- Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Guangdong Province Famous Fish Reproduction and Breeding Engineering Technology Research Center, Fisheries College of Guangdong Ocean University, Zhanjiang, China
42Sp50 is an isoform of the eukaryotic translation elongation factor 1 A (eEF1A) and is vital for fish ovarian development. Spotted scat (Scatophagus argus) is a popular marine cultured fish species in Southern Asia and China, and its artificial reproduction is complicated, with a relatively low success ratio in practice. In this study, the 42Sp50 gene was cloned from spotted scat. Tissue distribution analysis showed that 42Sp50 was mainly expressed in the ovary. qRT-PCR showed that 42Sp50 expression levels gradually decreased insignificantly in the ovaries from phase II to IV. Western blot analysis showed that 42Sp50 was highly expressed in the ovary, while it was almost undetectable in the testis. Immunohistochemistry analysis stained 42Sp50 mainly in the cytoplasm of the previtellogenic oocytes in ovaries of normal XX-female and sex-reversed XY-female. Aside from fish and amphibians, 42Sp50 was also identified in some reptile species using genomic database searching. Analyses of the transcriptome data from four different fish species (Hainan medaka (Oryzias curvinotus), silver sillago (Sillago sihama), Nile tilapia (Oreochromis niloticus), and Hong Kong catfish (Clarias fuscus)) revealed ovaries biased expression of 42Sp50 in all, similar to spotted scat. While the neighbor genes of 42Sp50 did not show ovary biased expression in the fish species analyzed. Bisulfite Sequencing PCR (BSP) results showed that the DNA methylation level of 42Sp50 promoter was low in ovaries, testes, and muscles. The luciferase reporter assay demonstrated that Dmrt4 activated 42Sp50 expression in the presence of Sf1 or Foxh1. These results suggest that 42Sp50 may be involved in regulating the early phase oocytes development of spotted scat.
Introduction
Translation elongation factor (EF) plays an essential role in protein synthesis by transporting the aminoacylated-tRNAs to the ribosome. There are three types of EF in prokaryotes, namely elongation factor thermo unstable (EF-TU), elongation factor thermo stable (EF-TS), and elongation factor G (EF-G). However, two types of EF exist in eukaryotes, including eukaryotic translation elongation factor 1 (eEF1) and eukaryotic translation elongation factor 2 (eEF2). eEF1 is composed of two types of proteins in vertebrates, which include eEF1A and eEF1B. eEF1A, and eEF1B are respectively homologues of prokaryotes EF-TU and EF-TS, while eEF2 is homologue of prokaryotes EF-G. eEF1A forms a compound with GTP and aminoacylated tRNA (aa-tRNA) to transport aa-tRNA to ribosomal site A (Browne and Proud, 2002). eEF1B binds to eEF1A-GDP, accelerating the exchange of GDP bound to eEF1A with exogenous GTP and prompting the formation of eEF1A-GTP (Janssen and Möller, 1988). When peptide bonds are formed, eEF2 promotes the movement of peptide chains from site A to site P of ribosome (Djumagulov et al., 2021).
The number of eEF1A subtypes varies among vertebrates species. Two eEF1A genes, eEF1A1 and eEF1A2, were reported in human (Homo sapiens) (Brands et al., 1986; Knudsen et al., 1993), mouse (Mus musculus) (Lee et al., 1994) and chicken (Gallus gallus) (Yoshikawa et al., 1984; Wang et al., 1994). Three eEF1A genes (eEF1S, eEF1O and 42Sp50) were identified in frog (Xenopus laevis) (Viel et al., 1991). The number of eEF1A gene subtypes is relatively high in fish. In Senegalese sole (Solea senegalensis) and Nile tilapia (Oreochromis niloticus), five eEF1A genes (eEF1A1a, eEF1A1b, eEF1A3, eEF1A4, and 42Sp50) were reported (Infante et al., 2008; Wei, 2011). Four eEF1A genes (eEF1A1a, eEF1A1b, eEF1A2 and eEF1A4) were described in zebrafish (Danio rerio) (Idigo et al., 2020). More eEF1A genes in fish might be due to the teleosts specific genome duplication (3R) (Hoegg et al., 2004).
The eEF1A genes show differential tissue-specific expression patterns in different species. In mammals, eEF1A1 and eEF1A2 were differentially expressed in tissues at different development stages, and eEF1A1 was expressed in more tissue types than eEF1A2 (Knudsen et al., 1993; Chambers et al., 1998; Kahns et al., 1998). But in the non-mammalian vertebrates, the expression patterns of eEF1A genes have not been widely studied. In zebrafish, eEF1A1a, eEF1A1b, and eEF1A4 were expressed in the brain, muscle, spleen, intestine, liver, testis, and ovary, while eEF1A2 was not expressed in the liver (Idigo et al., 2020). In Nile tilapia, eEF1A1b was highly expressed in the brain, heart, liver, intestine, kidney and testis, but weakly expressed in the ovary and undetectable in the gill, spleen, head kidney, and muscle. Whereas eEF1A4 was widely expressed in all tissues, 42Sp50 was specifically expressed in the ovary (Wei, 2011; Chen, 2018). The diverse expression pattern of the eEF1A genes in vertebrates suggests differential functions.
42Sp50 is a special member of the eEF1A gene family, first reported in X. laevis (Viel et al., 1991). 42Sp50 protein is one component of the ribonucleoprotein particle isolated from the ovary of frog sedimenting at 42 S (Wegnez and Denis, 1979). 42Sp50 has a molecular weight of around 50 kDa, hence the name 42Sp50. In X. laevis and Nile tilapia, 42Sp50 was specifically expressed in the oocytes (Deschamps et al., 1991; Wei, 2011; Chen 2018). The ovarian 42Sp50 gradually increases during the development process, reaching a peak at 60 days after hatching, and then decreases thereafter in Nile tilapia (Wei, 2011). In Japanese medaka (Oryzias latipes), 42Sp50 is one of the earliest markers of sex differentiation specifically expressed in oocytes (Kinoshita et al., 2009). While the reason for the ovary biased expression of 42Sp50 remains unclear. Knockout of 42Sp50 blocks the development of oocytes at the primary growth stage leading to abnormal proliferation and differentiation of follicular cells in female Nile tilapia, indicating that 42Sp50 is indispensable for ovarian development in Nile tilapia (Chen, 2018). Further comparative studies are required to understand the transcriptional regulation and functional importance of 42Sp50 in fish.
Spotted scat (Scatophagus argus) is a euryhaline subtropical marine fish widely distributed in Indian-Pacific waters (Barry et al., 1993). Spotted scat’s market value is relatively high due to its delicious taste. Although there are reports of artificial reproduction in spotted scat, the efficiency is still low in practice (Mandal et al., 2021). The cultured spotted scat fry were mainly wildly caught in China. This threatened and destroyed the natural resource of spotted scat. Studies focused on the reproduction-related gene in spotted scat might help improve artificial reproduction efficiency. In the previous study, 42Sp50 was found to play an important role in the development of ovaries in Nile tilapia (Chen, 2018). In this study, 42Sp50 was isolated in spotted scat, and its expression was analyzed by quantitative real-time PCR (qRT-PCR), Western blotting, and immunohistochemistry (IHC) during ovary development. In addition, the transcriptional regulation of 42Sp50 was examined. 42Sp50 in spotted scat was specifically expressed in the primary oocytes and regulated by Dmrt4, Sf1, and Foxh1 in vitro. The exclusive expression pattern of 42Sp50 in the primary oocytes enabled it as a good marker gene for further reproduction-related studies in spotted scat. The results provide a basis for the functional study of 42Sp50 in ovarian development in fish.
Materials and methods
Experimental fish and sampling
Spotted scats (body weight: 109.0–234.4 g, length: 15.4–16.9 cm) purchased from the Dongfeng market (Zhanjiang, Guangdong, China) were used for cDNA cloning and expression analysis. All fish were anesthetized using 100 mg/L of tricaine methane sulfonate (MS 222, Sigma, St. Louis, MO, United States) and dissected. Tissues used for further studies were excised and snap-frozen in liquid nitrogen and stored at −80°C prior to use. Parts of the excised gonads were fixed in Bouin’s solution for gonad developmental stage characterization and IHC analysis. The tail fins were also collected, stored in 100% ethanol at −20°C for DNA extraction. All experimental fish protocols followed the guidelines and approval of the Administration of Affairs Concerning Experimental Animals for the Science and Technology Bureau of China, and Animal Research and Ethics Committees of Guangdong Ocean University.
