- 1Department of Urology, The First Clinical Medical College of Jinan University, Jinan University, Guangzhou, China
- 2Department of Urology, The People’s Hospital of Guangxi Zhuang Autonomous Region, Nanning, China
Background: Clear cell renal cell carcinoma (ccRCC) is the most common pathological type of renal cell carcinoma. Tetratricopeptide repeat domain 21A (TTC21A), known as a component of intraflagellar transport complex A which is essential for the function of cilia, However, the role of TTC21A remains unclear in ccRCC. For the first time, we explore the role and potential mechanism of TTC21A in ccRCC based on multiple databases.
Methods: TTC21A expression across all TCGA tumor was analyzed via Tumor Immune Estimation Resource (TIMER) site. The correlation between TTC21A and clinicopathologic characteristics of ccRCC was analyzed with TCGA database. The diagnostic and prognostic value of TTC21A was evaluated by receiver operation characteristic curve, Kaplan-Meier plotter and Cox regression respectively. Moreover, functional enrichment analysis of TTC21A and the co-expression genes were performed by Gene Set Enrichment Analysis. The correlation of TTC21A and immune infiltration were evaluated by single sample Gene Set Enrichment Analysis.
Results: Pan-cancer analysis indicated that TTC21A was highly expressed in ccRCC and other cancer. In addition, elevated expression of TTC21A was associated with worse overall survival in ccRCC patients. Functional enrichment analysis showed that TTC21A and the co-expressed genes enriched in glucose metabolism and energy metabolism. Moreover, TTC21A expression was associated with infiltrating levels of dendritic cell, nature killer cell and other immune marker sets.
Conclusion: The results of analysis indicate that expression of TTC21A is associated with poor prognosis and immune infiltrating in ccRCC, which suggested TTC21A might be used as a potential predictor and target of treatment in ccRCC.
Introduction
Renal cell carcinoma (RCC) ranked the third most common urologic cancers worldwide (Sung et al., 2021). Furthermore, it represents the rising and the first mortality rate in the common urologic cancers (Partin et al., 2015). Despite advances in surgery for localized RCC, the efficacy of treatment for advanced or metastatic RCC is inadequate (Barata and Rini, 2017). Anatomical factors, including distant metastasis and perinephric invasion, are the important prognostic factors of RCC (Ljungberg et al., 2015). Therefore, lots of researches focused on advanced or metastatic RCC. Previous study had reported that immune infiltration of tumor was correlated with prognosis in RCC (Zhang et al., 2019). Tumor-infiltration immune cells, including CD4+ T cells, CD8+ T cells and Tfh cells, could regulate cancer progression in RCC, which could be regarded as targets for novel drugs to improve outcome (Vesely et al., 2011; Wang et al., 2018).
Renal cell carcinoma is consisted of three primary histologic subtypes: clear cell RCC, papillary RCC (pRCC) and chromophobe RCC (chRCC). Clear cell renal cell carcinoma (ccRCC or KIRC) is the most common subtype of RCC, which account for 70–80% of all RCC (Partin et al., 2015). Compared to pRCC or chRCC, the prognosis of ccRCC is worse, and 5-year relative survival of metastatic ccRCC is less than 10% (Cohen and McGovern, 2005). With the novel immunotherapy, the patients with ccRCC could have a longer overall survival. Nowadays, immune checkpoint inhibitor has been recommended as a treatment for advanced or metastatic RCC. Programed death-1 (PD-1) and cytotoxic T lymphocyte associated antigen 4 (CTLA4) has been considered as second-line therapy for RCC (Ljungberg et al., 2015).
Tetratricopeptide repeat domain 21A (TTC21A), a gene with protein coding, is a component of intraflagellar transport complex A (IFT-A complex) which is essential for the function of cilia (Picariello et al., 2019). Some studies had indicated that cilia contributed to tumorigenesis. Downregulation of cilia expression could be observed in renal cell carcinoma, pancreatic cancer, cholangiocarcinoma, and melanoma (Schraml et al., 2009; Kim et al., 2011; Gradilone et al., 2013). Besides, there were some researches indicated that IFT complex was associated with different types of cancers (Pidugu et al., 2019; Jiang et al., 2021; Tan et al., 2021). However, the investigations of TTC21A in tumor progression were limited. Previous studies reported TTC21A was associated with the prognosis and immune infiltrating level in lung adenocarcinoma (LUAD) and colorectal cancer (Wang et al., 2020; Li et al., 2022). The purpose of present study is to explore the role of TTC21A in clear cell renal cell carcinoma.
The data of clear cell renal cell carcinoma was downloaded from The Cancer Genome Atlas (TCGA) database. The diagnostic capacity of TTC21A expression in ccRCC was evaluated by receiver operation characteristic (ROC) curve. Meanwhile, the influence of TTC21A on the prognosis of ccRCC was analyzed by Kaplan-Meier plotter and Cox regression. Furthermore, the single sample Gene Set Enrichment Analysis (ssGSEA) was used to examine the correlations between TTC21A and different kinds of tumor-infiltrating immune cells (TILCs) in ccRCC. The results of present study could improve the understanding of the role of TTC21A in ccRCC, which make it possible to become a novel biomarker to predict tumor prognosis and immune infiltration for ccRCC patients.
Materials and methods
TCGA datasets analysis
Transcriptional expression data of TTC21A in clear cell renal cell carcinoma was downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov) database, which included 539 ccRCC tissues and 72 normal tissues. The gene expression with FPKM type was converted into the type of TPM for further analysis. The result was visualize using GraphPad Prism software (version 6.0; GraphPad software, Inc.). Statistical software R (https://www.r-project.org; version 3.6.3) was used for further analysis of TCGA database. Transcript data and clinical data of 33 TCGA cancer types was also downloaded from TCGA database for univariate cox regression analysis, and the result was visualized with a forest plot by using the “forestplot” package of R (https://CRAN.R-project.org/package=forestplot; version 2.0.1). The discrimination ability of TTC21A in KIRC was evaluated by ROC curve, and ROC curve was performed by “pROC” package of R (https://CRAN.R-project.org/package=pROC; version 1.18.0) (Robin et al., 2011). Multivariate analysis of overall survival was achieved by “survival” package of R (https://CRAN.R-project.org/package=survival; version 3.2–13), and nomogram and calibration plot were constructed by “survival” package and “rms” package of R (https://CRAN.R-project.org/package=rms; version 6.2–0) (Iasonos et al., 2008).
Kaplan-meier plotter database analysis
Kaplan-Meier Plotter Database can be used to analyze the effect of genes on survival in different types of cancer (Nagy et al., 2021). The correlation between TTC21A expression and overall survival in ccRCC was analyzed by Kaplan-Meier plotter (http://www.kmplot.com/analysis/). The log-rank p value and the hazard ratio were calculated.
LinkedOmics database analysis
The LinkedOmics database (http://www.linkedomics.org/login.php) is a publicly available platform for analyzing multi-omics data of 32 TCGA cancer types (Vasaikar et al., 2018). TTC21A co-expression was analyzed by using Pearson’s test, and the results were presented by volcano plot and heatmap. Gene Ontology (GO) term enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of TTC21A co-expression genes was performed by using gene set enrichment analysis (GSEA).
