- 1Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Brisbane, Australia
- 2Agency of Science, Technology and Research, Singapore, Singapore
- 3School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia
The hypothalamus and the pituitary gland are directly involved in the complex systemic changes that drive the onset of puberty in cattle. Here, we applied integrated bioinformatics to elucidate the critical proteins underlying puberty and uncover potential molecular mechanisms from the hypothalamus and pituitary gland of prepubertal (n = 6) and postpubertal (n = 6) cattle. Proteomic analysis in the hypothalamus and pituitary gland revealed 275 and 186 differentially abundant (DA) proteins, respectively (adjusted p-value < 0.01). The proteome profiles found herein were integrated with previously acquired transcriptome profiles. These transcriptomic studies used the same tissues harvested from the same heifers at pre- and post-puberty. This comparison detected a small number of matched transcripts and protein changes at puberty in each tissue, suggesting the need for multiple omics analyses for interpreting complex biological systems. In the hypothalamus, upregulated DA proteins at post-puberty were enriched in pathways related to puberty, including GnRH, calcium and oxytocin signalling pathways, whereas downregulated proteins were observed in the estrogen signalling pathway, axon guidance and GABAergic synapse. Additionally, this study revealed that ribosomal pathway proteins in the pituitary were involved in the pubertal development of mammals. The reported molecules and derived protein-protein networks are a starting point for future experimental approaches that might dissect with more detail the role of each molecule to provide new insights into the mechanisms of puberty onset in cattle.
Introduction
Early puberty is essential for the lifetime reproductive performance of cattle (Lesmeister et al., 1973; Johnston et al., 2013). However, Brahman cattle, a breed of the Bos indicus sub-species, which can withstand hostile conditions in northern production systems, are often older and heavier at puberty than Bos taurus breeds (Chenoweth 1994). As reaching the age of puberty is an important event contributing significantly to lifetime productivity, reducing the age at puberty is a major aim for Bos indicus breeders for efficient herd productivity.
Like other vertebrates and humans, puberty in cattle is initiated when the hypothalamus-pituitary-ovary (HPO) axis loses its sensitivity to negative feedback effects of steroid hormones, allowing an increase in gonadotropin-releasing hormone (GnRH) secretion from GnRH neurons in the hypothalamus. The increase then stimulates gonadotropins’ secretion: luteinizing hormone (LH), and follicle-stimulating hormone (FSH) are produced and released. FSH and LH then regulate gonadal development (Day et al., 1984; Day et al., 1987; Schillo et al., 1992; Day and Anderson, 1998; Gasser et al., 2006). However, the process is controlled by multiple factors, and the complex interactions between environmental and genetic factors regulating the process are only now coming to light.
The complex genetic architecture of puberty–that is multiple variants of small effect–is probably underpinned by variants with transcriptional and post-transcriptional regulatory effects. Variations in the expression of genes or proteins in the hypothalamus or the pituitary gland, either alone or simultaneously, will affect the pubertal process. To date, several transcriptomic studies have focused on investigating candidate molecules involved in critical molecular mechanisms that drive puberty in Bos indicus in the HPO axis. Fortes et al., 2016 identified five transcription factors, E2F8, NFAT5, SIX5, ZBTB38, and ZNF605, with potential regulatory roles at puberty in the hypothalamus in Brahman heifers. Nguyen et al., 2017 confirmed the role of zinc finger genes in a co-expression network using ovarian data of the same Brahman heifers. Although these transcriptomic studies confirmed the complexity of puberty, the correlation between expression levels of mRNA and protein is renowned poor (Gygi et al., 1999; de Sousa Abreu et al., 2009; Carvalhais et al., 2015; Payne, 2015; Edfors et al., 2016). As such, proteomic studies are complementary information, required to advance knowledge beyond differential gene expression or co-expression analyses.
Most studies for understanding puberty-related proteins involved in Brahman heifers were performed in peripheral tissues such as the liver and adipose tissues (Nguyen et al., 2018b; 2018c). Also, Tahir and others (Tahir et al., 2019) have published on the ovarian protein abundances in pre versus post-pubertal Brahman heifers. Although a neuropeptidome was performed on the hypothalamus and pituitary gland of Brangus heifers before and after puberty (DeAtley et al., 2018), there is no previous proteome study in the hypothalamus and pituitary gland of pubertal Brahman heifers. Therefore, this present study was aimed at measuring the abundance of proteins in the hypothalamus and pituitary gland in pubertal Brahman heifers using liquid chromatography-electrospray ionisation tandem mass spectrometry (LC-ESI-MS/MS). Discovering protein abundance profiles at the different pubertal stages in the hypothalamus and pituitary gland could be useful for revealing key targets controlling the complex mechanism of puberty and complementing the previous transcriptomic studies.
