and were placed into centrifuge tubes containing an anticoagulating agent.
Genomic DNA was extracted from the 30 chickens using the standard phenol/chloroform method, and random interruptions were made using a Covaris ultrasonic crusher. We performed 150-bp paired-end re-sequencing using the Illumina HiSeq 2500/NovaSeq 6000 system according to the manufacturer’s protocols. We used fastp (v0.20.0) software to filter residual primers, adapters trimming, and lower-quality reads, and used the -q 20 -u 30 parameters (from the Fastp manual, q, --qualified_quality_phred indicate the quality value that a base is qualified, thus default 15 means phred quality ≥ Q15 is qualified) for the quality control of the 150-bp paired-end raw reads (Chen et al., 2018). If the quality value of the per-base or per-read is less than 20, this is considered to indicate low quality. If the percentage of low-quality bases is 30%, the reads with too many N bases are excluded. If the content of N bases is greater than n, the read/pair will be discarded, and the default value is 5, such as when one has a high quality score (>Q30) and the other has a low quality score (<Q15). To reduce false corrections, fastp only performs a correction if the total mismatch is below a given threshold T (T ¼ 5) (Chen et al., 2018). The clean reads were mapped to the chicken genome (https://www.ncbi.nlm.nih.gov/, assembly: GCA_016700215.2) using BWA-MEM (v0.7.17) with default parameters, except for -t 4 -R (-t is thread, -R is set the reads header, and it is split with \t) option (Li et al., 2008). We sorted the alignment bam files using the Samtools (v-1.11) software package, and duplicate reads were removed using the picard (http://broadinstitute.github.io/picard/) tools MarkDuplicates. Genome Analysis Toolkit (GATK,v-4.2) (https://gatk.broad institute.org/) was then used for downstream processing and variant calling. Using the HaplotypeCaller algorithm of the GATK pipeline in the genomic Variant Call Format (gVCF), we obtained the genotype likelihoods at each site in the reference genome for each individual. We called the SNPs and indels using the HaplotypeCaller algorithm. Individual gVCF files were then merged into a multi-individual VCF using the GenotypeGVCFs tool of the GATK pipeline. It is imperative to filter raw SNP candidates in the genotyping workflow because it allows the shrinking of false-positive calls due to biases in the sequencing data (Jiang et al., 2021; Xiangtao Liu et al., 2013). We called SNPs and indels using the SelectVariants algorithm. Next, these variants were used as input for hard filtering in the GATK pipeline based on six statistics to identify SNPs or indels: QUAL < 30.0; QualByDepth (QD) < 2.0; FisherStrand (FS) > 60.0; RMS MappingQuality (MQ) < 40.0; MappingQualityRankSunTest (MQRankSum) < -12.5; and ReadPosRankSumTest (ReadPosRankSum) < -8.0 (Li et al., 2008). ClusterWindowSize and clusterSize were set to 10 and 3, respectively. Generally, consecutive SNPs tend to provide a high proportion of false-positive results in terms of selection signatures associated with plumage color (Margot E. Bowen et al., 2011).
The cross-population composite likelihood ratio (XP-CLR) method is mainly based on the gene Site Frequency Spectrum (SFS) principle, which is based on the difference in multilocus allele frequency between two populations and is used in selection signal detection (Maria Ines Fariello 2013; Chen et al. 2010). These methods have been widely used to discover the loci and genes related to difference in traits, such as hair length in Tianzhu white yaks (Bao et al., 2022), body size in dogs (Lyu et al., 2021) and ponies (Asadollahpour et al., 2020), and meat quality in Angus cattle (Taye et al., 2018). We used XP-CLR (v1.1.2) (Chen et al., 2010) statistical methods to detect genetic differentiation in Baicheng You chicken plumage colors (Pavlidis et al., 2013). The values of XP-CLR were calculated using the python script XPCLR, downloaded from Github (https://github.com/hardingnj/xpclr). The corresponding parameters were set as follows: maximum SNPs = 600, ld values = 0.95, window size = 40,000 (Lee et al., 2018). The regions with XP-CLR values in the top 1% were considered candidate regions, and genes with overlapped candidate sweeps were considered candidate genes. Six feather color models were used to identify candidate direct selection regions: 1) black vs. lavender; 2) black vs. yellow; 3) yellow vs. lavender; 4) black vs. yellow + lavender; 5) lavender vs. black + yellow; and 6) yellow vs. black + lavender. In addition, the genes were annotated using the chicken genome (https://www.ncbi.nlm.nih.gov/, assembly: GCA_016700215.2) and NCBI databases. Variant effect predictor (VEP) was used to annotate gene variants according to their functional categorization, including the following categories: up- and downstream gene, frameshift, 3- and 5-prime UTR, synonymous, missense, start lost, intronic, and splice region. We used the R qqman package to create a Manhattan plot of XP-CLR for the association analysis.
