
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 26 September 2022
Sec. Epigenomics and Epigenetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.970097
This article is part of the Research TopicApplication of Epigenomics Data to Improve Human and Livestock HealthView all 14 articles
The aim of this study was to explore the potential biological function of circular RNAs (circRNAs) in the sperm motility traits of Xinjiang Yili geese, and to provide a reference for analyzing the mechanism of regulation of Yili geese sperm motility. The 10 selected Xinjiang Yili Geese with high or low sperm motility (five for each group) were 3 years old, in good health, and were kept in the same feeding conditions. Yili geese were slaughtered for the collection of testicular tissue and high-throughput sequencing technology was used to screen differentially expressed circRNAs for bioinformatics analysis. Combined with the previously screened miRNAs related to the sperm motility of Yili geese, the circRNAs miRNAs regulatory network was constructed. The results showed that a total of 26,311 circRNAs were obtained from testicular tissues with high and low sperm motility, and 173 DECs were screened between the two groups (p < 0.05, |log2Foldchange|>0), of which 82 were up-regulated and 91 were down-regulated. Functional analysis of the source genes of these DECs showed that the source genes were mainly involved in biological processes. KEGG enrichment analysis showed that the source genes of DECs were mainly enriched in autophagy-animal, ubiquinone and other terpenoid-quinone biosynthesis, progesterone-mediated oocyte maturation, regulation of the actin cytoskeleton and other pathways. Furthermore, the visual regulatory network of differential circRNA-miRNA-mRNA was constructed, including 20 circRNAs, 18 miRNAs and 177 mRNAs, and nine core regulatory circRNAs were screened, including novell_circ_0045314, novel_circ_0019994 and novel_circ_0020422, etc., targeting ppy-mir-16, hsa-mir-221–3p, gga-mir-499–5p, etc. The results suggest that circRNAs may interact with miRNAs to further regulate mRNA to regulate sperm motility in Yili geese, so as to provide a reference for analyzing the molecular mechanism of sperm motility regulation.
The Yili goose is the only local poultry variety in China that is derived from the herbivorous characteristics of gray geese. As a high-quality local characteristic poultry, the Yili goose has the characteristics of strong adaptability, good flight, heat resistance, cold resistance, rough feeding resistance, strong disease resistance and stress resistance, as well as good meat quality (Zhao et al., 2020). Reproductive traits are the most important economic traits in poultry production. Improving reproductive performance is always an important goal of poultry genetic improvement. The semen quality of male birds is directly related to the fertilization ability of sperm, which is one of the important indicators affecting reproductive efficiency. According to previous research, the low fertility of male Yili geese and the low fertilization rate of eggs limit the development of the Yili goose industry (Zhao et al., 2019). We also found that the sperm motility of yili geese was significantly different among individuals, and there was a positive correlation between sperm motility and fertilization rate (Wu et al., 2022). Therefore, there is an urgent need to use molecular biotechnology to improve the reproductive performance of male Yili geese and to explore the genetic mechanisms affecting their semen quality.
CircRNAs are closed circular RNA transcripts formed by reverse splicing from a single RNA precursor and are found in all higher eukaryotes, including mammals (Li et al., 2018). It has been found that circRNA plays an important role in organisms. It can be used as competitive endogenous RNA (CeRNA) or as an “miRNA sponge” to inhibit the activity of miRNA, weaken or even relieve the inhibitory effect of miRNA on downstream target genes, and promote the expression of target genes (Zhong et al., 2018). At present, there are many studies on the roles of circRNAs in various aspects of the mammalian reproductive system, such as testicular development (Li et al., 2021; Zhang et al., 2021), follicle development (Guo et al., 2020; Liang et al., 2020), embryonic development (Quan and Li, 2018), etc. However, studies on the molecular mechanisms of circRNAs in the poultry reproductive system have mainly focused on the follicle development of female poultry (Shen M. et al., 2019; Shen M. et al., 2020; Wu et al., 2020). Research on the mechanism of regulation of circRNA with regard to sperm motility in the Yili goose is still lacking. In this study, Yili geese with extreme differences in sperm motility were taken as the objects of research. The expression pattern of circRNA in the testicular tissue of Yili geese with high and low sperm motility was analyzed by RNA-seq technology, and the circRNA related to the sperm motility of male geese was screened and identified, so as to provide a theoretical basis for the genetic and breeding improvement of Yili geese.
The experimental animals used in this study were provided by Hengxin Industrial Co., Ltd. Emin County, Xinjiang. According to the data on sperm motility and egg fertilization rate, among the 3-year-old Xinjiang Yili goose with similar body weight (3.65±0.40 kg) and the same feeding conditions, 5 geese with high and low sperm motility were selected (p < 0.01) (Table 1). After the geese were sacrificed, their testis tissues were collected immediately, and stored in liquid nitrogen, rinsed with PBS 1 to 2 times, and immediately placed in a cryopreservation tube containing RNA preservation solution. The testis samples of the HFR geese (high sperm motility group) were labeled as HFR-1∼HFR-5, and the testis samples of the LFR geese (low sperm motility group) were labeled as LFR-1∼LFR-5. The samples were labeled and stored at 4 °C overnight and then stored at -80 °C the next day until they were used for the extraction of total RNA.
The total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, United States), following the manufacturer’s protocol, and agarose gels, a Nanodrop ND-1000 spectrophotometer (IMPLEN,CA, United States) and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States) were used to ensure that the quality of the samples was sufficient for transcriptome sequencing. After the samples passed quality control, the strand-specific library was constructed by removing ribosomal RNA (circRNA library building and the linear RNA process). After the libraries were qualified, the library preparations were sequenced on an Illumina Hiseq platform and 150 bp paired-end reads were generated.
Raw data (raw reads) in fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low-quality reads from raw data. At the same time, for Q20, Q30 and GC content the clean data were calculated. The index of the reference genome (Yan et al., 2020) was built using Bowtie2 (v2.2.8) (Langmead et al., 2009)and paired-end clean reads were aligned to the reference genome using Bowtie. The circRNA was detected and identified using find_circ (v1.2) (Memczak et al., 2013)and CIRI2 (v2.0.5) (Gao et al., 2015), and the intersection of the two types of software was also used to identify the circRNA.