Materials and Methods
Ethics Statement
Animal use in this study was approved under animal ethics number QAAFI/279/12. Animals were housed at the Gatton Campus facilities of the University of Queensland.
Animal and Tissue Collection
The heifers used in this study and the assessment of puberty have been described by (Fortes et al., 2016; Nguyen et al., 2017; Nguyen et al., 2018a; Lau et al., 2020). In brief, these heifers were of the same age with no difference in body weight or body condition scores. Pubertal status was defined by the presence of the first corpus luteum (CL) and progesterone concentration. Postpubertal heifers were euthanized by stunning with a nonpenetratng captive bolt through the parietal bone of the head around day 15 after observing the first CL. The nonpenetrating captive bolt methodology was used for protecting the integrity of the lower brain tissues as previously described (Cánovas et al., 2014). A pre-pubertal heifer was then randomly selected to pair with a postpubertal heifer on slaughter day. Plasma progesterone concentrations were 0.4 ± 0.2 ng/ml and 2.0 ± 0.7 ng/ml in pre and post-pubertal heifers, respectively.
After euthanasia, the hypothalamus tissue (spanning from the preoptic to the arcuate nucleus) and pituitary gland (including anterior and posterior pituitary gland) tissues were quickly harvested, frozen in liquid nitrogen, and stored at −80°C until tissue processing.
Protein Sample Preparation
Protein extraction and digestion of heifers’ hypothalamus and pituitary tissues were performed as previously described (Tan et al., 2014; Nguyen et al., 2018b; Nguyen et al., 2018c). Briefly, the hypothalamus and pituitary tissues were manually homogenized into smaller fragments using rigid aluminium foil to protect the tissue and a hammer to “pulverize” the sample. This procedure was performed over a dry-ice bed to preserve the sample and randomize the fragments, ensuring that each sample was representative of the entire tissue. The resulting tissue fragments were transferred into Eppendorf® lobind microcentrifuge tubes (Sigma-Aldrich) where lysis solution (7 M urea, 2 M thiourea, 4%SDS, 10 mM DTT and 1 mM PMSF) was added. The fragments in the solution were sonicated at power level 4 for 10 s. Subsequently, the homogenate was vortexed vigorously for 1 h at 30°C. Following that, 25 mM acrylamide was added to the samples and subsequently incubated for 1 h at 30°C. Next, 5 mM DTT was added to samples in order to quench excess acrylamide. Four volumes of methanol: acetone (1:1) were added, and the samples precipitated overnight at −20°C. The precipitates were subsequently dissolved by adding 50 mM ammonium acetate, and the protein concentrations were measured with a Nanodrop (Thermo Scientific). After measuring concentration, about 100 µg of protein were transferred into a 10 kDa Amicon Ultra 0.5 centrifugal filter device (Merck Millipore). This filter device was then inserted into a collection tube and centrifuged at maximum speed for at least 30 min. Protein was diluted in 50 mM ammonium bicarbonate and then again centrifuged at maximum speed for at least 30 min. Trypsin was used as a protease for digestion of protein solution followed by incubation overnight at 37°C. Peptides were desalted by C-18 Zip-tip (adapted from Millipore procedure) and stored at −20°C until analysis.
Mass Spectrometry and Data Analysis
The LC-ESI-MS/MS was performed using a Prominence nanoLC system (Shimadzu) and TripleToF 5600 mass spectrometer with a Nanospray III interface (SCIEX) as previously described (Xu et al., 2015). Peptides were separated using a 70-min LC gradient. MS-TOF scan was performed. We performed information-dependent acquisition (IDA) of top peptides for one randomly chosen pre-pubertal sample and one randomly chosen post-pubertal sample. Subsequently, sequential window acquisition of all theoretical mass spectra (SWATH) was performed on all samples using the prepared IDA library for each group.
