- 1National Institute for Plant Biotechnology, Indian Council of Agricultural Research, New Delhi, India
- 2Punjab Agricultural University, Ludhiana, Punjab, India
Cytokinins constitutes a vital group of plant hormones regulating several developmental processes, including growth and cell division, and have a strong influence on grain yield. Chemically, they are the derivatives of adenine and are the most complex and diverse group of hormones affecting plant physiology. In this review, we have provided a molecular understanding of the role of cytokinins in developing seeds, with special emphasis on pulses and oilseed crops. The importance of cytokinin-responsive genes including cytokinin oxidases and dehydrogenases (CKX), isopentenyl transferase (IPT), and cytokinin-mediated genetic regulation of seed size are described in detail. In addition, cytokinin expression in germinating seeds, its biosynthesis, source-sink dynamics, cytokinin signaling, and spatial expression of cytokinin family genes in oilseeds and pulses have been discussed in context to its impact on increasing economy yields. Recently, it has been shown that manipulation of the cytokinin-responsive genes by mutation, RNA interference, or genome editing has a significant effect on seed number and/or weight in several crops. Nevertheless, the usage of cytokinins in improving crop quality and yield remains significantly underutilized. This is primarily due to the multigene control of cytokinin expression. The information summarized in this review will help the researchers in innovating newer and more efficient ways of manipulating cytokinin expression including CKX genes with the aim to improve crop production, specifically of pulses and oilseed crops.
Introduction
The importance of oilseeds and pulses in the human diet cannot be overstated. Wherein oilseeds comprise high-energy food with double the amount of energy as carbohydrate and protein (AGRIS FAO, 2022), pulses fulfill protein demands of the majority of people, and both the crop varieties are important for agriculture and livestock. Hormones constitute a pivotal component of regulatory mechanisms directing plant development and have been extensively studied with respect to various seed attributes.
In India, pulses and oilseeds are vital components of the food and nutritional security. India’s dietary habits are still predominantly vegetarian, and the country relies primarily on plant-based sources to achieve its daily protein and other nutritional needs. In addition, according to the FAO, pulses are an important part of a balanced diet. Pulses have been linked to lower risk factors for chronic disease. Apart from being an important aspect of human nutrition, pulses also play a key role in sustainable agriculture and climate change mitigation (Fao 2017; Ha et al., 2014; Jayalath et al., 2014; Kim et al., 2016; Sievenpiper et al., 2009; Viguiliouk et al., 2017; Calles, 2016; Foyer et al., 2016). Over the last 15 years, India has made significant success in increasing pulse production. In 2005–06, India’s total pulse production was 13.38 million metric tonnes (MT), which rose to 25.58 million MT in 2020–21. This represents a 91% increase, or a compound annual growth rate (CAGR) of 4.42%. Regarding oilseeds production, India grows roughly 15%–20% of the world’s total output, produces 6%–7% of vegetable oils, and consumes 9%–10% of all edible oils. Oilseeds are only second to food grains in terms of acreage, production, and economic worth.
Cytokinins are one of the most well studied plant hormones, exercising huge physiological and molecular impact throughout the life cycle of a plant. Cytokinins regulate several functions, such as root development, formation and maintenance of shoot meristem, organ formation, seed germination, seed and fruit development, senescence delay, and response to abiotic and biotic stress. An overview about cytokinins in terms of their biosynthesis, types, bioavailability, and storage forms is described in Figure 1. Cytokinin homeostasis is maintained through action of various enzymes involved in their activation, irreversible conjugation, and degradation. Cytokinins are present throughout the parts of higher plants, though abundantly in the tips of roots, apical meristem of the shoots, and the immature seeds. Majority of higher plants have more than a dozen cytokinins forms which are capable of interconversion. A plethora of previously undertaken studies (Jameson, et al., 1987; Werner and Schmülling, 2009; Kudo et al., 2010; Spichal and Spichal 2012; Schaller et al., 2014; Zwack and Rashotte, 2015) have provided a detailed and extensive overview of cytokinin.
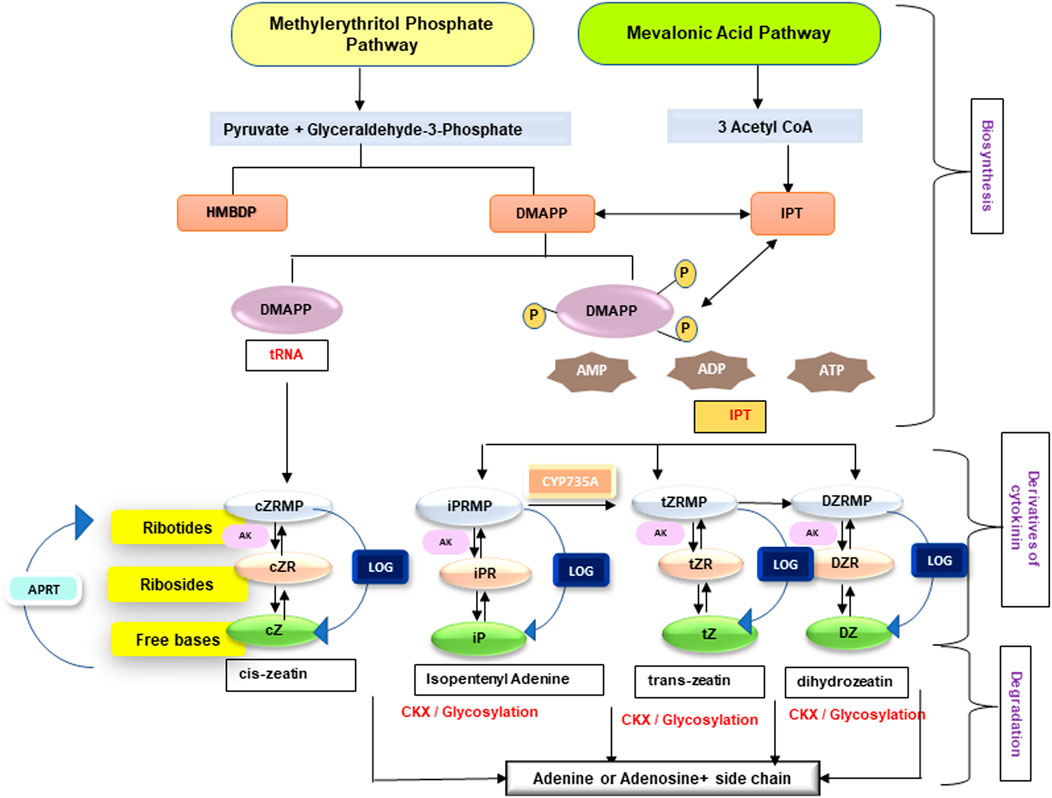
FIGURE 1. An overview on biosynthesis and enzymes of cytokinin homeostasis (iP, isopentenyladenine; DHZ, dihydrozeatin; cZ, cis-zeatin; tZ, trans-zeatin; DMAPP, dimethylallyl pyrophosphate; AMP, adenosine monophosphate) cytokinin biosynthetic pathways—mevalonic acid pathway (operates in cytosol and mitochondria) and methylerythritol pathway (operates in plastids). Enzymes involved are 1) IPT (isopentenyl transferase), which uses ATP, ADP, or ATP as acceptor and forms iPRTP (riboside 5′-diphosphate), iPRDP (riboside 5′-diphosphate), and iPRMP (riboside 5′-monophosphate); 2) CYP735A converts CK nucleotides in to tZ nucleotides; 3) AK (adenosine kinase) causes the phosphorylation of ribosides iPR, tZR, cZR, and DZR; 4) LOG (LONELY GUY) produces free bases from ribotides; 5) APRT (adenine phosphoribosyltransferase) catalyzes the conversion of CK bases to nucleotides; and 6) CKX (cytokinin oxidase/dehydrogenase) causes catabolism of free bases (iP, tZ, DZ, and cZ) to adenine or adenosine (for references, see Takei et al., 2004; Kudo et al., 2010; Sakakibara 2006; Kurakawa et al., 2007; Schoor et al., 2011).
Attempts have been made toward understanding the cytokinin-mediated molecular mechanism regulating important agronomic traits including plant height, plant density, date of flowering, number of primary and secondary branches, seed number per pod/silique, number of pod per plants, pod length, thousand seed weight, and seed size including seed length and seed width.
Cytokinin biosynthetic pathways and its regulation
Naturally occurring cytokinins are derivatives of adenine with an aromatic or an isoprenoid side chain of isopentenyladenine [N6-(D2-isopentenyl) adenine] and hydroxylated either at cis- or trans-terminal position, thereby forming zeatin, named after its discovery in maize (Strnad 1997; Mok and Mok 2001). Among these, cytokinins with the r side chains are widespread in nature. Later in mid-1970s, the presence of benzyladenine was reported in pea, a leguminous plant (Gaudinova’ et al., 2005). Numerous researchers in the past have reported the dominance of trans-zeatin and isopentenyladenine derivatives in nature, whereas cis-isomer is present in very low concentration and has very little or no activity. In contrast to this, recent studies on phytohormones revealed the abundance of cis-isomers in several legume species (Emery et al., 1998, 2000; Quesnelle and Emery, 2007) and other plants such as rice (Takagi et al., 1985) and maize (Veach et al., 2003; Vyroubalova’ et al., 2009). Naturally occurring derivatives of cytokinin encompass N6-(2-isopentenyl) adenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DHZ), which are a part of isoprenoid cytokinins, whereas benzyladenine (BA) along with its hydroxylated derivatives ortho- and metatopolin (oT and mT) and their methoxy-derivatives are included under the aromatic cytokinins group. Theoretically, 26 molecular species of cytokinin have been discovered (Murai, 2014). In total, seven distinct modifications at positions including, N9-position ribosides in adenine mono-, di-, and tri-phosphate ribonucleotides; at N7- and N9-positions glucosides; and O-glucoside in the isopentenyl side chain have been observed so far (Mok and Mok 1994; 2004; Sakakibara 2006; Kojima et al., 2009).
Essentially, two pathways are involved in the biosynthesis of cytokinin precursors, that is, isopentenyladenine-dependent pathway (mevalonic acid pathway) in the cytosol and mitochondria and isopentenyladenine-independent pathway (methylerythritol pathway) in the plastids in order to form isopentenyl transferase (IPT) and dimethylallyl pyrophosphate (DMAPP) as precursors in cytokinin biosynthesis (Figure 1). Subsequently, isopentenyl transferase (IPT) catalyzes transfer of isopentenyl moiety from DMAPP or hydroxymethyl butenyldiphosphate (HMBDP) to either AMP, ADP, or ATP for the formation of biologically active cytokinins and constitutes the rate limiting step. Furthermore, zeatin-type cytokinins are produced by the hydroxylation of the isopentenyl side chain. Alternatively, the hydroxylated side chains can be inserted straight to the N6-position of adenine moiety leading to formation of adenylate IPT. The isopentenyladenine derivative thus produced is then converted to trans-zeatin adenine by an action of a root localized cytochrome P450 monooxygenases (Takei et al., 2004; Kiba et al., 2013), thereby limiting trans-zeatin synthesis to roots but readily transported to other plant parts via xylem. The interconversion between the cis and the trans-isomer of zeatin is mediated by the enzyme cis-trans zeatin isomerase. Macro-concentration of cytokinin in plants is controlled by action of IPT and CKX enzymes; however, conversion of cytokinin nucleotides to cytokinin bases is catalyzed by enzymes, that is, cytokinin phosphoribosyl hydrolase (LOG) (Kurakawa et al., 2007; Kuroha et al., 2009; Tokunaga et al., 2012) and reverse action, that is, conversion from cytokinin bases to nucleotides is catalyzed by adenine phosphoribosyl transferases (APRTs) enzymes (Zhang et al., 2013). All the genes involved in cytokinin homeostasis exists as multi-gene families, with the individual members being differentially expressed in space and time. For instance, 8 and 9 IPT cDNAs and genes, respectively, have been reported in Arabidopsis genome. Of these, seven IPT genes use adenine nucleotides as substrate for transfer reaction, except IPT2. In addition, three of the IPT genes are expressed in the plastids (IPT1, 5, and 8), whereas the rest are localized to the cytoplasm. Furthermore, each gene has different spatial expression profile, such as IPT6 expresses in siliques, IPT4 in immature seeds, and IPT3 in phloem tissues.
