
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 09 September 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.945227
 Alessandra Modi1*†
Alessandra Modi1*† Maria Teresa Vizzari2†
Maria Teresa Vizzari2† Giulio Catalano3
Giulio Catalano3 Rajiv Boscolo Agostini2
Rajiv Boscolo Agostini2 Stefania Vai1
Stefania Vai1 Martina Lari1
Martina Lari1 Chiara Vergata1
Chiara Vergata1 Valentina Zaro1
Valentina Zaro1 Lucia Liccioli4
Lucia Liccioli4 Mariaelena Fedi4
Mariaelena Fedi4 Serena Barone4,5
Serena Barone4,5 Lorenzo Nigro6
Lorenzo Nigro6 Hovirag Lancioni7
Hovirag Lancioni7 Alessandro Achilli8
Alessandro Achilli8 Luca Sineo3
Luca Sineo3 David Caramelli1
David Caramelli1 Silvia Ghirotto2*
Silvia Ghirotto2*Sicily is one of the main islands of the Mediterranean Sea, and it is characterized by a variety of archaeological records, material culture and traditions, reflecting the history of migrations and populations’ interaction since its first colonization, during the Paleolithic. These deep and complex demographic and cultural dynamics should have affected the genomic landscape of Sicily at different levels; however, the relative impact of these migrations on the genomic structure and differentiation within the island remains largely unknown. The available Sicilian modern genetic data gave a picture of the current genetic structure, but the paucity of ancient data did not allow so far to make predictions about the level of historical variation. In this work, we sequenced and analyzed the complete mitochondrial genomes of 36 individuals from five different locations in Sicily, spanning from Early Bronze Age to Iron Age, and with different cultural backgrounds. The comparison with coeval groups from the Mediterranean Basin highlighted structured genetic variation in Sicily since Early Bronze Age, thus supporting a demic impact of the cultural transitions within the Island. Explicit model testing through Approximate Bayesian Computation allowed us to make predictions about the origin of Sicanians, one of the three indigenous peoples of Sicily, whose foreign origin from Spain, historically attributed, was not confirmed by our analysis of genetic data. Sicilian modern mitochondrial data show a different, more homogeneous, genetic composition, calling for a recent genetic replacement in the Island of pre-Iron Age populations, that should be further investigated.
Sicily is the largest island of the Mediterranean Sea, and it is widely known for its key role as a crossroad for human colonization from the early Holocene to the Middle Age. Indirect evidence of human presence in Sicily, based on lithic surface finds (Bianchini, 1972, 1971, 1969), could suggest an early settlement, but the evidence is insufficient and therefore it is thought that the human presence in the island does not pre-date the Last Glacial Maximum (Sineo et al., 2015) (LGM, for a commentary Sineo et al., 2015); however, it is only from the Late Glacial—Epigravettian period that a clear large-scale human presence in the island is documented, with the oldest human remains dated about 14,000 cal BCE (Addaura 1, Mannino et al., 2011). Since the middle Holocene, Sicily was settled by Neolithic Farmers from Anatolia and the Near East, as documented by the presence of Impressed Ware (Stentinello culture) dated about 5,800–5,500 cal BCE (Pessina and Tiné, 2008; Natali and Forgia, 2018). Historical and archaeological data offered a detailed description of more recent population settlements within the island, as the Italic people from Italian peninsula, the Phoenicians, the Greeks, the Romans, the Byzantines, the Arab and the Normans (Finley, 1968; Romano et al., 2021). In the pre-Hellenistic period, three distinct groups of people occupied the island: the Sicels (in the East), the Sicanians (in the West) and the Elymians (in the extreme West). The Sicels spoke an Indo-European dialect of the Italic group, as the Elymians, although ascribed to Trojan origin (Finley, 1968). No trace of the Sicanian language remains, and also their origin is extremely controversial. The Athenian historian Thucydides (VI, 2, 2) indeed postulated they came from Iberia (Finley 1968), while other authors proposed a local origin (Moscati, 1980). Toward the end of the second millennium BCE, Sicily was reached by Phoenician settlers who journeyed from Lebanon across the Mediterranean and established trading stations on promontories and on small islands around the coast. When the Greek colonization began (around the eighth century BCE) the Phoenicians withdrew toward the western corner of the island, possibly causing internal east-west cultural differentiation subsequently reshaped by later conquerors (Byzantines, Islamic and Normans) (Sarno et al., 2017).
Complex demographic and cultural dynamics should have affected the genomic landscape of Sicily at different levels; however, the geographical origin of some of the cultures dwelling in the Island, as well as the relative impact of these migrations on the genomic structure and differentiation of current Sicilian populations is actually largely unknown. The available genetic data are indeed mainly from modern populations, both uniparental (mitochondrial DNA and Y chromosome) (di Gaetano et al., 2009; Romano et al., 2003; Sarno et al., 2014) and autosomal (SNPs; Sarno et al., 2017), and generally only show that present-day Sicilian populations are characterized by shared genetic affinity with the modern inhabitants of Southern Italy and a large portion of eastern Mediterranean shores, have a predominant Neolithic-like component, and a substantial homogenous composition of paternal and maternal genetic pools. When analyzing modern genetic data, however, our ability to infer the genetic impact of different past migration processes is challenged by the number of demographic events and admixture layers involving ancestral populations, and by the level of genetic affinity among the putative sources of admixture. In this case, direct genomic evidence from key ancient populations would hence be fundamental to successfully quantify the historical presence of genetic structure, and to assess the genetic impact of past population dynamics and of the complex colonization histories. Unfortunately, the amount of ancient Sicilian genetic data is limited and scattered over time periods. Up to date, paleogenetics studies have focused mainly on the first colonization of the Island. Recently, the complete mitochondrial genome from Upper Paleolithic San Teodoro two and Mesolithic Oriente B individuals (Modi et al., 2021, 2020) revealed a low genetic diversity of the Sicilian hunter-gatherers presumably because of genetic drift processes during the early stages of colonization. Genome wide data from Late Epigravettian Oriente C individual (Mathieson et al., 2018; Catalano et al., 2020) confirmed the Western hunter-gatherer (WHG) affiliation of the specimen, suggesting a substantial genetic homogeneity among the Mediterranean Epigravettian population. Other whole-genome SNPs from ancient Sicilian individuals from Middle-Neolithic to Late Bronze Age has been recently produced (Fernandes et al., 2020), but the analyses presented in the paper were specifically conducted to extract information about the impact of the Bronze Age migration from Steppe in the islands of Western Mediterranean. Diroma et al., 2021 produced 27 complete mitochondrial sequences from an Iron Age necropolis in Polizzello but did not perform a comparison with other ancient Sicilian genomes, nor with other continental populations.
In this work, we reconstructed 43 complete or nearly complete mitochondrial genomes from individuals retrieved in five necropolises located in the island of Motya (western coast of Sicily) and in the archaeological sites of Baucina, Mokarta, Lilibeo and Ispica (see Figure 1), with the final aim of exploring the ancestral genetic structure of the Island and contextualizing this variation with that present in coeval groups from the Mediterranean area, as well as with that of its current inhabitants. To this scope we combined the newly generated sequences with other Sicilian mitochondrial genomes available from literature (Fernandes et al., 2020; Diroma et al., 2021), and with an extended dataset of about 300 complete mitochondrial sequences from the Mediterranean basin (Supplementary Table S1). To make up for the lack of unbiased modern mito-genetic data from Sicily, often focusing on specific macro-haplogroups and hence not being representative of the population’s genetic variability, we obtained a new mito-haplogroup assignment from 175 modern individuals. These samples come from all extant administrative provinces in Sicily, and may be properly compared with the haplo group variation retrieved in the ancient samples. Our results indicate a notable genetic structure within Sicily since the Early Bronze Age, in agreement with the complex historical cultural background of the Island. Approximate Bayesian Computation (ABC) model testing further allowed us to make explicit predictions about the origin of one of the most impressive cultures of Sicily, the Sicanian culture. Finally, the comparison with modern data advocates for a recent homogenization of the genetic variation, possibly resulting from a recent genetic turnover of pre Iron-Age populations whose dynamics should be further investigated.