After quality control, a total of ∼8.9 million high-quality paired-end reads were obtained from the 30 chickens (267.73 Gb clean base). The average sequencing depth per individual was 8.11×, and the average genome coverage was 98.62%.
A total of 18.37 million SNPs and 2.61 million indels were retained for the association analysis. In addition, where more than 3 SNPs were clustered within a 10-bp window, they were all considered false positives and deleted. In the subsequent analyses, we used only biallelic SNPs on the chromosomes.
The candidate genes for the three kinds of feather colors were identified using XP-CLR statistical methods in Baicheng You chicken. The total numbers of genes with positive selection signatures detected in the six groups were as follows: 934 genes (black vs. lavender), 3620 genes (black vs. yellow), 989 genes (yellow vs. lavender), 685 genes (black vs. yellow + lavender), 836 genes (lavender vs. black + yellow), and 820 genes (yellow vs. black + lavender), including cell growth, differentiation, migration, and immune regulation (Supplementary Table S2–S7). XP-CLR scores are presented in Figure 2.
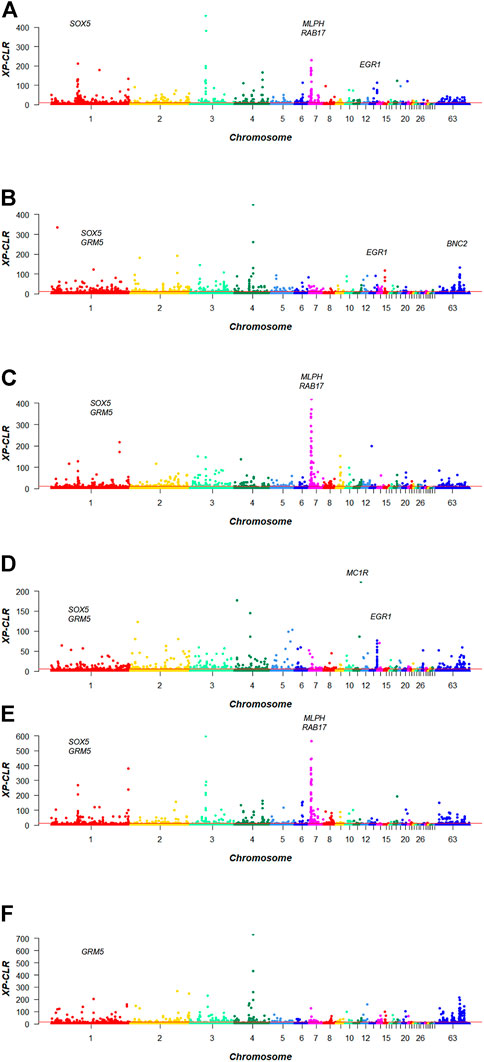
FIGURE 2. XP-CLR Mahattan plot of plumage colors in Baicheng You chicken. (A) Black vs. lavender; (B) black vs. yellow; (C) lavender vs. yellow; (D) black vs. lavender + yellow; (E) lavender vs. black + yellow; (F) yellow vs. black + lavender.
First, to obtain the genes associated with black plumage, the scanning regions of top 1% of black vs. lavender, black vs. yellow, and black vs. yellow + lavender models were extracted, respectively (Figures 2A,B,D), and the intersection of the three sets of data was taken. We obtained the overlap windows at chromosome 13: 18940001–18980000 bp, which overlapped with EGR1 gene.
Second, we extracted top 1% of sweep regions of black vs. lavender, lavender vs. yellow, and lavender vs. black + yellow models, respectively (Figures 2A,C,E), and the same method was used to obtain the intersection of the three groups of data. One window was found at chromosome 7: 4800001–4840000 bp, harboring RAB17, and we also identified a window at chromosome 7: 4820001–4860000 bp, containing MLPH. Another window was located at chromosome 1: 65940001–65980000 bp, which overlapped with the SOX5 gene. We thus obtained MLPH, RAB17, and SOX5 as genes that could be associated with lavender plumage color.
