
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 08 August 2022
Sec. Pharmacogenetics and Pharmacogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.949241
This article is part of the Research Topic Molecular Targets for Anti-Cancer Drug Discovery and Development View all 5 articles
Cancer is a complex disease whereby multiple genetic aberrations, epigenetic modifications, metabolic reprogramming, and the microenvironment contribute to the development of a tumor. In the traditional anticancer drug discovery pipeline, drug candidates are usually screened in vitro using two-dimensional or three-dimensional cell culture. However, these methods fail to accurately mimic the human disease state. This has led to the poor success rate of anticancer drugs in the preclinical stages since many drugs are abandoned due to inefficacy or toxicity when transitioned to whole-organism models. The common fruit fly, Drosophila melanogaster, has emerged as a beneficial system for modeling human cancers. Decades of fundamental research have shown the evolutionary conservation of key genes and signaling pathways between flies and humans. Moreover, Drosophila has a lower genetic redundancy in comparison to mammals. These factors, in addition to the advancement of genetic toolkits for manipulating gene expression, allow for the generation of complex Drosophila genotypes and phenotypes. Numerous studies have successfully created Drosophila models for colorectal, lung, thyroid, and brain cancers. These models were utilized in the high-throughput screening of FDA-approved drugs which led to the identification of several compounds capable of reducing proliferation and rescuing phenotypes. More noteworthy, Drosophila has also unlocked the potential for personalized therapies. Drosophila ‘avatars’ presenting the same mutations as a patient are used to screen multiple therapeutic agents targeting multiple pathways to find the most appropriate combination of drugs. The outcomes of these studies have translated to significant responses in patients with adenoid cystic carcinoma and metastatic colorectal cancers. Despite not being widely utilized, the concept of in vivo screening of drugs in Drosophila is making significant contributions to the current drug discovery pipeline. In this review, we discuss the application of Drosophila as a platform in anticancer drug discovery; with special focus on the cancer models that have been generated, drug libraries that have been screened and the status of personalized therapies. In addition, we elaborate on the biological and technical limitations of this system.
Cancer is one of the leading causes of death worldwide, claiming approximately 10 million lives in 2020. The disease represents a major global burden with 19.3 million new cases reported in 2020 (Sung et al., 2021). With these numbers expected to increase, it demonstrates the urgent need for intervention in detection and treatment strategies. Cancer is a complex disease whereby multiple genetic aberrations, epigenetic modifications, metabolic reprogramming, and the microenvironment contribute to the development of a tumor. Despite advances in understanding the molecular drivers of cancer, our ability to translate anticancer research into clinical success has been poor. The progression of new anticancer drugs through the drug discovery pipeline has one of the highest failure rates in comparison to other diseases. In effect, less than 5% of anticancer drugs that are developed ultimately reach the market (Moreno and Pearson, 2013; Liu et al., 2017). Several factors contribute to this high failure rate, most notably is the complexity of the disease and limitations in preclinical screening tools which fail to simulate these complexities.
In the traditional anticancer drug discovery pipeline, in vitro screening has become a standard tool used in the preclinical stages to identify candidate drugs with clinical potential (Figure 1). Small-molecule candidates are typically screened for activity using two-dimensional or three-dimensional cell culture. Two-dimensional (2-D) cell culture offers high-throughput screening with extensive panels of cell lines representing various cancer types. However, these cultures which are often found in monolayer, do not adequately represent key characteristics of human tumors such as the three-dimensional (3-D) architecture of tumors and the host-tumor environment. Furthermore, prolonged culturing of cancer cell lines leads to genetic drift. Consequently, cell lines may exhibit substantial genetic, epigenetic, and phenotypic variations induced by an artificial environment and therefore may not reflect the original tumor (Hoelder et al., 2012; Verjans et al., 2018). Three-dimensional cell culture, in the form of spheroids and organoids, was thus designed to resemble in vivo tumors more closely. These cell cultures offer the opportunity for a more in-depth study of characteristics such as cellular contact, the extracellular matrix, drug penetration, nutrient distribution, and aspects of drug resistance (Richardson et al., 2015; Horvath et al., 2016). Several factors, including the cost and reproducibility, limit the application of three-dimensional cell culture. For instance, different culture techniques often give rise to spheroids with varying sizes and shapes. This has been found to influence drug efficacy and toxicity (Zanoni et al., 2016). In addition, it is challenging to utilize this culture technique in a high-throughput manner (Verjans et al., 2018). Both 2-D and 3-D cell culture screening methods fail to accurately mimic the host-tumor microenvironment and recapitulate the complex mechanisms found in vivo. Consequently, many positive hits identified using these types of in vitro screens are abandoned in the preclinical stages due to inefficacy or toxicity when transitioned to whole-organism models (Horvath et al., 2016; Adams et al., 2021). This highlights the shortcomings of in vitro cell culture in the evaluation of anticancer drugs and emphasizes the need for more physiologically relevant screening tools that capture the properties of the cancer cell as well as the host-tumor microenvironment.
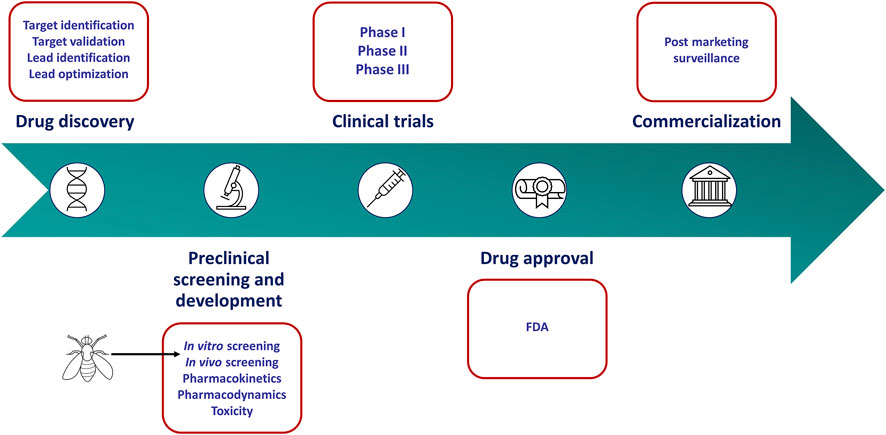
FIGURE 1. Drug discovery and development pipeline. In vitro culture screens have become a standard tool used in the preclinical stages to identify anticancer agents. However, these models fail to mimic key characteristics of human tumors. Drosophila melanogaster can recapitulate these characteristics more accurately and may serve as a more appropriate screening tool in the preclinical stages of the drug discovery pipeline.
The common fruit fly, Drosophila melanogaster, has emerged as a beneficial platform for investigating cancer. Drosophila has typically been utilized as a pathway discovery platform, enabling the successful identification of key components in cancer-related pathways such as WNT, HIPPO, JAK/STAT, RAS, NOTCH, HEDGEHOG, BMP and TGF-β (Rudrapatna et al., 2012; Cagan et al., 2019). However, apart from being a powerful pathway identification tool, the organism has also attracted attention as an anticancer drug screening tool. Conservation of key genes, a low genetic redundancy, development of genetic manipulation toolkits and the rapid life cycle of Drosophila have provided a selective advantage for modeling human tumors as well as whole-organism anticancer drug screening (Cagan et al., 2019). It has been estimated that approximately 75% of genes affiliated with a disease in humans, have functional homologs in Drosophila (Wangler et al., 2017). In a cancer-specific context, these include key genes involved in the cell cycle, differentiation, cell migration, cell polarity, cell adhesion and apoptosis. It is significant to point out that these genes appear at a lower frequency in Drosophila due to the lower genetic redundancy observed in comparison to mammals. This feature has proven advantageous for drug discovery since fewer genes need to be manipulated in order to set up a sensitized condition for drug screening (Su, 2019). These factors, in addition to the advancement of genetic toolkits for manipulating gene expression allow for the generation of complex Drosophila genotypes and phenotypes (Cagan et al., 2019; Su, 2019). Therefore, Drosophila can depict the cancer state more accurately than traditional in vitro cell culture systems, and has the added advantage of the host-tumor environment. Drosophila also offers many practical advantages. The organism has a rapid life cycle (∼ 10 days) with the ability to produce large numbers of offspring and is relatively cheap and easy to maintain. Moreover, both larvae and mature adults are small enough to fit into 96-well microtiter plates. This makes the organism suitable for high-throughput screening of large drug libraries (Richardson et al., 2015).
The translation of anticancer drug activity, determined in the preclinical screening stages, into efficacy in the clinic remains a major hurdle. The factors associated with the failure of such anticancer drugs have been reviewed elsewhere (Jardim et al., 2017). The burden of wasted money and resources applied into development of a drug that fails to show therapeutic relevance provides a strong motivation to re-consider our preclinical screening practices (Hoelder et al., 2012). Drosophila has the potential to recapitulate some hallmarks of cancer unerringly and the ability to be utilized in a high-throughput screening manner. Moreover, screening using Drosophila provides information on drug bioavailability, toxicity to the organism and host-tumor interactions. As such, Drosophila represents a far more valuable screening tool in the preclinical stages of anticancer drug discovery. Given the anatomical differences between Drosophila and humans, this system cannot be used to study all cancer types. Therefore, Drosophila may not replace whole-organism mammalian models but can serve as a more appropriate screening tool than in vitro cultures for particular cancer types (Figure 1). The application of this organism in the anticancer drug discovery pipeline is still in its infancy, and greater awareness and utilization should be encouraged. Herein, we discuss the application of Drosophila as a platform in anticancer drug discovery; with special focus on the cancer models that have been generated, drug libraries that have been screened and the status of personalized therapies. In addition, we elaborate on the biological and technical limitations of this system, placing it in context as an amenable tool for drug discovery.
The homologous nature of Drosophila and humans allows for the study of many cancer types in Drosophila. The systems used to study these cancers are not based on identical structural representation but rather genetic and molecular similarities between Drosophila and humans (Figure 2). Selected regions of Drosophila are modeled to represent the target cancer type. These regions are indicative of the genes and underlying mechanisms that are homologous in humans. Cancer represented in the Drosophila models is also applied to distinct stages in the life-cycle stage of the fly; larvae or adult, depending on the research particularities. The current cancer types modeled in Drosophila include, but is not limited to, colorectal, lung, thyroid, and brain cancer, which will be addressed in this article. Furthermore, molecular elements associated with several cancer types are also studied in these Drosophila models including tumor suppressors, chromatin regulators, cellular growth control, tumor microenvironment, tumor invasion, and metastasis (Rudrapatna et al., 2012).
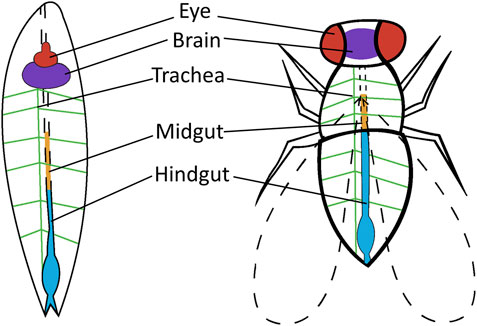
FIGURE 2. Drosophila tissue or organs used for cancer research. Tissue or organs are shown in the Drosophila larvae and Drosophila adult that are utilized in human cancer research. Thyroid cancer is targeted to the eye of adult Drosophila (red). Brain cancer is targeted to the brain of adult and larvae Drosophila (purple). Lung cancer is targeted to the trachea of larvae Drosophila (green) and colorectal cancer is targeted to the midgut (orange) and hindgut (blue) of adult Drosophila.
Transgenic Drosophila strains are typically modeled for cancer through the use of the GAL4/UAS system. Transgenes are carried by crossing parent flies transferring either the Enhancer-GAL4 or UAS-target gene that produces offspring with the induced target gene (Figure 3). Target gene selection is based on gene and pathway similarities corresponding to Drosophila and humans. The expression of the target protein generates a cascade of reactions specific to the cancer type. These reactions are observed and studied in conjunction with anticancer therapeutics (Table 1). Drosophila cancer models successfully identified several compounds capable of reducing cell proliferation from various Food and Drug Administration (FDA) approved chemical compounds, indicating the strong cancer-related chemical screening potential of using Drosophila models in anticancer research.
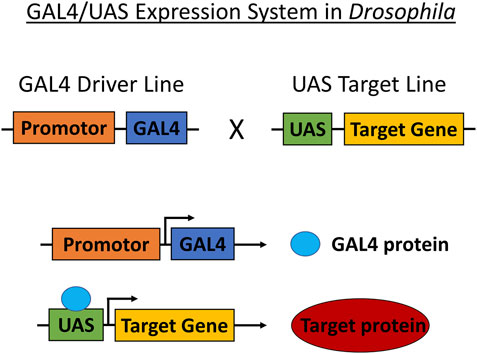
FIGURE 3. GAL4/UAS expression system in Drosophila. The GAL4/UAS system containing the GAL4 transcription factor (driven by cell-or tissue-specific enhancer/promoter) and UAS target used for targeting genetic manipulation in Drosophila. Transgenes are carried by crossing parent flies transferring either Enhancer-GAL4 or UAS-target gene that produces offspring translating the target protein.
