
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 10 August 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.983564
This article is part of the Research TopicEpigenetic Regulation and Non-histone Post-translational Modification in CancerView all 15 articles
Breast cancer (BC) has continued to be the leading cause of cancer deaths in women, accompanied by highly molecular heterogeneity. N6-methyladenosine (m6A), a methylation that happens on adenosine N6, is the most abundant internal mRNA modification type in eukaryotic cells. Functionally, m6A methylation is a reversible modification process and is regulated by 3 enzymes with different functions, namely “writer”, “reader”, and “eraser”. Abnormal m6A modifications trigger the expression, activation, or inhibition of key signaling molecules in critical signaling pathways and the regulatory factors acting on them in BC. These m6A-related enzymes can not only be used as markers for accurate diagnosis, prediction of prognosis, and risk model construction, but also as effective targets for BC treatment. Here, we have emphasized the roles of different types of m6A-related enzymes reported in BC proliferation, invasion, and metastasis, as well as immune regulation. The comprehensive and in-depth exploration of the molecular mechanisms related to m6A will benefit in finding effective potential targets and effective stratified management of BC.
Breast cancer (BC) has continued to be the leading cause of cancer deaths in women, and its incidence rates are still increasing globally (Sung et al., 2021). BC is a highly molecularly heterogeneous tumor type and is continually associated with headaches, such as recurrence, metastasis, and drug resistance (Yau et al., 2022) (Derose et al., 2011). These features cause certain bottlenecks in the diagnosis and treatment of BC. Therefore, exploring the molecular mechanism of the occurrence and development of BC, as well as the capability for the early diagnosis of BC, treatment monitoring, or the search for effective potential targets, is of great significance for the effective stratified management of BC and the development of new diagnosis and treatment methods (Ellis and Perou, 2013).
Epigenetic regulation, represented by N6-methyladenosine (m6A) modification, histone modification, DNA methylation, chromatin remodeling, and non-coding RNA (ncRNA) regulation, plays an overwhelming role in almost all biological behaviors, including cell differentiation and tissue development, and tumor progression (Wiener and Schwartz, 2021). M6A, a methylation that happens on adenosine N6, is the most abundant internal mRNA modification type in eukaryotic cells (Hu et al., 2022). There is a lot of evidence that m6A modification is an emerging important molecular modulation for tumors. In mammals, m6A modifications are known as a reversible dynamic process to influence different dimensions of RNA expression, including regulation of mRNA stability, splicing, translation efficiency, nuclear export, and degradation (Dierks et al., 2021). Functionally, this reversible m6A methylation modification is regulated by 3 enzymes with different functions, namely “writer”, “reader”, and “eraser” (Uddin et al., 2021). Among them, m6A can be installed by the methyltransferase complex, namely, writers, which include METTL3, METTL14, WTAP, RBM15/RBM15B, HAKAI, ZC3H13, and VIRMA/KIAA1429 (Huang et al., 2020). M6A eraser is able to remove methylation from m6A-modified RNA, mainly including FTO, ALKHB5, and ALKHB3 (Zhou et al., 2020). M6A reader is a selective RNA-binding protein that is responsible for catalyzing for recognizing m6A to activate downstream pathways, including YTH domain family 1-3 (YTHDF1, YTHDF1-2, YTHDF1), eIF3, IGF2BP1-3, hnRNPC, and hnRNPA2B1 (Dai et al., 2021) (Figure 1).
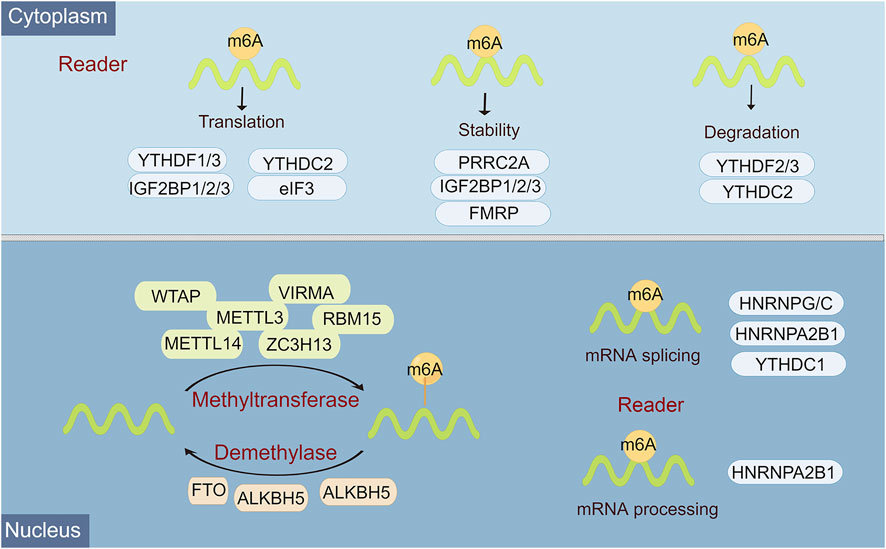
FIGURE 1. Overview of the classification and molecular mechanisms of m6A methylation. m6A RNA methylation is regulated by 3 different key enzymes, corresponding writers, erasers, and readers, which perform the functions of adding, deleting, or recognizing m6A, respectively. The consequences of m6A methylation lead to multiple processes in RNA metabolism and expression, including RNA splicing, miRNA processing, nuclear export, translation, stability, and RNA decay.
The enzymes involved in m6A modifications have been implicated in the regulation of gene expression and tumor evolution, including carcinogenesis, metastasis, and progression, especially in BC. For example, YTHDF1 overexpression is a not desirable signature for BC patients and is linked to lower immune infiltrate and poor clinical outcomes, while YTHDF1 inhibition promotes the proliferation, migration, and invasion in BC cell lines (Li et al., 2022). The results of sequencing data mining showed that m6A and its target genes have corresponding changes at the gene and protein levels in tumors, endowing the potential to indicate the prognosis of BC. In a bioinformatics analysis, high expression of IGF2BP1, a key m6A regulator, was often associated with shorter overall survival (OS) in BC patients. This suggests that IGF2BP1 is an independent prognostic factor in BC (Zhong et al., 2021). Besides, Chen et al. confirmed that METTL3 methylation is involved in KRT7-mediated m6A-induced BC lung metastasis (Chen et al., 2021). Furthermore, the expression profile of m6A regulators in BC is also prominently related to tumor malignancy, tumor immune score, anti-tumor immune response, and therapeutic effect (He et al., 2021). Gong et al. reported that METTL14 and ZC3H13 were positively correlated with the abundance of CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs) in BC.
