
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 09 September 2022
Sec. Epigenomics and Epigenetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.946524
This article is part of the Research Topic Epigenetic Regulation in Cardiovascular Diseases, Volume II View all 6 articles
 Xiao-Wen Meng1,2†
Xiao-Wen Meng1,2† Mian Zhang1,2†
Mian Zhang1,2† Jun-Kai Hu1,2†
Jun-Kai Hu1,2† Xin-Yu Chen1,2†
Xin-Yu Chen1,2† Yu-Qin Long1,2
Yu-Qin Long1,2 Hong Liu3
Hong Liu3 Xiao-Mei Feng4
Xiao-Mei Feng4 Fu-Hai Ji1,2*
Fu-Hai Ji1,2* Ke Peng1,2*
Ke Peng1,2*Background: The mechanisms underlying myocardial ischemia/reperfusion (I/R) injury are not fully understood. This study aims to explore key candidate genes and potential therapeutic targets for treatment of myocardial I/R injury.
Methods: The transcriptional profiles of ventricular myocardium during cardiac arrest, ischemia, and reperfusion were obtained from the Gene Expression Omnibus database. Based on the transcriptional data of GSE6381, functional pathway and process enrichment analyses, protein–protein interaction network, and gene set enrichment analyses were conducted. In the animal experiments, we established the myocardial I/R injury model in mice. We validated the mRNA and protein expression of the key genes using the qPCR and western blots. We further assessed the expression and localization of CCL21 and its receptors using immunofluorescence staining experiments.
Results: The microarray analyses identified five key genes (CCL21, XCR1, CXCL13, EDN1, and CASR). Myocardial I/R process in mice resulted in significant myocardial infraction, histological damage, and myocardial apoptosis. The results of qPCR and western blots showed that the expression of CCL21 and CXCL13 were increased following myocardial I/R injury in mice. Furthermore, the immunofluorescence staining results revealed that the expression of GPR174/CCR7 (CCL21 receptors), but not CXCR5 (CXCL13 receptor), was elevated following myocardial I/R injury. Moreover, the activated CCL21-GPR174/CCR7 signaling was located on the cardiac fibroblasts of the myocardium with I/R injury.
Conclusion: This study revealed several key factors underlying myocardial I/R injury. Of these, the activation of CCL21-GPR174/CCR7 signaling on cardiac fibroblasts was highlighted, which provides potential therapeutic targets for cardioprotection.
Timely restoration of blood flow is the most effective therapeutic strategy to salvage the viable myocardium for patients with acute myocardial infarction (Moran et al., 2014). Nevertheless, reperfusion per se leads to myocardial ischemia/reperfusion (I/R) injury, a complex pathophysiological process contributing up to 50% of final myocardial infarct size (Boag et al., 2015). Several characteristics of myocardial I/R injury are inflammatory responses, reactive oxygen species accumulation, cell apoptosis, and intracellular calcium overload (Hausenloy and Yellon, 2013; Zhang et al., 2017; Ge et al., 2019; Yuan et al., 2019). Despite recent progresses attempting to attenuate myocardial I/R injury, it remains an unsolved problem.
During cardiac surgery under cardiopulmonary bypass, myocardial I/R injury is predictable and inevitable (Bulluck et al., 2016). Compared with an unprotected myocardial I/R process, blood cardioplegic arrest and protective hypothermia are utilized to limit myocardial injury. A previous study assessed the gene expression profiles of myocardium in the early I/R stages in cardiac surgery using the microarray approach (Arab et al., 2007). Their results suggested that multiple signaling pathways such as PI3K signaling and hemoglobin synthesis were activated during the process (Arab et al., 2007). That study showed gene expression profiles only, without any further analysis or experimental validation. To date, the mechanisms of myocardial I/R injury are not fully understood.
This study aimed to explore the key genes and pathways in myocardial I/R injury. Through analyzing the transcriptional data, differentially expressed genes (DEGs) were identified from the Gene Expression Omnibus (GEO) database, and functional pathway and process enrichment analyses were performed. For the key genes, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein–protein interaction (PPI) were investigated. Gene set enrichment analysis (GSEA) was used to reveal enriched pathways in the stages of ischemia and reperfusion. Finally, the expression of key candidate genes was assessed in myocardial I/R injury of mice, and the activation of CCL21 with its receptors GPR174/CCR7 was highlighted.
The gene expression profile dataset GSE6381 was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE6381 presented early gene expression profiles during intraoperative myocardial I/R in cardiac surgery under cardiopulmonary bypass. Specifically, human right ventricular samples were obtained during surgical repair of ventricular septal defect and resection of right ventricular outflow obstruction, with four replicates at each stage (immediately post-cardioplegic arrest, end-ischemia, and 5 min after reperfusion). Data were detected on the Affymetrix Human Genome U133A Array platform (GPL96). The limma package in R language was used to identify DEGs for end-ischemia vs. cardiac arrest, reperfusion vs. end-ischemia, and reperfusion vs. cardiac arrest, respectively (Ritchie et al., 2015). The thresholds for DEGs selection were set as p < 0.05 and log2 FC (fold change) > 1. Volcano plots and heatmaps were generated to overview the gene expression profiles by using the R package.
For the DEGs from each comparison, pathway and process enrichment analyses were performed by using the Metascape (http://metascape.org/) (Zhou et al., 2019). The analyses were based on the following databases: GO Biological Processes, KEGG Pathways, Reactome Gene Sets, and Canonical Pathways. Terms with count ≥3, p-value < 0.01, and enrichment factor (observed counts/counts expected by chance) >1.5 were grouped in clusters based on the similarities. Specifically, p-values were calculated based on the accumulative hypergeometric distribution, and q-values were calculated by using the Banjamini-Hochberg procedure for multiple testing (Hochberg and Benjamini, 1990). Hierarchical clustering was performed on the enriched terms with the kappa scores, and sub-trees with a similarity >0.3 were considered a cluster (Roberts, 2008). The most statistically significant term within a cluster was used to represent the cluster. The overlaps between three gene lists were shown in a Circos plot (Krzywinski et al., 2009). Heatmaps of the top 20 and 100 enriched terms were generated by using the JTreeView (Saldanha, 2004).
To further investigate the relationships among the enriched terms, network plots of the top 20 clusters were generated, where terms with a similarity >0.3 were connected by edges. The networks were visualized by using the Cytoscape software v3.7.2 (https://cytoscape.org/), where each node represented an enriched term and was colored by its cluster ID and then by p-value (Shannon et al., 2003). In addition, the nodes were represented as pie charts, where the size of a pie was proportional to the total number of hits belonging to the specific term. The pie charts were color-coded based on the gene list identities, and the size of a slice showed the percentage of genes under the term which originated from the specific gene list.
For all DEGs, PPI network analysis was carried out by using the Metascape. The analyses were based on several databases, including BioGrid, InWeb_IM, and OmniPath (Stark et al., 2006; Turei et al., 2016; Li et al., 2017). The resultant network contained the subset of proteins with at least one physical interaction with one other member. The Molecular Complex Detection (MCODE) algorithm was applied to identify highly connected network components (Bader and Hogue, 2003). The PPI network plots were visualized by using the Cytoscape, where each node represented a hub protein and was colored by the cluster ID and gene list identity.
For the 26 key genes identified in MOCDE 1, GO enrichment analyses of biological process (BP), molecular function (MF), and cellular component (CC) were conducted by using the gProfiler (https://biit.cs.ut.ee/gprofiler/) (Raudvere et al., 2019). In addition, KEGG pathway analysis was performed to analyze significantly enriched signaling pathways. A p-value < 0.05 was applied as the threshold value.
By using the GeneMANIA (http://genemania.org/), a flexible and user-friendly web interface, PPI network and functional assays of 26 key genes were generated and visualized (Zuberi et al., 2013). Several types of relationships among proteins included co-expressions, physical interactions, shared protein domains, pathways, website predictions, co-localizations, and genetic interactions. GeneMANIA also reported weights that indicated the predictive value of each selected data set.
GSEA is a computational method used to determine whether a predefined set of genes shows significant and concordant differences between the two study groups. GSEA software v3.0 (http://www.gsea-msigdb.org/gsea/) was used to perform KEGG pathway enrichment for two comparisons: end-ischemia vs. cardiac arrest and reperfusion vs. cardiac arrest (Subramanian et al., 2005). A p-value < 0.05 was considered as statistically significant. The “c2.cp.kegg.v6.2.symbols.gmt” gene set was downloaded from the Molecular Signatures Database and used as the reference gene set.