Role of kinases in cytokinin signaling
Cytokinins act by regulating the expression of several genes downstream of the signaling cascade. The signaling mechanism of cytokinins in plants is unique and is very similar to the bacterial two component system (Stock et al., 2000; Cheung and Hendrickson, 2010). It involves a transmembrane histidine kinase receptor which dimerizes on ligand binding followed by autophosphorylation of the receptor. This leads to recruitment of an intermediate histidine phosphotransfer protein (HP) which in turns causes phosphorylation of downstream proteins known as nuclear response regulators in the nucleus which then execute cytokinin action. They either regulate the expression of cytokinin response genes as transcription factors or activate different downstream proteins as protein kinases. A detailed characterization of these response regulators in Arabidopsis, which in conjugation, led to their classification in four different classes: type-A, type-B, type-C, and pseudo response regulators (PRRs). The type-B response regulators are DNA-binding transcription factors that promote the expression of cytokinin primary response genes.
Recent evidences suggest the presence of cytokinin response factors in Arabidopsis and in conjugation with response regulators mediate the expression of cytokinin-responsive genes. The mode of signaling involved is either paracrine (local signal in meristematic tissues) or distal signaling (for signaling of availability of nutrients). Furthermore, the selective transport of the two most common cytokinins-trans-zeatin (tz) and isopentenyladenine (iP) is mediated specifically by xylem and phloem, respectively. The histidine kinases involved in the cytokinin signaling cascade are the transmembrane receptors that comprises of an extracellular CHASE domain (cyclases/histidine kinases–associated sensory extracellular) on which the ligand binds leading to the dimerization and hence activation of the receptor (Inoue et al., 2001; Nishimura et al., 2004, Nishimura et al., 2004 C.; Higuchi et al., 2004). The conserved histidine kinase domain is essential for the activation through autophosphorylation. The receiver domain comprises of a conserved aspartate domain which plays a key role in the transfer of phosphate group from HK domain to the histidine phosphotransfer protein (HP) that further conveys the signal specifically to the type-B response regulator (Appleby et al., 1996; Schaller et al., 2011) in the nucleus. The type-B response regulators possess a DNA-binding domain, thereby acting as transcription factors. In contrast to this, type-A response regulators do not have a DNA-binding domain and their downstream proteins are still undetermined. Once activated, the type-B response regulators are able to activate all the cytokinin-responsive genes including the type-A response regulators, which in turn suppress cytokinin signaling, thereby providing a negative feedback loop during the signaling pathway (Pareek et al., 2006; Du et al., 2007; Pils and Heyl, 2009; Kieber and Schaller, 2014).
Cytokinin-mediated genetic and epigenetic regulation of seed size
One of the key aspects of increasing crop productivity is the seed size. The major emphasis of crop production or improvement since time immemorial has been the selection of crops with bigger seed size (Song et al., 2007; Shomura et al., 2008; Fan et al., 2009). Seed size varies dramatically between species. The endosperm makes up the majority of the mature seed in monocots like rice and wheat. Majority of the dicots, for e.g., Arabidopsis thaliana and Brassica napus develop their endosperm rapidly at the initial stage; as a consequence, the embryo occupies the larger part of the developed mature seed. In flowering plants, the development of seeds is influenced by complex interactions between maternal tissues, embryo, and endosperm. It has been observed that the endosperm has a major role in regulating seed size. The endosperm exhibits more expeditive growth as compared to embryo during the early phase of seed development, and the seed volume also increases vis a vis the endosperm’s growth (Sundaresan, 2005). Several genes and transcription factors regulating growth of endosperm have also been reported to regulate the seed size in Arabidopsis. For example, HAIKU1 (IKU1), HAIKU2 (IKU2), and MINISEED3 (MINI3) have been reported to function synergistically in the same genetic pathway to enhance the endosperm size and embryo development (Garcia et al., 2003; Luo et al., 2005; Zhou et al., 2009). Promoters for MINI3 and IKU2 also associate with Short Hypocotyl Under Blue 1 (SHB1) transcription factor (recruited by WRKY10) to promote endosperm development (Zhou et al., 2009). A number of reviews have detailed out the role of these genes in the genetic regulation of seed size (Sundaresan, 2005; Sun et al., 2010; Kesavan et al., 2013). In addition, several maternally derived factors have also been shown to regulate seed size in various plants (Jofuku et al., 2005; Li et al., 2008; Adamski et al., 2009; Ohto et al., 2009; Xia et al., 2013; Singh et al., 2017). Recent studies have highlighted the involvement of various transcription factors, G protein-coupled hormone signaling and ubiquitin-mediated pathway in maternal control of seed size in Arabidopsis. The cytokinin oxidase 2 (CKX2) gene produces a protein that destroys active cytokinin present in the cell in an irreversible manner. The IKU pathway controls seed size via regulating endosperm growth, and CKX2 has been identified as a direct transcriptional target of the IKU system (Li et al., 2013). Overexpression of CKX2 resulted in the recovery from the decrease in seed size phenotypes, indicating the involvement of CKX2 in regulating seed size in a positive way. DNA methyltransferase 1 (MET1) (causes methylation of cytosine in CG) regulates CKX2 as well as epigenetic maternal imprinting (Li et al., 2013). Membrane-bound cytokinin receptors are encoded by the arabidopsis histidine kinases (AHK) family, and AHK2, AHK3, and CRE1 (cytokinin response 1)/AHK4 are the three histidine kinases that bind cytokinin. Although the deletion of one or both of these receptors had no effect on seed size, ahk1 ahk2, and ahk3 triple mutant seeds exhibited a 250% higher volume, with embryo cell number and size increasing by 15% and 30%, respectively (Riefler et al., 2006). Furthermore, it was suggested that the cytokinin-mediated regulation of seed size mostly occurs due to maternal and/or zygotic tissues. Exogenous application of cytokinin has also been proven beneficial in improving the yield-related traits in pulses and oil seed crops. Application of 100–200 µM BAP (6-benzylaminopurine) increased ovule and seed number in Brassica napus and also restored the replum development in wild-type B. napus and in the A. thaliana rpl ntt double mutant (Zuniga-Mayo et al., 2018). Moreover, when different forms of cytokinin viz., [6-benzyladenine (BA), N-(2-chloro-4-pyridyl)-n-phenylurea (CPPU), 6-furfurylaminopurine (KT), and thidiazuron (TDZ)] were applied on B. juncea, highest frequency of shoot regeneration was noticed in combinatorial treatment of thidiazuron (TDZ)] and NAA (Guo et al., 2005). Similarly, combination of BAP and IAA resulted in significantly high biomass and seed yield in Guizotia abyssinica (L.f.) Cass. (niger seed plant); a multipurpose oil seed crop (Talukdar et al., 2022). Application of 3.4 × 10–7 mol of 6-benzylaminopurine (BA) resulted in a 79% increase in soybean seed yield compared with controls (Nagel et al., 2001).
Cytokinin-induced/boosting number of seed per pod/pod set/pod development
Among yield-related traits, the number of seeds per/pod and/or number of seed per unit area determines the overall yield (Schou et al., 1978; Kokubun, 1988). The number of seeds per pod or number of seeds produced per unit area is directly proportional to the number of flowers that further develop in to mature pod. Leguminous plants, such as soybean, pigeonpea, and chickpea, produce a higher number of flowers, but most of them abort before reaching maturity (Abernethy et al., 1977). Likely causes include the lack of nutrition, vascular constrictions, and certain hormones (Heindl et al., 1982; Antos and Wiebold, 1984; Brun and Betts, 1984; Kokubun and Honda, 2000). Competition for photosynthesis among seeds and organs is also thought to be a major cause of abortion (Shibles et al., 1975). There are various reports highlighting the exogenous and/or endogenous cytokinin-mediated boost in flower and pod formation (Crosby, 1981; Peterson et al., 1984; Dyer et al., 1987; Mosjidis et al., 1993; Nagel et al., 2001; Yashima et al., 2015).
Figure 2 highlights various aspects of yield and yield-related traits as mediated by cytokinins. Increase in pod number was observed after application of exogenous BA4 to the floral raceme of mung bean (Clifford 1981) and two soybean cultivars (Crosby, 1981; Peterson, et al., 1984) The cultivar Shore, exhibited significant increases in pod number than Essex soybean cultivar. It was speculated that the difference in response was due to Shore ovules having a lower endogenous level of cytokinin-like activity than that in Essex ovules at the time of BA application. Similarly, the administration of three BA on the top of the nodes of field grown soybean resulted in an increase in total number of pod per plant by 27% and seed weight by 18% (Carlson et al., 1986). Furthermore, an increase in the total number of flowers, pods, and number of ovules per gynoecium observed in soybean and oilseed rape after exogenous application of cytokinin (Dyer et al., 1987; Honig et al., 2018) showed that the cytokinin system could be effectively utilized as a target for improving yield and yield-related traits in dicots as well. Bartrina et al., 2011 and Gailbiati et al., 2013 reported that an increased cytokinin levels in the ckx3 ckx5 double mutant result in a larger gynoecium and production of more ovules. Similarly, sextuple ckx3 ckx5 mutants were observed to have higher cytokinin concentrations with larger and highly active inflorescence meristems. They also produced up to 72% more flowers on the main stem, with the gynoecia had 32% and 54% more ovules pods, respectively. In addition, the weight of seeds extracted from the main stem of plants was found to be heavier by 20–32% (Schwarz et al., 2020). Furthermore, cytokinins have been shown to increase ovule quantity in other Brassicaceae species, implying that genetic manipulation of cytokinin metabolism could be an effective technique for increasing seed yield (Cucinotta et al., 2016; Zuniga-Mayo et al., 2018) Surprisingly, it was known recently that high amounts of cytokinin hindered fruit elongation in ckx7 mutants (Di Marzo et al., 2020). Niemann et al., 2015 highlighted the formation of more number of flowers and siliques as a result of mutation in the ROCK1 gene (REPRESSOR OF CYTOKININ DEFICIENCY1), which is essential for full CKX function. Furthermore, in the ugt85a3 (UDP-GLUCOSYL TRANSFERASE 85A3) mutant, lower cytokinin inactivation resulted in the development of more ovules per gynoecium (Cucinotta et al., 2018). In soybean, higher levels of cytokinin were found in the reproductive tissues during the pod set and seed filling phases (Jameson and Song, 2016; Liu et al., 2018; Zuniga-Mayo et al., 2018). Metabolite profiling of 27 cultivars of field-grown soybeans (pod and seed tissues) revealed that high producing varieties maintained a constant supply of cytokinins via de novo biosynthesis into later stages of development as compared to low yielding soybean genotypes. In addition, zeatin-type cytokinins are required for pod/seed set, whereas isopentenyladenine-type cytokinins have a role in seed filling (Kambhampati et al., 2017). Numerous studies in various crops have reported the significant impact of cytokinins in mediating yield and its related traits through a range of studies, such as exogenous cytokinin application, NGS, transgenic expression studies, advanced chromatographic techniques, mass spectrometry, and many others (Table 1).