Therefore, the systematic elucidation of the exact molecular mechanisms of m6A epigenetic regulation in BC progression is highly warranted. Here, we have reviewed and highlighted the roles of different types of m6A-related enzymes reported in BC proliferation, invasion, and metastasis, as well as immune regulation. Continued focus on the molecular mechanisms associated with m6A will benefit in finding effective potential targets and effective stratified management of BC.
At present, METTL3 is the most studied methylation-modified protein that plays a broad regulatory role in BC progression. METTL3 mainly participates in the biogenesis, decay, and translation control of mRNA through m6A modification (Lin et al., 2016). Both METTL3 and METTL14 have methyltransferase activity, and the methyltransferase complex formed by the two performs catalytic function during the m6A process (Yankova et al., 2021).
In TNBC, METTL3 is an important collaborator in regulating metastasis, and low expression of METTL3 is implicated in the poor prognosis of triple-negative breast cancer (TNBC) (Shi et al., 2020). As Shi et al. confirmed, this metastasis-suppressing function of METTL3 was achieved by suppressing the expression of COL3A1 and its m6A function (Shi et al., 2020). METTL3 could accelerate the protein levels of SOX2, CD133, and CD44 to maintain or promote BC cell stemness, which was triggered by the m6A modification of SOX2 mRNA by METTL3, ultimately leading to the alteration in enhanced BC invasion and migration capabilities (Xie et al., 2021). In BC lung metastasis cell lines, m6A and methyltransferase METTL3 expression was enhanced, while the expression level of demethylase FTO was reduced (Chen et al., 2021). M6A was capable of regulating lung metastasis in BC cells by regulating m6A/KRT7/KRT7-AS. The study by Wang et al. revealed a similar conclusion that METTL3 was a tumor promoter and its knockdown could inhibit tumor progression by reducing methylation levels (Wang H. et al., 2020). This mechanism of action was achieved through the Bcl-2 pathway targeted by METTL3. In addition, hepatitis B X-interacting protein (HBXIP) was identified to promote METTL3 expression by repressing miRNA let-7g, while METTL3 was simultaneously able to induce HBXIP expression (Cai et al., 2018). This mechanism caused a positive correlation between the expression of METTL3 and HBXIP in BC tissues and a positive feedback regulation phenomenon.
In the process of BC cell behavior, ncRNAs represented by miRNAs, lncRNAs, and circRNAs have been identified as very important direct regulators of METTL3. Therefore, interactions mediated by METTL3 and ncRNAs regulate the expression levels of post-transcriptionally regulated genes that determine tumor fate. METTL3 was capable of influencing the malignant behavior of BC EMT. Specifically, inhibition of METTL3 diminished the m6A modification of MALAT1, subsequently downregulated the MALAT1 expression to suppress EMT in BC by sponging miR-26b to reduce the expression of HMGA2 (Zhao et al., 2021). Fan et al. demonstrated that LINC00675 was a tumor protective factor, and its low expression was associated with higher tumor grade, lymphovascular invasion, and shorter survival (Fan and Wang, 2021). Furthermore, in vitro studies indicated that LINC00675 inhibited BC progression by suppressing miR-513b-5p in a METTL3-related m6A-dependent manner. From clinical, cellular, and tumor-bearing mouse levels, Xu et al. demonstrated that zinc finger protein 217 (ZNF217) silencing or miR-135 elevation inhibited BC cell migration, invasion, and EMT initiation (Xu et al., 2022). This was mediated by a mechanism that, ZNF217 could upregulate NANOG by reducing m6A levels through METTL3, thereby forming a miR-135/ZNF217/METTL3/NANOG axis.
LINC00958 was an overexpressed lncRNA that promoted the malignant progression of BC tumors (Rong et al., 2021). And, LINC00958 bound to miR-378a-3p to regulate YY1 expression, on the other hand, METTL3-mediated m6A modification promoted LINC00958 expression upregulation. In TNBC cell lines, the overexpressed METTL3 was an accelerator to suppress the proliferation and invasion (Ruan et al., 2021). Further validation showed that circMETTL3 served as a sponge for miR-34c-3p and exerted tumor-promoting functions by upregulating the expression of METTL3. METTL3-derived circRNAs contributed to the proliferation and invasion of BC cells, through the competitive endogenous RNA (ceRNA) effect of miR-31–5p with upregulated CDK1 (Li et al., 2021). This function was also affected by the m6A modification mechanism of circMETTL3, which included METTL3.
METTL14 is an important RNA methyltransferase that serves an essential and significant role in the growth of tumors by regulating RNA expression. METTL14 has been demonstrated to be a core component of the m6A methyltransferase complex and is implicated in the dynamic and reversible process of m6A modification (Zhu X. et al., 2021).
LINC00942 (LNC942) might function as an oncogene that promotes BC cell proliferation, and colony formation and inhibits apoptosis (Sun et al., 2020). In BC cells, LINC00942 increased the METTL14-mediated m6A methylation and its associated mRNA stability, as well as the CXCR4 and CYP1B1 expression of CXCR4 and CYP1B1, revealing a novel LNC942-METTL14-CXCR4/CYP1B1 regulatory axis. Zhao et al. demonstrated that silencing of lncRNA UCA1 suppressed DNA methylation of RNA methyltransferase METTL14 (Zhao et al., 2022). This event promoted m6A modification of miR-375, leading to reduced SOX12 expression and eventually restrained BC proliferation and invasion. METTL14 is also an m6A methyltransferase that is significantly elevated in BC tissues. When METTL14 was overexpressed or its activity was inhibited, the invasive ability of tumor cells becomes enhanced or weakened, accordingly (Yi et al., 2020). The abnormal expression of METTL14 reconstructed the miRNA expression profile of BC cells, and mainly regulated the cell adhesion and invasion ability by regulating the expression of Hsa-miR-146A-5p.