The induction of sex reversed XY fish was described in our previous study (Mustapha et al., 2021). Briefly, a batch of about 2-month-old spotted scat (average total length 2 cm) were obtained from the wild in August 2019 from Beihai-China and cultured at the Zhanjiang Donghai Island Cultivation Base (Zhanjiang, Guangdong, China). After 1 week, the fish were equally divided into two groups: the control group and the Estrogen group (E2 group). The E2 group was fed diets containing Estrogen (300 μg/g) while the control group was fed with common diets. Fish were fed three times daily for 3 months, and treatment terminated. The fish were subsequently fed with common diet until sampling. Before sampling, spotted scat were about 2.5 years old. Spotted scat were anesthetized, and the gonads were quickly dissected. Parts were fixed in Bouin’s solution for IHC, and the other part stored in 100% ethanol for DNA extraction. The tail fins were also collected, stored in 100% ethanol at −20°C for DNA extraction.
Identification of genetic sex of spotted scat
The DNA of spotted scats was extracted by a nucleic acid purification kit (N1173, DONGSHENG BIOTECON, China). After the quality and concentration were detected by 1% agarose gel and Nanodrop 2000 ultramicro nucleic acid protein analyzer (Thermo Scientific), DNA was stored at −20°C until use. The genetic sex of spotted scats was determined by using the sex-linked DNA Marker (Dmrt1-Marker-4-F/R) (Huang et al., 2022). Primer sequences are listed in Supplementary Table S1 and the PCR conditions were the same as Huang et al. (2022). Dmrt1-Marker-4-F/R amplified only one DNA fragment of 593 bp in female, while two DNA fragments of 593 bp and 693 bp were amplified in male.
cDNA cloning, gene identification and bioinformatics analysis
Total RNA was extracted from testes and ovaries of adult fish using TRIzol reagent (Invitrogen). The quality and concentration of RNA were detected by 1% agarose gel and Nanodrop 2000 ultramicro nucleic acid protein analyzer (Thermo Scientific). First-strand cDNA synthesis was performed using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, China) following the manufacturer’s protocol.
Local BLAST was used to search for the 42Sp50 transcripts from transcriptome sequence data of spotted scat gonads (He et al., 2019) using the 42Sp50 mRNA sequence from Nile tilapia (NCBI accession number: XM_019363060.2) as a query. Primers were subsequently designed from the 42Sp50 sequence obtained from the transcriptome data to amplify the partial cDNA fragment, including the open reading frame (ORF) in spotted scat (Supplementary Table S1). The PCR conditions were as follows: 4 min at 94°C; 30 s at 94°C, 30 s at 53°C, and 2 min at 72°C for 35 cycles; 10 min at 72°C. PCR products were examined on 1.5% agarose gel stained with ethidium bromide. Amplification bands were excised from the gel, purified and cloned into the pEasy-T3 vector (TransGen Biotech, China). Positive clones were screened by colony PCR and sequenced by Sanger sequencing (Sangon, Shanghai, China). The cDNA sequences of 42Sp50 cloned in this study was analyzed using DNASTAR software (http://www.dnastar.com). The ORF and protein sequences were predicted using ORF Finder software (https://www.ncbi.nlm.nih.gov). Sequence homology alignments were performed using MegAlign (part of the DNASTAR software package), and multiple alignments were carried out using ClustalX (http://www.clustal.org/). The whole protein sequences of 42Sp50 from other vertebrate species (the accession numbers are listed in Supplementary Table S2) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). The phylogenetic tree was constructed using the neighbor-joining method implemented in MEGA X (https://www.megasoftware.net/). Syntenic analysis was done by using data on Ensembl (https://asia.ensembl.org/index.html), NCBI (https://www.ncbi.nlm.nih.gov/) and Genomicus (https://www.genomicus.bio.ens.psl.eu/genomicus-101.01/cgi-bin/search.pl).
Tissue distribution of 42Sp50 mRNA
The hypothalamus, pituitary, gill, liver, heart, spleen, kidney, stomach, intestine, gonad, and muscle were dissected from adult male and female spotted scats and were used for tissue distribution analysis. Tissue distribution was performed by reverse transcription-polymerase chain reaction (RT-PCR). The cDNA was diluted 5-fold and subjected to amplification. β-Actin was used as an internal control. The PCR condition was as follow: 5 min at 94°C; 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C for 32 cycles (β-actin 28 cycles); 10 min at 72°C. The PCR products were separated by electrophoresis on 1.5% agarose gel and visualized with ethidium bromide.
Expression of 42Sp50 mRNA in ovaries at different stages
Ovaries from adult females at different stages of gonadal development were collected. The gonad developmental phases were determined by histological sectioning and staining, according to Cui et al. (2013). Total RNA was extracted from the gonads and reverse transcription was carried out, as mentioned above. The mRNA levels of 42Sp50 at different stages of gonadal development were detected by qRT-PCR and the conditions were as follows: 1 min at 95°C, 30 s at 95°C, 30 s at 60°C, 30 s at 72°C for 40 cycles (fluorescent data collection). The relative abundance of 42Sp50 mRNA transcripts was evaluated using the formula, R = 2−ΔΔCt (Livak and Schmittgen, 2001). β-Actin was used as a reference gene. All determinations were performed in triplicate. Primer sequences used for qPCR are listed in Supplementary Table S1. The amplified fragment length of 42Sp50 and β-actin were around 183 and 147 bp, respectively. The specificity of the primers were confirmed by cloning and sequencing.
Western blot
The whole protein sequence of spotted scat 42Sp50 was used as antigen to prepare the polyclonal antibody in this study. The recombinant 42Sp50 constructs were prepared by cloning the ORF into the expression vector pET16b. With MBP-tag at its N-terminus and His-tag at its C-terminus, the recombinant plasmid was expressed in Escherichia coli with isopropyl β-D-l-thiogalactopyranoside (IPTG, 500 μM) induction. Next, the recombinant protein (25–30 μg) was purified using a Ni-NTA super flow cartridge (Qiagen, Germany) and used as an antigen to immunize female rabbits (Japanese white rabbit) three times at 15- day intervals. Ten days after the last immunization, rabbit serum was collected and purified by affinity chromatography on Sepharose 4B Fast Flow Resin (Sigma, Germany). Polyclonal antibodies were confirmed by Western blot analysis. Briefly, total protein was extracted from male (n = 3) and female (n = 3) gonads (both at phase III) from adult spotted scat and diluted to a final concentration of 20 mg/ml. Western blotting was carried out as described previously (Ru et al., 2020). Antibody against 42Sp50 was diluted at 1:2,500 and the abundance of β-actin (rabbit anti-β-actin, 1:1,000 dilution of AA002, Abiotech co., Ltd. (Jinan, China)) was examined as a loading control. Goat anti-rabbit antibodies conjugated to horseradish peroxidase (1:1,000 dilution of YKCP-202-01, Youke, China) were used as secondary antibodies. Immunoreactive signals were detected with the BeyoECL Plus Kit (Beyotime, China) and then visualized on Tanon 5,200 (China). According to a previous report (Holmseth et al., 2012), an antigen pre-adsorption test was performed as a negative control.
Immunohistochemistry analysis
As described above, the gonads of the spotted scat at different gonad developmental phases and estradiol (E2) treated fish were collected. Gonads were fixed in Bouin’s solution for 24 h at room temperature, then dehydrated and embedded in paraffin. Tissue blocks were sectioned at 5 μm thickness. IHC was carried out according to the method used by Ru et al. (2020). Antibody against 42Sp50 was diluted to 1:500 and detected by the rabbit anti-goat antibody conjugated with horseradish peroxidase (1:1,000 dilution of YKCP-202-01, Youke, China). The pre-adsorption primary anti-42Sp50 antibody was also used as a negative control, according to a previous report (Holmseth et al., 2012).
Transcriptome analysis of the expression of 42Sp50 gene and its adjacent genes in different species
The sequences of 42Sp50 gene and its collinear genes of Nile tilapia and medaka were obtained from NCBI. Through local blast, the expression levels of 42Sp50 gene and its collinear genes were searched in the transcriptome data of spotted scat (He et al., 2019), Nile tilapia (Tao et al., 2013), Hainan medaka (O. curvinotus) (Dong et al., 2021), Hong Kong catfish (Clarias fuscus) (Lin et al., 2021) and silver sillago (Sillago sihama) (Tian et al., 2019). Transcriptome samples of Nile tilapia, medaka and silver sillago were all one female and one male, and the transcriptome samples of spotted scat and Hong Kong catfish were three females and three males, and the average value of the data was taken.