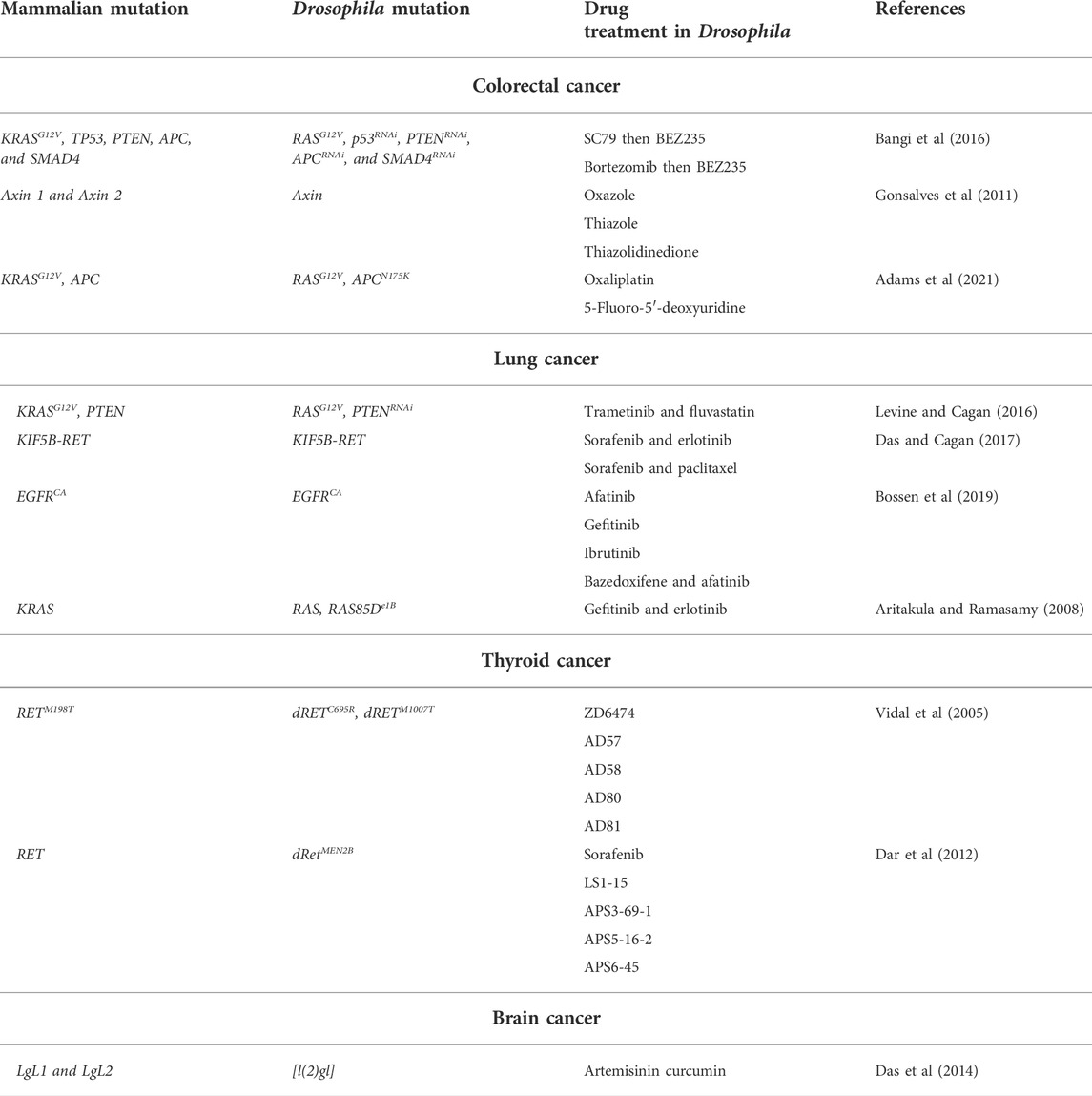
TABLE 1. Drosophila cancer models and drug treatment. Drosophila cancer models are described based on cancer type, mammalian mutation, corresponding Drosophila mutation and drug treatment of the Drosophila model.
Colorectal cancer (CRC) has the second-highest cancer-related mortality rate, with 1.9 million new cases and 935,000 deaths reported globally in 2020 (Sung et al., 2021). The development of CRC is caused by chromosomal abnormalities, gene mutations, and epigenetic alterations of genes regulating apoptosis, angiogenesis, proliferation, and differentiation (Vogelstein et al., 1988). In particular, the activation of oncogenes (KRAS and BRAF) and the inactivity of tumor suppressors (APC, p53, DCC, and p16), as well as mismatches during gene repair (MLH1 and MSH2), contribute to the development of the disease (Tanaka et al., 2006). The RAS gene family is especially well studied due to reports that 30–50% of colorectal tumors contain mutant KRAS oncogenes, while mutations in oncogenic RAS isoforms, such as NRAS and HRAS, are responsible for an estimated additional 6% of CRC tumors (Valtorta et al., 2013; Chang et al., 2016). Current therapeutic approaches targeting RAS-related tumors show limited efficacy, particularly in patients with KRAS-mutant metastatic CRC (Nazarian et al., 2010; Misale et al., 2012; Stephen et al., 2014). These poor therapeutic results are due to the rising resistance of late-stage CRC tumors to targeted therapies.
Safeguarded genes and pathways associated with CRC have been analyzed extensively in cell cultures and mouse modules (Sancho et al., 2004; Markowitz and Bertagnolli, 2009). The Drosophila hindgut distinctly retained these CRC-related pathways, presenting a near-identical composition of the cell populous to the mammalian colon (Takashima et al., 2008; Fox and Spradling, 2009). Similarly, the Drosophila midgut represents functions and molecular characteristics exhibited in the human intestine (Sadaqat et al., 2021). The intestinal maintenance of both Drosophila and mammals is controlled by proliferating cells that produce post-mitotic secretory cells and absorptive enterocytes (Casali and Batlle, 2009). These genetic similarities between Drosophila and mammals allow for the successful modeling of CRC in Drosophila. The Drosophila models demonstrate key hallmarks of human CRC, such as disruption of cellular differentiation, amplified cell proliferation, and decreased intestinal homeostasis (Gonzalez, 2013; Martorell et al., 2014; Villegas, 2019). The models also permit a whole-organism assessment of various CRC anticancer therapies.
Data analyzed from the Cancer Genome Atlas (TCGA) shows that 90% of CRC patient samples present mutations in two or more signaling pathways. Recurrent mutations specifically affect TGF-β, WNT, PI3K, RAS/MAPK, and p53 pathways (Bangi et al., 2016). Drosophila was effectively used by Bangi and colleagues (2016) to model 4 or 5 recurrent mutations based on CRC genotypes. The mutations reflected patient-specific genes identified through TCGA analysis corresponding to the 212 human colon tumors. The GAL4/UAS expression system allowed for the selected genes to be altered via transgene expression and tissue-specific RNAi (Figure 3). The authors combined active RASG12V with RNA interference (RNAi) knockdown of tumor suppressors p53, APC, PTEN, and SMAD4 (Bangi et al., 2016). Transgenes were targeted to the adult Drosophila hindgut epithelium. Principle colon cancer pathologies were cataloged, including cellular proliferation, basement membrane disruption, apoptosis and senescence circumvention, distant metastasis, and epithelial-mesenchymal transition (EMT). Specifically, RASG12V, p53RNAi, PTENRNAi, and APCRNAi mutations produced more peracute phenotypes.
Furthermore, the study by Bangi and colleagues (2016) accentuates the potential use of Drosophila in personalized medicine. These selectively mutated flies were used to test the effect of FDA-approved cancer drugs in CRC. The Drosophila models showed responses to oncogenic reagents based on grouped patient genotypes. The authors reported inhibition of the tumor suppressor mTORC1 with the activation of the RAS oncogene and simultaneous loss of the tumor suppressor PTEN (Bangi et al., 2016). This indicates possible CRC resistance to PI3K/mTOR inhibitors. Particular interest was set on BEZ235, the first PI3K/mTOR inhibitor drug to enter into clinical trials (Peyton et al., 2011). Mimicking clinic trial results, BEZ235 failed to diminish the spread of CRC in the Drosophila models. The efficacy of BEZ235 was, however, considerably enhanced with the addition of another FDA-approved cancer drug. Pretreatment of the CRC Drosophila models with SC79 (AKT-activating compound) followed by BEZ235 successfully reduced the spread of CRC. This two-step therapy also proved effective with the pretreatment of bortezomib (a proteasome inhibitor) followed by BEZ235. Interestingly reversing the drug order rendered the treatment ineffective, demonstrating the importance of the action mechanisms of drugs. Many genetically manipulated Drosophila model combinations were tested using sixteen anticancer drug compounds specific to RAS mutations (Bangi et al., 2016). Synergistically acting compounds demonstrated restored sensitivity against PI3K pathway inhibitors and an increase in mTORC1 activity. This was observed in Drosophila CRC models, mammalian CRC cells, and a CRC GEMM (genetically engineered mouse model) (Bangi et al., 2016).
Simpler gene families in Drosophila reduce potential complexities in loss-of-function studies, as fewer genes are modulated. Specifically, Drosophila offers a unique experimental state as it has only one Axin, as appose to the Axin 1 and Axin 2 present in mammalian cells. In a study by Gonsalves and colleagues (2011), the authors focused on the Drosophila loss-of-function mutation on this Wg/Wnt pathway inhibitor, namely Axin. The Drosophila Wg (wingless) gene, is a homolog of the mammalian WNT (wingless-related integration site) gene. These are structurally related genes that encode proteins implicated in developmental processes, such as patterning during embryogenesis and cell fate regulation, as well as oncogenesis (Miller, 2001; Clevers, 2006). In the absence of a Wg/Wnt ligand, Axin (forming part of a protein complex) controls β-catenin degradation. The Wg/Wnt ligand segregates Axin, causing β-catenin stabilization and, subsequently, Wg/Wnt transcription activation. The authors observed continuous Wg signal activation by depleting Drosophila cells of Axin, thereby detecting β-catenin stabilization. Stabilization of β-catenin is essential to CRC-linked tumorigenesis (Morin et al., 1997; Johnson et al., 2005). Mutations related to the regulation of this β-catenin transcriptional activator led to the accumulation of β-catenin in the nucleus, where the molecule causes uncontrolled gene transcription. The build-up of β-catenin in the nucleus is reported in 80% of CRC tumors (White et al., 2012).
Gonsalves and colleagues (2011) screened a small-molecule library to identify three small-molecule inhibitors of the Wg/Wnt pathway, particularly β-catenin responsive transcription inhibitors. The inhibitors (oxazole, thiazole, and thiazolidinedione) modeled in the Drosophila cells were shown to stabilize β-catenin by inhibiting β-catenin/TCF complex formation. These three drugs were tested in the transgenic Drosophila cells, human colon cancer patient biopsies, and human colon cancer cells (HCT-116 and HT29). The inhibitors proved cytotoxic in human colon cancer cells, non-toxic to the Drosophila cells, and when tested in human colon cancer patient biopsies, they indicated an efficacy comparable to FDA-approved anticancer drugs (Gonsalves et al., 2011). The study by Gonsalves and colleagues (2011) imitates the constitutive Wnt activity presented in cancers absent of tumor suppressor adenomatous polyposis coli (APC) (Rowan et al., 2000; Fearnhead et al., 2001). Similar to Axin, APC also acts to inhibit Wg/Wnt pathways. However, APC is a negative regulator of Wnt that controls Wnt signaling by blocking β-catenin in the nucleus. The oncogenic applications of APC inhibitor-related research are paramount, with 80–85% of sporadic colorectal tumors being exclusively associated with APC mutations (Zhang and Shay, 2017).
Accurate modeling of CRC in Drosophila models was also shown by clonal activation of RAS and Wg/WNT pathways in the adult Drosophila midgut (Wang et al., 2013; Martorell et al., 2014). Similarly, other studies have shown that Drosophila CRC models with these RAS and Wg/WNT mutations effectively identify cancer-triggering mutations (Gonzalez, 2013; Villegas, 2019). Combined RASV12 and WNT signaling pathway mutations are known as habitual CRC initiators in the Drosophila midgut (Jackstadt and Sansom, 2016). Mutations of the APC disrupt the Wg/Wnt pathways in the intestinal epithelia, while overexpression of the RAS oncogene causes tumor-like overgrowths (Martorell et al., 2014). The analysis of these genetic mutations typically includes dissection followed by the manual joining of midgut tumor images. The clonal area is then quantified with the number of clones and other required parameters by software-dependent image analysis (Martorell et al., 2014; Suijkerbuijk et al., 2016; Ngo et al., 2020). This method is effective; however, it is also tedious and time-consuming, limiting large-scale experimentation and rapid results.