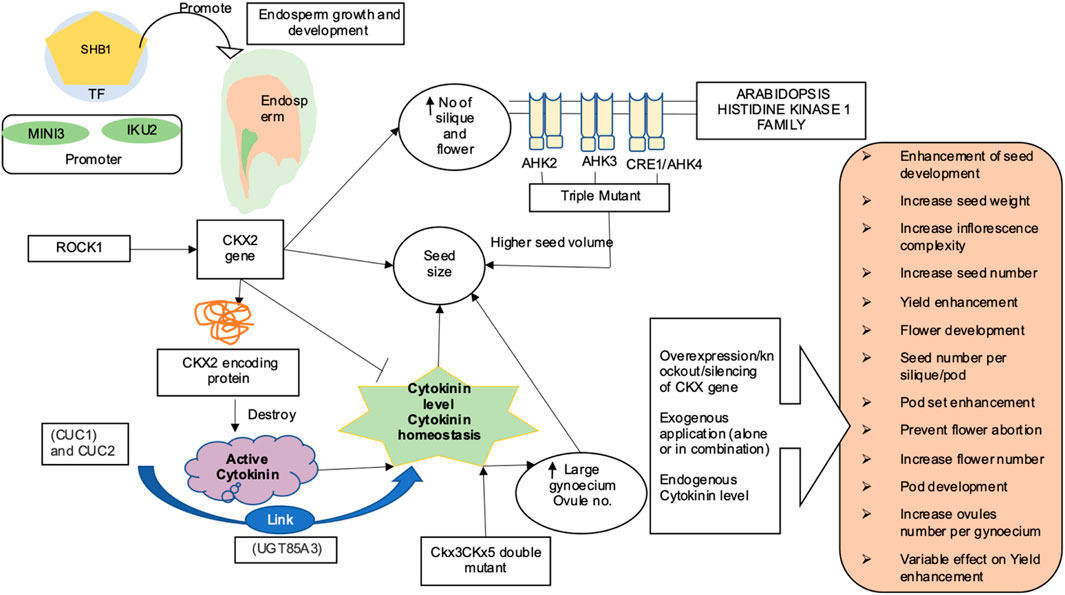
FIGURE 2. Cytokinin-mediated regulation of yield and yield-related traits (SHB1, Short Hypocotyl Under Blue1; ROCK1, Repressor of cytokinin deficiency 1; AHK1, Arabidopsis histidine kinase1 family; CKX2, Cytokinin oxidase/dehydrogenase; CUC, Cup-shaped cotyledon; MINI3, MINISEED 3; IKU2, HAIKU2) (Riefler et al., 2006; Zhou et al., 2009; Li et al., 2013). Various strategies such as overexpression of gene CKX/IPT (Werner et al., 2003), knockdown of CKX gene family (Ashikari et al., 2005), gene silencing (Heyl et al., 2008; Schoor et al., 2011), and exogenous application of CK (Nagel et al., 2001; Liu et al., 2018; Talukdar et al., 2022) have been attempted to enhance yield-related trait in pulses and oilseed crop. Dynamic role of cytokinin in orange box-in Arabidopsis; CKs are involved in formation of more number of flower, silique, seed (Bartrina et al., 2011; Niemann et al., 2015), ovule number, and seed yield (Werner et al., 2003; Cucinotta et al., 2018; Nguyen et al., 2021b; Zu et al., 2022) In soybean, CKs are involved in preventing flower abortion and in pod setting (Nonokawa et al., 2012), associated with seed development and fatty acid biosynthesis (Nguyen et al., 2016), increase seed yield (Kambhampati et al., 2017), pod set and seed filling (Jameson and Song 2016; Kambhampati et al., 2017; Cai et al., 2018; Liu et al., 2018), and reduction in floral abscission and increase pod number and seed weight (Carlson et al., 1986; Nagel et al., 2001) In Brassica increase in seed number per ovule number, pod number, seed weight (Zuniga-Mayo et al., 2018; Schwarz et al., 2020), more pod number, and increase in seed yield was observed (Atkins et al., 2011)
Source-sink pathways are always in as dynamic state during the entire life cycle of a plant. Initially starting their life cycle as sinks, leaves mature into source for the seeds, with the latter acted upon as a source to begin with, that is, providing energy and nutrients during germination. Central to these relationships are the availability and partitioning of two major resources: carbon and nitrogen. Sucrose and amino acid mobilization are controlled by SWEET (sugars will eventually be exported) and SUT (sucrose transporters)/SUCs (sucrose carriers) transporters and cell wall invertases for the former and primarily AAPs (amino acid permease) for the latter. Cytokinins have also been observed to regulate sink number as well as sink size in various legumes, cereals, and Arabidopsis. These signaling molecules are reported to increase the strength of sink tissues and magnetize the assimilates through either influencing sucrose metabolism and transport or promoting cell division (Brenner and Chiekh 1995; Emery and Atkins 2006) through upregulation of cyclins controlling check points of cell cycle (Riou-Khanlichi et al., 1999) and increased phloem unloading in seed coat (Brenner and Chiekh 1995). The insight into cytokinin-mediated influence on source-sink relations and nutrient allocation is depicted in Figure 3. Developing seeds have been documented with the highest cytokinins level among all plant tissues; thus, they are rich source of the former metabolites (Emery et al., 2000). As reported in Arabidopsis, IPT gene expression in the endosperm continues till early heart stage of its developing seeds, making them sites of cytokinin biosynthesis (Miyawaki et al., 2004), with minor quantities being translocated from xylem or phloem (Emery et al., 2000). Jameson et al., 2016 conducted expression profiling of SWEET, SUT, AAP, CWINV (cell wall invertases), IPT, LOG (Lonely Guy/cytokinin phosphoribosyl hydrolases) and CKX genes to ascertain source-sink dynamics in germinating seeds of P. sativum. They reported an active expression of cytokinins in imbibing seeds and its biosynthesis in germinating seedlings as well as strong expression of specific genes regulating source-sink dynamics in plants.
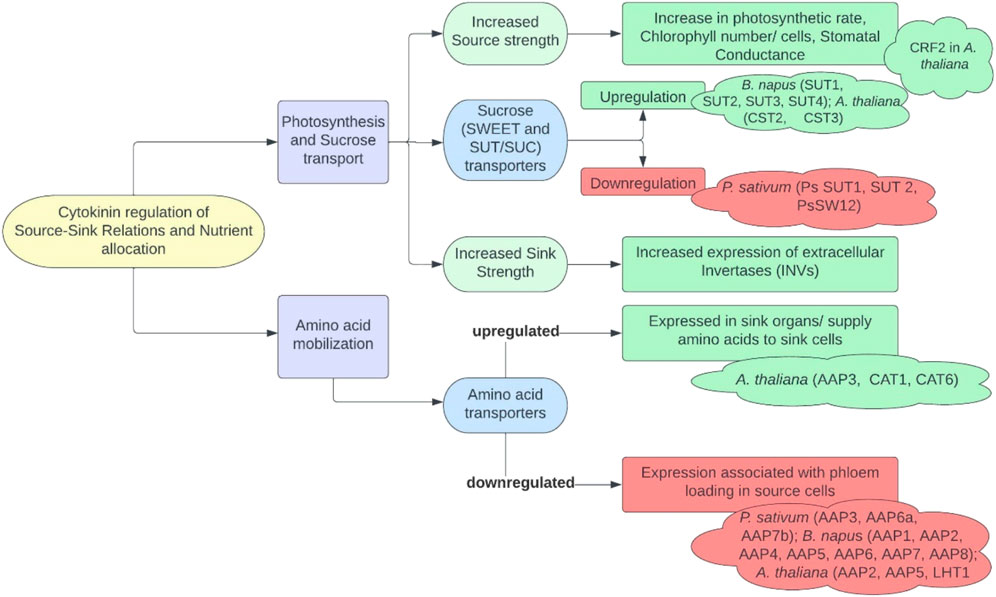
FIGURE 3. Insights into cytokinin-mediated source-sink relationship and nutrient allocation (SWEET, SUGARS WILL EVENTUALLY be EXPORTED TRANSPORTERS; SUT/SUCs, SUCROSE TRANSPORTERS/CARRIERS; AAP, amino acid permease; LHT, lysine and histidine transporters; CAT, cationic amino acid transporters) (for references, see McIntyre et al., 2021; Harms et al., 1994; Ninan et al., 2017; Song et al., 2015; Ehness and Roitsch, 1997; Godt and Roitsch, 1997; Yang et al., 2014; Chernyad’ev, 2000; Ahanger et al., 2020; Boasson and Laetsch, 1969; Okazaki et al., 2009; Lee et al., 2007; Kiba et al., 2011; Kiba et al., 2005; Yokoyama et al., 2007; and Jian et al., 2016)
Ectopic expression of an IPT gene (isopentenyltransferase) has been observed to increase seed yield. In addition, the inherent expression of cytokinins is highly dynamic and changes rapidly over time, as observed in developing cereal grains (Morris et al., 1993). In wheat, expression profile of specific members of the cytokinin biosynthesis (IPT), degradation (CKX), O-glucosylation, and ß-glucosidase gene families have been implicated in the changing cytokinins levels (Song et al., 2012). In legumes, the level of cytokinins also influence the pod set. Application of cytokinins in lupin has been observed to prevent abortion (Aitkins and Pigeaire, 1993). Emery et al. (2000) investigated correlation of cytokinins with abortion in developing flowers and pod set by increasing an IPT gene expression (Atkins et al., 2011). A detailed GC-MS analysis on developing white lupin revealed the expression nature of cytokinins and showed that the peak and transient cytokinin expression occurs in the liquid endosperm of developing seeds (Emery et al., 2000). Several evidences have pointed out the fact that in legumes maternally derived cytokinins are restricted to the pod and seed set (Emery et al., 2000), but in the developing embryo cytokinin biosynthesis is active in the filial tissues (Singh et al., 1988; Emery et al., 2000). In Arabidopsis, Day et al. (2008) observed that the cell cycle genes and cytokinin biosynthesis (IPT8) genes play a crucial role in the syncytial endosperm development.
Spatial expression of cytokinin gene regulating seed/seed number per pod development
Active sites for cytokinin biosynthesis include developing seeds, pod walls and seed coats. Cytokinins have been demonstrated to influence yield by inducing flowering, increasing silique/seed number/pod, and seed size. Various members of the cytokinin gene family viz cytokinin biosynthesis related gene family (BnIPT1, 2, 3, 5, 7, 8, and 9), cytokinin degradation gene family (BnCKX1 to BnCKX7), cell wall invertase gene family (BnCWINV1 to BnCWINV6), sugar transporter gene family (BnSUT1 to BnSUT6), and amino acid permease–related gene family (BnAAP1 to BnAAP8) have been identified in Brassica napus as a target for breeding (Song et al., 2015; Ninan et al., 2017). It has been reported that developing seeds are the major site of cytokinins and the filial tissues of developing legume seeds have been shown to rely on cytokinin biosynthesis (Singh, 1988; Jameson et al., 2016), whereas pod walls and seed coats have been observed to have significant amounts as well as different forms of cytokinin (Davey and van Staden, 1977, 1978, 1979; Zhang and Letham 1990; Emery et al., 2000; Song et al., 2015). In Arabidopsis, many studies have observed dynamic and differential spatiotemporal expression patterns of IPT gene family members (Miyawaki et al., 2004; Belmonte et al., 2013) as well as in other members of Brassicaceae family (O’Keefe et al., 2011; Liu et al., 2013). After profiling all seven AtIPT gene family members using RT-PCR, Miyawaki et al. (2004) observed the presence of the cytokinin biosynthesis pathway in most plant organs. However, distinct tissue specificity was observed in reporter gene (GUS) constructs used. But still, each family member was found to be expressed at several regions. Liu Z. et al. (2013) observed BrIPT1, 3, 5, and 7 genes to be strongly expressed in the roots, whereas BrIPT8-1 was largely confined to immature siliques and BrIPT8-2 in stamens of Brassica rapa. These evidences match well with the data on the developing seeds of the Arabidopsis, where expression of genes like AtIPT8 and AtIPT4 is localized to the chalazal region (Miyawaki et al., 2004; Day et al., 2008; Belmonte et al., 2013).
Cytokinin and seed germination.
Crop stand and productivity is a manifestation of seed germination and seedling establishment, with the germination process categorized into three phases (water uptake by seeds; mobilization of food reserves and reactivation of metabolism; and radical protrusion) and involves numerous physiological, morphological, and biochemical changes upon favorable conditions, which are regulated by endogenous and exogenous factors. One of the important internal components affecting germination is hormones. Emergence of seeds and buds from dormancy involves reduction in the levels of inhibitors and gradual buildup of growth promoters; thus, extensive changes in seed metabolome repertoire. Role of hormones in mediating a shift of an inert quiescent embryo to a rapidly metabolizing system, that is, seed germination and post germinative seedling growth is well elucidated, particularly that of abscisic acid (ABA) and gibberellins. However, several studies have implicated the regulatory role of cytokinins in seed germination. Different forms and activity of cytokinin are a function of developmental stages, tissues, and plant species in question (Kieber and Eric Schaller, 2010). Cytokinin application was observed to revert the ABA-induced inhibition of seed germination in Brassica oleraceae (Khan, 1971), suggesting permissive role of these hormones in removing the blocks (inhibitors) present in seeds. However, the study also suggested that if these inhibitors are absent, role of cytokinins to mediate germination becomes redundant. Water imbibition by seeds exclusively determines radical emergence; however, seedling growth is a feature of remobilization of stored food reserves to zones of growth and mitosis. Several research studies have implicated both of the latter processes to be possibly regulated by cytokinins (Gepstein and Ilan, 1980; Hepler, 1986; MacGaw and Borch, 1995). For instance, reserve material mobilization in Cicer arietinum seeds coincides with the period of supply of cytokinin from the embryonic axis to the cotyledons, that is, first 12 h s after the start of imbibition (Martin et al., 1987; Pino et al., 1991). Exogenous cytokinin application has been known to influence the development of embryonic axis as cited by several researchers (Tzou et al., 1973; Alvarez et al., 1987). Gallego et al., 1991 revealed variation in the endogenous level and compartmentalization of different cytokinin groups in embryonic axis of C. arientinum L. in response to exogenous cytokinin application and calcium treatment during germination. Zeatin, 2-isopentyl adenosine, and 2-isopentyl adenine application induces germinative changes peculiar to those under normal conditions, such as delay in epicotyl emergence and short and thick embryonic axis with reduced dry weight, whereas application of zeatin riboside and dihydro derivatives did not induce such changes. Furthermore, controlled conditions revealed high amounts of conjugated cytokinins (storage and inactive forms) in the basal regions of epicotyl and hypocotyl of embryonic axis, which are hydrolyzed to free bases followed by their transportation to apical zones, wherein their transformation to dihydro derivatives (most stable form) takes place, as the later form is resistant to the action of cytokinin oxidase enzyme present in the embryonic axis (MacGaw and Burch, 1995). However, a germination medium supplemented with calcium increased the level of dihydro derivatives, whereas exogenous cytokinin application leads to faster appearance but lower levels of these derivatives in embryonic axis segments displaying maximum growth. The later could be attributed to the fact that exogenous cytokinin could lead to stress or produce compounds resistant to enzymes (Whitty and Hall 1974).