Methyltransferase-like 5 (METTL5), can catalyze mA modification of 18S rRNA at adenosine 1832 (mA) in a critical position in the decoding center, possing the ability in regulating mRNA translation for impacting on cell growth (Sepich-Poore et al., 2022). In BC, METTL5 also exhibited a pattern of elevated expression and was required for the maintenance of BC cell lineage growth, reproduction, and S6K activation (Rong et al., 2020). The study by Rong et al. demonstrated that METTL5 was an 18S rRNA A1832-specific methyltransferase and was capable of regulating ribosome function via multiple models.
KIAA1429, also known as VIRMA, is recognized as the largest m6A methyltransferase and is employed as a scaffold for the catalytic core component of the bridging m6A methyltransferase complex (Lan et al., 2019). KIAA1429 plays an instrumental function in m6A modification and has previously been found to be dysregulated in a variety of cancer types. KIAA1429 is considered to be involved in BC carcinogenesis and progression. Zhang et al. showed that KIAA1429 was a significant promoter of tumor invasion and metastasis in vitro and in vivo, and affected the course of BC in a non-m6A-regulated manner (Zhang et al., 2022). KIAA1429 failed to interfere with m6A levels of SMC1A mRNA, implying that m6A modifications did not affect the interplay between KIAA1429 and SMC1A mRNA. KIAA1429 directly bound to the 3′-UTR of SMC1A mRNA, leading to the stability enhancement of SMC1A mRNA. KIAA1429 showed an interesting expression pattern with high expression in tumor entities but low expression in nontumorous tissues (Qian et al., 2019). In terms of prognosis, the high expression of KIAA1429 was often associated with a lower OS. Mechanistically, KIAA1429 plays a carcinogenic role in BC progression by regulating CDK1 in an m6A-independent manner. The detailed mechanisms of m6A writer in regulating BC progression could be seen in Table 1.
The fat mass and obesity-associated (FTO) gene is a well-known prominent factor in predicting obesity and is the first m6A eraser to be discovered in eukaryotic cells. FTO is responsible for controlling fatty acid transport, adipogenesis, fat metabolism, and obesity susceptibility. Single nucleotide polymorphisms (SNPs) of the FTO gene might be associated with various functions in different BC subtypes (Montazeri et al., 2022). It has been demonstrated that FTO expression is deregulated in a variety of tumors, including acute myeloid leukemia (AML), gastric cancer (GC), cervical squamous cell carcinoma (CSCC), ovarian cancer (OC), and BC (Deng et al., 2018).
As a key m6A demethylase, FTO is usually and aberrantly expressed up-regulated in BC tissues. High expression of FTO implies poor BC patient prognosis. Niu et al. determined that FTO remarkably contributed to BC cell proliferation and metastasis via the downregulation of tumor suppressor BNIP3, which involved FTO-mediated m6A demethylation in the 3′UTR of BNIP3 mRNA. Xu et al. demonstrated that in HER2-positive BC, the high FTO expression was linked to tumor progression, lymph node metastasis, TNM staging, and poor prognosis (Xu et al., 2020). In vitro experiments have similarly shown that FTO is a tumor-promoting factor that activates miR-181b-3p/ARL5B signaling leading to tumor migration.
AlkB homolog 5 (ALKBH5) is another key m6A demethylase for gene transcription, translation, and metabolism, and is regarded as an effective biomarker for various diseases, especially cancers (Zhang et al., 2017). ALKBH3 preferentially acts on m6A in tRNA sites.
For instance, Wang et al. have previously reported that ALKBH5 could specifically regulate the function of AML leukemia stem cells without affecting normal hematopoietic stem cells (HSCs) (Wang J. et al., 2020). This highlighted the crucial role of ALKBH5 in maintaining cancer stem cell (CSC) renewal and cancer occurrence and development through the KDM4C-ALKBH5-AXL Signaling Axis. Hypoxia induces a series of stress changes to support the stable viability of cells, including changes in cytokine secretion profiles, and post-transcriptional and translational regulation. Reprogramming of the mA epitranscriptome is vital for the formation of the transcriptome and proteome in the setting of hypoxia (Wang et al., 2021). Similarly, Zhang et al. showed that hypoxia-induced an enhancement of NANOG mRNA and protein expression and breast CSC phenotype in a HIF- and ALKBH5-dependent manner, and that ALKBH5 deficiency was a debilitating factor for the hypoxia-induced BC CSC enrichment (Zhang et al., 2016).
YTHDF1-3 are the three major m6A binding proteins and the most widely studied and versatile m6A readers (Chen et al., 2019). YTHDF1-3, containing special YTH domains, possess the capabilities of targeting and recognizing m6A-modified RNAs and mediating their degradation. YTHDF1 is a typical and highly expressed m6A reader protein in BC tissues and cell lines, and its high expression status is thought to be indicative of tumor size, metastasis, poor prognosis, and chemotherapy resistance (Anita et al., 2020). YTHDF1 is involved in almost the whole process of tumor biological behavior, and plays an important role in regulating transcription, translation, protein synthesis, angiogenesis, and EMT. Intriguingly, YTHDF1 as a target of tumor immune regulation has also attracted much attention.
Chen et al. showed that YTHDF1 promoted YTHDF1/FOXM1 to enhance FOXM1 expression, which in turn intensified the proliferation, invasion, and EMT phenotype of BC cells (Chen et al., 2022). Another 2022 similar study showed that a series of cascade reactions triggered by YTHDF1 were important molecular events in driving BC. Specifically, HIF1α expression could be induced and miR-16-5p levels were suppressed in a hypoxic microenvironment, resulting in upregulated YTHDF1 expression (Yao et al., 2022). Also, inhibition of YTHDF1 was able to promote the down-regulation of the glycolytic gene PKM2 to reduce BC glycolytic activity and lead to tumorigenicity and metastasis inhibition. These studies suggest that YTHDF1 can be used in a variety of regulatory pathways to modulate BC progression, and that depletion or targeted inhibition of YTHDF1 is a potentially efficient BC therapeutic strategy.