Alternatively, Adams and colleagues (2021) developed an innovative, fast, and simple method for the quantification of tumor formations. The Drosophila CRC models used by the authors contained APCN175K-RASG12V mutated tumor cells that were marked with two reporter genes: GFP and luciferase. The clones were generated using the GAL4/UAS expression system. The introduction of a second UAS-luciferase transgene reporter, in addition to UAS-GFP, allowed for two distinct screening methods. The first method relies on the sensitive luciferase-based assay used to test tumor formation. The luciferase activity is reported using a microplate reader, providing rapid batch processing of whole organisms. The second method uses the GFP reporter for clone visualization and quantification. Images are created with custom-designed macros used in combination with Fiji, an extensively available imaging software. This method provides an automated analysis of the total Drosophila midgut presented with clones. Particular attention was set on clone size, which was determined by the authors as the most pragmatic method for identifying the probability of clones conforming to tumors.
Following genetic alterations, the Drosophila CRC models were screened and tested with CRC-related drugs; oxaliplatin and 5-fluoro-5′-deoxyuridine. The current systems employed in identifying new CRC therapies are traditionally small molecule screening techniques based on computational software or cell culture followed by enzymatic assays. These systems present favorably during in vitro testing; however, they proved ineffective for toxicity screening in the proceeding whole-organism mammalian model experiments (Horvath et al., 2016). The Drosophila CRC models designed by Adam and colleagues (2021) also assisted in the development of a rapid drug-and genetic screening system. Quantified data collected from the Drosophila CRC models were used successfully to validate the model’s rapid response to the two standard CRC drug treatments. The introduction of a second UAS-luciferase transgene reporter, in addition to UAS-GFP, enabled the high-throughput screening tool in Drosophila.
Statistically, lung cancer has the highest mortality rate among all cancer types, with 1.8 million deaths and an additional 2.2 million new cases reported in 2020 (Sung et al., 2021). Approximately 85% of lung cancer cases present with non-small-cell lung carcinoma (NSCLC) (Griffin and Ramirez, 2017). Conventional lung cancer treatments generally include chemoradiotherapy with complimenting targeted therapies. However, the current drugs targeting lung cancer have proven ineffective in tumor suppression while inducing toxicity and drug resistance. The homologous molecular factors and epithelial cellular similarities between the Drosophila tracheal and vertebrate lung development are significant (Andrew and Ewald, 2010; Behr, 2010; Roeder et al., 2012). The multi-branched tubular structure of the Drosophila tracheal system consists of larger primary tubes that branch out into smaller diameter branches and end in terminal sectors. This forms an interconnected hierarchy of tubes, analogous to vertebrate lungs, with both providing oxygen to the organism. The development of the Drosophila tracheal branching system is distinctly dependent on Fibroblast Growth Factor (FGF) signaling (Ghabrial et al., 2003; Grifoni et al., 2015). Tracheal branching in Drosophila is distinctly triggered by the FGF receptor homolog breathless (btl). The FGF signaling also initiates the development of the vertebrate lung branching system (Bellusci et al., 1997; Park et al., 1998).
Drosophila was used by Levine and Cagan (2016) to model lung cancer with mutated RASG12V and PTEN (PI3K negative regulator) knockdown. The Drosophila model targeted the btl gene using the GAL4/UAS expression system (Figure 3). Genetic alterations to the trachea caused tumor-like growth, tracheal cell proliferation, and early larval stage model death. Mutation selection was based on the association of active oncogenic RAS isoforms, commonly affiliated with active PI3K pathway signals (Cancer Genome Atlas Research Network, 2012; Kandoth et al., 2013). Chemical screening of 1192 FDA-approved drugs (chosen to reduce model lethality) identified trametinib, the MEK1/2 kinases inhibitor drug, and fluvastatin, the HMG-CoA reductase inhibitor drug used in cardiovascular treatment. Synergistically, these two compounds respectively inhibited RAS and PI3K pathway activity. This led to decreased over-proliferation, reduced whole-organism toxicity, and inhibited the growth of A549 human NSCLC cells with activated RASG12V (Levine and Cagan, 2016). Trametinib is currently the only MEK inhibitor with FDA approval for NSCLC treatment as a monotherapy or in combination with dabrafenib. Clinical treatment of trametinib and dabrafenib in NSCLC patients indicates survival benefits and continued response to therapy (Planchard et al., 2022). While fluvastatin has been indicated in various cancer research, the drug is not FDA approved for anticancer treatment. Consequently, the synergistic treatment of trametinib and fluvastatin in NSCLC patients is unknown.
Additionally, Drosophila facilitated the creation of novel therapeutic strategies focused on the multiple gene fusion oncogene KIF5B-RET (Das and Cagan, 2017). Identified as a critical lung cancer driver, the structure of this KIF5B-RET oncoprotein suggests that simultaneous multi-kinase activation is therapeutically required (Ju et al., 2012; Kohno et al., 2012; Lipson et al., 2012; Takeuchi et al., 2012). Reported primarily in non-smoker patients with few genetic alterations in notable cancer agents, KIF5B-RET account for an estimated 2% of NSCLC cases (Takeuchi et al., 2012). Previous research done by Levinson and Cagan (2016) optimized CCDC6-RET and NCOA4-RET genes to generate transgenic Drosophila models. They used the GAL4/UAS expression system in a thyroid-linked cancer study, concluding with an unknown pathway of RET fusion activation (Levinson and Cagan, 2016). Accordingly, Das and Cagan (2017) focused on activation pathways associated with the RET fusions to generate a patient-derived KIF5B-RET mutation. This selected mutation was also expressed in Drosophila epithelium through the binary GAL4/UAS system. The study identified canonical signaling pathway activation through the C-terminal RET kinase domain of KIF5B-RET. The N-terminal domain of KIF5B also activated multiple receptor tyrosine kinases (RTKs), including fibroblast growth factor receptor (FGFR) and epidermal growth factor receptor (EGFR) signaling (Das and Cagan, 2017). These findings offer valuable insight into pathway vulnerabilities requiring multi-targeted agents of selected therapeutic cocktails. Administration of drugs designed singularly for RET inhibition proved ineffective in KIF5B-RET transformed cells. However, in combining the RET inhibitor sorafenib, with the EGFR inhibitor erlotinib or microtubule inhibitor paclitaxel, high efficacy was demonstrated in human cell line KIF5B-RET models and Drosophila KIF5B-RET models alike (Das and Cagan, 2017). While these KIF5B-RET-positive NSCLC therapies await patient study validation, therapeutics of other fusion kinases may advance through Drosophila signal pathway activation investigations.
Effective NSCLC treatment is frequently associated with simultaneously activated pathway inhibitors. However, the activation and preservation of NSCLC are limited to relatively few oncogenic driver mutations, with EGFR as the most notable lung cancer oncogene (Kandoth et al., 2013; Rotow and Bivona, 2017). Oncogenic KRAS mutations are established prime promoters of NSCLC and are known to incite resistance to secondary cancer therapies and EGFR inhibitors (Riely et al., 2009). The KRAS proto-oncogene functions in cascade signal transductions initiated by EGFR binding (Graziani et al., 1993). EGFR participates in a complex of signaling pathways and developmental processes. Structurally, EGFR contains an intracellular tyrosine kinase (TK), with the Drosophila TK domain presenting distinct similarities to humans (Bier, 2005). Additionally, EGFR signaling in Drosophila is vital to the organism’s eye structure, throughout developmental stages, and to the wing development, during the late third instar larval stage (Guichard et al., 1999). Overexpression of EGFR accounts for up to 80% of NSCLC cases (Molina et al., 2008).
A Drosophila lung tumor model was designed to target ectopic expression of constitutively active EGFR isoform (EGFRCA) in the airway epithelium of Drosophila (Bossen et al., 2019). Extensive phenotyping of the NSCLC model induced massive hyper- and metaplasia. Modified blt-GAL4>UAS-EGFRCA expression promoted death in later larval development stages. Furthermore, ppk4-GAL4>UAS-EGFRCA expression prompts early third instar stage death. Model death caused by alterations to the pickpocket (ppk) gene is presumably from oxygen deficiency. The authors quantified these Drosophila larval mortality readouts to screen numerous possible combinations of NSCLC therapies from 1000 FDA-approved compounds (Bossen et al., 2019). Drug screening identified TK inhibitor (TKI) compounds afatinib, gefitinib, and ibrutinib. These compounds proved capable of rescuing lethality in the Drosophila whole-organism model. They also reversed structural trachea phenotypes with afatinib and gefitinib, showing impressive survival rates (Bossen et al., 2019). Furthermore, secondary screening of the pharmacologic FDA-approved library was performed. Bazedoxifene and afatinib were identified during this drug screening as potential synergistically acting compounds. Combined, these two compounds were able to rescue EGFR-induced lethality by reducing hypoxia-inducing JAK/STAT signals.
Another study used Drosophila lung cancer models as an in vivo drug screening system to analyze EGFR-associated TKI pathways via enhancer-suppressor assays (Aritakula and Ramasamy, 2008). Mutations and transgenics relating to EGFR pathways are well documented, providing an optimal system for drug target identification, target validation, and secondary effect determination research. The lung cancer Drosophila model was developed using the GAL4/UAS system. The gain-of-function analysis included upstream activation sequences: UAS–EGFRλtop, UAS-Argos, and UAS-CycE. Simultaneously, tissue-specific GAL4 gene drivers included vg-GAL4, ey-GAL4, and omb-GAL5 with a lethal recessive allele of RAS and RAS85De1B used for loss-of-function RAS-related pathways (Aritakula and Ramasamy, 2008). Model evaluations focused on mechanisms by which gefitinib and erlotinib (TKIs) block Drosophila EGFR signaling. Enhancer-suppressor analysis and in silico analysis were used for evaluations. The selection of the FDA-approved gefitinib and erlotinib TKIs was based on the reported efficacy of these drugs in chemotherapeutic treatments of cancer patients with EGFR-induced tumors (Maemondo et al., 2010; Ciuleanu et al., 2012). Gefitinib and erlotinib combined showed suppression of EGFR-induced eye phenotypes in Drosophila. On its own, gefitinib suppressed EGFR-induced wing phenotypes. Both drugs act to inhibit diphosphorylated forms of the extracellular signal-regulated kinase (dp-ERK1/2) in both the eye and wing imaginal discs of wild-type larvae models (Aritakula and Ramasamy, 2008).
In 2020, thyroid cancer was responsible for an estimated 44,000 deaths worldwide and 586,000 new cases (Sung et al., 2021). Globally, a rise in thyroid cancer cases is reported yearly, particularly from developed countries (Kim et al., 2020). Thyroid cancer presents a wide range of complexities and mutations (Vidal et al., 2005; Das and Cagan, 2017). Mutations in the RET receptor tyrosine kinase (RTC), a 120 kDa transmembrane protein, are responsible for the development of thyroid cancer in Drosophila and humans. Additionally, mutations that trigger RET activity led to multiple endocrine neoplasia type 2A and 2B (MEN2A and MEN2B) and familial medullary thyroid carcinoma (FMTC) thyroid cancers (Vidal et al., 2005).
Vidal and colleagues (2005) developed transgenic Drosophila models expressing mutated RET (dRET) isoforms using the GAL4/UAS system (Figure 3). This thyroid cancer model was created to mimic FMTC, MEN2A, and MEN2B- like isoforms. The mutated dRET isoforms were targeted to the developing eye in adult Drosophila, as flies do not have a thyroid (Figure 2). The authors successfully generated the Drosophila genotypes (GMR-dRetM559T and ptc-dRetM955T) to mimic thyroid cancer in Drosophila (Vidal et al., 2005). The selection of the RETM198T mutation was based on known medullary thyroid carcinoma (MTC) patient genetics. The MEN2A-associated mutation was mimicked by replacing cysteine with arginine at position 695 (C695R). The MEN2B-associated mutation was mimicked by engineering a methionine-to-threonine point mutation into a full-length dRET cDNA at codon 1,007. Altered dRET isoforms were each cloned behind the eye-specific glass multiple reporter (GMR) promoter. The stable transgenic fly lines were generated by standard methods to create GMRd-RET, GMR-dRETC695R, and GMR-dRETM1007T Drosophila thyroid cancer models. These newly generated models expressed wild-type, C695R, or M1007T dRET isoforms, respectively, in their developing eyes.