DNA bisulfite treatment and sequencing
The 3 kb sequence before the 42Sp50 start codon was found in the spotted scat genome as its promoter sequence. The CpG Islands of 42Sp50 promoter were predicted with MethPrimer (https://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). Selected fragment within the 3 kb region was used to design a pair of primers (Supplementary Table S1). The gonads of three wild XY individuals, three wild XX individuals and two XY individuals treated with E2 were selected for DNA extraction, referring to the previous method. Bisulfite modification of spotted scat DNA was performed using the EZ DNA Methylation-Gold™ Kit (D5005, Murphy Ave, Irvine, CA, United States), following the manufacturer’s instructions. The bisulfate-treated DNA was amplified by PCR using the previously mentioned primers, and then the PCR product was ligated into the pEasy-T3 vector (TransGen Biotech, China). For each individual, 7–10 positive clones were sequenced. The Sanger sequencing was carried out by Sangon (Shanghai, China). The sequencing results were compared with spotted scat 42Sp50 gene sequence.
Transient transfections and luciferase assays
The possible binding sites of 42Sp50 promoter were predicted by PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). The 42Sp50 promoter of spotted scat (3 kb, ligation into Mlu I/Xho I sites) was generated by PCR and subcloned into the pGL3-basic vector (Promega Corp., Madison, WI). Sf1, Foxh1, and Dmrt4 sequence was synthesized by Sangon Biotech and ligated to pcDNA3.1 vector (Invitrogen). The dual-luciferase assay was carried out according to a previous report (Wang et al., 2007). HEK 293 cells cultured at a density of 2 × 104 cells per well were used for transfection. The reagents used in the experiment include FBS (16140071, Gibco, America), DMEM (SH30022.01, Hyclone, America), streptomycin/penicillin (15070063, Thermofisher, America), Dual-Luciferase Reporter Gene Assay Kit (K801, Biovision, America) and lipo2000 (Invitrogen, America).
Statistical analysis
All data are expressed as means ± standard deviation (SD). Significant differences in the data among the groups were tested by one-way analysis of variance (ANOVA) with Duncan’s post-hoc test or independent-samples t-test using a threshold of p < 0.05. All statistical calculations were performed using SPSS 19.0 (SPSS, Chicago, IL, United States).
Results
cDNA cloning and sequence analysis of 42Sp50
The partial cDNA of spotted scat 42Sp50 was 1543 bp with 1368 bp ORF which encodes 455 amino acids (NCBI accession number: MT774144) (Figure 1A). The predicted molecular mass was 50.05 kDa, and the isoelectric point was 9.63. Spotted scat 42Sp50 contains seven exons and six introns, similar to other fish (Figure 1B). The sequence analysis showed that 42Sp50 has no transmembrane domain and signal peptide, but with one N-link glycosylation site, nine protein kinase C phosphorylation sites, three casein kinase II phosphorylation sites and six N-myristoylation sites. Additionally, GTP binding domain and EF-TU binding domain were predicted in the sequence (Figure 1A). The structure and sequence of spotted scat 42Sp50 are conserved with other fish species (Supplementary Figure S1).
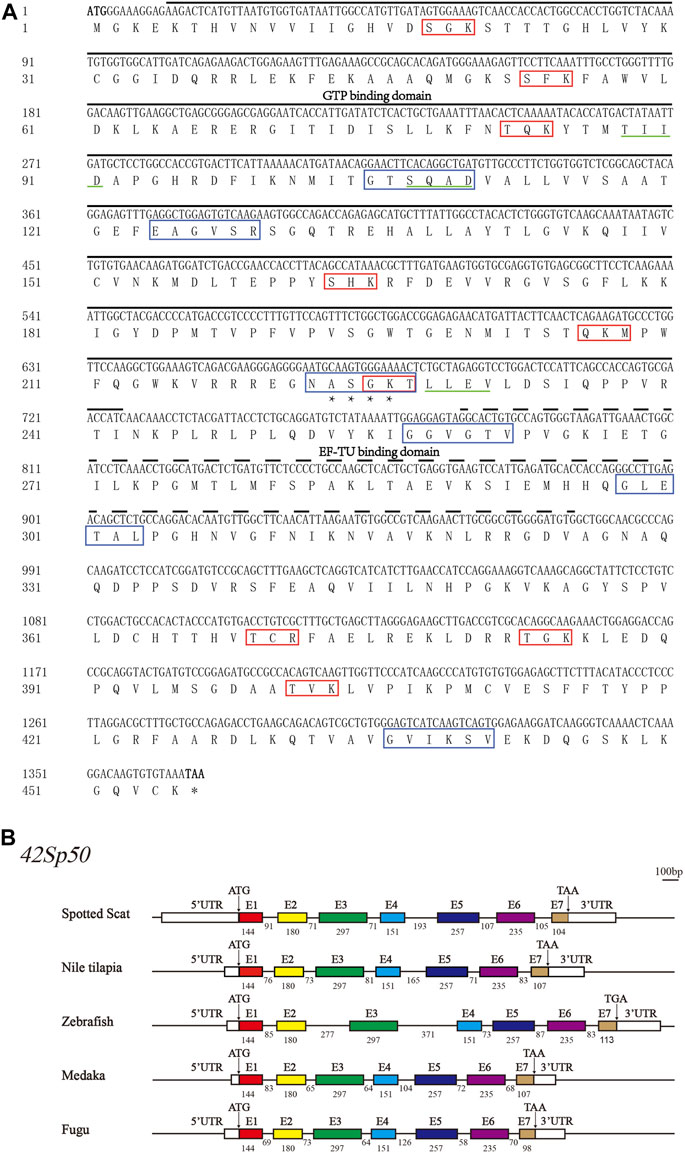
FIGURE 1. The gene sequence and structure of 42Sp50 in spotted scat (Scatophagus argus). (A) Nucleic acid sequence and amino acid sequence of spotted scat 42Sp50. Black solid line, the GTP domain; Dotted line, EF-TU domain; “*”, N-glycosylation site; Red box, Protein kinase C phosphorylation site; Green solid line, Casein kinase II phosphorylation site; Blue box, N-myristoylation sites. (B) Schematic diagram of the exon–intron structure of 42Sp50 in bony fish. The white rectangles represent the 5′ and 3′ UTR. Colored boxes represent exons, and horizontal lines between exons represent introns. ATG stands for the start codon and TAA/TGA stands for the stop codon. The number represents the base length. The short solid lines at the top right are scale bars, representing 100 bp.
In this study, 42Sp50 was isolated from the genome database of fish, amphibia and some reptiles. A phylogenetic tree based on the amino acid of 42Sp50 sequences of vertebrates was constructed (Figure 2A) (the accession numbers are listed in Supplementary Table S2). The analysis showed that the sequence found in reptiles was indeed 42Sp50. It was observed that 42Sp50 of sauropsida, amphibia, chondrichthyes, and osteichthyes were clustered into four distinct branches. 42Sp50 of spotted scat was most closely related to Fugu (Takifugu rubripes). However, 42Sp50 was not isolated in all reptile genomes. 42Sp50 was found in chelonia and crocodilia, but not in lecertifromes and serpentiformes. Due to the lack of data, the presence of 42Sp50 is unknown in rhynchocephalia and amphisbaeniformes. In addition, 42Sp50 was not found in aves and mammalia (Figure 2B).
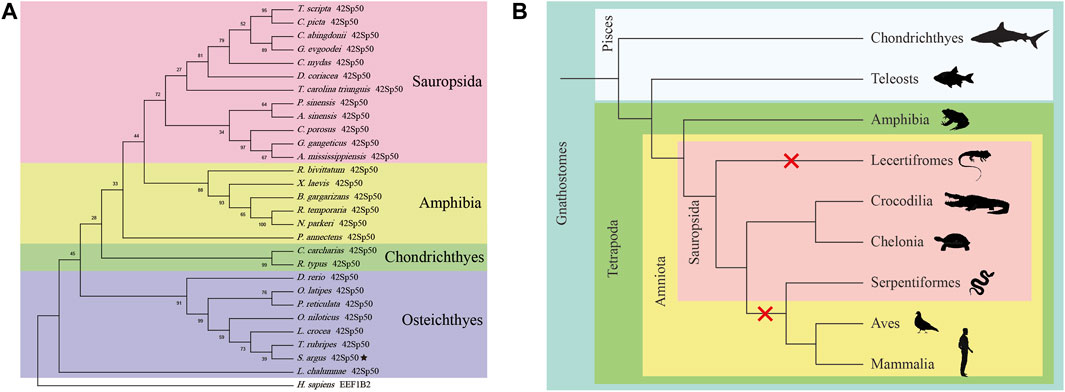
FIGURE 2. Phylogenetic analysis of 42Sp50 in vertebrates. (A) Phylogenetic tree of the 42Sp50 in selected vertebrate species. The tree was constructed using MEGA X and the Neighbor-joining approach. Bootstrap testing was based on 500 replicates. The 42Sp50 genes of spotted scat are denoted by pentagram. Genbank accession numbers of these are provided in Supplementary Table S2. (B) The evolution of 42Sp50 in Gnathostomes. The red X signs indicate the events of 42Sp50 losses during the evolution of Lecertifromes, Serpentiformes, Aves and Mammalia.