HTSeq v0.6.0 (Anders et al., 2015)was used to count the reads numbers mapped to each gene, and the expression amount was normalized with TPM (Zhou et al., 2010). The input data of circRNA differential expression is readcounts data obtained from circrna expression level analysis. Differential expression analysis of the two groups was performed using the DESeq2 R package (1.10.1) (Michael et al., 2014). circRNAs with a p < 0.05 and |log2Foldchange|>0 found by DESeq2 were set as the threshold for significantly differential expression. The GOseq (Release 2.1.2) (Young et al., 2010) software was used for the GO enrichment analysis of differentially expressed circRNA-derived genes, and KOBAS (2.0) (Mao et al., 2005; Kanehisa et al., 2008) was used for Paththe way enrichment analysis. A P of <0.05 was set as the threshold for statistically significant results.
miRNA target sites in the exons of circRNA loci were identified using miRanda (Enright et al., 2003). miRNA target genes were predicted to be the intersection of miRanda (Enright et al., 2003) and RNAhybrid (Kruger and Rehmsmeier, 2006). Cytoscape (v3.7.1) (Paul et al., 2003) software was used to construct the circRNA-miRNA-mRNA networks.
Randomly selected from the transcriptome sequencing results, 8 DECs related to the HFR and LFR of Xinjiang Yili geese were used for fluorescence-based quantitative validation. Oligo 7.0 software was used to design primers (Supplementary Table S1). SYBR GREEN reagent (TaKaRa) was used to amplify the target gene and internal reference gene (beta-actin) mRNAs on a ROCHE 480 quantitative PCR instrument (Eppendorf, Germany). The PCR reaction system (20 μl) included the 10 μl AceQ Universal SYBR qPCR Master Mix, 0.4 μl upstream primer (10 μmol L/L), 0.4 μl downstream primer (10 μmol L/L), 6.7 μl ddH2O, and 2.5 μl cDNA. The reaction conditions were as follows: 95°C for 5 min, 95°C for 10 s, 60°C for 30 s, 40 cycles; dissolution curve: 60°C→95°C, with a temperature increase of 0.3°C every 15 s. Quantitative expression results were calculated according to the cross point (CP) values, and the relative expression levels were calculated according to the 2−∆∆Ct method (Bustin et al., 2009).
Based on the semen quality data from six instances and the fertilization rate data from five instances, five individuals in the high sperm motility group and five individuals in the low sperm motility group were screened out. An independent t-test showed that the sperm motility and fertilization rate of the individuals in the high sperm motility group were significantly higher than in the low sperm motility group (p < 0.01, Table 1).
It can be seen from Table 2 that 495,401,294 clean reads and 475,402,620 reads were obtained by sequencing Yili geese with high and low sperm motility, respectively. The Q20 and Q30 of each sample were at least 97.49% and 93.06%, respectively, and the GC contents were between 45.40% and 47.40%. These results indicated that the quality of transcriptome sequencing results met the needs of subsequent analyses. According to the statistical results, 91.29%–92.37% of the clean reads were mapped to the reference genome, of which 86.06%–89.40% of the clean reads were uniquely mapped. The results showed that there were enough reads of each sample mapped to the reference genome, and the selected reference genome was suitable.
A total of 26,311 circRNAs were identified in testicular tissues (Figure 1), of which 6,520 and 6,032 were unique of the Xinjiang Yili geese with HFR and LFR, respectively. According to the annotation of the reference genome, it was found that many of the host genes of circRNAs are related to male reproduction (Table 3, Supplementary Table S2), such as spermatogenesis-related SPA5L, SPAT5, SPAT6, SPT17, SPT48, etc., testis-specific genes TESK1, TEX10, TSG10 etc., and flagellar motility genes SPEF2, CF206, IF172, etc., showing that it is feasible to sequence testicular tissue by RNA-seq technology and to screen for biomarkers related to geese reproduction.
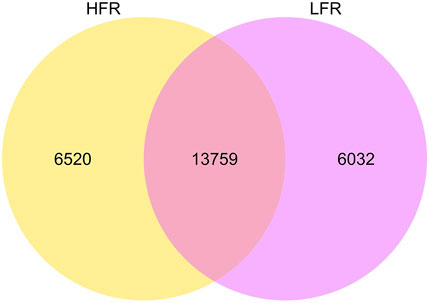
FIGURE 1. circRNAs difference between HFR and LFR. The big circle represents each comparison combination, the sum of the numbers in each big circle represents the total number of circrnas identified by the comparison combination, and the overlapping part of the circle represents the number of circrnas common between the combinations.
According to the comparison results of circRNAs and the reference genome, there were 14,232 circRNAs without corresponding host genes, and other circRNAs were mapped to 12,080 genes. The lengths of these circRNAs ranged from 23 bp to 1,511 bp, with an average length of 357 bp (Figure 2). Most circRNAs were distributed on chromosome 2, accounting for 8.43%, followed by chromosomes 1, 7, and 10, accounting for 6.38%, 5.83%, and 4.87%, respectively (Figure 3). According to their location in the genome, most circRNAs were derived from intergenic regions (34.19%), followed by exonic regions (33.45%) and intronic regions (32.36%) (Figure 4).
Compared with the low sperm motility group, a total of 173 DECs (differentially expressed circRNAs) were screened in the high sperm motility group, of which 82 were up-regulated and 91 were down-regulated (p < 0.05, |log2Foldchange|>0) (Figure 5), and 77 circRNAs had the source genes. Among them, the circRNAs with the smallest p values were novel_circ_0013769 and novel_circ_0007998, both located in the intergenic region, and both being up-regulated circRNAs. The fold difference was the largest in novel_circ_0042868 (source gene PDE6C), novel_circ_0018689 (intergenic_region) and novel_circ_0030568 (source gene ADAM9), with a fold difference of 7.6687, 7.1261 and 7.0849, respectively. There were 87 circRNAs with more than a 4-fold difference, accounting for 50.29% of the DECs. Hierarchical clustering analysis was performed on the circRNAs that were significantly differentially expressed in the testes of the two groups, as shown in the heat map (Figure 6), red color and blue color respectively represent the significant increase or decrease in the HFR group compared with the LFR group. It can be seen that the repeatability within the sample group was good, and the difference between the groups was large, which can better reflect the differences between different treatments.