ABI5 (abscisic acid insensitive 5) encodes for a basic leucine zipper transcription factor that is best characterized as a key component involved in ABA signaling and early seedling development. ABI5 transcripts buildup during germination and degrade to basal amount post seed germination (Lopez-Molina et al., 2001; Cho et al., 2002). A total of two mechanisms have been proposed wherein cytokinin targets ABI5 to regulate seed germination, as shown in the Figure 4. One route explains marginal regulation by cytokinin through repression of ABI5 expression in an ABA-independent manner (Wang et al., 2011). Second mechanism pertains to the role of cytokinin in promoting plastid differentiation. In Arabidopsis, Guan et al. (2014) demonstrated that cytokinin mediates ABI5 protein degradation, thereby alleviating ABA-induced inhibition of cotyledon greening, a focal point during post germination growth and marking the transition from heterotrophy to autotrophy and establishing photosynthetic capacity. In cytokinin signaling, hybrid histidine protein kinases (AHKs) autophosphorylate upon sensing cytokinins through receptor binding. This phosphorelay system comprises of transfer of the phosphate group from the receptors to the downstream components, initially to AHPs (Arabidopsis histidine phosphotransfer proteins), followed by phosphorylation and activation of Myb transcription factor encoding type B ARRs (response regulators). The latter induces expression of cytokinin response gene, including type A ARRs as characterized in Arabidopsis (Nishimura et al., 2004; Riefler et al., 2006). When phosphorylated, type A ARRs, through unknown mechanism, negatively regulates cytokinin signaling, thus forming a feedback regulatory loop (Muller and Sheen 2007; To and Kieber 2008; Hwang et al., 2012). To et al. (2007) and Ren et al. (2009) reported cytokinin to positively regulate steady level of type A- ARRs mRNA by increasing the stability of protein in a phosphorylation-dependent manner and further postulated type A ARR–ABI5 complex to inhibit ABI5 protein interaction with proteasome degradation. Cytokinin promotes seed germination by mimicking the action of auxin, stimulating ABI5 protein degradation and thus uplifting the abscisic acid inhibitory effect on post germination growth of Arabidopsis. Thereby, degradation of ABI5 protein rather than its mRNA is a major step in cytokinin-mediated ABA signaling.
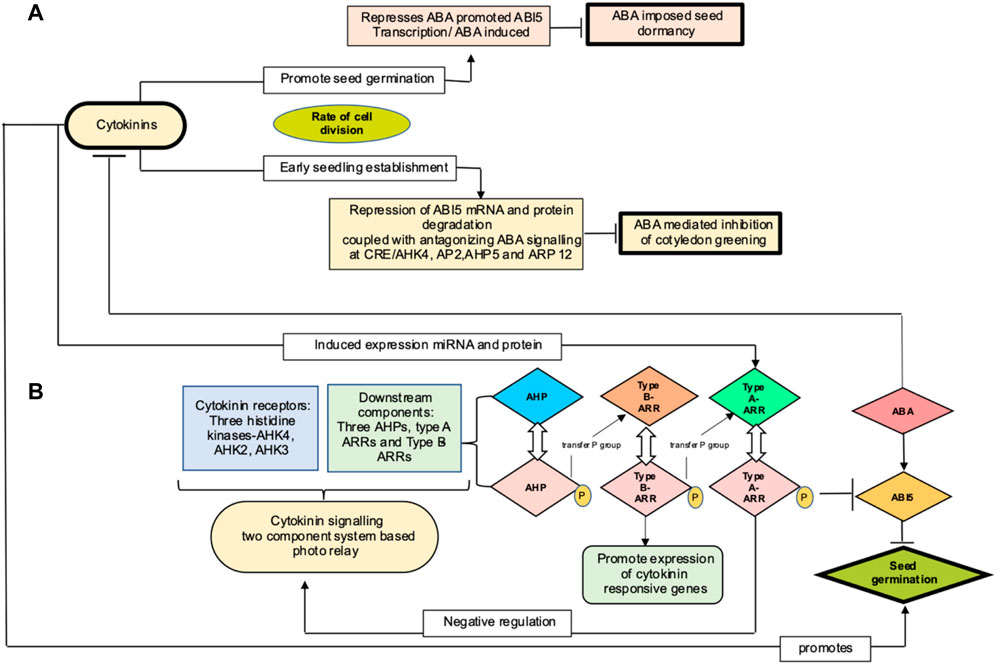
FIGURE 4. (A) Proposed mechanism for cytokinin action in regulating seed germination in Arabidopsis by Wang et al. (2011) and Guan et al. (2014), represented at top (light pink (marginal mechanism) and yellow (major mechanism) color boxes, respectively). Cytokinin influences cell division to promote seed germination and seedling establishment and thereby activates ABA-imposed seed dormancy and inhibition of cotyledon greening. (B) represents cytokinin signaling components and their interaction with ABI5 to mediate seed germination (AHK, Arabidopsis histidine kinases, ARRs, Arabidopsis response regulators; ABA, abscisic acid; ABI5, abscisic acid insensitive protein 5)
Convergence of cytokinin with light signaling has been demonstrated through interaction of type-A ARRs with phytochrome b (phyb) (Sweere et al., 2001) and a bZIP transcription factor, HY5, in Arabidopsis (Ang et al., 1998). Hutchison et al. (2006) and Riefler et al. (2006) emphasized role of cytokinin receptor genes and AHPs in far red light regulated seed germination, implicating cytokinin repressed ABA signaling as an important regulatory mechanism to coordinate early seedling establishment. An enhanced cell division rate as a function of seed priming with cytokinin, particularly kinetin has also been related to improved germination and robustness of seeds (Sawan et al., 2000; Tahaei et al., 2016). Wang et al., 2011 characterized Arabidopsis gim1 (germination insensitive to ABA mutant 1) mutants’ deficit for AtIPT8 gene encoding for isopentenyl transferases, catalyzing a rate limiting step in the cytokinin biosynthetic pathway (Sun et al., 2003; Miyawaki et al., 2006). These mutants were characterized for reduced expression of ABI5 gene, and the expression could not be restored with exogenous ABA treatment. However, ectopic expression of AtIPT8 (ecotypic expression) in OE-2/Com1 transgenic plants was observed to raise the cytokinin level and ABA insensitive seed germination features of gim1 mutants was observed.
Several advanced studies have also been conducted to elucidate the role and establish mechanism of cytokinin in germination of oil seed crops and legumes. For instance, first study underpinning legume metabolism in relation to seed germination was conducted by Araujo et al. (2019). Priming of Medicago truncatula seeds (50 mM) kinetin throughout germination followed by detailed metabolome and physiological characterization was observed to speed up radical protrusion caused an impairment of seedling growth at the root level. Kinetin affected content of 27 metabolites at radicle emergence stage, chiefly associated with rapid decline of metabolites linked to germination and stress indicating the role of kinetin as both stress agent and inducer of seed germination. Such targeted studies hold potential in identifying the point, wherein priming of cytokinin needs to be stopped for preventing genotoxicity. However, Riefler et al. (2006) demonstrated role of cytokinin in seed germination, shoot and root development, seed size, and senescence through loss of function mutants for cytokinin receptors (AHK2, AHK3, and CRE1/AHK4) in Arabidopsis. Rapid germination, decreased sensitivity to far-red light and increased dark germination were observed in the mutants, revealing functions of these cytokinin receptors in regulating these processes. Cytokinin, thus, was observed to negatively regulate light-dependent seed germination in Arabidopsis. Similarly, Oh et al. (2009) conducted genome wide chromatin immunoprecipitation (ChIP)—chip analysis in Arabidopsis targeting role of phytochrome interacting factor 3–LIKE5 (PIL5), a basic helix-loop-helix transcription factor (TF) in seed germination. It was observed that phytochromes when activated mediated degradation of PIL5 and enabled seed germination. PIL5 was identified to directly or indirectly regulate gene expression of several hormonal signaling networks, including GA, ABA, JA, ethylene, and cytokinins, and also impacting the expressions of various genes encoding cell enzymes involved in cell wall modification. Of interest, PIL 5 was observed to directly downregulate expression of cytokinin response factors, that is, CRF1, CRF2, and CRF3 genes as well as to upregulate the expression of AHP5 gene. CRFs (AP2 domain TFs) and AHP5 are known to positively upregulate subset of cytokinin responses (Rashotte et al., 2006) and cytokinin signaling, respectively; therefore, PIL5-mediated reverse regulation of these cytokinin positive signaling genes.
Future perspectives
It has been observed that when cytokinin levels rise, their degradation also kicks in. This is the one of the most important considerations for researchers aiming to target increased cytokinin content as plants will respond to it by accelerating the cytokinin degradation as a result of active homeostasis. Key participants to cytokinin degradation are the CKX catalyzing degradation of several cytokinin forms (Werner et al., 2006; Gajdošová et al., 2011). Table 2 shows the expression of the members of IPT and CKX gene family in different tissues of oilseed and pulses. These gene families have been the preferred targets for yield improvement in several crops. It has been shown that natural or artificial induction of cytokinins result in increased activity of CKX (Brugière et al., 2008; Motyka et al., 2003; Liu et al., 2013a, Liu et al., 2013b). Pertinently, a detailed characterization of the spatiotemporal expression of cytokinin gene family need to be performed in oilseeds and pulse crops for shortlisting significant genes that can be further targeted for genome engineering with the aim to boost the yield. However, appropriate genome editing strategies need to be developed as genes involved in cytokinin biosynthesis and its catabolism belong to multigene family and are pleiotropic in nature.
In the past decade, several reviews have focussed on summarizing the role of cytokinins in biotic and abiotic stresses (Cortleven et al., 2019), nitrogen nutrition (Gu et al., 2018), senescence (Zwack and Rashotte, 2013; Honig et al., 2018), and seed yield (Jameson et al., 2016). In addition, the significance of cytokinin dehydrogenase: a genetic target for yield improvement in wheat crop (Chen et al., 2020) has also been extensively discussed. However, the role of cytokinins in oilseeds and pulses has not been reviewed so far. In this review, we have tried to gather all the relevant information concerning cytokinin-regulated yield traits in these crops. Nevertheless, information on cytokinin metabolomics also needs to be reviewed which might open new avenues for cytokinin-targeted research for improving crop yield and quality.
Quantification and qualitative analysis of cytokinins is pivotal to determine their association with agronomically important traits. Several studies have substantiated the relation between yield, associated parameters, and cytokinins and described them as evolutionary conserved and of utmost functional significance. However, advanced multidisciplinary approaches targeting cytokinin purification, profiling, quantification, and underlying molecular mechanisms are required to be undertaken in pulses and legumes, specifically. Omics tools and techniques provides a peek through in cytokinin gene families, spaciotemporal gene expression patterns, and metabolite characterization, to further dissect cytokinin-mediated source-sink relations and associated metabolic pathways in details. Identifying and targeting candidate genes will further pave the road to enhance the endogenous cytokinins level using transgenic and breeding approaches. Cytokinin homeostasis and its regulatory networking could also be fine-tuned in pulses and oilseeds through detailed characterization of its gene families-, tissue-, and stage-specific expression data, detecting novel mutations and their applicability to harness the potential of techniques such as MAS, TILING, and CRISPR and many more for developing new germplasm, breeding lines, and varieties with maintained quality traits. Comprehensive depiction of the inhibitors and activators of key genes of cytokinins homeostasis, such as CKX and IPT, and standardization of studies conducting their exogenous applications could also be utilized for enhancing abiotic stress tolerance as well as in vitro organogenesis and holds immense potential toward micropropagation in numerous agricultural, horticultural crops, and in forestry.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
Authors are grateful to the Director ICAR- NIPB, New Delhi for providing funding and laboratory facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abernethy, R. H., Wright, L. N., and Matsuda, K. (1977). Association of seedling respiratory metabolism and adenylate energy charge with seed weight of Panicum antidotale Retz.1. Crop Sci. 17 (4), 563–566. doi:10.2135/CROPSCI1977.0011183X001700040021X
Adamski, N. M., Anastasiou, E., Eriksson, S., O’Neill, C. M., and Lenhard, M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U. S. A. 106 (47), 20115–20120. doi:10.1073/PNAS.0907024106
agris.fao (2022). Application of cytokinins to flowers to increase pod set in Lupinus angustifolius L. [lupins]. (n.d.). Retrieved May 10, 2022, fromAvailable at: https://agris.fao.org/agris-search/search.do?recordID=AU9301192.
