- 1Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, Jiangxi Agricultural University, Nanchang, China
- 2College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
Plants usually respond to the external environment by initiating a series of signal transduction processes mediated by protein kinases, especially calcineurin B-like protein–interacting protein kinases (CIPKs). In this study, 54 CIPKs were identified in the peanut genome, of which 26 were from cultivated species (named AhCIPKs) and 28 from two diploid progenitors (Arachis duranensis—AdCIPKs and Arachis ipaensis—AiCIPKs). Evolution analysis revealed that the 54 CIPKs were composed of two different evolutionary branches. The CIPK members were unevenly distributed at different chromosomes. Synteny analysis strongly indicated that whole-genome duplication (allopolyploidization) contributed to the expansion of CIPK. Comparative genomics analysis showed that there was only one common collinear CIPK pairs among peanut, Arabidopsis, rice, grape, and soybean. The prediction results of cis-acting elements showed that AhCIPKs, AdCIPKs, and AiCIPKs contained different proportions of transcription factor binding motifs involved in regulating plant growth, abiotic stress, plant hormones, and light response elements. Spatial expression profiles revealed that almost all AhCIPKs had tissue-specific expression patterns. Furthermore, association analysis identified one polymorphic site in AdCIPK12 (AhCIPK11), which was significantly associated with pod length, seed length, hundred seed weight, and shoot root ratio. Our results provide valuable information of CIPKs in peanut and facilitate better understanding of their biological functions.
Introduction
Calcium is an important second messenger in plants (Feng et al., 2017), and the signal transduction pathway mediated by calcium plays an important role in plant growth, development, and stress (Mao et al., 2016). Calcium signals formed during plant growth are further transmitted by calcium sensor proteins. Common calcium sensor proteins include calmodulins (CaMs), calmodulin-like proteins (CMLs), calcium-dependent protein kinases (CDPK), calcineurin B-like proteins (CBL), and CBL-interacting protein kinase (CIPK) (Zhao et al., 2009). CIPK, the interacting protein kinase of CBL, is a kind of protein that interacts with activated CBL as a downstream protein and is also a kind of Ca2+-dependent serine/threonine protein kinase (Kim et al., 2000), which contains a conserved catalytic kinase domain at its N-terminal (Albrecht et al., 2001). The C-terminal is an NAF regulatory control domain composed of 24 amino acids, which is highly conserved and mediates the interaction between CBL and CIPK protein (Kolukisaoglu et al., 2004).
Up to now, 25, 30, 16, and 43 CIPK genes have been identified in Arabidopsis thaliana (Yu et al., 2007), Oryza sativa (Kolukisaoglu et al., 2004), Vitis vinifera (Lu et al., 2017), and Zea mays (Chen et al., 2011), respectively. CIPK genes were widely involved in stress response, growth, and development regulation of plants (Hu et al., 2015). Studies of Arabidopsis have shown that the protein encoded by AtCIPK23 played an important role in potassium metabolism (Wang and Wu, 2009). The combination of AtCBL1, AtCBL9, and AtCIPK23 activated the potassium transport channel (Cheong et al., 2007). AtCIPK15 interacted with PP2C phosphatase ABI2 to form a complex, which controlled the expression of ABA-related genes (Li et al., 2006). AtCIPK3 responded to various abiotic stresses through ABA-dependent or ABA-independent pathway (Xu et al., 2006). It was found that overexpression of OsCIPK3, OsCIPK12, and OsCIPK15 improved cold, drought, and salt stress tolerance of rice, respectively (Xiang et al., 2007). Overexpression of wheat CIPK24 increased the contents of Na+ and antioxidant protective enzymes (Hrabak et al., 2003) and further improved the tolerance of A. thaliana to high salt stress (Deng et al., 2013). Apple CIPK6 interacted with AtCBL4 protein after transferring A. thaliana AtCBL4 gene into apple and improved the resistance of apple seedlings to low temperature, drought, and high salt stress (Wang and Liu, 2018). With the completion of large-scale plant genome sequencing, the research on CIPK genes in soybean (Feng et al., 2015), poplar (Sun et al., 2015), and many other plants received considerable attention, but research on CIPK genes in peanut has not been reported yet.
Peanut is one of the major oil and cash crops in China (Pandey et al., 2020). Calcium is the second largest nutrient element in peanut (Rui, 2015). In recent years, frequent occurrence of land drought, cold damage, and soil salinization has been a serious impact on the increase of peanut yield and the improvement of peanut quality (Lu et al., 2017). Under the pressure of reducing production cost and protecting the environment, screening peanut varieties with low calcium tolerance and exploring multistress response proteins and stress resistance mechanisms in an adverse environment have become the top priority in studying the stress resistance breeding of peanut (Sanders et al., 2002). To deal with abiotic stresses such as drought, cold, and salt damage and improve the yield and quality of peanut, it is of great significance to explore peanut CIPK genes and reveal their role in the calcium signaling pathway. In this study, the CIPK genes of cultivated peanut and its two diploid progenitors were comprehensively analyzed. At last, a total of 54 CIPK genes were explicitly identified. Their basic protein information, exon–intron structure, phylogeny, and cis-acting elements were systematically analyzed, which provided valuable theoretical basis and genetic resources for the high-yield breeding of peanut.
Materials and methods
Genome-wide identification of the CIPK genes in peanut
To identify CIPK genes in peanut, the protein sequences of three peanut genomes were downloaded from PeanutBase (http://www.peanutbase.org/). The conserved domains of all proteins encoded by peanut genome were analyzed using the HMMER 3.0 software, and genes including both the Pkinase (PF00069.24) and NAF (PF03822.13) domains were selected as peanut CIPK candidates. The PROSTIE and SMART software were used to verify the 54 CIPKs as calcineurin B-like protein–interacting protein kinases. Candidates with PROTEIN_KINASE_DOM (PS50011) and NAF (PS50816) in PROSTIE and the S_TKc (SM00220) domain in SMART were selected as CIPKs. The physicochemical data such as gene number, coding sequence (CDS) length, amino acid number, isoelectric point, molecular weight, and EF hand structure number were obtained from PeanutBase or analyzed using the ExPASy Proteomics Sever online tool (Gasteiger et al., 2005).
Evolution and structure analysis of CIPK genes in peanut
The gene structure diagram was drawn using the GSDS 2.0 mapping software based on the genome annotation information of CIPK genes with the GFF format, which was downloaded from PeanutBase (http://www.peanutbase.org/). The phylogenetic tree using 179 CIPK proteins from peanut, rice, grape, Arabidopsis, and soybean was constructed using the MEGA 5.2 software with the neighbor-joining method (Bootstrap value 1,000, Poisson model, uniform rates, pairwise deletion). The analyses of the composition of conserved motifs were conducted using MEME (http://meme-suite.org/tools/meme) with the maximum number 20 (classic mode, zero or one occurrence per sequence).
Cis-acting elements analysis of peanut CIPKs
We defined the 2-kb upstream sequence of the initiation codon as the promoter of the peanut CIPKs and downloaded it from PeanutBase to search for cis-acting regulatory elements through PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Then, only the cis-acting elements related to adversity stress were screened out statistically.
Expression profiles of AhCIPK genes in different tissues and treatments
RNA-seq datasets of 22 peanut tissues were downloaded from PeanutBase (http://www.peanutbase.org/) and NCBI SRA (https://www.ncbi.nlm.nih.gov/sra/), and the expression levels of AhCIPK genes in different tissues were obtained (Clevenger et al., 2016) with all raw data deposited as BioSamples SAMN03944933–SAMN03944990. The expression data (FPKM value) of peanut CIPK genes were normalized and output using the TBtools software (Chen et al., 2020). Ralstonia solanacearum infection was carried out as described before according to (Zhang et al. 2017). Submergence treatment followed the method described by (Zeng et al. 2021).
Candidate gene association mapping
The genotype data of the CIPK genes used for association analyses were obtained from the transcriptome sequencing data of a peanut germplasm population with 146 accessions (unpublished data). The phenotypes were collected from five environments (Wuhan 2016, Wuhan 2017, Yangluo 2016, Yangluo 2017, Zhanjiang 2016). Three replicates were randomly planted in each environment, with 12 plants in each row.
Results
Genome-wide identification of the CIPK genes in cultivated peanut and its diploid progenitors
To systematically determine CIPK genes in peanut, genes containing both the conserved Pkinase (PF00069.24) and NAF (PF03822.13) domains were searched through the whole peanut genome. The SMART and PROSTIE software tools were used to verify the Pkinase domains. A total of 54 CIPK candidates were identified from the peanut genome of cultivated species Arachis hypogaea (26, namely AhCIPK1–AhCIPK26) and its two wild species Arachis duranensis (13, namely, AdCIPK1–AdCIPK13) and Arachis ipaensis (15, namely, AiCIPK1–AiCIPK15) (Table 1). Then, we determined their chromosome locations, mRNA length, number of amino acids (aa), MW, theoretical pI, and transmembrane domain (TMD) (Table 1). AhCIPK genes were distributed on chromosomes 1, 2, 3, 7, 8, 9, and 10 (A genome) and 11, 12, 13, 17, 19, and 20 (B genome). Two wild species–specific CIPK genes (AdCIPK7 and AiCIPK5) were uncovered (Figure 1). The gene length of the peanut CIPKs ranged from 1,146 to 3,177 bps, of which the shortest was AiCIPK4 with 1,146 bps and the longest length was AhCIPK22 with 3,177 bps. The amino acid length of CIPKs varied from 381 to 570. The isoelectric point ranged from 5.8 (AdCIPK11) to 9.79 (AiCIPK8), and the molecular weight ranged from 45,067.06 to 64,189.82 Da. All CIPKs do not contain a TMD.
