
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 02 September 2022
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.956866
This article is part of the Research TopicGenomic alteration landscapes of aging, metabolic disorders, and cancer: emerging overlaps and clinical importanceView all 12 articles
Background: The association between autophagy, structural alterations of the aortic wall, and endothelial dysfunction in humans has yet to be fully elucidated. The family of ULK (UNC51-like) enzymes plays critical roles in autophagy and development. This study aimed to evaluate the association between ULK gene family members and patient age of first type B aortic dissection (TBAD) onset.
Methods: The genotype data in a TBAD cohort from China and the related summary-level datasets were analyzed. We applied the sequence kernel association test (SKAT) to test the association between single-nucleotide polymorphisms (SNPs) and age of first onset of TBAD controlling for gender, hypertension, and renal function. Next, we performed a 2-sample Mendelian randomization (MR) to explore the potential causal relationship between ULK4 and early onset of TBAD at the level of gene expression coupled with DNA methylation with genetic variants as instrumental variables.
Results: A total of 159 TBAD patients with 1,180,097 SNPs were included. Concerning the association between the ULK gene family and the age of first onset of the TBAD, only ULK4 was found to be significant according to SKAT analysis (q-FDR = 0.0088). From 2-sample MR, the high level of ULK4 gene expression was related to a later age of first onset of TBAD (β = 4.58, p = 0.0214).
Conclusion: This is the first study of the ULK gene family in TBAD, regarding the association with the first onset age. We demonstrated that the ULK4 gene is associated with the time of onset of TBAD based on both the SKAT and 2-sample MR analyses.
Type B aortic dissection (TBAD) is a rare while life-threatening condition, in which a tear occurs in the descending part of the aorta and may extend into the abdomen (Prêtre and Von Segesser, 1997; Hagan et al., 2000; Nienaber and Clough, 2015). Prevention of premature death from TBAD depends on the early identification of high-risk individuals, careful monitoring of the dissected aorta for aneurysmal dilations, medications to slow the rate of growth of aneurysms, and timely surgical repair of aneurysms (Mokashi and Svensson, 2019).
Aortic expansion is one of the risk factors associated with the need for intervention or adverse outcomes in patients with TBAD. It was reported that younger age at presentation was a clinical predictor of aortic expansion. Patient age <60 years was significantly associated with increasing aortic diameter, which was thought to be due to a less rigid aortic wall, making the aorta more prone to dilation in younger patients (Kamman et al., 2017). However, the essential reasons for this finding deserve further research, and the association of genetic variants with the age of first onset of type B aortic dissections is a valuable research direction.
Autophagy is a process in which intracellular components and dysfunctional organelles are delivered to the lysosome for degradation and recycling. Therefore, autophagy has various connections to many human diseases, as its functions are essential for cell survival, bioenergetic homeostasis, organism development, and cell death regulation. The association between autophagy and structural alterations of the aortic wall and endothelial dysfunction in humans has yet to be fully elucidated (Yang and Klionsky, 2020). Previous studies have shown that more than 20% of individuals with thoracic aortic aneurysms and dissections have a family history of disease that may be caused by a genetic syndrome, resulting from a single-gene mutation such as Marfan syndrome (MFS [MIM:154700]) arising from a fibrillin-1 (FBN1 [MIM:134797]) mutation (Biddinger et al., 1997). However, autophagy-related biomarkers studies for aortic dissection diseases are still rare. According to existing research, AMPK increases the process of autophagy after its activation. Although the mechanisms of AD and autophagy have not been fully elucidated, autophagy has been observed to be activated in impaired vascular smooth muscle cells (VSMCs). Excessive or impaired autophagy may lead to VSMC death or dysfunction, which is thought to promote aneurysm and AD (Clément et al., 2019).