YTHDF2 is an N-methyladenosine-binding protein and can modulate mRNA stability, thus impacting central nervous system responses, embryonic development, and tumor evolution. Tumor biology studies have shown that YTHDF2 can modulate m6A modification to regulate downstream signaling molecules to regulate tumor cell proliferation, invasion, and migration (Shen et al., 2021). For instance, SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity to m6A-modified mRNA (Hou et al., 2021). Einstein et al. uncovered a mechanism associated with RNA-binding proteins (RBPs), that the suppression of YTHDF2 initiated proteotoxic cell death pattern in MYC-driven TNBC (Einstein et al., 2021). This work not only demonstrated that YTHDF2, aberrantly expressed RBP and its mode of interaction with RNA were essential for BC cell growth, but that targeting YTHDF2 and specific RBP possessed outstanding BC therapeutic potential. The important mechanism of post-translational protein modification of YTHDF2, fully confirmed that YTHDF2 has a delicate manipulation between the regulation of protein post-translational modification and RNA chemical modification.
YTHDF3, in combination with YTHDF1 and YTHDF2, has a crucial effect in enhancing the synthesis of m6A-modified mRNAs in the cytoplasm (Shi et al., 2017). In TNBC subtypes, YTHDF3 expression was associated with poorer disease-free survival (DFS) and overall survival (OS) in patients (Lin et al., 2022). YTHDF3 could intensify the ZEB1 mRNA stability in an m6A-dependent manner, consequently leading to BC cell growth and EMT (Lin et al., 2022).
Intriguingly, the high expression level of YTHDF3 was also closely related to the prognosis of patients with breast cancer brain metastases (BCBMs). By enhancing the translation of m6A-enriched transcripts of ST6GALNAC5, GJA1, and EGFR, YTHDF3 promoted the communication between BC cells, endothelial cells, astrocytes, and tumor metastasis phenotypes represented by angiogenesis (Chang et al., 2020). Therefore, YTHDF3 could affect cascade steps in the BCBM, and then domesticate the evolution of BC cell changes in TME toward inducing brain metastatic polarity.
Totally, different binding proteins selectively recognize m6A-modified RNAs for achieving gene expression regulation. YTHDF1 is conducive to the enhanced mRNA translation, YTHDF2 is responsible for mRNA degradation, and YTHDF3 precipitates in the translation and degradation via the reciprocity actions with YTHDF1 and YTHDF2. This means that YTHDF3 can both collaborate with YTHDF1 to catalyze the translation of methylated RNAs, or directly engage with YTHDF2 to accelerate the decay of mRNAs. Thus, YTHDF1, YTHDF2, and YTHDF3 exert irreplaceable functions to foster BC progression and potentially even become robust therapeutic targets for prognostic stratification and effective treatment of BC.
As a post-transcriptional fine regulator, IGF2BP1 plays a role in remodeling tumor growth, chemotherapy resistance, and macroscopically, OS and recurrence of tumor patients. GF2BP1 potentiates tumor malignant progression in a variety of solid tumors and exhibits a poor prognostic indicative value (Glaß et al., 2021). The principal action of IGF2BP1 in oncogenic cells is to stabilize mRNA encoding oncogenic factors. In pan-cancer studies, the high expression and tumor-promoting characteristics of IGF2BP1 in specific tumors make it a promising therapeutic target, but it is also inhibitory in some tumors (Huang et al., 2018).
Zhu et al. identified a hypoxia-induced lncRNA KB-1980E6.3, that exhibited abnormal BC tissue upregulation and was associated with a poor prognosis (Zhu P. et al., 2021). LncRNA KB-1980E6.3 increased the stability of c-Myc mRNAs by binding to m6A reader IGF2BP1 and consequently maintained the stemness of BCSCs. Interrupting this mechanism was of the potential to provide a therapeutic strategy for hypoxic tumors. MIR210HG acts as an oncogenic lncRNA highly expressed in BC tissue, and could promote BC metastasis, by inhibiting its encoded miR-210 (Shi et al., 2022). Moreover, MYCN directly activated IGF2BP1, and both IGF2BP1 and ELAVL1 strengthened the MIR210HG stability, resulting in a MYCN/IGF2BP1/MIR210HG regulatory axis.
Immune cells, secreted factors, and the tumor immune microenvironment in which they are intertwined are indispensable key links in the anti-tumor response (Liu et al., 2021). Considerable evidence suggests that m6A is involved in processes that regulate innate and adaptive immune cells, which in turn have been assigned roles in anti-inflammatory, anti-infective, and anti-tumor immunity (Ma et al., 2021). There is also much literature based on the existing reported m6A regulators, mining the correlation and prognostic scoring model of m6A regulators and immune infiltrating cells, for providing a novel evaluation tool for the diagnosis, prognosis, and immune status of BC (Yuan et al., 2022).
The immunomodulatory role of METTL3 in BC has been frequently reported. Yin et al. found that knockdown of METTL3 in bone marrow cells triggered malignant tumor proliferation and metastasis and exhibited an elevated abundance of M1/M2-like tumor-associated macrophages (TAMs) and Treg infiltration (Yin et al., 2021). Mechanistic studies suggested that deletion of METTL3 disrupted YTHDF1-mediated SPRED2 translation, thereby enhancing NF-kB and STAT3 activation via the ERK pathway, leading to tumor progression. Meanwhile, as the therapeutic benefit of programmed cell death receptor 1 (PD-1) inhibitor was weakened in Mettl3−/− mice, METTL3 could be a potential target for tumor immunotherapy. In addition, it has been reported that the expression of programmed cell death 1 ligand (PD-L1) was positively linked to the expression of METTL3 and IGF2BP3 in BC tissues (Wan et al., 2022). Since METTL3-mediated m6A modification could enhance PD-L1 mRNA stability through the METTL3-IGF2BP3 axis, tumor immune cell infiltrations and CD8+ T cell functions were enhanced forcefully when METTL3 or IGF2BP3 is inhibited. Ou et al. identified a specific C5aR1+ neutrophil subpopulation that potentiated BC cell glycolysis through ERK1/2-WTAP-ENO1 signaling, indicating that C5aR1+ neutrophils and the associated WTAP-ENO1 axis contribute to potential BC therapeutic target (Yin et al., 2021).