Ahanger, M. A., Aziz, U., Sahli, A. A., Alyemeni, M. N., and Ahmad, P. (2020). Combined kinetin and spermidine treatments ameliorate growth and photosynthetic inhibition in Vigna angularis by up-regulating antioxidant and nitrogen metabolism under cadmium stress. Biomolecules 10, 147. doi:10.3390/biom10010147
Alvarez, J., Guerra, H., Calvarro, L. M., and Villalobos, N. (1987). The effect of embryo Axis, 6-benzylaminopurine and kinetin on protein metabolism in protein bodies during germination of lentil seeds. J. Plant Physiology 126, 319–328. doi:10.1016/s0176-1617(87)80016-0
Ang, L. H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 (2), 213–222. doi:10.1016/S1097-2765(00)80022-2
Antos, M., and Wiebold, W. J. (1984). Abscission, total soluble sugars, and starch profiles within a soybean Canopy1. Agron. J. 76 (5), 715–719. doi:10.2134/AGRONJ1984.00021962007600050002X
Appleby, J. L., Parkinson, J. S., and Bourret, R. B. (1996). Signal transduction via the multi-step phosphorelay: Not necessarily a road less traveled. Cell 86 (6), 845–848. doi:10.1016/S0092-8674(00)80158-0
Araújo, S., Pagano, A., Dondi, D., Lazzaroni, S., Pinela, E., Macovei, A., et al. (2019). Metabolic signatures of germination triggered by kinetin in Medicago truncatula. Sci. Rep. 9 (1), 10466. doi:10.1038/S41598-019-46866-6
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Sci. (New York, N.Y.) 309 (5735), 741–745. doi:10.1126/science.1113373
Atkins, C. A., and &Pigeaire, A. (1993). Application of cytokinins to flowers to increase pod set in Lupinus angustifolius L. Aust. J. Agric. Res. 44 (8), 1799–1819. doi:10.1071/ar9931799
Atkins, J. P., Gregory, A. J., Burdon, D., and Elliott, M. (2011). Managing the marine environment: Is the DPSIR framework holistic enough? Syst. Res. Behav. Sci. 28 (5), 497–508. doi:10.1002/SRES.1111
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23 (1), 69–80. doi:10.1105/TPC.110.079079
Belmonte, M. F., Kirkbride, R. C., Stone, S. L., Pelletier, J. M., Bui, A. Q., Yeung, E. C., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. U. S. A. 110 (5), E435./-/DCSUPPLEMENTAL/SD01.XLS. doi:10.1073/PNAS.1222061110
Boasson, R., and Laetsch, W. M. (1969). Chloroplast replication and growth in tobacco. Science 166, 749–751. doi:10.1126/science.166.3906.749
Brenner, M. L., and Cheikh, N. (1995). “The role of hormones in photosynthate partitioning and seed filling,” in Plant hormones. Editor P. J. Davies (Dordrecht: Springer). doi:10.1007/978-94-011-0473-9_30
Brugière, N., Humbert, S., Rizzo, N., Bohn, J., and Habben, J. E. (2008). A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development. Cytokinin biosynthesis in maize. Plant Mol. Biol. 67 (3), 215–229. doi:10.1007/s11103-008-9312-x
Brun, W. A., and Betts, K. J. (1984). Source/Sink relations of abscising and nonabscising soybean flowers. Plant Physiol. 75 (1), 187–191. doi:10.1104/PP.75.1.187
Cai, L., Zhang, L., Fu, Q., and Xu, Z. F. (2018). Identification and expression analysis of cytokinin metabolic genes IPTs, CYP735A and CKXs in the biofuel plant Jatropha curcas. PeerJ 2018, e4812. doi:10.7717/PEERJ.4812/SUPP-4
Calles, T. (2016). The international year of pulses: What are they and why are they important. Agric. Dev. 26, 40–42.
Carlson, G. A., Kingsbury, D. T., Goodman, P. A., Coleman, S., Marshall, S. T., DeArmond, S., et al. (1986). Linkage of prion protein and scrapie incubation time genes. Cell 46 (4), 503–511. doi:10.1016/0092-8674(86)90875-5
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2020). Cytokinin dehydrogenase: A genetic target for yield improvement in wheat. Plant Biotechnol. J. 18 (3), 614–630. doi:10.1111/pbi.13305
Cherniad’ev, I. (2000). [Ontogenetic changes in the photosynthetic apparatus and effect of cytokinins]. Prikl. Biokhim. Mikrobiol. 36, 611–625. doi:10.1023/A:1026628119243
Cheung, J., and Hendrickson, W. A. (2010). Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 13 (2), 116–123. doi:10.1016/J.MIB.2010.01.016
Cho, Y., Suh, S. K., Park, H. K., and Wood, A. (2002). Impact of 2, 4-DP and BAP upon pod set and seed yield in soybean treated at reproductive stages. Plant Growth Regul. 36 (3), 215–221. doi:10.1023/A:1016590505449
Clifford, P. E. (1981). Control of reproductive sink yield in mung beans. Z. Für Pflanzenphysiol. 102 (2), 173–181. doi:10.1016/S0044-328X(81)80196-1
Cortleven, A., Leuendorf, J. E., Frank, M., Pezzetta, D., Bolt, S., and Schmülling, T. (2019). Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 42 (3), 998–1018. doi:10.1111/pce.13494
Crosby, D. G. (1981). Environmental chemistry of pentachlorophenol. Pure Appl. Chem. 53 (5), 1051–1080. doi:10.1351/pac198153051051
Cucinotta, M., Manrique, S., Cuesta, C., Benkova, E., Novak, O., and Colombo, L. (2018). CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in arabidopsis. J. Exp. Bot. 69 (21), 5169–5176. doi:10.1093/JXB/ERY281
Cucinotta, M., Manrique, S., Guazzotti, A., Quadrelli, N. E., Mendes, M. A., Benkova, E., et al. (2016). Cytokinin response factors integrate auxin and cytokinin pathways for female reproductive organ development. Dev. Camb. Engl. 143 (23), 4419–4424. doi:10.1242/DEV.143545
D’Agostino, I. B., Deruère, J., and Kieber, J. J. (2000). Characterization of the response of the arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124 (4), 1706–1717. doi:10.1104/PP.124.4.1706
Davey, J. E., and Van Staden, J. (1977). A cytokinin complex in the developing fruits of lupinus albus. Physiol. Plant. 39 (3), 221–224. doi:10.1111/J.1399-3054.1977.TB04040.X
Davey, J. E., and van Staden, J. (1978). Cytokinin activity in Lupinus albus: III. Distribution in fruits. Physiol. Plant. 43, 87–93. doi:10.1111/j.1399-3054.1978.tb01572.x
Davey, J. E., and van Staden, J. (1979). Cytokinin activity in Lupinus albus: IV. Distribution in seeds. Plant Physiol. 63, 873–877. doi:10.1104/pp.63.5.873
Day, T. A., Ruhland, C. T., and Xiong, F. S. (2008). Warming increases aboveground plant biomass and C stocks in vascular‐plant‐dominated Antarctic tundra. Glob. Chang. Biol. 14 (8), 1827–1843. doi:10.1111/j.1365-2486.2008.01623.x
de Bouillé, P., Sotta, B., Miginiac, E., and Merrien, A. (1989). Hormones and inflorescence development in oilseed rape. Plant Physiol. Biochem. 27, 443–450.
De Bouille, P., Sotta, B., Miginiac, E., and Merrien, A. (1989). Hormones and pod development in oilseed rape (Brassica napus). Plant Physiol. 90 (3), 876–880. doi:10.1104/PP.90.3.876
Di Marzo, M., Herrera-Ubaldo, H., Caporali, E., Novák, O., Strnad, M., Balanzà, V., et al. (2020). SEEDSTICK controls arabidopsis fruit size by regulating cytokinin levels and FRUITFULL. Cell Rep. 30 (8), 2846–2857. e3. doi:10.1016/J.CELREP.2020.01.101
Du, C., Bollen, M. H. J., Agneholm, E., and Sannino, A. (2007). A new control strategy of a VSC-HVDC system for high-quality supply of industrial plants. IEEE Trans. Power Deliv. 22 (4), 2386–2394. doi:10.1109/TPWRD.2007.899622
Dyer, D. J., Carlson, D. R., Cotterman, C. D., Sikorski, J. A., and Ditson, S. L. (1987). Soybean pod set enhancement with synthetic cytokinin analogs. Plant Physiol. 84 (2), 240–243. doi:10.1104/PP.84.2.240
Ehness, R., and Roitsch, T. (1997). Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 11, 539–548. doi:10.1046/j.1365-313x.1997.11030539.x
Emery, R. J. N., and Atkins, C. A. (2006). “Cytokinins and seed development,” in Seed science and technology: Trends andadvances. Binghamton. Editor A. S. Basra (New York: Haworth Press Inc), 63–93.
Emery, R. J. N., Leport, L., Barton, J. E., Turner, N. C., and Atkins, C. A. (1998). cis-isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol. 117 (4), 1515–1523. doi:10.1104/PP.117.4.1515
Emery, R. J. N., Ma, Q., and Atkins, C. A. (2000). The forms and sources of cytokinins in developing white lupine seeds and fruits. Plant Physiol. 123 (4), 1593–1604. doi:10.1104/PP.123.4.1593
Fan, C., Yu, S., Wang, C., and Xing, Y. (2009). A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 118 (3), 465–472. doi:10.1007/S00122-008-0913-1
Fao (2017). Food and agricultural organisation of the united nations nutritional benefits of pulses. Available at: Published http://www.fao.org/resources/infographics/infographics-details/en/c/412361/(Accessed October 10, 2017).