Protein Pilot v5.0.1 (ABSCIEX) was then utilized for peptide identification. The bovine protein database was used for peptide mapping and retrieved from Uniprot (www.uniprot.org; 43,813 entries assigned to Bos taurus). For subsequent analyses, identified peptides with more than 99% confidence and a false discovery rate (FDR) of less than 1% were used. Ion libraries were used for SWATH analyses. The abundance of proteins and peptides was computed by PeakView v2.1 software (ABSCIEX). The differential expression analysis between the two grops of pre and post-pubertal heifers as performed using MSstats (v2.6) in R (Choi et al., 2014). For controlling the false discovery rate, the p-values were adjusted using the Benjamini and Hochberg’s approach. Proteins with an adjusted p-value < 0.01 were assigned as differentially abundant.
Transcriptomic Data
The hypothalamus and pituitary gland transcriptomic data were obtained from Fortes et al., 2016 and Nguyen et al., 2017. These transcriptomic studies used the same tissues harvested from the same heifers at pre- and post-puberty. The mRNA and protein pairs were identified from sets of expressed genes (Fortes et al., 2016; Nguyen et al., 2017) and set of abundant proteins (current study). The Pearson’s correlation coefficient (r) was used to compute the correlations between expression levels of mRNAs and the abundance of proteins in the hypothalamus and pituitary gland from pre- and postpubertal Brahman heifers. The cor() function in R was used in this calculation. The unmatched mRNA-proteins were not included in this analysis.
Functional Enrichment Analysis of DA Proteins
The gene ontology analysis of differentially abundant (DA) proteins was obtained through STRINGv10 system (http://string-db.org). The DA proteins were classified according to biological process (BP), cellular component (CC), molecular function (MF) and pathways. Further, Cytoscape software (http://cytoscape.org/) was used to retrieve biological terms for the protein interaction networks. In order to determine significant terms and pathways, the terms having a corrected p-value (FDR) < 0.05 were considered.
Results
Protein Identification, Quantification, and Differential Abundance
In total, 765 proteins were identified and quantified in the hypothalamus. Of this total, 275 differentially abundant (DA) proteins were identified (adjusted p-value < 0.01) in the hypothalamus libraries, of which 120 were significantly downregulated and 155 were significantly upregulated at post-puberty (adjusted p-value < 0.01) (Figures 1A, 2A and Supplementary Table S1).
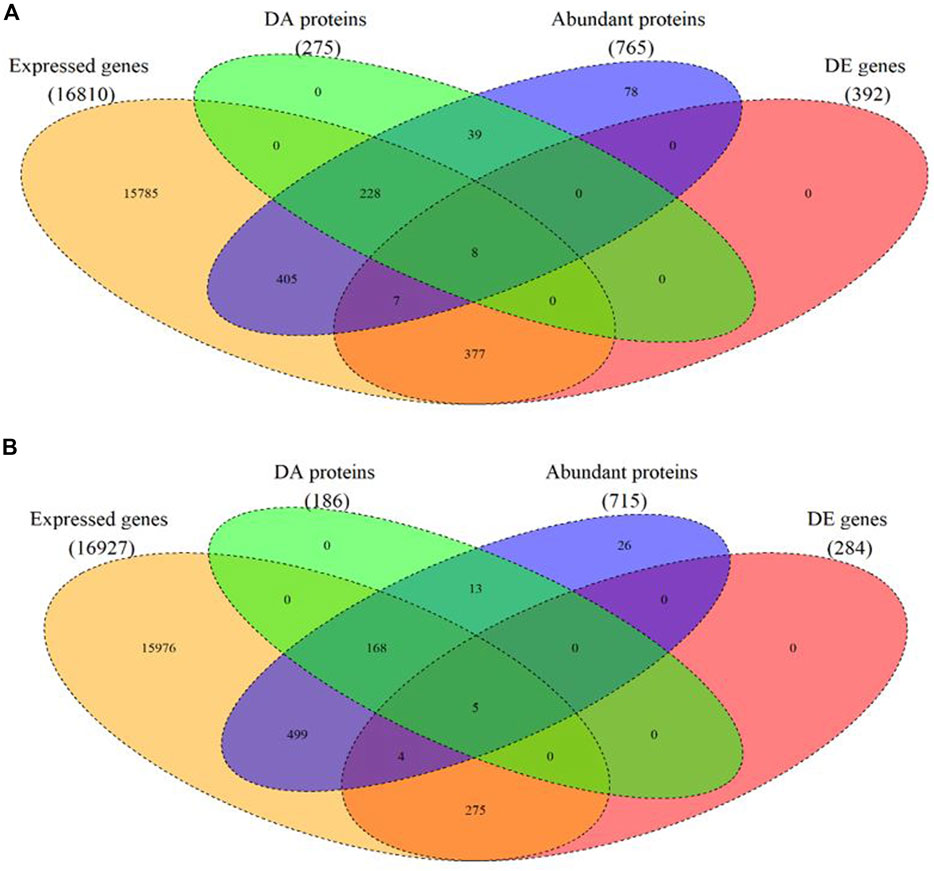
FIGURE 1. Venn diagram of expressed mRNAs and proteins as well as differentially expressed (DE) mRNA and differentially abundant (DA) protein in each tissue. (A) in the hypothalamus tissue, (B) in the pituitary gland.