Vandetanib, also known as ZD6474, was originally identified as a chemical inhibitor of the vascular endothelial growth factor receptor 2 (VEGFR2) RTK, with additional activity against EGFR (Vidal et al., 2005). ZD6474 is an anticancer medication used for the treatment of thyroid gland tumors. The drug acts as a kinase inhibitor of cell receptors such as the VEGFR, EGFR, and the RET-tyrosine kinase. Drosophila thyroid cancer models were given different concentrations of ZD6474. An increase in mortality of MR-dRET was observed in higher concentrations of ZD6474 administration. Low concentrations of ZD6474 led to a partial rescue of GMR-dRET, GMR-dRETC695R, and GMR-dRETM1007T models. Following treatment with higher doses of ZD6474, there were no phenotypically observable differences in the eyes of the thyroid cancer and wild-type Drosophila models (Vidal et al., 2005).
The authors could not establish the ability of the drug to rescue different isoforms, as the exact concentration of ZD6474 that entered the fly system was not obtained (Vidal et al., 2005). However, the initial phenotypes were comparable. ZD6474 is less effective in Drosophila isoforms of EGFRs namely, RAS and RAF. Phenotypes presented in the Drosophila models were not rescued by ZD6474. According to Vidal and colleagues (2005), ZD6474 either suppresses dRET activity independent of the RAS pathway signaling or the drug acts upstream of dRAS1 and dRAD. These results support previous studies that ZD6474 acts directly on the receptor; consequently, inhibition of downstream cytoplasm kinases linked to RAS signaling is likely minor. The study concluded that ZD6474 acts as an in vivo inhibitor of the RET signaling pathway. RET-dependent phenotypes observed in Drosophila are strongly suppressed by ZD6474. Targeting chemical kinase inhibitors to tissues with oncogenic RET may offer a viable treatment for RET-dependent cancers (Vidal et al., 2005; Das and Cagan, 2017). ZD6474 completed phase three clinical trials in 2010 and became the first FDA-approved chemotherapy for RET-based thyroid tumors in 2011 (Wells et al., 2010). The approval of this drug validated the potential use of Drosophila cancer models as a powerful tool for anticancer drug discovery. However, it should be noted that ZD6474 has been reported to have high toxicity and resistance issues (Dar et al., 2012; Das and Cagan, 2017).
Furthermore, Dar and colleagues (2012) utilized the transgenic dRetMEN2B Drosophila model for a whole-organism efficacy validation of ZD6474 in the treatment of medullary thyroid carcinoma (MTC) patients. The authors improved the efficacy of ZD6474 for drug screening by developing a quantitative viability assay that uses the GAL4-UAS expression system. The cancer models targeted dRetMEN2B in developing Drosophila eye, wing, and leg. The ptc˃dRetMEN2B assay results indicated that the patched promoter is responsible for oncogene expression of developing epithelial and other tissues. The dRetMEN2B Drosophila model was used to structurally modify ZD6474. The modified ZD6474 generated improved anticancer targets, including AD57, AD58, AD80, and AD81 (Dar et al.,2012). The alteration showed minimal toxicity and improved efficacy (Dar et al., 2012; Das and Cagan, 2017).
Another study developed Drosophila MTC models to access novel targets and chemical space from the clinical kinase inhibitor sorafenib (Sonoshita and Cagan, 2017). The combination of chemical and genetic modification screening with computational modeling showed that kinases strongly enhance or limit the activity of sorafenib. Previous studies found that sorafenib inhibits RET, BRAF, and KDR/VEGFR19 (Wilhelm et al., 2004). However, sorafenib was progressively refined, and the results were a new class of kinase inhibitors (LS1-15, APS3-69-1, APS5-16-2, and APS6-45) with distinct polypharmacology and an improved therapeutic index in Drosophila and human MTC xenograft models.
Cancer of the brain and nervous system accounted for approximately 308,000 new cases and 251,000 deaths in 2020 (Sung et al., 2021). Brain cancer affects young and old individuals and contributes to high mortality rates worldwide with limited therapeutic options (Sampson et al., 2020). The central nervous system (CNS) shows remarkable evolutionary conservation in cellular composition and neuro-developmental mechanisms (Bilen and Bonini, 2005). Gliomas account for almost 80% of all malignant primary CNS tumors (Hanif et al., 2017). Glioblastoma (GBM) tumors are the most common and aggressive CNS tumors. GBM tumors infiltrate the brain and proliferate rapidly with limited therapeutical therapies currently available (Read et al., 2009; Sonoshita and Cagan, 2017; Yamamura et al., 2021). Understanding the genetic and molecular rationale underpinning gliomagenesis can assist the development of effective anticancer therapeutics.
The mutation of the EGFR tyrosine kinase is the most common genetic lesion in gliomas. Glioma-associated EGFR mutant forms have constitutive kinase activity, which drives cellular proliferation and migration by persistently stimulating Ras signaling (Maher et al., 2001; Furnari et al., 2007). Loss of the lipid phosphatase PTEN, which inhibits the phosphatidylinositol-3 kinase (PI3K) signaling pathway, also causes glioma formation. Furthermore, activating mutations in PIK3CA, which encodes the p110 catalytic subunit of PI3K, are another prevalent genetic disease. Akt, a key PI3K effector, is frequently constitutively activated in gliomas (Maher et al., 2001; Furnari et al., 2007). Multiple mutations that coactivate the EGFR-Ras and PI3K-Akt pathways are required to cause glioma. EGFR-Ras or PI3K mutations alone are not sufficient to convert glial cells (Holland et al., 2000; Maher et al., 2001). Understanding how these mutations interact with the tumors’ neurodevelopmental origins could lead to new insights into the mechanisms of gliomagenesis. Multiple glial cell types maintain proliferative capacity in the mammalian brain, including differentiated astrocytes, glial progenitors, and multipotent neural stem cells. Many developmental processes in these cell types are regulated by EGFR-Ras and PTEN-PI3K signaling, including proliferation and self-renewal, which are also characteristics of glioma cells (Furnari et al., 2007). Drosophila has multiple cell types that require EGFR pathway signaling for normal development. Brain cancer can be studied using glia cells in the instar larvae or adult fly. These cells are homologous to mammalian glia units of development with comparable gene expression and function (Freeman and Doherty, 2006).
Read and colleagues (2009) developed a glioma Drosophila model by generating mutant phenotypes through hyperactivation of these pathways in Drosophila glia and glia precursors. EGFR (dEGFR), Raf(dRaf), PIK3CA(dp110), PTEN (dPTEN), and Akt (dAkt) each have a single functioning ortholog in Drosophila, and Ras(dRas85D, dRas64B) has two functional orthologs (Read et al., 2009). The authors used the GAL4/UAS expression system (Figure 3) to undertake glial overexpression tests using the repo-Gal4 driver. The repo-Gal4 driver provides continuous UAS-transgene expression in almost all glia from embryos through adulthood (Brand and Perrimon, 1993). The authors used the UAS-dsRNA constructs for glial-specific RNAi, which they confirmed with phenotypic testing and/or antibody labeling (Dietzl et al., 2007; Read et al., 2009). Constitutive co-activation of the EGFR-RAS and PI3K pathways in Drosophila brain cancer models stimulated glial neoplasia, which resulted in highly proliferative and invasive neoplastic cells. The co-overexpression of activated dEGFR (dEGFRλ) and dp110 (dp110CAAX) via repo-Gal4 led to a 50-fold increase in glia. Co-overexpression of dEGFR and PI3K pathway core components, such as dAkt, resulted in phenotypes comparable to repo > dEGFRλ;dp110CAAX. The phenotypes did; however, differ depending on the pathway activation level and transgene expression. Co-overexpression of constitutively active dRas (dRas85DV12) or its effector dRaf (dRafgof) with dp110CAAX, dAkt, or a dPTENdsRNA, which partially knocked-down dPTEN, resulted in dramatic glial overgrowth. Finally, co-overexpression of dPTEN or dominant-negative dRas85D (dRas85DN17) inhibited glial outgrowth in repo > dEGFRλ;dp110CAAX larvae. This demonstrates that Ras activity and excess phospho-inositols are required for neoplasia.
Furthermore, when the EGFR and PI3K pathways are activated together, they induce far more severe phenotypes than would be expected if individually active (Read et al., 2009). Excess glia appeared in early larval stages in repo > dEGFRλ;dp110CAAX brains and accumulated during 5–7 days. The dEGFRλ;dp110CAAX glia disrupts the normal cellular architecture of the brain consequently concluding that the dEGFRλ;dp110CAAX glia are neoplastic. Read and colleagues (2009) demonstrated that Drosophila glia and glial precursors, constitutive coactivation of EGFR-Ras and PI3K signaling leads to neoplastic, invasive cells that form transplantable tumor-like growths, replicating human glioma and animal glioma models. This strong organotypic and cell-type-specific Drosophila cancer model showed malignant cells produced by mutations in hallmark genes and pathways known to be driving forces in analogous human cancer. These effectors interact together in a network to promote improper cellular growth and migration by coordinating cell cycle entry and progression, blocking cell cycle exit, and stimulating cell cycle entry and progression. While the pathways in this network are interrelated, they behave in a synergistic rather than additive manner.
Four pathway circuits were found to be necessary for glial neoplasia formation. The dRas and dMyc pathways caused dCyclinE and dCyclinD to drive cell cycle entry. Phosphorylation and inactivation of the retinoblastoma protein triggered E2F activators to stimulate cell cycle entry. The pointed (Pnt) pathway causes the string gene (Stg) to promote cell cycle progression through the activation of EGFR-Ras-Erk. Glial-specific RNAi knockdown of Pnt reduced stg expression and completely suppressed dEGFRλ;dp110CAAX neoplasia. Furthermore, pnt proteins are important for stg expression and over-proliferation in the dEGFRλ;dp110CAAX glia. The Tor-elF4E-S6K pathway increased protein translation mediated by dS6K and deIF4E. Particularly, the expression of cell cycle regulators and ribosomal components, through dMyc, the single Drosophila ortholog of the Myc and bHLH transcription factors. The authors presume that dMyc mediates signal integration between EGFR-Ras and PI3K pathways. Drosophila orthologs of CyclinE, Cdc25, and Myc were shown to be key rate-limiting genes required for glia neoplasia. Furthermore, orthologs of Sin1, Rictor, and Sdk4 are genes required only for abnormal neoplastic glia proliferation.
PTEN loss occurs in 90% of primary glioblastomas, as a consequence of increased signaling of the PI3-kinase pathway. This pathway is also activated by mutations in the EGFR, PIK3CA, and PIK3R1 genes. Activation of this pathway in turn activates atypical protein kinase C (PKC), which phosphorylates and inactivates the lethal giant larvae (Lgl) gene (Verhaak et al., 2010). Inactivity of the Lgl gene causes downstream PTEN loss and leads to lgl inactivation (Plant et al., 2003; Betschinger et al., 2005; Verhaak et al., 2010; Killela et al., 2014). Humans have two genes that are homologous to Drosophila Lgl gene, the LgL1, and LgL2 (Klezovitch et al., 2004). Similar to Drosophila, the LgL1 gene is phosphorylated and inactivated by downstream PTEN loss in human glioblastoma.
Das and colleagues (2014) developed Drosophila brain cancer models targeting the loss of lethal(2) giant [L(2)gl] gene. The P127 protein is encoded by l(2)gl, and has a human homolog LLGL-1 (previously known as HUGL), which codes for LLGL. Low expression of LLGL is a known cause of colorectal cancer (Schimanski et al.,2005; Zimmermann et al., 2008). Lgl1 knockout animal models reportedly develop severe brain dysplasia, in which neural progenitor cells fail to differentiate but continue to proliferate uncontrollably (Klezovitch et al., 2004). Mutation in the lgl gene promotes brain tumor formation in Drosophila at the larval stage of development and brain dysplasia in mice (Grifoni et al., 2013; Das et al., 2014). During the extended life of the larvae, the cerebral hemispheres and imaginal discs become several times larger than their typical size. Overgrowth of the brain tissue is caused by the over-proliferation of neuroblasts, leading to brain cancer (Woodhouse and Liotta, 2004; Gont et al., 2014). The tumor cells of l(2)gl mutants have many of the same characteristics as human cancers, such as loss of cellular form, tissue architecture, and differentiation ability (Gateff, 1978). Drosophila, that is homozygous for the recessive fatal gene l(2)gl, produces pupae considerably later than the wild type, yet most of them are unable to metamorphose and perish as third instar larvae (Gateff, 1978).