It is known that the ULK (UNC51-like) enzymes are a family of mammalian kinases and play critical roles in autophagy and development. The mammalian ULK family of kinases comprises 5 genes: ULK1 to ULK4 and STK36 (Chan and Tooze, 2009). These enzymes share a conserved N-terminal kinase domain, which is homologous to C. elegans UNC51 and yeast Atg1, the original kinase identified in the autophagy pathway. ULK kinases can be found in all observed eukaryotes. ULK1 kinases are involved in autophagy (Young et al., 2009), ULK3 is also implicated in hedgehog signaling and in autophagy-mediated senescence (Maloverjan et al., 2010), and several genome-wide association studies (GWASs) show linkage to blood pressure (Levy et al., 2009). ULK4 is a pseudo kinase in all species and may be linked to neurogenesis, brain function (Liu et al., 2017), and blood pressure (Levy et al., 2009). It has also been reported that ULK4 is potentially associated with acute aortic dissections (Guo et al., 2016). As can be seen, there is demonstrable link between the ULK family, autophagy, and acute aortic dissection, and thus an assessment of the effect of the ULK family on aortic dissection is warranted.
We performed an association analysis between ULK gene family members and the age of first onset of Chinese TBAD patients. The association study was performed in Chinese TBAD patients. The causal effect of ULK genes was subsequently verified by 2-sample Mendelian randomization (MR) at the gene expression and DNA methylation levels.
We obtained genotype data from a TBAD cohort enrolled through the Vascular Surgery Department of Zhongshan Hospital Fudan University, which included 162 Chinese patients with TBAD from January 2018 to June 2019. Each participant signed a consent form. The study was approved by the relevant ethics committees (Ethical approval No. B2019-110R) and was administered by trained personnel.
In association analysis, we obtained genotype data using whole genome sequencing, using the Illumina NovaSeq platform (Illumina, San Diego, CA, United States) in a paired-end 150 bp mode on 162 TBAD patients. Briefly, the samples were excluded if they met any of the following quality control (QC) criteria (Supplementary Figure S1): 1) overall genotype completion rate <95%, 2) unexpected duplicates or probable relatives, 3) heterozygosity rates more than six times the SD from the mean, or 4) gender discrepancies. SNPs were excluded if they met any of the following QC criteria: 1) SNPs had a low call rate of <95% in all samples, 2) the genotype distributions of SNPs deviated from those expected by the Hardy–Weinberg equilibrium (p < 0.000001), or 3) single-nucleotide variants (SNVs) with minor allele frequencies were less than 1%.
Methylation quantitative trait loci (mQTL) data were obtained from the Brisbane Systems Genetics Study (n = 614) and Lothian Birth Cohorts of 1921 and 1936 (n = 1366) (Wu et al., 2018). Details of the QC procedures were described in a previous study. Briefly, all the individuals were of European descent. Only the DNA methylation probes with at least a cis-mQTL at p < 0.0001 and only SNPs within 500 kb distance from each probe were included in the analysis. As for the summary statistics of expression quantitative trait loci (eQTL), we used the cis-eQTL in the prefrontal cortex from the PsychENCODE project (n = 1387) (Gandal et al., 2018; Wang et al., 2018). The eQTL analyses of PsychENCODE were performed by including 100 hidden covariate factors as covariates. Only the data of SNPs in a 500 kb window around the ULK4 gene were included in the subsequent analysis.
Continuous variables are summarized as mean ± SD and categorical variables are described as numbers and percentages. The sequence kernel association test (SKAT), which is a supervised, flexible, and computationally efficient regression method was used to test for the association between a set of genetic variants and a continuous or dichotomous trait with adjustments made for relevant covariates (Ionita-Laza et al., 2013). A total of 421 variants from the ULK family passed QC and were included in SKAT analysis with controlling for gender, hypertension, and smoking status and renal function (Wu et al., 2011). Furthermore, separate analyses were conducted for all variants (n = 421) and rare variants with minor allele frequency (MAF) < 0.05 (n = 290). We used COXPRESdb (http://coxpresdb.jp) to drawing the co-expressed gene network with pathway and protein–protein interaction information. COXPRESdb was first released for human and mouse models in 2007 (Takeshi et al., 2007). One characteristic feature of COXPRESdb is its ability to compare multiple co-expression data derived from different transcriptomics technologies and different species, which strongly reduces false-positive relationships in individual gene co-expression data (Takeshi et al., 2012; Takeshi et al., 2019). To clarify the molecular mechanisms, we used COXPRESdb to perform co-expression analysis on those genes that exerted significant effect on aortic dissection.