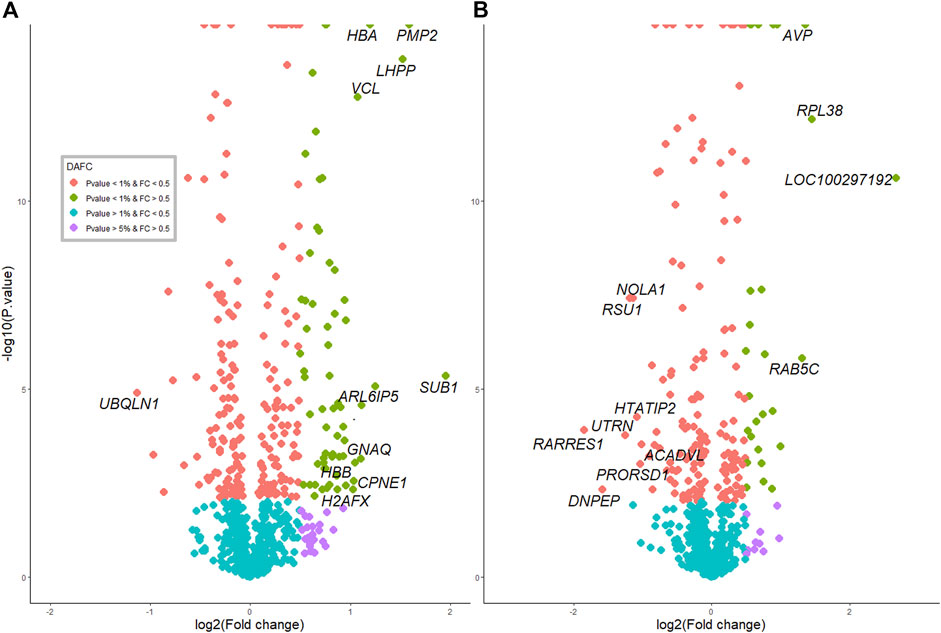
FIGURE 2. Protein profiles and overlap with RNAseq between pre- and postpubertal heifers in each tissue. (A) in the hypothalamus tissue. (B) in the pituitary gland.
In the pituitary libraries, a total of 715 proteins were identified and quantified. Of these, 186 were DA proteins, where 96 proteins were significantly decreased, and 90 proteins were significantly increased at post-puberty (adjusted p-value < 0.01) (Figures 1B, 2B and Supplementary Table S1).
The extent of DA proteins between pre and post-puberty in each tissue was visualized in volcano plots (Figures 2A,B). Protein IDs, corresponding gene names and fold change (log2FC) information are listed in Supplementary Table S1.
Derived Correlations Between mRNA Levels and Protein Abundances
The matching pairs of expressed mRNA and proteins in each tissue were used for comparison because of the relatively small number of matched DE mRNA and DA proteins. In the hypothalamus, 641 out of 765 identified proteins corresponded to mRNA transcripts discovered in the hypothalamic transcriptome data (Fortes et al., 2016) (Figure 1A and Supplementary Table S2). The correlation between 641 protein and transcript pairs in the hypothalamus was positive but insignificant, with a correlation coefficient of 0.05 (p-value = 0.16). Among these pairs, only eight DE genes were DA proteins, namely TUBB2B, OMG, SPTAN1, RAP1A, SSBP1, PLCB1, EIF3J and H2AFX (Table 1). Six of these eight DA proteins showed the same direction of regulation as their mRNAs.