In order to further understand 42Sp50, synteny analysis was performed. The 42Sp50 gene of spotted scat is located at 11311436-11314216 bp of LG8 chromosome. The results indicated that the adjacent genes of 42Sp50 have a high degree of consistency in fish. Even though 42Sp50 was not found in human, mouse, chicken and lizard (Anolis carolinensis), its upstream and downstream genes (e.g., Myc, Fam49b and Asap1) exist in these species (Figure 3). The accession number of genes used in syntenic analysis are listed in Supplementary Table S3.
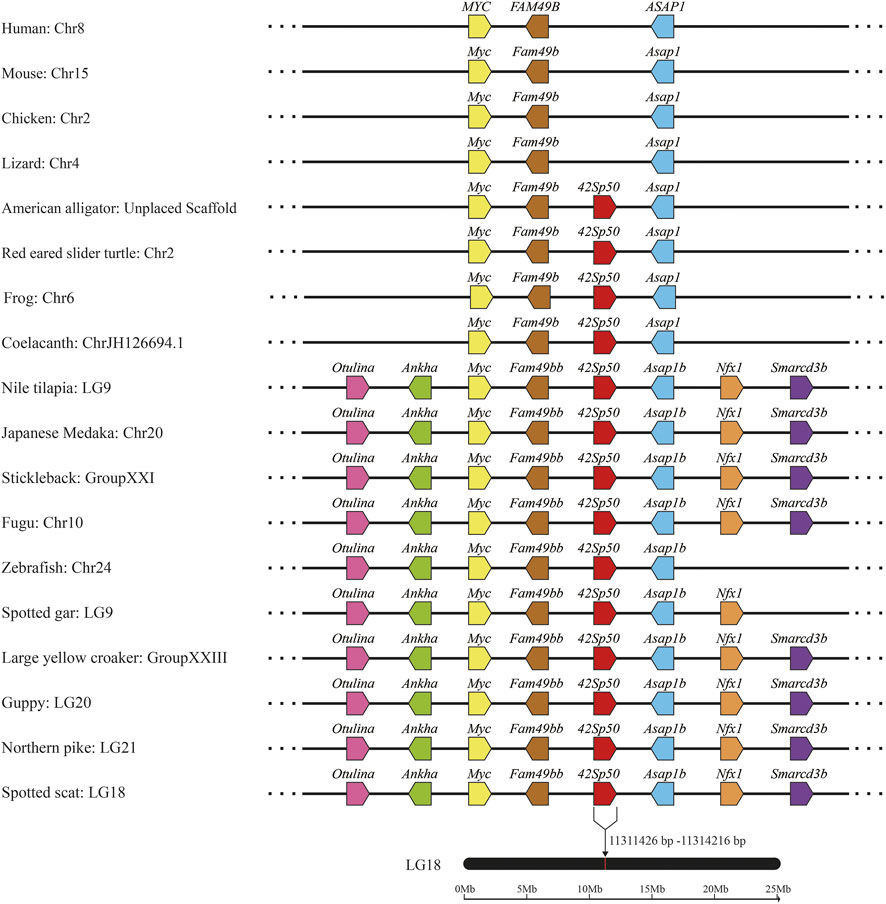
FIGURE 3. Syntenic analysis of 42Sp50 genes from spotted scat using the genomes of 18 selected vertebrate species (human, mouse, chicken, lizard, American alligator, red eared slider turtle, frog, coelacanth, Nile tilapia, Japanese medaka, stickleback, Fugu, zebrafish, spotted gar, large yellow croaker, guppy and northem pike). Genes are represented by pentagons, and the directions of the reading frames are represented by the direction that the pentagon is pointing. Genbank accession numbers of these gene are provided in Supplementary Table S3.
Tissue distribution and expression of 42Sp50 mRNA at different gonad developmental phases
Tissue expression results showed that 42Sp50 was highly expressed in the female ovaries and was weakly expressed in the testis (Figure 4A). The distinction of ovarian stages is detailed in Cui et al. (2013). The ovaries of stage II, stage III and stage IV of spotted scat were collected, and the 42Sp50 mRNA expression levels were examined by qRT-PCR. The highest expression level of 42Sp50 mRNA was observed in stage II and the lowest in stage IV, but there was no significant difference between the expression levels in each stage (Figure 4B).
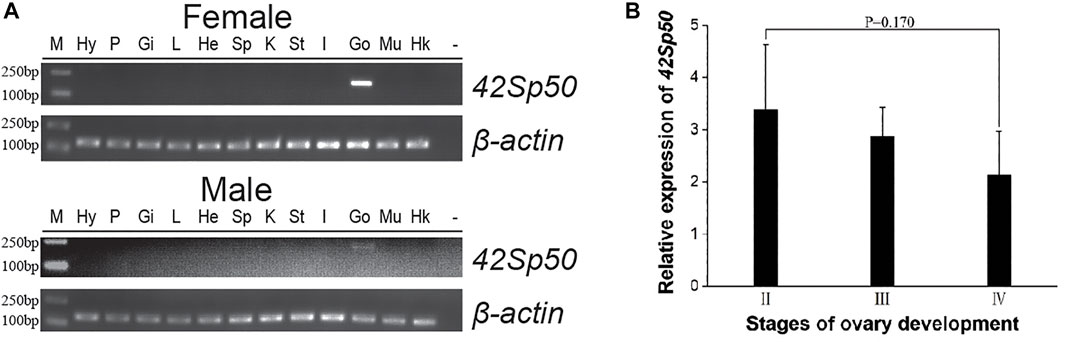
FIGURE 4. Tissue distribution and expression of 42Sp50 mRNA at different gonad developmental phases. (A) Tissue distribution of 42Sp50 in spotted scat. The PCR products for spotted scat 42Sp50 and β-actin were 183 bp and 145 bp, respectively. Hy, hypothalamus; P, pituitary; Gi, gill; L, liver; He, heart; Sp, spleen; K, kidney; St, stomach; I, intestines; Go, gonad; Mu, muscle; “−”, negative control; Marker, DS™2000. (B) The expression of 42Sp50 in the ovary at different development stages in spotted scat using qRT-PCR. Data are presented as mean ± SEM (n = 3).
42Sp50 protein expression in gonads by western blot
In this study, spotted scat 42Sp50 polyclonal antibody was produced. The anti-42Sp50 antibody detected a single 50.0 kDa band in the protein prepared from the spotted scat stage III ovary (Figure 5). The detected band size is similar to the calculated 42Sp50 protein size of 50.0 kDa. While 42Sp50 was almost undetectable in the testis. When the pre-adsorbed primary antibody was used, the 42Sp50 specific band of the ovary could not be detected (Figure 5).
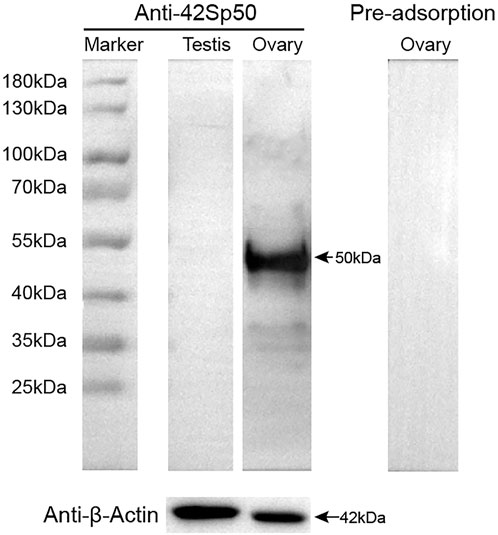
FIGURE 5. Specificity of anti-42Sp50 antibody analyzed by Western blotting. β-Actin abundance was examined as the loading control. The primary antibody which was preadsorbed with its respective recombination protein was used as negative controls.