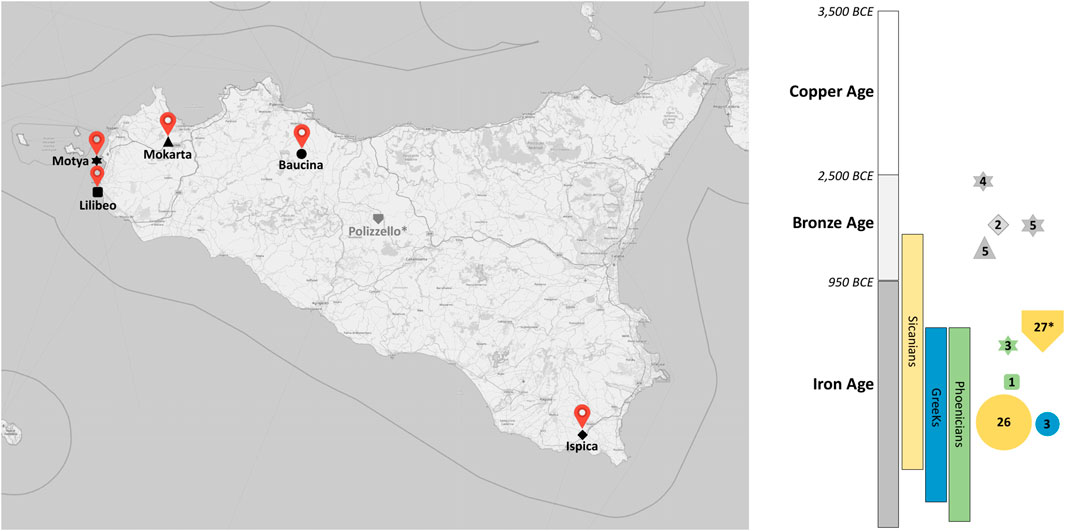
FIGURE 1. Samples geographical and chronological distribution. Map of Sicily showing the location of the archaeological sites from which individuals were sampled. Time period and civilizations covered in this study are shown by color blocks with collected samples represented on the right side. The size of the symbols is proportional to the number of individuals. *Samples from Diroma et al., 2021.
Ancient sample preparation and sequencing. We collected petrous bones and teeth of 58 individuals from five different archeological sites: 16 specimens from Motya, 34 from Baucina, five from Mokarta, one from Lilibeo and two from Ispica, spanning from the Bronze Age to the Hellenistic period (Figure 1 and Supplementary Table S2). For Baucina, we analyzed samples from two cultural contexts classified on the basis of the burial rites: 31 Sicanian and three Greeks (Belvedere et al., 2017). Sampling location and archaeological context of the samples we generated are detailed in Supplementary Data. Molecular analysis was carried out at the Laboratory of Anthropology (University of Florence) in exclusively dedicated ancient DNA facilities and following strict guidelines to avoid contaminations (Gilbert et al., 2005; Willerslev and Cooper, 2005). Moreover, blank controls were included during all DNA extraction and library preparation experiments to monitor for contamination of the reagents. Before powder sampling, the outer layer of temporal bones and teeth was brushed with disposable tools and irradiated by ultraviolet light (254 nm) for 30 min to remove external contaminants. For petrous bones, powder was collected from the densest part of the inner ear as described in Pinhasi et al., 2015 using a dentist microdrill with disposable tips. For teeth, powder was recovered from the inner dentine in root canals, taking care to preserve the crown morphology. For each sample, approximately 50 mg of bone or dentine powder were used for DNA extraction, applying a protocol optimized to retain short DNA fragments (Dabney et al., 2013). A double-strand and double indexed library was generated from 20 µL of each extract (Meyer and Kircher, 2010). No uracil-DNA glycosylase (UDG) treatment was performed for 29 libraries of Baucina samples, while all the other libraries were processed with partial UDG treatment (Rohland et al., 2015) for reducing the rate of deamination (Supplementary Table S2). Each library was then enriched for mtDNA following a multiplexed capture protocol as described in Maricic et al., 2010. After quality control, libraries were sequenced on Illumina MiSeq in paired-end mode, setting 2 × 75 + 8 + 8 run parameters.
Mitochondrial consensus sequences reconstruction and contamination estimation. Sequencing data were demultiplexed and sorted according to the individual sample barcodes added to each DNA fragment during library preparation. Raw reads were then processed using the pipeline described in Peltzer et al., 2016. Adapters were trimmed and reads with a minimum overlap of 10 bp were merged in a single sequence using Clip & Merge v1.7.4. Merged reads were mapped on the revised Cambridge Reference Sequence (rCRS, NC_012920) using CircularMapper and BWA v.0.6.2 (Li and Durbin, 2009), setting -l 16,500 -o two and -n 0.01 parameters to optimize alignment performance with degraded DNA (Schubert et al., 2012). Reads with mapping quality below 30 were discarded. PCR duplicates were removed using DeDup v0.12.1, a tool that takes both mapping positions (start and end) into account to identify duplicates. Degradation patterns (i.e fragment length and deamination rates at the ends of the molecules) were estimated using MapDamage 2.0 (Jónsson et al., 2013). Schmutzi software (Renaud et al., 2015) was used for endogenous consensus calling and for estimating present-day human contamination (details in Supplementary Data). Bases with individual likelihood <20 were considered as missing positions (Ns) in the reconstructed mitogenome sequences (see Supplementary Tables S3, S4). For each sample, mitochondrial haplogroup was determined with Haplogrep2 (van Oven, 2015; Weissensteiner et al., 2016) on the command line, using fasta file as input.
Radiocarbon dating. Radiocarbon measurements were performed on collagen extracted from petrous bones according to the modified-Longin method. After reducing to graphite the collected material, C-14 concentration in unknown samples was measured by Accelerator Mass Spectrometry (Fedi et al., 2007). Conventional radiocarbon ages were converted to calibrated ages using OxCal 4.3 by the intCal13 calibration curve (Reimer et al., 2013).
Data collection and processing. We collected 300 high quality complete mitochondrial genomes from the literature accessible on public databases such as The European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) and GenBank (https://www.ncbi.nlm.nih.gov/genbank). The samples from Fernandes et al. (2020) were mapped to the reconstructed human mtDNA consensus sequence (RSRS, Behar et al., 2012) so we decided to re-align these sequences to the Revised Cambridge Reference Sequence (rCRS, GenBank Accession Number NC_012920) in order to make them comparable to the other collected sequences. We mapped the raw reads using BWA-aln v.0.7.15 (Li and Durbin, 2009), with parameters -n 0.01 and, -o 2 -L 16,500 and used picard-tools-1.98 (http://picard.sourceforge.net/) to add read groups (AddOrReplaceReadGroups) and to remove duplicated reads from the obtained BAM files (MarkDuplicates).
Reads showing a mapping quality below 30 were filtered out from all the alignment files collected in our datasets. We then used the schmutzi pipeline described above to estimate the endogenous mitochondrial consensus sequence for each sample.
Finally, we performed a multiple sequence alignment of our dataset exploiting the software MAFFT (Katoh and Standley, 2013) with the following options FFT-NS-2 and-maxiterate 100.
Modern dataset. The modern collection consisted of 175 biological samples from unrelated subjects with a Sicilian maternal grandmother as a terminal maternal ancestor (TMA) from all administrative provinces of Sicily. After signing an informed consent, each volunteer provided a saliva sample, through mouthwash rinse, and a pedigree chart with known genealogical data. This genealogical information was analyzed to reallocate the samples based on their terminal maternal ancestor (TMA), as follows: Agrigento (n = 18), Caltanissetta (n = 29), Catania (n = 10), Enna (n = 5), Messina (n = 6), Palermo (n = 81), Ragusa (n = 8), Siracusa (n = 5) and Trapani (n = 13), thus representing all the nine administrative provinces of the Island. An anonymous code was assigned to each participant and mitochondrial control region sequence was obtained, as previously described (Modi et al., 2020).
Genetic structure and differentiation. All mtDNA haplotypes were classified into haplogroups using Haplogrep 2.0 (Weissensteiner et al., 2016) according to the most up-to-date mtDNA phylogeny, as reported in PhyloTree build 17 (http://www.phylotree.org/) (van Oven and Kayser, 2009). The consistency of haplogroup classifications based on control-region and full mitogenome data in Eurasian populations have been confirmed in recent papers (Modi et al., 2020; Cardinali et al., 2022). A diachronic comparison between modern and ancient individuals was achieved through a principal component analysis (PCA) based on haplogroup frequencies (Supplementary Table S5) and conducted by employing the function fviz_pca_biplot from the R-package factoextra (Kassambara and Mundt, 2017).
We also calculated FST values between groups with Arlequin v.3.5 (Excoffier and Lischer, 2010) and visualized them by plotting a heatmap using the function levelplot from the R-package Lattice (Sarkar, 2008). The FST values were also visualized by Multidimensional Scaling (MDS), using the cmdscale function in the R environment.