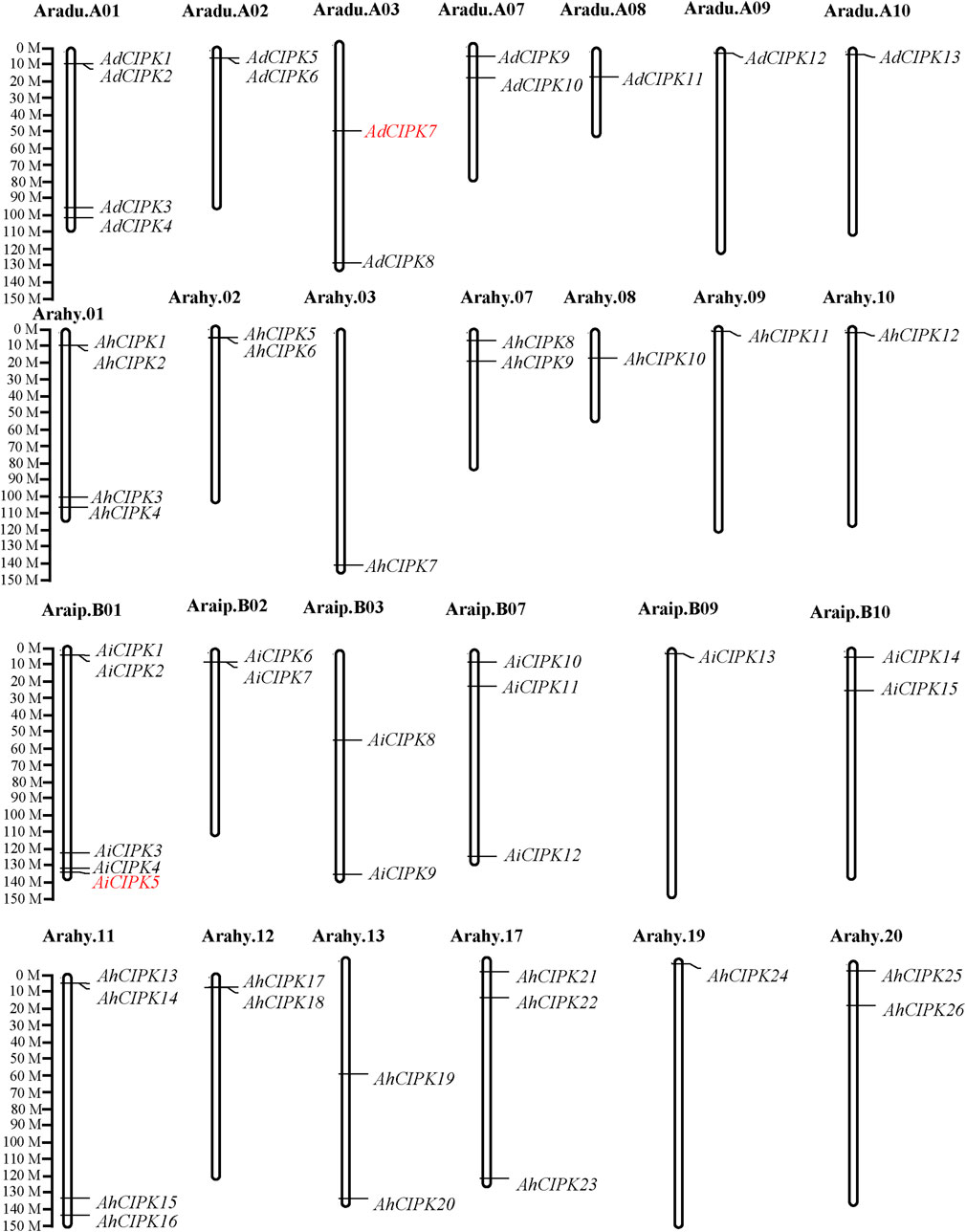
FIGURE 1. Chromosomal locations of peanut calcineurin B-like protein–interacting protein kinase (CIPK) genes. Chromosomal positions of the peanut CIPK genes were mapped based on GFF data downloaded from PeanutBase. The chromosome number is indicated above each chromosome. Genes in red mean wild species specific.
Phylogenetic and gene structure analysis of CIPKs in peanut
To determine the evolutionary relationship of CIPKs among A. hypogaea, A. duranensis, and A. ipaensis, the phylogenetic tree of the 54 CIPKs was constructed. The results indicated that the CIPKs can be classified into two clades (I and Ⅱ) (Figure 2A). Clades I and II consisted of 31 CIPKs (15 AhCIPKs, 8 AdCIPKs, and 8 AiCIPKs) and 23 CIPKs (11 AhCIPKs, 5 AdCIPKs, and 7 AiCIPKs), respectively. It is interesting that the results of the gene structure based on the genome annotations also showed that the CIPK genes can be divided into two groups, corresponding to the two phylogenetic families (the intron-rich group corresponded to phylogenetic clade I, and the intron-less group corresponded to phylogenetic clade Ⅱ). The intron numbers of the intron-less group were less than 3 (0 to 2), while those of the intron-rich group were more than 10 (Figure 2B). Further, the phylogenetic relationship and classification of peanut CIPKs were supported by motif analysis. A total of 20 motifs were identified (Figure 2C); in general, peanut CIPKs had 11–17 motifs. Motif 1, Motif 2, and Motif 4 were the most common, present in all CIPK proteins. Otherwise, the vast majority of CIPKs included Motifs 3, 5, 6, 7, and 8, which covered more than 50 CIPK members. Motifs 13 and 19 were clade-specific elements in clade II, and Motif 17 only existed in AhCIPK15 of clade I but 13 CIPKs in clade II.
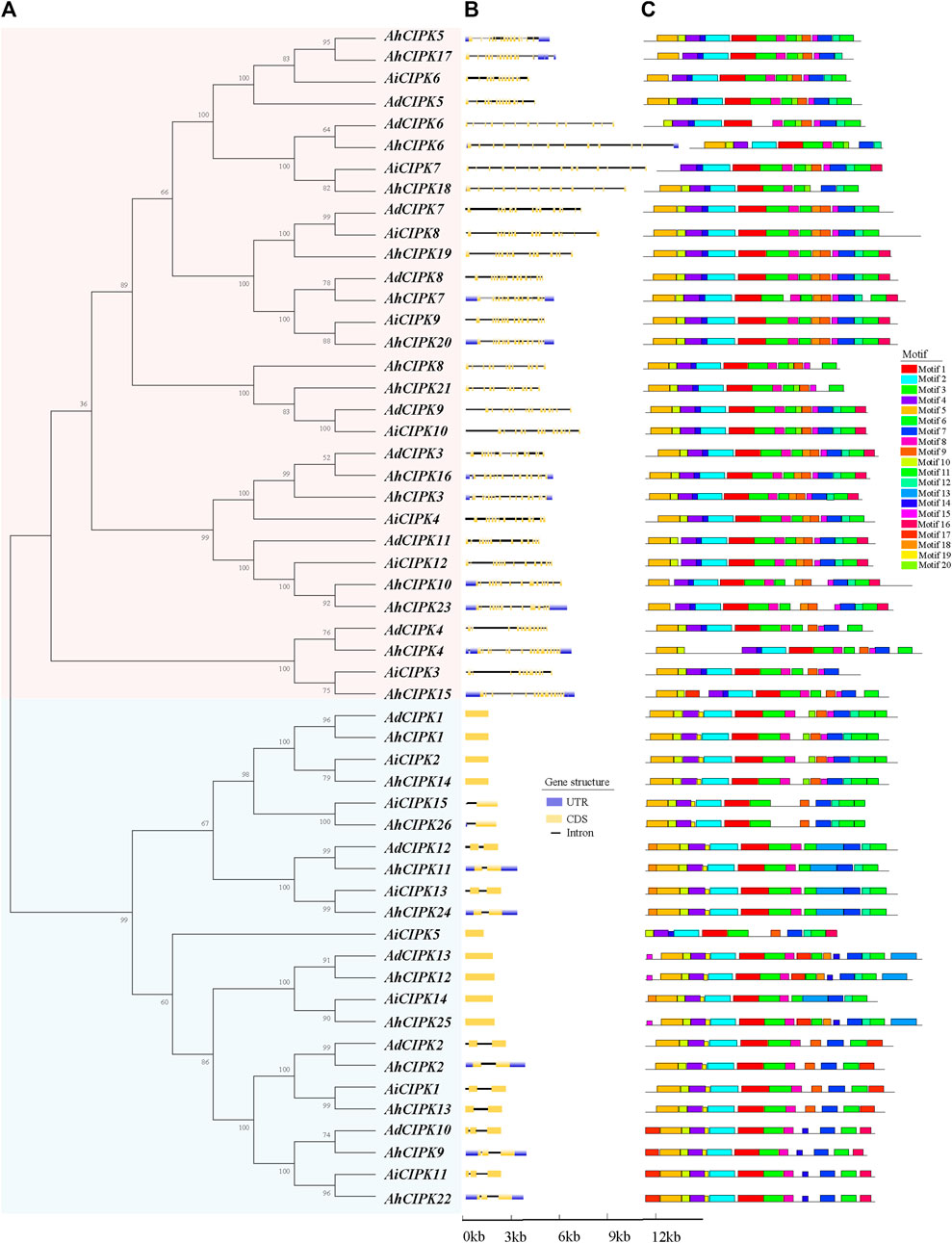
FIGURE 2. Comparison of the gene structure and motif of 54 CIPK genes in peanut. (A) unrooted phylogenetic tree with over 50% bootstrap value above the branch. Clades I and II are displayed in pink and blue colors, respectively. The names of the species are abbreviated to two letters, named as Arachis duranensis (Ad), Arachis ipaensis (Ai), and Arachis hypogaea L. Tifrunner (Ah). (B) exon/introns and untranslated regions (UTRs) of CIPKs. Blue boxes denote UTR (untranslated region); yellow boxes denote coding sequence (CDS); and black lines denote introns. The length of protein can be estimated using the scale at the bottom. (C) motif architectures of all CIPK genes. Each motif is illustrated with a specific color, and the distribution of identified motifs corresponds to their positions.
