- 1Centre de Recherche CERVO, Département de Psychiatrie et de Neurosciences, Faculté de Médecine, Université Laval, Québec City, QC, Canada
- 2Département des Sciences Animales, Faculté des Sciences de l’agriculture et de l’alimentation, Université Laval, Québec City, QC, Canada
- 3Centre de Recherche en Reproduction, Développement et Santé Intergénérationnelle (CRDSI), Université Laval, Québec City, QC, Canada
- 4Université Côte d’Azur, CNRS, Institut de Pharmacologie Moléculaire et Cellulaire, Valbonne, France
Introduction
In the early days of gene discoveries, the denominations in initial reports were often imprecise, restrictive or uninformative. Indeed, genes were usually originally named in reference to the context of their discovery, according to the specific tissue, cell type, cellular function, disease or phenotypic outcome under scrutiny. As a result, it was not uncommon that the same gene would have several names, simply because different researchers were working on the same gene, albeit in distinct contexts. Consequently, despite gene names harmonization, which coincided with the adoption of official gene symbols used in databases, most genes still exhibit a handful of aliases. Also, the official alias was generally chosen on historical grounds rather than as a reflection of the actual functions of the gene. More recently, several genes were renamed as their historical name was either unprecise and/or vector of negative connotations. Renaming the FMR1 gene has recently been the object of such debate in the community (Herring et al., 2020).
The specific case of renaming the FMR1 gene involved in fragile X syndrome
Since its cloning in 1991 (Verkerk et al., 1991), the official name of the gene mutated in Fragile X Syndrome (FXS) has been Fragile X Mental Retardation 1 (official gene symbol FMR1), as it was the first of a long series of X-linked genes associated with intellectual deficiency, termed at that time mental retardation. FXS is a leading cause of developmental delay, inherited intellectual disability, and the most frequent monogenic cause of autism spectrum disorder (ASD) (Kaufmann et al., 2017). FXS is caused by abnormal expansions of CGG triplets (n > 200 repeats) in the untranslated region (5′-UTR) of the X-linked FMR1 gene, which turn off its expression (Verkerk et al., 1991). Over the past two decades, while the general population exhibit in average 30 CGG repeats, premutations between 50 and 200 repeats in the FMR1 gene have been associated with Fragile X Premutation Associated Conditions (FXPAC), a set of conditions with no intellectual disability (Hagerman and Hagerman, 2016; Johnson et al., 2020). Also, it is important to note that mutations in the FMR1 gene are incompletely penetrant, and some individuals, in particular females bearing abnormal CGG repeat expansions will exhibit no signs of FXS or FXPAC (Johnson et al., 2020). As our society becomes more inclusive and recommends not to stigmatize individuals with intellectual deficiency, the term mental retardation has been progressively banned from scientific or clinical reports. Also, since FMR1 mutations are not necessarily accompanied by intellectual deficiency, calling it based on this single phenotypic trait was judged non-inclusive (Herring et al., 2020). Upon a suggestion of the European Fragile X Network that received support from Fragile X Societies, it was petitioned to change the definition of the FMR1 acronym as well as of its encoded protein FMRP, by modifying the meaning of the last letters (Herring et al., 2020). As a result, recently, the HUGO Gene Nomenclature Committee agreed to rename the FMR1 gene with the more inclusive name of Fragile X Messenger Ribonucleoprotein 1 (HGNC, 2021; Bruford and On behalf of the Hugo Gene Nomenclature Committee, 2022).