The protocol of animal study was approved by the Ethic Committee for Animal Care and Use at Soochow University, Suzhou, Jiangsu, China. The animal experimental procedures followed the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication No. 85-23, revised in 1996). Adult male C57BL/6J mice (8 weeks old, 20–25 g) were provided by the Experimental Animal Center of Soochow University. Mice were housed in a controlled condition (temperature of 24–26°C, 12-h light/dark cycle, and free access to food and water).
We induced myocardial I/R injury in mice as previously reported (Peng et al., 2021). First, the mice were anesthetized with intraperitoneal sodium pentobarbital (50 mg/kg). Next, the mice were endotracheally intubated and received mechanical ventilation using a rodent respirator (RWD, Shenzhen, China). A left thoracotomy was performed in the left fourth intercostal space. The left anterior descending coronary artery (LAD) was temporarily ligated at ∼2 mm under the lower edge of the left auricle. Observation of tissue blanching in the left ventricular myocardium and elevation in the ST-segment of electrocardiography indicated that the myocardial I/R model was successfully established. The LAD was occluded for 30 min to induce myocardial ischemia, and the ligature was removed for reperfusion of 30 min. For mice in the sham surgery group, the same surgical procedure was performed except that the suture placed around the LAD was not tied. At the end of the reperfusion, the hearts were harvested.
Evans blue staining and 2,3,5-triphenyltetrazolium chloride (TTC) staining were used to assess the area at risk and infarct size of myocardium, as previously described (Peng et al., 2020; Song et al., 2020; Yang et al., 2021). Briefly, the LAD was re-ligated following 30 min of reperfusion, and 1 ml of 2% Evans blue dye (Sigma, St. Louis, MO) was perfused into the coronary artery. The heart was then immediately excised and quickly frozen at −80°C. The heart samples below the ligature were sliced into five transverse sections, and the slices were incubated with 1% TTC (Sigma, St. Louis, MO) at 37°C for 15 min in a dark chamber and fixed with 10% formaldehyde overnight. The non-ischemic area (blue), the myocardium area at risk (AAR, red), and infarct area (IA, pale) were measured using the Image Pro Plus software (Media Cybernetics, Silver Spring, MD, United States). The ratios of IA/AAR and AAR/left ventricular area (LV) were calculated.
Histological structure of the left ventricular myocardium samples was detected using the hematoxylin and eosin (HE) staining (Yang et al., 2021). Briefly, myocardial tissues were fixed with paraformaldehyde and embedded with paraffin. The samples were cut transversely into 5 µm-thick sections. After dewaxing with dimethylbenzene and dehydrating with graded ethanol series, the slices were stained with hematoxylin and eosin. Finally, histological damage of the myocardium was observed under a light microscope (Nikon Corporation; Tokyo, Japan).
Cell apoptosis in the left ventricular myocardium samples was assessed using the terminal deoxynucleotidyl transferase dUTP nick‐end labeling (TUNEL) analysis with the in situ Cell Death Detection kit (Roche Molecular Biochemicals, Mannheim, Germany) (Peng et al., 2020; Peng et al., 2021). The heart samples were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, and cut into 4 μm‐thick sections. The slices were incubated with TUNEL reaction mixture and then counterstained with the 4′,6-diamino-2-phenylindole (DAPI) to show the nuclei. At least four random and nonoverlapping of views in areas surrounding the infarct area in the I/R group or the corresponding area in the sham group were observed using a fluorescence microscope (Nikon Corporation; Tokyo, Japan). The apoptosis rate was expressed as a percentage of TUNEL-positive cells to total cell nuclei.
Total RNA in the left ventricular myocardium samples was extracted using the TRIZOL reagent (Thermo Fisher Scientific, Waltham, MA). After RNA quantification and purity evaluation, reverse transcription was conducted with a cDNA Synthesis Kit (abm, Richmond, BC, Canada), in a 20 μl reaction system containing 5× All-in-one, DEPC water, and RNA samples. Then, qPCR was performed with the EvaGreen (abm, Richmond, BC, Canada) on the Roche Light Cycler R480 System (Roche, Bedford, MA). The amplification conditions were: Pre-denaturation at 93°C for 10 s, denaturation at 55°C for 15 s, and annealing at 72°C for 20 s for 40 cycles. Three replicates were used for each sample. Gene expression were analyzed by using the 2−ΔΔCT method and normalized to β-tubulin. The sequences of primers (Sangon Biotech, Shanghai, China) are listed in Table 1.
The total protein of the left ventricular myocardium samples was extracted using the RIPA reagents (Beyotime, Shanghai, China). The protein concentrations were detected using a bicinchoninic acid protein assay kit (Beyotime, Shanghai, China). All proteins were separated with 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and transferred onto polyvinylidene fluoride membranes (Millipore Corp., Bedford, MA). The membranes were incubated with the primary antibodies overnight at 4°C, and then incubated with the horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. The primary antibodies were as follows: goat anti-CCL21 (1:1000; R&D Systems, Minneapolis, MN), rabbit anti-XCR1 (1:1000; Novus Biologicals, Littleton, CO), goat anti-CXCL13 (1:1000; R&D Systems, Minneapolis, MN), goat anti-CCR7 (1:1000; Novus Biologicals, Littleton, CO), rabbit anti-GPR174 (1:1000; Biorbyt, Cambridge, United Kingdom), rabbit anti-CXCR5 (1:1000; Absin, Shanghai, China), and rabbit anti-β-tubulin (1:5000; Cell Signaling Technology, Beverly, MA). Finally, the protein bands were detected using the Tanon 5200 luminescent workstation (Tanon, Shanghai, China). The protein expression was normalized to β-tubulin as control.
Following 30 min of reperfusion, the left ventricular myocardium samples were fixed with 10% formalin and embedded in paraffin. Then, the tissues were cut transversely into 5 µm-thick sections, deparaffinized, rehydrated, and subjected to antigen retrieval. For immunofluorescence staining, the slices were incubated with primary antibodies overnight at 4°C, and then incubated with secondary antibodies for 2 h at room temperature. The following primary antibodies were used: goat anti-CCR7 (1:200; Novus Biologicals, Littleton, CO), rabbit anti-GPR174 (1:200; Biorbyt, Cambridge, United Kingdom), mouse anti-α-Actinin (1:250; Santa Cruz Technology, CA), and mouse anti-Vimentin (1:200; Abcam, CA). The nuclei were counterstained with DAPI. At least four random and nonoverlapping of views in areas surrounding the infarct area in the I/R group or the corresponding area in the sham group were observed using the fluorescence microscope. Images were analyzed using the Image Pro Plus software.
Statistical analysis was performed by using the GraphPad Prism software (version 9.0, GraphPad, San Diego, CA). Data were expressed as mean ± standard error of the mean and compared using t-test. A two-tailed value of p < 0.05 was considered statistically significant.
The expression profiles of 12,549 genes in GSE6381 of human right ventricular samples during myocardial I/R process were analyzed. The DEGs were identified in three comparisons (Figure 1A): 309 DEGs (247 upregulated and 62 downregulated) in end-ischemia vs. cardiac arrest (Supplementary Table S1), 175 DEGs (90 upregulated and 85 downregulated) in reperfusion vs. end-ischemia (Supplementary Table S2), and 217 DEGs (165 up-regulated and 52 down-regulated) in reperfusion vs. cardiac arrest (Supplementary Table S3). These DEGs showed apparently differential expression profiles (Figure 1B). The top 10 up- and downregulated DEGs for each comparison are presented in Table 2. There were 40, 34, and 51 common DEGs between two comparisons, but no gene was found as common to the three (Figure 1C; Supplementary Table S4).