Biological evolution analysis of CIPKs in peanut and other plant species
To further understand the relationship of CIPK members among different species, the phylogenetic tree of CIPK proteins of Arabidopsis, rice, grape, soybean [AtCIPK (26), OsCIPK (31), VvCIPK(16), and GmCIPK (52)], and peanut was constructed using maximum parsimony (Figure 3). The results showed that CIPK proteins of these species can be divided into two subfamilies (Ⅰ and Ⅱ). The analysis of the phylogenetic tree revealed that all the peanut CIPKs were clustered together (Figure 3). The relationships between the two wild species A. duranensis and A. ipaensis and cultivated species A. hypogaea were closer than that between the other four species. In addition, many CIPK members of peanut and soybean clustered together, suggesting that the two legume species were evolutionarily closer than others. TThe second evolutionary closest of peanut was Arabidopsis. There were more family members in peanut and soybean than in Arabidopsis, rice, and grape plants, suggesting a specific linear amplification of the gene family in legume plant. Whether these additional members of the genes have additional functions as well or whether they are produced only because functional redundancy requires further experimental verification.
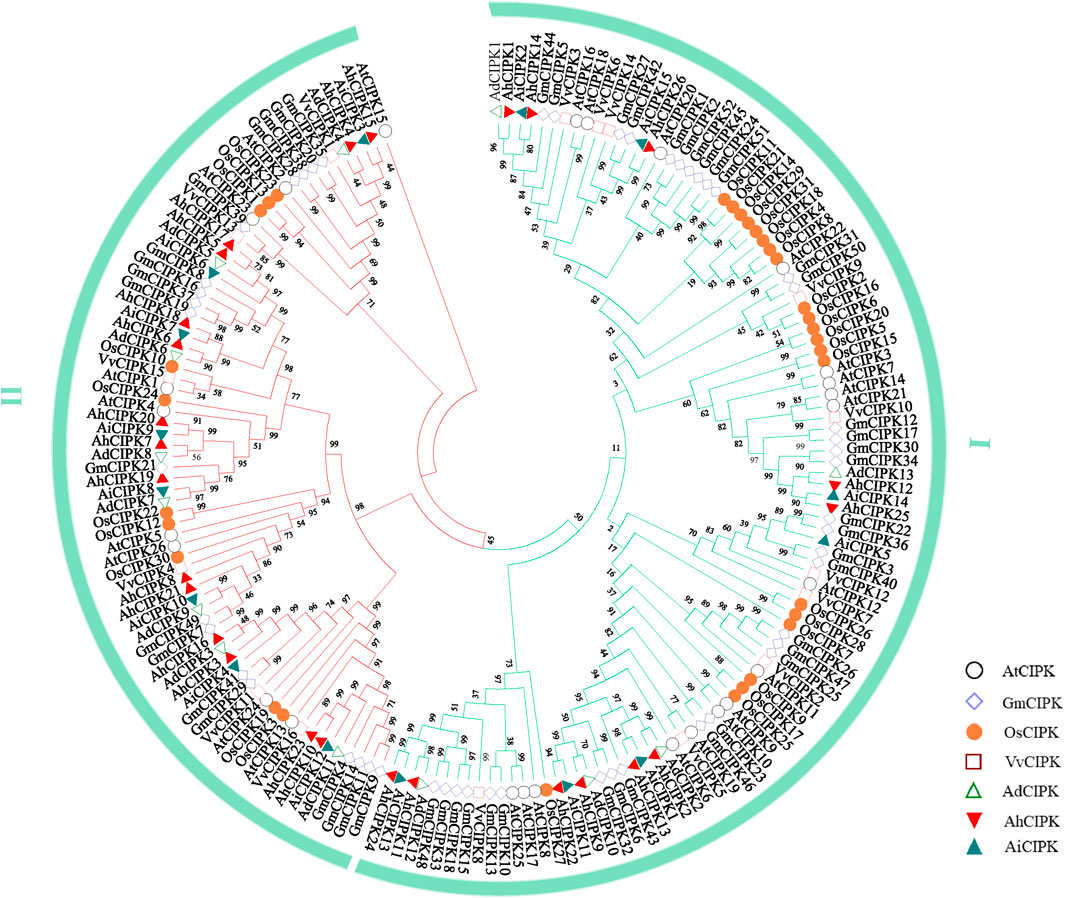
FIGURE 3. Phylogenetic tree of CIPKs in peanut and other plant species. The plants in the square frame indicate that the CIPK members outside of peanuts have the closest evolutionary relationship with soybean CIPKs.
Gene duplication and synteny analyses of peanut CIPKs
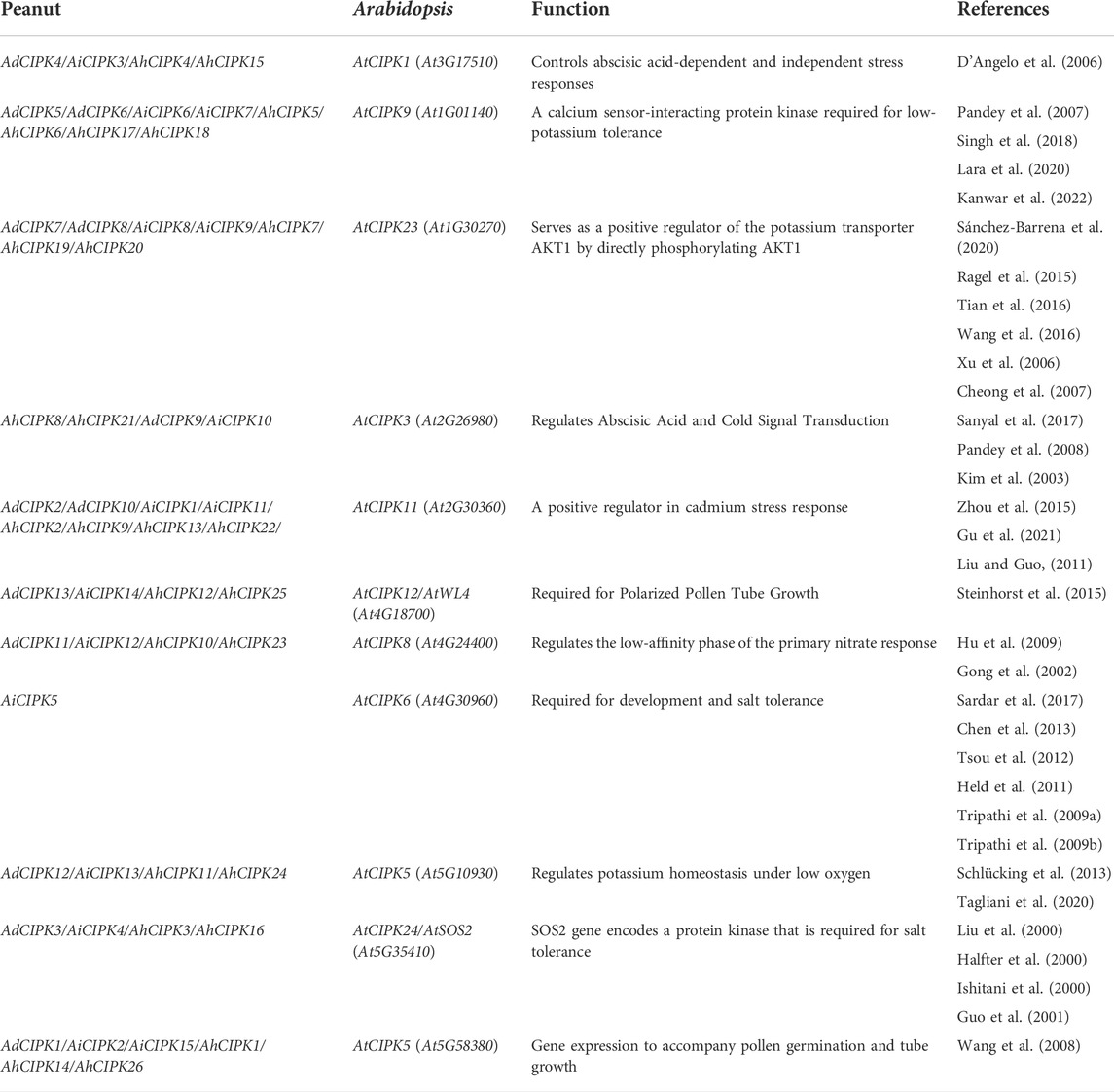
Chromosomal location analyses revealed that the 26 AhCIPKs distributed unevenly on 13 chromosomes (chromosomes 01, 02, 03, 07, 08, 09, 10, 11, 12, 13, 17, 19, and 20). The 13 AdCIPKs presented on chromosomes A01, A02, A03, A07, A08, A09, and A10, and the 15 AiCIPKs distributed on chromosomes B01, B04, B06, B09, and B10 (Figure 4). A total of 16 chromosomal fragment repeat gene