We detected the top 1% of sweep regions of black vs. yellow, lavender vs. yellow, and yellow vs. black + lavender models, respectively (Figures 2B,C,F), and again took the intersection of three sets of data. The overlap window at chromosome 1: 18240001–18280000 bp, containing the GRM5 gene.
The lavender phenotype has been found to be related to MLPH and Rab17 (Roulin and Ducres, 2013). SOX5, as a transcription factor, indirectly regulates the synthesis and transport of melanocytes (Min et al., 2022; Stolt et al., 2008; Wang et al., 2016). EGR1 is expressed in mouse hair follicles (Mcmahon et al., 1990). GRM5 is a candidate gene in the Polverara chicken (black vs. white) (Mastrangelo et al., 2020). Thus, the results showed that MLPH, RAB17, SOX5, EGR1, and GRM5 genes were associated with lavender, black, and yellow plumage colors in Baicheng You chicken.
A total of 258 SNPs and 28 indels were identified in MLPH (Supplementary Table S8). In the coding regions of the MLPH gene, we detected eight synonymous, two non-synonymous nucleotide substitutions, and one deletion. Interestingly, we found an A99G substitution in exon 1 and one deletion (AG861A) in exon 6 that were present in lavender feather chickens.
RAB17 possessed a total 179 SNPs and 6 indels (Supplementary Table S9). Five synonymous and four non-synonymous nucleotide substitutions were detected in the coding region of RAB17 gene, and G514A (Val172Ile) substitution in exon 5 appeared in lavender plumage color (Table 1).Three synonymous and two non-synonymous nucleotide substitutions were detected in exon 2 of the EGR1 gene (Supplementary Table S10).
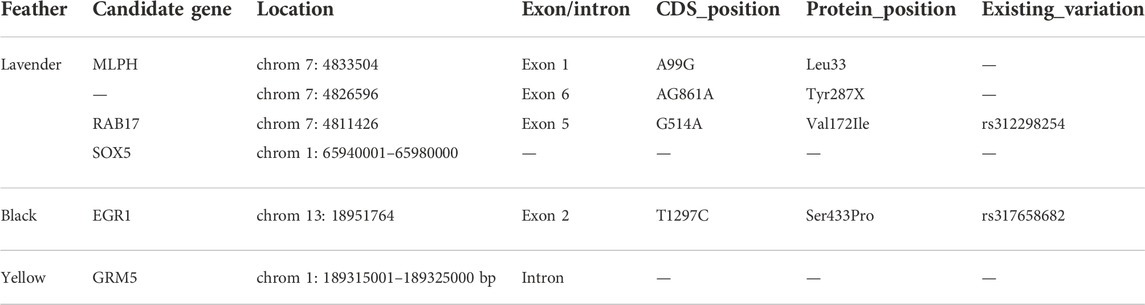
TABLE 1. Candidate genes based on the SNPs and amino acid substitutions in chicken with black/lavender/yellow plumage .
The remaining two plumage color loci at chromosome 1 exhibited the strongest association signals within the intronic regions of GRM5 and SOX5, and they are obvious candidates for controlling normal variation in chicken plumage colors (Supplementary Tables S11, S12). However, GRM5 lies 33.82 kb upstream of TYR, which is also involved in pigmentation, which could affect pigmentation by regulating the expression of TYR (Sandra Beleza et al., 2013; Smyth et al., 2006). Therefore, we hypothesized that the non-coding DNA variation of GRM5 and SOX5 might affect pigmentation by regulating the expression of neighboring genes or influence itself expression by regulating the enhancer activity of the genes themselves. Candidate genes based on the SNPs and amino acid substitutions in black, lavender, and yellow feathers in Baicheng You chicken are summarized in Table 1.