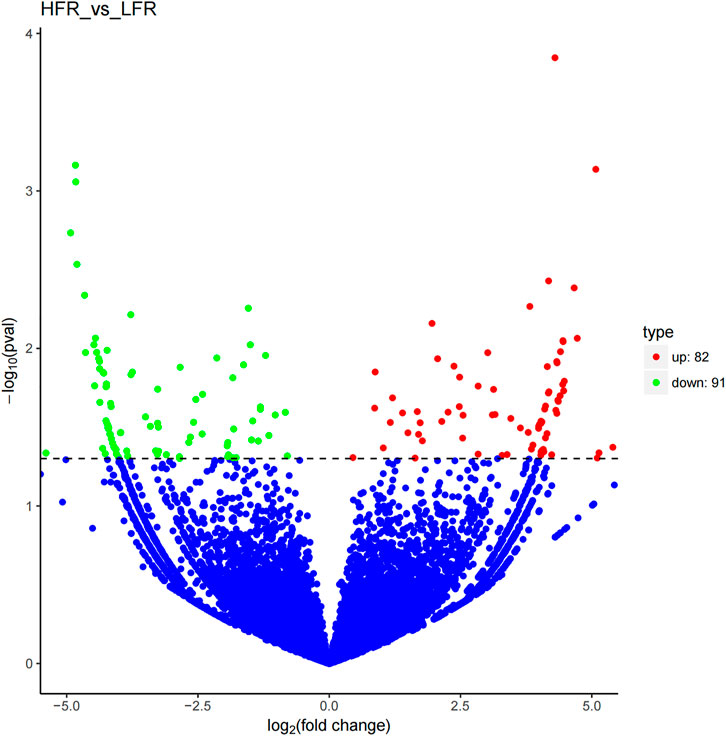
FIGURE 5. Differential circRNA volcano map. Differentially expressed circRNAs were filtered using a p < 0.05 as a threshold. Red spots represent up-regulated circRNAs, and green spots indicate down-regulated circRNAs. Blue spots represent circRNAs that did not show obvious changes between the HFR and LFR samples.
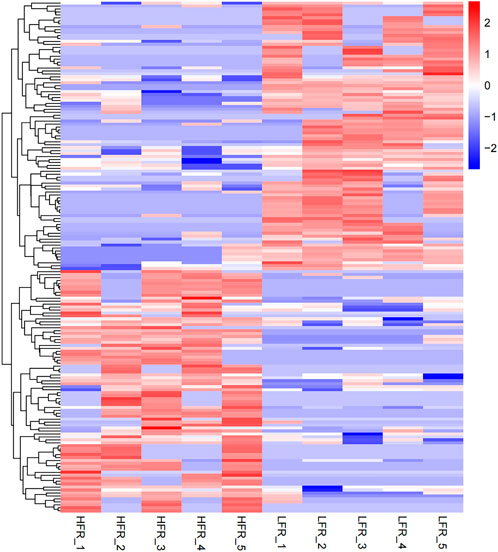
FIGURE 6. Differential circRNA cluster map. The horizontal axis is the sample, and the vertical axis is the differential gene. The left side clusters the genes according to the expression similarity, and the upper part clusters each sample according to the expression profile similarity. The expression level gradually increases from blue to red, and the number is the relative expression level after homogenization.
The functional analysis of the source genes of these DECs (Figure 7; Table 4) showed that 58 significant GO terms were enriched (p < 0.05), and biological processes (63.79%) were mainly enriched in cellular component organization, multi-organism processes, and the reproduction of a single-celled organism. The molecular function (31.03%) was mainly enriched in adenyl nucleotide binding, cytoskeletal protein binding, actin binding, etc., and the cellular component (5.17%) was mainly enriched in the microtubule organizing center, integral to the Golgi membrane. KEGG analysis was mainly enriched in autophagy-animal, ubiquinone and other terpenoid-quinone biosynthesis, progesterone-mediated oocyte maturation, oocyte meiosis and purine metabolism and other signaling pathways, of which metabolic pathways were enriched. The pathways with the most genes, autophagy-animal and ubiquinone and other terpenoid-quinone biosynthesis were the most significantly enriched pathways, and the source genes of differential circRNAs involved in these two pathways were Pdpk1 (phosphoinositide-dependent protein kinase 1) and COQ5 (methoxy-6-polypropylene-1,4-benzoquinone methylase) (Figure 8;Table 5).
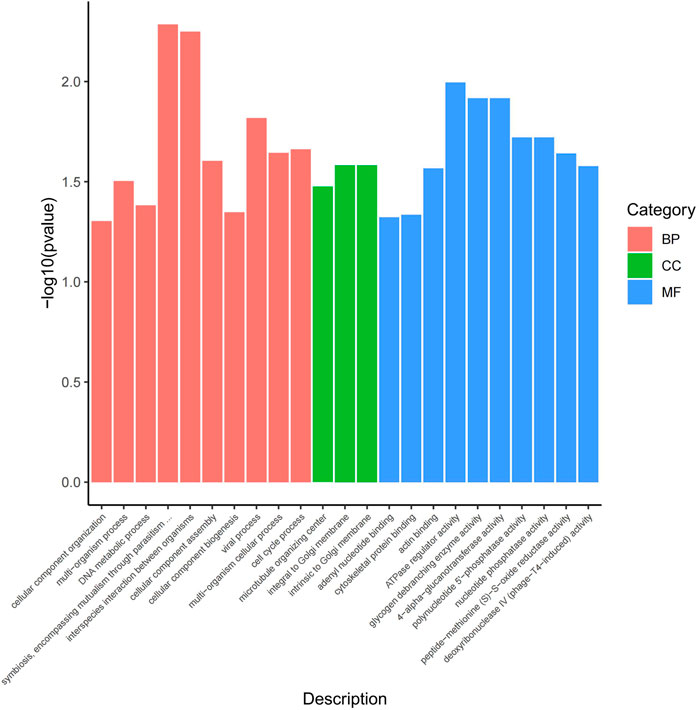
FIGURE 7. GO enrichment map. The horizontal axis represents the GO term of the next level of the three categories of GO, and the vertical axis represents the p value ranges.
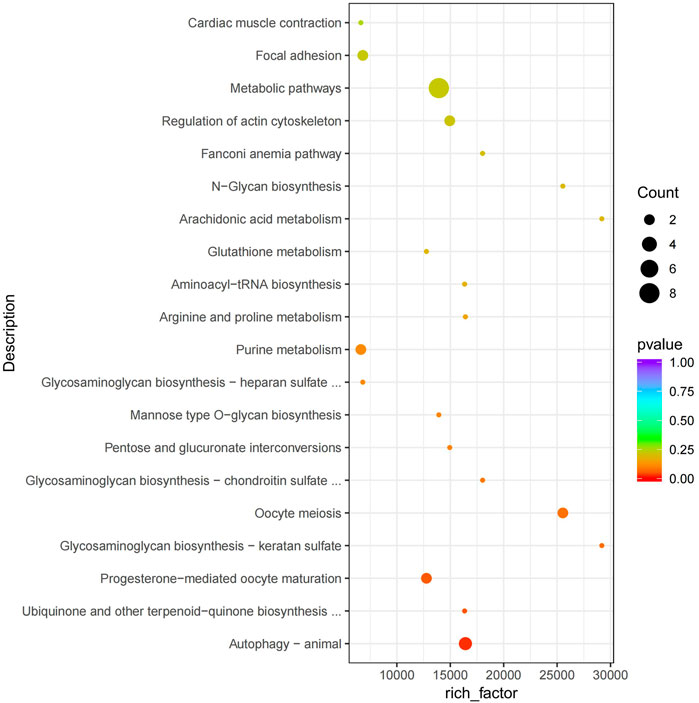
FIGURE 8. KEGG enrichment map. The vertical axis represents the pathway name, the horizontal axis represents rich factor, the size of the point represents the number of differentially expressed genes in this pathway, and the color of the point corresponds to different p value ranges.