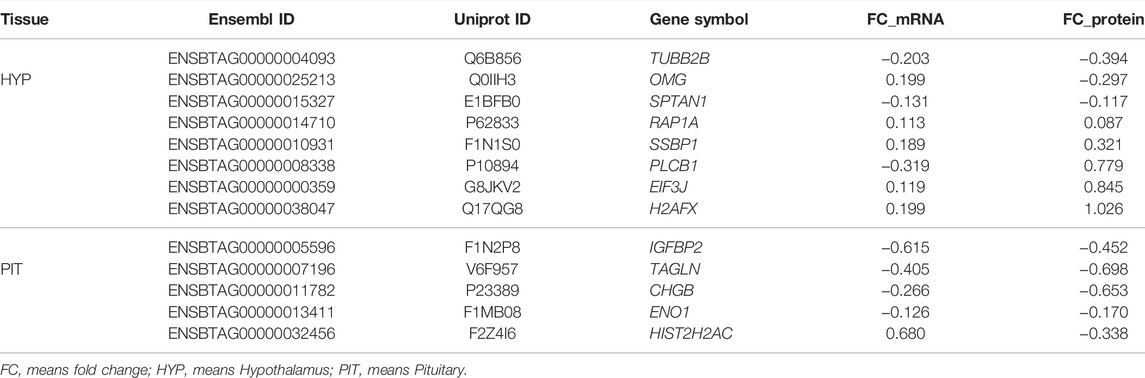
TABLE 1. List of common differentially expressed genes and abundant proteins in the hypothalamus and pituitary studies.
When compared the proteins in the pituitary gland with its corresponding mRNA data (Nguyen et al., 2017), 648 out of 715 mRNA-proteins pairs were identified (Fortes et al., 2016) (Figure 1B and Supplementary Table S2). The correlation between transcript and protein in pituitary for 648 proteins was minimal (r = 0.006; p-value = 0.8). Among these pairs, five DA proteins and DE genes were in common. They were: IGFBP2, TAGLN, CHGB, ENO1 and HIST2H2AC (Table 1). Four of these five proteins showed the same direction of regulations as their mRNAs.
Gene Ontology Enrichment Analysis
In order to evaluate the major biological processes influencing puberty onset in Brahman cattle, GO enrichment was performed to examine the functional characteristics of DA proteins between pre and post-pubertal samples in each tissue (Supplementary Tables S3, S4).
As shown in Figure 3, DA proteins in the hypothalamus were predominantly enriched for “organic substance metabolic process” (31%), “single-organism metabolic process” (27%), “oxidation-reduction process” (14%) and “nervous system development” (12%), in the BP category of GO terms. As expected, DA proteins were involved in the peptide metabolic process and brain development (including development of neurons, glial cells and axons). These DA proteins were also annotated to the terms “response to oxidative stress,” “glutamine metabolic and catabolic process,” “energy derivation by oxidation of organic compounds,” and “regulation of stress-activated MAPK cascade” (FDR <0.05). MF analysis revealed DA proteins were connected to 89 enriched GO terms, including proteins involved in binding (38%) and catalytic activity (34%). There were 116 enriched GO terms found in the CC category (FDR <0.05), and the majority of DA proteins were located in the cytoplasmic (51%) and intracellular (45%) space.