Expression of 42Sp50 in gonads at different developmental phases and E2 treated gonads by immunohistochemistry
IHC was carried out on ovaries of stage I-IV and the testis of stage II-V. The results showed positive signals in the ovaries of all stages. A strong anti-42Sp50 positive signals were only observed in the cytoplasm of the oocytes at phase I and II, weak signals were observed in phase Ⅲ, and no signal was observed in the oocytes of phase IV. No positive signal was observed in the testes, indicating that low expression level of 42Sp50 in the testis (Figure 6A). In addition, gonads of 2.5 years old spotted scat were collected from the E2 group. The genetic sex of this fish is identified as XY (Figure 6B), but with ovarian features. The IHC results showed a positive signal in the cytoplasm of the oocyte, which was consistent with the result observed in the ovary of a wild-type female individual (Figure 6C). No positive signal was observed in the negative control (Figure 6C).
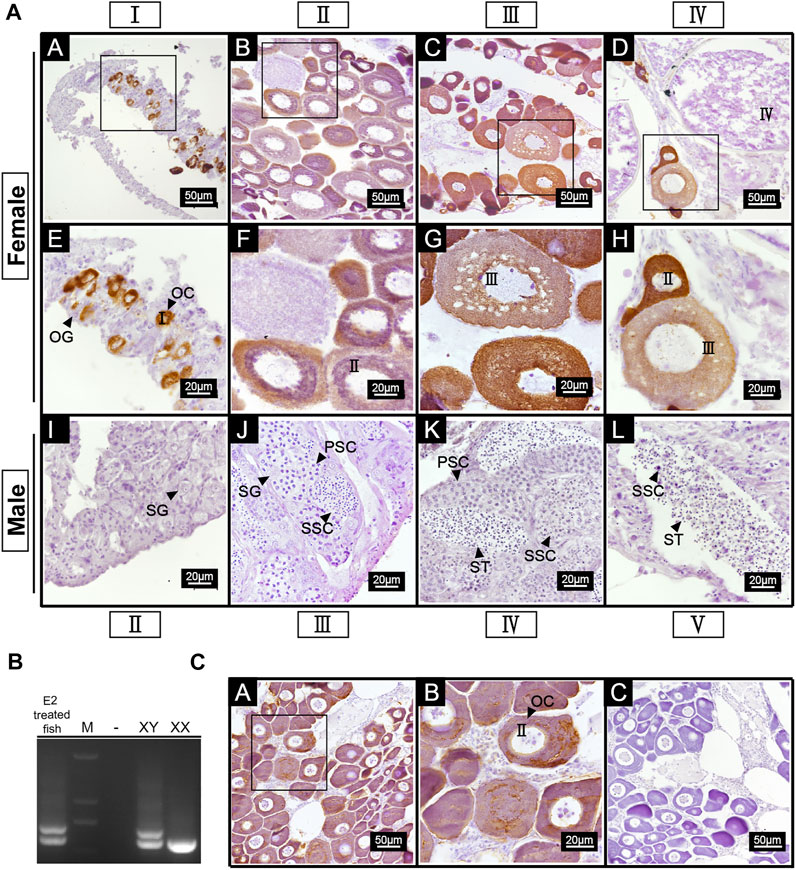
FIGURE 6. Cellular location of 42Sp50 in gonad of spotted scat by IHC. (A) A–H are ovaries at stage I, II, III and IV, respectively. E–H are the high magnifications of the boxed areas in A–D, respectively. I–L are testes at stage II, III, IV and V, respectively. (B) The identification of genetic sex of E2 treated fish. (C) A,B are the gonad of 2.5 years old E2 treated fish, B is the high magnification of the boxed area in (A) C is the negative control of E2 treated fish gonad. PBS was used instead of the primary antibody as the negative control.
Transcriptome analysis results
We obtained the mRNA expression data of 42Sp50 and its syntenic genes in five different teleosts (Supplementary Figure S2). The expression of Myc, 42Sp50, and Nfx1 was higher in females than in males, while Fam49b was higher in males than females. Different from them, the expressions of Ankha and Asap1 differ among different species.
Methylation levels of the 42Sp50 promoter
According to the predictions, the 42Sp50 promoter has one CpG island, located between −1000 bp and −1500 bp (Figure 7A). The fragment selected for this experiment has 7 CG sites (Figure 7B). The genetic sex of nine fish treated with E2 were identified (Figure 7C), there are two bands in XY individuals and only one band in XX individuals. XY individuals are selected for the subsequent experiments. The comparison results show that the chosen site in this study has a lower degree of methylation whether in the gonads of XY-male, XX-female and XY-E2-treated fish and the muscles of XY-male and XX-female (Figure 7D).
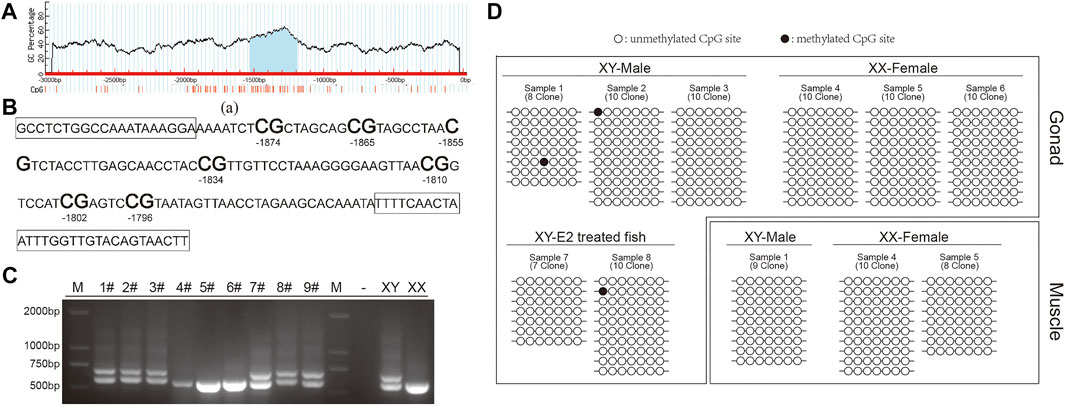
FIGURE 7. DNA methylation in the 42Sp50 promoter. (A) The prediction of CpG island in 42Sp50 promoter. (B) The fragment of 42Sp50 promoter selected in this study. The primers are marked with boxes. (C) The identification of genetic sex of E2 treated fish. (D) The site methylation level was analysed. Black solids and circles correspond to methylated and unmethylated sites, respectively.
Luciferase assays of 42Sp50
Figure 8A shows the binding sites of 42sp50 promoter with Sf1 and Fox gene family members predicted by PROMO. The result of luciferase assays showed that 42sp50 was not activated when Foxh1, Sf1, and Dmrt4 were alone, respectively (Figure 8B). In the combination of two transcription factors, Sf1 or Foxh1 could activate the transcription of 42Sp50 when they were present together with Dmrt4. Among the combinations of three transcription factors, the combination of Dmrt4, Foxh1, and Sf1 promoted the transcription of 42Sp50. Other combinations had no effect on the transcription of 42Sp50.
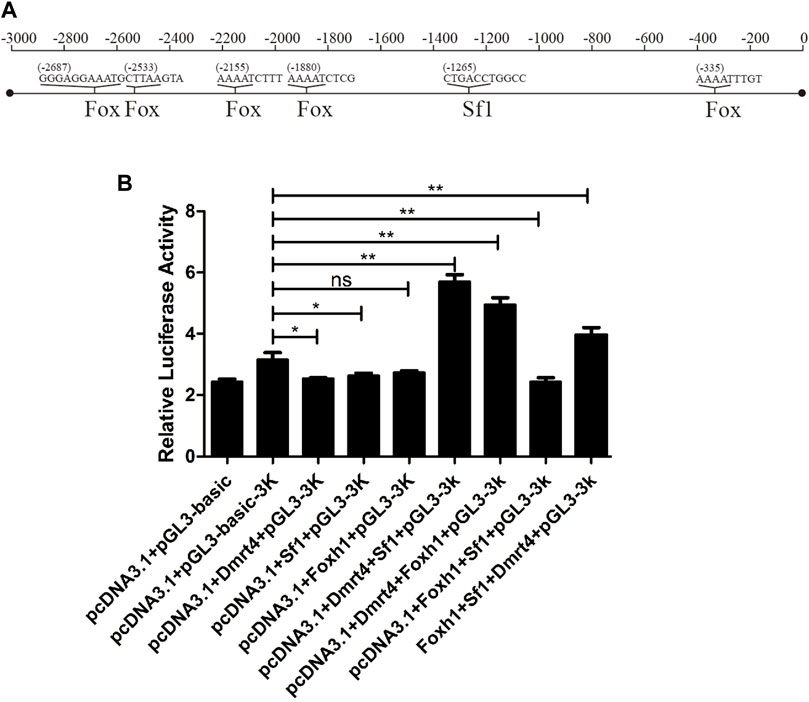
FIGURE 8. Regulation experiment of 42Sp50 promoter in vitro. (A) The binding sites of 42Sp50 promoter with Sf1 and Fox gene family members predicted by PROMO. (B) Luciferase analysis to detect the regulatory effects of Sf1, Foxh1, and Dmrt4 and their combinations on the promoter of 42Sp50.