Discussion
We agree that removing “mental retardation” was necessary and keeping the original acronym FMR1 was essential for database nomenclature issues. However, we wish to point that the new name “Fragile X Messenger Ribonucleoprotein” does not reflect the pleiotropic functions of FMR1 gene that three decades of research have contributed to unravel. The letter M now standing for “messenger” and the R for “ribonucleoprotein. First, the choice of “messenger” for M does not reflect the fact that FMRP, the FMR1 encoded protein, not only binds messenger RNA in the translation machinery to modulate translation (Corbin et al., 1997; Khandjian et al., 2004; Stefani et al., 2004; Darnell et al., 2011), but also micro-RNA, piwi-RNA, and lncRNA (Caudy et al., 2002; Guo et al., 2018; Lannom et al., 2021). Therefore, its principal functional category is RNA-binding protein, although FMRP not only binds RNA, but also chromatin where it intervenes in DNA repair processes (Alpatov et al., 2014; Chakraborty et al., 2020). Second, the abbreviation, MRP for “messenger-ribonucleoprotein” is insufficiently precise, a more adequate term would have been “messenger-binding”. Third, FMRP not only binds nucleic acids, but also a diversity of partner proteins within large protein complexes [Figure 10A in Dury et al. (2013), Brown et al. (2010)]. These multiple protein-protein interactions likely modulate FMRP affinity to RNA (Bechara et al., 2007). In addition, it should be recalled that FMRP, besides being a critical component of neuronal granules, which travel in the neuronal arborization to deliver RNA at the synapse (El Fatimy et al., 2016), is also a molecular adaptor or a bridge between these granules and microtubule elements (Davidovic et al., 2007; Dictenberg et al., 2008). For all these reasons, assigning the term “messenger ribonucleoprotein” in FMR1 full name de facto restricts its biological role and do not reflect the full spectrum of the 26 functional categories assigned to FMR1 in the Gene Ontology (GO) repository. As a matter of fact, GO terms associated to FMR1 notably encompass: Nucleic acid binding (GO:0003676), RNA-binding (GO:0003729), miRNA-binding (GO:0035198), siRNA binding (GO:0035197), ribosome binding (GO:0043022), methylated histone binding (GO:0035064), to microtubule binding (GO:0008017), protein binding (GO:0001948), and protein homodimerization activity (GO:0042803). These multifaceted functions should be reflected in FMR1 new alias.
As for the protein name, the UniProt database has chosen a name totally unrelated to the acronym as it is now termed “Synaptic functional regulator FMR1” (https://www.uniprot.org/uniprot/Q06787). This is also clearly misleading and restrictive in terms of functions for FMRP in other cell types than neurons and outside the brain. The synaptic compartment containing only a minor fraction of FMRP, and its functions are in no way restricted to the synapse. As a matter of fact, FMRP is expressed in all tissues of the body, albeit at different levels (Khandjian et al., 1998, see also Figure 1) to the exception of striated muscle (Khandjian et al., 1998). As a consequence, in light of the 30 years of research in the Fragile X field, neither the HUGO name for the FMR1 gene, nor the UniProt name for FMRP appear adequate since they do not actually reflect their pleiotropic functions.
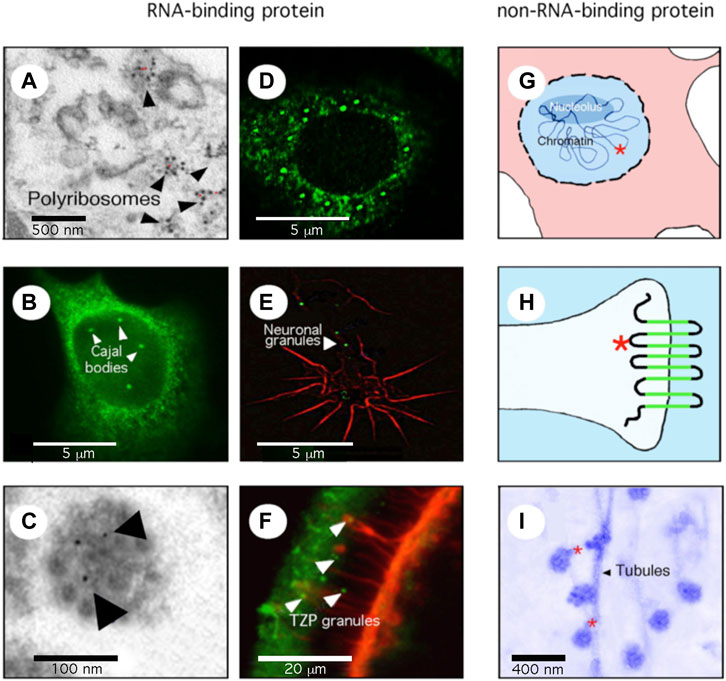
FIGURE 1. FMRP is a multifunctional protein. (A) the great majority of FMRP is associated with the translation machinery. Electron microscopy (EM) micrography showing FMRP (red dots) on polyribosomes. (B) A minor fraction of FMRP, in this case the nuclear isoform 6, is detected in Cajal Bodies, site of splicing of pre-messenger RNA. (C) FMRP is present in compacted granules containing mRNA, travelling in neuronal dendrites/axons as seen by EM and immuno-gold labelling (indicated by arrows). (D) FMRP relocalizes to stress granules after arsenite treatments. (E) FMRP (green) in compacted granules delivered locally at the axonal growth cone (red). (F) FMRP (green) is present in RNA-granules translocated from cumulus cells to the oocytes through transzonal projections (red). (G) FMRP (red star) is associated with chromatin and is involved in DNA repair. (H) FMRP (red star) associates with ion channels at the synapse. (I) FMRP (red dots) is a molecular adaptor between repressed granules and microtubules through kinesin interaction in the neuronal arborization.