circRNAs can adsorb miRNAs by combining with them, and can act as miRNA sponges (Li et al., 2018). Therefore, miRNA binding site analysis on the identified circRNAs is helpful to further study the function of circRNAs. In this study, the differentially expressed miRNAs in the testis of Yili geese with high and low sperm motility obtained in the previous stage were combined to construct a differential circRNA-miRNA-mRNA interaction network. As shown in Figure 9, the network was enriched with 20 circRNAs, 18 miRNAs and 177 mRNAs, and each circRNAs had at least 2 or more miRNA binding sites. Aca-mir-212–5p has 10 target circRNAs and 49 target mRNAs. Hsa-mir-221–3p has 3 target circRNAs and 10 target mRNAs. Bta-mir-221 has 3 target circRNAs and 11 target mRNAs. In addition, novel_ circ_ 0017590, novel_ circ_0018059 had four miRNA binding sites.
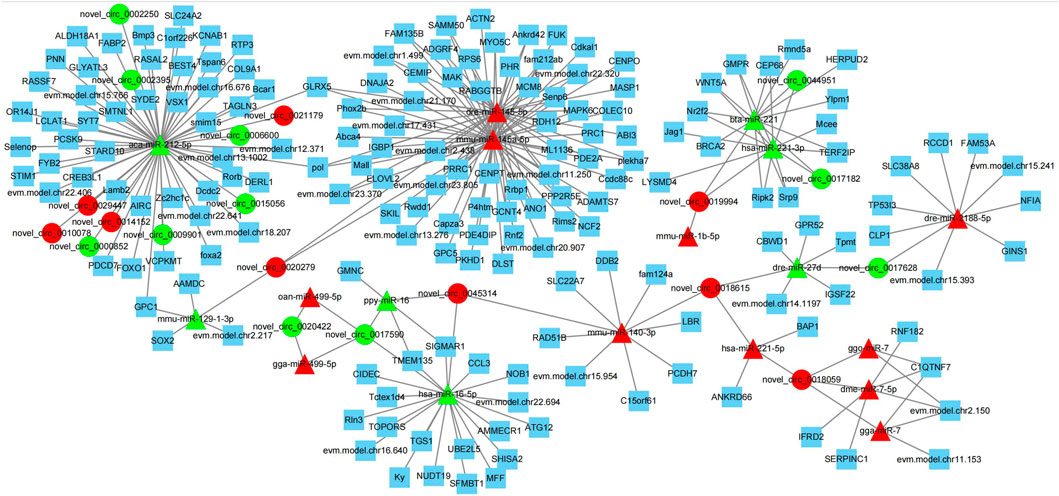
FIGURE 9. Interaction diagram of differential circRNA-miRNA-mRNA network. Triangle nodes represent the targeted miRNAs, circular nodes show circRNAs, and squares nodes show mRNA. Red color represent up-regulated, and green color indicate down-regulated.
novel_ circ_ 0045314, novel_ circ_ 0019994 and novel_ circ_ 0020422 has two negatively regulated target miRNAs, novell_ circ_0017628, novel_ circ_ 0018615, novel_ circ_ 0021179, novel_circ_ 0029447, novel_ circ_ 0014152 and novel_ circ_ 0010078 has one negatively regulated target miRNA. These results suggested that circRNAs in testis may regulate testicular development and sperm motility by actively participating in binding with miRNAs, acting as competitive endogenous RNAs, regulating the function of target miRNAs, and thus indirectly targeting mRNA levels.
In order to validate the differentially expressed genes identified by transcriptome sequencing, eight circRNAs were randomly selected and confirmed by qRT-PCR using GAPDH as the internal reference gene. The results showed that the expression trends of the eight circRNAs were consistent with those of the transcriptome sequencing results. Therefore, the transcriptome sequencing results are reliable and can be further studied and analyzed (Figure 10).
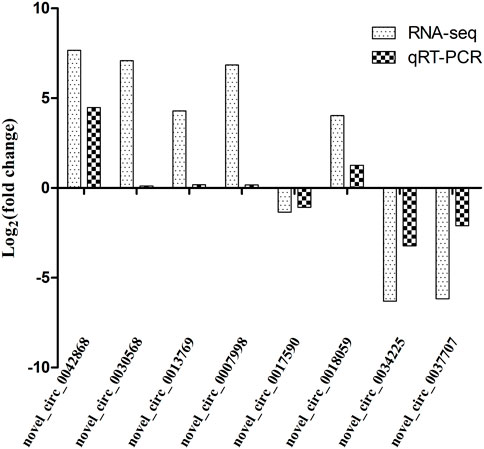
FIGURE 10. Comparison of qRT-PCR and RNA-seq results. Total RNA extracted from the hypothalami, pituitary glands, and ovaries tissues that were measured by qRT-PCR analysis; relative expression levels were calculated according to the 2−∆∆Ct method using GAPDH as an internal reference gene.