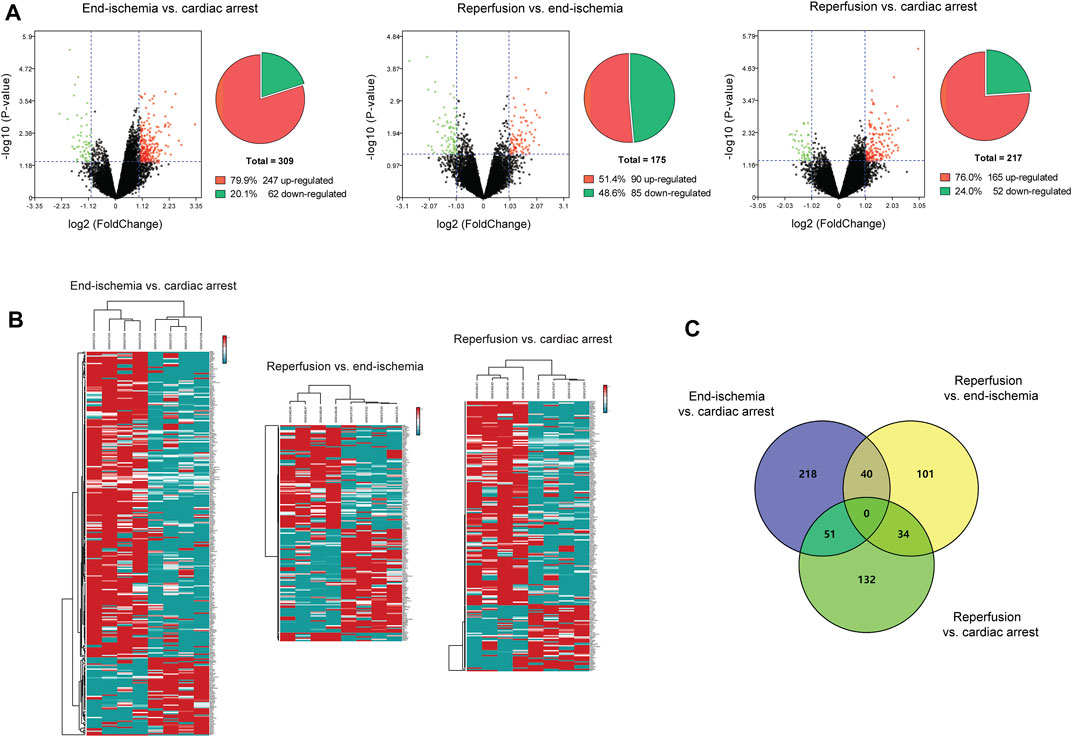
FIGURE 1. Identification of DEGs. (A) Volcano plots and pie charts showing DEGs for end-ischemia vs. cardiac arrest, reperfusion vs. end-ischemia, and reperfusion vs. cardiac arrest. (B) Heatmaps of DEGs. (C) Venn’s diagram showing common DEGs among comparisons.
The pathway and process enrichment analyses of DEGs for each comparison were conducted by using the Metascape. Figure 2A shows gene overlaps and genes that belong to the same enriched ontology term. The top 20 enriched terms are colored by p-values in the heatmap (Figure 2B), with the characteristics shown in Figure 2C. Of these, inorganic ion homeostasis, positive regulation of JNK cascade, and cytokine-mediated signaling pathway may be closely related to the myocardial I/R process. Neuroactive ligand-receptor interaction is the common KEGG pathway. The top 100 enriched terms are available in Supplementary Figure S1. Furthermore, the relationships of the significantly enriched terms are visualized in networks, colored by cluster IDs (Figure 2D), p-values (Figure 2E), and gene list identities (Figure 2F). All results of the enrichment analyses are listed in Supplementary Table S5.
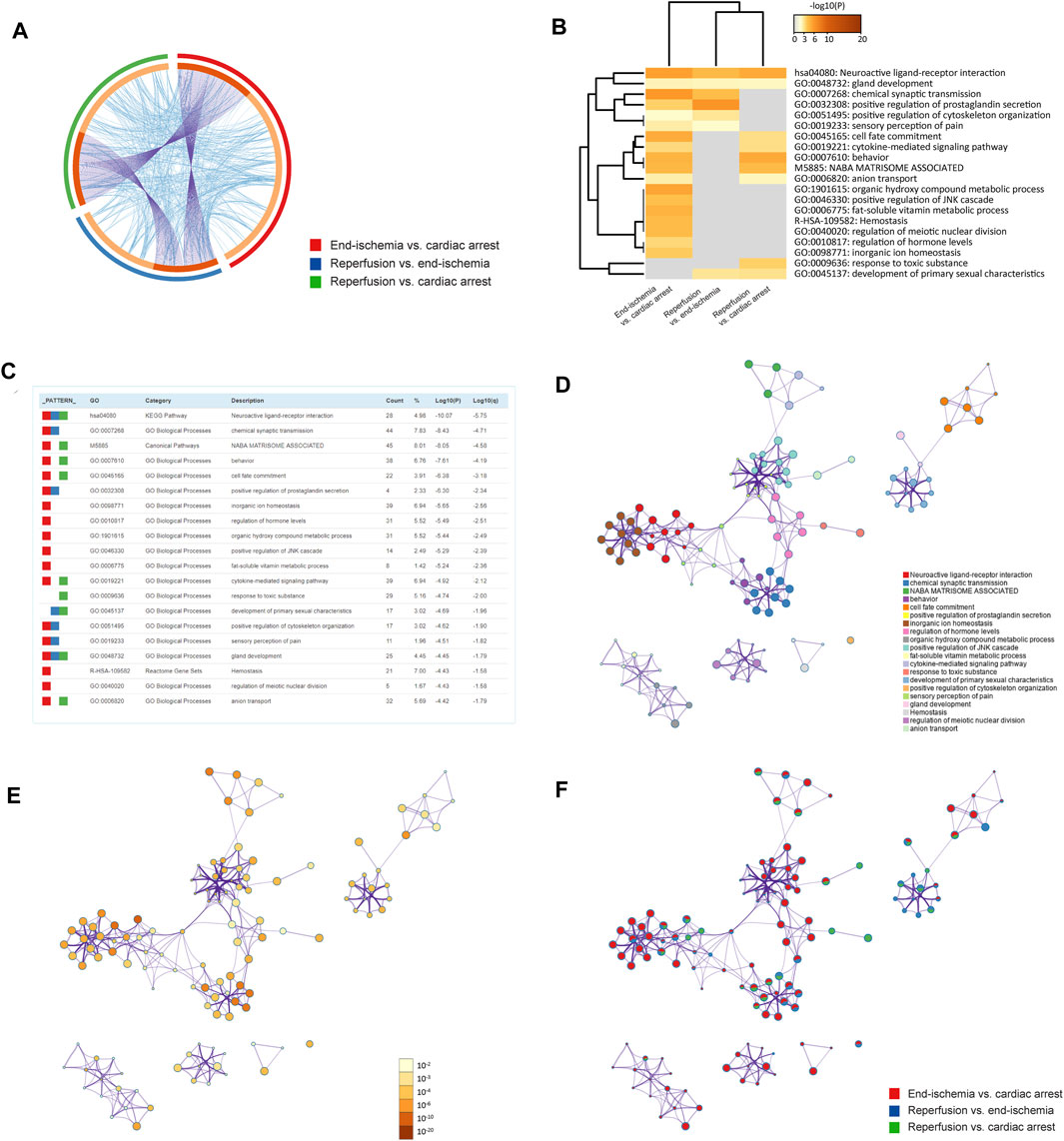
FIGURE 2. Functional enrichment analyses of DEGs. (A) Circos plot showing gene overlaps and genes of the same enriched ontology term. Purple curves link common genes, and blue curves link genes belonging to the same enriched term. Dark orange indicates genes that hit multiple lists, and light orange indicates genes unique to a list. (B) Heatmap showing top 20 enriched terms, colored by p-values. (C) Characteristics of top 20 enriched terms. Count (%) indicates the number (percentage) of genes in the given ontology term. (D) Networks colored by cluster IDs, where nodes with the same ID are close to each other. (E) Networks colored by p-values. (F) Networks with pie charts, color-coded by identities of the genes.
The PPI networks of DEGs were performed to identify key modules with highly clustered proteins using the MCODE functionality. All interactions of the proteins are presented in Supplementary Figure S2. The PPI analyses revealed a total of 10 key modules. The MCODE one contained 26 highly clustered proteins: C5AR1, CASR, CCL21, CHRM4, CNR1, CORT, CXCL13, EDN1, EDN3, GNRH2, GNRHR, GRM8, HCRTR1, HCRTR2, MTNR1A, NMBR, NPY1R, NTS, NTSR1, OPRD1, OPRK1, P2RY10, P2RY6, PPY, TACR3, and XCR1. In the MCODE networks, the interactions among the 26 hub proteins were colored by MCODE cluster IDs (Figure 3A) and identities of the proteins (Figure 3B). The annotations of the 26 genes are presented in Table 3.