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was conducted with a ccRCC cohort from TCGA database by using the GSEA software (www.gsea-msigdb.org/gsea/index.jsp) (Mootha et al., 2003; Subramanian et al., 2005). GSEA was performed for Gene Ontology (GO) term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment with 1000 permutations, and corresponding gene sets (C5. go. v7.5. symbols. gmt and C2. cp. kegg. v7.5. symbols. gmt) were used as reference gene sets (Liberzon et al., 2011; Liberzon et al., 2015). False discovery rate (FDR) q-value < 0.25, |normalized enrichment score (NES)| > 1 and adjust p-value < 0.05 were identified as significant.
TIMER database analysis
TIMER is a comprehensive web for analysis of immune infiltrates in different cancer types (http://timer.comp-genomics.org/) (Li et al., 2020). The TIMER database comprises 10897 samples from The Cancer Genome Atlas (TCGA). The expression of TTC21A in different cancer types was analyzed by TIMER database.
Immune infiltration analysis
The correlation between TTC21A and tumor-infiltrating immune cells was analyzed by ssGSEA with “GSVA” package of Bioconductor (https://bioconductor.org/packages/GSVA/) (Hänzelmann et al., 2013). The enrichment score of immunocyte was quantified based on the markers of 24 types immunocyte as reported previously (Bindea et al., 2013). The correlation between TTC21A and immune infiltrates cells was analyzed by Spearman correlation.
Statistical analysis
The TTC21A expression between tumor and normal tissues was analyzed with Wilcoxon rank sum test. The correlation between TTC21A expression and clinicopathological characteristics were compared with Pearson χ2 test and Wilcoxon rank sum test. Survival curves were generated in Kaplan-Meier Plotter Database, and were displayed hazard ratio (HR) and p - values. The correlation between TTC21A expression and immune infiltration were analyzed by Spearman correlation. p < 0.05 was considered significant.
Results
The expression of TTC21A in different types of cancers
The expression of TTC21A in different types of cancers was evaluated by using TERM database. The TTC21A expression between tumor and normal tissues in TCGA tumors was shown in Figure 1A. The expression of TTC21A was significantly higher in CHOL (Cholangiocarcinoma), KIRC (Kidney Renal Clear Cell Carcinoma), KIRP (Kidney Renal Papillary Cell Carcinoma), LIHC (Liver Hepatocellular Carcinoma), PCPG (Pheochromocytoma and Paraganglioma), PRAD (Prostate Adenocarcinoma), READ (Rectum Adenocarcinoma), STAD (Stomach Adenocarcinoma) compared with that of normal tissues. However, TTC21A expression was significantly lower in HNSC (Head and Neck Cancer), KICH (Kidney Chromophobe), LUAD (Lung Adenocarcinoma), LUSC (Lung Squamous Cell Carcinoma), THCA (Thyroid Carcinoma) compared with normal tissues. Analysis of ccRCC cohort in TCGA database revealed that TTC21A expression was significantly elevated in tumor tissues compared with adjacent normal tissues (Figure 1B).
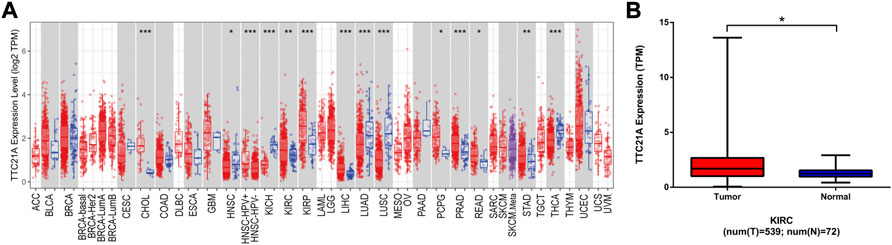
FIGURE 1. Differential expression of TTC21A. (A) Differential expression of TTC21A in 33 types cancers (*p < 0.05, **p < 0.01, ***p < 0.001) (B) Differential expression of TTC21A in ccRCC and normal tissues from TCGA database (*p < 0.05).
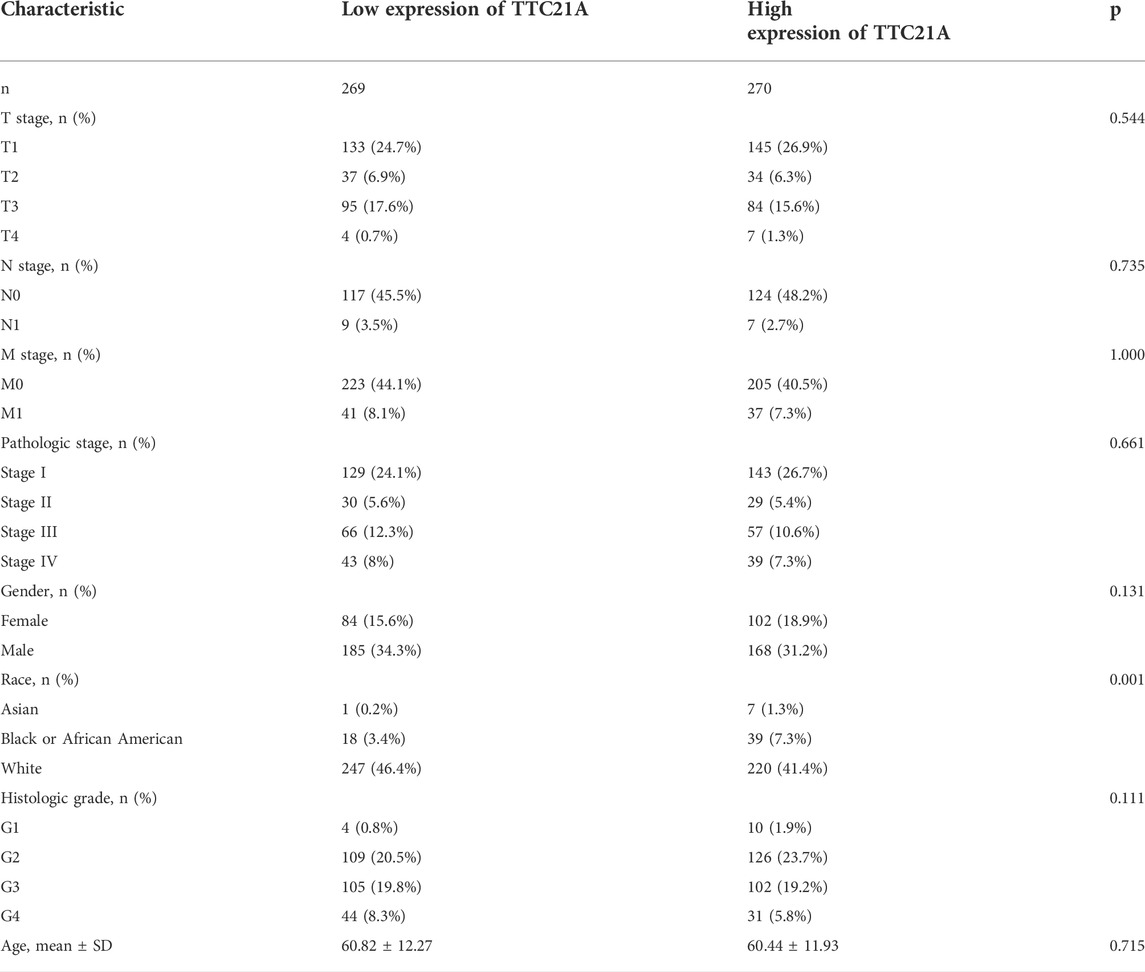
Correlation between TTC21A expression and clinicopathologic characteristics
539 KIRC samples were classified into two groups by the median value of TTC21A expression. As shown in Table 1, high expression of TTC21A was related with race (p = 0.001). Whereas, the other clinicopathologic characteristics had no different between high- and low-expression groups. Different races showed significant difference in the expression of TTC21A (Figure 2A). Besides, the area under the ROC curve (AUC) showed a diagnostic accuracy of TTC21A in KIRC, however, the accuracy was low (AUC = 0.649, 95%CI = 0.600–0.698) (Figure 2B).