pairs were identified without tandem repeats (Figure 4, Supplementary Table S1). Further, we calculated the Ks (synonymous) and Ka (nonsynonymous) values of the duplicated gene pairs and found that the Ka/Ks ratio for duplicated AhCIPK gene pairs ranged from 0.00 to 0.57 with an average of 0.17 (Supplementary Table S2). The ω values of all duplicated gene pairs were less than 1, showing that purifying selection occurred on these duplicated gene pairs. Synteny analysis with Arabidopsis, rice, grape, and soybean revealed one conserved CIPK gene (AhCIPK14) in these species (Figure 5, Supplementary Table S1). BLASTP methods were used to identify peanut CIPK gene orthologs between peanut and Arabidopsis. In total, we found 54 orthologous gene pairs between peanut and Arabidopsis (Table 2). The orthologs in Arabidopsis included AtCIPK12/AtWL4 and AtCIPK5 participating in pollen germination and tube growth (Wang et al., 2008; Steinhorst et al., 2015), AtCIPK24/AtSOS2 required for salt tolerance in A. thaliana (Halfter et al., 2000; Ishitani et al., 2000; Liu et al., 2000; Guo et al., 2001), and AtCIPK1 and AtCIPK3 relating to the ABA signal transduction (D'Angelo et al., 2006; Sanyal et al., 2017; Pandey et al., 2008; Kim et al., 2003). Therefore, we speculated that these AhCIPK homologous genes might play multiple roles not only in peanut growth and development but also in plant hormone and stress resistance.
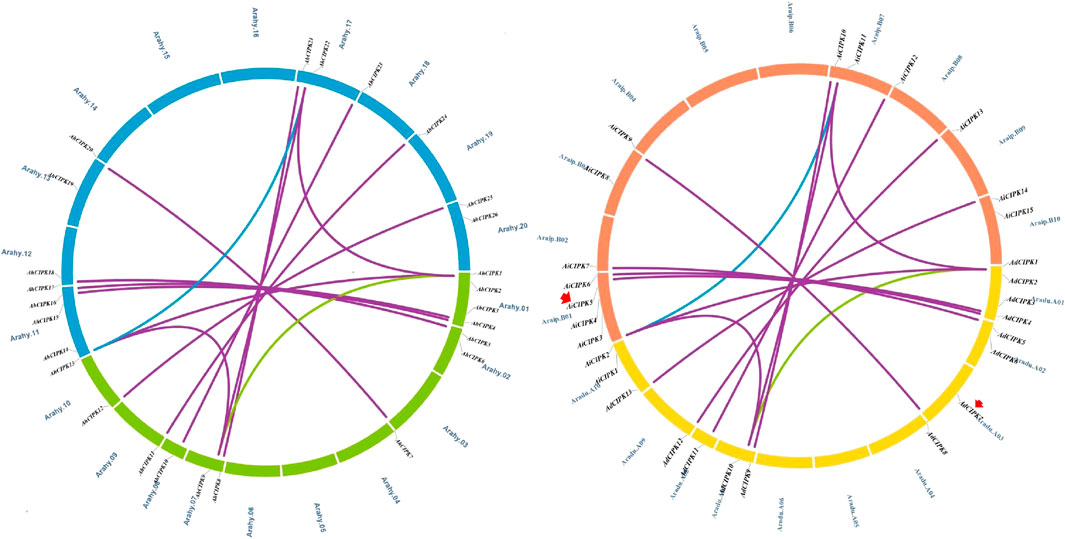
FIGURE 4. Chromosomal distribution and gene duplications of the AdCIPK, AiCIPK, and AhCIPK genes. The scales on the circle are in megabases. Each colored bar represents a chromosome as indicated. Gene IDs are labeled on the basis of their positions on the chromosomes. Red arrows indicate wild species–specific CIPK genes.
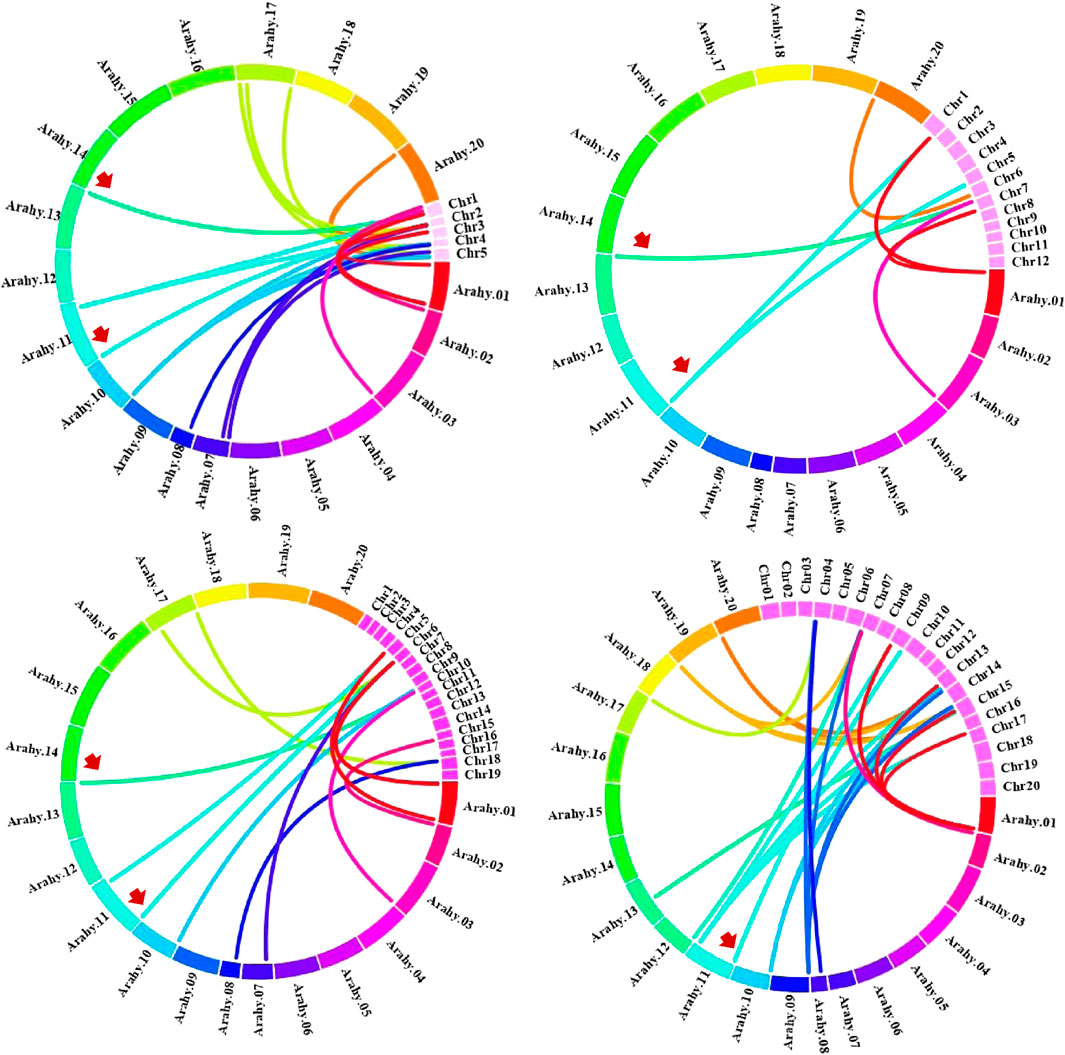
FIGURE 5. Comparative physical mapping showed orthologous relationships of AhCIPK genes with (A) Arabidopsis, (B) rice, (C) grape, and (D) soybean. Red arrows indicate common collinear CIPK orthologs.