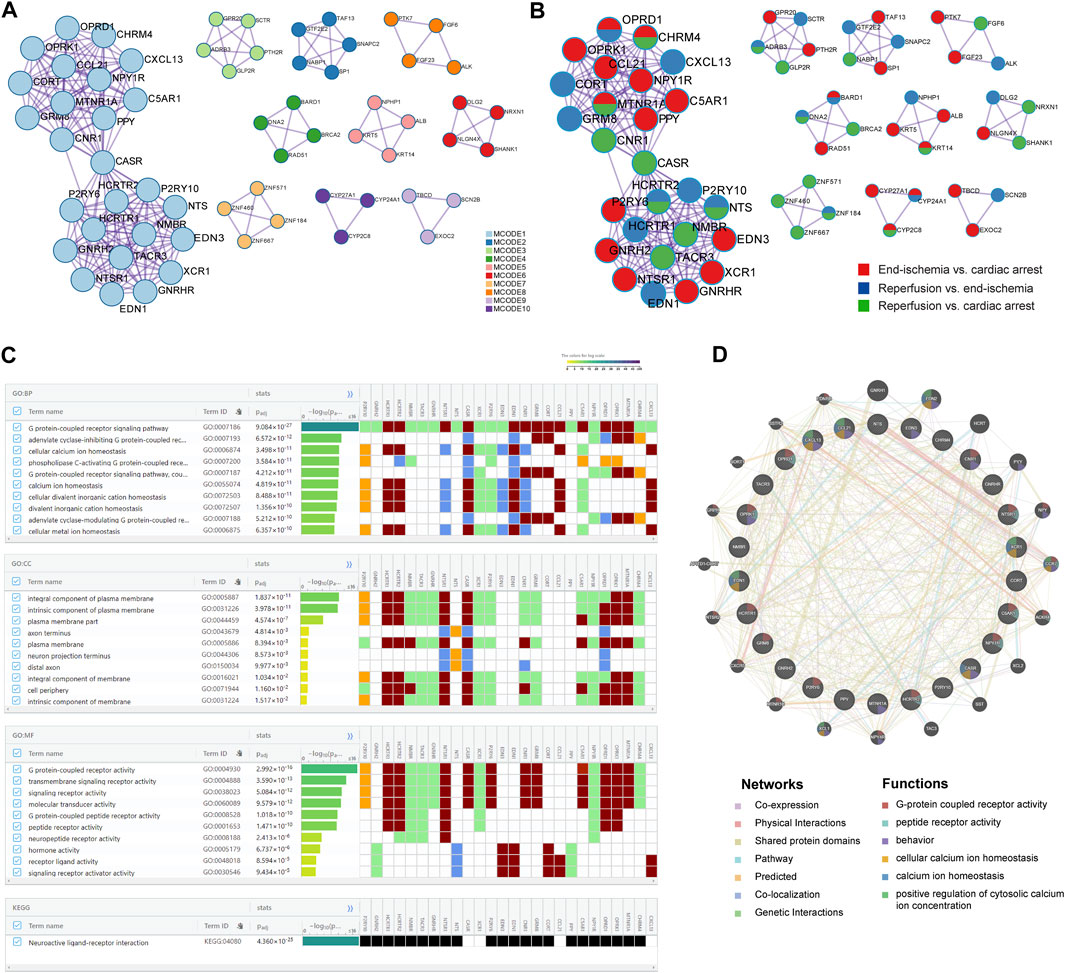
FIGURE 3. Key candidate genes and PPI network analysis. (A) Interactions among hub proteins in MCODE cluster 1. (B) Interactions among hub proteins, colored by identities of the proteins. (C) Top 10 enriched BP, CC, and MF terms, and enriched KEGG pathway. (D) Protein-protein interaction networks and functions.
Based on the 26 genes, GO and KEGG enrichment analyses were performed using the gProfiler. Of the top significantly enriched BP, CC, and MF terms, cellular calcium ion homeostasis (GO:0006874) and calcium ion homeostasis (GO:0055074) were noted in the process of myocardial I/R injury (Figure 3C). Thirteen genes were involved, including P2RY10, HCRTR1, HCRTR2, NTSR1, CASR, XCR1, P2RY6, EDN3, EDN1, CNR1, CCL21, C5AR1, and CXCL13. In addition, KEGG analysis showed one enriched pathway, neuroactive ligand-receptor interaction (Figure 3C), in line with the previous results (Figure 2C). All enrichment results are available in Supplementary Figure S3; Supplementary Table S6.
To further reveal the interactions of the 26 hub proteins, PPI network and functional analysis were conducted using the GeneMANIA (Figure 3D). There were substantial and complex relationships within these proteins, including co-expressions, physical interactions, shared protein domains, pathways, predicted interactions, co-localizations, and genetic interactions (Supplementary Table S7). In addition, the functional analysis revealed several gene functions (Supplementary Table S8). Of note, there were three functions of calcium regulation (cellular calcium ion homeostasis, calcium ion homeostasis, and positive regulation of cytosolic calcium ion concentration). In these calcium-related functions, five hub proteins (CCL21, XCR1, CXCL13, EDN1, and CASR) were revealed, which were significantly enriched in two calcium processes (Figure 3C).
Based on the ischemia samples, GSEA revealed several significantly enriched pathways, including neuroactive ligand receptor interaction, GNRH signaling pathway, calcium signaling pathway, WNT signaling pathway, VEGF signaling pathway, and JAK-STAT signaling pathway (Figure 4A). Based on the reperfusion samples, GSEA revealed several significantly enriched pathways, including neuroactive ligand receptor interaction, VEGF signaling pathway, JAK-STAT signaling pathway, and WNT signaling pathway (Figure 4B). All results of GSEA enriched pathways are available in Supplementary Tables S9, S10.
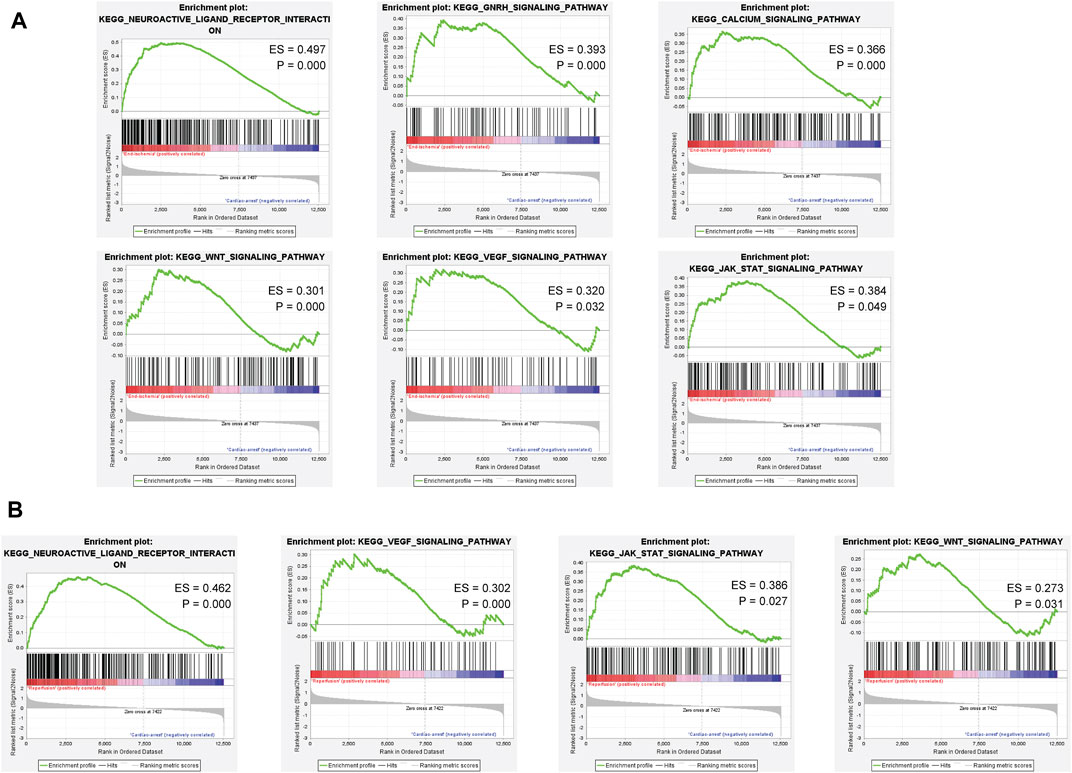
FIGURE 4. Gene set enrichment analysis of the ischemia and reperfusion samples. (A) Enriched pathways in ischemia samples. (B) Enriched pathways in reperfusion samples.