Phylogenetic inference. The phylogenetic network based on nucleotide variation in the complete mitochondrial genomes, was constructed using the Median-Joining algorithm (Bandelt et al., 1995) implemented in PopART software (http://popart.otago.ac.nz/). We built a Neighbor-Joining phylogenetic trees (Saitou and Nei, 1987) directly on mtDNA sequences using the software MEGA (Tamura et al., 2007) based on pairwise molecular distances corrected using Kimura 2 parameters model (bootstrap value = 500).
Model comparison through Approximate Bayesian Computation. In order to give insights into the origin of Sicanians from Baucina we designed two alternative models of evolution (Continuity and Discontinuity), detailed in Supplementary Table S6, that have been compared through an ABC approach (Supplementary Data for details of the methods and of the demographic models tested). We generated 50,000 simulations under each model, with parameters values drawn from prior distributions (Supplementary Table S6), using fastsimcoal2 (Excoffier et al., 2021) within the ABCToolbox suite (Wegmann et al., 2010). We summarized our data calculating different summary statistics with arlsumstat (Excoffier and Lischer, 2010): number of haplotypes (K), Heterozygosity (H), number of Segregating sites (S), Tajima’s D (TAJIMAD), Fixation index (Fst) and Pairwise differences (Pi). For the model comparison we exploited a machine-learning algorithm named Random-Forest (ABC-RF) (Pudlo et al., 2016). Under the ABC-RF approach the model selection analysis is treated as a classification problem; the classifier is constructed from simulations via the RF algorithm and hence applied to the observed data in order to estimate the posterior probability of the most supported model. To perform the model selection procedure, we used the function abcrf from the R package abcrf, employing a forest of 500 trees (Pudlo et al., 2016). Moreover, following Pudlo et al. (2016), we also included the linear discriminant analysis (LDA) axes as additional summary statistics, enabling the flag LDA = TRUE of the function abcrf. LDA is a method of dimensionality reduction that provides a linear projection of the simulated and observed dataset on a space of dimension M−1 (where M is the number of models tested) called the LDA axes, that optimize the differentiation between the models. The projection of the simulated reference table datasets and of the observed dataset on the first LDA axes gives a visual way to evaluate how much the models are differentiated and if the simulated genetic variation overlaps with the observed variation. Before calculating the posterior probabilities of the most supported model, we computed the confusion matrix and evaluated the out-of-bag classification error.
We obtained complete or nearly complete mitochondrial sequences from 43 individuals, with an average coverage between 6x and 511x and modern human contamination level lower than 6% for all the samples with the exception of the sample MOK2 that showed a contamination level of 32% (Supplementary Tables S3, S4, Supplementary Data for details). Radiocarbon dating was performed on collagen extracted from petrous bones of individuals from Motya according to the modified-Longin method (Fedi et al., 2007). Radiocarbon dates allowed us to identify samples from two different phases: Bronze Age individuals related to the prehistoric occupation of the island and three individuals from the Phoenician period.
The novel sequences have been deposited in GenBank under accession number: ON496938-ON496980.
For the population genetic analyses we subdivided the newly generated ancient samples into seven groups, considering geographic criteria (i.e., sampling locations) and/or historical and cultural information (i.e. Bronze Age and Phoenician individuals from Motya were treated as two distinct groups and the same was for Sicanian and Greeks from Baucina). Samples that showed a mean coverage <20x and a contamination level >6% were excluded (6 individuals from Baucina and one from Mokarta). A detailed description of the samples analyzed is shown in Table 1.
We combined the newly produced mitochondrial sequences with data collected from the literature (Lazaridis et al., 2017, 2016; Hofmanová et al., 2016; Lipson et al., 2017; Martiniano et al., 2017; Olivieri et al., 2017; Olalde et al., 2019, 2018; Emery et al., 2018; Fregel et al., 2018; Zalloua et al., 2018; González-Fortes et al., 2019; Villalba-Mouco et al., 2019; Fernandes et al., 2020; Marcus et al., 2020; Rivollat et al., 2020; Skourtanioti et al., 2020; Diroma et al., 2021). We included Sicilian individuals from Fernandes et al. (2020) as other samples from populations of the Mediterranean (Supplementary Table S1). In total we considered 336 individuals from 35 populations, spanning a time range from Neolithic to Iron Age.
Figure 2 panel A shows the distribution of mitochondrial haplogroup frequencies in the whole dataset we built. The haplogroup assignment is also reported in Table 1 and in Supplementary Table S1. In the newly sequenced Sicilian samples, the most frequent haplogroups observed are T and H which reached a frequency of 41% in the Sicanian group from Baucina (n = 17) and a frequency of 37,5% in the Motya Bronze Age (n = 8) group, respectively. The same haplogroups are represented at high frequencies in previously reported samples from Early Bronze Age Sicily and mainland Iron Age Italy. The Sicilian samples attributed to the Phoenician culture belong to HV and V haplogroups, today found predominantly in the Near East and Caucasus (HV) and Northern Europe (V) (Torroni et al., 2001; Shamoon-Pour et al., 2019). The Bronze Age sample of Motya does not show any HV and V haplogroups, and single individuals reporting T, K, I, and U5. In this group, we also found the presence of a L3 lineage, currently found at high frequency in Northeast Africa and possibly highlighting a genetic link with the African continent during the Bronze Age. The Baucina samples show a variety of haplogroups composition, with Sicanian individuals showing a high percentage of T lineages, followed in frequency by H and U5. The three Greek individuals are assigned to three different haplogroups, H, V, and U3. In the Bronze Age Ispica, we identified one X and 1 K individual. The haplogroups differentiation of the individuals in the dataset is also represented by the Neighbor-Joining (NJ) tree (Supplementary Figure S1), whereas the phylogenetic relationship among haplotypes is represented by the phylogenetic network (Figure 3). The NJ tree (Supplementary Figure S1) reflects the known phylogenetic relationship among mitochondrial haplogroups. Phoenicians in Motya and Lilibeo cluster together, close to Bronze Age and Iron Age samples from Iberian Peninsula, and a Sardinian and an Italian sequence. Sicanians of Baucina and Iron Age indigenous from Polizzello share closeness within the haplogroup T. Bronze Age Motya individuals are spread along the whole tree. The phylogenetic network (Figure 3) describes the genetic relationship among the 336 sequences in the dataset we analyzed. Most of the mitochondrial genomes are unique haplotypes, reported in the network as small circles of a single color. The closeness among sequences in the phylogenetic network mirrors the structure that emerged from the NJ tree.