In our study of the three feather colors of Baicheng You chicken, we focused specifically on the loci that may play a more important role in the evolution of feather color through natural, sexual, and artificial selection. Here, we used comparative genome analysis between black, yellow, and lavender sub-populations to identify the genetic basis underlying the variation in feather color among Baicheng You chicken. In this study, several previously reported genes were found to be involved in lavender feather-related traits, such as MLPH, RAB17, and SOX5. In previously published articles, non-synonymous mutation (c. 103 C>T) (Vaez et al., 2008) and (c. 1909 A>G) (Xu et al., 2016) of MLPH resulted in the development of the lavender or light-gray plumage color phenotype in chicken. In this study, we did not find SNP (c.1909 A>G), but a frameshift deletion (c. 861 indel G) was located at exon 6 of MLPH, which leads to a completely different protein. Vaez et al. found that the absence of exon 6 in MLPH gene transcript in lavender phenotype chickens (Vaez et al., 2008), and frameshift deletion in exon 5 of MLPH gene has similarly been reported in rabbit (Oryctolagus cuniculus) breeds (Fontanesi et al., 2014). Deletions have also been found in leaden mice and cat (Ishida et al., 2006). The variations in MLPH gene could cause transport defect melanosomes in melanocytes, resulting in the dilution of coat or plumage colors in cats (Ishida et al., 2006), dogs (Bauer et al., 2018; Drögemüller et al., 2007; Van Buren et al., 2020), cattle (Li et al., 2016), sheep (Posbergh et al., 2020), rabbits (Demars et al., 2018; Jia et al., 2021), and minks (Cirera et al., 2013), as well as Griscelli syndrome type-3 in humans (Al-Mousa et al., 2016; Çağdaş et al., 2012; Gironi et al., 2019; Jo et al., 2020; Ménasché et al., 2005). McMurtrie et al. reported that RAB17 is an attractive candidate for leaden mice, though failed to identify any RAB17-coding region mutations, because it was expressed in polarized epithelial cells (E. B. McMurtrie et al., 1997). A non-synonymous substitution (G514A) in exon 5 of RAB17 gene by whole genome re-sequencing was detected, resulting in change in the amino acid Val172Ile in this study.
The transcription factor SOX5 has been detected in the early migrating neural crest of chicken (Perez-Alcala et al., 2004). There are two explanations for the regulation of transcription factor SOX5 on melanocytes. A 7.6-kb non-coding deletion near SOX10 was found to be associated with pale yellow feathers (Zhu et al., 2022), and SOX5 played a role in melanocyte lineage by regulating the activity of transcription factor SOX10 (Min et al., 2022). The other was up-regulation of the expression of TYR through MITF, thereby reducing the synthesis of melanin (Wang et al., 2016).
In chicken, MC1R is a single exon gene without intron on chromosome 11, and it is associated with the traditional feather color Extension locus (E). Although MC1R is among the genes that have been extensively studied for their association with feather color, we did not identify it except for in the black vs. yellow + lavender model. We had the major gene EGR1 of black plumage by XP-CLR algorithm, and we hypothesized that the variation loci T1297C (Ser433Phr) in EGR1 could be associated with the synthesis of eumelanin. EGR1, a member of the immediate early family, is an important nuclear transcription factor (Gómez-Martín et al., 2010). EGR1 mRNA was highly expressed in hair follicles in the process of mouse embryogenesis (Mcmahon et al., 1990). EGR1 could also up-regulate the expression of TYR by binding MC1R, leading to the synthesis of large amounts of eumelanin.
In this study, GRM5 was associated with yellow plumage color. Our results coincided with those of Mastrangelo et al. (2020). In recent years, studies of human skin and eye color have found that the skin color loci at 11q14.3 is strongly correlated with the intronic regions of GRM5, and non-synonymous mutation in TYR (S192Y) is in linkage disequilibrium with the most significant SNPs in GRM5 (Sandra Beleza et al., 2013). Kaustubh Adhikari et al. have also demonstrated the presence of multiple independent signals of association in GRM5/TYR (Adhikari et al., 2019; Miao et al., 2017). GRM5 may affect pigmentation by regulating the expression of TYR.
In our study, we showed that MLPH, RAB17, SOX5, EGR1, and GRM5 were candidate pigmentation genes in Baicheng You chicken breed, using whole genome data. Notably, mutations of MLPH (AG861A), RAB17 (G583A), and SOX5 have been found to be related to lavender plumage. EGR1 (T1297C) was a most plausible candidate gene for black plumage, and GRM5 for yellow plumage in Baicheng You chickens. Overall, the candidate genes identified herein could help elucidate the structure and composition of the genome underlying plumage colors and provide novel insights into the regulation mechanisms of feather color development in Baicheng You chicken, as well as other chicken breeds.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Ministry of Agriculture of China (Beijing, China) and Animal Welfare Committee of China Agricultural University (Beijing, China). Written informed consent was obtained from the owners for the participation of their animals in this study. Written informed consent was obtained from the owners for the participation of their animals in this study.