Compared to the sham controls, myocardial I/R injury led to an obvious myocardial infract size (approximately 40%) in the Evans blue/TTC staining (Figure 5A). In addition, HE staining showed remarkable myocardial histological damage following myocardial I/R injury (Figure 5B). The structure of myocardial tissue in the sham group was clear and aligned, with intact myocardial fibers; however, the I/R group showed a damaged tissue structure, swollen cardiomyocytes, disarranged myofibrils, and leukocytes infiltration. The TUNEL assay was performed to assess the level of DNA fragmentation in myocardial apoptotic cells. The results showed a significantly increased TUNEL apoptosis rate induced by myocardial I/R injury (Figure 5C).
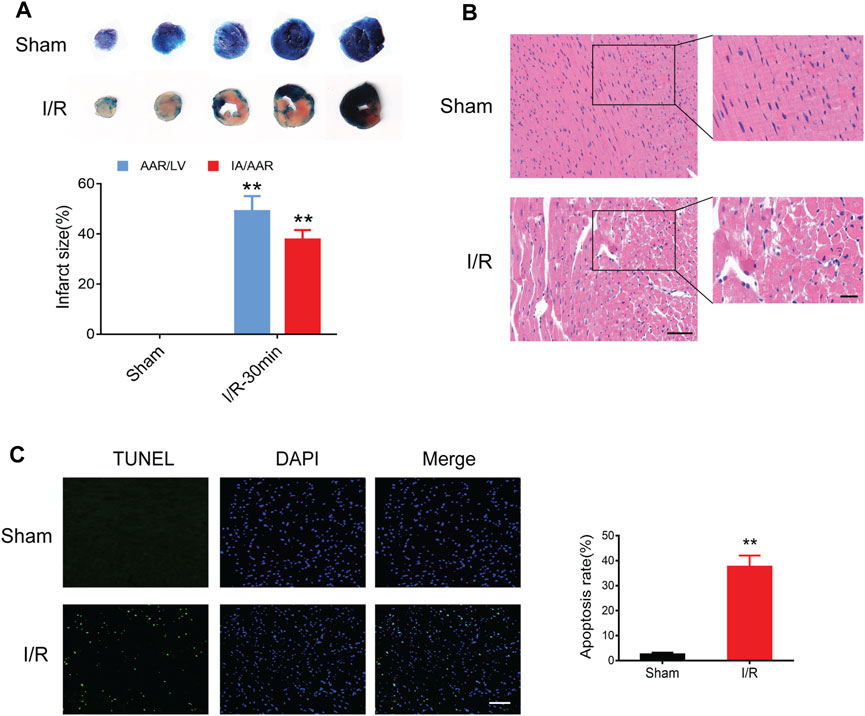
FIGURE 5. Myocardial I/R injury in mice. (A) Evans blue/TTC staining showing myocardial infract size in the sham and I/R groups. (B) HE staining showing myocardial histological damage. Scale bar = 100 μm (left) and 50 μm (right). (C) TUNEL staining showing the apoptosis rate in the myocardium. Scale bar = 100 μm n = 3. **p < 0.01 vs. the sham group. I/R, ischemia reperfusion; AAR, area at risk; IA, infarct area; LV, left ventricle.
Based on the PPI network and hub protein analyses, five key genes (CCL21, XCR1, CXCL13, EDN1, and CASR) were identified. To investigate the role of these genes at the early stage of myocardial I/R injury, the mRNA expression of these genes was determined using the qPCR. The results showed that the mRNA expression of CCL21, XCR1, CXCL13, and CASR were significantly up-regulated in the left ventricular samples of myocardial I/R injury (Figure 6A).
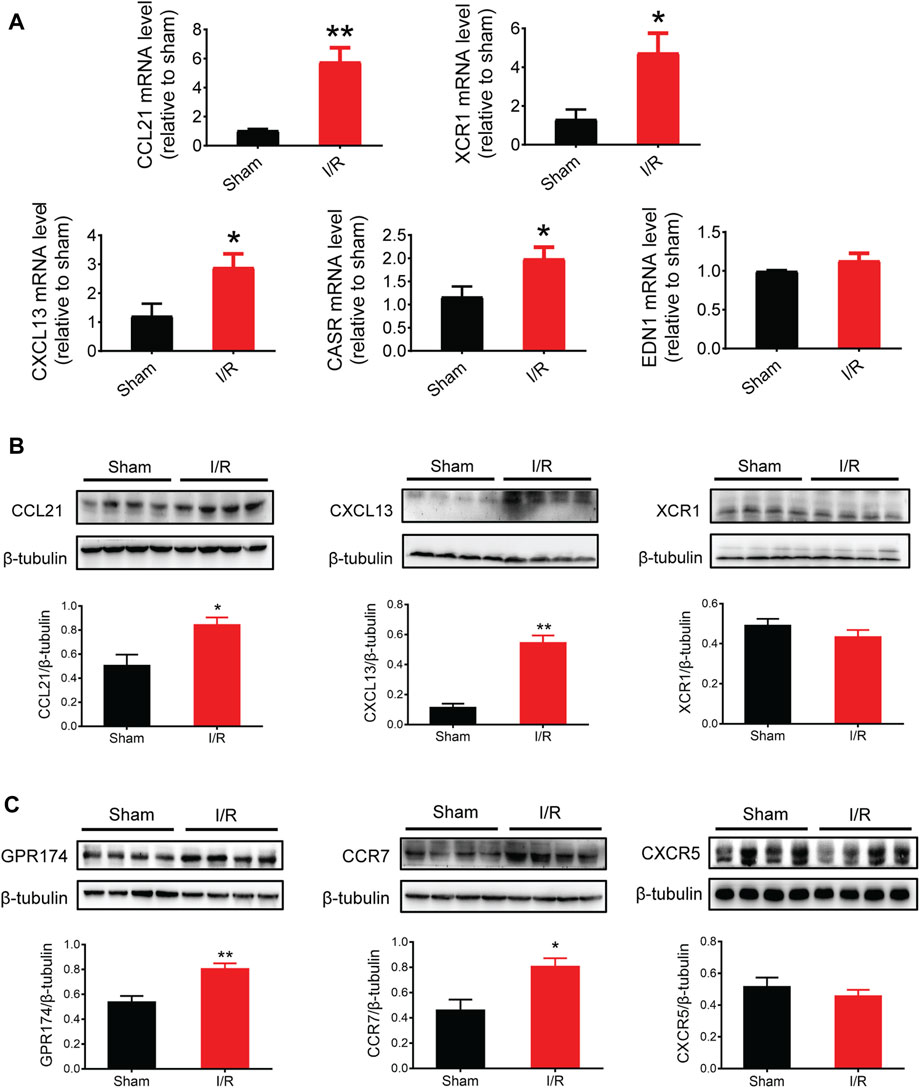
FIGURE 6. mRNA and protein expression of key genes and related receptors. (A) mRNA expression of CCL21, XCR1, CXCL13, CASR, and EDN1. (B) Protein expression of CCL21, CXCL13, and XCR1. (C) Protein expression of GPR174/CCR7 (the receptors of CCL21) and CXCR5 (the receptors of CXCL13). n = 3–6. *p < 0.05, **p < 0.01 vs. the sham group. I/R, ischemia reperfusion.
Given that chemokines are small secreted proteins with chemoattractant properties that play a key role in inflammation and cell apoptosis (Jarrah et al., 2018; Zhang et al., 2020), we assessed the protein expression of CCL21, XCR1, and CXCL13. The results showed that the protein expression of CCL21 and CXCL13 was significantly increased following myocardial I/R injury (Figure 6B). Next, we tested the protein expression of GPR174 and CCR7 (known receptors of CCL21) (Chen et al., 2020; Van Raemdonck et al., 2020; Zhao et al., 2020) and CXCR5 (a known receptor of CXCL13) (Hussain et al., 2019; Shen et al., 2020). We found that the protein expression of GPR174 and CCR7, but not CXCR5, was significantly elevated. These results suggested that the CCL21-GPR174/CCR7 chemokine axis was involved in the early stage of myocardial I/R injury (Figure 6C).