The analysis workflow is shown in Figure 1. In general, we adopted two analysis steps. First, we applied the SKAT to test the association between a set of SNP and patients’ age of first onset of TBAD, controlling for gender, hypertension, smoking status, and renal function. Then, linear regression models were applied to further detect significant SNPs in significant genes. Multiple comparisons were adjusted with the false discovery rate method (FDR) to control the overall false-positive rate at a 5% level. Biomarkers measured by a q-FDR value ≤0.05 were included in further study (Benjamini and Hochberg, 1995).
To further explore the role of ULK4 in different clinical features, we constructed a genetic score with the significant SNPs and conducted subgroup analysis to compare the differences between different categories of features. The genetic score (GS) was calculated based on a weighted linear combination of individual values of the significant SNPs, with weights derived from the stepwise linear regression model. As a result, three SNPs were finally included in the genetic score with the formula defined as follows: GS = (−25.326 × rs191792955) + (12.408 × rs142574024) + (−4.425 × rs74282513).
In the second step, we performed a 2-sample MR to explore the potential causal relationship between ULK4 and the early onset of TBAD (Y) at the level of gene expression and DNA methylation (X) using genetic variants (G) as instrumental variables (Evans and Davey Smith, 2015). As shown in Figure 4, SNPs with a q-FDR < 0.05 were regarded as candidate instrumental variables and linkage disequilibrium (LD) clumping with a window of 1 MB, and an r2 < 0.2 was applied to remove SNPs with high LD (Hemani et al., 2018). In addition, we performed sensitivity analyses using several approaches to investigate potential pleiotropic bias and verify the robustness of the results, including MR-Egger regression, weighted median MR, weighted mode MR, simple mode MR, funnel plots, and leave-1-variant-out analysis.
Statistical analyses were performed using R version 3.4.4 (The R Foundation of Statistical Computing, Vienna, Austria). A 2-sided p-value of less than 0.05 was considered to indicate a statistically significant difference.
A total of 162 Chinese TBAD patients from Zhongshan Hospital were genotyped with two patients being excluded because of gender mismatches, and one patient being excluded because of familial relationships. Finally, 159 TBAD patients with 1,180,097 SNPs after QC were included in further analyses (Supplementary Figure S1). Demographics and clinical characteristics are shown in Supplemetary Table S1. The mean age of the enrolled TBAD inpatients was 56.11 years (SD 14.33), and 78.62% of the patients were male. The mean age of the first onset of TBAD was 54.89 years with a range of 18–85 years. Most of the enrolled TBAD inpatients (88.68%) also had hypertension, and 10.06% had diabetes.
Concerning on the association between the ULK gene family and age of first onset of TBAD, only ULK4 was significant according to SKAT analysis (q-FDR = 0.0088) based on our TBAD samples. Among 361 variants in the ULK4 gene, there were 243 variants with MAF <0.05. In addition, SKAT analysis for variants with MAF <0.05 of ULK4 was also significant (p = 0.0015) (Table 1). Further linear regression analysis of SNPs in ULK4 revealed 17 SNPs, that were associated with age of first onset of TBAD, and the most strongly associated SNP in ULK4 was rs191792955 (Figure 2; Supplementary Figure S2). The details of the association results of the SNPs sites of ULK4 are shown in Supplementary Table S2.
Among the significant 17 SNPs, three SNPs were maintained for genetic score generation after stepwise regression with p < 0.05. As the genetic score increased, the age of first onset decreased for TBAD patients with well-controlled hypertension (coefficient 1.21, 95% CI: 0.677–1.751), and similar results were found for complex, male, and normal renal function TBAD patients. Heterogeneity details are shown in a forest plot (Supplementary Figure S3).