Various clinical attempts to improve brain cancer treatments including concurrent radiation, and chemotherapy treatments, as well as surgical interventions, have been unsuccessful. As a result, in vivo studies for screening possible harmless anticancer drugs were conducted. Das and colleagues (2014) created Drosophila brain cancer models by deleting the tumor suppressor gene, Igl, during larval development. The Drosophila early instar larvae were then fed with various concentrations of artemisinin and curcumin. Artemisinin, is an antimalarial medicine with possible antitumor qualities, and curcumin, is an excellent anticancer agent that has already been clinically proven to treat certain types of cancers (Grifoni et al., 2013). These cancers includes pancreatic cancer, colorectal carcinoma, and head and neck squamous cell carcinoma; however, the effectiveness of these molecules has not been studied intensively against brain cancer (LoTempio et al., 2005; Lu et al., 2011; Crespo-Ortiz and Wei, 2012). More specifically artemisinin is a sesquiterpene lactone with a 1,2,4-trioxane ring system extracted from Artemisia annua or annual wormwood. Artemisinin derivatives, such as dihydroartemisinin and arteether, have been used to treat certain cancers including leukemia, colon, melanoma, breast, ovarian, prostate, central nervous system (CNS), renal, pancreatic, osteosarcoma, and lung cancer cells (Crespo-Ortiz and Wei, 2012). On the other hand, curcumin is a polyphenol extracted from Curcuma longa. The anticancer properties of curcumin in preclinical and clinical studies are well documented (Reuter et al., 2011; Wilken et al., 2011; Aggarwal et al., 2013; Gupta et al., 2013; Kunnumakkara et al., 2017). The Drosophila model regarded 500 µM of artemisinin and 100 µM curcumin as optimum concentrations to induce apoptosis in brain cancer cells without being toxic to the normal cells. Additionally, effects on tumor inhibition with the combination treatment of artemisinin and curcumin showed positive results including an improvement in the median lifespan and locomotion response of the Drosophila models (Yadav et al., 2016).
Tumor heterogeneity poses one of the biggest challenges in cancer therapy; it causes patients suffering from the same type of cancer or even the primary and secondary tumors in an individual to respond differently to the same treatments. Heterogeneity limits the efficacy of monotherapeutic agents to entirely eliminate all the cells in a tumor (Chen and Lahav, 2016). Using monotherapeutic agents in cancer therapy frequently results in the development of drug resistance in the cancer cells (Chen and Lahav, 2016; Palmer and Sorger, 2017). As cancer results from so many aberrant signaling pathways, targeting a singular pathway results in ineffective treatment, even if there is an initial positive response. To overcome this, multiple therapeutic agents targeting multiple pathways simultaneously can significantly increase drug efficacy whilst decreasing the therapeutic dosage, which potentially results in fewer unwanted side effects (Chen and Lahav, 2016).
Restrictive combinations (RCs) of drugs are an area of cancer therapy that has been focused on recently, although not tested in humans yet. RCs focus on strategic dosing and administration of drugs that aim to help spare normal cells and target cancer cells with a cytotoxic effect (Mokhtari et al., 2017). This regimen utilizes slight differences between cancer cells and normal cells, for example, the lack of or presence of a target (Blagosklonny, 2008). Another approach is drug re-purposing, where FDA approved pharmaceutical agents currently used for non-cancerous diseases are re-purposed in cancer therapy (Ashburn and Thor, 2004; Chong and Sullivan, 2007). This approach is efficient as the FDA-approved drugs have already passed clinical safety protocols, had pharmacokinetic profiles, and helped reduce the financial burden of novel drug development (Ashburn and Thor, 2004). Drosophila cancer models have been beneficial in preclinical studies into high-efficacy synergistic drug combinations (Levine and Cagan, 2016; Bossen et al., 2019).
There are, however, challenges in combinatorial therapies, for example, multidrug resistance in chemotherapy (Gottesman et al., 2002), cross drug resistance (Aird et al., 2002; Yardley, 2013), adverse drug interactions, and even the need for increased therapeutic doses as treatment regimens continue over time (Murayama and Gotoh, 2019). In addition, drug specifics, such as time of administration and dosage, are mainly only known for administrating the drug alone. Therefore, trial and error is often needed when drugs are combined, as the dynamic response of the combined drugs is often not predictable (Chen and Lahav, 2016). Drosophila is a great tool to be used in the process of combinational drug testing, as they are an easy and cheap alternative to other animal models such as mice. Drosophila cannot replace higher animal models; however, they can narrow down the drugs that should be taken into other animal models for further investigation.
For example, multiple studies have found combinational therapy options to enhance radiation therapy in Drosophila, that have been found to be effective in humans as well (Edwards et al., 2011; Gladstone et al., 2012; Stickel et al., 2015). More specifically, Edwards and colleagues (2011) found that a microtubule depolymerizing agent, namely, maytansinol isobutyrate increased sensitivity to radiation therapy in Drosophila and later in human colon cancer cells. Next Gladstone and colleagues (2012) as well as Stickel and colleagues (2015) investigated a protein synthesis inhibitor, namely bouvardin and found increased sensitivity to radiation therapy in Drosophila which transferred to increased sensitively in human NSCLC, head and neck cancer (HNC) and glioma. Another example of this is the use of an EGFR-induced lung cancer Drosophila model to identify an alternative combinational therapy for lung cancer patients (Bossen et al., 2019). Bossen and colleagues (2019), when investigating the EGFR-induced lung cancer Drosophila model, found that targeting EGFR along with STAT-signaling is a promising strategy for lung cancer therapy. Additionally, Adams and colleagues (2021) investigated a Drosophila CRC model’s response to currently approved chemotherapy drugs for colorectal cancer in humans. They found that the Drosophila CRC models responded well to both oxaliplatin and 5-Fluoro-5′-deoxyuridine, two commonly used chemotherapy drugs for colorectal cancer in humans. This helps support that Drosophila models are a viable candidate for screening drugs against human CRC, a notoriously hard cancer to treat. Therefore, Drosophila studies play a role in optimization of the use of multi-therapeutic agents in cancer therapy in order for it to reach its potential in effectively treating cancer in humans.
One of the biggest problems with cancer treatment currently is that there is only a limited number of FDA approved regimens that display variable effectiveness amongst different patients and usually cause a host of severe side effects (Ocana et al., 2011; DiMasi et al., 2013). Lastly, a big issue is the development of drug resistance against the limited approved drug regimens (Gottesman et al., 2002). Personalized medicine aims to treat each individual patient with the best possible drug, where they receive the optimal therapeutic benefit with the lowest possible side effects. One of the primary areas being focused on to achieve this is understanding the genetic contribution to the diseased state and how this contributes to the toxicity and efficiency of potential drugs. One big hurdle in personalized therapy is understanding which candidate genes are of therapeutic significance. This is complicated by the complex genetic landscape within a living organism; this is where Drosophila is a powerful tool for understanding the complex relationships within a whole-organism.
The advantages of Drosophila in drug discovery and development have already been addressed in this review; however, some particularly apply to utilizing them for personalized medicine screening as they quickly develop and are relatively cheap and easy to use. This lowers costs, which is vital in making personalized therapy affordable (Kasai and Cagan, 2010). Drosophila’s most valuable characteristic for personalized therapy, however, is by far the ability to easily manipulate their genes, as it has been seen that the activity of almost all Drosophila genes can be increased or decreased within nearly all cell types at any stage of development (Kasai and Cagan, 2010). The permission for rapid and inexpensive manipulation of multiple genes in Drosophila, as well as the ability to perform high-throughput whole-organism screening (Adams et al., 2000; Kasai and Cagan, 2010), makes it popular amongst the other model animals. Manipulation of gene expression and generation of specific mutations are becoming more accessible, quicker, and more affordable as the years result in the development of these technologies. Significantly it has been estimated that 75% of genes involved in disease causation are conserved between humans and Drosophila (Reiter et al., 2001; Pandey and Nichols, 2011). For cancer, several studies have identified novel therapeutic targets in vivo using the Drosophila model system (Vidal et al., 2005; Bangi et al., 2011; Levine and Cagan, 2016; Bossen et al., 2019).
It is important to recognize the multigenic nature of cancers. Two types of Drosophila models have been produced; firstly, an in-silico Drosophila Patient Model (DPM), which models the biomolecular regulation in cancer utilizing patient-specific gene expression data to develop personalized cancer therapeutic strategies (Gondal et al., 2021). Secondly is an in vivo Drosophila Patient Model (DPM) where the use of Drosophila in personalized cancer therapy has come with the creation of cancer avatars. These cancer avatar flies are genetically mutated to contain several mutations that mimic the human patient. The Drosophila are then used to screen drugs, including multi-therapeutic approaches (Bangi et al., 2019). These models may help identify compounds with higher whole-organism efficacy and decreased toxicity.
Briefly, a specific example is the treatment of KRAS-mutant metastatic colorectal cancer with nodules in the lungs, where the development of a personalized Drosophila model resulted in the identification of a two-drug cocktail of trametinib and zoledronate. Overall, the patient was treated with trametinib and zoledronate for approximately 11 months, exhibiting a maximum of 45% reduction in tumor burden. One side effect that developed was a severe rash; however, it was controlled using antibiotics and antihistamines, allowing for the continued use of the treatment regimen. The patient had to be taken off treatment due to the emergence of previously unobserved lesions. When the lesions were genetically analyzed, two new mutations were found, suggesting resistance formation to treatment (Bangi et al., 2019).
A second example was the case of a patient with advanced adenoid cystic carcinoma cancer type that originates in the salivary glands. A Drosophila cancer avatar was created containing selected mutations mimicking the patient. Robotics-based screening identified a three-drug cocktail (namely vorinostat, pindolol and tofacitinib) that was able to rescue transgene-mediated lethality in the Drosophila patient-specific line. A partial response was seen for 12 months, after which relapse occurred. Subsequent resistance was due to new genomic amplifications and deletions (Bangi et al., 2021). However, it must be noted that the patients in these two cases had already been on several different therapy regimens, and their cancer had already metastasized by the time personalized therapy had started. The positive response seen, even if not followed through, encourages the use of personalized therapy for future cases.
Lastly, a clinical trial ran between 2015 and 2020, which investigated personalized cancer therapy in patients with metastatic medullary thyroid or metastatic colon cancer (ClinicalTrials.gov Identifier: NCT02363647). This trial aimed to identify tumor mutations and incorporate them into personalized Drosophila. These personalized Drosophila would then be screened with FDA approved drugs to identify a single drug or any drug combinations that are the most effective and least toxic. The identified drug or drug combinations would then be tested in xenograft models before a board of experts choose the best therapeutic option for each individual. To date, 10 individuals have been recruited into the trial; however, no further details are supplied as the trial was suspended in October 2020, due to lack of funding.
One of the biggest obstacles to overcome, even in multi-therapeutic personalized therapy, is the development of drug resistance. Therefore, identifying potential drug resistance could be important in optimizing personalized treatments. Again, personalized Drosophila comes into play as it was found that they could potentially be used to identify biomarkers of drug resistance (Bangi et al., 2014). As the use of Drosophila in multi-therapeutic personalized therapy is still in its infancy additional research is needed to fully reach the potential of multi-therapeutic personalized therapy in cancer treatment. As well as an increase in fly scientists trained in using Drosophila in personalized medicine.
Drosophila has emerged as an auspicious model for screening anticancer drugs. Notable characteristics of fruit flies such as 1) short life span and generation time, 2) low genetic redundancy, 3) high reproductive rates, 4) low maintenance costs and 5) genetic and functional similarities to humans make them attractive models for the discovery of anticancer therapeutics. However, some normal physiological and cancer-relevant mechanisms differ in Drosophila and humans due to several differences between the two organisms.
The most notable difference is the physiology and anatomy of the two organisms. Humans have a more complex physiology and anatomical organization compared to Drosophila. Tumors in humans are far more complex than those generated in Drosophila. Cancer fly models, therefore, produce a partial picture of the disease in humans (Willoughby et al., 2013; Richardson et al., 2015; Yadav et al., 2016). The consequence of such differences may produce pseudo-positive or pseudo-negative findings during drug screening. Drosophila particularly lacks the direct equivalent of the mammalian organs, such as the liver, pancreas, spleen, thymus, kidneys, lungs, and thyroid gland. However, cancers of the thyroid, lungs and brain can be modeled with Drosophila equivalent organs or using other organs such as the eye. A classic example is the modeling of thyroid cancer using the cells of the developing eye as described previously (Das and Cagan, 2017). Although not a perfect model for the study of thyroid cancer, it proved useful in understanding the mechanism of development and identification of potential anticancer therapeutics.
The current fly models do not adequately address tumor genetic heterogeneity and evolution observed in human patients. Tumor genetic heterogeneity and evolution play a key role in formation of aggressive and treatment resistant cancers (Pelham et al., 2020). Whilst there are significant strides made in replicating the genetic architecture of tumors in patients using fly “avatars” and improving treatment, an effort is required to create fly models that will address tumor evolution and heterogeneity in pursuit of improving cancer treatment outcomes.