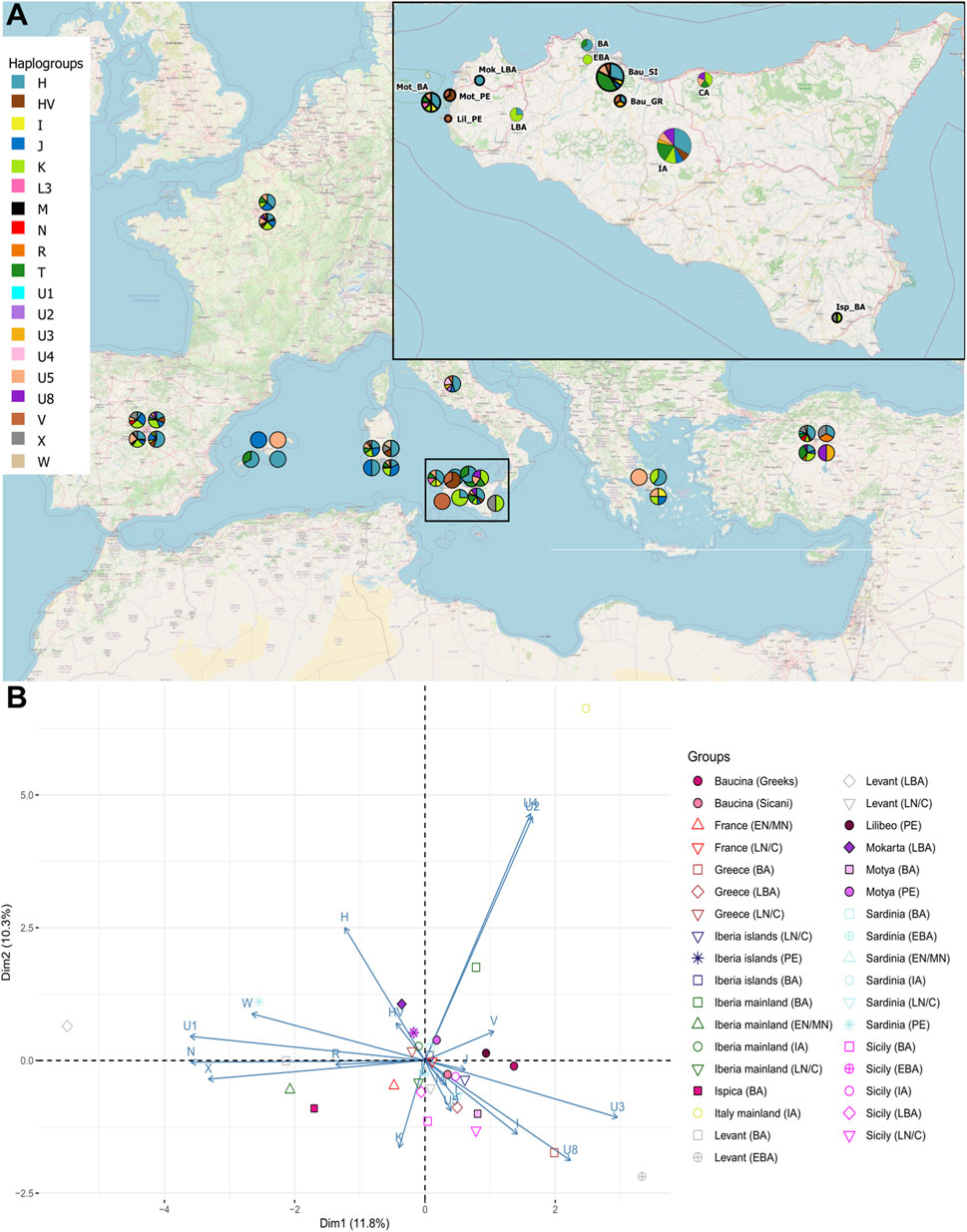
FIGURE 2. Mitochondrial genetic structure of ancient Sicily. (A) Geographical distribution of haplogroup frequency in the analyzed samples. (B) Principal Component Analysis (PCA) on the haplogroup frequencies within groups; geographical population are indicated with different colors, while cultures are indicated with different symbols and labelled as follow: EN/MN = Early/Middle Neolithic, LN/C= Late Neolithic/Chalcolithic, BA = Bronze Age, EBA = Early Bronze Age, LBA = Late Bronze Age, IA = Iron Age, PE = Phoenicians.
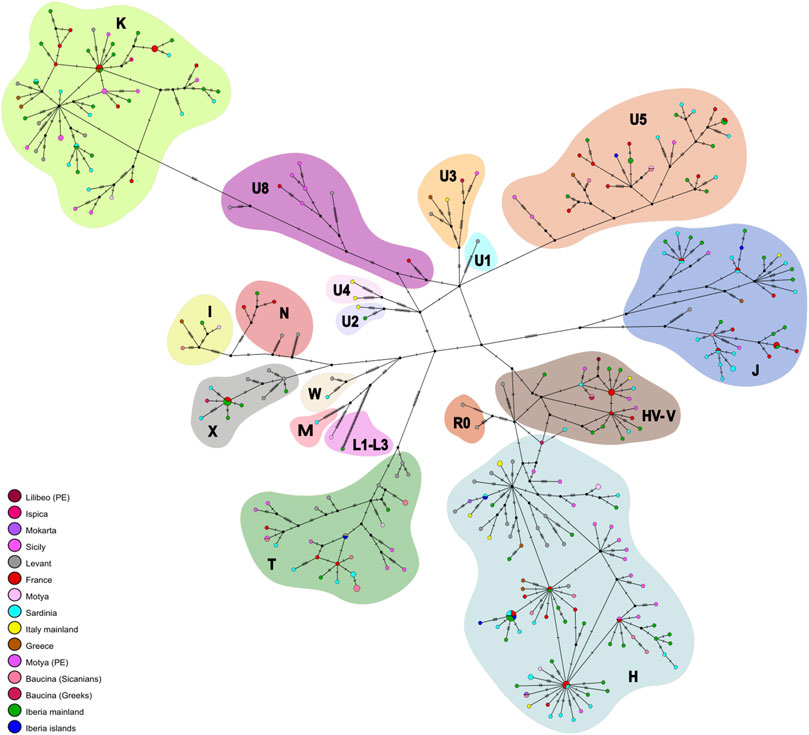
FIGURE 3. Mitochondrial genetic structure of ancient Sicily. Median joining network. Phylogenetic network based on nucleotide variation in the complete mitochondrial genomes.
To better explore the genetic relationship among Sicilian samples we run a Principal Component Analysis (PCA), on the haplogroup frequencies within groups (Figure 2B). In Figure 2B the Sicilian samples are compared with all the Mediterranean populations, from Early Neolithic to Iron Age. With respect to other samples from the Mediterranean Area, Sicilian group occupy a wider range of genetic variation, with the sample from Ispica falling far from other Sicilian individuals, and closer to Bronze Age samples from the Levant and Early Neolithics from mainland Iberia. Phoenicians of Motya and Lilibeo fall closer between each other than with the Bronze Age individuals of Motya. There is no particular closeness with other Phoenicians in Sardinia and Iberia islands. For the Baucina site, the two populations - Sicanians and Greeks - defined through the burial rituals (i.e., structure and design of graves) display genetic differences clearly evident in the PCA (Figure 2B). Moreover, Sicanians fall closer to the Iron Age Sicilian samples from Polizzello, which have also been attributed to the Sicanian culture. The same pattern is evident even taking into account molecular distance among groups instead of haplogroups frequency composition, as shown in the MDS analysis reported in Figure 4A This analysis further highlights the genetic divergence between Sicanians and Greeks in Baucina, with Sicanians falling again close to Iron Age Polizzello, and Baucina Greeks overlapping to the Late Bronze Age Greece and being close to Iron Age mainland Italy and Early Bronze Age Sardinia. The Bronze Age/Iron Age samples fall mainly together in the right portion of the MDS, while Neolithic/Chalcolithic samples occupy the left side of the plot. Exceptions to this pattern are Baucina Sicanians and Iron Age Polizzello, which are closer to Neolithic/Chalcolithic groups. Phoenicians in Motya, Sardinia and Iberia Islands continue to be well separated.
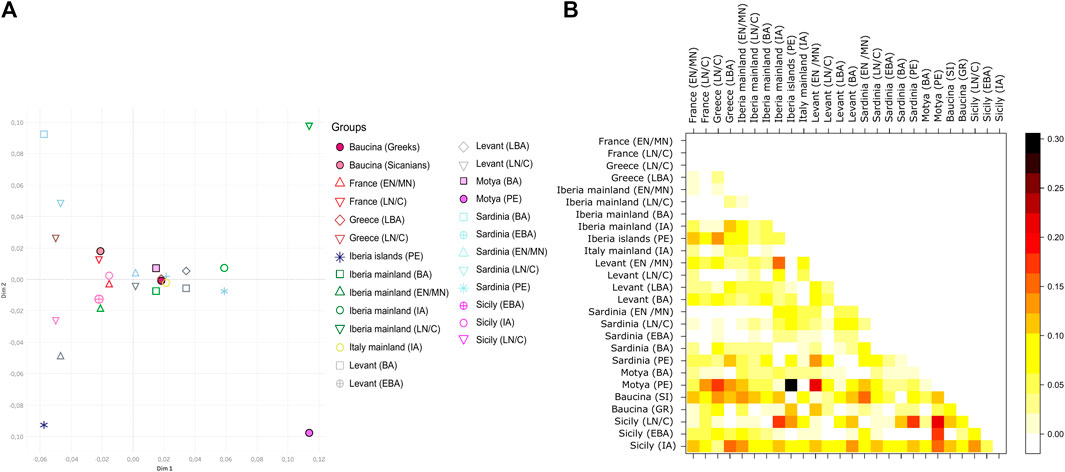
FIGURE 4. Pattern of similarities with populations of Mediterranean. (A) Multidimensional Scaling (MDS) based on a matrix of molecular distances (FST) among groups; geographical population are indicated with different colors, while cultures are indicated with different symbols. (B) Heatmap describing a matrix of molecular distances (FST) among groups, each group is indicated by a number. Cultures are labelled as follow: EN/MN = Early/Middle Neolithic, LN/C= Late Neolithic/Chalcolithic, BA = Bronze Age, EBA = Early Bronze Age, LBA = Late Bronze Age, IA = Iron Age, PE = Phoenicians, SI = Sicanians and GR = Greeks.