Another study reported that the co-expression network of YTHDF1 is critical in shaping immune responses, including antigen processing and presentation (Hu et al., 2021). YTHDF1 may act as a hopeful pan-cancer immune biomarker, as well as a novel promising marker for tumor immunotherapy. These results provide strong evidence that m6A modification is involved in the complex immune regulation of BC. M6A modifications can reshape antitumor immune responses by affecting immune cell state and function, and post-transcriptional regulation of specific cytokines and proteins.
Based on these current developments, it is evident that m6A plays a dual role in shaping tumor progression. Specifically, m6A regulates the expression of its target genes to influence tumor progression, and whether the target genes act as tumor promoters or tumor suppressors determines the tumor-promoting or tumor-suppressing function of m6A. The presence of m6A modifications contributes to the promotion/suppression of various cellular functions, such as precursor mRNA splicing, nuclear translocation, stability, translation, and microRNA biogenesis, as the modification represented by tumor cells and immune cells, thus remodeling the BC progression.
In terms of BC diagnosis, m6A enzymes have also demonstrated good predictive efficacy. This is due to the fact that m6A enzymes show a characteristic pattern of differential expression in different BC subtypes and BC staging classifications. There have been more than 20 risk models based on these screened m6A regulators in BC research. For example, the overexpression of YTHDF1, YTHDF3, and KIAA1429 predicted a poor prognosis in terms of overall survival (OS), and the upregulation of YTHDF3 was an independent prognostic factor for OS in BR patients (Liu et al., 2019). There is even a strong performance of m6A enzymes in the treatment efficacy and recurrent metastasis of BC.
Aberrant expression of m6A regulators, are potential indicators for BC prediction, individually or synergistically. Other reported factor-based (such as ferroptosis, autophagy, lncRNA, m6A regulator-mediated immune Genes) models are also of huge value that have been confirmed in multiple studies. He et al. constructed an m6A regulator pattern, which could be effective for predicting malignancy, outcomes, and antitumor immune response (He et al., 2021). Moreover, the established models based on both m6A and other factors are attracting more and more attention. For instance, we previously constructed a risk signature based on 6 screened m6A-related lncRNAs, including Z68871.1, AL122010.1, OTUD6B-AS1, AC090948.3, AL138724.1, EGOT (Lv et al., 2021). This model could identify the prognosis and immune state in BC. Zhang et al. also adopt 21 m6A-related lncRNAs to establish a predictive model for predicting prognostic situations and BC subtypes with different immunogenicity (Zhang et al., 2021). Most of these models tend to build more accurate and extensive prediction models based on the inclusion of M6A-related indicators, thus providing rich information on tumor malignancy, tumor metastasis and recurrence, immune microenvironment, efficacy monitoring, and drug resistance evaluation.
Moreover, m6A-related models may have superior diagnostic value by associating with other models, including death (necroptosis, autophagy, and ferroptosis) genes, immune genes, glycosylation genes, and so on. However, since most of these are studies based on database excavation, they are retrospective studies. There are still relatively few prospective studies that can actually be performed in a large sample of realistic cohorts, and need to be further corroborated in actual clinical practice to prove their credibility. Overall, as a single regulator included in the model can offer many disease information, more gene combination models potentially provide a more comprehensive for breast cancer diagnosis and therapeutic outcomes. We must first admit that the reported single or the risk models based on these screened m6A regulators, is not the replacement for traditional and classical methods such as serology, pathology, and imaging. To be more precise, the traditional multi-method routine is the gold standard for diagnosis and prediction, while the risk models based on these screened m6A regulators are potential multifactorial predictors, which can be used as a useful supplement to clinical routine evaluation methods.
In terms of m6A-targeted BC tumor therapy, the following points still deserve in-depth consideration. First, m6A can regulate tumor progression through multiple mechanisms, which has been confirmed in BC. How m6A modification dysregulation is regulated by affecting tumor stem cells, immune environment, tumor cell fate, and other multiple ways is an important theoretical basis for strengthening m6A as tumor therapy. However, the role of m6A is dual in tumors, and a comprehensive assessment of how to regulate m6A-related enzymes by inhibition or activation requires specific BC subtypes, tumor microenvironment, and other underlying diseases. Moreover, ncRNA is an important player involved in tumor regulation, and how m6A RNA modification affects ncRNA function deserves further exploration.
Secondly, the primary prerequisite for targeting m6A-modified enzymes is the resolution of the protein crystal complexes of these enzymes in order to mine and design docked high-affinity small molecules and antibodies by conformational relationships (Oerum et al., 2021). These inhibitors and antibodies, in turn, will be considered to provide a good pre-requisite for clinical targeting only after effective validation in cellular and animal experiments (You et al., 2022). Finally, numerous studies have explained the critical role of m6A recognition proteins in BC, but a considerable number of m6A regulators have not been fully validated, including RBM15/RBM15B, HAKAI, ZC3H13, ALKBH3, eIF3, hnRNPC, and hnRNPA2B1. Multi-omics information mining based on single-cell sequencing, proteomics, RNA-seq, and m6A methylation sequencing will provide comprehensive information mining for BC tumor sites and cellular models. This unreported regulator in BC is a research gap and therefore has considerable research value, including the relationship between expression abundance and prognosis, diagnostic and therapeutic potential. These can provide a more profound complement to the discovery of new molecules or previous regulatory networks. Thus, continued research is still needed to fully elucidate the role of m6A regulators in mRNA and ncRNA biology.