LQ, MG, and HL conceived and designed the experiments. JW, TZ, JZ, and XZ performed the experiments. HW and JW analyzed the data. HW wrote the manuscript. JW, CT, LQ, and MG revised the manuscript. All authors read and approved the final version of the manuscript.
This study was supported by Xinjiang Production and Construction Corps Key Laboratory of Protection and Utilization of Biological Resources in Tarim Basin Open Fund Project “Evaluation and Utilization of Genetic Resources in Baicheng You chicken by Using Genomic Information” (BRZD2104); the Graduate Research Innovation Project of Tarim University “MLPH gene variation and the formation mechanism of lavender feather in Baicheng You chicken” (TDBSCX202109); and the Beijing Innovation Team of the Modem Agro-industry Technology Research System (BAIC04-2022).
We appreciate the chicken farmers for chicken sampling.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.985228/full#supplementary-material
Adhikari, K., Mendoza-Revilla, J., Sohail, A., Fuentes-Guajardo, M., Lampert, J., Chacón-Duque, J. C., et al. (2019). A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat. Commun. 10 (1), 358. doi:10.1038/s41467-018-08147-0
Al-Mousa, H., Abouelhoda, M., Monies, D. M., Al-Tassan, N., Al-Ghonaium, A., Al-Saud, B., et al. (2016). Unbiased targeted next-generation sequencing molecular approach for primary immunodeficiency diseases. J. Allergy Clin. Immunol. 137 (6), 1780–1787. doi:10.1016/j.jaci.2015.12.1310
Alzahofi, N., Welz, T., Robinson, C. L., Page, E. L., Briggs, D. A., Stainthorp, A. K., et al. (2020). Rab27a co-ordinates actin-dependent transport by controlling organelle-associated motors and track assembly proteins. Nat. Commun. 11 (1), 3495. doi:10.1038/s41467-020-17212-6
Asadollahpour, N. H., Esmailizadeh, A., Ayatollahi, M. A., Han, J., Wu, D. D., Li, Y., et al. (2020). Comparative population genomic analysis uncovers novel genomic footprints and genes associated with small body size in Chinese pony. BMC Genomics 21 (1), 496. doi:10.1186/s12864-020-06887-2
Bao, Q., Ma, X., Jia, C., Wu, X., Wu, Y., Meng, G., et al. (2022). Resequencing and signatures of selective scans point to candidate genetic variants for hair length traits in long-haired and normal-haired Tianzhu white yak. Front. Genet. 13, 798076. doi:10.3389/fgene.2022.798076
Bauer, A., Kehl, A., Jagannathan, V., and Leeb, T. (2018). A novel MLPH variant in dogs with coat colour dilution. Anim. Genet. 49 (1), 94–97. doi:10.1111/age.12632
Baxter, L. L., Watkins Chow, D. E., Pavan, W. J., and Loftus, S. K. (2019). A curated gene list for expanding the horizons of pigmentation biology. Pigment. Cell Melanoma Res. 32 (3), 348–358. doi:10.1111/pcmr.12743
Çağdaş, D., Özgür, T. T., Asal, G. T., Tezcan, İ., Metin, A., Lambert, N., et al. (2012). Griscelli syndrome types 1 and 3: Analysis of four new cases and long-term evaluation of previously diagnosed patients. Eur. J. Pediatr. 171 (10), 1527–1531. doi:10.1007/s00431-012-1765-x
Chen, H., Patterson, N., and Reich, D. (2010). Population differentiation as a test for selective sweeps. Genome Res. 20 (3), 393–402. doi:10.1101/gr.100545.109
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi:10.1093/bioinformatics/bty560
Cieslak, M., Reissmann, M., Hofreiter, M., and Ludwig, A. (2011). Colours of domestication. Biol. Rev. Camb. Philos. Soc. 86 (4), 885–899. doi:10.1111/j.1469-185X.2011.00177.x
Cirera, S., Markakis, M. N., Christensen, K., and Anistoroaei, R. (2013). New insights into the melanophilin (MLPH) gene controlling coat color phenotypes in American mink. Gene 527 (1), 48–54. doi:10.1016/j.gene.2013.05.047