Cis-acting elements prediction of CIPK genes in peanut
Cis-acting elements in a promoter as the binding target of transcription factors are essential in the regulation of gene expression. In order to understand the regulation mechanisms of peanut CIPK genes, 2-kb upstream sequences of the peanut CIPK genes were analyzed via the PlantCARE database. In total, 54 cis-regulatory elements were detected. Four main categories were defined as the light responsiveness element, phytohormone responsiveness, abiotic stress responsiveness, and plant growth groups (Figures 6A–C, Supplementary Table S3). In the promoter region of the AdCIPKs, the largest subdivision was the light responsiveness group, containing 53.2% of the predicted cis-elements; phytohormone responsiveness elements ranked second (24.2%) (Figure 6A); abiotic stress response elements were 15.9%; and elements involved in plant growth accounted for 6.7% (Figure 6A). AdCIPK2 had the greatest number of elements with 38 in total, which contained six abscisic acid responsiveness elements (ABREs) (Supplementary Table S3). For AiCIPKs, the percentage of light, phytohormone, abiotic stress, and plant growth responsiveness cis-elements was 57.7, 21.5, 16.6, and 4.2% (Figure 6B). AiCIPK14 had the greatest number of elements at 40 in total, which also contained six ABREs. In AhCIPKs, the proportions were 56.0, 20.9, 16.0, and 7.1% (Figure 6C). In the light response category, Box 4 (light-responsive element) and GT1-motif (part of a module for light response) were the most dominant. Meanwhile, cis-acting elements responding to auxin, abscisic acid, gibberellin, flavonoids, methyl jasmonate, and salicylic acid were detected in the phytohormone responsiveness group.
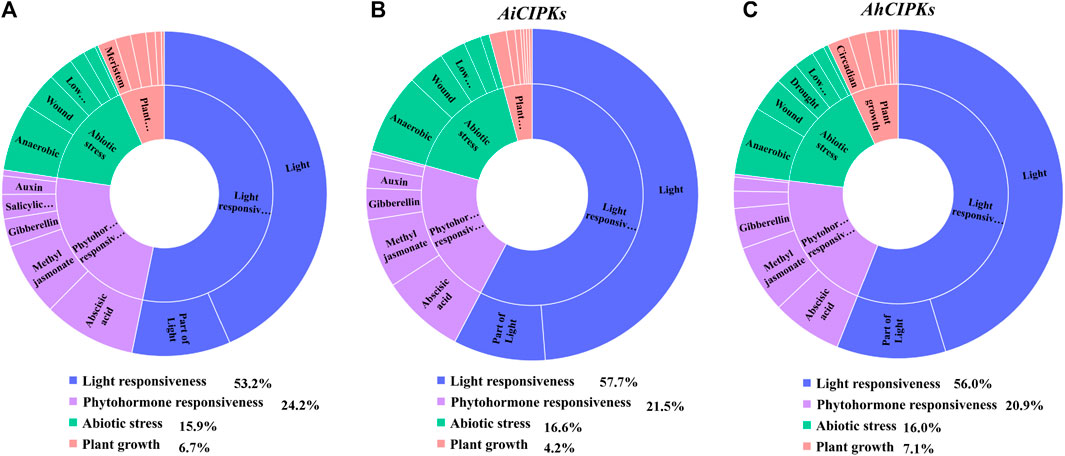
FIGURE 6. cis-acting elements in peanut CIPK genes. (A) assessment of AdCIPK different subclass and category proportions in sunburst chart. (B) assessment of AiCIPK different subclass and category proportions in sunburst chart. (C) assessment AhCIPK different subclass and category proportions in sunburst chart. The lengths of the petals are proportional to the number of elements in each subclass qualitatively. Blue, purple, green, and red represent light responsiveness, phytohormone responsiveness, abiotic stress responsiveness, and plant growth regulation, respectively.
Tissue expression profiles of AhCIPKs
To further study the expression pattern of peanut CIPKs in different tissues and explore its function in peanut growth and development, the tissue expression profiles of CIPK genes were analyzed by using the transcriptome data of 22 peanut tissues (Figure 7). The results showed that the 26 AhCIPK genes had distinct tissue-specific expression patterns across the 22 tissues (leaves, stem, roots, flower, pod, and seed). AhCIPK9 and AhCIPK25 showed a higher expression level in leaf. AhCIPK7 and AhCIPK20 were mostly expressed in reproductive shoot and pattee 1 stalk; interestingly, nearly half of AhCIPK genes were highly expressed in reproductive organs, among which AhCIPK2, AhCIPK3, AhCIPK10, AhCIPK14, AhCIPK15, AhCIPK18, and AhCIPK22 had strong expression in perianth; AhCIPK3, AhCIPK6, AhCIPK8, AhCIPK16, AhCIPK17, and AhCIPK21 were highly expressed in stamens; and AhCIPK12 and AhCIPK26 were obviously expressed in roots. AhCIPK11, AhCIPK23, and AhCIPK24 were highly expressed in nodules. AhCIPK4, AhCIPK12, AhCIPK19, and AhCIPK24 had strong expression during the relatively later pericarp developmental stage. In addition, AhCIPK1 and AhCIPK19 were enriched in the earlier seed developmental stage, while AhCIPK4 and AhCIPK13 were expressed highly in the later seed developmental stage (Figure 7).
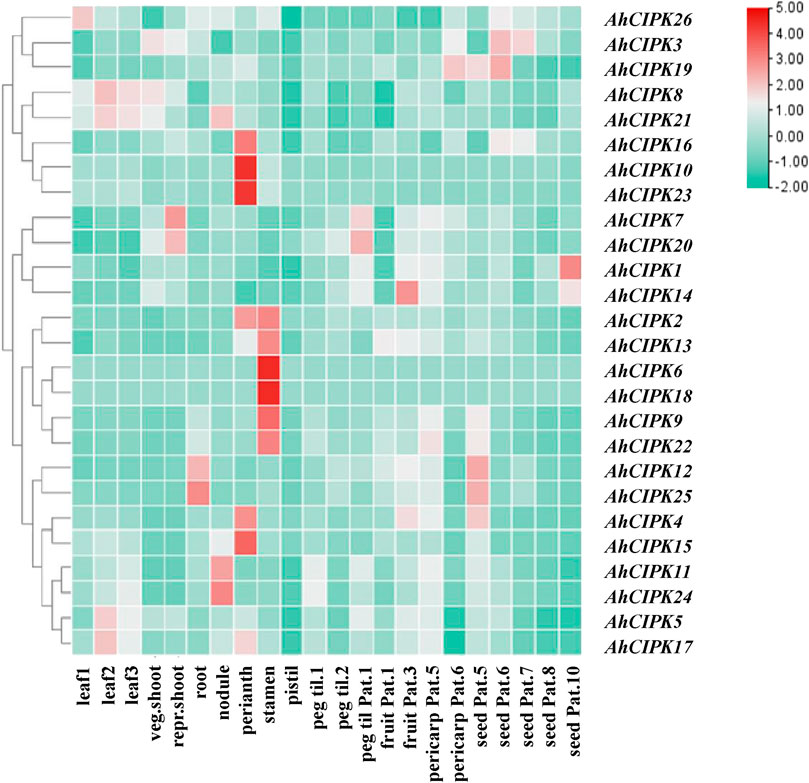
FIGURE 7. Expression profiles of AhCIPK genes. The heatmap of the AhCIPK gene expression levels was hierarchically clustered using TBtools with the data normalization method of Z-score standardization. The color scale bar from green to red represents low and high expressions, respectively. Abbreviations of the 22 tissues used in the expression profiles of AhCIPK genes were from Ren et al. (2022).
Expression pattern of the AhCIPKs under submergence and Ralstonia solanacearum infection
Plants suffer from a wide variety of environmental stressors under natural conditions. To determine the abiotic and biotic stress responses, we detected the expression of AhCIPK genes responding to submergence and R. solanacearum infection. The results showed that except the two unexpressed members AhCIPK6 and 20, almost all other AhCIPKs respond to submergence stress (Figure 8A, Supplementary Table S4). AhCIPK3, 5, 7, 8, 9, 17, 19, 20, 21, and 22 were upregulated rapidly after 6 h of the submergence treatment, while AhCIPK10, 11, 23, and 23 reached their highest expression at 24 h (Figure 8). By contrast, the expression levels of AhCIPK1, 2, 4, 14, 15, 16, and 26 were inhibited under the whole submergence treatment process. We found it interesting that AhCIPK12, 13, and 25 were first repressed under the earlier submergence treatment stages and were induced at the later stages. In addition, seven AhCIPKs (AhCIPK1, 5, 7, 11, 14, 19, and 20) were activated after 6 h R. solanacearum infection (Figure 8B), while six AhCIPK members (AhCIPK2, 9, 11, 12, 22, and 25) were obviously upregulated and three (AhCIPK3, 16, and 26) were depressed after 48 h of R. solanacearum infection (Figure 8B, Supplementary Table S4); In total, AhCIPK genes might have functioned differentially in both abiotic and biotic environmental stress regulation.