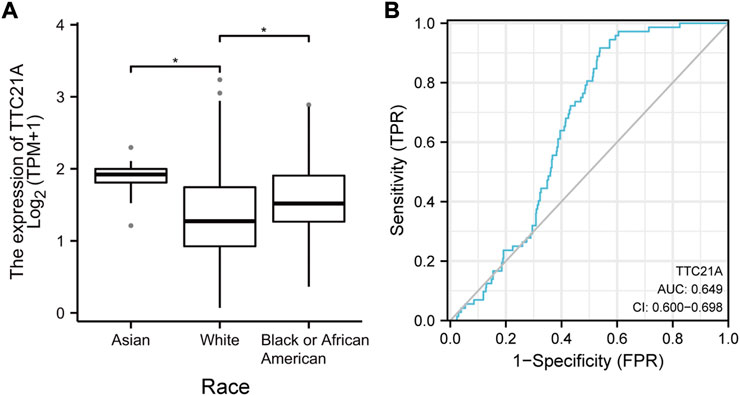
FIGURE 2. Correlation of TTC21A and clinicopathologic characteristics in ccRCC. (A) TTC21A expression was associated with different races (*p < 0.05) (B) Receiver operation characteristic (ROC) curve of TTC21A (TPR true positive rate, FPR false positive rate).
TTC21A expression is correlated with survival in ccRCC
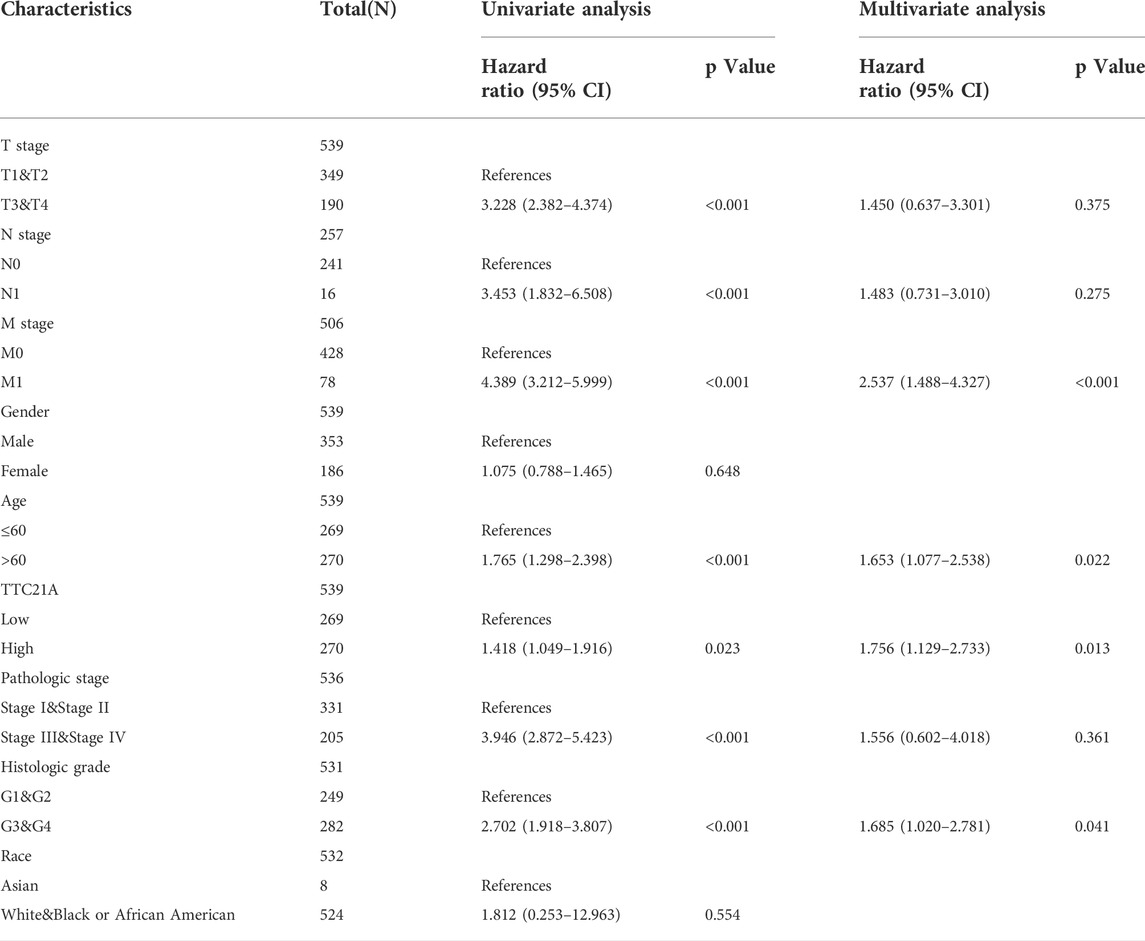
Univariate Cox regression indicated that high TTC21A expression was associated with overall survival in KIRC, HNSC, LUAD, READ and STAD (Figure 3A). Kaplan-Meier survival curve revealed that high expression of TTC21A was associated with worse prognosis in KIRC (OS HR = 1.86, 95%CI = 1.37 to 2.52, log-rank test, p < 0.05) (Figure 3B). In addition, the elevated TTC21A expression led to significantly shorter overall survival (OS) in different stage of ccRCC (Stage I HR = 2.05, 95%CI = 1.12 to 3.78, log-rank test, p < 0.05; Stage IV HR = 1.80, 95%CI = 1.10 to 2.95, log-rank test, p < 0.05) (Figures 3D,G). However, High TTC21A expression was not associated with relapse free survival (RFS) and stage II, Stage III disease in patients with ccRCC (Figures 3C,E,F).