Figure 4 panel B and Supplementary Figure S2 quantify the level of populations’ differentiation due to genetic structure of our dataset. From the heatmap plot of Supplementary Figure S2 it is evident that, among the Sicilian groups, Sicily Late Bronze Age is the most differentiated group, with FST values that reaches 0.5 with Phoenicians in Motya. Sicily Late Bronze Age rather shows genetic similarities with Late Neolithic/Chalcolithic European groups (FST values ranging from 0.07 to 0.12) and in particular with Late Neolithic/Chalcolithic and Bronze Age Greek groups (FST values below 0.05), as also highlighted in Fernandes et al. (2020). To better explore the genetic structure among other groups, we excluded Late Bronze age Sicily from the heatmap, and the resulting picture is shown in Figure 4B. The genetic differentiation among Sicilian groups is higher with respect to that observed in other regions of the Mediterranean. Compared with Sardinia (the other main island of the Mediterranean Sea), the genetic structure within Sicily is stronger, with pairwise populations FST values reaching 0.2–0.3, against a maximum of 0.1 among Sardinian groups. Among the Sicilian groups, the most differentiated sample is Phoenicians in Motya. Interestingly the same group, compared with other populations analyzed, shows the maximum level of genetic differentiation with Phoenicians in the Iberia islands, and, to a lesser extent, with Neolithic/Chalcolithic populations. The most similar group to the Phoenicians in Motya, other than their Bronze Age neighbors, is the Levant Late Bronze Age, with an FST value of 0.011. The Late Neolithic/Chalcolithic group in Sicily also have a high level of genetic differentiation with the other groups, thus highlighting a possible genetic structure since the first colonization by early farmers. The same group shows the lowest level of differentiation with Early/Late Neolithic groups in Sardinia. The Sicanians of Baucina do not show any particular resemblance with populations from Iberian Peninsula, as first postulated by Thucydides (VI, 2, 2); the groups with which they show the lowest differentiation are Neolithic and Late Bronze Age Levant.
We also compared the ancient mitochondrial genetic structure of Sicily with that present in modern populations of the Island. To this aim, we analyzed the mtDNA haplogroup distribution of a novel dataset of 175 modern individuals from different geographic areas of the region. The most common western Eurasian macro-haplogroups have been identified in modern Sicilians (Supplementary Table S5). The most frequent haplogroup in modern Sicilians is H (34.4%), followed by T (12.6%), L (12.0%), HV (7.4%) and J (6.3%). Haplogroup H is homogeneously distributed throughout the region. Instead, T mtDNAs are absent in the southern part of the region (Ragusa and Siracusa) and very rare (<1.0%) in the north-western provinces (Palermo and Trapani). The high incidence of L mtDNAs in Palermo (23.5%) could be explained by migrations from northern Africa in rather recent historical times (Cerezo et al., 2012). This is also confirmed by their absence in virtually all ancient individuals, with the only notable exception of an ancient L3 mtDNA from Motya. In general, the higher number of macro-haplogroups identified in modern individuals than in ancient ones (18 vs. 11; Supplementary Table S5) could be easily explained by their wider geographical origin. We further explored this evidence through a Principal Component Analysis based on haplogroup frequencies (Figure 5). The PCA plot emphasizes the differences between ancient and modern Sicily, with all the modern groups falling quite far from the variation generated by the ancient individuals. Enna and Siracusa show more resemblance with ancient groups, mostly due to an unexpected high incidence of haplogroup K in these two modern populations (20% in both), which should be further explored in a larger dataset.
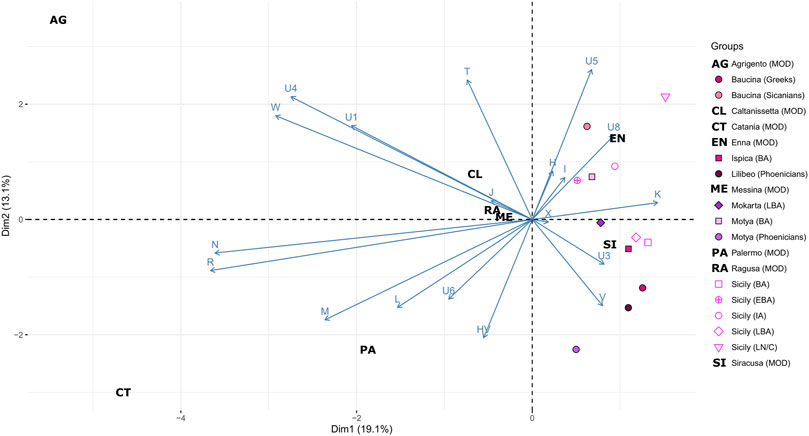
FIGURE 5. Mitochondrial genetic structure of ancient and modern Sicily. Principal Component Analysis (PCA) on the haplogroup frequencies within Sicilian groups including contemporary populations. Geographical ancient populations are indicated with different colors, while modern populations (MOD) are indicated with letters. Cultural information of the ancient population is represented using distinct symbols and labelled as follow: LN/C= Late Neolithic/Chalcolithic, BA = Bronze Age, EBA = Early Bronze Age, LBA = Late Bronze Age, IA = Iron Age, PE = Phoenicians.
Among the ancient populations dwelling in Sicily, the Sicanians are known to have the most controversial origin. Thucydides (VI, 2, 2) claims that they descended from Iberians, while other authors have described Sicanians as Illyrians descendants (Fine, 1985) or developed locally from Neolithic/Bronze Age people (Diodorus Siculus V, 6,1–3). We exploited the inferential framework of Approximate Bayesian Computation to investigate the origin of Sicanians from Baucina and their relationships with other Sicilian groups (details of the approach in Supplementary Data). For this analysis we decided to exclude the late Bronze Age Sicilians samples collected from Fernandes et al. (2020) due to their higher genetic affinities with Neolithic/Chalcolithic Mediterranean groups rather than with coeval Sicilians groups.
We defined two alternative demographic models as shown in Figure 6. The first model, “Continuity”, considers that Sicanians from Baucina are direct descendants of early Bronze Age Sicilian populations, thus assuming a local development of Sicanian culture. The second model, “Discontinuity”, assumed that Sicanians descent from a Mediterranean population that diverged from the ancestral population of Bronze Age Sicilians during the Neolithic (8,000–6,000 BP), thus accounting for an external migration that gave rise to the Sicanian culture in Sicily. We first evaluated the accuracy of our inferential ABC procedure in discriminating between the two models considered, performing a power test computing the confusion matrix and the classification errors. For the Continuity model we obtained a classification error of 30%, while for the Discontinuity model we obtained a classification error of 28%, showing that our procedure is sufficiently able to discriminate between the two models, as also confirmed by the LDA plot (Supplementary Figure S3). We then performed the model selection analysis, which selected the Continuity model as the most supported by the observed data, with a posterior probability of 63%. The results of our ABC analysis are summarized in Table 2.
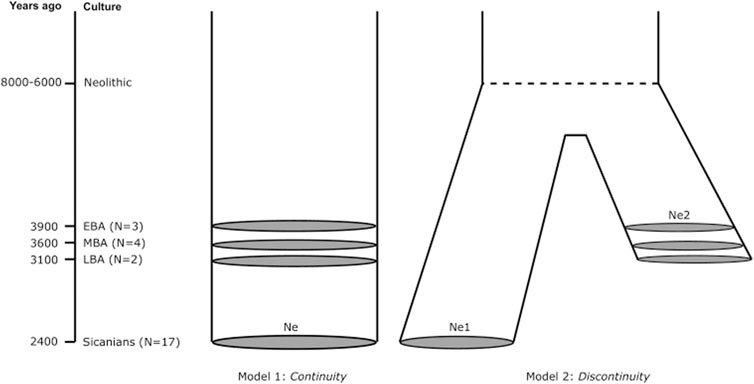
FIGURE 6. Origin of Sicanian culture. Demographic models describing hypotheses on the origin of Sicanians in Baucina.
The study of ancient complete mitochondrial genomes from Sicily presented in this paper has generated insight into the diachronic level of population genetic structure of one of the main islands of the Mediterranean Sea. The analysis of mitogenomes from the Mediterranean allowed us to contextualize the Sicilian genetic variation with that of possibly related groups from Early Neolithic to Iron Age. Whereas the comparison with modern Sicilian variation marked the first step towards the understanding the genetic impact of ancient cultures on the modern inhabitants of the Island.