Consequently, we wish to propose here a more inclusive full name for FMR1. Keeping intact the FMR1 acronym, we propose the F to remain as in Fragile X, but that M stands for multifunctional, R for RNA-binding, the main functional category assigned to FMRP, and obviously in the case of its encoded product, P for protein. We believe that “Fragile X Multifunctional RNA-binding Protein” is a more naturalistic full name which captures best the multifaceted functions of FMRP unravelled in the course of 30 years of research in the field.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alpatov, R., Lesch, B. J., Nakamoto-Kinoshita, M., Blanco, A., Chen, S., Stutzer, A., et al. (2014). A chromatin dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell 157, 869–881. doi:10.1016/j.cell.2014.03.040
Bechara, E., Davidovic, L., Melko, M., Bensaid, M., Tremblay, S., Grosgeorge, J., et al. (2007). Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Res. 35, 299–306. doi:10.1093/nar/gkl1021
Brown, M. R., Kronengold, J., Gazula, V. R., Chen, Y., Strumbos, J. G., Sigworth, F. J., et al. (2010). Fragile X Mental Retardation Protein controls gating of the sodium- activated potassium channel Slack. Nat. Neurosci. 13, 819–821. doi:10.1038/nn.2563
Bruford, E.On behalf of the Hugo Gene Nomenclature Committee (Hgnc) (2022). Comment on Herring et al. The Use of “Retardation” in FRAXA, FMRP,FMR1 and Other Designations. Cells 2022, 11, 1044. Cells 11, 1937. doi:10.3390/cells11121937
Caudy, A. A., Myers, M., Hannon, G. J., and Hammond, S. M. (2002). Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16, 2491–2496. doi:10.1101/gad.1025202
Chakraborty, A., JenjaroenpunLi, J., El Hilali, S., McCulley, A., Haarer, B., Hoffman, E. A., et al. (2020). Replication stress induces global chromosome breakage in the fragile X genome. Cell Rep. 32, 108179. doi:10.1016/j.celrep.2020.108179
Corbin, F., Bouillon, M., Fortin, A., Morin, S., Rousseau, F., and Khandjian, E. W. (1997). The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 6, 1465–1472. doi:10.1093/hmg/6.9.1465
Darnell, J. C., Van Driesche, S. J., Zhang, C., Hung, K. Y., Mele, A., Fraser, C. E., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi:10.1016/j.cell.2011.06.013
Davidovic, L., Jaglin, X. H., Lepagnol-Bestel, A. M., Tremblay, S., Simonneau, M., Bardoni, B., et al. (2007). The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 16, 3047–3058. doi:10.1093/hmg/ddm263
Dictenberg, J. B., Swanger, S. A., Antar, L. N., Singer, R. H., and Bassell, G. J. (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939. doi:10.1016/j.devcel.2008.04.003
Dury, A. Y., El Fatimy, R., Tremblay, S., Rose, T. M., Côté, J., De Koninck, P., et al. (2013). Nuclear fragile X mental retardation protein is localized to cajal bodies. PLoS Genet. 9, e1003890. doi:10.1371/journal.pgen.1003890
El Fatimy, R., Davidovic, L., Tremblay, S., Jaglin, X., Dury, A., Robert, C., et al. (2016). Tracking the fragile X mental retardation protein in a highly ordered neuronal RiboNucleoParticles population: A link between stalled polyribosomes and RNA granules. PLoS Genet. 12 (7), e1006192. doi:10.1371/journal.pgen.1006192