Semen quality is one of the important indicators to measure the breeding value of male birds. The evaluation indicators of semen quality mainly include semen color, sperm motility, sperm viability, sperm deformity rate, semen volume, pH, etc (Jerysz and Lukaszewicz, 2013; Lukaszewicz et al., 2021). Sperm motility is a reflection of metabolic capacity, and is an important factor in ensuring that the sperm and egg meet and complete fertilization. Studies have shown that sperm motility traits are highly positively correlated with fertilization rate (Nguyen, 2019). Moreover, the evaluation of sperm motility traits is relatively simple compared with other semen quality traits, and is easy to generalize. Therefore, sperm motility is the most suitable indicator to measure the reproductive performance of breeding poultry. The testis is an important reproductive organ of male birds, and its main functions are spermatogenesis and endocrine function (Iddrriss et al., 2018). The seminiferous tubules are distributed inside the testis and are the main location of spermatogenesis, making them the most suitable organ for determining male sperm motility.
circRNAs are a new class of RNAs that are different from traditional linear RNAs. They are conserved in different species and are specifically expressed in tissues at different developmental stages. circRNAs can be regulated by affecting the transcription, mRNA conversion and translation of RNA-binding proteins and miRNAs(Panda, 2018; Kristensen et al., 2019). So far, research on circRNAs has mainly focused on humans and model animals, and less research has been carried out on testicular development. Like other vertebrates, the avian testis is the site of spermatogenesis and androgen production, and research on testicular biology in avians mainly focuses on chickens and ducks (Estermann et al., 2021). Research on circRNA in poultry mainly focuses on muscle cell proliferation and differentiation (Ouyang et al., 2018; Shen X. X. et al., 2019), follicle development (Shen M. M. et al., 2020; Wu et al., 2020), disease and immunity (Qiu et al., 2018; Wang et al., 2020) and intramuscular fat deposition (Wang, 2018). However, in the study of the genetic mechanisms regulating sperm motility, the mechanism of generation and the downstream functions of circRNAs remain unclear.
Studies have shown that in mammals, the brain and testis are the tissues with the highest expression levels of circRNAs(Zhou et al., 2018; Mahmoudi and Cairns, 2019). The normal progression of testicular development and spermatogenesis depends on the precise regulation of related genes at the transcriptional and post-transcriptional levels, and ncRNAs can be temporally and spatially refined. At present, research on sperm motility regulation, especially non-coding RNA-mediated gene regulation, is relatively scarce. In this study, based on the data on semen quality and egg fertilization rate, male Yili geese with extreme differences in sperm motility were strictly screened. High-throughput sequencing technology was used for the first time to study the circRNA expression profile of the testicular tissue of Yili geese with high and low sperm motility, and a total of 26,312 circRNAs were identified. Most of these circRNAs were 23 bp-1,511 bp in length, with an average length of 357 bp, which is similar to the length of circRNAs identified in other animals (Liang et al., 2017; Yuan et al., 2018). It is worth noting that in this study, circRNAs were relatively evenly distributed in intergenic, exonic and intronic regions, and most of them were derived from intergenic regions (34.19%). Tang et al, (2020) and other studies have shown that circRNAs are abundantly expressed in male germ cells during spermatogenesis, and circRNA levels increase with the progress of spermatogenesis. This study identified a higher number of circRNAs in high sperm motility testes, indicating that there are more sperm cells in testes with high sperm motility, that is, the spermatogenesis ability is stronger. In addition, compared with the low sperm motility group, a total of 173 DECs were screened, of which 82 were up-regulated and 91 were down-regulated. The source genes of these DECs are widely involved in biological processes such as growth and development, reproduction, and metabolism.
To explore the relationship between these DECs and sperm motility in Yili geese, GO enrichment analysis was performed on the host genes of these circRNAs, and it was found that they were mainly involved in biological processes (63.79%), and were significantly enriched in the reproduction of a single-celled organism, cytoskeletal protein binding, actin binding, adenyl nucleotide binding, microtubule organizing center and other terms. Signaling pathways that play an important role in sperm motility are included in the significantly enriched signaling pathways. Among them, actin is an important part of the cytoskeleton (Hohmann and Dehghani, 2019), and is distributed in mammalian cells, including Sertoli cells in the testis, and actin polymerization in the sperm tail during capacitation regulates motility (Finkelstein et al., 2013). The core of sperm flagella is made up of microtubules, along with tens of thousands of tiny molecular motors called dyneins, which bend these microtubules rhythmically, creating wave after wave of motion, resulting in steering. Sudarshan (Gadadhar et al., 2021) and others found that a special enzymatic modification of tubulin, called glycylation, is the key to keeping sperm moving in a straight line. It is suggested that these circRNAs may be involved in the regulation of sperm motility.
KEGG enrichment analysis showed that autophagy-animal and ubiquinone and other terpenoid-quinone biosynthesis were the most significantly enriched pathways (p < 0.05), and the source genes of differential circRNAs involved in these two pathways were Pdpk1 (phosphomuscle alcohol-dependent protein kinase 1) and COQ5 (methoxy-6-polyacryl-1,4-benzoquinone methylase). The PDPK1 gene is expressed in spermatogenic cells, and PDPK1 is localized in the post-acrosomal region of mouse epididymal caudate sperm (Vadnais et al., 2013). COQ5 is a methyltransferase with adenopicromethionine as a methyl donor. It is located on the stromal side of the inner mitochondrial membrane and plays a crucial role in the stability and activity of the CoQ protein complex (Nguyen et al., 2014). CoQ may not only participate in electron transfer in the mitochondrial respiratory chain of cells, maintaining the functions of cellular respiration and energy metabolism, but also participate in the redox reaction of the body as an antioxidant (Zhong and Zhong, 2014).
Autophagy is a basic process that exists in all eukaryotes, and is involved in the life processes of various cells in the male reproductive system, as well as being involved in the key pathophysiological processes of many diseases of the male reproductive system, such as azoospermia, oligospermia, asthenozoospermia, cryptorchidism, and orchitis (Zhu et al., 2019). Lei et al, (2021) studied the initiation of autophagy in mouse testis and found that after autophagy was blocked in sperm cells, the assembly of a series of key structures during sperm differentiation was disrupted. For example, the deformed scaffold of the sperm–sperm collar (manchette) and the tail shaft (axoneme) to ensure sperm movement at the same time lead to a large amount of cytoplasm to remain in the sperm head that ought to have been removed, resulting in morphological deformities and movement disorders. Yefimova et al, (2013)abolished autophagy by germ-cell-specific knockout of Atg7, resulting in reduced testicular weight, sperm deformities, and significantly reduced fertility in male mice. Liu et al, (2016) found that Sertoli cell-specific knockout of Atg5 or Atg7 disrupted autophagy, resulting in disorders of the vas deferens and deformed sperm heads, which in turn affected the reproductive performance of male mice.