Ferreira, F. J., and Kieber, J. J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8, 518–525. doi:10.1016/j.pbi.2005.07.013
Foyer, C. H., Lam, H., Nguyen, H. T., Siddique, K. H. M., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2, 1–10. doi:10.1038/nplants.2016.112
Gajdošová, S., Spíchal, L., Kamínek, M., Hoyerová, K., Novák, O., Dobrev, P. I., et al. (2011). Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 62 (8), 2827–2840. doi:10.1093/jxb/erq457
Galbiati, F., Sinha Roy, D., Simonini, S., Cucinotta, M., Ceccato, L., Cuesta, C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant J. 76 (3), 446–455. doi:10.1111/TPJ.12309
Gallego, P., Hernandez-Nistal, J., Martin, L., Nicolas, G., and Villalobos, N. (1991). Cytokinin levels during the germination and seedling growth of cicer arietinum L.: Effect of exogenous application of calcium and cytokinins. Plant Sci. 77 (2), 207–221. doi:10.1016/0168-9452(91)90090-U
Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jürgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131 (4), 1661–1670. doi:10.1104/PP.102.018762
Garcia, S. M., Garcia, K. L. C., Cochrane, S. M., Garcia, S. M., and Cochrane, K. L. (2005). Ecosystem approach to fisheries: A review of implementation guidelines1. ICES J. Mar. Sci. 62 (3), 311–318. doi:10.1016/J.ICESJMS.2004.12.003
Gaudinova, A., Dobrev, P. I., Solcova, B., Novak, O., Strand, M., Friedecky, D., et al. (2005). The involvement of cytokinin oxidase/dehydrogenase and zeatin reductase in regulation of cytokinin levels in pea (Pisum sativum L.) leaves. J. Plant Growth Regul. 24, 188–200. doi:10.1007/s00344-005-0043-9
Gepstein, S., and Ilan, I. (1980). Evidence for the involvement of cytokinins in the regulation of proteolytic activity in cotyledons of germinating beans. Plant Cell Physiology 21 (1), 57–63. doi:10.1093/OXFORDJOURNALS.PCP.A075990
Godt, D. E., and Roitsch, T. (1997). Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol. 115, 273–282. doi:10.1104/pp.115.1.273
Grant, J. E., Ninan, A., Cripps-Guazzone, N., Shaw, M., Song, J., Petřík, I., et al. (2021). Concurrent overexpression of amino acid permease AAP1(3a) and SUT1 sucrose transporter in pea resulted in increased seed number and changed cytokinin and protein levels. Funct. Plant Biol. 48 (9), 889–904. doi:10.1071/FP21011
Gu, J., Li, Z., Mao, Y., Struik, P. C., Zhang, H., Liu, L., et al. (2018). Roles of nitrogen and cytokinin signals in root and shoot communications in maximizing of plant productivity and their agronomic applications. Plant Sci. 274, 320–331. doi:10.1016/j.plantsci.2018.06.010
Guan, C., Wang, X., Feng, J., Hong, S., Liang, Y., Ren, B., et al. (2014). Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of abscisic acid Insensitive5 protein in arabidopsis. Plant Physiol. 164 (3), 1515–1526. doi:10.1104/PP.113.234740
Guo, D. P., Zhu, Z. J., Hu, X. X., and Zheng, S. J. (2005). Effect of cytokinins on shoot regeneration from cotyledon and leaf segment of stem mustard (Brassica juncea var. tsatsai). Plant Cell Tissue Organ Cult. 83 (1), 123–127. doi:10.1007/S11240-005-3799-5
Ha, V., Sievenpiper, J. L., De Souza, R. J., Jayalath, V. H., Mirrahimi, A., Agarwal, A., et al. (2014). Effect of dietary pulse intake on established therapeutic lipid targets for cardiovascular risk reduction: A systematic review and meta-analysis of randomized controlled trials. Cmaj 186 (8), E252–E262. doi:10.1503/cmaj.131727
Harms, K. V. W. R., Schulz, B., and Frommer, W. B. (1994). Isolation and characterization of P-type H+-ATPase genes from potato. Plant Mol. Biol. 26, 979–988. doi:10.1007/BF00028864
Heindl, J. C., Carlson, D. R., Brun, W. A., and Brenner, M. L. (1982). Ontogenetic variation of four cytokinins in soybean root pressure exudate. Plant Physiol. 70 (6), 1619–1625. doi:10.1104/PP.70.6.1619
Hepler, P. K. (1986). Calcium regulation of mitosis: The metaphase/anaphase transition. Mol. Cell. Aspects Calcium Plant Dev., 167–174. doi:10.1007/978-1-4613-2177-4_21
Heyl, A., Ramireddy, E., Brenner, W. G., Riefler, M., Allemeersch, J., and Schmülling, T. (2008). The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 147 (3), 1380–1395. doi:10.1104/PP.107.115436
Higuchi, M., Pischke, M. S., Mähönen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. U. S. A. 101 (23), 8821–8826. doi:10.1073/PNAS.0402887101
Hönig, M., Plíhalová, L., Husičková, A., Nisler, J., and Doležal, K. (2018). Role of cytokinins in senescence, antioxidant defence and photosynthesis. Int. J. Mol. Sci. 19 (12), 4045. doi:10.3390/ijms19124045
Honig, M., Plihalova, L., Husickova, A., Nisler, J., and Dolezal, K. (2018). Role of cytokinins in senescence, antioxidant defence and photosynthesis. Int. J. Mol. Sci. 19, E4045. doi:10.3390/ijms19124045
Horgan, R., Hewett, E. W., Horgan, J. M., Purse, J., and &Wareing, P. F. (1975). A new cytokinin from Populus x robusta. Phytochemistry 14 (4), 1005–1008. doi:10.1016/0031-9422(75)85176-4
Hutchison, C. E., Li, J., Argueso, C., Gonzalez, M., Lee, E., Lewis, M. W, et al. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 (11), 3073–3087. doi:10.1105/TPC.106.045674
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi:10.1146/annurev-arplant-042811-105503
Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., et al. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 (6823), 1060–1063. doi:10.1038/35059117
Jameson, P. E., Dhandapani, P., Novak, O., and Song, J. (2016). Cytokinins and expression of SWEET, SUT, CWINV and AAP genes increase as pea seeds germinate. Int. J. Mol. Sci. 17 (12), E2013. doi:10.3390/IJMS17122013
Jameson, P. E., and Song, J. (2016). Cytokinin: A key driver of seed yield. J. Exp. Bot. 67 (3), 593–606. doi:10.1093/JXB/ERV461
Jameson, P., Letham, D., Zhang, R., Parker, C., and Badenoch-Jones, J. (1987). Cytokinin translocation and metabolism in lupin species. I. Zeatin riboside introduced into the xylem at the base of lupinus angustifolius stems. Funct. Plant Biol. 14 (6), 695. doi:10.1071/PP9870695
Jayalath, V. H., De Souza, R. J., Sievenpiper, J. L., Ha, V., Chiavaroli, L., Mirrahimi, A., et al. (2014). Effect of dietary pulses on blood pressure: A systematic review and meta-analysis of controlled feeding trials. Am. J. Hypertens. 27 (1), 56–64. doi:10.1093/ajh/hpt155
Jian, H., Lu, K., Yang, B., Wang, T., Zhang, L., Zhang, A., et al. (2016). Genome-wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L.). Front. Plant Sci. 7, 1464. doi:10.3389/fpls.2016.01464
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U. S. A. 102 (8), 3117–3122. doi:10.1073/PNAS.0409893102
jstor (2022). Factors affecting shedding of flowers in soybean (Glycine max (L.) merrill) on JSTOR. Retrieved May 10, 2022, fromAvailable at: https://www.jstor.org/stable/23688254.
Jung, H. W., Kim, K. D., and Hwang, B. K. (2005). Identification of pathogen-responsive regions in the promoter of a pepper lipid transfer protein gene (CALTPI) and the enhanced resistance of the CALTPI transgenic Arabidopsis against pathogen and environmental stresses. Planta 221 (3), 361–373. doi:10.1007/s00425-004-1461-9
Kakimoto, T. (2001). Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 42 (7), 677–685. doi:10.1093/PCP/PCE112
Kambhampati, S., Kurepin, L. V., Kisiala, A. B., Bruce, K. E., Cober, E. R., Morrison, M. J., et al. (2017). Yield associated traits correlate with cytokinin profiles in developing pods and seeds of field-grown soybean cultivars. Field Crops Res. 214, 175–184. doi:10.1016/J.FCR.2017.09.009
Kant, S., Burch, D., Badenhorst, P., Palanisamy, R., Mason, J., and Spangenberg, G. (2015). Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLOS ONE 10 (1), e0116349. doi:10.1371/JOURNAL.PONE.0116349
Kariuki, M. (2002). Height estimation in complete stem analysis using annual radial growth measurements. Forestry 75 (1), 63–74. doi:10.1093/FORESTRY/75.1.63
Kempster, R., Barat, M., Bishop, L., Rufino, M., Borras, L., and Dodd, I. C. (2021). Genotype and cytokinin effects on soybean yield and biological nitrogen fixation across soil temperatures. Ann. Appl. Biol. 178 (2), 341–354. doi:10.1111/AAB.12652
Kesavan, M., Song, J. T., and Seo, H. S. (2013). Seed size: A priority trait in cereal crops. Physiol. Plant. 147 (2), 113–120. doi:10.1111/J.1399-3054.2012.01664.X
Khan, A. A. (1971). Cytokinins: Permissive role in seed germination. Science 171 (3974), 853–859. doi:10.1126/SCIENCE.171.3974.853
Kiba, T., Kudo, T., Kojima, M., and Sakakibara, H. (2011). Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409. doi:10.1093/jxb/erq410
Kiba, T., Naitou, T., Koizumi, N., Yamashino, T., Sakakibara, H., and Mizuno, T. (2005). Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His->Asp phosphorelay circuitry. Plant Cell Physiol. 46, 339–355. doi:10.1093/pcp/pci033
Kiba, T., Takei, K., Kojima, M., and Sakakibara, H. (2013). Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27 (4), 452–461. doi:10.1016/J.DEVCEL.2013.10.004
Kiba, T., Yamada, H., and Mizuno, T. (2002). Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol. 43 (9), 1059–1066. doi:10.1093/PCP/PCF121
Kieber, J. J., and Eric Schaller, G. (2010). The perception of cytokinin: A story 50 Years in the making. Plant Physiol. 154 (2), 487–492. doi:10.1104/PP.110.161596
Kim, S. J., De Souza, R. J., Choo, V. L., Ha, V., Cozma, A. I., Chiavaroli, L., et al. (2016). Effects of dietary pulse consumption on body weight: A systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 103 (5), 1213–1223. doi:10.3945/ajcn.115.124677
Kojima, M., Kamada-Nobusada, T., Komatsu, H., Takei, K., Kuroha, T., Mizutani, M., et al. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: An application for hormone profiling in oryza sativa. Plant Cell Physiol. 50 (7), 1201–1214. doi:10.1093/pcp/pcp057
Kokubun, M. (1988). Design and evaluation of soybean ideotypes. Bull. Tohoku Natl. Agric. Exp. Stn. 77, 77–142.
Kokubun, M., and Handa, I. (2000). Intra-raceme variation in pod-set probability is associated with cytokinin content in soybeans. Plant Prod. Sci. 3, 354–359. doi:10.1626/pps.3.354
Kokubun, M., and Honda, I. (2015). Intra-raceme variation in pod-set probability is associated with cytokinin content in soybeans. Plant Prod. Sci. 3 (4), 354–359. doi:10.1626/PPS.3.354
Kudo, T., Kiba, T., and Sakakibara, H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52 (1), 53–60. doi:10.1111/J.1744-7909.2010.00898.X
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 (7128), 652–655. doi:10.1038/nature05504
Kuroha, T., Tokunaga, H., Kojima, M., Ueda, N., Ishida, T., Nagawa, S., et al. (2009). Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21 (10), 3152–3169. doi:10.1105/tpc.109.068676
Le, D. T., Nishiyama, R., Watanabe, Y., Vankova, R., Tanaka, M., Seki, M., et al. (2012). Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLOS ONE 7 (8), e42411. doi:10.1371/JOURNAL.PONE.0042411
Lee, D. J., Park, J. Y., Ku, S. J., Ha, Y. M., Kim, S., Kim, M. D., et al. (2007). Genome-wide expression profiling of Arabidopsis Response Regulator 7(ARR7) overexpression in cytokinin response. Mol. Genet. Genomics 277, 115–137. doi:10.1007/s00438-006-0177-x
Letham, D. S. (2019). Cytokinins as phytohormones — sites of biosynthesis, translocation, and function of translocated cytokinin. Cytokinins, 57–80. doi:10.1201/9781351071284-5
Li, J., Nie, X., Tan, J. L. H., and Berger, F. (2013). Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 110 (38), 15479–15484. doi:10.1073/pnas.1305175110
Li, N., Xu, R., and Li, Y. (2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. doi:10.1146/annurev-arplant-050718-095851
Li, S., Gu, S., Liu, W., Han, H., and Zhang, Q. (2008). Water quality in relation to land use and land cover in the upper Han River Basin, China. Catena 75 (2), 216–222. doi:10.1016/J.CATENA.2008.06.005
Li, S. S., Kendall, C. W., de Souza, R. J., Jayalath, V. H., Cozma, A. I., Ha, V.., et al. (2014). Dietary pulses, satiety and food intake: A systematic review and meta-analysis of acute feeding trials. Obes. (Silver Spring) 8, 1773–1780. doi:10.1002/oby.20782
Liu, F., Xing, S., Ma, H., Du, Z., and Ma, B. (2013a). Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl. Microbiol. Biotechnol. 97 (20), 9155–9164. doi:10.1007/S00253-013-5193-2
Liu, H., Mi, Z., Lin, L., Wang, Y., Zhang, Z., Zhang, F., et al. (2018). Shifting plant species composition in response to climate change stabilizes grassland primary production. Proc. Natl. Acad. Sci. U. S. A. 115 (16), 4051–4056. doi:10.1073/PNAS.1700299114
Liu, L. M., Zhang, H. Q., Cheng, K., and Zhang, Y. M. (2021). Integrated bioinformatics analyses of PIN1, CKX, and yield-related genes reveals the molecular mechanisms for the difference of seed number per pod between soybean and cowpea. Front. Plant Sci. 12, 749902. doi:10.3389/fpls.2021.749902