Drosophila have a simple innate immune system whilst mammals possess complex adaptive and innate immune systems. Moreover, blood or lymphatic vessels are absent in Drosophila. For these reasons, the direct and indirect effects of drugs on the immune system and tumor neo-vasculature cannot be assessed (Gao et al., 2015; Yadav et al., 2016; Yamamura et al., 2021). In addition, the lack of an adaptive immune system further limits the testing of newer therapies such as immune checkpoint inhibitors (ICIs) which have shown greater success than small molecule/chemotherapeutic drugs.
The limitation posed by the physiological and anatomical differences between flies and humans, therefore, warrants additional preclinical testing in mammals such as mice prior to testing in humans (clinical trials). Drug screening using Drosophila can help verify the suitability of drug candidates in a whole-organism prior to testing in costly rodent experiments and clinical trials. However, Drosophila cannot replace rodent models. Mammalian models are still required to establish the pharmacokinetics and pharmacodynamics to validate the pharmacological activity of the drug candidate (Pandey and Nichols, 2011; Su, 2019). Although Drosophila may not replace mouse models, it is a more appropriate screening tool than in vitro cell culture and can narrow down the potential drugs to be screened in the more expensive mouse models.
The route of administration of a drug is important in the drug discovery process. Drug candidates are often diluted in Drosophila culture media for larvae (and adults), and the drug is ingested orally. Limitations of oral administration are that drug concentrations in the food may differ significantly from actual physiological concentrations. Other issues with this route of administration also include the taste of a drug: if a drug tastes bad, a fly is likely to not eat it. In addition, significant variability in dosage among flies as a result of various body sizes and amounts of drug ingested, as well as relatively low throughput, may further pose a limitation. It may be necessary to examine in vivo concentrations using high-performance liquid chromatography or mass spectrometry (Kuklinski et al., 2010). In adult flies, several routes of administration may be tested. These include vapor (e.g. ethanol) via injection in the abdomen or dropped directly onto the exposed nerve cord of decapitated flies (Moore et al., 1998; Torres and Horowitz, 1998). Following administration, drug candidates are often metabolized before making their way to target sites. The difference in the metabolism between humans and Drosophila can further limit the use of drugs that are toxic to Drosophila and not humans and vice versa. However, there is a strong correlation of toxicity between the two organisms for most drugs (Rand, 2010; Pandey and Nichols, 2011).
All of these factors, and more, accentuate the use of Drosophila as a screening platform for target discovery, primary small-molecule screening, or post-screening validation to narrow down a large pool of potential drug candidates prior to screening in costly mammalian models. Nonetheless, the use of Drosophila in target discovery and high-throughput screening remains a relevant whole-organism drug screening approach. The use of Drosophila improves the rate of discovery by reducing the time necessary to identify a small collection of potentially more effective lead compounds for final validation. Although Drosophila models can be informative in the discovery process, having a well-defined hypothesis and a thorough understanding of the limitations of the fly are absolutely critical for success.
Current anticancer therapies are often ineffective or result in the development of drug-resistance, especially when administered as a monotherapy. The discovery of new anticancer drugs involves complex experimental procedures, extensive assessment processes and high implementation costs. Generally, initial drug discovery relies on target identification using computational software and in vitro drug screening; however, these have been found to be frequently inefficient when applied to whole-organism models. The common fruit fly Drosophila melanogaster presents a valuable tool for screening and testing anticancer drugs in the preclinical stages of drug discovery. Drosophila present a high conservation of key genes, a low genetic redundancy, easy genetic manipulation and a rapid life cycle. In addition, Drosophila are easy to maintain and produce large amounts of offspring making them suitable for cost-effective high-throughput screening of anticancer therapies. Generated Drosophila cancer models, either specified per cancer type or personalized to patient genotype, can mediate high-throughput screening of FDA-approved non-cancer and anticancer drugs adapted to specified genetic requirements. In addition, Drosophila can provide valuable information on drug bioavailability and toxicity. In this way Drosophila can be used in personalized therapy to study a cancer model with multiple genetic mutations. They can also be used to screen for combinational therapies in a multi-therapeutic approach. These models are inexpensive and efficient, thus reducing the cost and time of anticancer drug discovery. However, the application of this organism in the anticancer drug discovery pipeline is still in its infancy, and greater awareness, skill development and application are key to fully utilizing the potential of Drosophila in drug discovery and development.
MX, CM, BR, SM and SS wrote and edited the manuscript. CM, BR and SS created the figures. BR, SM and SS conceptualized the article. SS is the corresponding author. All authors contributed to the article and approved the submitted version.
BR is funded by The Technology and Human Resources for Industry programme (THRIP) and UNISA. SM is funded by the National Research Foundation (NRF) GUN: 121878 and the UNISA Academic Qualification Improvement Programme (AQIP). MX is funded by NRF through the grant linked scholarship associated with SM NRF funding. CM is funded by UNISA.
BR is funded by the company Buboo (Pty) Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, J., Casali, A., and Campbell, K. (2021). Sensitive high-throughput assays for tumour burden reveal the response of a Drosophila melanogaster model of colorectal cancer to standard chemotherapies. Int. J. Mol. Sci. 22 (10), 5101. doi:10.3390/ijms22105101
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287 (5461), 2185–2195. doi:10.1126/science.287.5461.2185
Aggarwal, B. B., Yuan, W., Li, S., and Gupta, S. C. (2013). Curcumin‐free turmeric exhibits anti‐inflammatory and anticancer activities: Identification of novel components of turmeric. Mol. Nutr. Food Res. 57 (9), 1529–1542. doi:10.1002/mnfr.201200838
Aird, R. E., Cummings, J., Ritchie, A. A., Muir, M., Morris, R. E., Chen, H., et al. (2002). In vitro and in vivo activity and cross resistance profiles of novel ruthenium (II) organometallic arene complexes in human ovarian cancer. Br. J. Cancer 86 (10), 1652–1657. doi:10.1038/sj.bjc.6600290
Andrew, D. J., and Ewald, A. J. (2010). Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev. Biol. 341 (1), 34–55. doi:10.1016/j.ydbio.2009.09.024
Aritakula, A., and Ramasamy, A. (2008). Drosophila-based in vivo assay for the validation of inhibitors of the epidermal growth factor receptor/Ras pathway. J. Biosci. 33 (5), 731–742. doi:10.1007/s12038-008-0093-9
Ashburn, T. T., and Thor, K. B. (2004). Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3 (8), 673–683. doi:10.1038/nrd1468
Bangi, E., Garza, D., and Hild, M. (2011). In vivo analysis of compound activity and mechanism of action using epistasis in Drosophila. J. Chem. Biol. 4 (2), 55–68. doi:10.1007/s12154-010-0051-5
Bangi, E., Ang, C., Smibert, P., Uzilov, A. V., Teague, A. G., Antipin, Y., et al. (2019). A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci. Adv. 5 (5), eaav6528. doi:10.1126/sciadv.aav6528
Bangi, E., Murgia, C., Teague, A. G., Sansom, O. J., and Cagan, R. L. (2016). Functional exploration of colorectal cancer genomes using Drosophila. Nat. Commun. 7 (1), 13615. doi:10.1038/ncomms13615
Bangi, E., Murgia, C., Teague, A., Sansom, O., and Cagan, R. (2015). Abstract PR03: Identifying biomarkers of drug response and resistance using personalized Drosophila models of colorectal cancer. Clin. Cancer Res. 21, PR03. doi:10.1158/1557-3265.PMS14-PR03
Bangi, E., Smibert, P., Uzilov, A. V., Teague, A. G., Gopinath, S., Antipin, Y., et al. (2021). A Drosophila platform identifies a novel, personalized therapy for a patient with adenoid cystic carcinoma. iScience 24 (3), 102212. doi:10.1016/j.isci.2021.102212
Behr, M. (2010). Molecular aspects of respiratory and vascular tube development. Respir. Physiol. Neurobiol. 173, S33–S36. doi:10.1016/j.resp.2010.04.011
Bellusci, S., Furuta, Y., Rush, M. G., Henderson, R., Winnier, G., Hogan, B. L., et al. (1997). Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Dev. Camb. Engl. 124 (1), 53–63. doi:10.1242/dev.124.1.53
Betschinger, J., Eisenhaber, F., and Knoblich, J. A. (2005). Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr. Biol. 15 (3), 276–282. doi:10.1016/j.cub.2005.01.012
Bier, E. (2005). Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6 (1), 9–23. doi:10.1038/nrg1503
Bilen, J., and Bonini, N. M. (2005). Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 39, 153–171. doi:10.1146/annurev.genet.39.110304.095804
Blagosklonny, M. V. (2008). Targeting the absence” and therapeutic engineering for cancer therapy. Cell Cycle 7 (10), 1307–1312. doi:10.4161/cc.7.10.6250
Bossen, J., Uliczka, K., Steen, L., Pfefferkorn, R., Mai, M. M., Burkhardt, L., et al. (2019). An EGFR-induced Drosophila lung tumor model identifies alternative combination treatments. Mol. Cancer Ther. 18 (9), 1659–1668. doi:10.1158/1535-7163.MCT-19-0168
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. development 118 (2), 401–415. doi:10.1242/dev.118.2.401
Cagan, R. L., Zon, L. I., and White, R. M. (2019). Modeling cancer with flies and fish. Dev. Cell 49 (3), 317–324. doi:10.1016/j.devcel.2019.04.013
Cancer Genome Atlas Research Network (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489 (7417), 519–525. doi:10.1038/nature11404
Casali, A., and Batlle, E. (2009). Intestinal stem cells in mammals and Drosophila. Cell stem cell 4 (2), 124–127. doi:10.1016/j.stem.2009.01.009
Chang, Y. Y., Lin, P. C., Lin, H. H., Lin, J. K., Chen, W. S., Jiang, J. K., et al. (2016). Mutation spectra of RAS gene family in colorectal cancer. Am. J. Surg. 212 (3), 537–544. doi:10.1016/j.amjsurg.2016.02.013
Chen, S. H., and Lahav, G. (2016). Two is better than one; toward a rational design of combinatorial therapy. Curr. Opin. Struct. Biol. 41, 145–150. doi:10.1016/j.sbi.2016.07.020
Chong, C. R., and Sullivan, D. J. (2007). New uses for old drugs. Nature 448 (7154), 645–646. doi:10.1038/448645a
Ciuleanu, T., Stelmakh, L., Cicenas, S., Miliauskas, S., Grigorescu, A. C., Hillenbach, C., et al. (2012). Efficacy and safety of erlotinib versus chemotherapy in second-line treatment of patients with advanced, non-small-cell lung cancer with poor prognosis (TITAN): A randomised multicentre, open-label, phase 3 study. Lancet. Oncol. 13 (3), 300–308. doi:10.1016/S1470-2045(11)70385-0
Clevers, H. (2006). Wnt/beta-catenin signaling in development and disease. Cell 127 (3), 469–480. doi:10.1016/j.cell.2006.10.018
Crespo-Ortiz, M. P., and Wei, M. Q. (2012). Antitumor activity of artemisinin and its derivatives: From a well-known antimalarial agent to a potential anticancer drug. J. Biomed. Biotechnol. 2012, 247597. doi:10.1155/2012/247597
Dar, A. C., Das, T. K., Shokat, K. M., and Cagan, R. L. (2012). Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486 (7401), 80–84. doi:10.1038/nature11127
Das, S. S., Nanda, G. G., and Alone, D. P. (2014). Artemisinin and curcumin inhibit Drosophila brain tumor, prolong life span, and restore locomotor activity. Iubmb Life 66 (7), 496–506. doi:10.1002/iub.1284
Das, T. K., and Cagan, R. L. (2017). KIF5B-RET oncoprotein signals through a multi-kinase signaling hub. Cell Rep. 20 (10), 2368–2383. doi:10.1016/j.celrep.2017.08.037
Dietzl, G., Chen, D., Schnorrer, F., Su, K. C., Barinova, Y., and Fellner, M. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 (7150), 151–156. doi:10.1038/nature05954
DiMasi, J. A., Reichert, J. M., Feldman, L., and Malins, A. (2013). Clinical approval success rates for investigational cancer drugs. Clin. Pharmacol. Ther. 94 (3), 329–335. doi:10.1038/clpt.2013.117
Edwards, A., Gladstone, M., Yoon, P., Raben, D., Frederick, B., Su, T. T., et al. (2011). Combinatorial effect of maytansinol and radiation in Drosophila and human cancer cells. Dis. Model. Mech. 4 (4), 496–503. doi:10.1242/dmm.006486
Fearnhead, N. S., Britton, M. P., and Bodmer, W. F. (2001). The ABC of APC. Hum. Mol. Genet. 10 (7), 721–733. doi:10.1093/hmg/10.7.721
Fox, D. T., and Spradling, A. C. (2009). The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell stem cell 5 (3), 290–297. doi:10.1016/j.stem.2009.06.003
Freeman, M. R., and Doherty, J. (2006). Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 29 (2), 82–90. doi:10.1016/j.tins.2005.12.002
Furnari, F. B., Fenton, T., Bachoo, R. M., Mukasa, A., Stommel, J. M., Stegh, A., et al. (2007). Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 21 (21), 2683–2710. doi:10.1101/gad.1596707
Gao, G., Chen, L., and Huang, C. (2015). Anti-cancer drug discovery: Update and comparisons in yeast, Drosophila, and zebrafish. Curr. Mol. Pharmacol. 7 (1), 44–51. doi:10.2174/1874467207666140702113629
Gateff, E. (1978). The genetics and epigenetics of neoplasms in Drosophila. Biol. Rev. Camb. Philos. Soc. 53 (1), 123–168. doi:10.1111/j.1469-185x.1978.tb00994.x
Ghabrial, A., Luschnig, S., Metzstein, M. M., and Krasnow, M. A. (2003). Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19, 623–647. doi:10.1146/annurev.cellbio.19.031403.160043
Gladstone, M., Frederick, B., Zheng, D., Edwards, A., Yoon, P., Stickel, S., et al. (2012). A translation inhibitor identified in a Drosophila screen enhances the effect of ionizing radiation and taxol in mammalian models of cancer. Dis. Model. Mech. 5 (3), 342–350. doi:10.1242/dmm.008722
Gondal, M. N., Butt, R. N., Shah, O. S., Sultan, M. U., Mustafa, G., Nasir, Z., et al. (2021). A personalized therapeutics approach using an in silico Drosophila patient model reveals optimal chemo-and targeted therapy combinations for colorectal cancer. Front. Oncol. 11, 692592. doi:10.3389/fonc.2021.692592
Gonsalves, F. C., Klein, K., Carson, B. B., Katz, S., Ekas, L. A., Evans, S., et al. (2011). An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 108 (15), 5954–5963. doi:10.1073/pnas.1017496108
Gont, A., Hanson, J. E., Lavictoire, S. J., Daneshmand, M., Nicholas, G., Woulfe, J., et al. (2014). Inhibition of glioblastoma malignancy by Lgl1. Oncotarget 5 (22), 11541–11551. doi:10.18632/oncotarget.2580
Gonzalez, C. (2013). Drosophila melanogaster: A model and a tool to investigate malignancy and identify new therapeutics. Nat. Rev. Cancer 13 (3), 172–183. doi:10.1038/nrc3461
Gottesman, M. M., Fojo, T., and Bates, S. E. (2002). Multidrug resistance in cancer: Role of ATP–dependent transporters. Nat. Rev. Cancer 2 (1), 48–58. doi:10.1038/nrc706
Graziani, A., Gramaglia, D., Dalla Zonca, P., and Comoglio, P. M. (1993). Hepatocyte growth factor/scatter factor stimulates the Ras-guanine nucleotide exchanger. J. Biol. Chem. 268 (13), 9165–9168. doi:10.1016/S0021-9258(18)98327-4
Grifoni, D., Sollazzo, M., Fontana, E., Froldi, F., and Pession, A. (2015). Multiple strategies of oxygen supply in Drosophila malignancies identify tracheogenesis as a novel cancer hallmark. Sci. Rep. 5 (1), 9061. doi:10.1038/srep09061
Griffin, R., and Ramirez, R. A. (2017). Molecular targets in non-small cell lung cancer. Ochsner J. 17 (4), 388–392.