For 2-sample MR, the individual instrument-gene expression and instrument-age of first onset for TBAD are shown in Supplementary Table S3. Among the 19 SNPs associated with gene expression (q-FDR < 0.05), three were left after LD clumping. A similar process was followed for the instrumental variable determination of ULK4 methylation, and there were 10 CpG probes with 25 SNPs found (Supplementary Table S4). The details of the instrumental variable selections are shown in Figure 3A. The intercept of MR-Egger for ULK4 gene expression and DNA methylation indicated that there was no potential horizontal pleiotropy, and the inverse variance weighted (IVW) analysis with fixed effects was conducted for two sample MR analyses. The high ULK4 gene expression was related to a later age of onset for TBAD (β = 4.58, p = 0.0214) (Figures 3B, 4A). Among MR analyses of the 10 CpG probes, eight SNPs in cg25209153 were significant (p = 0.0155) (Figures 3B, 4A), and the higher cg25209153 was related to the earlier age of first onset (β = −4.02). In addition, several MR methods were used to evaluate the robustness of results, and the direct of the causal effects of both ULK4 and cg25209153 were consistent with IVW, although the p values were not significant (Figures 4B, C). The details of sensitively analyses are shown in Supplementary Table S5.
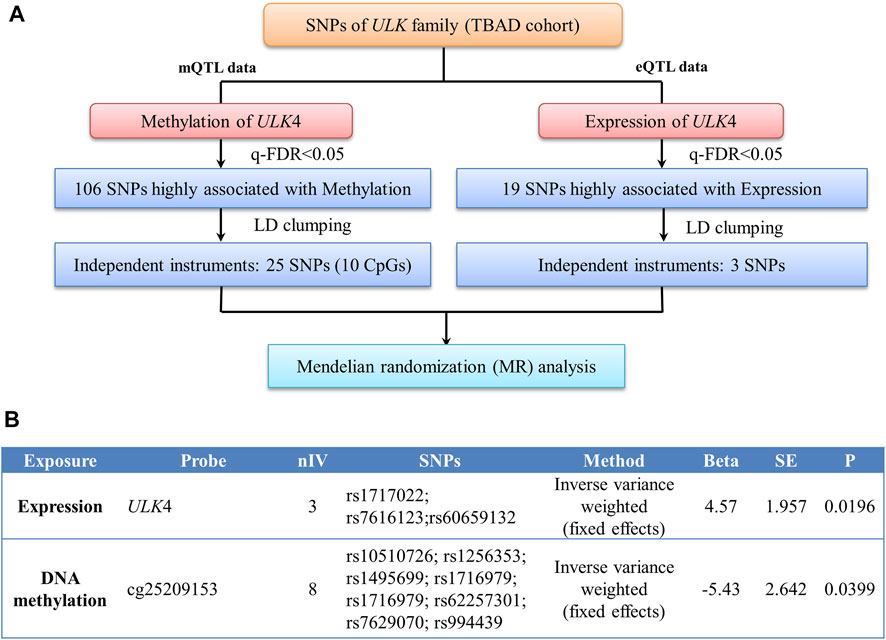
FIGURE 3. Two sample Mendelian randomization analyses. (A) Diagram of instrumental variable (IV) selection. (B) Results of Mendelian randomization (MR) between gene expression, DNA methylation, and onset age of TBAD.
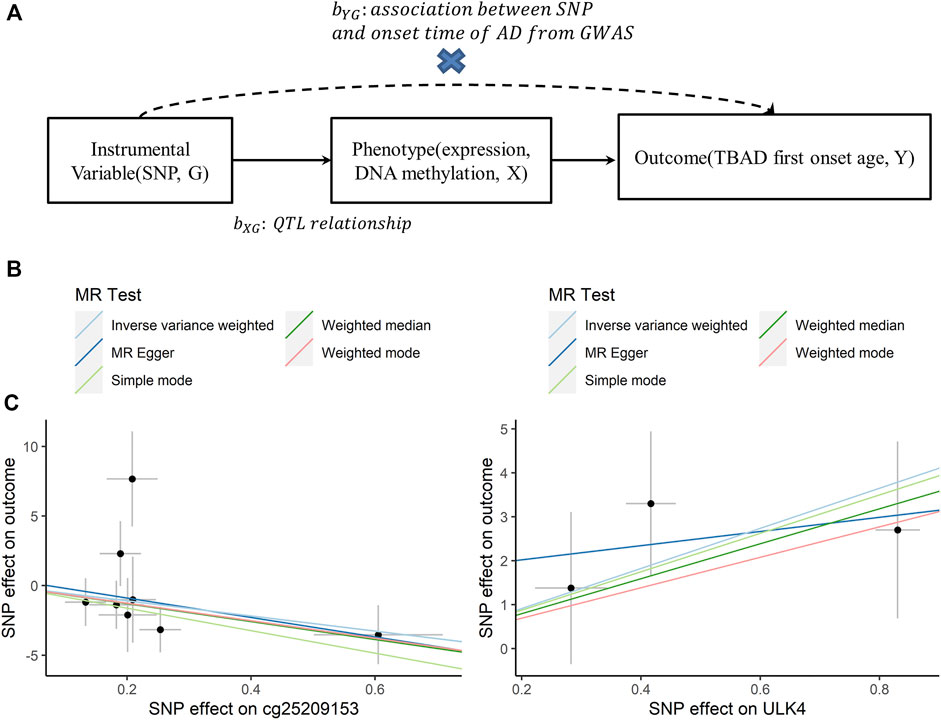
FIGURE 4. Association plots between ULK4 or cg25209153 and first onset age. (A) Diagram of Mendelian randomization (MR) analysis. (B) Scatter plots for association between cg25209153 and first onset age by different MR analytical methods. (C) Scatter plots for association between ULK4 and first onset age by different MR analytical methods.