Guo, Y., Chen, X., Xing, R., Wang, M., Zhu, X., and Guo, W. (2018). Interplay between FMRP and lncRNA TUG1 regulates axonal development through mediating SnoN-Ccd1 pathway. Hum. Mol. Genet. 27, 475–485. doi:10.1093/hmg/ddx417
Hagerman, R. J., and Hagerman, P. (2016). Fragile X-associated tremor/ataxia syndrome - features, mechanisms and management. Nat. Rev. Neurol. 12 (7), 403–412. doi:10.1038/nrneurol.2016.82
Herring, J., Johnson, K., and Richstein, J. (2020). The use of “retardation” in FRAXA, FMRP, FMR1 and other designations. Cells 11, 1044. doi:10.3390/cells11061044
HGNC (2021). Symbol report for FMR1. Available at: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:3775.
Johnson, K., Herring, J., and Richstein, J. (2020). Fragile X premutation associated conditions (FXPAC). Front. Pediatr. 8, 266. doi:10.3389/fped.2020.00266
Kaufmann, W. E., Kidd, S. A., Andrews, H. F., Budimirovic, D. B., Esler, A., Haas-Givler, B., et al. (2017). Autism spectrum disorder in fragile x syndrome: Cooccurring conditions and current treatment. Pediatrics 139 (3), S194–S206. doi:10.1542/peds.2016-1159F
Khandjian, E. W., Huot, M. E., Tremblay, S., Davidovic, L., Mazroui, R., and Bardoni, B. (2004). Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. U. S. A. 101, 13357–13362. doi:10.1073/pnas.0405398101
Khandjian, E. W., Bardoni, B., Corbin, F., Sittler, A., Giroux, S., Heitz, D., et al. (1998). Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet. 7, 2121–2128. doi:10.1093/hmg/7.13.2121
Khandjian, E. W., Fortin, A., Thibodeau, A., Tremblay, S., Côté, F., Devys, D., et al. (1995). A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum. Mol. Genet. 4, 783–789. doi:10.1093/hmg/4.5.783
Lannom, M. C., Nielsen, J., Nawaz, A., Shilikbay, T., and Ceman, S. (2021). FMRP and MOV10 regulate Dicer1 expression and dendrite development. PLoS One 16 (11), e0260005. doi:10.1371/journal.pone.0260005
Stefani, G., Fraser, C. E., Darnell, J. C., and Darnell, R. B. (2004). Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276. doi:10.1523/JNEUROSCI.2306-04.2004
Verkerk, A. J. M. H., Pieretti, M., Sutcliffe, J. S., Fu, Y-H., Kuhl, D. P. A., Pizzuti, A., et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. doi:10.1016/0092-8674(91)90397-h
Keywords: FMR1, FMRP, fragile X syndrome, RNA binding protein, gene name
Citation: Khandjian EW, Robert C and Davidovic L (2022) FMRP, a multifunctional RNA-binding protein in quest of a new identity. Front. Genet. 13:976480. doi: 10.3389/fgene.2022.976480
Received: 23 June 2022; Accepted: 11 July 2022;
Published: 10 August 2022.
Edited by:
Daman Kumari, National Institutes of Health (NIH), United StatesReviewed by:
Anita Bhattacharyya, University of Wisconsin-Madison, United StatesWilliam T. Brown, AOL, United States
Copyright © 2022 Khandjian, Robert and Davidovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edouard W. Khandjian, RWR3YXJkLmtoYW5kamlhbkBjcnVscmcudWxhdmFsLmNh
 Edouard W. Khandjian
Edouard W. Khandjian Claude Robert2,3
Claude Robert2,3 Laetitia Davidovic
Laetitia Davidovic