Demars, J., Iannuccelli, N., Utzeri, V., Auvinet, G., Riquet, J., Fontanesi, L., et al. (2018). New insights into the melanophilin (MLPH) gene affecting coat color dilution in rabbits. Genes 9 (9), 430. doi:10.3390/genes9090430
Drögemüller, C., Philipp, U., Haase, B., Günzel-Apel, A., and Leeb, T. (2007). A noncoding melanophilin gene (MLPH) SNP at the splice donor of exon 1 represents a candidate causal mutation for coat color dilution in dogs. J. Hered. 98 (5), 468–473. doi:10.1093/jhered/esm021
Fang, M. F. M., Larson, G. L. G., Ribeiro, H. R. H. S., Li, N. L. N., and Andersson, L. A. L. (2014). Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5, e1000341. doi:10.1371/journal.pgen.1000341
Fontanesi, L., Scotti, E., Allain, D., and Dall'Olio, S. (2014). A frameshift mutation in themelanophilin gene causes the dilute coat colour in rabbit (Oryctolagus cuniculus) breeds. Anim. Genet. 45 (2), 248–255. doi:10.1111/age.12104
Gironi, L. C., Zottarelli, F., Savoldi, G., Notarangelo, L. D., Basso, M. E., Ferrero, I., et al. (2019). Congenital hypopigmentary disorders with multiorgan impairment: A case report and an overview on gray hair syndromes. Medicina 55 (3), 78. doi:10.3390/medicina55030078
Gómez-Martín, D., Díaz-Zamudio, M., Galindo-Campos, M., and Alcocer-Varela, J. (2010). Early growth response transcription factors and the modulation of immune response: Implications towards autoimmunity. Autoimmun. Rev. 9 (6), 454–458. doi:10.1016/j.autrev.2009.12.006
Gross, S. P., Tuma, M. C., Deacon, S. W., Serpinskaya, A. S., Reilein, A. R., and Gelfand, V. I. (2002). Interactions and regulation of molecular motors in Xenopus melanophores. J. Cell Biol. 156 (5), 855–865. doi:10.1083/jcb.200105055
Ishida, Y., David, V. A., Eizirik, E., Schäffer, A. A., Neelam, B. A., Roelke, M. E., et al. (2006). A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics 88 (6), 698–705. doi:10.1016/j.ygeno.2006.06.006
Jia, X., Ding, P., Chen, S., Zhao, S., Wang, J., and Lai, S. (2021). Analysis of MC1R, MITF, TYR, TYRP1, and MLPH genes polymorphism in four rabbit breeds with different coat colors. Animals. 11 (1), 81. doi:10.3390/ani11010081
Jiang, S. V. A. E., David, G., Li, V. S. L. B., Wang, H., and Balaji, A. (2020). Microtubule motor transport in the delivery of melanosomes to the actin-rich apical domain of the retinal pigment epithelium. J. Cell Sci. 133. doi:10.1242/jcs.242214
Jiang, F., Lin, R., Xiao, C., Xie, T., Jiang, Y., Chen, J., et al. (2021). Analysis of whole-genome re-sequencing data of ducks reveals a diverse demographic history and extensive gene flow between Southeast/South Asian and Chinese populations. Genet. Sel. Evol. 53 (1), 35. doi:10.1186/s12711-021-00627-0
Jo, C. S., Park, H. I., Jung, H. J., Park, J. I., Lee, J. E., Myung, C. H., et al. (2020). A novel function of Prohibitin on melanosome transport in melanocytes. Theranostics 10 (9), 3880–3891. doi:10.7150/thno.41383
Kerje, S., Lind, J., Schutz, K., Jensen, P., and Andersson, L. (2003). Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim. Genet. 34 (4), 241–248. doi:10.1046/j.1365-2052.2003.00991.x
Lee, W., Park, K., Taye, M., Lee, C., Kim, H., Lee, H., et al. (2018). Analysis of cross-population differentiation between Thoroughbred and Jeju horses. Asian-Australas. J. Anim. Sci. 31 (8), 1110–1118. doi:10.5713/ajas.17.0460
Li, H., Ruan, J., and Durbin, R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 (11), 1851–1858. doi:10.1101/gr.078212.108