According to the gene co-expression network of ULK4 (Supplementary Figure S4), it was found that OSCP1 acts on smooth muscle cells in the tunica media layers of artery walls, participates in the regulation of the extracellular matrix (ECM) related to intimal proliferation after endothelial injury, and is related to restenosis after vascular injury (Martí‐Pàmies et al., 2017). WDPCD is believed to be related to the development of arteries. In the process of coronary vasculature development, WDPCD participates in the regulation of epithelial–mesenchymal transition (EMT) to enable migration that gives rise to smooth muscle cells (Liu et al., 2018).
Acute aortic dissection may be fatal without early diagnosis and appropriate management, and thus biomarker tests play an important role in preventing and diagnosing aortic dissection disease (Ranasinghe and Bonser, 2010). Several epigenetic studies of TBAD have identified potential biomarkers relevant to the etiology of TBAD (Wang et al., 2012; Erhart et al., 2020). Multiple GWAS studies have identified a significant association of the ULK4 SNPs with hypertension. Genetic variants in ULK4 have also been reported to be associated with the pathogenesis of sporadic thoracic aortic dissection (STAD) (Guo et al., 2016). To the best of our knowledge, this is the first study of the ULK gene family in TBAD to focus on the association with age of first onset.
The ULK (UNC51-like) enzymes play critical roles in autophagy and development. While ULK1, ULK2, and ULK3 have been characterized, and very little is known about ULK4. Recently, deletions in ULK4 have genetically linked to increased susceptibility to developing schizophrenia, which is a devastating neuropsychiatric disease with high heritability (Khamrui et al., 2019). Similarly, TBAD has also been identified as a suspected heritable characteristic. In their single-institute study, Shalhub et al. (2021) found that heritable TBAD was the cause of TBAD in one of four patients, and familial TBAD was presented at an early age. Finally, it has been established that hypertension is an essential component of both familial TBAD and sporadic TBAD (Shalhub et al., 2020).
Most patients in our TBAD cohort were male, most had hypertension, and few of them had diabetes. These population characteristics are consistent with the reported TBAD in the Chinese population (Huang et al., 2021). Based on whole genome sequencing data, we demonstrated that the ULK4 gene is associated with the age of TBAD onset based on both the SKAT and 2-sample MR analyses. Furthermore, we found that DNA methylation of cg25209153ULK4 and expression of ULK4 were associated with the age of first onset. Furthermore, high DNA methylation of cg25209153ULK4 was negatively correlated with age of first onset. Inversely, high expression of ULK4 was positively correlated with age of first onset.