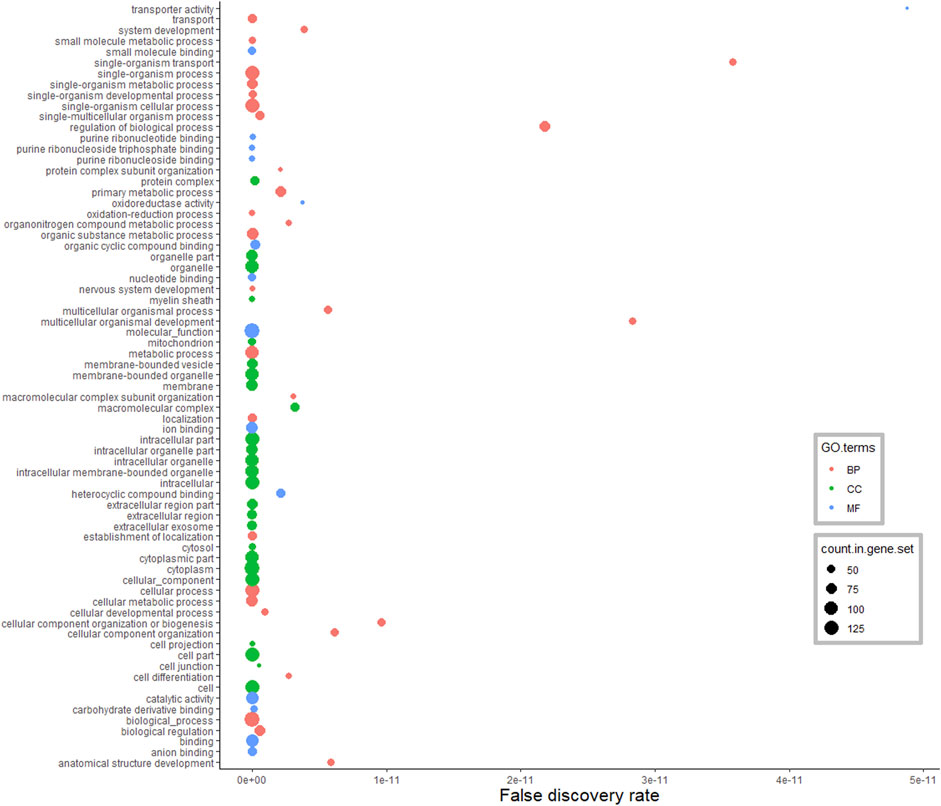
FIGURE 3. Functional classification of differentially abundant proteins in the hypothalamus between pre-versus postpubertal heifers. Only highly significant GO terms were shown (FDR <1.0E−10).
According to the GO annotations of DA proteins from the pituitary gland (Figure 4), among 214 enriched BP terms, the majority of DA proteins were classified in the single-organism cellular process (39%), cellular process (38%), metabolic process (35%) and biological regulation (30%). Enriched MF terms were 39 in the analysis of the pituitary gland (FDR <0.05). Hydrolases, oxidoreductases and endopeptidase were the main protein classes identified in this MF anotation. Moreover, the CC terms of pituitary DA proteins were assigned to the organelle, membrane-bounded organelle, cytoplasm, intracellular and extracellular regions.
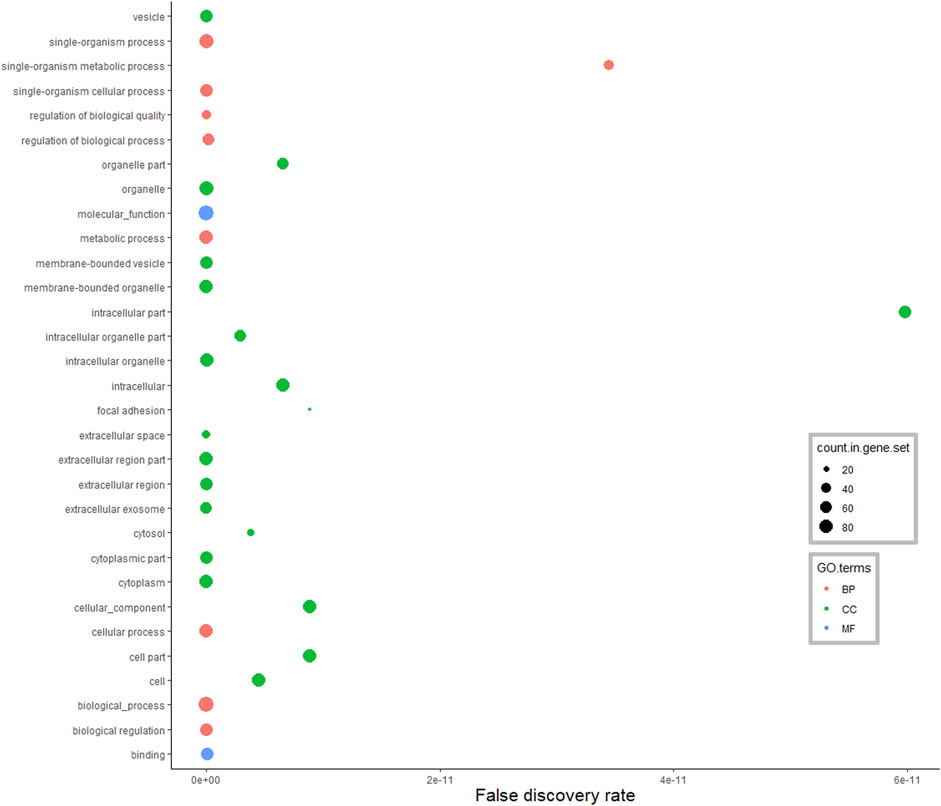
FIGURE 4. Functional classification of differentially abundant proteins in the pituitary between pre-versus postpubertal heifers. Only highly significant GO terms were shown (FDR <1.0E−10).