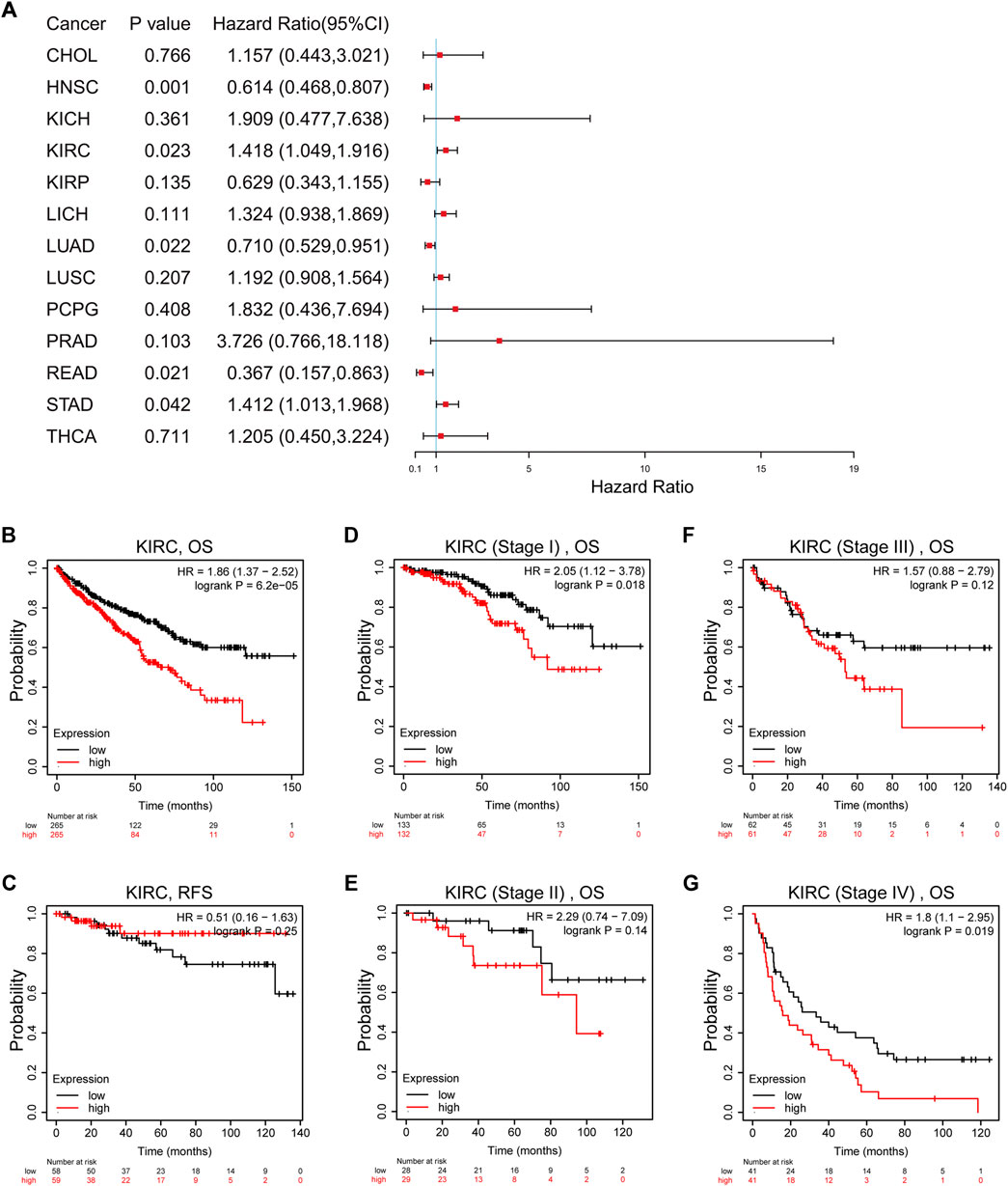
FIGURE 3. Survival analysis of TTC21A in ccRCC. (A) Correlation between TTC21A expression and overall survival of different types cancers. (B) Overall survival was significantly higher in patients with low TTC21 expression (HR = 1.86, 95%CI = 1.37 to 2.52, log-rank test, p < 0.05) (C) Relapse free survival was not significantly higher in patients with low TTC21 expression (HR = 0.51, 95%CI = 0.16 to 1.63, log-rank test, p > 0.05). (D–G) Overall survival in stage I and stage IV of ccRCC were significantly higher in patients with low TTC21 expression, while overall survival in stage II and stage III were not significant.
Univariate and multivariate Cox analyses indicated that M stage (p < 0.001), age (p < 0.05), histologic grade (p < 0.05) and TTC21A expression (p < 0.05) were independently associated with overall survival (Table 2). Based on multivariate Cox regression, the nomogram was conducted (Figure 4A). Potential covariates included M stage, age, histologic grade and TTC21A expression. The concordance index (C-index) was used to evaluate the predictive ability of the prognostic model (C-index = 0.741, 95%CI = 0.722–0.760). Calibration curve of the nomogram was made, and the bias-corrected lines were approaching the ideal line (Figure 4B).
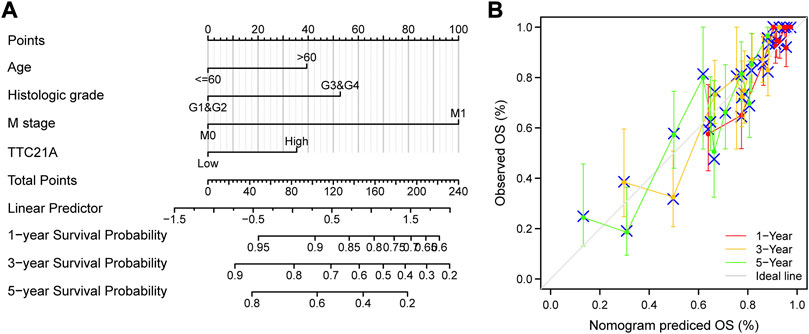
FIGURE 4. Predictive ability of TTC21A in ccRCC. (A) Nomogram for predicting overall survival of ccRCC. (B) Calibration curve of the nomogram.
Biological functions of TTC21A in ccRCC
GO term enrichment analysis and KEGG pathway enrichment analysis between low and high TTC21A expression were performed by GSEA. GO term enrichment analysis in biological process (BP) revealed that high expression of TTC21A was associated with ‘ATP synthesis coupled electron transport’, “electron transport chain”, oxidative phosphorylation’ and “tricarboxylic acid cycle” (Figures 5A–D). KEGG pathway enrichment analysis revealed that high expression of TTC21A was associated with signaling pathway of mTOR, Notch, VEGF and oxidative phosphorylation (Figures 5E–H).