The 36 sequences we analyzed belong to five different archaeological sites, and the haplogroups composition revealed a wide range of variation, even within the same necropolis. The high mitochondrial genetic variation of ancient Sicily is evident also when compared with other ancient European populations, as shown by the PCA in Figure 2B and by MDS plot in Figure 4A. The FST heatmap in Figure 4B further confirms this pattern, highlighting a higher differentiation among Sicilian groups with respect to what is present in other Mediterranean islands (e.g., Sardinia), or in Continental Europe. Indeed, we observed a statistically significant genetic structure within Sicilian groups (AMOVA FST: 0.08947; p-value: 0.00139 + −0.00035) whereas in Sardinia, the other main island in the Mediterranean basin, the genetic differentiation is not significant (AMOVA FST: 0.00158; p-value:0.42158 + −0.00459). The mitochondrial genetic structure we identify in Sicily is in agreement with the documented historical migrations of populations belonging to different cultures, which made the Island a major Mediterranean crossroad for different populations from Europe, North Africa, and the Levant for a long time (Sarno et al., 2017), and deserves to be further investigated. It also supports that these cultural exchanges were actually accompanied by movements of people, with a possible following exchange of genes.
The eleven individuals sequenced in the necropolis of Motya clustered in two different subgroups, with the three individuals attributed to the Phoenician culture that genetically clustered together and separated from the Bronze Age samples from the same necropolis. These three samples also show similarities with the Phoenician individual from Lilibeo, whose sampling location in the main Island postulates a migration between the two areas or a recent shared ancestry, and further confirms the Phoenician presence within the island of Motya. Another noteworthy pattern of genetic similarities come from the analysis of Baucina and Polizzello samples. In Baucina the individuals have been attributed to the Sicanian culture (26 individuals) and to the Greek culture (3 individuals); the haplogroups compositions and the FST heatmap (Figures 2, 4) actually show the presence of genetic differentiation between these two groups, thus reflecting their different cultural attribution. The observed genetic structure within the Baucina settlement could suggest that a certain social and ethnic distinction was maintained during the Greek colonization. This scenario is also supported by the archaeological records that show a strong Hellenization of the settlements and necropolises starting from the second half of the sixth century BCE, as a consequence of the arrival and establishment of Greeks in the region living alongside the local people (Lyons, 1996; Morgan, 1999).
As highlighted in Figures 2B, 4A, the Sicanians of Baucina show rather more genetic links with the Iron Age samples from Polizzello (Diroma et al., 2021), coming from a necropolis located in the heart of Sicania, and dated ninth-seventh century BCE, than with their Greek neighbors. The genetic closeness of mitochondrial genomes of Sicanians from Baucina and Iron Age indigenous from Polizzello may support the attribution of Polizzello individuals to the Sicanian culture (de Miro, 1988). Additionally, these results reveal a certain genetic homogeneity of the inhabitants of central and western Sicily associated with the same culture.
When ancient Sicilians were contextualized within the Mediterranean domain, we did not find any genetic link between Sicanians individuals (both from Baucina and Iron Age Polizzello) and other Iberian populations. Fernandes et al. (2020) identified Iberia as a key ancestry source for Bronze Age people of Sicily, but the explicit demographic analyses of Sicanian sequences show that this ancestry may not be directly linked to the origin of the Sicanian culture, as originally postulated by Thucydides (VI, 2, 2). The resemblance between Iberia and Sicily seems instead to trace back to Late Neolithic, as emerging also from the low FST values reported in Figure 4B. Our inferential model-based analysis through Approximate Bayesian Computation further supports these results, favoring a local development of Sicanian individuals with a genetic continuity in Central/Western Sicily at least since the Early Bronze Age. Bearing in mind that we are only considering the evolution of the maternal lineage and cannot test other models that may be compatible with the observed genetic variation (such as demographic scenarios that account for sex-biased migrations), our model-based analysis still represents a first step toward a comprehensive and inferential reconstruction of past evolutionary and demographic dynamics in Sicily.
Another interesting similarity pattern came from the Phoenicians in Motya, that showed large resemblance with Levant Late Bronze Age, and the highest FST values (about 0.3) with Phoenicians from Iberian islands. Among the haplogroups identified in the new sequenced samples the most notable is undoubtedly the L3, currently present at high frequency in Northeast Africa (Soares et al., 2012). This L3 sequence found in a Bronze Age individual of Motya explicitly confirmed that the widespread human mobility from North Africa to Europe during the Chalcolithic and Bronze Age, involved also the most remote part of the Island, as emerged also in Fernandes et al. (2020).
Finally, we compared the ancient Sicilian genetic structure with that of modern individuals with known Sicilian ancestry, coming from nine cities around the Island. From the comparison of frequency distribution of mitochondrial haplogroups of ancient and modern Sicilian populations, as well as from the structure emerging from the PCA of Figure 5, we cannot exclude the possibility that Bronze/Iron Age Sicilians made a modest ancestry contribution to modern Sicilians, at least for the maternal lineage. The Y chromosome variation, indeed, has proven to overlap between current and Bronze Age inhabitants of Sicily (Fernandes et al., 2020), postulating a different demographic and evolutionary history for the females and males inhabitants of the Island. A more comprehensive analysis of sex biased processes and of the underlying demographic and evolutionary forces would benefit from an increased and extensive sequencing of modern populations, that would allow to perform explicit comparison between continuity/isolation models from Bronze Age to current time. While our data are indeed consistent with a nearly complete replacement (at least for the mitochondrial lineage) of the pre-Iron Age populations of Sicily by modern inhabitants of the Island, we cannot exclude the hypothesis that locally we may still find a degree of continuity that deserves to be investigated.
This study is restricted to the analysis of a uniparental marker, the mitochondrial genome. Focusing on this marker gave us the opportunity to extend the sampling and the sequencing to a higher number of individuals, so as to adequately representing different cultures dwelling in Sicily in different time periods, and allowing us to identify a structured genetic variation and quantify genetic distances among groups. Albeit limited to the maternal lineage, the present study indeed emphasizes the complex genetic scenario of Sicily since its colonization. The structured genetic variation in culturally defined groups actually supports that cultural processes and exchanges within the Island have been accompanied and promoted by movement of people, and that these dynamics left a footprint on the genetic background of ancient individuals. Modern populations present a rather different pattern of maternal genetic variation; the more homogeneous composition of contemporary uniparental gene pool within Sicily (also reported by Sarno et al., 2014) points towards a recent genetic replacement of pre-Iron Age populations that should be further explicitly addressed.
We acknowledge that the amount of genetic information as well as the inferential power of this uniparental marker is limited with respect to genome-wide ancient DNA data. The analysis of whole-genome variation of different ancient populations from Sicily would provide a more accurate and comprehensive source of information to make inference about past dynamics, such as the time and the origin of principal migration events within the island, the extent of genetic links among contemporary and diachronic groups, and will allow us to explicitly test the hypothesis of a genetic turnover within the island in the last two to three thousand years.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON496938-ON496980, https://www.ncbi.nlm.nih.gov/genbank/, OP081220—OP081394.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
SG, AM, SV, and DC conceived the project; AM, GC, CV, and VZ performed laboratory experiments; ML contributed reagents/materials for libraries preparation and sequencing; AM and GC performed the bioinformatic analysis for mitochondrial consensus sequences reconstruction; HL and AA performed analyses on modern samples; SG, MTV, and RBA performed population genetic analyses; LL, MF, and SB performed radiocarbon dating on samples from Motya; LS and LN provided skeletal material; SG, AM, MTV, GC, RBA, and LS wrote the manuscript. All authors read the manuscript and provided critical input.
This research was supported by the PRIN 2017 grant 20177PJ9XF, PRIN 2017 grant 20174BTC4R_002, and PRIN 2020 grant 2020TACEZR from the Italian Ministry of Education, University and Research (MIUR).
The skeletal samples from the sites of Baucina, Ispica, Lilibeo, Polizzello and Mokarta were studied at the Anthropology laboratory of the STEBICEF Department of Palermo. LS and the authors thank M.G. Griffo of the Baglio Anselmi Archaeological Museum of Marsala, Vincenza Forgia, Aurelio Burgio of the University of Palermo, Giuseppe Bordonaro, archaeologist, Rossella Giglio of the Archaeological Park of Segesta (TP), the beloved Anna Maria Sammito of the Superintendency of Ragusa, that recently passed away, and Arch. Caruso of the Superintendency of Caltanissetta.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.945227/full#supplementary-material
Bandelt, H. J., Forster, P., Sykes, B. C., and Richards, M. B. (1995). Mitochondrial portraits of human populations using median networks. Genetics 141, 743–753. doi:10.1093/genetics/141.2.743
Behar, D. M., van Oven, M., Rosset, S., Metspalu, M., Loogväli, E.-L., Silva, N. M., et al. (2012). A “copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684. doi:10.1016/j.ajhg.2012.03.002
Belvedere, O., Burgio, A., Bordonaro, G., and Forgia, V. (2017). Baucina (Pa)–Monte falcone 2014 indagini nella necropoli. FOLD&R FastiOnLine documents Res. 380, 1–7.