Liu, P., Zhang, C., Ma, J. Q., Zhang, L. Y., Yang, B., Tang, X. Y., et al. (2018). Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development and stress responses in oilseed rape (Brassica napus L.). Genes 9 (3), 168. doi:10.3390/genes9030168
Liu, Z., Chen, Y., Han, J., Chen, D., Yang, G., Lan, T., et al. (2021). Determination, dissipation dynamics, terminal residues and dietary risk assessment of thiophanate-methyl and its metabolite carbendazim in cowpeas collected from different locations in China under field conditions. J. Sci. Food Agric. 101 (13), 5498–5507. doi:10.1002/JSFA.11198
Liu, Z., Lv, Y., Zhang, M., Liu, Y., Kong, L., Zou, M., et al. (2013b). Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14 (1), 1–20. doi:10.1186/1471-2164-14-594
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 98 (8), 4782–4787. doi:10.1073/PNAS.081594298
Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32 (3), 317–328. doi:10.1046/J.1365-313X.2002.01430.X
Lorenzi, R., Horgan, R., and Wareing, P. F. (1975). Cytokinins in picea sitchensis carriere: Identification and relation to growth. Biochem. Und Physiol. Der Pflanz. 168 (1–4), 333–339. doi:10.1016/S0015-3796(17)30130-0
Luo, J. J., Masson, S., Behera, S., Shingu, S., and Yamagata, T. (2005). Seasonal climate predictability in a coupled OAGCM using a different approach for ensemble forecasts. J. Clim. 18 (21), 4474–4497. doi:10.1175/JCLI3526.1
Martin, L., Diez, A., Nicolas, G., and Villalobos, N. (1987). Variation of the levels and transport of cytokinins during germination of chick-pea seeds. J. Plant Physiology 128 (1–2), 141–151. doi:10.1016/S0176-1617(87)80188-8
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., et al. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in arabidopsis. Plant Cell 17 (11), 3007–3018. doi:10.1105/TPC.105.035451
McGaw, B. A., and Burch, L. R. (1995). Cytokinin biosynthesis and metabolism. Plant Horm., 98–117. doi:10.1007/978-94-011-0473-9_5
McIntyre, K. E., Bush, D. R., and Argueso, C. T. (2021). Cytokinin regulation of source-sink relationships in plant-pathogen interactions. Front. Plant Sci. 12, 1618. doi:10.3389/fpls.2021.677585
Mens, C., Li, D., Haaima, L. E., Gresshoff, P. M., and Ferguson, B. J. (2018). Local and systemic effect of cytokinins on soybean nodulation and regulation of their Isopentenyl Transferase (IPT) biosynthesis genes following Rhizobia inoculation. Front. Plant Sci. 9, 1150. doi:10.3389/fpls.2018.01150
Miyawaki, K., Matsumoto-Kitano, M., and Kakimoto, T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37 (1), 128–138. doi:10.1046/J.1365-313X.2003.01945.X
Miyawaki, K., Tarkowski, P., Matsumato-Kitano, M., Kato, T., Sato, S., Tarkowska, D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 103 (44), 16598–16603. doi:10.1073/pnas.0603522103
Mok, D. W. S., and Mok, M. C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118. doi:10.1146/ANNUREV.ARPLANT.52.1.89
Morris, R. O., Blevins, D. G., Dietrich, J. T., Durley, R. C., Gelvin, S. B., Gray, J., et al. (1993). Cytokinins in plant pathogenic bacteria and developing cereal grains. Funct. Plant Biol. 20 (5), 621–637. doi:10.1071/PP9930621
Mosjidis, C. O., Peterson, C. M., Truelove, B., and Dute, R. R. (1993). Stimulation of pod and ovule growth of soybean, Glycine max (L.) Merr. by 6-benzylaminopurine. Ann. Bot. 71, 193–199. doi:10.1006/anbo.1993.1024
Motyka, V., Vaňková, R., Čapková, V., Petrášek, J., Kamínek, M., and Schmülling, T. (2003). Cytokinin‐induced upregulation of cytokinin oxidase activity in tobacco includes changes in enzyme glycosylation and secretion. Physiol. Plant. 117 (1), 11–21. doi:10.1034/j.1399-3054.2003.1170102.x
Müller, B., and Sheen, J. (2007). Arabidopsis cytokinin signaling pathway. Science’s STKE Signal Transduct. Knowl. Environ. 2007 (407), Cm5. doi:10.1126/STKE.4072007CM5
Murai, N. (2014). Plant growth hormone cytokinins control the crop seed yield. Am. J. Plant Sci. 2014, 14. doi:10.4236/ajps.2014.514231
Nagel, L., Brewster, R., Riedell, W. E., and Reese, R. N. (2001). Cytokinin regulation of flower and pod set in soybeans ( Glycine max (L.) merr.). Ann. Bot. 88 (1), 27–31. doi:10.1006/ANBO.2001.1423
Nguyen, H. N., Kambhampati, S., Kisiala, A., Seegobin, M., and Emery, R. J. N. (2021a). The soybean (Glycine max L.) cytokinin oxidase/dehydrogenase multigene family; Identification of natural variations for altered cytokinin content and seed yield. Plant Direct 5 (2), e00308. doi:10.1002/PLD3.308
Nguyen, H. N., Lai, N., Kisiala, A. B., and Emery, R. N. (2021b). Isopentenyltransferases as master regulators of crop performance: Their function, manipulation, and genetic potential for stress adaptation and yield improvement. Plant Biotechnol. J. 19 (7), 1297–1313. doi:10.1111/pbi.13603
Nguyen, H. N., Perry, L., Kisiala, A., Olechowski, H., and Emery, R. J. N. (2020). Cytokinin activity during early kernel development corresponds positively with yield potential and later stage ABA accumulation in field-grown wheat (Triticum aestivum L.). Planta 252 (5), 76. doi:10.1007/S00425-020-03483-2
Nguyen, T., Kisiala, A., Andreas, P., Emery, N., and Narine, S. (2016). Soybean seed development: Fatty acid and phytohormone metabolism and their interactions. Curr. Genomics 17, 241–260. doi:10.2174/1389202917666160202220238
Niemann, M. C. E., Bartrina, I., Ashikov, A., Weber, H., Novák, O., Spíchal, L., et al. (2015). Arabidopsis ROCK1 transports UDP-GlcNAc/UDP-GalNAc and regulates ER protein quality control and cytokinin activity. Proc. Natl. Acad. Sci. U. S. A. 112 (1), 291–296. doi:10.1073/pnas.1419050112
Ninan, A. S., Shah, A., Song, J., and Jameson, P. E. (2017). Differential gene expression in the meristem and during early fruit growth of pisum sativum L. Identifies potential targets for breeding. Int. J. Mol. Sci. 18 (2), E428. doi:10.3390/IJMS18020428
Nishimura, A. L., Mitne-Neto, M., Silva, H. C. A., Richieri-Costa, A., Middleton, S., Cascio, D., et al. (2004a). A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 75 (5), 822–831. doi:10.1086/425287
Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., and Ueguchi, C. (2004b). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in arabidopsis. Plant Cell 16 (6), 1365–1377. doi:10.1105/TPC.021477
Nonokawa, K., Nakajima, T., Nakamura, T., and Kokubun, M. (2012). Effect of synthetic cytokinin application on pod setting of individual florets within raceme in soybean. Plant Prod. Sci. 15 (2), 79–81. doi:10.1626/pps.15.79
Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., et al. (2009). Genome-wide analysis of genes targeted by phytochrome interacting factor 3-like5 during seed germination in arabidopsis. Plant Cell 21 (2), 403–419. doi:10.1105/TPC.108.064691
O’Hara Mosjidis, C., Peterson, C. M., Truelove, B., and Dute, R. R. (1993). Stimulation of pod and ovule growth of soybean, glycine max (L.) merr. by 6-benzylaminopurine. Ann. Bot. 71 (3), 193–199. doi:10.1006/ANBO.1993.1024
Ohto, M. aki, Floyd, S. K., Fischer, R. L., Goldberg, R. B., and Harada, J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 22 (4), 277–289. doi:10.1007/s00497-009-0116-1
Okazaki, K., Kabeya, Y., Suzuki, K., Mori, T., Ichikawa, T., Matsui, M., et al. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21, 1769–1780. doi:10.1105/tpc.109.067785
O’Keefe, D., Song, J., and Jameson, P. E. (2011). Isopentenyl transferase and cytokinin oxidase/dehydrogenase gene family members are differentially expressed during pod and seed development in rapid-cycling Brassica. J. Plant Growth Regul. 30 (1), 92–99. doi:10.1007/S00344-010-9171-Y
Pareek, A., Singh, A., Kumar, M., Kushwaha, H. R., Lynn, A. M., and Singla-Pareek, S. L. (2006). Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 142 (2), 380–397. doi:10.1104/PP.106.086371
Park, J., Lee, S., Park, G., Cho, H., Choi, D., Umeda, M., et al. (2021). C Yto K Inin-Responsive G Rowth Regulator regulates cell expansion and cytokinin-mediated cell cycle progression. Plant Physiol. 186 (3), 1734–1746. doi:10.1093/plphys/kiab180
Peterson, C. H., Summerson, H. C., and Duncan, P. B. (1984). The influence of seagrass cover on population structure and individual growth rate of a suspension-feeding bivalve, Mercenaria mercenaria. J. Mar. Res. 42 (1), 123–138. doi:10.1357/002224084788506194
Peterson, C. M., Williams, J. C., and Kuang, A. (2015). Increased pod set of determinate cultivars of soybean, Glycine max, with 6-benzylaminopurine. Bot. Gaz. 151 (3), 322–330. doi:10.1086/33783210.1086/337832
Pils, B., and Heyl, A. (2009). Unraveling the evolution of cytokinin signaling. Plant Physiol. 151 (2), 782–791. doi:10.1104/PP.109.139188
Pino, E., Martin, L., Guerra, H., Nicolas, G., and Villalobos, N. (1991). Effect of dihydrozeatin on the mobilization of protein reserves in protein bodies during the germination of chick-pea seeds. J. Plant Physiology 137 (4), 425–432. doi:10.1016/S0176-1617(11)80311-1
Prerostova, S., Dobrev, P. I., Knirsch, V., Jarosova, J., Gaudinova, A., Zupkova, B., et al. (2021). Light quality and intensity modulate cold acclimation in Arabidopsis. Int. J. Mol. Sci. 22 (5), 2736. doi:10.3390/ijms22052736
Quesnelle, P. E., and Emery, R. J. N. (2007). cis-cytokinins that predominate in Pisum sativum during early embryogenesis will accelerate embryo growth in vitro. Can. J. Bot. 85 (1), 91–103. doi:10.1139/B06-149
Rashotte, A. M., Mason, M. G., Hutchison, C. E., Ferreira, F. J., Schaller, G. E., and Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathwayProc. Natl. Acad. Sci. U. S. A. J. Exp. Bot. 103 (29), 11081–11085. doi:10.1073/PNAS.0602038103/SUPPL_FILE/02038FIG11
Reid, D. E., Heckmann, A. B., Novák, O., Kelly, S., and Stougaard, J. (2016). CYTOKININ OXIDASE/DEHYDROGENASE3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol. 170 (2), 1060–1074. doi:10.1104/PP.15.00650
Ren, B., Liang, Y., Deng, Y., Chen, Q., Zhang, J., Yang, X., et al. (2009). Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 19 (1010), 1178–1190. doi:10.1038/cr.2009.88
Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 (1), 40–54. doi:10.1105/TPC.105.037796
Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J. A. H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Sci. (New York, N.Y.) 283 (5407), 1541–1544. doi:10.1126/SCIENCE.283.5407.1541
Roeckel, P., Oancia, T., and Drevet, J. (1997). Effects of seed-specific expression of a cytokinin biosynthetic gene on canola and tobacco phenotypes. Transgenic Res. 6, 133–141. doi:10.1023/a:1018425720949
Romanov, G. A., Kieber, J. J., and Schmülling, T. (2002). A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signalling. FEBS Lett. 515 (1–3), 39–43. doi:10.1016/S0014-5793(02)02415-8
S Singh, J. P. (1988). A rapid method for determination of nitrate in soil and plant extracts. Plant Soil 110 (1), 137–139. doi:10.1007/bf02143549
Sakakibara, H. (2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 (1), 431–449. doi:10.1146/annurev.arplant.57.032905.105231
Sawan, Z. M., Mohamed, A. A., Sakr, R. A., and Tarrad, A. M. (2000). Effect of kinetin concentration and methods of application on seed germination, yield components, yield and fiber properties of the Egyptian cotton (Gossypium barbadense). Environ. Exp. Bot. 44 (1), 59–68. doi:10.1016/S0098-8472(00)00054-X
Schaller, G. E., Shiu, S. H., and Armitage, J. P. (2011). Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 21 (9), R320–R330. doi:10.1016/J.CUB.2011.02.045
Schaller, G. E., Street, I. H., and Kieber, J. J. (2014). Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 21, 7–15. doi:10.1016/J.PBI.2014.05.015
Schoor, S., Farrow, S., Blaschke, H., Lee, S., Perry, G., von Schwartzenberg, K., et al. (2011). Adenosine kinase contributes to cytokinin interconversion in arabidopsis. Plant Physiol. 157 (2), 659–672. doi:10.1104/PP.111.181560
Schou, J. B., Jeffers, D. L., and Streeter, J. G. (1978). Effects of reflectors, black boards, or shades applied at different stages of plant development on yield of Soybeans1. Crop Sci. 18 (1), 29–34. doi:10.2135/CROPSCI1978.0011183X001800010009X
Schwarz, I., Scheirlinck, M. T., Otto, E., Bartrina, I., Schmidt, R. C., and Schmülling, T. (2020). Cytokinin regulates the activity of the inflorescence meristem and components of seed yield in oilseed rape. J. Exp. Bot. 71 (22), 7146–7159. doi:10.1093/JXB/ERAA419
semanticscholar (2022). Design and evaluation of soybean ideotypes | Semantic Scholar. Retrieved May 10, 2022, from Available at: https://www.semanticscholar.org/paper/Design-and-evaluation-of-soybean-ideotypes-Kokubun/c313a8a8f90f4f9852ae730dd8e1a9ee7b260694.