KEGG Pathway Analysis
In order to investigate the biological pathways enriched in response to puberty onset in Brahman heifers, a KEGG pathway enrichment analysis was performed.
In the hypothalamus, comparison of pre- and post-pubertal samples, yielded a total of 275 DA proteins mapped to 67 enriched pathways (Supplementary Table S5). Among enriched pathways, known pathways that are related to puberty were also identified. The enriched pathways included the estrogen signalling pathway (FDR = 0.002), axon guidance (FDR = 0.006), glutamatergic synapse (FDR = 0.0001), GABAergic synapse (FDR = 0.005), GnRH signalling pathway (FDR = 0.02), oxytocin signalling pathway (FDR = 0.001), calcium signalling (FDR = 0.01) and PPAR signalling pathway (FDR = 0.05). DA proteins involved in the GnRH signalling pathway were highlighted in Figure 5. In addition, DA proteins were also involved in other pathways such as oxidative phosphorylation (FDR = 6.6 × 10−12), the tricarboxylic acid cycle (TCA, FDR = 3.58 × 10−9), non-alcoholic fatty liver disease (NAFLD, FDR = 1.08 × 10−5), glycolysis and gluconeogenesis (FDR <0.05), thyroid hormone synthesis (FDR = 0.05), metabolism of amino acids and carbohydrate metabolism (FDR <0.05). The DE gene and corresponding DA protein, PLCB1, was annotated to four enriched pathways: GnRH signalling, estrogen signalling, calcium signalling and oxytocin signalling. In short, pathways related to neuronal-hormonal signalling and metabolism were significant for the DA proteins in the hypothalamus (Table 2).
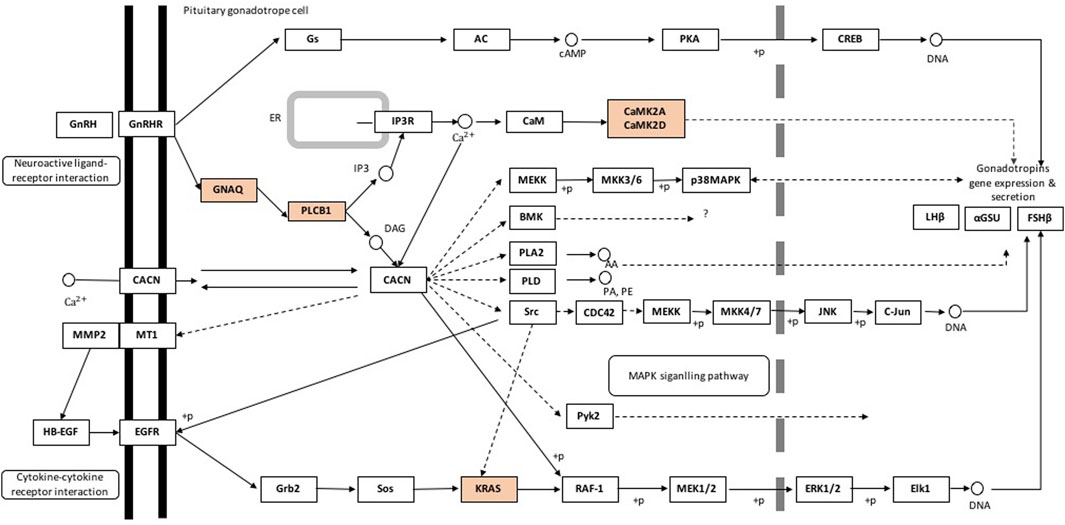
FIGURE 5. Gonadotropin-releasing hormone signalling pathway containing differentially abundant proteins in the hypothalamus of pre- and postpubertal brahman heifers. Pathway components representing differentially abundant proteins are in orange. Pathway was adapted from the KEGG database (http://www.genome.jp/kegg-bin/show_pathway?bta04912, accessed: 11/03/2018).