Bianchini, G. (1969). Manufatti della «pebble culture» in sicilia. Riv. Sci. Preistoriche 24, 171–180.
Bianchini, G. (1972). “Nuovi rinvenimenti della “Pebble Culture” nei terrazzi del quaternario antico della Sicilia Sud Occidentale,” in Studi preliminari e comparativi sulla tipologia e l’evoluzione delle industrie litiche (Florence: Istituto Italiano di Preistoria e Protostoria), 89–109.
Bianchini, G. (1971). “Risultati delle ricerche sul Paleolitico inferiore in Sicilia e la scoperta di industrie del gruppo delle Pebble-Culture nei terrazzi quaternari di Capo Rossello in territorio di Realmonte,” in Atti Della XIII Riunione Scientifica Dell’Istituto Italiano Di Preistoria e Protostoria, Siracusa–Malta 1968 (Florence: Istituto Italiano di Preistoria e Protostoria), 89–109.
Cardinali, I., Bodner, M., Capodiferro, M. R., Amory, C., Rambaldi Migliore, N., Gomez, E. J., et al. (2022). Mitochondrial DNA footprints from western eurasia in modern Mongolia. Front. Genet. 12, 819337. doi:10.3389/fgene.2021.819337
Catalano, G., lo Vetro, D., Fabbri, P. F., Mallick, S., Reich, D., Rohland, N., et al. (2020). Late upper palaeolithic hunter-gatherers in the central mediterranean: New archaeological and genetic data from the late epigravettian burial Oriente C (favignana, sicily). Quat. Int. 537, 24–32. doi:10.1016/j.quaint.2020.01.025
Cerezo, M., Achilli, A., Olivieri, A., Perego, U. A., Gómez-Carballa, A., Brisighelli, F., et al. (2012). Reconstructing ancient mitochondrial DNA links between Africa and Europe. Genome Res. 22 (5), 821–826. doi:10.1101/gr.134452.111
Dabney, J., Knapp, M., Glocke, I., Gansauge, M.-T., Weihmann, A., Nickel, B., et al. (2013). Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U. S. A. 110, 15758–15763. doi:10.1073/pnas.1314445110
di Gaetano, C., Cerutti, N., Crobu, F., Robino, C., Inturri, S., Gino, S., et al. (2009). Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome. Eur. J. Hum. Genet. 17, 91–99. doi:10.1038/ejhg.2008.120
Diroma, M. A., Modi, A., Lari, M., Sineo, L., Caramelli, D., and Vai, S. (2021). New insights into mitochondrial DNA reconstruction and variant detection in ancient samples. Front. Genet. 12, 619950. doi:10.3389/fgene.2021.619950
Emery, M. v., Duggan, A. T., Murchie, T. J., Stark, R. J., Klunk, J., Hider, J., et al. (2018). Ancient Roman mitochondrial genomes and isotopes reveal relationships and geographic origins at the local and pan-Mediterranean scales. J. Archaeol. Sci. Rep. 20, 200–209. doi:10.1016/j.jasrep.2018.04.036
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 10, 564–567. doi:10.1111/j.1755-0998.2010.02847.x
Excoffier, L., Marchi, N., Marques, D. A., Matthey-Doret, R., Gouy, A., and Sousa, V. C. (2021). fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics 37, 4882–4885. doi:10.1093/bioinformatics/btab468
Fedi, M. E., Cartocci, A., Manetti, M., Taccetti, F., and Mandò, P. A. (2007). The 14C AMS facility at LABEC, Florence. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 259, 18–22. doi:10.1016/j.nimb.2007.01.140
Fernandes, D. M., Mittnik, A., Olalde, I., Lazaridis, I., Cheronet, O., Rohland, N., et al. (2020). The spread of steppe and Iranian-related ancestry in the islands of the Western Mediterranean. Nat. Ecol. Evol. 4, 334–345. doi:10.1038/s41559-020-1102-0
Fine, J. (1985). The ancient Greeks: A critical history. Massachusetts, United States: Harvard University Press, 72.
Fregel, R., Méndez, F. L., Bokbot, Y., Martín-Socas, D., Camalich-Massieu, M. D., Santana, J., et al. (2018). Ancient genomes from north Africa evidence prehistoric migrations to the maghreb from both the levant and Europe. Proc. Natl. Acad. Sci. U. S. A. 115, 6774–6779. doi:10.1073/pnas.1800851115
Gilbert, M. T. P., Bandelt, H.-J., Hofreiter, M., and Barnes, I. (2005). Assessing ancient DNA studies. Trends Ecol. Evol. 20, 541–544. doi:10.1016/j.tree.2005.07.005
González-Fortes, G., Tassi, F., Trucchi, E., Henneberger, K., Paijmans, J. L. A., Díez-del-Molino, D., et al. (2019). A Western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc. Biol. Sci. 286, 20182288. doi:10.1098/rspb.2018.2288
Hofmanová, Z., Kreutzer, S., Hellenthal, G., Sell, C., Diekmann, Y., Díez-del-Molino, D., et al. (2016). Early farmers from across Europe directly descended from neolithic aegeans. Proc. Natl. Acad. Sci. U. S. A. 113, 6886–6891. doi:10.1073/pnas.1523951113
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F., and Orlando, L. (2013). mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29 (13), 1682–1684. doi:10.1093/bioinformatics/btt193
Kassambara, A., and Mundt, F., 2017. Package ‘factoextra.’ Extract and visualize the results of multivariate data analyses 76.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi:10.1093/molbev/mst010
Lazaridis, I., Mittnik, A., Patterson, N., Mallick, S., Rohland, N., Pfrengle, S., et al. (2017). Genetic origins of the minoans and mycenaeans. Nature 548, 214–218. doi:10.1038/nature23310
Lazaridis, I., Nadel, D., Rollefson, G., Merrett, D. C., Rohland, N., Mallick, S., et al. (2016). Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424. doi:10.1038/nature19310
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Lipson, M., Szécsényi-Nagy, A., Mallick, S., Pósa, A., Stégmár, B., Keerl, V., et al. (2017). Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372. doi:10.1038/nature24476
Lyons, C. L. (1996). Sikel burials at morgantina: Defining social and ethnic identities. Early Soc. Sicily new Dev. Archaeol. Res., 177–188.
Mannino, M. A., di Salvo, R., Schimmenti, V., di Patti, C., Incarbona, A., Sineo, L., et al. (2011). Upper palaeolithic hunter-gatherer subsistence in mediterranean coastal environments: An isotopic study of the diets of the earliest directly-dated humans from sicily. J. Archaeol. Sci. 38 (11), 3094–3100. doi:10.1016/j.jas.2011.07.009
Marcus, J. H., Posth, C., Ringbauer, H., Lai, L., Skeates, R., Sidore, C., et al. (2020). Genetic history from the middle neolithic to present on the mediterranean island of Sardinia. Nat. Commun. 11, 939. doi:10.1038/s41467-020-14523-6
Maricic, T., Whitten, M., and Pääbo, S. (2010). Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE 5 (11), e14004. doi:10.1371/journal.pone.0014004
Martiniano, R., Cassidy, L. M., Ó’Maoldúin, R., McLaughlin, R., Silva, N. M., Manco, L., et al. (2017). The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 13, e1006852. doi:10.1371/journal.pgen.1006852
Mathieson, I., Alpaslan-Roodenberg, S., Posth, C., Szécsényi-Nagy, A., Rohland, N., Mallick, S., et al. (2018). The genomic history of southeastern Europe. Nature 555, 197–203. doi:10.1038/nature25778
Meyer, M., and Kircher, M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, prot5448. doi:10.1101/pdb.prot5448
Modi, A., Catalano, G., D’Amore, G., di Marco, S., Lari, M., Sineo, L., et al. (2020). Paleogenetic and morphometric analysis of a Mesolithic individual from Grotta d’Oriente: An oldest genetic legacy for the first modern humans in Sicily. Quat. Sci. Rev. 248, 106603. doi:10.1016/j.quascirev.2020.106603
Modi, A., Vai, S., Posth, C., Vergata, C., Zaro, V., Diroma, M. A., et al. (2021). More data on ancient human mitogenome variability in Italy: New mitochondrial genome sequences from three upper palaeolithic burials. Ann. Hum. Biol. 48 (3), 213–222. doi:10.1080/03014460.2021.1942549
Morgan, C. (1999). “The archaeology of ethnicity in the colonial world of the eighth to sixth centuries BC: Approaches and prospects,” in Confini e Frontiera Nella Grecità d’Occidente. Atti Del Trentasettesimo Convegno Di Studi Sulla Magna Grecia (Taranto, 85–145.