Finally, aberrant regulation of m6A regulatory proteins is involved in BC drug resistance and tumor immune response. Targeted m6A-based therapies will contribute to the oncological treatment of BC. However, it is worth mentioning that because the regulatory network of m6A modifications is complex and involves multiple signaling molecules and pathways, inhibition of a single molecule may lead to unintended responses. Therefore, for targeted therapies of m6A, or combination strategies with other tumor-targeting drugs, the optimal combination will lead to the best efficacy.
Overall, m6A is an important mechanism for epigenetic modifications that regulate BC progression. Abnormal m6A modifications trigger the expression, activation, or inhibition of key signaling molecules in critical signaling pathways and the regulatory factors acting on them in BC. M6A-related proteins and their targets show differentially expressed patterns in BC tissue and blood. These m6A-related genes can not only be used as markers for accurate diagnosis, prediction of prognosis, and risk model construction, but also as effective targets for BC treatment.
All authors contributed to the design of the study and the writing of the manuscript. MZ, MD, and XY designed the project and revised the manuscript. XL, QZ, and ZL performed the literature search and wrote the manuscript. JG helped revise the manuscript. All authors reviewed the manuscript and approved the final version.
This work was supported by the National Natural Science Foundation of China (81900518).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AML, acute myeloid leukemia; ALKBH5, AlkB homolog 5; BC, breast cancer; BCBMs, breast cancer brain metastases ; CSC, cancer stem cell; CSCC, cervical squamous cell carcinoma; ceRNA, competitive endogenous RNA; FTO, fat mass and obesity-associated; GC, gastric cancer; HSCs, hematopoietic stem cells; HBXIP, hepatitis B X-interacting protein; METTL5, methyltransferase-like 5; m6A, N6-methyladenosine; OC, ovarian cancer; OS, overall survival; PD-L1, programmed cell death 1 ligand; PD-1, programmed cell death receptor 1; RBPs, RNA-binding proteins; SNPs, single nucleotide polymorphisms; TNBC, triple-negative breast cancer; TAMs, tumor-associated macrophages.
Anita, R., Paramasivam, A., Priyadharsini, J. V., and Chitra, S. (2020). The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am. J. Cancer Res. 10, 2546–2554.
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 415, 11–19. doi:10.1016/j.canlet.2017.11.018
Chang, G., Shi, L., Ye, Y., Shi, H., Zeng, L., Tiwary, S., et al. (2020). YTHDF3 induces the translation of m6A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell 38, 857–871. e7. doi:10.1016/j.ccell.2020.10.004
Chen, F., Chen, Z., Guan, T., Zhou, Y., Ge, L., Zhang, H., et al. (2021). N6 -methyladenosine regulates mRNA stability and translation efficiency of KRT7 to promote breast cancer lung metastasis. Cancer Res. 81, 2847–2860. doi:10.1158/0008-5472.CAN-20-3779
Chen, H., Yu, Y., Yang, M., Huang, H., Ma, S., Hu, J., et al. (2022). YTHDF1 promotes breast cancer progression by facilitating FOXM1 translation in an m6A-dependent manner. Cell Biosci. 12, 19. doi:10.1186/s13578-022-00759-w
Chen, X.-Y., Zhang, J., and Zhu, J.-S. (2019). The role of m6A RNA methylation in human cancer. Mol. Cancer 18, 103. doi:10.1186/s12943-019-1033-z
Dai, X.-Y., Shi, L., Li, Z., Yang, H.-Y., Wei, J.-F., and Ding, Q. (2021). Main N6-methyladenosine readers: YTH family proteins in cancers. Front. Oncol. 11, 635329. doi:10.3389/fonc.2021.635329
Deng, X., Su, R., Stanford, S., and Chen, J. (2018). Critical enzymatic functions of FTO in obesity and cancer. Front. Endocrinol. 9, 396. doi:10.3389/fendo.2018.00396
Derose, Y. S., Wang, G., Lin, Y. C., Bernard, P. S., Buys, S. S., Ebbert, M. T. W., et al. (2011). Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 17, 1514–1520. doi:10.1038/nm.2454
Dierks, D., Garcia-Campos, M. A., Uzonyi, A., Safra, M., Edelheit, S., Rossi, A., et al. (2021). Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 18, 1060–1067. doi:10.1038/s41592-021-01242-z
Einstein, J. M., Perelis, M., Chaim, I. A., Meena, J. K., Nussbacher, J. K., Tankka, A. T., et al. (2021). Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell 81, 3048–3064. e9. doi:10.1016/j.molcel.2021.06.014
Ellis, M. J., and Perou, C. M. (2013). The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 3, 27–34. doi:10.1158/2159-8290.CD-12-0462
Fan, S., and Wang, L. (2021). N 6 -Methyladenosine-regulated LINC00675 suppress the proliferation, migration and invasion of breast cancer cells via inhibiting miR-513b-5p. Bioengineered 12, 10690–10702. doi:10.1080/21655979.2021.2001905
Glaß, M., Misiak, D., Bley, N., Müller, S., Hagemann, S., Busch, B., et al. (2021). IGF2BP1, a conserved regulator of RNA turnover in cancer. Front. Mol. Biosci. 8, 632219. doi:10.3389/fmolb.2021.632219
He, X., Tan, L., Ni, J., and Shen, G. (2021). Expression pattern of m6A regulators is significantly correlated with malignancy and antitumor immune response of breast cancer. Cancer Gene Ther. 28, 188–196. doi:10.1038/s41417-020-00208-1
Hou, G., Zhao, X., Li, L., Yang, Q., Liu, X., Huang, C., et al. (2021). SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 49, 2859–2877. doi:10.1093/nar/gkab065
Hu, J., Qiu, D., Yu, A., Hu, J., Deng, H., Li, H., et al. (2021). YTHDF1 is a potential pan-cancer biomarker for prognosis and immunotherapy. Front. Oncol. 11, 607224. doi:10.3389/fonc.2021.607224
Hu, L., Liu, S., Peng, Y., Ge, R., Su, R., Senevirathne, C., et al. (2022). m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. [Epub ahead of print]. doi:10.1038/s41587-022-01243-z