We detected the expression and cellular localization of GPR174 and CCR7 using the immunofluorescence staining. The double-labeling immunofluorescence results showed that there was no GPR174 positive cells (green) co-localized with the α-Actinin positive cells (red) in the sham or myocardial I/R mice, suggesting that GPR174 did not present on cardiomyocytes (Figure 7A). Therefore, we further investigated whether GPR174 was expressed in the cardiac fibroblasts. Following myocardial I/R injury, GPR174 positive cells (green) and Vimentin positive cells (red) co-localized in the left ventricular myocardial tissues, and the expression of GPR174 on cardiac fibroblasts was significantly increased (Figure 7B). Similarly, we also found that myocardial I/R-induced activation of CCR7 was also evident on cardiac fibroblasts (Figure 7C). Taken together, these results suggested that the up-regulated GPR174 and CCR7 on the cardiac fibroblasts underlie the pathological course of myocardial I/R injury.
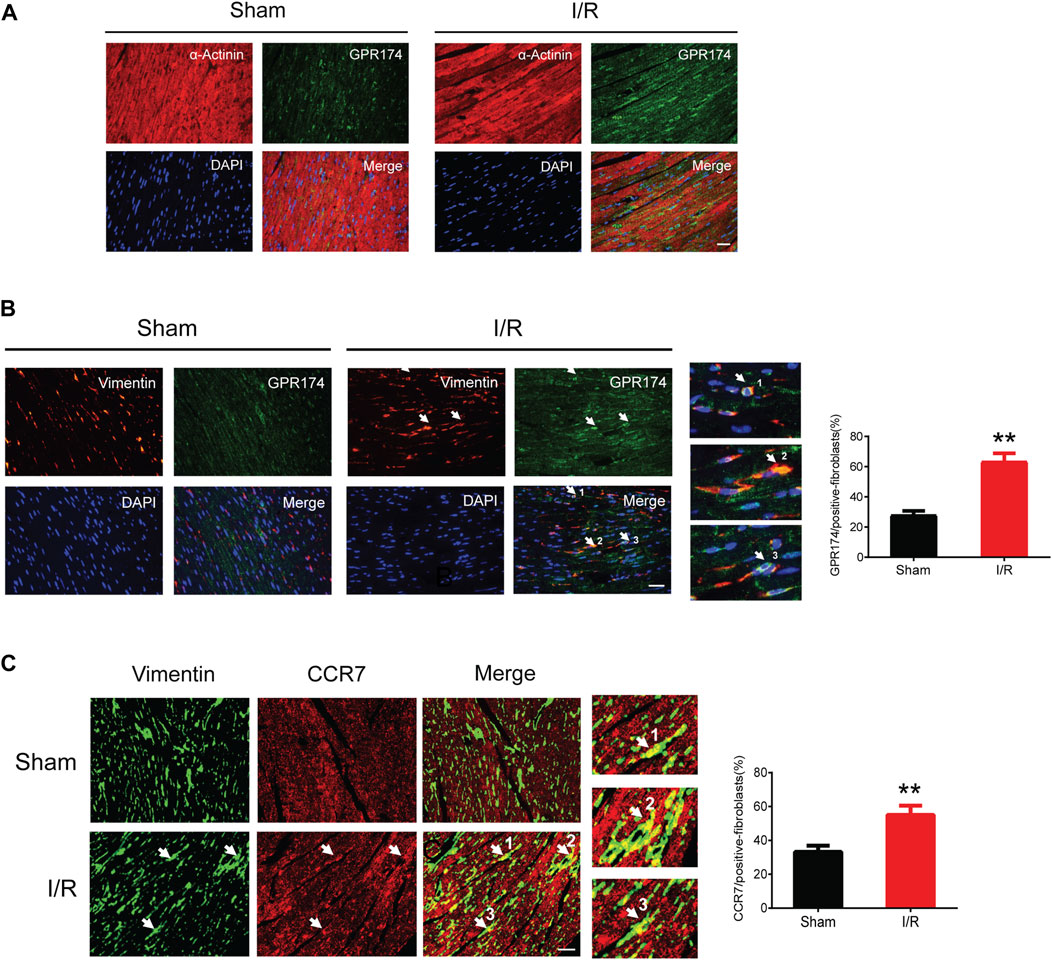
FIGURE 7. Expression of GPR174 and CCR7 on cardiac fibroblasts during myocardial I/R injury. (A) Immunofluorescence staining showing that GPR174 did not present on cardiomyocytes in the sham or I/R groups. (B) Immunofluorescence staining showing the expression of GPR174 on cardiac fibroblasts. (C) Immunofluorescence staining showing the expression of CCR7 on cardiac fibroblasts in the sham and I/R groups. Scale bar = 50 μm n = 3. **p < 0.01 vs. the sham group. I/R, ischemia reperfusion.
In this study, the gene expression profiles of human right ventricular samples during myocardial I/R process in cardiac surgery was explored with the use of a comprehensive bioinformatics approach. We identified 309 DEGs for end-ischemia vs. cardiac arrest, 175 DEGs for reperfusion vs. end-ischemia, and 217 DEGs for reperfusion vs. cardiac arrest. Based on pathway and process enrichment analyses, PPI analysis, and MCODE assay, several key modules with hub proteins were revealed. Further enrichment and network analyses of the highly clustered proteins highlighted five key candidate genes (CCL21, XCR1, CXCL13, EDN1, and CASR) related to calcium homeostasis and regulation. In addition, GSEA confirmed several significantly enriched pathways in the ischemia and reperfusion stages. Next, we performed the validation experiments in animal studies using qPCR and Western blots. The results confirmed that the mRNA and protein expression of CCL21 and CXCL13 were elevated during myocardial I/R injury in mice. Furthermore, myocardial I/R injury induced the activation of GPR174/CCR7 (the CCL21 receptors), but not CXCR5 (the CXCL13 receptor). Notably, the increased expression of GPR174/CCR7 was located within cardiac fibroblasts.
The major difference between our study and the previous one (Arab et al., 2007) is the discovery of novel key candidate genes associated with calcium signaling pathway during the myocardial I/R process in cardiac surgery. Of note, these key genes (CCL21, XCR1, CXCL13, EDN1, and CASR) were significantly enriched in the processes of calcium homeostasis and regulation, including cellular calcium ion homeostasis, calcium ion homeostasis, and positive regulation of cytosolic calcium ion concentration. Given that intracellular calcium overload is a critical mechanism for cardiomyocyte apoptosis and cell death during myocardial I/R injury (Garcia-Dorado et al., 2012; Mattiazzi et al., 2015), the identification of these novel genes may provide a new preventive and therapeutic basis for intraoperative cardioprotection.
Of these five genes, CCL21, XCR1, CXCL13, and EDN1 have yet to be evaluated in the context of myocardial I/R injury. CCL21, chemokine (C-C motif) ligand 21, is a small cytokine belonging to the CC chemokine family. A previous study showed that CCL21 was responsible for ischemic brain damage, while CD93 mediated neuroprotection via suppressing neuroinflammation through downregulation of CCL21 (Harhausen et al., 2010). XCR1, X-C motif chemokine receptor 1, is a chemokine receptor belonging to the G protein-coupled receptor superfamily, which induces calcium mobilization (Yoshida et al., 1998). CXCL13, chemokine (C-X-C motif) ligand 13, has recently been found to play an important role in spinal cord I/R injury (Chen et al., 2019). Specifically, knockdown of CXCL13 by small interfering RNA protected the rats against spinal cord I/R injury via suppressing the activity of relevant signaling pathways and proinflammatory cytokines (Chen et al., 2019). EDN1, endothelin-1, is a potent vasoconstrictor produced by vascular endothelial cells. In a porcine liver transplant mode, improved viability of damaged liver grafts by oxygenated hypothermic machine perfusion was associated with a reduced EDN1 expression (Vekemans et al., 2011). CASR, a calcium-sensing receptor, has been investigated in myocardial I/R injury in recent studies, suggesting that inhibition of CASR attenuated myocardial I/R injury through suppressing endoplasmic reticulum stress-associated apoptotic signaling pathways (Sun et al., 2017; Liu et al., 2019; Yin et al., 2019).