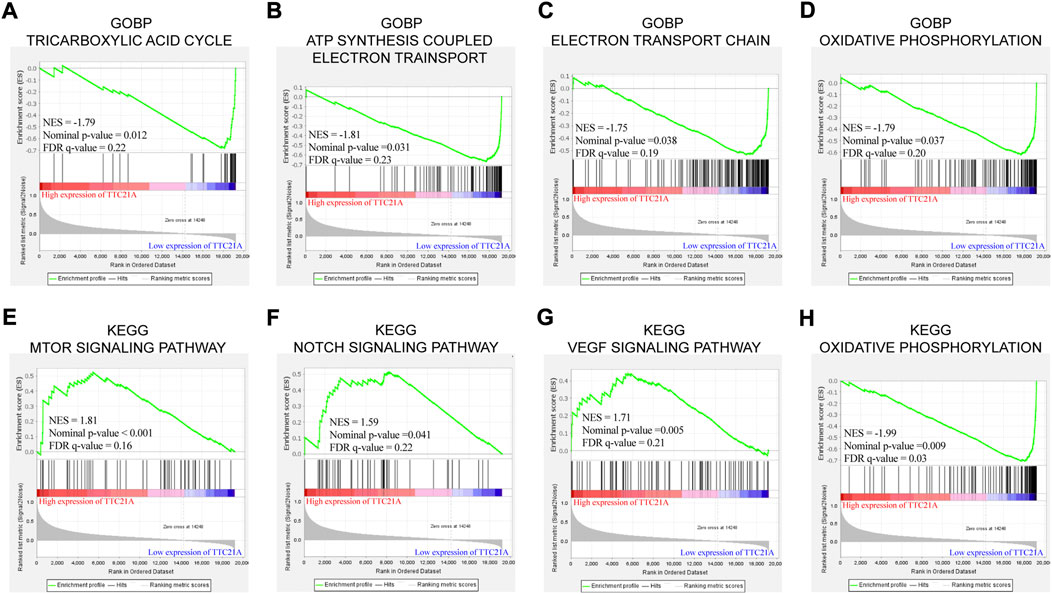
FIGURE 5. Enrichment analysis of TTC21A in ccRCC. Tricarboxylic acid cycle (A), ATP synthesis coupled electron transport (B), electron transport chain (C) and oxidative phosphorylation (D) were enriched in biological process of ccRCC. MTOR signaling pathway (E), NOTCH signaling pathway (F), VEGF signaling pathway (G) and oxidative phosphorylation (H) were enriched in KEGG pathway.
The co-expression networks of TTC21A in ccRCC
The database of LinkedOmics was performed to evaluate the co-expression networks of TTC21A in ccRCC. 8463 genes (red dots) were positively correlated with TTC21A significantly, whereas 4634 genes (green dots) were negatively correlated with TTC21A (p < 0.05) (Figure 6A). The top 50 genes positively or negatively correlated with TTC21A were presented in the heatmap respectively (Figures 6B,C). Furthermore, GO term enrichment analysis of TTC21A co-expression genes was performed by gene set enrichment analysis (GSEA). The results of GO enrichment analysis in biological processes (BP) indicated that the activities of peroxisome organization, peptidyl-asparagine modification, nucleobase metabolic process, tricarboxylic acid metabolic process, and peroxisomal transport were inhibited by co-expression genes (Figure 6D). KEGG pathway enrichment suggested that TTC21A co-expression genes were negatively associated with glycosphingolipid biosynthesis, prion diseases, terpenoid backbone biosynthesis, biosynthesis of unsaturated fatty acids, adherens junction, ferroptosis, protein processing in endoplasmic reticulum, oxidative phosphorylation, peroxisome, carbon metabolism, valine, leucine and isoleucine degradation, and citrate cycle (TCA cycle) (Figure 6E).
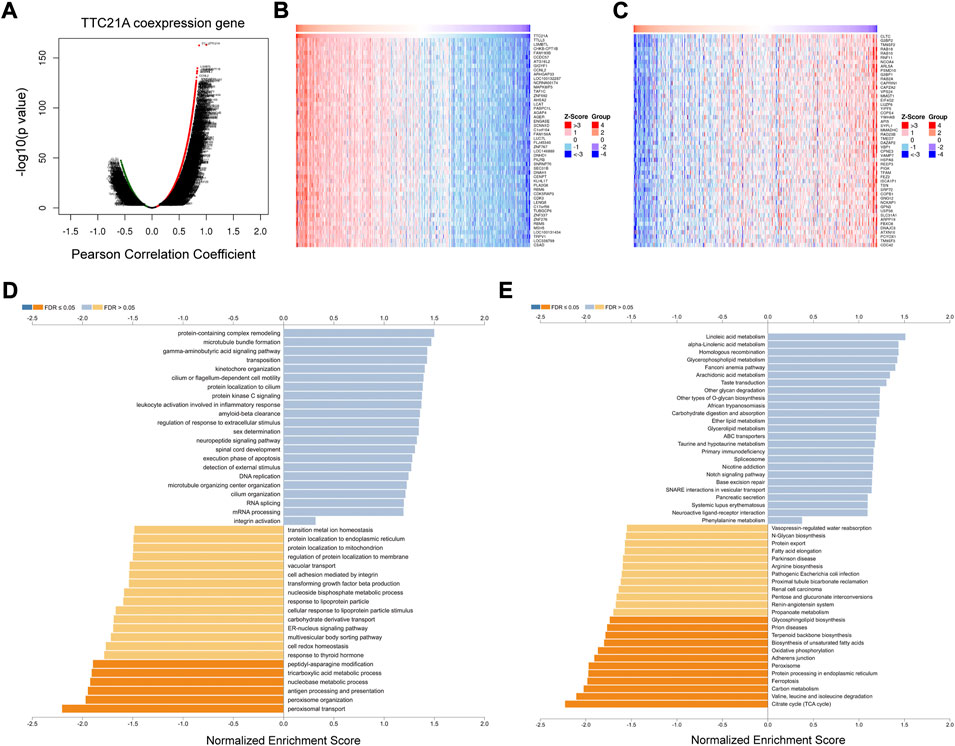
FIGURE 6. Analysis of the co-expression genes of TTC21A in ccRCC. (A) The co-expression genes of TTC21A in ccRCC. (B) Heat map of top 50 genes positively correlated with TTC21A (C) Heat map of top 50 genes negatively correlated with TTC21A. (D) Enrichment analysis of the co-expression genes in biological process of ccRCC. (E) Enrichment analysis of the co-expression genes in KEGG pathway of ccRCC.
Correlation analysis between TTC21A and immune infiltration in ccRCC
We investigated the correlation between TTC21A and tumor-infiltrating immune cells in ccRCC. The results of analysis indicated that TTC21A correlated with most tumor-infiltrating immune cells in ccRCC (Figure 7A). In addition, TTC21A was negatively correlated with B cells, NK CD56dim cells, T follicular helper (TFH), dendritic cells (DC), macrophages, immature dendritic cells (iDC), Type 2 T helper (Th2) cells and T gamma delta (Tgd) (Figures 7B–I).