Several GWASs have reported significant associations of ULK4 SNPs with hypertension in individuals of European (particularly those with high diastolic blood pressure), African American, and East Asian descent (Guo et al., 2016). It is also known that poorly controlled hypertension is a major risk factor for TBAD, and our study demonstrated that the ULK4 gene, involved in endocytosis and axon growth (Ostberg et al., 2020), is associated with age of first onset of TBAD, suggesting that in addition to the association with the control of hypertension, genomic variants in ULK4 have a potential mechanism for contributing to the early onset of TBAD. As autophagy is a highly conserved catabolic process and a major cellular pathway for the degradation of long-lived proteins and cytoplasmic organelles, ULK4 plays critical roles in autophagy and dysregulation of autophagy may lead to the early onset of TBAD. Further studies are needed to validate the link between ULK4 and the age of first onset of TBAD. ULK4 may be an effective companion diagnostic target in TBAD if it is confirmed by further fundamental and clinical studies.
In addition, our study will follow up the prognosis of the existing cases so as to suggest early detection and early treatments for at-risk patients. At the same time, we will also consider measuring different omics data such as proteomics from the same batch to further validate the importance of the ULK4 gene in TBAD.
Our study also has some limitations. First, the sample size was limited, which may impact the robustness of the 2-sample MR analyses. Second, because only TBAD patients were included in our study, the association of ULK4 with type A dissections could not be investigated; moreover, as a case-only study, which did not involve the control group, the current study cannot conclude the effective companion diagnostic target in TBAD. Third, the validated DNA samples were not collected. In addition, the clinical information was not comprehensive, such as concomitant medication of antiplatelet and statin, the potential bias risk may impact the robustness of the results.
In conclusion, this is the first study of the ULK gene family in TBAD to focus on an analysis of the association with the first onset age. We demonstrated that the ULK4 gene is associated with the age of first onset of TBAD based on both the SKAT and 2-sample MR analyses. ULK4 may be an effective companion diagnostic target in TBAD.
The data presented in the study are deposited through these following websites. The eQTL summary data can be found in http://www.psychENCODE.org/. The mQTL data from the meta-analyses of Brisbane Systems Genetics Study and the Lothian Birth Cohorts of 1921 and 1936 are available at http://cnsgenomics.com/software/smr/#Download. The whole genome sequencing data of TBAD cohort was provided by Yi Si, et al. Requests to access this dataset should be directed to c3lzaXlAeWFob28uY29t.
The studies involving human participants were reviewed and approved by the relevant ethics committees of Zhongshan Hospital (Ethical approval No. B2019-110R) and was administered by trained personnel. The patients/participants provided their written informed consent to participate in this study.
LH, YS, and WF designed the research, wrote the manuscript, and responsible for final content. LH, YW, YS, and FC conceived and designed the research. LH, JT, LL, and RW conducted the research. LH, JT, and YW analyzed the data. FC and WF reviewed and edited the manuscript. All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis and for the decision to submit the manuscript for publication.
This work was supported by the National Natural Science Foundation of China (Grant 81903407 to LH).
We are grateful to all patients who participated in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.956866/full#supplementary-material