According to the GSEA results, several calcium-related KEGG pathways were significantly enriched in the human right ventricular samples during the ischemia and reperfusion stages, including calcium signaling pathway, GNRH signaling pathway, WNT signaling pathway, VEGF signaling pathway, and JAK-STAT signaling pathway. These findings suggest their important contribution to the cellular calcium homeostasis and regulation in the myocardial I/R process. It has been highlighted that disorder of calcium homeostasis is a pivotal mechanism underlying myocardial I/R injury (Garcia-Dorado et al., 2012; Mattiazzi et al., 2015; Kristiansen et al., 2017). Our recent study also showed that H9c2 cardiomyocytes exposed to oxygen-glucose deprivation/reoxygenation had decreased cell viability, elevated intracellular calcium levels, changes in calcium-related signaling activity, and increased cell apoptosis, which was alleviated by pretreatment with dexmedetomidine, a selective α2-adrenoceptor agonist offering protective effects against I/R injury (Yuan et al., 2019).
Until recently, the relation between CCR7 and cardiac fibroblasts in context of myocardial injury has not been well elucidated. A previous study suggested the redistribution of chemokine receptor CCR7 from peripheral blood to the infarcted heart during myocardial I/R following primary percutaneous coronary intervention procedures (Hoffmann et al., 2012). Another study suggested that CCL21/CCR7 interactions was involved in left ventricular remodeling during pressure overload in patients with aortic stenosis (Finsen et al., 2014). Based on the existing literature, we assume that CCR7 might migrate from peripheral blood to cardiac fibroblasts with an interaction of CCL21, which plays a role in the etiology of myocardial I/R injury.
This study has several limitations. First, the bioinformatic analyses were based on a relatively small number of human ventricular samples from a single microarray data. An integrated analysis of several datasets might have provided more information and insight. Second, the GEO dataset (GSE6381) and ensuring animal studies were based on different species and different parts of heart: right ventricular of human from the GEO dataset, and left ventricular of mice from the animal studies. Although we performed the qPCR, western blots, and immunofluorescence staining experiments to validate the expression of the key genes, the myocardial I/R injury model in mice was different from the human heart samples during cardiac surgery. Hence, the differences in methodology and species should be taken into consideration when interpreting these data, and more studies are still needed to collaborate our findings. Third, the expression profiles of myocardium during intraoperative myocardial I/R obtained in this study was only on the gene level, without assessment and relationships of non-coding RNAs, such as micro RNAs, circular RNAs, or long non-coding RNAs. Last, we showed that myocardial I/R injury increased the expression of CCL21, along with the activation of GPR174/CCR7 within cardiac fibroblasts, but further functional experiments are still needed for mechanistic explorations.
In conclusion, the present study revealed five key genes (CCL21, XCR1, CXCL13, EDN1, and CASR) and calcium homeostasis and regulation pathways during myocardial I/R injury. The expression of CCL21 and CXCL13 were validated in myocardial I/R injury of mice. We highlighted the increased expression of CCL21 with the activation of receptors GPR174/CCR7 on cardiac fibroblasts underlying myocardial I/R injury. Our findings may provide insights into development of preventive and therapeutic targets for intraoperative cardioprotection.
The data presented in the study are deposited in the Jianguoyun/Nutstore repository, available at: https://www.jianguoyun.com/p/Dd2OkgkQxvzPChiIpcMEIAA.
The animal study was reviewed and approved by Ethic Committee for Animal Care and Use of Soochow University.
All authors made substantial contributions to the design of this study, data acquisition and interpretation, statistical plan, drafting the manuscript, or revising the manuscript critically. All authors agreed to be accountable for all aspects of the work and gave their final approval of this version to be published.
This study was supported by National Natural Science Foundation of China (82072130 and 81873925), Natural Science Foundation of Jiangsu Province (BK20191171), Jiangsu Government Scholarship for Overseas Studies (JS-2018-178), 333 High-level Talent Training Project in Jiangsu Province (BRA2020089), and Six Talent Peaks Project in Jiangsu Province (WSN-022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.946524/full#supplementary-material
Arab, S., Konstantinov, I. E., Boscarino, C., Cukerman, E., Mori, A., Li, J., et al. (2007). Early gene expression profiles during intraoperative myocardial ischemia-reperfusion in cardiac surgery. J. Thorac. Cardiovasc. Surg. 134 (74-81), 74–81. doi:10.1016/j.jtcvs.2007.01.025
Bader, G. D., and Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 4, 2. doi:10.1186/1471-2105-4-2
Boag, S. E., Das, R., Shmeleva, E. V., Bagnall, A., Egred, M., Howard, N., et al. (2015). T lymphocytes and fractalkine contribute to myocardial ischemia/reperfusion injury in patients. J. Clin. Invest. 125, 3063–3076. doi:10.1172/JCI80055
Bulluck, H., Yellon, D. M., and Hausenloy, D. J. (2016). Reducing myocardial infarct size: Challenges and future opportunities. Heart 102, 341–348. doi:10.1136/heartjnl-2015-307855
Chen, F., Li, X., Li, Z., Zhou, Y., Qiang, Z., and Ma, H. (2019). The roles of chemokine (C-X-C motif) ligand 13 in spinal cord ischemia-reperfusion injury in rats. Brain Res. 1727, 146489. doi:10.1016/j.brainres.2019.146489
Chen, Y., Shao, Z., Jiang, E., Zhou, X., Wang, L., Wang, H., et al. (2020). CCL21/CCR7 interaction promotes EMT and enhances the stemness of OSCC via a JAK2/STAT3 signaling pathway. J. Cell. Physiol. 235, 5995–6009. doi:10.1002/jcp.29525
Finsen, A. V., Ueland, T., Sjaastad, I., Ranheim, T., Ahmed, M. S., Dahl, C. P., et al. (2014). The homeostatic chemokine CCL21 predicts mortality in aortic stenosis patients and modulates left ventricular remodeling. PLoS One 9, e112172. doi:10.1371/journal.pone.0112172
Garcia-Dorado, D., Ruiz-Meana, M., Inserte, J., Rodriguez-Sinovas, A., and Piper, H. M. (2012). Calcium-mediated cell death during myocardial reperfusion. Cardiovasc. Res. 94, 168–180. doi:10.1093/cvr/cvs116
Ge, L., Cai, Y., Ying, F., Liu, H., Zhang, D., He, Y., et al. (2019). miR-181c-5p exacerbates hypoxia/reoxygenation-induced cardiomyocyte apoptosis via targeting PTPN4. Oxid. Med. Cell. Longev. 2019, 1957920. doi:10.1155/2019/1957920
Harhausen, D., Prinz, V., Ziegler, G., Gertz, K., Endres, M., Lehrach, H., et al. (2010). CD93/AA4.1: A novel regulator of inflammation in murine focal cerebral ischemia. J. Immunol. 184, 6407–6417. doi:10.4049/jimmunol.0902342
Hausenloy, D. J., and Yellon, D. M. (2013). Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J. Clin. Invest. 123, 92–100. doi:10.1172/JCI62874
Hochberg, Y., and Benjamini, Y. (1990). More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818. doi:10.1002/sim.4780090710
Hoffmann, J., Fiser, K., Weaver, J., Dimmick, I., Loeher, M., Pircher, H., et al. (2012). High-throughput 13-parameter immunophenotyping identifies shifts in the circulating T-cell compartment following reperfusion in patients with acute myocardial infarction. PLoS One 7, e47155. doi:10.1371/journal.pone.0047155