Grifoni, D., Froldi, F., and Pession, A. (2013). Connecting epithelial polarity, proliferation and cancer in Drosophila: The many faces of lgl loss of function. Int. J. Dev. Biol. 57 (9-10), 677–687. doi:10.1387/ijdb.130285dg
Guichard, A., Biehs, B., Sturtevant, M. A., Wickline, L., Chacko, J., Howard, K., et al. (1999). Rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Dev. Camb. Engl. 126 (12), 2663–2676. doi:10.1242/dev.126.12.2663
Gupta, S. C., Patchva, S., and Aggarwal, B. B. (2013). Therapeutic roles of curcumin: Lessons learned from clinical trials. AAPS J. 15 (1), 195–218. doi:10.1208/s12248-012-9432-8
Hanif, F., Muzaffar, K., Perveen, K., Malhi, S. M., and Simjee, S. U. (2017). Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian pac. J. Cancer Prev. 18 (1), 3–9. doi:10.22034/APJCP.2017.18.1.3
Hoelder, S., Clarke, P. A., and Workman, P. (2012). Discovery of small molecule cancer drugs: Successes, challenges and opportunities. Mol. Oncol. 6 (2), 155–176. doi:10.1016/j.molonc.2012.02.004
Holland, E. C., Celestino, J., Dai, C., Schaefer, L., Sawaya, R. E., Fuller, G. N., et al. (2000). Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat. Genet. 25 (1), 55–57. doi:10.1038/75596
Horvath, P., Aulner, N., Bickle, M., Davies, A. M., Del Nery, E., Ebner, D., et al. (2016). Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 15 (11), 751–769. doi:10.1038/nrd.2016.175
Jackstadt, R., and Sansom, O. J. (2016). Mouse models of intestinal cancer. J. Pathol. 238 (2), 141–151. doi:10.1002/path.4645
Jardim, D. L., Groves, E. S., Breitfeld, P. P., and Kurzrock, R. (2017). Factors associated with failure of oncology drugs in late-stage clinical development: A systematic review. Cancer Treat. Rev. 52, 12–21. doi:10.1016/j.ctrv.2016.10.009
Johnson, V., Volikos, E., Halford, S. E., Sadat, E. E., Popat, S., Talbot, I., et al. (2005). Exon 3 β-catenin mutations are specifically associated with colorectal carcinomas in hereditary non-polyposis colorectal cancer syndrome. Gut 54 (2), 264–267. doi:10.1136/gut.2004.048132
Ju, Y. S., Lee, W. C., Shin, J. Y., Lee, S., Bleazard, T., Won, J. K., et al. (2012). A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 22 (3), 436–445. doi:10.1101/gr.133645.111
Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B., Lu, C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502 (7471), 333–339. doi:10.1038/nature12634
Kasai, Y., and Cagan, R. (2010). Drosophila as a tool for personalized medicine: A primer. Per. Med. 7 (6), 621–632. doi:10.2217/pme.10.65
Killela, P. J., Pirozzi, C. J., Reitman, Z. J., Jones, S., Rasheed, B. A., Lipp, E., et al. (2014). The genetic landscape of anaplastic astrocytoma. Oncotarget 5 (6), 1452–1457. doi:10.18632/oncotarget.1505
Kim, J., Gosnell, J. E., and Roman, S. A. (2020). Geographic influences in the global rise of thyroid cancer. Nat. Rev. Endocrinol. 16, 17–29. doi:10.1038/s41574-019-0263-x
Klezovitch, O., Fernandez, T. E., Tapscott, S. J., and Vasioukhin, V. (2004). Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 18 (5), 559–571. doi:10.1101/gad.1178004
Kohno, T., Ichikawa, H., Totoki, Y., Yasuda, K., Hiramoto, M., Nammo, T., et al. (2012). KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 18 (3), 375–377. doi:10.1038/nm.2644
Kuklinski, N. J., Berglund, E. C., and Ewing, A. G. (2010). Micellar capillary electrophoresis - electrochemical detection of neurochemicals from Drosophila. J. Sep. Sci. 33 (3), 388–393. doi:10.1002/jssc.200900634
Kunnumakkara, A. B., Bordoloi, D., Harsha, C., Banik, K., Gupta, S. C., Aggarwal, B. B., et al. (2017). Curcumin mediates anticancer effects by modulating multiple cell signaling pathways. Clin. sci. 131 (15), 1781–1799. doi:10.1042/CS20160935
Levine, B. D., and Cagan, R. L. (2016). Drosophila lung cancer models identify trametinib plus statin as candidate therapeutic. Cell Rep. 14 (6), 1477–1487. doi:10.1016/j.celrep.2015.12.105
Levinson, S., and Cagan, R. L. (2016). Drosophila cancer models identify functional differences between ret fusions. Cell Rep. 16 (11), 3052–3061. doi:10.1016/j.celrep.2016.08.019
Lipson, D., Capelletti, M., Yelensky, R., Otto, G., Parker, A., Jarosz, M., et al. (2012). Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 18 (3), 382–384. doi:10.1038/nm.2673
Liu, Z., Delavan, B., Roberts, R., and Tong, W. (2017). Lessons learned from two decades of anticancer drugs. Trends Pharmacol. Sci. 38 (10), 852–872. doi:10.1016/j.tips.2017.06.005
LoTempio, M. M., Veena, M. S., Steele, H. L., Ramamurthy, B., Ramalingam, T. S., Cohen, A. N., et al. (2005). Curcumin suppresses growth of head and neck squamous cell carcinoma. Clin. Cancer Res. 11 (19), 6994–7002. doi:10.1158/1078-0432.CCR-05-0301
Lu, J. J., Cai, Y. J., and Ding, J. (2011). Curcumin induces DNA damage and caffeine-insensitive cell cycle arrest in colorectal carcinoma HCT116 cells. Mol. Cell. Biochem. 354 (1), 247–252. doi:10.1007/s11010-011-0824-3
Maemondo, M., Inoue, A., Kobayashi, K., Sugawara, S., Oizumi, S., Isobe, H., et al. (2010). Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 362 (25), 2380–2388. doi:10.1056/NEJMoa0909530
Maher, E. A., Furnari, F. B., Bachoo, R. M., Rowitch, D. H., Louis, D. N., Cavenee, W. K., et al. (2001). Malignant glioma: Genetics and biology of a grave matter. Genes Dev. 15 (11), 1311–1333. doi:10.1101/gad.891601
Markowitz, S. D., and Bertagnolli, M. M. (2009). Molecular origins of cancer: Molecular basis of colorectal cancer.. N. Engl. J. Med. 361 (25), 2449–2460. doi:10.1056/NEJMra0804588
Martorell, Ò., Merlos-Suárez, A., Campbell, K., Barriga, F. M., Christov, C. P., Miguel-Aliaga, I., et al. (2014). Conserved mechanisms of tumorigenesis in the Drosophila adult midgut. PLoS ONE 9 (2), e88413. doi:10.1371/journal.pone.0088413
Miller, J. R. (2001). The wnts. Genome Biol. 3 (1), REVIEWS3001. doi:10.1186/gb-2001-3-1-reviews3001
Misale, S., Yaeger, R., Hobor, S., Scala, E., Janakiraman, M., Liska, D., et al. (2012). Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486 (7404), 532–536. doi:10.1038/nature11156
Mokhtari, R. B., Homayouni, T. S., Baluch, N., Morgatskaya, E., Kumar, S., Das, B., et al. (2017). Combination therapy in combating cancer.. Oncotarget 8, 38022–38043. doi:10.18632/oncotarget.16723
Molina, J. R., Yang, P., Cassivi, S. D., Schild, S. E., and Adjei, A. A. (2008). Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 83 (5), 584–594. doi:10.4065/83.5.584
Morin, P.J., Sparks, A.B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B., et al. (1997). Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science, 275 (5307), 1787-1790. doi:10.1126/science.275.5307.1787
Moore, M. S., DeZazzo, J., Luk, A. Y., Tully, T., Singh, C. M., Heberlein, U., et al. (1998). Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell 93 (6), 997-1007. doi:10.1016/s0092-8674(00)81205-2
Moreno, L., and Pearson, A. D. (2013). How can attrition rates be reduced in cancer drug discovery? Expert Opin. Drug Discov. 8 (4), 363–368. doi:10.1517/17460441.2013.768984
Murayama, T., and Gotoh, N. (2019). Patient-derived xenograft models of breast cancer and their application. Cells 8 (6), 621. doi:10.3390/cells8060621
Nazarian, R., Shi, H., Wang, Q., Kong, X., Koya, R. C., Lee, H., et al. (2010). Melanomas acquire resistance to B-RAF (V600E) inhibition by RTK or N-RAS upregulation. Nature 468 (7326), 973–977. doi:10.1038/nature09626
Ngo, S., Liang, J., Su, Y. H., and O'Brien, L. E. (2020). Disruption of EGF feedback by intestinal tumors and neighboring cells in Drosophila. Curr. Biol. 30 (8), 1537–1546. e3. doi:10.1016/j.cub.2020.01.082
Ocana, A., Pandiella, A., Siu, L. L., and Tannock, I. F. (2011). Preclinical development of molecular-targeted agents for cancer. Nat. Rev. Clin. Oncol. 8 (4), 200–209. doi:10.1038/nrclinonc.2010.194
Palmer, A. C., and Sorger, P. K. (2017). Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 171 (7), 1678–1691. doi:10.1016/j.cell.2017.11.009
Pandey, U. B., and Nichols, C. D. (2011). Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 63 (2), 411–436. doi:10.1124/pr.110.003293
Park, W. Y., Miranda, B., Lebeche, D., Hashimoto, G., and Cardoso, W. V. (1998). FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev. Biol. 201 (2), 125–134. doi:10.1006/dbio.1998.8994
Pelham, C. J., Nagane, M., and Madan, E. (2020). Cell competition in tumor evolution and heterogeneity: Merging past and present. Semin. Cancer Biol. 63, 11–18. doi:10.1016/j.semcancer.2019.07.008
Peyton, J. D., Ahnert, J. R., Burris, H., Britten, C., Chen, L. C., TaberneroV Duval, J., et al. (2011). A dose-escalation study with the novel formulation of the oral pan-class I PI3K inhibitor BEZ235, solid dispersion system (SDS) sachet, in patients with advanced solid tumors. J. Clin. Oncol. 29 (15), 3066. doi:10.1200/jco.2011.29.15_suppl.3066
Planchard, D., Besse, B., Groen, H. J., Hashemi, S. M., Mazieres, J., Kim, T. M., et al. (2022). Phase 2 study of dabrafenib plus trametinib in patients with BRAF V600E-mutant metastatic NSCLC: Updated 5-year survival rates and genomic analysis. J. Thorac. Oncol. 17 (1), 103–115. doi:10.1016/j.jtho.2021.08.011
Plant, P. J., Fawcett, J. P., Lin, D. C., Holdorf, A. D., Binns, K., Kulkarni, S., et al. (2003). A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5 (4), 301–308. doi:10.1038/ncb948