Coenzyme Q (coenzyme Q, CoQ) is a class of quinone substances widely distributed in living organisms. Benzoquinone, also known as ubidearenone and ubiquinone, is a lipid-soluble antioxidant mainly present in the mitochondria, and is also a natural antioxidant and free radical scavenger in mammals (Lafuente et al., 2013; Liu et al., 2017). As an energy promoter and antioxidant, coenzyme Q10 is mainly distributed in the mitochondria in the midsection of sperm, and the availability of the energy required for sperm motility and other energy-dependent processes depends on the availability of coenzyme Q10 (Aby and Lavon, 1997). Studies have shown that sperm concentration, motility and semen parameters are related to CoQ10 concentration, as CoQ10 can reduce stress oxidation, increase antioxidant enzyme activity, and improve overall antioxidant capacity (Balercia et al., 2009; Lafuente et al., 2013). Studies have confirmed that ubiquinone (Q10) coenzyme is more active in the biosynthesis of the male testis and female follicular fluid, and confirmed that coenzyme Q10 is inseparable from mammalian reproduction (Shetty et al., 2013; Varela-Lopez et al., 2017). Huo Min (Huo, 2018) added different concentrations of CoQ10 exogenously to a frozen dilution solution, and detected various indicators of cashmere goat sperm after thawing. The results show that compared with the control group, the sperm quality after freezing and thawing was significantly improved (p < 0.05). When the concentration of coenzyme Q10 was 400 μg/ml, the sperm motility rate was 60.2%, which was significantly higher than that of the control group (p < 0.05), and the intracellular ROS content was significantly decreased (p < 0.05); when the concentration of coenzyme Q10 was 40 μg/ml, the sperm plasma membrane integrity rate was 63.0%, the acrosome integrity rate was 76.5%, and the mitochondrial membrane potential was 2.9, which were significantly higher than the levels found in the control group (p < 0.05). Hamed et al, (2019) explored the protective effect of coenzyme Q10 (CoQ10) and berberine (BB) combined with and without varicocelectomy on sperm parameters in rats with postoperative varicocele. They showed that BB and CoQ10 alone and/or together could improve sperm parameters and reduce sperm DNA damage in varicocele-induced rats. It is worth noting that although the metabolic pathways in the KEGG enrichment were not significantly enriched, the pathways with the most enrichment of source genes were identified. These results indicate that there are cellular differences in testicular tissue in Yili geese with high and low sperm motility, and many of the DECs identified may play a roles in regulating autophagy, anti-oxidation and cell metabolism, thereby participating in the regulation of sperm motility in Yili geese.
The circRNA-miRNA-mRNA interaction network further revealed that cirRNA may regulate the expression of mRNA through the interaction with miRNA to regulate the sperm movement ofYili geese. In the interaction network, miR-212–5p, which has the most binding sites for circRNAs, was reported to be developmentally related to the follicles of ewe during FSH stimulation (Hya et al., 2022). Spermatogonial stem-cell-specific miR-221 is involved in the regulation of spermatogenesis in mice (Smorag et al., 2012). Qin et al, (2019) confirmed that circRNA-9119 is a regulatory circRNA involved in testicular inflammation and acts as a sponge for miR26a and miR-136 in Sertoli cells and Leydig cells in response to mimic RNAs, and that the stimulation of viral replication produces poly I:C. These results indicate that circRNAs have potential roles in the regulation of testicular development or sperm motility in Yili geese, and whether these circRNAs really affect testicular development or sperm motility requires further experiments in order to study and verify their mechanisms.
In summary, this study analyzed the expression profile of circRNAs in the testis of Yili geese with high and low sperm motility, identified a large number of circRNAs in the testis, revealed the genomic characteristics and length distribution of circRNAs, and constructed the circRNA-miRNA interaction network. The results suggest that circRNAs may regulate the sperm motility of Yili geese through interaction with miRNAs, which provides a solid foundation for identifying and characterizing the key circRNAs involved in testis development or the regulation of sperm motility.
The data presented in the study are deposited in the BioProjectin NCBI repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA856143), accession number is PRJNA856143.
The animal study was reviewed and approved by Animal Welfare and Ethics Committee of XinJiang Agricultural University (Animal Protocol number:2020035).
Conceptualization, HL, and YW; methodology, YW; validation, YW and XZ; investigation, YW, XZ, GB, CM, and YY; resources, HL; data curation, YW, JL, YY, LW and GB; writing—original draft preparation, YW and XZ; writing-review and editing, YW; visualization, HL; supervision, HL; project administration, HL; funding acquisition, HL.
This study was supported by the National Natural Science Foundation of China (Grant No. 32160785), 2022 Xinjiang Agricultural University Graduate Research Innovation Project (XJAUGRI2022004); and 2022 Xinjiang poultry breeding technology integration demonstration and promotion project (2022xjjq-z-01).
We highly acknowledge Hengxin Industrial Co., Ltd. of Ermin County, Xinjiang, China, who provided the animals and physical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.970097/full#supplementary-material
Aby, Lewin., and Lavon., Haim (1997). The effect of coenzyme Q10 on sperm motility and function. Mol. Asp. Med. 18, s213–s219. doi:10.1016/s0098-2997(97)00036-8
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31 (2), 166–169. doi:10.1093/bioinformatics/btu638
Balercia, G., Buldreghini, E., Vignini, A., Tiano, L., Paggi, F., Amoroso, S., et al. (2009). Coenzyme Q10 treatment in infertile men with idiopathic asthenozoospermia: A placebo-controlled, double-blind randomized trial. Fertil. Steril. 91 (5), 1785–1792. doi:10.1016/j.fertnstert.2008.02.119
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi:10.1373/clinchem.2008.112797
Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5 (1), R1. doi:10.1186/gb-2003-5-1-r1
Estermann, M. A., Major, A. T., and Smith, C. A. (2021). Genetic regulation of avian testis development. Genes 12 (9), 1459. doi:10.3390/genes12091459
Finkelstein, M., Megnag, B., Ickowicz, D., and Breitbart, H. (2013). Regulation of sperm motility by PIP2(4, 5) and actin polymerization. Dev. Biol. 381 (1), 62–72. doi:10.1016/j.ydbio.2013.06.014
Gadadhar, S., Viar, G. A., Hansen, J. N., Gong, A., Kostarev, A., Ialy-Radio, C., et al. (2021). Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science 371 (6525), eabd4914. doi:10.1126/science.abd491410.1126/science.abd4914
Gao, Y., Wang, J., and Zhao, F. (2015). CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16 (1), 4. doi:10.1186/s13059-014-0571-3
Guo, T. Y., Huang, L., Yao, W., Du, X., Li, Q. Q., Ma, M. L., et al. (2020). The potential biological functions of circular RNAs during the initiation of atresia in pig follicles. Domest. Anim. Endocrinol. 72, 106401. doi:10.1016/j.domaniend.2019.106401
Hamed, N., Hamid, S., Mohammadi, A., Soleimanifar, F., Izadpanah, F., Haddad Kashani, H., et al. (2019). The protective effect of coenzyme Q10 and berberine on sperm parameters, with and without varicocelectomy in rats with surgically induced varicoceles. Comp. Clin. Path. 28 (2), 479–485. doi:10.1007/s00580-018-2850-y
Hohmann, T., and Dehghani, F. (2019). The cytoskeleton-A complex interacting meshwork. Cells 8 (4), 362. doi:10.3390/cells8040362
Huo, min. (2018). Effects of Antioxidants on Cryopreservation of Cashmere Semen. Chinese Dissertation of Inner Mongolia Agricultural University.