Discussion
42Sp50 is a member of eEF1A family, which is lost during evolution in some vertebrates, including mammals and birds. Expression and functional analysis of 42Sp50 showed it is highly expressed in the ovary and necessary for oocyte development. While its transcriptional regulation is rarely reported. The partial cDNA of spotted scat 42Sp50 was cloned in the present study. Genome-wide searching of 42Sp50 in vertebrates confirmed that it was lost in some tetrapod groups. Spotted scat 42Sp50 showed a similar expression pattern with other fish, indicating its conserved role of it in ovary development. DNA methylation and in vitro promoter analysis of 42Sp50 were carried out to reveal the possible mechanisms for its ovary specific expression in spotted scat.
42Sp50 was evolutionarily lost in some tetrapod species
Gene duplication usually happens during evolution and is important for acquiring novel genes promoting the evolution of organisms (Magadum et al., 2013). There are three rounds of genome duplication during the evolution of vertebrates, named 1, 2, and 3R. 1R happened before and 2R after Cyclostomata formation (Steinke et al., 2006). While the 3R, or fish-specific genome duplication (FSGD), specifically occurred in the ancestor of teleosts, and most of the duplicated genes were secondarily lost (Hoegg et al., 2004; Meyer and Van de Peer, 2005). Herein we identified 42Sp50, which is located in a conserved gene cluster in fish and some tetrapods. Meanwhile, 42Sp50 could not be found in lamprey, indicating that 42Sp50 appears after the 2R before 3R in the vertebrates. Only one 42Sp50 was found in the analyzed teleosts indicating that this gene did not duplicate during the 3R or its duplicates were secondarily lost after 3R. As 42Sp50 is a reproduction-related gene, its disappearance in some tetrapods might be caused by the change of reproduction strategy, which makes this gene redundant and thereafter lost. There are a lot of reproduction-related genes lost during species diversification and evolution. Dmrt6 exists in Nile tilapia and stickleback, while it is not found in zebrafish, medaka, spot gar, and mammals (Zhang et al., 2014). Similar with 42Sp50, Gsdf exists in most fish, including spotted scat and some tetrapods (Hsu and Chung, 2021). Interestingly, protein interaction analysis showed that Gsdf could interact with eEF1a3, which is homologous to 42Sp50 in medaka (Zhang et al., 2021). It is interesting to test if 42Sp50 could also interact with Gsdf in fish. If 42Sp50 and Gsdf are functionally related and evolutionary lost in similar species, we can propose that functionally related genes will be lost together during evolution. Genes like 42Sp50 and Gsdf are very good models for evolutionary gene analysis. They could increase our knowledge of the rule of gene loss in vertebrates.
The conservation of 42Sp50 expression
In the present study, RT-PCR, qPCR, Western blot and IHC results showed that 42Sp50 was predominantly and highly expressed in the ovaries of spotted scat, showing a sex-dimorphic expression pattern. This expression pattern is identical to that observed in Nile tilapia (Wei, 2011; Chen, 2018). Consistently, the transcriptome of gonads in several unrelated fish confirmed that 42Sp50 also expressed much higher in ovary than that of testis. In X. laevis, 42Sp50 was expressed abundantly in previtellogenic oocytes (Viel et al., 1991). Similarly, 42Sp50 was highly expressed in the phase I and II oocytes in spotted scat and Nile tilapia indicating that 42Sp50 might be critical for early oocytes development (Chen, 2018). In XX 42Sp50−/− Nile tilapia, the development of the oocytes was blocked at the early stages, confirming that 42Sp50 is necessary for ovary development (Chen, 2018). However, the detailed mechanism of 42Sp50 in oocyte development remains unclear. 42Sp50 might act as its family member`s classical function as protein synthesis factor to transfer aminoacyl tRNA to the ribosomes during elongation. 42Sp50 is present in the 42 S particles and binds with tRNA transcribed in oocytes at the early stage of vitellogenesis, thus 42Sp50 might be important for long-term storage of aminoacyl tRNA in early oocyte development stages (Viel et al., 1987). It is well documented that the location and transcription of the RNA in the oocytes is delicately regulated, and the maternal RNA is critical for embryos development (Miao et al., 2017). The expression of 42Sp50 is deceased in the mature oocyte and lowly expressed in the embryo, indicating that it did not act as maternal protein (Supplementary Figure S3). Alternatively, 42Sp50 might participate in the protection, location and transcription regulation of RNA in the oocytes at the early stages. In future studies, the detailed mechanism of 42Sp50 in oocyte development should be addressed.
Considering the special expression pattern of 42Sp50, it could be taken as the oocytes marker gene in fish (Kinoshita et al., 2009; Chen, 2018). Recently, some gonads of XY male spotted scat individuals were found to develop abnormally with occurrence of primary oocytes (PO) in testes and was referred to as ectopic primary oocytes (Ecto-PO), whereas 42Sp50 was found to be expressed in the Ecto-PO (Mustapha et al., 2022). It is still unknown why and how the appearance of the Ecto-PO, and whether the fertility of the individuals containing Ecto-PO would be influenced. 42Sp50 is a good marker gene which could be used for tracing the Ecto-PO in the intersex male fish. There will be a broad application of 42Sp50 antibody prepared in the present study in future artificial reproduction related studies in spotted scat.
The reason for the ovary special expression of 42Sp50
Numerous genes show sexually biased expression patterns in gonads, and the detailed transcription regulation mechanisms for those genes are not widely studied. Through RNA-Seq, a total of 32,122 differentially expressed genes (DEGs) were identified in the gonads of spotted scat, of which 11,156 were highly expressed in the testis and 20,966 were highly expressed in the ovary (He et al., 2019). Some gene clusters containing many genes showed similar expression patterns, e.g., the genes located near Gsdf were highly expressed in fish ovaries (Gautier et al., 2011). However, the adjacent genes of 42Sp50 are not consistently highly expressed in ovaries, so 42Sp50 is not located within a gene cluster which consists of several ovary biased expressed genes. DNA Methylation is a chemical modification of DNA that can regulate gene expression. DNA methylation may be one reason for genes’ sexual dimorpic expression pattern. In European sea bass (Dicentrarchus labrax), the testicular DNA methylation level of Cyp19a promoter is higher than that of ovary, resulting in the higher expression of Cyp19a in ovaries than that of testes (Navarro-Martín et al., 2011). In barramundi (Lates calcarifer), the promoter DNA methylation degree of sex biased genes Dmrt1 and NR5A2 was higher in females, and the gene expression was also negatively correlated with their promoter DNA methylation level (Domingos et al., 2018). In this study, the DNA methylation levels of 42Sp50 promoter were all very low in testes, ovaries and muscles. Thus it is speculated that the expression of 42Sp50 does not depend on the DNA methylation regulation of its promoter. Transcription factors can regulate gene expression by interacting with transcription factor binding sites of the target gene promoter. Sexual biased expression of genes can be attributed to their transcription factors. Herein, 42Sp50 was found to be regulated by Sf1 or Foxh1 in the presence of Dmrt4 in vitro in spotted scat. Dmrt4 seems to be essential for 42Sp50 transcription. In blue tilapia (Oreochromis aurea), IHC analysis showed that strong Dmrt4 signals were observed in the cytoplasm of early previtellogenic oocytes and male germ cells at stages from A-type spermatogonia to primary spermatocytes (Cao et al., 2007). In spotted scat, Dmrt4 is highly expressed in the ovaries and testes (Jiang et al., 2019). While Dmrt4 is relatively expressed higher in testis than that of ovary in spotted scat. This seems to conflict with our results that 42Sp50 was only weakly expressed in the testis. To interpret this, we can deduce that Dmrt4 alone can not activate 42Sp50 expression in the testis, and this speculation is consistent with the in vitro analysis. Alternatively, some transcription factors could restrict the 42Sp50 expression in the testis in spotted scat. In Nile tilapia, IHC analysis showed that Sf1 was expressed in the oogonia, interstitial cells, granulosa cells, theca cells of the ovary, and spermatogonia Leydig cells of the testis at 90 and 180 days after hatching (Xie et al., 2016). The cellular location of 42Sp50 and Sf1 are not overlapped, indicating that our in vitro results might be artifact. While we still can not exclude that Sf1 might be expressed in the oocyte at phase I and II at lower level, which was undetectable by IHC. In Nile tilapia, ISH showed that Foxh1 has a sexually dimorphic expression pattern in gonads, with extremely high expression in phase I and II oocytes (Yuan et al., 2014). Herein, tissue distribution analysis showed that Foxh1 was specifically expressed in the ovary (Supplementary Figure S4). So female-specific Foxh1 might be the reason for the higher expression of its target gene 42Sp50 in the ovary. In addition, the Foxh1−/− XX Nile tilapia also showed oogenesis arrest similar to that of 42Sp50 knockout XX fish (Tao et al., 2019). Foxh1 and 42Sp50 might also be functionally related in fish.