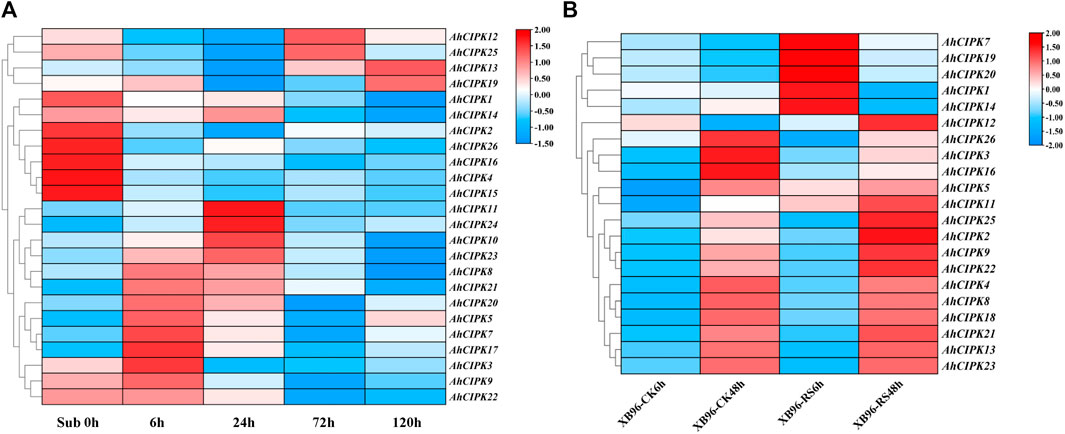
FIGURE 8. Relative expression of AhCIPKs under submergence and Ralstonia solanacearum infection. (A) expression characteristics of AhCIPKs in response to submergence at five time points (00, 06, 24, 72, and 120 h). (B) expression of AhCIPKs in response to R. solanacearum infection at two time points (6 and 48 h). The heatmap of the AhCIPK gene expression levels was hierarchically clustered using TBtools with the data normalization method of Z-score standardization. Legend at the right of the heatmap: red means upregulation and blue shows downregulated expression.
Candidate gene association of peanut CIPKs polymorphisms with 104 traits
To further uncover the roles of CIPK genes in peanut development and stress response, we performed candidate gene association analysis using 22 single-nucleotide polymorphisms in CIPKs from transcriptome data of 146 peanut lines and 104 phenotypes related to peanut development and stress response collected in five environments. The results indicated that one polymorphic site [A09_903480(G/K/T)] was significantly associated with pod length (PL), seed length (SL), hundred seed weight (HSW), and shoot root ratio (SR) traits (Figure 9A, Supplementary Tables S5–S7). Site B09_903480 mainly formed three haplotypes [B09_903480(G/K/T)] (Figure 9B) in the population and was located in the predicted exon region of AiCIPK10 (Figure 9C). Results showed that PL, SL, HSW, and SR in haplotype G were significantly higher than those in haplotype T (Figure 9D).
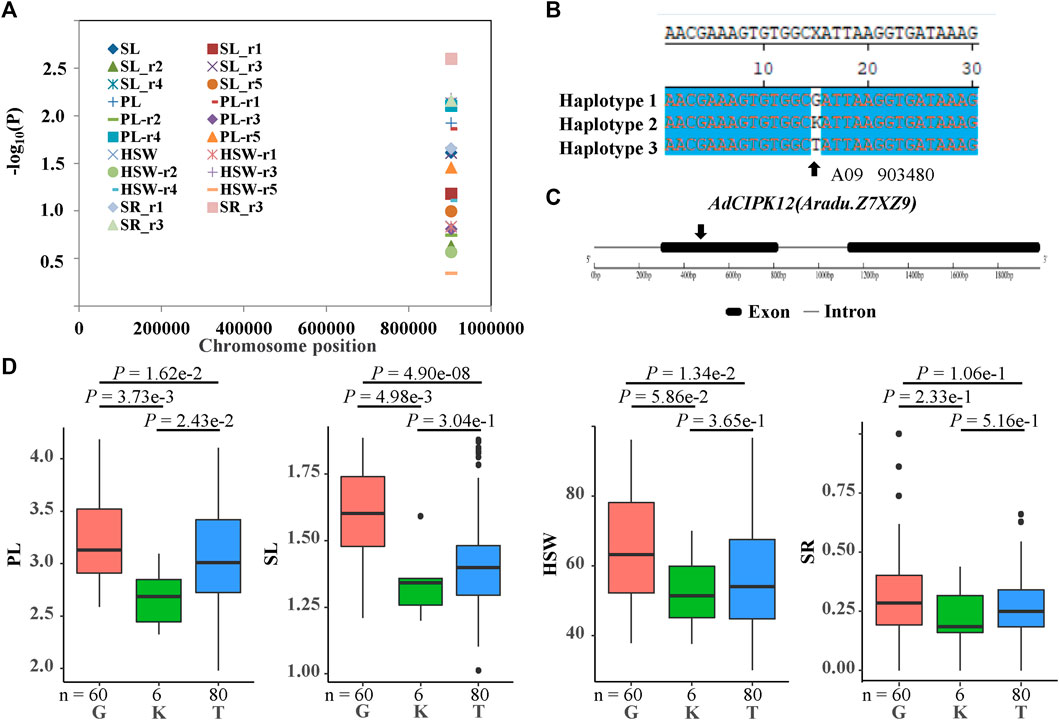
FIGURE 9. Association results and the phenotypes of the polymorphic sites of the peanut CIPKs associated with hundred seed weight (HSW)/pod length (PL)/ seed length (SL)/shoot root ratio (SR) variation. (A) association results between HSW/PL/SL/SR and the polymorphisms in peanut CIPKs. (B) sequences of the site significantly associated with HSW/PL/SL/SR variation. (C) gene structures of AdCIPK12(Aradu.Z7XZ9). (D) phenotypic comparison of haplotypes of the associated site with HSW/PL/SL/SR in five environments of the population.
Discussion
Peanut CIPKs did not expand with genome duplication
CIPK genes are widely distributed widely in the biological world; however, their number varies greatly among different species. One, two, and seven CIPK genes were found in green algae, Chlorella, and Physcomitrella patens, respectively (Cheong et al., 2007; Weinl and Kudla, 2009). According to previous studies, 25, 30, 27, 43, and 79 CIPK genes were identified in A. thaliana (125–155 Mb), rice (389 Mb), poplar (416 Mb), corn (2,400 Mb), and wheat (14,500 Mb) (Kolukisaoglu et al., 2004; Xiang et al., 2007; Chen et al., 2011; Sun et al., 2015; Zhu et al., 2021). Among Solanaceae plants, there were 21 and 22 CIPK members in tomato (Liu et al., 2017; Wang and Liu, 2018). Woody plants had 27 members (Zhang et al., 2008). This study uncovered 54 CIPK genes in three peanut genomes, among which 26 were from cultivated peanut (2,540 Mb). Our results support the hypothesis that the number of CIPK members in monocotyledonous plants is more than that in dicotyledonous plants. However, the number of peanut CIPK members did not expand with peanut genome size expansion.
The phylogenetic analysis revealed that CIPK members in Arabidopsis, rice, soybean, grape, and peanut could be coincidentally clustered into two distinct groups with different numbers of introns. This cluster pattern is the same as that in the previously reported CIPK-phylogenetic trees of Arabidopsis and rice (Kolukisaoglu et al., 2004). Our results further supported the hypothesis that the ancestor of CIPKs evolutionarily formed in the plant genome prior to the separation of the lineages of monocotyledons and dicotyledons (Kolukisaoglu et al., 2004; Yu et al., 2007).
Peanut CIPK genes functioned in stress response
The prediction of cis-acting elements can provide important clues for the study of gene expression regulation (Zhu et al., 2021). Cis-acting elements of biotic and abiotic stresses presented ubiquitously in the promoter region of peanut CIPKs, indicating these impact factors may interact to act on the CIPK regulatory mechanism (Figure 6). Compared to those in the AhCIPKs, the defense, stress, low-temperature response element TC-rich repeats and LTR were distributed concentratedly in several AdCIPKs or AiCIPKs in the two diploid progenitors; for example, four and three LTRs were identified in the promoter region of AdCIPK4 and AiCIPK11, and four TC-rich repeats were found in the promoter region of AdCIPK4, indicating that the regulatory mechanism might be different between the cultivated peanut and the two diploid progenitors. Further the stress-induced expression data also elucidated the potential functions of peanut CIPKs in stress.