Hya, B., Jla, B., Syxa, B., Han, X. Y., Song, X. T., Qi, M. Y., et al. (2022). miRNA expression analysis of the sheep follicle during the prerecruitment, dominant, and mature stages of development under FSH stimulation. Theriogenology 181, 161–169. doi:10.1016/j.theriogenology.2022.01.001
Iddrriss, A. R. I., Obese, F. Y., and Robinson, J. E . (2018). Testis size and asymmetry in the Guinea fowl (Numida meleagris): A test of the compensation hypothesis. Avian Biol. Res. 11 (2), 123–131. doi:10.3184/175815618X15203333340470
Jerysz, A., and Lukaszewicz, E. (2013). Effect of dietary Selenium and Vitamin E on ganders' response to semen collection and Ejaculate characteristics. Biol. Trace Elem. Res. 153 (1-3), 196–204. doi:10.1007/s12011-013-9652-5
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi:10.1093/nar/gkm882
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Kruger, J., and Rehmsmeier, M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, 451–454. doi:10.1093/nar/gkl243
Lafuente, R., Gonzalez-Comadran, M., Sola, I., Lopez, G., Brassesco, M., Carreras, R., et al. (2013). Coenzyme Q10 and male infertility: A meta-analysis. J. Assist. Reprod. Genet. 30 (9), 1147–1156. doi:10.1007/s10815-013-0047-5
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10 (3), R25–R10. doi:10.1186/gb-2009-10-3-r25
Lei, Y., Zhang, X., Xu, Q., Liu, S., Li, C., Jiang, H., et al. (2021). Autophagic elimination of ribosomes during spermiogenesis provides energy for flagellar motility. Dev. Cell 56 (16), 2313–2328.e7. doi:10.1016/j.devcel.2021.07.015
Li, T. T., Luo, R. R., Wang, X., Wang, H., Zhao, X., Guo, Y., et al. (2021). Unraveling stage-dependent expression patterns of circular RNAs and their related ceRNA modulation in ovine postnatal testis development. Front. Cell Dev. Biol. 9, 627439. doi:10.3389/fcell.2021.627439
Li, X., Yang, L., and Chen, L. L. (2018). The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71 (3), 428–442. doi:10.1016/j.molcel.2018.06.034
Liang, G., Yan, J., Guo, J., and Tang, Z. (2020). Identification of ovarian circular RNAs and differential expression analysis between Meishan and Large White pigs. Animals. 10 (7), 1114. doi:10.3390/ani10071114
Liang, G., Yang, Y., Niu, G., Tang, Z., and Li, K. (2017). Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 24 (5), 523–535. doi:10.1093/dnares/dsx022
Liu, C., Wang, H. N., Shang, Y. L., Liu, W., Song, Z., Zhao, H., et al. (2016). Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy 12 (5), 814–832. doi:10.1080/15548627.2016.1159377
Liu, H. T., Cheng, S. B., Huang, Y. C., and Lin, P. T. (2017). Coenzyme Q10 and oxidative stress: Inflammation Status in Hepatocellular carcinoma Patients after Surgery. Nutrients 9 (1), 29. doi:10.3390/nu9010029
Lukaszewicz, E., Kowalczyk, A., and Jerysz, A. (2021). Characteristics of semen collected from gander included in the genetic resources conservation program. Poult. Sci. 100 (9), 101314. doi:10.1016/j.psj.2021.101314
Mahmoudi, E., and Cairns, M. J. (2019). Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci. Rep. 9, 2564. doi:10.1038/s41598-019-38860-9
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21 (19), 3787–3793. doi:10.1093/bioinformatics/bti430
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 (7441), 333–338. doi:10.1038/nature11928
Michael, I. L., Wolfgang, H., and Simon, A. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Nguyen, T. M. D. (2019). Main signaling pathways involved in the control of fowl sperm motility. Poult. Sci. 98, 1528–1538. doi:10.3382/ps/pey465
Nguyen, T. P., Casarin, A., Desbats, M. A., Doimo, M., Trevisson, E., Santos-Ocana, C., et al. (2014). Molecular characterization of the human COQ5 C -methyltransferase in coenzyme Q10 biosynthesis. Biochim. Biophys. Acta 1841, 1628–1638. doi:10.1016/j.bbalip.2014.08.007
Ouyang, H. J., Chen, X. L., Wang, Z. J., Yu, J., Jia, X., Li, Z., et al. (2018). Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 25 (1), 71–86. doi:10.1093/dnares/dsx039
Panda, A. C. (2018). “Circular RNAs act as miRNA sponges,” in Circular RNAs. Advances in experimental medicine and biology, vol 1087. Editor J. Xiao (Singapore: Springer). doi:10.1007/978-981-13-1426-1_6
Paul, S., Andrew, M., Owen, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Qin, L., Lin, J., and Xie, X. (2019). CircRNA-9119 suppresses poly I:C induced inflammation in Leydig and Sertoli cells via TLR3 and RIG-I signal pathways. Mol. Med. 25, 28. doi:10.1186/s10020-019-0094-1
Qiu, L., Chang, G., Bi, Y., Liu, X., and Chen, G. (2018). Circular RNA and mRNA profiling reveal competing endogenous RNA networks during avian leukosis virus, subgroup J-induced tumorigenesis in chickens. PLoS One 13 (10), e0204931. doi:10.1371/journal.pone.0204931
Quan, G., and Li, J. (2018). Circular RNAs: Biogenesis, expression and their potential roles in reproductionCircular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 11 (1), 9. doi:10.1186/s13048-018-0381-4
Shen, M., Li, T., Zhang, G., Wu, P., Chen, F., Lou, Q., et al. (2019). Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genomics 20 (1), 96. doi:10.1186/s12864-019-5462-2
Shen, M. M., Wu, P., Li, T. T., Wu, P. F., Chen, F. X., Chen, L., et al. (2020). Transcriptome analysis of circRNA and mRNA in Theca cells during follicular development in chickens. Genes 11 (5), 489. doi:10.3390/genes11050489
Shen, M., Wu, P., Li, T., Wu, P., Chen, F., Chen, L., et al. (2020). Transcriptome analysis of circRNA and mRNA in Theca cells during follicular development in chickens. Genes 11 (5), 489. doi:10.3390/genes11050489