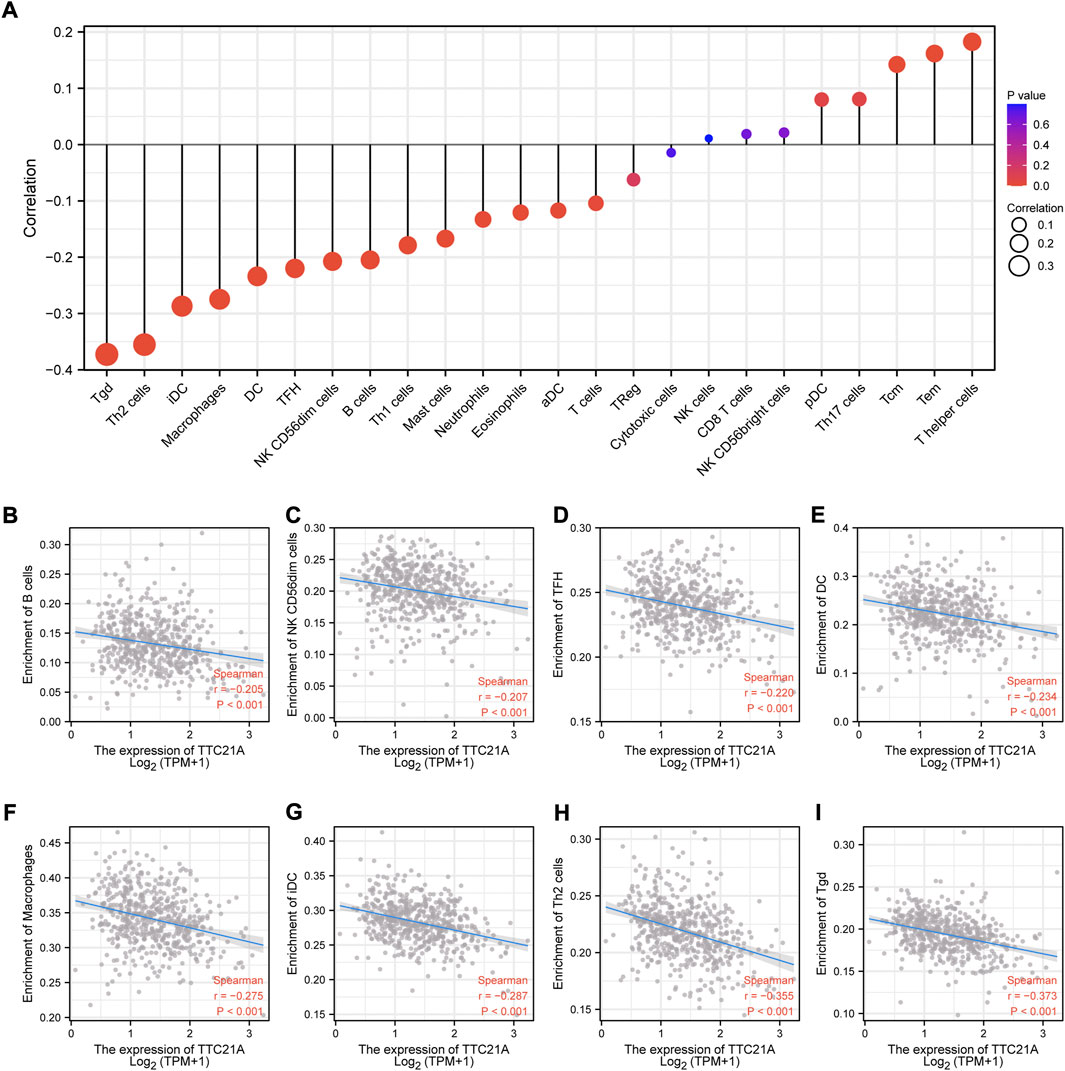
FIGURE 7. Association of TTC21A with immune infiltration. (A) Association of TTC21A with immune infiltration (Tem T effector memory, Tcm T central memory, Th helper T cells, pDC plasmacytoid DC, NK natural killer, Treg regulatory T cells, aDC activated DC, TFH T follicular helper, DC dendritic cells, iDC immature DC, Tgd T gamma delta). (B–I) TTC21A was negatively correlated with infiltration of B cells, NK CD56dim cells, TFH, DC, macrophages, iDC, Th2 cells and Tgd.
Discussion
TTC21A is a protein-coding gene associated with male infertility, and might involve in reduced motility of spermatozoa and multiple morphological abnormalities of the sperm flagella (MMAF) (Liu et al., 2019). In addition, a recent study reported high expression of TTC21A predicts favorable prognosis in lung adenocarcinoma (Wang et al., 2020). In our study, we found that TTC21A was over-expressed in ccRCC, which correlated with poor prognosis. Furthermore, our analysis indicated that immune infiltration levels were correlated with the expression of TTC21A in ccRCC. Thus, TTC21A might be a potential biomarker for ccRCC.
In our study, the expression of TTC21A was calculated in different types of cancer. The result revealed that TTC21A was highly expressed in ccRCC tissues compared to normal tissues. The analysis of clinicopathologic features showed that TTC21A expression was correlated with race. Whereas, other clinicopathologic features were not correlated with high expression of TTC21A. In addition, the ROC analysis suggested that TTC21A might be a diagnostic marker for ccRCC, although the diagnostic power was relatively low. The prognostic value of TTC21A in ccRCC was also evaluated based on the Kaplan-Meier survival analysis and multivariate Cox analysis, which showed that TTC21A was an independent risk factor for ccRCC, and high TTC21A expression was associated with poor OS. All these results suggested that TTC21A might be a potential prognostic marker of ccRCC.
Gene set enrichment analysis and co-expression analysis were performed to explore the possible mechanism of TTC21A in ccRCC. The enrichment analysis of biological process revealed that TTC21A was involved in tricarboxylic acid cycle (TCA cycle), oxidative phosphorylation (OxPhos) and electron transport in ccRCC, which were related to glucose and energy homeostasis. Otherwise, the enrichment analysis of pathway showed that TTC21A was associated with mTOR, Notch, VEGF signaling pathway and oxidative phosphorylation. Glucose homeostasis disorder is an essential part of the Warburg effect, and it was widely accepted the Warburg effect contributes to tumorigenesis, proliferation, angiogenesis, metastasis and immune evasion in tumor (Vaupel et al., 2019). The Warburg effect is mainly characterized by enhanced aerobic glycolysis and mitochondrial dysfunction. Mitochondrial dysfunction can lead to inhibition of OxPhos and TCA cycle via mutation of mitochondrial enzymes (King et al., 2006; Ratcliffe, 2007; Rasheed and Tarjan, 2018; Schmidt et al., 2020; Vaupel and Multhoff, 2021). Research on the mechanism reveal that the Warburg effect is triggered by activation of hypoxia-inducible factor-1 (HIF-1), oncogene activation (e.g. K-Ras, Akt, mTORC1), altered signaling pathways (e.g. PI3K-Akt-mTORC1 signaling pathway and Ras-Raf-MEK-ERK-cMyc signaling pathway) and inactivation of tumor suppressors (e.g. p53 and PTEN) (Senyilmaz and Teleman, 2015; Srinivasan et al., 2016; Cassim et al., 2020; Grasso et al., 2020). Based on the results of enrichment analysis, we hypothesized TTC21A may affect progression of ccRCC by regulating glucose homeostasis via mTOR pathway. However, the hypothesis needs further experiments in vivo for validation.