Natali, E., and Forgia, V. (2018). The beginning of the neolithic in southern Italy and sicily. Quat. Int. 470, 253–269. doi:10.1016/j.quaint.2017.07.004
Olalde, I., Brace, S., Allentoft, M. E., Armit, I., Kristiansen, K., Booth, T., et al. (2018). The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196. doi:10.1038/nature25738
Olalde, I., Mallick, S., Patterson, N., Rohland, N., Villalba-Mouco, V., Silva, M., et al. (2019). The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234. doi:10.1126/science.aav4040
Olivieri, A., Sidore, C., Achilli, A., Angius, A., Posth, C., Furtwängler, A., et al. (2017). Mitogenome diversity in sardinians: A genetic window onto an island’s past. Mol. Biol. Evol. 34, 1230–1239. doi:10.1093/molbev/msx082
Peltzer, A., Jäger, G., Herbig, A., Seitz, A., Kniep, C., Krause, J., et al. (2016). Eager: Efficient ancient genome reconstruction. Genome Biol. 17, 60. doi:10.1186/s13059-016-0918-z
Pessina, A., and Tiné, V. (2008). Archeologia del neolitico: l’Italia tra VI e IIV millennio AC. Rome, Italy: Carocci.
Pinhasi, R., Fernandes, D., Sirak, K., Novak, M., Connell, S., Alpaslan-Roodenberg, S., et al. (2015). Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLOS ONE 10, e0129102. doi:10.1371/journal.pone.0129102
Pudlo, P., Marin, J. M., Estoup, A., Cornuet, J. M., Gautier, M., and Robert, C. P. (2016). Reliable ABC model choice via random forests. Bioinformatics 32, 859–866. doi:10.1093/bioinformatics/btv684
Reimer, P. J., Bard, E., Bayliss, A., Beck, J. W., Blackwell, P. G., Ramsey, C. B., et al. (2013). IntCal13 and Marine13 radiocarbon age calibration curves 0–50, 000 Years cal BP. Radiocarbon 55, 1869–1887. doi:10.2458/azu_js_rc.55.16947
Renaud, G., Slon, V., Duggan, A. T., and Kelso, J. (2015). Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 16, 224. doi:10.1186/s13059-015-0776-0
Rivollat, M., Jeong, C., Schiffels, S., Küçükkalıpçı, İ., Pemonge, M.-H., Rohrlach, A. B., et al. (2020). Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers. Sci. Adv. 6, eaaz5344. doi:10.1126/sciadv.aaz5344
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S., and Reich, D. (2015). Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370 (1660), 20130624. doi:10.1098/rstb.2013.0624
Romano, V., Cali, F., Ragalmuto, A., D’Anna, R. P., Flugy, A., de Leo, G., et al. (2003). Autosomal microsatellite and mtDNA genetic analysis in sicily (Italy). Ann. Hum. Genet. 67, 42–53. doi:10.1046/j.1469-1809.2003.00007.x
Romano, V., Catalano, G., Bazan, G., Calì, F., and Sineo, L. (2021). Archaeogenetics and landscape dynamics in sicily during the Holocene: A review. Sustainability 13, 9469. doi:10.3390/su13179469
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Sarkar, D. (2008). Lattice: Multivariate data visualization with R. Berlin, Germany: Springer Science & Business Media.
Sarno, S., Boattini, A., Carta, M., Ferri, G., Alù, M., Yao, D. Y., et al. (2014). An ancient mediterranean melting pot: Investigating the uniparental genetic structure and population history of sicily and southern Italy. PLoS ONE 9, e96074. doi:10.1371/journal.pone.0096074
Sarno, S., Boattini, A., Pagani, L., Sazzini, M., de Fanti, S., Quagliariello, A., et al. (2017). Ancient and recent admixture layers in Sicily and Southern Italy trace multiple migration routes along the Mediterranean. Sci. Rep. 7, 1984. doi:10.1038/s41598-017-01802-4
Schubert, M., Ginolhac, A., Lindgreen, S., Thompson, J. F., al-Rasheid, K. A., Willerslev, E., et al. (2012). Improving ancient DNA read mapping against modern reference genomes. BMC Genomics 13 (1), 178. doi:10.1186/1471-2164-13-178
Shamoon-Pour, M., Li, M., and Merriwether, D. A. (2019). Rare human mitochondrial HV lineages spread from the Near East and Caucasus during post-LGM and Neolithic expansions. Sci. Rep. 9, 14751. doi:10.1038/s41598-019-48596-1
Sineo, L., Petruso, D., Forgia, V., Messina, A., and D’Amore, G. (2015). Human peopling of Sicily during quaternary. Cheyenne, USA: AcademyPublish.org.
Skourtanioti, E., Erdal, Y. S., Frangipane, M., Balossi Restelli, F., Yener, K. A., Pinnock, F., et al. (2020). Genomic history of neolithic to Bronze age Anatolia, northern levant, and southern Caucasus. Cell. 181, 1158–1175. e28. doi:10.1016/j.cell.2020.04.044
Soares, P., Alshamali, F., Pereira, J. B., Fernandes, V., Silva, N. M., Afonso, C., et al. (2012). The expansion of mtDNA haplogroup L3 within and out of Africa. Mol. Biol. Evol. 29, 915–927. doi:10.1093/molbev/msr245
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi:10.1093/molbev/msm092
Torroni, A., Bandelt, H.-J., Macaulay, V., Richards, M., Cruciani, F., Rengo, C., et al. (2001). A signal, from human mtDNA, of postglacial recolonization in Europe. Am. J. Hum. Genet. 69, 844–852. doi:10.1086/323485
van Oven, M., and Kayser, M. (2009). Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394. doi:10.1002/humu.20921
van Oven, M. (2015). PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci. Int. Genet. Suppl. Ser. 5, e392–e394. doi:10.1016/j.fsigss.2015.09.155
Villalba-Mouco, V., van de Loosdrecht, M. S., Posth, C., Mora, R., Martínez-Moreno, J., Rojo-Guerra, M., et al. (2019). Survival of late pleistocene hunter-gatherer ancestry in the iberian peninsula. Curr. Biol. 29, 1169–1177. e7. doi:10.1016/j.cub.2019.02.006
Wegmann, D., Leuenberger, C., Neuenschwander, S., and Excoffier, L. (2010). ABCtoolbox: A versatile toolkit for approximate bayesian computations. BMC Bioinforma. 11, 116. doi:10.1186/1471-2105-11-116
Weissensteiner, H., Pacher, D., Kloss-Brandstätter, A., Forer, L., Specht, G., Bandelt, H.-J., et al. (2016). HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63. doi:10.1093/nar/gkw233
Willerslev, E., and Cooper, A. (2005). Ancient DNA. Proc. Biol. Sci. 272, 3–16. doi:10.1098/rspb.2004.2813
Keywords: ANCIENT DNA, mitochondrial genomes, genetic structure, coalescent simulations, approximate bayesian computation
Citation: Modi A, Vizzari MT, Catalano G, Boscolo Agostini R, Vai S, Lari M, Vergata C, Zaro V, Liccioli L, Fedi M, Barone S, Nigro L, Lancioni H, Achilli A, Sineo L, Caramelli D and Ghirotto S (2022) Genetic structure and differentiation from early bronze age in the mediterranean island of sicily: Insights from ancient mitochondrial genomes. Front. Genet. 13:945227. doi: 10.3389/fgene.2022.945227
Received: 16 May 2022; Accepted: 18 August 2022;
Published: 09 September 2022.
Edited by:
Charleston Chiang, University of Southern California, United StatesReviewed by:
Francesco Montinaro, University of Tartu, EstoniaCopyright © 2022 Modi, Vizzari, Catalano, Boscolo Agostini, Vai, Lari, Vergata, Zaro, Liccioli, Fedi, Barone, Nigro, Lancioni, Achilli, Sineo, Caramelli and Ghirotto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandra Modi, YWxlc3NhbmRyYS5tb2RpQHVuaWZpLml0; Silvia Ghirotto, c2lsdmlhLmdoaXJvdHRvQHVuaWZlLml0
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.