Shen, X. X., Liu, Z. H., Cao, X. N., He, H., Han, S., Chen, Y., et al. (2019). Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int. J. Biol. Sci. 15 (10), 2265–2281. doi:10.7150/ijbs.36412
Shetty, R. A., Forster, M. J., and Sumien, N. (2013). Coenzyme Q(10) supplementation reverses age-related impairments in spatial learning and lowers protein oxidation. Age 35 (5), 1821–1834. doi:10.1007/s11357-012-9484-9
Smorag, L., Zheng, Y., Nolte, J., Zechner, U., Engel, W., and Pantakani, D. V. K. (2012). MicroRNA signature in various cell types of mouse spermatogenesis: Evidence for stage-specifically expressed miRNA-221, -203 and -34b-5p mediated spermatogenesis regulation. Biol. Cell 104 (11), 677–692. doi:10.1111/boc.201200014
Tang, C., Xie, Y., Yu, T., Liu, N., Wang, Z., Woolsey, R. J., et al. (2020). m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 30 (3), 211–228. doi:10.1038/s41422-020-0279-8
Vadnais, M. L., Aghajanian, H. K., Lin, A., and Gerton, G. L. (2013). Signaling in sperm: Toward a molecular understanding of the acquisition of sperm motility in the mouse epididymis. Biol. Reprod. 89 (5), 127. doi:10.1095/biolreprod.113.110163
Varela-Lopez, A., Ochoa, J. J., Llamas-Elvira, J. M., Lopez-Frias, M., Planells, E., Ramirez-Tortosa, M., et al. (2017). Age-related Loss in Bone Mineral density of rats fed Lifelong on a fish Oil-based diet is Avoided by coenzyme Q10 addition. Nutrients 9 (3), 176. doi:10.3390/nu9020176
Wang, L. L., You, Z., Wang, M. Y., Yuan, Y., Liu, C., Yang, N., et al. (2020). Genome-wide analysis of circular RNAs involved in Marek's disease tumourigenesis in chickens. RNA Biol. 17 (4), 517–527. doi:10.1080/15476286.2020.1713538
Wang, S. S. (2018). Integrated analysis to identify candidate functional genes for fat formation and deposition in ducks. Yangzhou: Yangzhou University, 43–53. doi:10.1038/s41598-017-04178-7
Wu, Y. P., Ding, Y. W., Li, H. Y., Zhao, X. Y., Cao, Y., Pan, L., et al. (2022). Comparative study on semen quality, reproductive performance and testicular development of Yili geese with high and low sperm motility. Chin. J. Animal Sci., 1–9. doi:10.19556/j.0258-7033.20211202-01
Wu, Y., Xiao, H., Pi, J., Zhang, H., Pan, A., Pu, Y., et al. (2020). The circular RNA aplacirc_13267 upregulates duck granulosa cell apoptosis by the apla-miR-1-13/THBS1 signaling pathway. J. Cell. Physiol. 235 (18), 5750–5763. doi:10.1002/jcp.29509
Yan, Li., Gao., Guangliang, Yu, Lin., Hu, S., Luo, Y., Wang, G., et al. (2020). Pacific Biosciences assembly with Hi-C mapping generates an improved, chromosome-level goose genome. GigaScience 9 (10), giaa114. doi:10.1093/gigascience/giaa114
Yefimova, M. G., Messaddeq, N., Harnois, T., Meunier, A. C., Clarhaut, J., Noblanc, A., et al. (2013). A chimerical phagocytosis model reveals the recruitment by Sertoli cells of autophagy for the degradation of ingested illegitimate substrates. Autophagy 9 (5), 653–666. doi:10.4161/auto.23839
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11 (2), R14. doi:10.1186/gb-2010-11-2-r14
Yuan, G., Wu, M., Fan, Y., Li, S., Lai, Z., Huang, Y., et al. (2018). Identification and characterization of circular RNAs in Qinchuan cattle testis. R. Soc. Open Sci. 5 (7), 180413. doi:10.1098/rsos.180413
Zhang, F., Zhang, X., Ning, W., Zhang, X., Ru, Z., Wang, S., et al. (2021). Expression analysis of circular RNAs in Y oung and Sexually mature boar testes. Animals. 11 (5), 1430. doi:10.3390/ani11051430
Zhao, X. Y., Shaershanbieke, A., Ma, C., Wu, Y. P., Wang, J. H., Li , H. Y., et al. (2020). Effects of dietary Crude protein I evel on Laying raws, reproductive Hormone concentrations and related gene mRNA expression in reproductive Axis of yili geese. China Anim. Husb. & Veterinary Med. 47 (06), 1729–1738. doi:10.16431/j.cnki.1671-7236.2020.06.011
Zhao, X. Y., Wu, Y. P., Peng, X., Wang, J. H., Duan, Y. Q., Li, Z. P., et al. (2019). Effects of dietary Crude protein level on performance, Hatching performance and Serum Biochemcial Indexes of yili geese. Chin. J. Animal Nutr. 31 (04), 1630–1636.
Zhong, X. L., and Zhong, C. G. (2014). Role of coenzyme Q in xenobiotics-induced mitochondrial damage:its advances[J]. Chin. J. Pharmacol. Toxicol. 28 (2), 302–308. doi:10.3867/j.issn.1000-3002.2014.02.027
Zhong, Y. X., Du, Y. J., Yang, X., Mo, Y., Fan, C., Xiong, F., et al. (2018). Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 17 (1), 79. doi:10.1186/s12943-018-0827-8
Zhou, L., Chen, J., Li, Z., Li, X., Hu, X., Huang, Y., et al. (2010). Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One 305 (12), e15224. doi:10.1371/journal.pone.0015224
Zhou, T., Xie, X., Li, M., Shi, J., Zhou, J., Knox, K., et al. (2018). Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA 24, 1443–1456. doi:10.1261/rna.067132.118
Keywords: yili geese, sperm motility, testis, circrnas, transcriptomic
Citation: Wu Y, Li H, Zhao X, Baki G, Ma C, Yao Y, Li J, Yao Y and Wang L (2022) Differential expression of circRNAs of testes with high and low sperm motility in Yili geese. Front. Genet. 13:970097. doi: 10.3389/fgene.2022.970097
Received: 15 June 2022; Accepted: 07 September 2022;
Published: 26 September 2022.
Edited by:
Xiao Wang, Konge Larsen ApS, DenmarkReviewed by:
Md Zubbair Malik, Jawaharlal Nehru University, IndiaCopyright © 2022 Wu, Li, Zhao, Baki, Ma, Yao, Li, Yao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiying Li, bGh5LTNAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.