Hussain, M., Adah, D., Tariq, M., Lu, Y., Zhang, J., and Liu, J. (2019). CXCL13/CXCR5 signaling axis in cancer. Life Sci. 227, 175–186. doi:10.1016/j.lfs.2019.04.053
Jarrah, A. A., Schwarskopf, M., Wang, E. R., LaRocca, T., Dhume, A., Zhang, S., et al. (2018). SDF-1 induces TNF-mediated apoptosis in cardiac myocytes. Apoptosis 23, 79–91. doi:10.1007/s10495-017-1438-3
Kristiansen, S. B., Haanes, K. A., Sheykhzade, M., and Edvinsson, L. (2017). Endothelin receptor mediated Ca(2+) signaling in coronary arteries after experimentally induced ischemia/reperfusion injury in rat. J. Mol. Cell. Cardiol. 111, 1–9. doi:10.1016/j.yjmcc.2017.07.013
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi:10.1101/gr.092759.109
Li, T., Wernersson, R., Hansen, R. B., Horn, H., Mercer, J., Slodkowicz, G., et al. (2017). A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods 14, 61–64. doi:10.1038/nmeth.4083
Liu, C., Li, H., Zheng, H., Zhai, M., Lu, F., Dong, S., et al. (2019). CaSR activates PKCδ to induce cardiomyocyte apoptosis via ER stress‑associated apoptotic pathways during ischemia/reperfusion. Int. J. Mol. Med. 44, 1117–1126. doi:10.3892/ijmm.2019.4255
Mattiazzi, A., Argenziano, M., Aguilar-Sanchez, Y., Mazzocchi, G., and Escobar, A. L. (2015). Ca2+ Sparks and Ca2+ waves are the subcellular events underlying Ca2+ overload during ischemia and reperfusion in perfused intact hearts. J. Mol. Cell. Cardiol. 79, 69–78. doi:10.1016/j.yjmcc.2014.10.011
Moran, A. E., Forouzanfar, M. H., Roth, G. A., Mensah, G. A., Ezzati, M., Flaxman, A., et al. (2014). The global burden of ischemic heart disease in 1990 and 2010: The global burden of disease 2010 study. Circulation 129, 1493–1501. doi:10.1161/CIRCULATIONAHA.113.004046
Peng, K., Chen, W. R., Xia, F., Liu, H., Meng, X. W., Zhang, J., et al. (2020). Dexmedetomidine post-treatment attenuates cardiac ischaemia/reperfusion injury by inhibiting apoptosis through HIF-1α signalling. J. Cell. Mol. Med. 24, 850–861. doi:10.1111/jcmm.14795
Peng, K., Liu, H., Yan, B., Meng, X. W., Song, S. Y., Ji, F. H., et al. (2021). Inhibition of cathepsin S attenuates myocardial ischemia/reperfusion injury by suppressing inflammation and apoptosis. J. Cell. Physiol. 236, 1309–1320. doi:10.1002/jcp.29938
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. doi:10.1093/nar/gkz369
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi:10.1093/nar/gkv007
Roberts, C. (2008). Modelling patterns of agreement for nominal scales. Stat. Med. 27, 810–830. doi:10.1002/sim.2945
Saldanha, A. J. (2004). Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi:10.1093/bioinformatics/bth349
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shen, Y., Zhang, Y., Chen, L., Du, J., Bao, H., Xing, Y., et al. (2020). Chemokine CXCL13 acts via CXCR5-ERK signaling in hippocampus to induce perioperative neurocognitive disorders in surgically treated mice. J. Neuroinflammation 17, 335. doi:10.1186/s12974-020-02013-x
Song, N., Ma, J., Meng, X. W., Liu, H., Wang, H., Song, S. Y., et al. (2020). Heat shock protein 70 protects the heart from ischemia/reperfusion injury through inhibition of p38 MAPK signaling. Oxid. Med. Cell. Longev. 2020, 3908641. doi:10.1155/2020/3908641
Stark, C., Breitkreutz, B. J., Reguly, T., Boucher, L., Breitkreutz, A., and Tyers, M. (2006). BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539. doi:10.1093/nar/gkj109
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi:10.1073/pnas.0506580102
Sun, J., Wei, T., Bai, S., Zhao, H., Liu, X., Yu, J., et al. (2017). Calcium-sensing receptor-mediated mitogen-activated protein kinase pathway improves the status of transplanted mouse embryonic stem cells in rats with acute myocardial infarction. Mol. Cell. Biochem. 431, 151–160. doi:10.1007/s11010-017-2987-z
Turei, D., Korcsmaros, T., and Saez-Rodriguez, J. (2016). OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 13, 966–967. doi:10.1038/nmeth.4077
Van Raemdonck, K., Umar, S., Palasiewicz, K., Volkov, S., Volin, M. V., Arami, S., et al. (2020). CCL21/CCR7 signaling in macrophages promotes joint inflammation and Th17-mediated osteoclast formation in rheumatoid arthritis. Cell. Mol. Life Sci. 77, 1387–1399. doi:10.1007/s00018-019-03235-w
Vekemans, K., van Pelt, J., Komuta, M., Wylin, T., Heedfeld, V., Detry, O., et al. (2011). Attempt to rescue discarded human liver grafts by end ischemic hypothermic oxygenated machine perfusion. Transpl. Proc. 43, 3455–3459. doi:10.1016/j.transproceed.2011.09.029
Yang, Y. F., Wang, H., Song, N., Jiang, Y. H., Zhang, J., Meng, X. W., et al. (2021). Dexmedetomidine attenuates ischemia/reperfusion-induced myocardial inflammation and apoptosis through inhibiting endoplasmic reticulum stress signaling. J. Inflamm. Res. 14, 1217–1233. doi:10.2147/JIR.S292263
Yin, B., Hou, X. W., and Lu, M. L. (2019). Astragaloside IV attenuates myocardial ischemia/reperfusion injury in rats via inhibition of calcium-sensing receptor-mediated apoptotic signaling pathways. Acta Pharmacol. Sin. 40, 599–607. doi:10.1038/s41401-018-0082-y
Yoshida, T., Imai, T., Kakizaki, M., Nishimura, M., Takagi, S., and Yoshie, O. (1998). Identification of single C motif-1/lymphotactin receptor XCR1. J. Biol. Chem. 273, 16551–16554. doi:10.1074/jbc.273.26.16551
Yuan, M., Meng, X. W., Ma, J., Liu, H., Song, S. Y., Chen, Q. C., et al. (2019). Dexmedetomidine protects H9c2 cardiomyocytes against oxygen-glucose deprivation/reoxygenation-induced intracellular calcium overload and apoptosis through regulating FKBP12.6/RyR2 signaling. Drug Des. devel. Ther. 13, 3137–3149. doi:10.2147/DDDT.S219533
Zhang, J. J., Peng, K., Zhang, J., Meng, X. W., and Ji, F. H. (2017). Dexmedetomidine preconditioning may attenuate myocardial ischemia/reperfusion injury by down-regulating the HMGB1-TLR4-MyD88-NF-кB signaling pathway. PLoS One 12, e0172006. doi:10.1371/journal.pone.0172006
Zhang, W., Zhu, T., Chen, L., Luo, W., and Chao, J. (2020). MCP-1 mediates ischemia-reperfusion-induced cardiomyocyte apoptosis via MCPIP1 and CaSR. Am. J. Physiol. Heart Circ. Physiol. 318, H59–H71. doi:10.1152/ajpheart.00308.2019
Zhao, R., Chen, X., Ma, W., Zhang, J., Guo, J., Zhong, X., et al. (2020). A GPR174-CCL21 module imparts sexual dimorphism to humoral immunity. Nature 577, 416–420. doi:10.1038/s41586-019-1873-0
Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A. H., Tanaseichuk, O., et al. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. doi:10.1038/s41467-019-09234-6
Keywords: myocardial ischemia/reperfusion injury, differentially expressed genes, CCL21, GPR174/CCR7, cardiac fibroblasts, transcriptome
Citation: Meng X-W, Zhang M, Hu J-K, Chen X-Y, Long Y-Q, Liu H, Feng X-M, Ji F-H and Peng K (2022) Activation of CCL21-GPR174/CCR7 on cardiac fibroblasts underlies myocardial ischemia/reperfusion injury. Front. Genet. 13:946524. doi: 10.3389/fgene.2022.946524
Received: 17 May 2022; Accepted: 15 August 2022;
Published: 09 September 2022.
Edited by:
Suowen Xu, University of Science and Technology of China, ChinaReviewed by:
Zhengyuan Xia, The University of Hong Kong, Hong Kong, SAR ChinaCopyright © 2022 Meng, Zhang, Hu, Chen, Long, Liu, Feng, Ji and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fu-Hai Ji, amlmdWhhaXN1ZGFAMTYzLmNvbQ==; Ke Peng, cGVuZ2tlMDQyMkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.