Huang, H., Weng, H., and Chen, J. (2020). The biogenesis and precise control of RNA m6A methylation. Trends Genet. 36, 44–52. doi:10.1016/j.tig.2019.10.011
Huang, X., Zhang, H., Guo, X., Zhu, Z., Cai, H., and Kong, X. (2018). Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J. Hematol. Oncol. 11, 88. doi:10.1186/s13045-018-0628-y
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18, 186. doi:10.1186/s12943-019-1106-z
Li, C., Zhang, C., Zhang, G., Chen, B., Li, X., Li, K., et al. (2022). YTHDF1 amplification is correlated with worse outcome and lower immune cell infiltrations in breast cancer. Cancer Biomark. 1, 1–16. doi:10.3233/CBM-203103
Li, Z., Yang, H., Dai, X., Zhang, X., Huang, Y., Shi, L., et al. (2021). CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int. J. Biol. Sci. 17, 1178–1190. doi:10.7150/ijbs.57783
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. doi:10.1016/j.molcel.2016.03.021
Lin, Y., Jin, X., Nie, Q., Chen, M., Guo, W., Chen, L., et al. (2022). YTHDF3 facilitates triple-negative breast cancer progression and metastasis by stabilizing ZEB1 mRNA in an m6A-dependent manner. Ann. Transl. Med. 10, 83. doi:10.21037/atm-21-6857
Liu, C., Yang, Z., Li, R., Wu, Y., Chi, M., Gao, S., et al. (2021). Potential roles of N6-methyladenosine (m6A) in immune cells. J. Transl. Med. 19, 251. doi:10.1186/s12967-021-02918-y
Liu, L., Liu, X., Dong, Z., Li, J., Yu, Y., Chen, X., et al. (2019). N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J. Cancer 10, 5447–5459. doi:10.7150/jca.35053
Lv, W., Wang, Y., Zhao, C., Tan, Y., Xiong, M., Yi, Y., et al. (2021). Identification and validation of m6A-related lncRNA signature as potential predictive biomarkers in breast cancer. Front. Oncol. 11, 745719. doi:10.3389/fonc.2021.745719
Ma, Z., Gao, X., Shuai, Y., Xing, X., and Ji, J. (2021). The m6A epitranscriptome opens a new charter in immune system logic. Epigenetics 16, 819–837. doi:10.1080/15592294.2020.1827722
Montazeri, F., Hatami, H., Fathi, S., Hasanpour Ardekanizadeh, N., Bourbour, F., Rastgoo, S., et al. (2022). FTO genotype was associated with breast cancer in HER2 negative patients. Clin. Nutr. ESPEN 49, 495–498. doi:10.1016/j.clnesp.2022.02.122
Oerum, S., Meynier, V., Catala, M., and Tisne, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49, 7239–7255. doi:10.1093/nar/gkab378
Qian, J. Y., Gao, J., Sun, X., Cao, M. Da, Shi, L., Xia, T. S., et al. (2019). KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 38, 6123–6141. doi:10.1038/s41388-019-0861-z
Rong, B., Zhang, Q., Wan, J., Xing, S., Dai, R., Li, Y., et al. (2020). Ribosome 18S m6A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth. Cell Rep. 33, 108544. doi:10.1016/j.celrep.2020.108544
Rong, D., Dong, Q., Qu, H., Deng, X., Gao, F., Li, Q., et al. (2021). m6A-induced LINC00958 promotes breast cancer tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Discov. 7, 27. doi:10.1038/s41420-020-00382-z
Ruan, H., Gu, W., Xia, W., Gong, Y., Zhou, X., Chen, W., et al. (2021). METTL3 is suppressed by circular RNA circMETTL3/miR-34c-3p signaling and limits the tumor growth and metastasis in triple negative breast cancer. Front. Oncol. 11, 778132. doi:10.3389/fonc.2021.778132
Sepich-Poore, C., Zheng, Z., Schmitt, E., Wen, K., Zhang, Z. S., Cui, X.-L., et al. (2022). The METTL5-TRMT112 N6-methyladenosine methyltransferase complex regulates mRNA translation via 18S rRNA methylation. J. Biol. Chem. 298, 101590. doi:10.1016/j.jbc.2022.101590
Shen, X., Zhao, K., Xu, L., Cheng, G., Zhu, J., Gan, L., et al. (2021). YTHDF2 inhibits gastric cancer cell growth by regulating FOXC2 signaling pathway. Front. Genet. 11, 592042. doi:10.3389/fgene.2020.592042
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N 6-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi:10.1038/cr.2017.15
Shi, W., Tang, Y., Lu, J., Zhuang, Y., and Wang, J. (2022). MIR210HG promotes breast cancer progression by IGF2BP1 mediated m6A modification. Cell Biosci. 12, 38. doi:10.1186/s13578-022-00772-z
Shi, Y., Zheng, C., Jin, Y., Bao, B., Wang, D., Hou, K., et al. (2020). Reduced expression of METTL3 promotes metastasis of triple-negative breast cancer by m6A methylation-mediated COL3A1 up-regulation. Front. Oncol. 10, 1126. doi:10.3389/fonc.2020.01126
Sun, T., Wu, Z., Wang, X., Wang, Y., Hu, X., Qin, W., et al. (2020). LNC942 promoting METTL14-mediated m6A methylation in breast cancer cell proliferation and progression. Oncogene 39, 5358–5372. doi:10.1038/s41388-020-1338-9
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Uddin, M. B., Wang, Z., and Yang, C. (2021). The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 20, 61. doi:10.1186/s12943-021-01356-0
Wan, W., Ao, X., Chen, Q., Yu, Y., Ao, L., Xing, W., et al. (2022). METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer 21, 60. doi:10.1186/s12943-021-01447-y
Wang, H., Xu, B., and Shi, J. (2020a). N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene 722, 144076. doi:10.1016/j.gene.2019.144076
Wang, J., Li, Y., Wang, P., Han, G., Zhang, T., Chang, J., et al. (2020b). Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling Axis. Cell Stem Cell 27, 81–97. e8. doi:10.1016/j.stem.2020.04.001
Wang, Y. J., Yang, B., Lai, Q., Shi, J. F., Peng, J. Y., Zhang, Y., et al. (2021). Reprogramming of m6A epitranscriptome is crucial for shaping of transcriptome and proteome in response to hypoxia. RNA Biol. 18, 131–143. doi:10.1080/15476286.2020.1804697
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m6A. Nat. Rev. Genet. 22, 119–131. doi:10.1038/s41576-020-00295-8
Xie, J., Ba, J., Zhang, M., Wan, Y., Jin, Z., and Yao, Y. (2021). The m6A methyltransferase METTL3 promotes the stemness and malignant progression of breast cancer by mediating m6A modification on SOX2. J. B.U.ON. 26, 444–449.