It is well appreciated that immune infiltration is critical for the progression of diverse tumors (Gajewski et al., 2013). Clear cell renal cell carcinoma is characterized with enrich immune cell infiltrate, including CD4+ cells, CD8+ cells and nature killer (NK) cells (Kopecký et al., 2007; Komohara et al., 2011). Accumulated evidence shown metabolic reprogramming was correlated with tumor microenvironments (TMEs) (Vaupel, 2004; Vaupel and Multhoff, 2018). Metabolic disorder in tumor can shape tumor microenvironments, and lead to immunosuppression and immune evasion (Li and Simon, 2020). Taken together, TTC21A was related to some metabolic processes, which arise the possibility that TTC21A may be associated with immune infiltration. The analysis of the interrelation between TTC21A expression and diverse immune infiltration cell in ccRCC showed the expression of TTC21A was negatively correlated with infiltration level of B cells, NK CD56dim cells, TFH, DC, macrophages, iDC, Th2 cells and Tgd. Accumulated lactate produced by aerobic glycolysis could inhibit NK cells, T cells and M1 state macrophages. In addition, lactate could prohibit the presentation of tumor antigens from DCs to other immune cells (Pavlova and Thompson, 2016). Based on our results, we hypothesized that immune infiltration correlated with TTC21A may contribute to the poor prognosis of ccRCC.
In conclusion, high expression of TTC21A is associated with poor prognosis in ccRCC. Moreover, the metabolic reprogramming and immune infiltration may be the possible mechanism of TTC21A. Obviously, all of the conclusions still need well designed experiments to further confirm. Another limitation was the clinical information was not comprehensive in TCGA database. The significance of present study was to provide a novel predictor and target for local advanced or metastatic RCC.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
Conception and design, WL and JL. Data curation, WL and GH. Analysis and interpretation of data, JL, WW, DN, and CL. Visualization, HW, ZC, XL, and BL. Original draft, JL and XG. Review of the manuscript, All authors.
Funding
This work was funded by National Natural Science Foundation of China (NO: 81760456), the key Research and Development Program of Guangxi (Guike AB 18294027) and the Research Funds of Guangxi Zhuang Autonomous Region Health Commission (NO: Z202000666).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barata, P. A.-O., and Rini, B. I. (2017). Treatment of renal cell carcinoma: Current status and future directions. Ca. Cancer J. Clin. 67, 507–524. (1542-4863 (Electronic)). doi:10.3322/caac.21411
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795. (1097-4180 (Electronic)). doi:10.1016/j.immuni.2013.10.003
Cassim, S., Vucetic, M., Zdralevic, M., and Pouyssegur, J. (2020). Warburg and beyond: The power of mitochondrial metabolism to collaborate or replace fermentative glycolysis in cancer. Cancers (Basel) 12, E1119. [doi] LID - 1119. (2072-6694 (Print)). doi:10.3390/cancers12051119
Cohen, H. T., and McGovern, F. J. (2005). Renal-cell carcinoma. N. Engl. J. Med. 353, 2477–2490. (1533-4406 (Electronic)). doi:10.1056/NEJMra043172
Gradilone, S. A., Radtke, B. N., Bogert, P. S., Huang, B. Q., Gajdos, G. B., and LaRusso, N. F. (2013). HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer Res. 73, 2259–2270. (1538-7445 (Electronic)). doi:10.1158/0008-5472.CAN-12-2938
Grasso, D., Zampieri, L. X., Capeloa, T., Van de Velde, J. A., and Sonveaux, P. (2020). Mitochondria in cancer. Cell Stress 4, 114–146. (2523-0204 (Electronic)). doi:10.15698/cst2020.06.221
Hänzelmann, S., Castelo R Fau - Guinney, J., and Guinney, J. (2013). GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 14, 7. (1471-2105 (Electronic)). doi:10.1186/1471-2105-14-7
Iasonos, A., Schrag, D., Raj, G. V., and Panageas, K. S. (2008). How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 26, 1364–1370. (1527-7755 (Electronic)). doi:10.1200/JCO.2007.12.9791
Jiang, Y., Zhang, C., Zhang, J., Han, D., and Shi, X. (2021). Comprehensive analysis of the prognosis and biological significance for IFIT family in skin cutaneous melanoma. Int. Immunopharmacol. 101, 108344. (1878-1705 (Electronic)). doi:10.1016/j.intimp.2021.108344
Kim, J., Dabiri S Fau - Seeley, E. S., and Seeley, E. S. (2011). Primary cilium depletion typifies cutaneous melanoma in situ and malignant melanoma. PLoS One 6 (11), e27410. (1932-6203 (Electronic)). doi:10.1371/journal.pone.0027410
King, A., Selak Ma Fau - Gottlieb, E., and Gottlieb, E. (2006). Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 25 (34), 4675–4682. (0950-9232 (Print)). doi:10.1038/sj.onc.1209594
Komohara, Y., Hasita, H., Ohnishi, K., Fujiwara, Y., Suzu, S., Eto, M., et al. (2011). Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci. 102, 1424–1431. (1349-7006 (Electronic)). doi:10.1111/j.1349-7006.2011.01945.x
Kopecký, O., Lukesova, S., Vroblova, V., Vokurkova, D., Moravek, P., Safranek, H., et al. (2007). Phenotype analysis of tumour-infiltrating lymphocytes and lymphocytes in peripheral blood in patients with renal carcinoma. Acta Med. 50, 207–212. (1211-4286 (Print)). doi:10.14712/18059694.2017.84
Li, F., and Simon, M. C. (2020). Cancer cells don't live alone: Metabolic communication within tumor microenvironments. Dev. Cell 54, 183–195. (1878-1551 (Electronic)). doi:10.1016/j.devcel.2020.06.018
Li, N., Zhou, Z., Zhang, L., Tang, H., Chen, X., and Zhou, H. (2022). High expression of TTC21A predict poor prognosis of colorectal cancer and influence the immune infiltrating level. Transl. Cancer Res. 11, 981–992. (2219-6803 (Electronic)). doi:10.21037/tcr-21-2674
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514. (1362-4962 (Electronic)). doi:10.1093/nar/gkaa407
Liberzon, A., Birger, C., Thorvaldsdottir, H., Ghandi, M., Mesirov, J. P., and Tamayo, P. (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. (2405-4712 (Print)). doi:10.1016/j.cels.2015.12.004
Liberzon, A., Subramanian, A., Pinchback, R., Thorvaldsdottir, H., Tamayo, P., and Mesirov, J. P. (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. (1367-4811 (Electronic)). doi:10.1093/bioinformatics/btr260
Liu, W., He, X., Yang, S., Zouari, R., Wang, J., Wu, H., et al. (2019). Bi-Allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am. J. Hum. Genet. 104, 738–748. (1537-6605 (Electronic)). doi:10.1016/j.ajhg.2019.02.020
Ljungberg, B., Bensalah, K., Canfield, S., Dabestani, S., Hofmann, F., Hora, M., et al. (2015). EAU guidelines on renal cell carcinoma: 2014 update. Eur. Urol. 67, 913–924. (1873-7560 (Electronic)). doi:10.1016/j.eururo.2015.01.005
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. (1061-4036 (Print)). doi:10.1038/ng1180
Nagy, Á., Munkácsy, G., and Győrffy, B. (2021). Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 11, 6047. (2045-2322 (Electronic)). doi:10.1038/s41598-021-84787-5
Partin, A. W., Wein, A. J., Kavoussi, L. R., and Peters, C. A. (2015). Campbell-walsh urology E-book. New York: Elsevier Health Sciences.