eQTL, expression quantitative trait loci; KAT, sequence kernel association test; MAF, minor allele frequency; MFS, Marfan syndrome; mQTL, methylation quantitative trait loci; MR, Mendelian randomization; STBAD, type B aortic dissection; VSMCs, vascular smooth muscle cells.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 (1), 289–300. doi:10.1111/j.2517-6161.1995.tb02031.x
Biddinger, A., Rocklin, M., Coselli, J., and Milewicz, D. M. (1997). Familial thoracic aortic dilatations and dissections: A case control study. J. Vasc. Surg. 25 (3), 506–511. doi:10.1016/s0741-5214(97)70261-1
Chan, E. Y., and Tooze, S. A. (2009). Evolution of Atg1 function and regulation. Autophagy 5 (6), 758–765. doi:10.4161/auto.8709
Clément, M., Chappell, J., Raffort, J., Lareyre, F., Vandestienne, M., Taylor, A. L., et al. (2019). Vascular smooth muscle cell plasticity and autophagy in dissecting aortic aneurysms. Arterioscler. Thromb. Vasc. Biol. 39 (6), 1149–1159. doi:10.1161/ATVBAHA.118.311727
Erhart, P., Gieldon, L., Ante, M., Körfer, D., Strom, T., Grond-Ginsbach, C., et al. (2020). Acute stanford type B aortic dissection—Who benefits from genetic testing? J. Thorac. Dis. 12 (11), 6806–6812. doi:10.21037/jtd-20-2421
Evans, D. M., and Davey Smith, G. (2015). Mendelian randomization: New applications in the coming age of hypothesis-free causality. Annu. Rev. Genomics Hum. Genet. 16, 327–350. doi:10.1146/annurev-genom-090314-050016
Gandal, M. J., Zhang, P., Hadjimichael, E., Walker, R. L., Chen, C., Liu, S., et al. (2018). Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362 (6420), eaat8127. doi:10.1126/science.aat8127
Guo, D-c., Grove, M. L., Prakash, S. K., Eriksson, P., Hostetler, E. M., LeMaire, S. A., et al. (2016). Genetic variants in LRP1 and ULK4 are associated with acute aortic dissections. Am. J. Hum. Genet. 99 (3), 762–769. doi:10.1016/j.ajhg.2016.06.034
Hagan, P. G., Nienaber, C. A., Isselbacher, E. M., Bruckman, D., Karavite, D. J., Russman, P. L., et al. (2000). The international registry of acute aortic dissection (IRAD): New insights into an old disease. Jama 283 (7), 897–903. doi:10.1001/jama.283.7.897
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. elife 7, e34408. doi:10.7554/eLife.34408
Huang, L., Kan, Y., Zhu, T., Chen, B., Xu, X., Dong, Z., et al. (2021). Ten-year clinical characteristics and early outcomes of type B aortic dissection patients with thoracic endovascular aortic repair. Vasc. Endovasc. Surg. 55 (4), 332–341. doi:10.1177/1538574420983652
Ionita-Laza, I., Lee, S., Makarov, V., Buxbaum, J. D., and Lin, X. (2013). Sequence kernel association tests for the combined effect of rare and common variants. Am. J. Hum. Genet. 92 (6), 841–853. doi:10.1016/j.ajhg.2013.04.015
Kamman, A. V., Brunkwall, V., Eric, L., Heijmen, R. H., and Trimarchi, S. (2017). Predictors of aortic growth in uncomplicated type B aortic dissection from the acute dissection stent grafting or best medical treatment (ADSORB) database. J. Vasc. Surg. 65 (4), 964–971. doi:10.1016/j.jvs.2016.09.033
Khamrui, S., Ung, P. M., Secor, C., Schlessinger, A., and Lazarus, M. B. (2019). High-resolution structure and inhibition of the schizophrenia-linked pseudokinase ULK4. J. Am. Chem. Soc. 142 (1), 33–37. doi:10.1021/jacs.9b10458
Levy, D., Ehret, G. B., Rice, K., Verwoert, G. C., Launer, L. J., Dehghan, A., et al. (2009). Genome-wide association study of blood pressure and hypertension. Nat. Genet. 41 (6), 677–687. doi:10.1038/ng.384
Liu, M., Xu, P., O'Brien, T., and Shen, S. (2017). Multiple roles of Ulk4 in neurogenesis and brain function. Neurogenesis 4 (1), e1313646. doi:10.1080/23262133.2017.1313646
Liu, X., Wang, Y., Liu, F., Zhang, M., Song, H., Zhou, B., et al. (2018). Wdpcp promotes epicardial EMT and epicardium-derived cell migration to facilitate coronary artery remodeling. Sci. Signal. 11 (519), eaah5770. doi:10.1126/scisignal.aah5770
Maloverjan, A., Piirsoo, M., Michelson, P., Kogerman, P., and Østerlund, T. (2010). Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp. Cell Res. 316 (4), 627–637. doi:10.1016/j.yexcr.2009.10.018