Li, W., Sartelet, A., Tamma, N., Coppieters, W., Georges, M., and Charlier, C. (2016). Reverse genetic screen for loss-of-function mutations uncovers a frameshifting deletion in themelanophilin gene accountable for a distinctive coat color in Belgian Blue cattle. Anim. Genet. 47 (1), 110–113. doi:10.1111/age.12383
Ludwig, A., Pruvost, M., Reissmann, M., Benecke, N., Brockmann, G. A., Castaños, P., et al. (2009). Coat color variation at the beginning of horse domestication. Science 324 (5926), 485. doi:10.1126/science.1172750
Lyu, G., Feng, C., Zhu, S., Ren, S., Dang, W., Irwin, D. M., et al. (2021). Whole genome sequencing reveals signatures for artificial selection for different sizes in Japanese primitive dog breeds. Front. Genet. 12, 671686. doi:10.3389/fgene.2021.671686
MargotBowen, E. E. D. B. I., Luisa, B., Superti-Furga, A., Shiro, I., Valerie, C., Judith, V. B. E., et al. (2011). Loss-of-Function mutations in PTPN11Cause metachondromatosis, but not ollier disease or maffucci syndrome. PLoS Genet. 7, e1002050. doi:10.1371/journal.pgen.1002050
María Inés Fariello, S. B. H. N., Boitard, S., Naya, H., SanCristobal, M., and Servin, B. (2013). Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 3 (193), 929–941. doi:10.1534/genetics.112.147231
Mastrangelo, S., Cendron, F., Sottile, G., Niero, G., Portolano, B., Biscarini, F., et al. (2020). Genome-Wide analyses identifies known and new markers responsible of chicken plumage color. Animals. 10 (3), 493. doi:10.3390/ani10030493
Mcmahon, A. P., Champion, J. E., Mcmahon, J. A., and Sukhatme, V. P. (1990). Developmental expression of the putative transcription factor Egr-1 suggests that Egr-1 and c-fos are coregulated in some tissues. Dev. Camb. 108 (2), 281–287. doi:10.1242/dev.108.2.281
McMurtrie, E. B., Barbosa, M. D. F. S., S, M. Z. A., and Kingsmore, S. F. (1997). Rab17 and rab18, small GTPases with specificity for polarized epithelial cells: Genetic mapping in the mouse. Genomics 45, 623–625. doi:10.1006/geno.1997.4959
Ménasché, G., Hsuan Ho, C., Sanal, O., Feldmann, J., Tezcan, I., Ersoy, F., et al. (2005). Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1). J. Clin. Invest. 115 (4), 1100. doi:10.1172/JCI18264C1
Miao, Y., Soudy, F., Xu, Z., Liao, M., Zhao, S., and Li, X. (2017). Candidate gene identification of feed efficiency and coat color traits in a C57BL/6J × kunming f2 mice population using Genome-Wide association study. Biomed. Res. Int. 2017, 7132941. doi:10.1155/2017/7132941
Min, Y., Li, Q., and Yu, H. (2022). Heme-peroxidase 2 modulated by POU2F1 and SOX5 is involved in pigmentation in pacific oyster (Crassostrea gigas). Mar. Biotechnol. 24 (2), 263–275. doi:10.1007/s10126-022-10098-z
Moreiras, H., Seabra, M. C., and Barral, D. C. (2021). Melanin transfer in the epidermis: The pursuit of skin pigmentation control mechanisms. Int. J. Mol. Sci. 22 (9), 4466. doi:10.3390/ijms22094466
Nadeau, N. J., Minvielle, F., and Mundy, N. I. (2006). Association of a Glu92Lys substitution in MC1R with extended Brown in Japanese quail (Coturnix japonica). Anim. Genet. 37 (3), 287–289. doi:10.1111/j.1365-2052.2006.01442.x
Pavlidis, P., Živković, D., Stamatakis, A., and Alachiotis, N. (2013). SweeD: Likelihood-Based detection of selective sweeps in thousands of genomes. Mol. Biol. Evol. 30 (9), 2224–2234. doi:10.1093/molbev/mst112
Perez-Alcala, S., Nieto, M. A., and Barbas, J. A. (2004). LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Dev. Camb. 131 (18), 4455–4465. doi:10.1242/dev.01329
Posbergh, C. J., Staiger, E. A., and Huson, H. J. (2020). A Stop-Gain mutation within MLPH is responsible for the lilac dilution observed in jacob sheep. Genes (Basel) 11, 618. doi:10.3390/genes11060618