Conclusion
Herein, 42Sp50 was cloned from the gonads of spotted scat. 42Sp50 gene was identified in some reptile species, while it was evolutionarily lost in all birds and mammals. 42Sp50 was expressed higher in ovaries than that of testis in spotted scat and mainly expressed in the cytoplasm of phase I and II oocytes indicating that it is critical for ovarian development in spotted scat. The expression of 42Sp50 might not be regulated by the DNA methylation modification of its promoter. The luciferase reporter assay demonstrated that Dmrt4 activated 42Sp50 expression in the presence of Sf1 or Foxh1 in spotted scat. This is the first report of the transcription regulation study of 42Sp50. 42Sp50 is proved to be a good oocytes marker gene in fish.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MT774144.
Ethics statement
The animal study was reviewed and approved by Animal Research and Ethics Committees of the Institute of Aquatic Economic Animals of Guangdong Ocean University (201903004). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
D-NJ and FZ conducted investigation. FZ, D-NJ, UFM, and S-XL conceptualized the data. FZ, H-JS, and D-NJ performed formal analysis. FZ, D-NJ, and UFM wrote the original draft. FZ, D-NJ, UFM, and C-HZ performed review and editing. G-LL administered the project. D-NJ and C-HZ acquired fund.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant number: 32172971); Guangdong Basic and Applied Basic Research Foundation (grant number: 2021A1515010430); Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (grant number: 2019KJ149).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.964150/full#supplementary-material
References
Barry, T. P., Castanos, M. T., Macahilig, M. P. S. C., and Fast, A. W. (1993). Gonadal maturatio and spawning induction in female spotted scat (Scatophagus argus). J. Aquac. Trop. 8, 121–130. doi:10.1016/j.anireprosci.2020.106273
Brands, J. H., Maassen, J. A., Van Hemert, F. J., Amons, R., and Möller, W. (1986). The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur. J. Biochem. 155 (1), 167–171. doi:10.1111/j.1432-1033.1986.tb09472.x
Browne, G. J., and Proud, C. G. (2002). Regulation of peptide-chain elongation in mammalian cells. Eur. J. Biochem. 269 (22), 5360–5368. doi:10.1046/j.1432-1033.2002.03290.x
Cao, J., Cao, Z., and Wu, T. (2007). Generation of antibodies against DMRT1 and DMRT4 of Oreochromis aurea and analysis of their expression profile in Oreochromis aurea tissues. J. Genet. Genomics 34 (6), 497–509. doi:10.1016/S1673-8527(07)60055-1
Chambers, D. M., Peters, J., and Abbott, C. M. (1998). The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc. Natl. Acad. Sci. U. S. A. 95 (8), 4463–4468. doi:10.1073/pnas.95.8.4463
Chen, J. L. (2018). The role of elongation factor eEF1A1b and 42Sp50 in gametogenesis in Nile tilapia. Doctor Thesis. Chongqing, China: Southwest University, 1–125. Available at: https://kns.cnki.net/kcms/detail/detail.aspx? (Supervisor: Wang D.S).
Cui, D., Liu, Z. W., Liu, N. X., Zhang, Y. Y., and Zhang, J. B. (2013). Histological study on the gonadal development of Scatophagus argus. J. Fish. China 7 (05), 696. doi:10.3724/SP.J.1231.2013.38442
Deschamps, S., Morales, J., Mazabraud, A., Le Maire, M., Denis, H., and Brown, D. D. (1991). Two forms of elongation factor 1 alpha (EF-1 alpha O and 42Sp50), present in oocytes, but absent in somatic cells of Xenopus laevis. J. Cell Biol. 114 (6), 1109–1111. doi:10.1083/jcb.114.6.1109
Djumagulov, M., Demeshkina, N., Jenner, L., Rozov, A., Yusupov, M., Yusupova, G., et al. (2021). Accuracy mechanism of eukaryotic ribosome translocation. Nature 600 (7889), 543–546. doi:10.1038/s41586-021-04131-9
Domingos, J. A., Budd, A. M., Banh, Q. Q., Goldsbury, J. A., Zenger, K. R., Jerry, D. R., et al. (2018). Sex-specific dmrt1 and cyp19a1 methylation and alternative splicing in gonads of the protandrous hermaphrodite barramundi. PLoS One 13 (9), e0204182. doi:10.1371/journal.pone.0204182
Dong, Z. D., Li, X. Y., Yao, Z. B., Wang, C., Guo, Y. S., Wang, Q., et al. (2021). Oryzias curvinotus in Sanya does not contain the male sex-determining gene dmy. Animals. 11 (5), 1327. doi:10.3390/ani11051327
Gautier, A., Le Gac, F., and Lareyre, J. J. (2011). The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 472 (1-2), 7–17. doi:10.1016/j.gene.2010.10.014
He, F. X., Jiang, D. N., Huang, Y. Q., Mustapha, U. F., Yang, W., Cui, X. F., et al. (2019). Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat (Scatophagus argus). Fish. Physiol. Biochem. 45 (6), 1963–1980. doi:10.1007/s10695-019-00693-8
Hoegg, S., Brinkmann, H., Taylor, J. S., and Meyer, A. (2004). Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 59 (2), 190–203. doi:10.1007/s00239-004-2613-z
Holmseth, S., Zhou, Y., Follin-Arbelet, V. V., Lehre, K. P., Bergles, D. E., Danbolt, N. C., et al. (2012). Specificity controls for immunocytochemistry: the antigen preadsorption test can lead to inaccurate assessment of antibody specificity. J. Histochem. Cytochem. 60 (3), 174–187. doi:10.1369/0022155411434828
Hsu, C. W., and Chung, B. C. (2021). Evolution, expression, and function of gonadal somatic cell-derived factor. Front. Cell Dev. Biol. 9, 684352. doi:10.3389/fcell.2021.684352
Huang, Y., Huang, Y. Q., Deng, Q. M., Mustapha, U. F., Peng, Y. X., Li, G. L., et al. (2022). A rapid method for genetic sex identification in the spotted scat (Scatophagus argus). J. Fish. Sci. China 29 (04), 515–524. doi:10.12264/JFSC2021-0359
Idigo, N. J., Soares, D. C., and Abbott, C. M. (2020). Translation elongation factor 1A2 is encoded by one of four closely related eef1a genes and is dispensable for survival in zebrafish. Biosci. Rep. 40 (1), BSR20194191. doi:10.1042/BSR20194191
Infante, C., Asensio, E., Cañavate, J. P., and Manchado, M. (2008). Molecular characterization and expression analysis of five different elongation factor 1 alpha genes in the flatfish Senegalese sole (Solea senegalensis kaup): differential gene expression and thyroid hormones dependence during metamorphosis. BMC Mol. Biol. 30 (9), 19. doi:10.1186/1471-2199-9-19
Janssen, G. M., and Möller, W. (1988). Kinetic studies on the role of elongation factors 1 beta and 1 gamma in protein synthesis. J. Biol. Chem. 263 (4), 1773–1778. doi:10.1016/s0021-9258(19)77943-5
Jiang, D. N., Peng, Y. X., Mustapha, U. K., Gu, H. T., Deng, S. P., Chen, H. P., et al. (2019). Molecular cloning and expression profile of Dmrt4 in spotted scat (Scatophagus argus). J. Guangdong Ocean. Univ. 39 (01), 7–13. doi:10.3969/j.issn.1673-9159.2019.01.002
Kahns, S., Lund, A., Kristensen, P., Knudsen, C. R., Clark, B. F., Cavallius, J., et al. (1998). The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res. 26 (8), 1884–1890. doi:10.1093/nar/26.8.1884
Kinoshita, M., Okamoto, G., Hirata, T., Shinomiya, A., Kobayashi, T., Kubo, Y., et al. (2009). Transgenic medaka enables easy oocytes detection in live fish. Mol. Reprod. Dev. 76 (2), 202–207. doi:10.1002/mrd.20942