Pavlova, N. N., and Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47. (1932-7420 (Electronic)). doi:10.1016/j.cmet.2015.12.006
Picariello, T. A., Brown, J. M., Hou, Y., Swank, G., Cochran, D. A., King, O. D., et al. (2019), A global analysis of IFT-A function reveals specialization for transport of membrane-associated proteins into cilia. J. Cell Sci. 132, jcs220749, (1477-9137 (Electronic)), doi:10.1242/jcs.220749
Pidugu, V. K., Pidugu, H. B., Wu, M. M., Liu, C. J., and Lee, T. C. (2019). Emerging functions of human IFIT proteins in cancer. Front. Mol. Biosci. 6, 148. (2296-889X (Print)). doi:10.3389/fmolb.2019.00148
Rasheed, M., and Tarjan, G. (2018). Succinate dehydrogenase complex: An updated review. Arch. Pathol. Lab. Med. 142, 1564–1570. (1543-2165 (Electronic)). doi:10.5858/arpa.2017-0285-RS
Ratcliffe, P. J. (2007). Fumarate hydratase deficiency and cancer: Activation of hypoxia signaling? Cancer Cell 11, 303–305. (1535-6108 (Print)). doi:10.1016/j.ccr.2007.03.015
Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J. C., et al. (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 12, 77. (1471-2105 (Electronic)). doi:10.1186/1471-2105-12-77
Schmidt, C., Sciacovelli, M., and Frezza, C. (2020). Fumarate hydratase in cancer: A multifaceted tumour suppressor. Semin. Cell Dev. Biol. 98, 15–25. (1096-3634 (Electronic)). doi:10.1016/j.semcdb.2019.05.002
Schraml, P., Frew, I. J., Thoma, C. R., Boysen, G., Struckmann, K., Krek, W., et al. (2009). Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Mod. Pathol. 22, 31–36. (1530-0285 (Electronic)). doi:10.1038/modpathol.2008.132
Senyilmaz, D., and Teleman, A. A. (2015). Chicken or the egg: Warburg effect and mitochondrial dysfunction. F1000Prime Rep. 7, 41. (2051-7599 (Print)). doi:10.12703/P7-41
Gajewski, T. F., Schreiber, H., and Fu, Y. X. (2013).Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14 (10), 1014–1022. doi:10.1038/ni.2703
Srinivasan, S., GuhaM., , Dong, D. W., Whelan, K. A., Ruthel, G., Uchikado, Y., et al. (2016). Disruption of cytochrome c oxidase function induces the Warburg effect and metabolic reprogramming. Oncogene 35, 1585–1595. (1476-5594 (Electronic)). doi:10.1038/onc.2015.227
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. (0027-8424 (Print)). doi:10.1073/pnas.0506580102
Sung, H. A., Ferlay, J., Siege, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. (1542-4863 (Electronic)). doi:10.3322/caac.21660
Tan, X. F., Chen, Q., Hua, S. H., and Yip, G. W. (2021). Roles of interferon induced protein with Tetratricopeptide repeats (IFIT) family in cancer. Curr. Med. Chem. 28, 5034–5047. (1875-533X (Electronic)). doi:10.2174/0929867328666210617105209
Vasaikar, S. V., Straub, P., Wang, J., and Zhang, B. (2018). LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. (1362-4962 (Electronic)). doi:10.1093/nar/gkx1090
Vaupel, P. A.-O., and Multhoff, G. (2021). Revisiting the Warburg effect: Historical dogma versus current understanding. J. Physiol. 599, 1745–1757. (1469-7793 (Electronic)). doi:10.1113/JP278810
Vaupel, P., and Multhoff, G. (2018). Hypoxia-/HIF-1α-Driven factors of the tumor microenvironment impeding antitumor immune responses and promoting malignant progression. Adv. Exp. Med. Biol. 1072, 171–175. (0065-2598 (Print)). doi:10.1007/978-3-319-91287-5_27
Vaupel, P., Schmidberger, H., and Mayer, A. A.-O. (2019). The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Biol. 95, 912–919. (1362-3095 (Electronic)). doi:10.1080/09553002.2019.1589653
Vaupel, P. (2004). Tumor microenvironmental physiology and its implications for radiation oncology. Semin. Radiat. Oncol. 14, 198–206. (1053-4296 (Print)). doi:10.1016/j.semradonc.2004.04.008
Vesely, M. D., Kershaw, M. H., Schreiber, R. D., and Smyth, M. J. (2011). Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271. (1545-3278 (Electronic)). doi:10.1146/annurev-immunol-031210-101324
Wang, W., Ren, S., Wang, Z., Zhang, C., and Huang, J. (2020). Increased expression of TTC21A in lung adenocarcinoma infers favorable prognosis and high immune infiltrating level. Int. Immunopharmacol. 78, 106077. (1878-1705 (Electronic)). doi:10.1016/j.intimp.2019.106077
Wang, Y., Xu, L., Lu, X., Fu, D., Su, J., Geng, H., et al. (2018). CD4 + T cells promote renal cell carcinoma proliferation via modulating YBX1. Exp. Cell Res. 363, 95–101. (1090-2422 (Electronic)). doi:10.1016/j.yexcr.2017.12.026
Keywords: tetratricopeptide repeat domain 21A, prognosis, immune infiltrating, clear cell renal cell carcinoma, bioinformactics analysis
Citation: Lin J, Nong D, Wang W, Guo X, Li C, Li B, Wang H, Chen Z, Li X, Huang G and Li W (2022) High expression of TTC21A predicts unfavorable prognosis and immune infiltrates in clear cell renal cell carcinoma. Front. Genet. 13:967378. doi: 10.3389/fgene.2022.967378
Received: 12 June 2022; Accepted: 12 October 2022;
Published: 03 November 2022.
Edited by:
Bing Han, Shanghai Jiao Tong University, ChinaReviewed by:
Liangyou Gu, People’s Liberation Army General Hospital, ChinaXiaojian Zhu, Second Affiliated Hospital of Nanchang University, China
Copyright © 2022 Lin, Nong, Wang, Guo, Li, Li, Wang, Chen, Li, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Li, bGl3ZWk5NV8yMDAwQDE2My5jb20=
 Junhao Lin
Junhao Lin DeYong Nong2
DeYong Nong2