Rand, M. D. (2010). Drosophotoxicology: The growing potential for Drosophila in neurotoxicology. Neurotoxicol. Teratol. 32 (1), 74–83. doi:10.1016/j.ntt.2009.06.004
Read, R. D., Cavenee, W. K., Furnari, F. B., and Thomas, J. B. (2009). A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 5 (2), e1000374. doi:10.1371/journal.pgen.1000374
Reiter, L. T., Potocki, L., Chien, S., Gribskov, M., and Bier, E. (2001). A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11 (6), 1114–1125. doi:10.1101/gr.169101
Reuter, S., Gupta, S. C., Park, B., Goel, A., and Aggarwal, B. B. (2011). Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 6 (2), 93–108. doi:10.1007/s12263-011-0222-1
Richardson, H. E., Willoughby, L., and Humbert, P. O. (2015). Screening for anti‐cancer drugs in drosophila. eLS 2015, 1–14. doi:10.1002/9780470015902.a0022535
Riely, G. J., Marks, J., and Pao, W. (2009). KRAS mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 6 (2), 201–205. doi:10.1513/pats.200809-107LC
Roeder, T., Isermann, K., Kallsen, K., Uliczka, K., and Wagner, C. (2012). A Drosophila asthma model - what the fly tells us about inflammatory diseases of the lung. Adv. Exp. Med. Biol. 710, 37–47. doi:10.1007/978-1-4419-5638-5_5
Rotow, J., and Bivona, T. G. (2017). Understanding and targeting resistance mechanisms in NSCLC. Nat. Rev. Cancer 17 (11), 637–658. doi:10.1038/nrc.2017.84
Rowan, A. J., Lamlum, H., Ilyas, M., Wheeler, J., Straub, J., Papadopoulou, A., et al. (2000). APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc. Natl. Acad. Sci. U. S. A. 97 (7), 3352–3357. doi:10.1073/pnas.97.7.3352
Rudrapatna, V. A., Cagan, R. L., and Das, T. K. (2012). Drosophila cancer models. Dev. Dyn. 241 (1), 107–118. doi:10.1002/dvdy.22771
Sadaqat, Z., Kaushik, S., Kain, P., Sampson, J. H., Gunn, M. D., Fecci, P. E., et al. (20212020). Gut feeding the brain: Drosophila gut an animal model for medicine to understand mechanisms mediating food preferences animal models in medicine. Rijeka, Croatia: Intech OpenBrain immunology and immunotherapy in brain tumours. Nat. Rev. Cancer 20 (1), 12–25. doi:10.1038/s41568-019-0224-7
Sampson, J. H., Gunn, M. D., Fecci, P. E., and Ashley, D. M. (2020). Brain immunology and immunotherapy in brain tumours. Nature reviews. Cancer 20 (1), 12–25. doi:10.1038/s41568-019-0224-7
Sancho, E., Batlle, E., and Clevers, H. (2004). Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20, 695–723. doi:10.1146/annurev.cellbio.20.010403.092805
Schimanski, C. C., Schmitz, G., Kashyap, A., Bosserhoff, A. K., Bataille, F., Schäfer, S. C., et al. (2005). Reduced expression of Hugl-1, the human homologue of Drosophila tumour suppressor gene lgl, contributes to progression of colorectal cancer. Oncogene 24 (19), 3100–3109. doi:10.1038/sj.onc.1208520
Sonoshita, M., and Cagan, R. L. (2017). Modeling human cancers in Drosophila. Curr. Top. Dev. Biol. 121, 287–309. doi:10.1016/bs.ctdb.2016.07.008
Stephen, A. G., Esposito, D., Bagni, R. K., and McCormick, F. (2014). Dragging ras back in the ring. Cancer cell 25 (3), 272–281. doi:10.1016/j.ccr.2014.02.017
Stickel, S. A., Gomes, N. P., Frederick, B., Raben, D., and Su, T. T. (2015). Bouvardin is a radiation modulator with a novel mechanism of action. Radiat. Res. 184 (4), 392–403. doi:10.1667/RR14068.1
Su, T. T. (2019). Drug screening in Drosophila; why, when, and when not? Wiley Interdiscip. Rev. Dev. Biol. 8 (6), e346. doi:10.1002/wdev.346
Suijkerbuijk, S. J., Kolahgar, G., Kucinski, I., and Piddini, E. (2016). Cell competition drives the growth of intestinal adenomas in Drosophila. Curr. Biol. 26 (4), 428–438. doi:10.1016/j.cub.2015.12.043
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Takashima, S., Mkrtchyan, M., Younossi-Hartenstein, A., Merriam, J. R., and Hartenstein, V. (2008). The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature 454 (7204), 651–655. doi:10.1038/nature07156
Takeuchi, K., Soda, M., Togashi, Y., Suzuki, R., Sakata, S., Hatano, S., et al. (2012). RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18 (3), 378–381. doi:10.1038/nm.2658
Tanaka, H., Deng, G., Matsuzaki, K., Kakar, S., Kim, G. E., Miura, S., et al. (2006). BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. Int. J. Cancer 118 (11), 2765–2771. doi:10.1002/ijc.21701
Torres, G., and Horowitz, J. M. (1998). Activating properties of cocaine and cocaethylene in a behavioral preparation of Drosophila melanogaster. Synapse 29, 148–161.
Valtorta, E., Misale, S., Sartore-Bianchi, A., Nagtegaal, I. D., Paraf, F., Lauricella, C., et al. (2013). KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. Int. J. Cancer 133 (5), 1259–1265. doi:10.1002/ijc.28106
Verhaak, R. G., Hoadley, K. A., Purdom, E., Wang, V., Qi, Y., Wilkerson, M. D., et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell 17 (1), 98–110. doi:10.1016/j.ccr.2009.12.020
Verjans, E. T., Doijen, J., Luyten, W., Landuyt, B., and Schoofs, L. (2018). Three‐dimensional cell culture models for anticancer drug screening: Worth the effort? J. Cell. Physiol. 233 (4), 2993–3003. doi:10.1002/jcp.26052
Vidal, M., Wells, S., Ryan, A., and Cagan, R. (2005). ZD6474 suppresses oncogenic RET isoforms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Res. 65 (9), 3538–3541. doi:10.1158/0008-5472.CAN-04-4561
Villegas, S. N. (2019). One hundred years of Drosophila cancer research: No longer in solitude. Dis. Model. Mech. 12 (4), dmm039032. doi:10.1242/dmm.039032
Vogelstein, B., Fearon, E. R., Hamilton, S. R., Kern, S. E., Preisinger, A. C., Leppert, M., et al. (1988). Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319 (9), 525–532. doi:10.1056/NEJM198809013190901
Wang, C., Zhao, R., Huang, P., Yang, F., Quan, Z., Xu, N., et al. (2013). APC loss-induced intestinal tumorigenesis in Drosophila: Roles of Ras in Wnt signaling activation and tumor progression. Dev. Biol. 378 (2), 122–140. doi:10.1016/j.ydbio.2013.03.020
Wangler, M. F., Yamamoto, S., Chao, H. T., Posey, J. E., Westerfield, M., Postlethwait, J., et al. (2017). Model organisms facilitate rare disease diagnosis and therapeutic research.. Genetics 207, 9–27. doi:10.1534/genetics.117.203067
Wells, S. A., Gosnell, J. E., Gagel, R. F., Moley, J., Pfister, D., Sosa, J. A., et al. (2010). Vandetanib for the treatment of patients with locally advanced or metastatic hereditary medullary thyroid cancer. J. Clin. Oncol. 28 (5), 767–772. doi:10.1200/JCO.2009.23.6604
White, B. D., Chien, A. J., and Dawson, D. W. (2012). Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology 142 (2), 219–232. doi:10.1053/j.gastro.2011.12.001
Wilhelm, S. M., Carter, C., Tang, L., Wilkie, D., McNabola, A., Rong, H., et al. (2004). BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 64 (19), 7099–7109. doi:10.1158/0008-5472.CAN-04-1443
Wilken, R., Veena, M. S., Wang, M. B., and Srivatsan, E. S. (2011). Curcumin: A review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma. Mol. Cancer 10 (1), 12. doi:10.1186/1476-4598-10-12
Willoughby, L. F., Schlosser, T., Manning, S. A., Parisot, J. P., Street, I. P., Richardson, H. E., et al. (2013). An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Dis. Model. Mech. 6 (2), 521–529. doi:10.1242/dmm.009985
Woodhouse, E. C., and Liotta, L. A. (2004). Drosophila invasive tumors: A model for understanding metastasis. Cell cycle 3 (1), 37–39. doi:10.4161/cc.3.1.636
Yadav, A. K., Srikrishna, S., and Gupta, S. C. (2016). Cancer drug development using drosophila as an in vivo tool: From bedside to bench and back. Trends Pharmacol. Sci. 37 (9), 789–806. doi:10.1016/j.tips.2016.05.010
Yamamura, R., Ooshio, T., and Sonoshita, M. (2021). Tiny Drosophila makes giant strides in cancer research. Cancer Sci. 112 (2), 505–514. doi:10.1111/cas.14747
Yardley, D. A. (2013). Drug resistance and the role of combination chemotherapy in improving patient outcomes. Int. J. Breast Cancer 2013, 137414. doi:10.1155/2013/137414
Zanoni, M., Piccinini, F., Arienti, C., Zamagni, A., Santi, S., Polico, R., et al. (2016). 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 6 (1), 19103. doi:10.1038/srep19103
Zhang, L., and Shay, J. W. (2017). Multiple roles of APC and its therapeutic implications in colorectal cancer. J. Natl. Cancer Inst. 109 (8), djw332. doi:10.1093/jnci/djw332
Zimmermann, T., Kashyap, A., Hartmann, U., Otto, G., Galle, P. R., Strand, S., et al. (2008). Cloning and characterization of the promoter of Hugl-2, the human homologue of Drosophila lethal giant larvae (lgl) polarity gene. Biochem. Biophys. Res. Commun. 366 (4), 1067–1073. doi:10.1016/j.bbrc.2007.12.084
Keywords: Drosophila melanogaster, cancer models, high-throughput screening, drug discovery, personalized therapy
Citation: Munnik C, Xaba MP, Malindisa ST, Russell BL and Sooklal SA (2022) Drosophila melanogaster: A platform for anticancer drug discovery and personalized therapies. Front. Genet. 13:949241. doi: 10.3389/fgene.2022.949241
Received: 20 May 2022; Accepted: 06 July 2022;
Published: 08 August 2022.
Edited by:
Zodwa Dlamini, Pan African Cancer Research Institute (PACRI), South AfricaReviewed by:
Andreu Casali, University of Lleida, SpainCopyright © 2022 Munnik, Xaba, Malindisa, Russell and Sooklal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Selisha A. Sooklal, c29va2xzYUB1bmlzYS5hYy56YQ==
†These authors have contributed equally to the work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.