Roulin, A., and Ducrest, A. (2013). Genetics of colouration in birds. Semin. Cell Dev. Biol. 24 (6-7), 594–608. doi:10.1016/j.semcdb.2013.05.005
Sandra Beleza, N. A. J. S., Jailson Lopes, J. C. I. I., Magnus Nordborg, A. N. C. E., Tang, H., and Coram, M. A. (2013). Genetic architecture of skin and eye color in an african- European admixed population. PLoS Genet. 9 (3), 1003372. doi:10.1371/journal.pgen.1003372
Schiöth, H. B., Raudsepp, T., Ringholm, A., Fredriksson, R., Takeuchi, S., Larhammar, D., et al. (2003). Remarkable synteny conservation of melanocortin receptors in chicken, human, and other vertebrates. Genomics 81 (5), 504–509. doi:10.1016/S0888-7543(03)00028-4
Smyth, I. M., Wilming, L., Lee, A. W., Taylor, M. S., Gautier, P., Barlow, K., et al. (2006). Genomic anatomy of the tyrp1 (Brown) deletion complex. Proc. Natl. Acad. Sci. U. S. A. 103 (10), 3704–3709. doi:10.1073/pnas.0600199103
Stolt, C. C., Lommes, P., Hillgartner, S., and Wegner, M. (2008). The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res. 36 (17), 5427–5440. doi:10.1093/nar/gkn527
Takeuchi, S., Suzuki, H., Hirose, S., Yabuuchi, M., Sato, C., Yamamoto, H., et al. (1996). Molecular cloning and sequence analysis of the chick melanocortin 1-receptor gene. Biochim. Biophys. Acta 1306 (2), 122–126. doi:10.1016/0167-4781(96)00026-7
Taye, M., Yoon, J., Dessie, T., Cho, S., Oh, S. J., Lee, H., et al. (2018). Deciphering signature of selection affecting beef quality traits in Angus cattle. Genes Genomics 40 (1), 63–75. doi:10.1007/s13258-017-0610-z
Vaez, M., Follett, S. A., Bed'Hom, B., Gourichon, D., Tixier-Boichard, M., and Burke, T. (2008). A single point-mutation within the melanophilin gene causes the lavender plumage colour dilution phenotype in the chicken. BMC Genet. 9, 7. doi:10.1186/1471-2156-9-7
Van Buren, S. L., Minor, K. M., Grahn, R. A., Mickelson, J. R., Grahn, J. C., Malvick, J., et al. (2020). A third MLPH variant causing coat color dilution in dogs. Genes (Basel) 11 (6), E639. doi:10.3390/genes11060639
Wang, P., Zhao, Y., Fan, R., Chen, T., and Dong, C. (2016). MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting sox5 in mouse skin melanocytes. Int. J. Mol. Sci. 17 (7), 959. doi:10.3390/ijms17070959
Xiangtao Liu, S. H. Z. W., Joel, G. B. Y., Wang, Z., Gelernter, J., and Yang, B. Z. (2013). Variant callers for next-generation sequencing data: A comparison study. PLoS One 8 (9), 75619. doi:10.1371/journal.pone.0075619
Xu, J. G., Xie, M. G., Zou, S. Y., Liu, X. F., Li, X. H., Xie, J. F., et al. (2016). Interactions of allele E of the MC1R gene with FM and mutations in the MLPH gene cause the five-gray phenotype in the Anyi tile-like gray chicken. Genet. Mol. Res. 15 (2). doi:10.4238/gmr.15027633
Keywords: plumage colors, genome, chicken, pigmentation genes, genetic marker
Citation: Wang H, Wen J, Li H, Zhu T, Zhao X, Zhang J, Zhang X, Tang C, Qu L and Gemingguli M (2022) Candidate pigmentation genes related to feather color variation in an indigenous chicken breed revealed by whole genome data. Front. Genet. 13:985228. doi: 10.3389/fgene.2022.985228
Received: 03 July 2022; Accepted: 10 October 2022;
Published: 21 November 2022.
Edited by:
Salvatore Mastrangelo, University of Palermo, ItalyReviewed by:
Filippo Biscarini, National Research Council (CNR), ItalyCopyright © 2022 Wang, Wen, Li, Zhu, Zhao, Zhang, Zhang, Tang, Qu and Gemingguli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lujiang Qu, cXVsdWpAMTYzLmNvbQ==; M. Gemingguli, Z21nbC0xMTNAZm94bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.