Among the orthologs between peanut and Arabidopsis (Table 2), the functions of the corresponding ortholog genes in Arabidopsis have been determined, and they functioned in influencing plant stress response (Table 2). AdCIPK3, AiCIPK4, AhCIPK3, and AhCIPK16 were identified as the orthologous genes of the famous salt tolerance gene AtCIPK24/AtSOS2 in A. thaliana (Halfter et al., 2000; Ishitani et al., 2000; Liu et al., 2000; Guo et al., 2001), and AhCIPK8, AhCIPK21, AdCIPK9, and AiCIPK10 were found as the orthologs of AtCIPK3 relating to the cold signal transduction (Kim et al., 2003; Pandey et al., 2008; Sanyal et al., 2017). Many other orthologs were also uncovered between peanut and the Arabidopsis abscisic acid-dependent and Arabidopsis abscisic acid-independent stress response gene AtCIPK1 (D'Angelo et al., 2006), low-potassium tolerance gene AtCIPK9 (Pandey et al., 2007; Singh et al., 2018; Lara et al., 2020; Kanwar et al., 2022), and cadmium stress response gene AtCIPK11 (Liu and Guo, 2011; Zhou et al., 2015; Gu et al., 2021). Therefore, these peanut CIPK orthologous genes may also play multiple roles in peanut stress response, especially in salt response.
Peanut CIPK genes play important roles in growth and development
Many important genes were selectively expressed in specific tissues during various physiological and developmental processes (Wan et al., 2014). Our results showed that the 26 AhCIPK genes had distinct tissue-specific expression patterns, and several AhCIPKs showed higher expression level in the leaf, reproductive shoot, root, nodule, pod, and seed. We found it interesting that nearly half of AhCIPK genes are highly expressed in reproductive organs (Figure 7), indicating AhCIPKs play important roles in multiple tissue growth and development, especially the reproductive organs.
The single-nucleotide polymorphic sites in AdCIPK12 (corresponding to AhCIPK11) were significantly associated with PL, SL, HSW, and SR variation. The polymorphic site in AdCIPK12 [A09_ 903,480 (G/K/T)], located in the predicted first exon region of the gene, A09_903480(G/K/T), led no transition (synonymous mutation) in the peanut population. Recent studies proved that synonymous mutations also have dramatic effects on protein output (Gillen et al., 2021). These results indicated that the B09_903480 (G/K/T) sequence polymorphisms might be the actual functional sites. Further, AhCIPK11 was mainly expressed in the nodule, peg, pericarp, and seed, especially in the middle pericarp and seed development stages, which provided additional evidence for its function in peanut pod development (Figure 6). Further investigation was needed to confirm the roles of AdCIPK12 (AhCIPK11) in the pod and seed development of peanut.
Conclusions
We definitely identified 54 CIPK members in cultivated and wild peanut for the first time and determined their chromosomal locations, gene structures, evolution, and expression patterns under biotic and abiotic conditions. We also focused on one gene, AiCIPK10/AhCIPK21, which was involved in pod and seed development. Our results provide valuable information for understanding the functions of the peanut CIPK gene family in regulating yield, quality, and stress responses in peanut.
Data availability statements
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
WR and JZ carried out all the experiments and data analyses. WR and JH prepared the figures and tables. LW, JF, and JZ made modifications to the article. All authors read and approved the final manuscript.
Funding
This research was funded by grants from the Jiangxi Agriculture Research System (No. JCARS-18) and Scientific Research Foundation for Scholars of HZNU (2022QDL034). Funders did not participate in the design of the study, analysis of the results and writing of the manuscript, but provided financial support for the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.939255/full#supplementary-material
References
Albrecht, V., Ritz, O., Linder, S., Harter, K., and Kudla, J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+regulated kinases. Embo J. 2 (5), 1051–1063. doi:10.1093/emboj/20.5.1051
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, L., Wang, Q. Q., Zhou, L., Ren, F., Li, D. D., and Li, X. B. (2013). Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 40 (8), 4759–4767. doi:10.1007/s11033-013-2572-9
Chen, X., Gu, Z., Xin, D., Hao, L., Liu, C., Huang, J., et al. (2011). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 38 (2), 77–87. doi:10.1016/j.jcg.2011.01.005
Cheong, Y. H., Pandey, G. K., Grant, J. J., Batistic, O., Li, L., Kim, B. G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52 (2), 223–239. doi:10.1111/j.1365-313X.2007.03236.x
Clevenger, J., Chu, Y., Scheffler, B., and Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7, 1446. doi:10.3389/fpls.2016.01446
D'Angelo, C., Weinl, S., Batistic, O., Pandey, G. K., Cheong, Y. H., Schültke, S., et al. (2006). Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48 (6), 857–872. doi:10.1111/j.1365-313X.2006.02921.x
Deng, X., Zhou, S., Hu, W., Feng, J., Zhang, F., Chen, L., et al. (2013). Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant. 149 (3), 367–377. doi:10.1111/ppl.12046
Feng, Z. J., Cui, X. Y., Cui, X. Y., Chen, M., Yang, G. X., Ma, Y. Z., et al. (2015). The soybean GmDi19-5 interacts with GmLEA3.1 and increases sensitivity of transgenic plants to abiotic stresses. Front. Plant Sci. 6, 179–186. doi:10.3389/fpls.2015.00179
Feng, Z. J., Xu, S. C., Liu, N., Zhang, G. W., Hu, Q. Z., and Gong, Y. M. (2017). Expression character of vegetable soybean CIPKs in response to abiotic stresses and hormones. J. Plant Genet. Resour. 18 (6), 1168–1178. doi:10.13430/j.cnki.jpgr.2017.06.019
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identifification and analysis tools on the ExPASy server,” in The Proteomics protocols handbook (New Jersey: Humana Press), 571–607. doi:10.1385/1-59259-890-0:571
Gillen, S. L., Waldron, J. A., and Bushell, M. (2021). Codon optimality in cancer. Oncogene 40 (45), 6309–6320. doi:10.1038/s41388-021-02022-x
Gong, D., Gong, Z., Guo, Y., Chen, X., and Zhu, J. K. (2002). Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J. Biol. Chem. 277 (31), 28340–28350. doi:10.1074/jbc.M107719200
Gu, S., Wang, X., Bai, J., Wei, T., Sun, M., Zhu, L., et al. (2021). The kinase CIPK11 functions as a positive regulator in cadmium stress response in Arabidopsis. Gene 772, 145372. doi:10.1016/j.gene.2020.145372
Guo, Y., Halfter, U., Ishitani, M., and Zhu, J. K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13 (6), 1383–1400. doi:10.1105/tpc.13.6.1383
Halfter, U., Ishitani, M., and Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U. S. A. 97 (7), 3735–3740. doi:10.1073/pnas.040577697
Held, K., Pascaud, F., Eckert, C., Gajdanowicz, P., Hashimoto, K., Corratgé-Faillie, C., et al. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21 (7), 1116–1130. doi:10.1038/cr.2011.50
Hrabak, E. M., Chan, C. W. M., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK super family of protein kinases. Plant Physiol. 132 (2), 666–680. doi:10.1104/pp.102.011999
Hu, H. C., Wang, Y. Y., and Tsay, Y. F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57 (2), 264–278. doi:10.1111/j.1365-313X.2008.03685.x
Hu, W., Xia, Z., Yan, Y., Ding, Z., Tie, W., Wang, L., et al. (2015). Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front. Plant Sci. 30 (6), 914–929. doi:10.3389/fpls.2015.00914
Ishitani, M., Liu, J., Halfter, U., Kim, C. S., Shi, W., and Zhu, J. K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12 (9), 1667–1678. doi:10.1105/tpc.12.9.1667
Kanwar, P., Sanyal, S. K., Mahiwal, S., Ravi, B., Kaur, K., Fernandes, J. L., et al. (2022). CIPK9 targets VDAC3 and modulates oxidative stress responses in Arabidopsis. Plant J. 109 (1), 241–260. doi:10.1111/tpj.15572
Kim, K. N., Cheong, Y. H., Grant, J. J., Pandey, G. K., and Luan, S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15 (2), 411–423. doi:10.1105/tpc.006858
Kim, K. N., Cheong, Y. H., Gupta, R., and Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124 (4), 1844–1853. doi:10.1104/pp.124.4.1844
Kolukisaoglu, Ü., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks.:genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134 (1), 43–58. doi:10.1104/pp.103.033068
Lara, A., Ródenas, R., Andrés, Z., Martínez, V., Quintero, F. J., Nieves-Cordones, M., et al. (2020). Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9. J. Exp. Bot. 71 (16), 5053–5060. doi:10.1093/jxb/eraa212
Li, L., Kim, B. G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 103 (33), 12625–12630. doi:10.1073/pnas.0605129103
Liu, J., and Guo, Y. (2011). The alkaline tolerance in Arabidopsis requires stabilizing microfilament partially through inactivation of PKS5 kinase. J. Genet. Genomics 38 (7), 307–313. doi:10.1016/j.jgg.2011.05.006
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., and Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U. S. A. 97 (7), 3730–3734. doi:10.1073/pnas.060034197
Liu, S. Y., Huang, H. F., Wang, X. D., Gong, C., Song, H. H., Chen, X. L., et al. (2017). Identification and characterization of CBL and CIPK gene families in pepper genome analyse. Mol. Plant Breed. 15 (8), 2977–2985. doi:10.1186/s12864-019-6125-z
Lu, Q., Li, S. X., Chen, X. P., Zhou, G. Y., Hong, Y. B., Li, H. F., et al. (2017). Current situation, problems and suggestions of peanut breeding in southern China. Chin. J. Oil Crop Sci. 39 (4), 556–566. doi:10.7505/j.issn.1007-9084.2017.04.019
Mao, J., Manik, S. M., Shi, S., Chao, J., Jin, Y., Wang, Q., et al. (2016). Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes 7 (9), 62–72. doi:10.3390/genes7090062
Pandey, G. K., Cheong, Y. H., Kim, B. G., Grant, J. J., Li, L., and Luan, S. (2007). CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res. 17 (5), 411–421. doi:10.1038/cr.2007.39
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, le.G., and Luan, S. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1 (2), 238–248. doi:10.1093/mp/ssn003
Pandey, M. K., Pandey, A. K., Kumar, R., Nwosu, C. V., Guo, B., Wright, G. C., et al. (2020). Translational genomics for achieving higher genetic gains in groundnut. Theor. Appl. Genet. 133 (5), 1679–1702. doi:10.1007/s00122-020-03592-2
Ragel, P., Ródenas, R., García-Martín, E., Andrés, Z., Villalta, I., Nieves-Cordones, M., et al. (2015). The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 169 (4), 2863–2873. doi:10.1104/pp.15.01401
Ren, W., Zeng, Z., Wang, S., Zhang, J., Fang, J., Wan, L., et al. (2022). Global Survey, Expressions and Association Analysis of CBLL Genes in Peanut. Front. genet. 13, 821163. doi:10.3389/fgene.2022.821163
Rui, Y. (2015). Calcium availability to runner -type peanut (Arachis hypogaea L.) in the southeastern United States. Auburn: Graduate Faculty of Auburn University.