Xu, L., Zhang, J., Ma, Y., Yuan, Y., Yu, H., Wang, J., et al. (2022). MicroRNA-135 inhibits initiation of epithelial-mesenchymal transition in breast cancer by targeting ZNF217 and promoting m6A modification of NANOG. Oncogene 41, 1742–1751. doi:10.1038/s41388-022-02211-2
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020). The FTO/miR‐181b‐3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun. 40, 484–500. doi:10.1002/cac2.12075
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601. doi:10.1038/s41586-021-03536-w
Yao, X., Li, W., Li, L., Li, M., Zhao, Y., Fang, D., et al. (2022). YTHDF1 upregulation mediates hypoxia-dependent breast cancer growth and metastasis through regulating PKM2 to affect glycolysis. Cell Death Dis. 13, 258. doi:10.1038/s41419-022-04711-1
Yau, C., Osdoit, M., van der Noordaa, M., Shad, S., Wei, J., de Croze, D., et al. (2022). Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet. Oncol. 23, 149–160. doi:10.1016/S1470-2045(21)00589-1
Yi, D., Wang, R., Shi, X., Xu, L., Yilihamu, Y., and Sang, J. (2020). METTL14 promotes the migration and invasion of breast cancer cells by modulating N6–methyladenosine and hsa–miR–146a–5p expression. Oncol. Rep. 43, 1375–1386. doi:10.3892/or.2020.7515
Yin, H., Zhang, X., Yang, P., Zhang, X., Peng, Y., Li, D., et al. (2021). RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 12, 1394. doi:10.1038/s41467-021-21514-8
You, Y., Fu, Y., Huang, M., Shen, D., Zhao, B., Liu, H., et al. (2022). Recent advances of m6A demethylases inhibitors and their biological functions in human diseases. Int. J. Mol. Sci. 23, 5815. doi:10.3390/ijms23105815
Yuan, B., Liu, W., Huo, M., Zhang, J., Yang, Y., Gao, T., et al. (2022). The hub ten gene-based risk score system using RNA m6A methylation regulator features and tumor immune microenvironment in breast cancer. Breast Cancer 29, 645–658. doi:10.1007/s12282-022-01341-5
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113, E2047–E2056. doi:10.1073/pnas.1602883113
Zhang, J., Shan, B., Lin, L., Dong, J., Sun, Q., Zhou, Q., et al. (2021). Corrigendum: Dissecting the role of N6-methylandenosine-related long non-coding RNAs signature in prognosis and immune microenvironment of breast cancer. Front. Cell Dev. Biol. 9, 812770. doi:10.3389/fcell.2021.812770
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606. e6. doi:10.1016/j.ccell.2017.02.013
Zhang, X., Dai, X. Y., Qian, J. Y., Xu, F., Wang, Z. W., Xia, T., et al. (2022). SMC1A regulated by KIAA1429 in m6A-independent manner promotes EMT progress in breast cancer. Mol. Ther. Nucleic Acids 27, 133–146. doi:10.1016/j.omtn.2021.08.009
Zhao, C., Ling, X., Xia, Y., Yan, B., and Guan, Q. (2022). LncRNA UCA1 promotes SOX12 expression in breast cancer by regulating m6A modification of miR-375 by METTL14 through DNA methylation. Cancer Gene Ther. 29, 1043–1055. doi:10.1038/s41417-021-00390-w
Zhao, C., Ling, X., Xia, Y., Yan, B., and Guan, Q. (2021). The m6A methyltransferase METTL3 controls epithelial-mesenchymal transition, migration and invasion of breast cancer through the MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int. 21, 441. doi:10.1186/s12935-021-02113-5
Zhong, S., Lin, Z., Chen, H., Mao, L., Feng, J., and Zhou, S. (2021). The m 6 A-related gene signature for predicting the prognosis of breast cancer. PeerJ 9, e11561. doi:10.7717/peerj.11561
Zhou, Z., Lv, J., Yu, H., Han, J., Yang, X., Feng, D., et al. (2020). Mechanism of RNA modification N6-methyladenosine in human cancer. Mol. Cancer 19, 104. doi:10.1186/s12943-020-01216-3
Zhu, P., He, F., Hou, Y., Tu, G., Li, Q., Jin, T., et al. (2021a). A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 40, 1609–1627. doi:10.1038/s41388-020-01638-9
Keywords: breast cancer, M6A, methylation, gene expression, miRNA, metastasis
Citation: Zhou M, Dong M, Yang X, Gong J, Liao X, Zhang Q and Liu Z (2022) The emerging roles and mechanism of m6a in breast cancer progression. Front. Genet. 13:983564. doi: 10.3389/fgene.2022.983564
Received: 01 July 2022; Accepted: 18 July 2022;
Published: 10 August 2022.
Edited by:
Zhongxiao Wang, Biogen Idec (United States), United StatesReviewed by:
Yawen Guo, Zhejiang University, ChinaCopyright © 2022 Zhou, Dong, Yang, Gong, Liao, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinghua Liao, eGluZ2h1YWxpYW9Ad3VzdC5lZHUuY24=; Qi Zhang, emhhbmdxaTA2MTcyQDE2My5jb20=; Zeming Liu, Nm15dEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.