Knudsen, S. M., Frydenberg, J., Clark, B. F., and Leffers, H. (1993). Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms:isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1 alpha. Eur. J. Biochem. 215 (3), 549–554. doi:10.1111/j.1432-1033.1993.tb18064.x
Lee, S., Ann, D. K., and Wang, E. (1994). Cloning of human and mouse brain cDNAs coding for S1, the second member of the mammalian elongation factor-1 alpha gene family: analysis of a possible evolutionary pathway. Biochem. Biophys. Res. Commun. 203 (3), 1371–1377. doi:10.1006/bbrc.1994.2336
Lin, X. H., Zhou, D. Y., Zhang, X. M., Li, G. L., Zhang, Y. L., Huang, C. L., et al. (2021). A first insight into the gonad transcriptome of Hong Kong catfish (Clarias fuscus). Animals. 11 (4), 1131. doi:10.3390/ani11041131
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., and Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92 (1), 155–161. doi:10.1007/s12041-013-0212-8
Mandal, B., Kailasam, M., Bera, A., Sukumaran, K., Hussain, T., Biswas, G., et al. (2021). Standardization of oocyte size during artificial fertilization and optimization of stocking density during indoor larval and outdoor nursery rearing of captive spotted scat (Scatophagus argus) for a viable juvenile production system. Aquaculture 534, 736262. doi:10.1016/j.aquaculture.2020.736262
Meyer, A., and Van de Peer, Y. (2005). From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27, 937–945. doi:10.1002/bies.20293
Miao, L. Y., Yuan, Y., Cheng, F., Fang, J. J., Zhou, F., Ma, W. R., et al. (2017). Translation repression by maternal RNA binding protein Zar1 is essential for early oogenesis in zebrafish. Development 144 (1), 128–138. doi:10.1242/dev.144642
Mustapha, U. F., Huang, Y., Huang, Y. Q., Assan, D., Shi, H. J., Jiang, M. Y., et al. (2021). Gonadal development and molecular analysis revealed the critical window for sex differentiation, and E2 reversibility of XY-male spotted scat, Scatophagus argus. Aquaculture 544, 737147. doi:10.1016/j.aquaculture.2021.737147
Mustapha, U. F., Zhi, F., Huan, Y. Q., Assan, D., Li, G. L., Jiang, D. N., et al. (2022). First account of a transient intersex in spotted scat, Scatophagus argus: a marine gonochoristic fish. Fish. Physiol. Biochem. doi:10.1007/s10695-022-01097-x
Navarro-Martin, L., Vinas, J., Ribas, L., Diaz, N., Gutierrez, A., Di Croce, L., et al. (2011). DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 7 (12), e1002447. doi:10.1371/journal.pgen.1002447
Ru, X. Y., Shi, H. J., Wang, T., Liu, Q. Q., Jiang, D. N., Peng, Y. H., et al. (2020). Effects of 17β-Estradiol on growth-related genes expression in female and male spotted scat (Scatophagus argus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 250, 110492. doi:10.1016/j.cbpb.2020.110492
Steinke, D., Hoegg, S., Brinkmann, H., and Meyer, A. (2006). Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol. 4, 16. doi:10.1186/1741-7007-4-16
Tao, W. J., Shi, H. J., Yang, J., Diakite, H. D., Kocher, T. D., Wang, D. S., et al. (2019). Homozygous mutation of foxh1 arrests oogenesis causing infertility in female Nile tilapia. Biol. Reprod. 102 (3), 758–769. doi:10.1093/biolre/ioz225
Tao, W., Yuan, J., Zhou, L., Sun, L., Sun, Y., Yang, S., et al. (2013). Characterization of Gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS One 8, 63604. doi:10.1371/journal.pone.0063604
Tian, C. X., Li, Z. Y., Dong, Z. D., Huang, Y., Du, T., Chen, H. P., et al. (2019). Transcriptome analysis of male and female mature gonads of silver sillago (Sillago sihama). Genes 10 (2), 129. doi:10.3390/genes10020129
Viel, A., Djé, M. K., Mazabraud, A., Denis, H., and le Maire, M. (1987). Thesaurin a, the major protein of Xenopus laevis previtellogenic oocytes, present in the 42 S particles, is homologous to elongation factor EF-1 alpha. FEBS Lett. 223 (2), 232–236. doi:10.1016/0014-5793(87)80295-8
Viel, A., Le Maire, M., Philippe, H., Morales, J., Mazabraud, A., Denis, H., et al. (1991). Structural and functional properties of thesaurin a (42Sp50), the major protein of the 42 S particles present in Xenopus laevis previtellogenic oocytes. J. Biol. Chem. 266 (16), 10392–10399. doi:10.1016/S0021-9258(18)99238-0
Wang, H., Parent, M., and Morais, R. (1994). Cloning and characterization of a cDNA encoding elongation factor 1 alpha from chicken cells devoid of mitochondrial DNA. Gene 140 (2), 155–161. doi:10.1016/0378-1119(94)90539-8
Wang, D. S., Kobayashi, T., Zhou, L. Y., Paul-Prasanth, B., Ijiri, S., Sakai, F., et al. (2007). Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 21 (3), 712–725. doi:10.1210/me.2006-0248
Wegnez, M., and Denis, H. (1979). Biochemical research on oogenesis. Transfer RNA is fully charged in the 42-S storage particles of Xenopus laevis oocytes. Eur. J. Biochem. 98 (1), 67–75. doi:10.1111/j.1432-1033.1979.tb13161.x
Wei, Y. Y. (2011). Molecular cloning and expression of translation elongation fascors (eEF1A) in the Nile tilapia. Master Thesis. Chongqing, China: Southwest University, 1–51. Available at: https://kns.cnki.net/kcms/detail/detail.aspx? (Supervisor: Wang D. S).
Xie, Q. P., He, X., Sui, Y. N., Chen, L. L., Sun, L. N., Wang, D. S., et al. (2016). Haploinsufficiency of SF-1 causes female to male sex reversal in nile Tilapia, Oreochromis niloticus. Endocrinology 157 (6), 2500–2514. doi:10.1210/en.2015-2049
Yoshikawa, A., Takano-ohmuro, H., and Masaki, T. (1984). Increase in the amount of elongation factor 2 in chicken muscular dystrophy. Muscle Nerve 7 (9), 733–740. doi:10.1002/mus.880070907
Yuan, J., Tao, W. J., Cheng, Y. Y., Huang, B. F., and Wang, D. S. (2014). Genome-wide identification, phylogeny, and gonadal expression of fox genes in Nile tilapia, Oreochromis niloticus. Fish Physiol. Biochem. 40 (4), 1239–1252. doi:10.1007/s10695-014-9919-6
Zhang, X. B., Wang, H., Li, M. H., Cheng, Y. Y., Jiang, D. N., Sun, L. N., et al. (2014). Isolation of doublesex- and mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia. Biol. Reprod. 91 (6), 136. doi:10.1095/biolreprod.114.121418
Zhang, X. T., Chang, Y. Y., Zhai, W. Y., Qian, F., Zhang, Y. Q., Xu, S. M., et al. (2021). A potential role for the gsdf-eEF1α complex in inhibiting germ cell proliferation: A protein-interaction analysis in medaka (Oryzias latipes) from a proteomics perspective. Mol. Cell Proteomics 20, 100023. doi:10.1074/mcp.RA120.002306
Keywords: Scatophagus argus, 42Sp50, 17β-estradiol (E2), promoter analysis, DNA methylation
Citation: Zhi F, Jiang D-N, Mustapha UF, Li S-X, Shi H-J, Li G-L and Zhu C-H (2022) Expression and regulation of 42Sp50 in spotted scat (Scatophagus argus). Front. Genet. 13:964150. doi: 10.3389/fgene.2022.964150
Received: 08 June 2022; Accepted: 14 July 2022;
Published: 11 August 2022.
Edited by:
Xiangshan Ji, Shandong Agricultural University, ChinaReviewed by:
Wenjing Tao, Southwest University, ChinaPrapansak Srisapoome, Kasetsart University, Thailand
Copyright © 2022 Zhi, Jiang, Mustapha, Li, Shi, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Hua Zhu, Y2h6NDE2QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Fei Zhi†
Fei Zhi† Dong-Neng Jiang
Dong-Neng Jiang Umar Farouk Mustapha
Umar Farouk Mustapha Hong-Juan Shi
Hong-Juan Shi Guang-Li Li
Guang-Li Li Chun-Hua Zhu
Chun-Hua Zhu