Shibles, R., Anderson, I. C., and Gibson, A. H. (1975). “Soybean,” in Crop physiology. Editor L. T. Evans (London: Cambridge Univ. Press), 151–189.
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40 (8), 1023–1028. doi:10.1038/NG.169
Sievenpiper, J. L., Kendall, C. W. C., Esfahani, A., Wong, J. M. W., Jiang, H. Y., Bazinet, R. P., et al. (2009). Effect of non-oil-seed pulses on glycaemic control: A systematic review and meta-analysis of randomised controlled experimental trials in people with and without diabetes. Diabetologia 52, 1479–1495. doi:10.1007/s00125-009-1395-7
Singh, J., Clavijo Michelangeli, J. A., Gezan, S. A., Lee, H., and Vallejos, C. E. (2017). Maternal effects on seed and seedling phenotypes in reciprocal F 1 hybrids of the common bean (Phaseolus vulgaris L.). Front. Plant Sci. 8, 42. doi:10.3389/fpls.2017.00042
Singh, S., Letham, D. S., Jameson, P. E., Zhang, R., Parker, C. W., Bandenoch-Jones, J., et al. (1988). Cytokinin biochemistry in relation to leaf senescence: IV. Cytokinin metabolism in soybean explants. Plant Physiol. 88 (3), 788–794. doi:10.1104/pp.88.3.788
Song, J., Jiang, L., and Jameson, P. E. (2012). Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol. 12, 78. doi:10.1186/1471-2229-12-78
Song, J., Jiang, L., and Jameson, P. E. (2015). Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase, and amino acid permease gene family members in leaf, flower, silique, and seed development. J. Exp. Bot. 66 (16), 5067–5082. doi:10.1093/JXB/ERV133
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 (5), 623–630. doi:10.1038/ng2014
Spíchal, L., and Spíchal, L. (2012). Cytokinins – recent news and views of evolutionally old molecules. Funct. Plant Biol. 39 (4), 267–284. doi:10.1071/FP11276
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi:10.1146/ANNUREV.BIOCHEM.69.1.183
Strnad, M. (1997). The aromatic cytokinins. Physiol. Plant. 101 (4), 674–688. doi:10.1111/J.1399-3054.1997.TB01052.X
Sun, J., Li, L., Liu, M., Wang, M., Ding, M., Deng, S., et al. (2010). Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl-stressed callus cells of two contrasting poplars. Plant Cell Tissue Organ Cult. 103 (2), 205–215. doi:10.1007/S11240-010-9768-7
Sun, J., Niu, Q. W., Tarkowski, P., Zheng, B., Tarkowska, D., Sandberg, G., et al. (2003). The arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol. 131 (1), 167–176. doi:10.1104/PP.011494
Sundaresan, V. (2005). Control of seed size in plants. Proc. Natl. Acad. Sci. U. S. A. 102 (50), 17887–17888. doi:10.1073/PNAS.0509021102
Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Bäurle, I., Kudla, J., et al. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Sci. (New York, N.Y.) 294 (5544), 1108–1111. doi:10.1126/SCIENCE.1065022
Tahaei, A., Soleymani, A., and Shams, M. (2016). Seed germination of medicinal plant, fennel (Foeniculum vulgare Mill), as affected by different priming techniques. Appl. Biochem. Biotechnol. 180 (1), 26–40. doi:10.1007/S12010-016-2082-Z
Tajima, Y., Imamura, A., Kiba, T., Amano, Y., Yamashino, T., and Mizuno, T. (2004). Comparative studies on the type-B response regulators revealing their distinctive properties in the his-to-asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 45 (1), 28–39. doi:10.1093/PCP/PCG154
Takagi, M., Yokota, T., Murofushi, N., Ota, Y., and Takahashi, N. (1985). Fluctuation of endogenous cytokinin contents in rice during its life cycle—quantification of cytokinins by selected ion monitoring using deuterium-labelled internal standards. Agric. Biol. Chem. 49 (11), 3271–3277. doi:10.1080/00021369.1985.10867233
Takei, K., Yamaya, T., and Sakakibara, H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 279 (40), 41866–41872. doi:10.1074/JBC.M406337200
Talukdar, M., Swain, D. K., and Bhadoria, P. B. S. (2022). Effect of IAA and BAP application in varying concentration on seed yield and oil quality of Guizotia abyssinica (L.f.) Cass. Ann. Agric. Sci. 67 (1), 15–23. doi:10.1016/J.AOAS.2022.02.002
Taylor, J. S., Harker, K. N., Robertson, J. M., and Foster, K. R. (1990). The effect of A seaweed extract containing cytokinin on the growth and yield of barley. Can. J. Plant Sci. 70 (4), 1163–1167. doi:10.4141/CJPS90-140
To, J. P. C., Deruère, J., Maxwell, B. B., Morris, V. F., Hutchison, C. E., Ferreira, F. J., et al. (2007). Cytokinin regulates type-A arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19 (12), 3901–3914. doi:10.1105/TPC.107.052662
To, J. P. C., and Kieber, J. J. (2008). Cytokinin signaling: Two-components and more. Trends Plant Sci. 13 (2), 85–92. doi:10.1016/J.TPLANTS.2007.11.005
Tokunaga, H., Kojima, M., Kuroha, T., Ishida, T., Sugimoto, K., Kiba, T., et al. (2012). Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG‐dependent pathway in cytokinin activation. Plant J. 69 (2), 355–365. doi:10.1111/j.1365-313X.2011.04795.x
Tzou, D.-S., Galson, E. C., and Sondheimer, E. (1973). The metabolism of hormones during seed germination and release from dormancy: III. The effects and metabolism of zeatin in dormant and nondormant ash embryos. Plant Physiol. 51 (5), 894–897. doi:10.1104/PP.51.5.894
Veach, Y. K., Martin, R. C., Mok, D. W. S., Malbeck, J., Vankova, R., and Mok, M. C. (2003). O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 131 (3), 1374–1380. doi:10.1104/PP.017210
Viguiliouk, E., Blanco Mejia, S., Kendall, C. W., and Sievenpiper, J. L. (2017). Can pulses play a role in improving cardiometabolic health? Evidence from systematic reviews and meta‐analyses. Ann. N. Y. Acad. Sci. 1392 (1), 43–57. doi:10.1111/nyas.13312
Viguiliouk, E., Stewart, S. E., Jayalath, V. H., Ng, A. P., Mirrahimi, A., De Souza, R. J., et al. (2015). Effect of replacing animal protein with plant protein on glycemic control in diabetes: A systematic review and meta-analysis of randomized controlled trials. Nutrients 7 (12), 9804–9824. doi:10.3390/nu7125509
Vyroubalová, Š., Václavíková, K., Turečková, V., Novák, O., Šmehilová, M., Hluska, T., et al. (2009). Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol. 151 (1), 433–447. doi:10.1104/PP.109.142489
Wang, C., Wang, H., Zhu, H., Ji, W., Hou, Y., Meng, Y., et al. (2021). Genome-wide identification and characterization of cytokinin oxidase/dehydrogenase family genes in Medicago truncatula. J. Plant Physiol. 256, 153308. doi:10.1016/j.jplph.2020.153308
Wang, Y., Li, L., Ye, T., Zhao, S., Liu, Z., Feng, Y. Q., et al. (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 68 (2), 249–261. doi:10.1111/J.1365-313X.2011.04683.X
Werner, S., Bartrina, I., Novák, O., Strnad, M., Werner, T., and Schmülling, T. (2021). The cytokinin status of the epidermis regulates aspects of vegetative and reproductive development in Arabidopsis thaliana. Front. Plant Sci. 12, 613488. doi:10.3389/fpls.2021.613488
Werner, T., Köllmer, I., Bartrina, I., Holst, K., and Schmülling, T. (2006). New insights into the biology of cytokinin degradation. Plant Biol. 8 (3), 371–381. doi:10.1055/S-2006-923928
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 (11), 2532–2550. doi:10.1105/TPC.014928
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12 (5), 527–538. doi:10.1016/j.pbi.2009.07.002
Whitty, C. D., and Hall, R. H. (1974). A cytokinin oxidase in Zea mays. Can. J. Biochem. 52 (9), 789–799. doi:10.1139/O74-112
Wu, A., Su, C., Wang, D., Peng, Y., Liu, M., Hua, S., et al. (2013). Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe 14 (4), 446–452. doi:10.1016/J.CHOM.2013.09.001
Xia, N., Zhang, L., Wang, G., Feng, Q., and Liu, L. (2013). Label-free and sensitive strategy for microRNAs detection based on the formation of boronate ester bonds and the dual-amplification of gold nanoparticles. Biosens. Bioelectron. 47, 461–466. doi:10.1016/J.BIOS.2013.03.074
Yang, H., Postel, S., Kemmerling, B., and Ludewig, U. (2014). Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ. 37, 1404–1414. doi:10.1111/pce.12244
Yashima, Y., Kaihatsu, A., Nakajima, T., and Kokubun, M. (2015). Effects of source/sink ratio and cytokinin application on pod set in soybean. Plant Prod. Sci. 8 (2), 139–144. doi:10.1626/PPS.8.139
Ye, X. L., Yeung, E. C., and Zee, S. Y. (2002). Sperm movement during double fertilization of a flowering plant, Phaius tankervilliae. Planta 215 (1), 60–66. doi:10.1007/S00425-002-0736-2
Yokoyama, A., Yamashino, T., Amano, Y. I., Tajima, Y., Imamura, A., Sakakibara, H., et al. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48 (1), 84–96. doi:10.1093/PCP/PCL040
Zhang, R., and Letham, D. S. (1990). Cytokinin translocation and metabolism in lupin species. III. Translocation of xylem cytokinin into the seeds of lateral shoots of Lupinus angustifolius. Plant Sci. 70 (1), 65–71. doi:10.1016/0168-9452(90)90033-K
Zhang, X., Chen, Y., Lin, X., Hong, X., Zhu, Y., Li, W., et al. (2013). Adenine phosphoribosyl transferase 1 is a key enzyme catalyzing cytokinin conversion from nucleobases to nucleotides in Arabidopsis. Mol. Plant 6 (5), 1661–1672. doi:10.1093/mp/sst071
Zhou, Y., Zhang, X., Kang, X., Zhao, X., Zhang, X., and Ni, M. (2009). Short hypocotyl under Blue1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate arabidopsis seed Development.MINISEED3 and HAIKU2 promoters in vivo to regulate arabidopsis seed development. Plant Cell 21, 106–117. doi:10.1105/tpc.108.064972
Zu, S. H., Jiang, Y. T., Chang, J. H., Zhang, Y. J., Xue, H. W., and Lin, W. H. (2022). Interaction of brassinosteroid and cytokinin promotes ovule initiation and increases seed number per silique in Arabidopsis. J. Integr. Plant Biol. 64 (3), 702–716. doi:10.1111/JIPB.13197
Zu, S. H., Jiang, Y. T., Chang, J. H., Zhang, Y. J., Xue, H. W., and Lin, W. H. (2022). Interaction of brassinosteroid and cytokinin promotes ovule initiation and increases seed number per silique in Arabidopsis. J. Integr. Plant Biol. 64 (3), 702–716. doi:10.1111/jipb.13197
Zuñiga-Mayo, V. M., Baños-Bayardo, C. R., Díaz-Ramírez, D., Marsch-Martínez, N., and De Folter, S. (2018). Conserved and novel responses to cytokinin treatments during flower and fruit development in Brassica napus and Arabidopsis thaliana. Sci. Rep. 6836 (1), 1–10. doi:10.1038/s41598-018-25017-3
Zwack, P. J., and Rashotte, A. M. (2013). Cytokinin inhibition of leaf senescence. Plant Signal. Behav. 8 (7), e24737. doi:10.4161/psb.24737
Keywords: cytokinin, yield contributing traits, pulses and oilseeds, seed development, CKX, cytokinin oxidase/dehydrogenase
Citation: Sharma S, Kaur P and Gaikwad K (2022) Role of cytokinins in seed development in pulses and oilseed crops: Current status and future perspective. Front. Genet. 13:940660. doi: 10.3389/fgene.2022.940660
Received: 10 May 2022; Accepted: 11 July 2022;
Published: 12 October 2022.
Edited by:
Santosh Kumar Gupta, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Pranav Pankaj Sahu, Max Planck Institute for Chemical Ecology, GermanyBhawana Israni, Max Planck Institute for Chemical Ecology, Germany
Rupesh K. Deshmukh, Plaksha University, India
Copyright © 2022 Sharma, Kaur and Gaikwad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kishor Gaikwad, a2lzaDIwMTJAZ21haWwuY29t
 Sandhya Sharma
Sandhya Sharma Parampreet Kaur
Parampreet Kaur Kishor Gaikwad
Kishor Gaikwad