Martí‐Pàmies, I., Cañes, L., Alonso, J., Rodríguez, C., and Martínez‐González, J. (2017). The nuclear receptor NOR‐1/NR4A3 regulates the multifunctional glycoprotein vitronectin in human vascular smooth muscle cells. FASEB J. 31 (10), 4588–4599. doi:10.1096/fj.201700136RR
Mokashi, S. A., and Svensson, L. G. (2019). Guidelines for the management of thoracic aortic disease in 2017. Gen. Thorac. Cardiovasc. Surg. 67 (1), 59–65. doi:10.1007/s11748-017-0831-8
Nienaber, C. A., and Clough, R. E. (2015). Management of acute aortic dissection. Lancet 385 (9970), 800–811. doi:10.1016/S0140-6736(14)61005-9
Ostberg, N. P., Zafar, M. A., Ziganshin, B. A., and Elefteriades, J. A. (2020). The genetics of thoracic aortic aneurysms and dissection: A clinical perspective. Biomolecules 10 (2), 182. doi:10.3390/biom10020182
Prêtre, R., and Von Segesser, L. K. (1997). Aortic dissection. Lancet 349 (9063), 1461–1464. doi:10.1016/S0140-6736(96)09372-5
Ranasinghe, A. M., and Bonser, R. S. (2010). Biomarkers in acute aortic dissection and other aortic syndromes. J. Am. Coll. Cardiol. 56 (19), 1535–1541. doi:10.1016/j.jacc.2010.01.076
Shalhub, S., Rah, J. Y., Campbell, R., Sweet, M. P., Quiroga, E., and Starnes, B. W. (2021). Characterization of syndromic, nonsyndromic familial, and sporadic type B aortic dissection. J. Vasc. Surg. 73 (6), 1906–1914.e2. doi:10.1016/j.jvs.2020.10.080
Shalhub, S., Roman, M. J., Eagle, K. A., LeMaire, S. A., Zhang, Q., Evangelista, A., et al. (2020). Type B aortic dissection in young individuals with confirmed and presumed heritable thoracic aortic disease. Ann. Thorac. Surg. 109 (2), 534–540. doi:10.1016/j.athoracsur.2019.07.004
Takeshi, O., Shinpei, H., Masayuki, S., Motoshi, S., Hiroyuki, O., and Kengo, K. (2007). COXPRESdb: A database of coexpressed gene networks in mammals. Nucleic Acids Res. 36, D77–D82. doi:10.1093/nar/gkm840
Takeshi, O., Yasunobu, O., Satoshi, I., and Shu, T. (2012). Motoike IN, kengo K: COXPRESdb: A database of comparative gene coexpression networks of eleven species for mammals. Nucleic Acids Res. 41 (D1), 1014–1020.
Takeshi, O., Yuki, K., Yuichi, A., Shu, T., and Kengo, K. (2019). COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 47 (D1), D55-D62. doi:10.1093/nar/gky1155
Wang, D., Liu, S., Warrell, J., Won, H., Shi, X., Navarro, F. C., et al. (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science 362 (6420), eaat8464. doi:10.1126/science.aat8464
Wang, L., Yao, L., Guo, D., Wang, C., Wan, B., Ji, G., et al. (2012). Gene expression profiling in acute Stanford type B aortic dissection. Vasc. Endovasc. Surg. 46 (4), 300–309. doi:10.1177/1538574412443315
Wu, M. C., Lee, S., Cai, T., Li, Y., Boehnke, M., and Lin, X. (2011). Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89 (1), 82–93. doi:10.1016/j.ajhg.2011.05.029
Wu, Y., Zeng, J., Zhang, F., Zhu, Z., Qi, T., Zheng, Z., et al. (2018). Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 9 (1), 918–1014. doi:10.1038/s41467-018-03371-0
Yang, Y., and Klionsky, D. J. (2020). Autophagy and disease: Unanswered questions. Cell Death Differ. 27 (5), 858–871. doi:10.1038/s41418-019-0480-9
Keywords: type B aortic dissection, ULK4, age first onset, genetic variants, gene expression
Citation: Huang L, Tang J, Lin L, Wang R, Chen F, Wei Y, Si Y and Fu W (2022) Association of genetic variants in ULK4 with the age of first onset of type B aortic dissection. Front. Genet. 13:956866. doi: 10.3389/fgene.2022.956866
Received: 30 May 2022; Accepted: 28 July 2022;
Published: 02 September 2022.
Edited by:
Jaspreet Kaur Dhanjal, Indraprastha Institute of Information Technology Delhi, IndiaReviewed by:
Aditya Yashwant Sarode, Columbia University, United StatesCopyright © 2022 Huang, Tang, Lin, Wang, Chen, Wei, Si and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Si, c3lzaXlAeWFob28uY29t; Weiguo Fu, ZnUud2VpZ3VvQHpzLWhvc3BpdGFsLnNoLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.