Sánchez-Barrena, M. J., Chaves-Sanjuan, A., Raddatz, N., Mendoza, I., Cortés, Á., Gago, F., et al. (2020). Recognition and activation of the plant AKT1 potassium channel by the kinase CIPK23. Plant Physiol. 182 (4), 2143–2153. doi:10.1104/pp.19.01084
Sanders, D., Pelloux, J., Brownlee, C., and Harper, J. F. (2002). Calcium at the crossroads of signaling. Plant Cell 14, S401–S417. doi:10.1105/tpc.002899
Sanyal, S. K., Kanwar, P., Samtani, H., Kaur, K., Jha, S. K., and Pandey, G. K. (2017). Alternative splicing of CIPK3 results in distinct target selection to propagate ABA signaling in Arabidopsis. Front. Plant Sci. 8, 1924. doi:10.3389/fpls.2017.01924
Sardar, A., Nandi, A. K., and Chattopadhyay, D. (2017). CBL-interacting protein kinase 6 negatively regulates immune response to Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 68 (13), 3573–3584. doi:10.1093/jxb/erx170
Schlücking, K., Edel, K. H., Köster, P., Drerup, M. M., Eckert, C., Steinhorst, L., et al. (2013). A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol. Plant 6 (6), 1814–1829. doi:10.1093/mp/sst065
Singh, A., Yadav, A. K., Kaur, K., Sanyal, S. K., Jha, S. K., Fernandes, J. L., et al. (2018). A protein phosphatase 2C, AP2C1, interacts with and negatively regulates the function of CIPK9 under potassium-deficient conditions in Arabidopsis. J. Exp. Bot. 69 (16), 4003–4015. doi:10.1093/jxb/ery182
Steinhorst, L., Mähs, A., Ischebeck, T., Zhang, C., Zhang, X., Arendt, S., et al. (2015). Vacuolar CBL-CIPK12 Ca(2+)-sensor-kinase complexes are required for polarized pollen tube growth. Curr. Biol. 25 (11), 1475–1482. doi:10.1016/j.cub.2015.03.053
Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y., Wang, X., et al. (2015). Identification and comprehensive analysis of the CBL and CIPK gene family in Wheat (Triticum aestivum L). BMC Plant Biol. 15, 269. doi:10.1186/s12870-015-0657-4
Tagliani, A., Tran, A. N., Novi, G., Di, Mambro.R., Pesenti, M., Sacchi, G. A., et al. (2020). The calcineurin β-like interacting protein kinase CIPK25 regulates potassium homeostasis under low oxygen in Arabidopsis. J. Exp. Bot. 71 (9), 2678–2689. doi:10.1093/jxb/eraa004
Tian, Q., Zhang, X., Yang, A., Wang, T., and Zhang, W. H. (2016). CIPK23 is involved in iron acquisition of Arabidopsis by affecting ferric chelate reductase activity. Plant Sci. 246, 70–79. doi:10.1016/j.plantsci.2016.01.010
Tripathi, V., Parasuraman, B., Laxmi, A., and Chattopadhyay, D. (2009a). CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 58 (5), 778–790. doi:10.1111/j.1365-313X.2009.03812.x
Tripathi, V., Syed, N., Laxmi, A., and Chattopadhyay, D. (2009b). Role of CIPK6 in root growth and auxin transport. Plant Signal. Behav. 4 (7), 663–665. doi:10.1111/j.1365-313-X.2009.03812.x
Tsou, P. L., Lee, S. Y., Allen, N. S., Winter-Sederoff, H., and Robertson, D. (2012). An ER-targeted calcium-binding peptide confers salt and drought tolerance mediated by CIPK6 in Arabidopsis. Planta 235 (3), 539–552. doi:10.1007/s00425-011-1522-9
Wan, L., Wu, Y., Huang, J., Dai, X., Lei, Y., Yan, L., et al. (2014). Identification of ERF genes in peanuts and functional analysis of AhERF008 and AhERF019 in abiotic stress response. Funct. Integr. Genomics 14 (3), 467–477. doi:10.1007/s10142-014-0381-4
Wang, A. X., and Liu, S. Y. (2018). Identification and bioinformatics analysis on CIPK gene family in tomato. J. Northeast Agric. Univ. 49 (2), 31–38. doi:10.3969/j.issn.1005-9369.2018.02.004
Wang, X. P., Chen, L. M., Liu, W. X., Shen, L. K., Wang, F. L., Zhou, Y., et al. (2016). AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in Arabidopsis. Plant Physiol. 170 (4), 2264–2277. doi:10.1104/pp.15.01493
Wang, Y., and Wu, W. H. (2004). Molecular genetic mechanism of high efficient potassium up take in plants. Chin. Bull. Bot. 44 (1), 27–36. doi:10.3969/j.issn.1674-3466.2009.01.003
Wang, Y., Zhang, W. Z., Song, L. F., Zou, J. J., Su, Z., and Wu, W. H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148 (3), 1201–1211. doi:10.1104/pp.108.126375
Weinl, S., and Kudla, J. (2009). The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi:10.1111/j.1469-8137.2009.02938.x
Xiang, Y., Huang, Y., and Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144 (3), 1416–1428. doi:10.1104/pp.107.101295
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 (7), 1347–1360. doi:10.1016/j.cell.2006.06.011
Yu, Y. H., Xia, X. L., Yin, W. L., and Zhang, H. C. (2007). Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 52 (2), 101–110. doi:10.1007/s10725-007-9165-3
Zeng, R., Chen, T., Wang, X., Cao, J., Li, X., Xu, X., et al. (2021). Physiological and expressional regulation on photosynthesis, starch and sucrose metabolism response to waterlogging stress in peanut. Front. Plant Sci. 12, 601771. doi:10.3389/fpls.2021.601771
Zhang, C., Chen, H., Cai, T., Deng, Y., Zhuang, R., Zhang, N., et al. (2017). Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 15 (1), 39–55. doi:10.1111/pbi.12589
Zhang, H., Yin, W., and Xia, X. (2008). Calcineurin B-like family in populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 56 (2), 129–140. doi:10.1007/s10725-008-9293-4
Zhao, J. F., Sun, Z. F., Zheng, J., Guo, X., Dong, Z., Huai, J., et al. (2009). Cloning and characterization of a novel CBL-interacting protein kinase from maize. Plant Mol. Biol. 69 (6), 661–674. doi:10.1007/s11103-008-9445-y
Zhou, X., Hao, H., Zhang, Y., Bai, Y., Zhu, W., Qin, Y., et al. (2015). SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 168 (2), 659–676. doi:10.1104/pp.114.255455
Keywords: peanut, expression, adversity response, association analysis, CIPK genes
Citation: Ren W, Zhang J, He J, Fang J and Wan L (2022) Identification, expression, and association analysis of calcineurin B-like protein–interacting protein kinase genes in peanut. Front. Genet. 13:939255. doi: 10.3389/fgene.2022.939255
Received: 09 May 2022; Accepted: 21 July 2022;
Published: 05 September 2022.
Edited by:
Ajay Kumar, North Dakota State University, United StatesCopyright © 2022 Ren, Zhang, He, Fang and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liyun Wan, d2FubGl5dW4yMDE5QDE2My5jb20=
†These authors have contributed equally to this work
 Weifang Ren
Weifang Ren Juncheng Zhang
Juncheng Zhang Jie He1
Jie He1 Liyun Wan
Liyun Wan