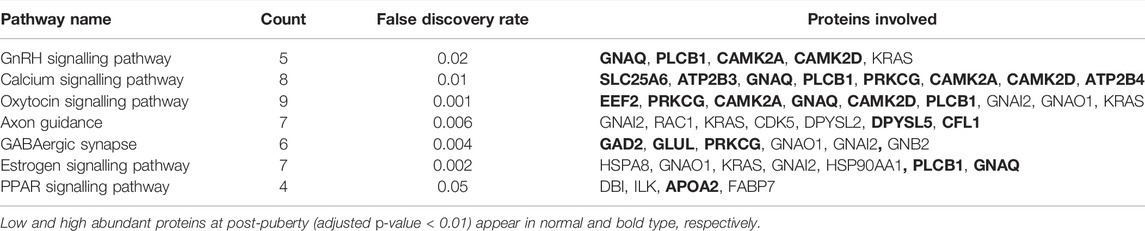
TABLE 2. Enriched pathways related to differentially abundant proteins in heifer puberty in the hypothalamus.
In the pituitary gland, a total of 186 DA proteins were assigned to 12 KEGG pathways (FDR <0.05). Among these enriched pathways, the ribosome pathway represented the largest number of DA proteins (FDR = 0.0002). Complement and coagulation cascades, glycolysis and gluconeogenesis, biosynthesis of amino acids, and focal adhesion were also overrepresented (Figure 6).
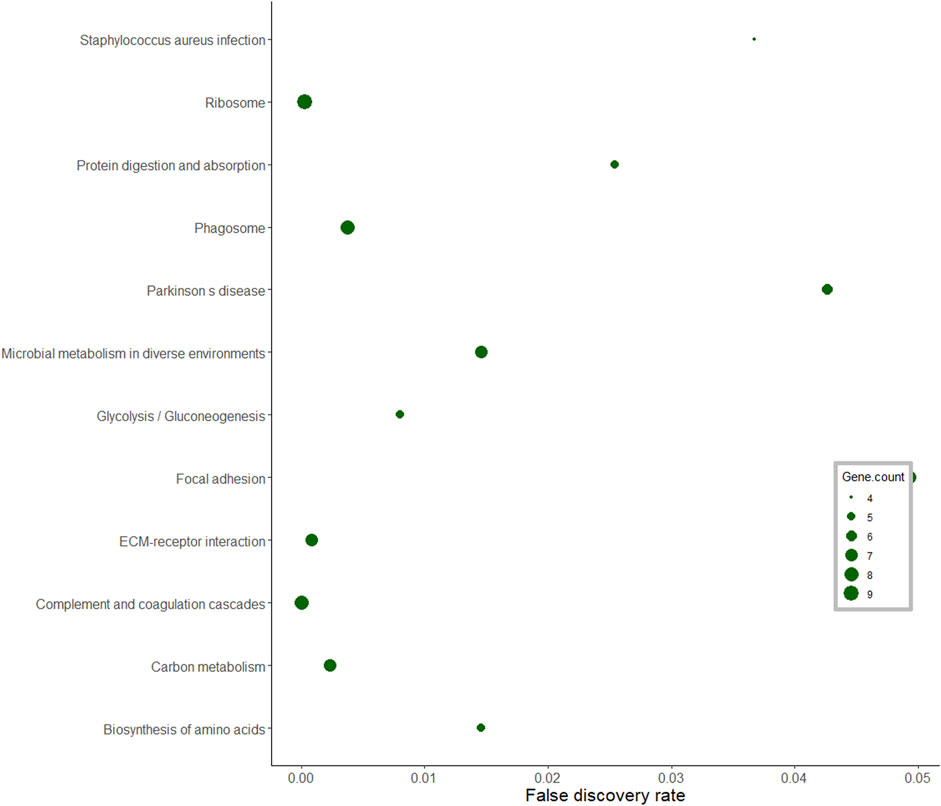
FIGURE 6. KEGG pathway analysis of differentially abundant proteins in the pituitary gland of pubertal Brahman heifers.
Protein -Protein Interaction Network Analysis
The relationship among DA proteins was further investigated in a network analysis. The hypothalamus network contained 181 nodes (proteins) with 486 edges (interactions) (Figure 7). Heat shock protein 90 alpha family class A member 1 (HSP90AA1) had the highest number of connections in the network (34 edges), followed by malate dehydrogenase 2 (MDH2) and albumin (ALB) with 23 and 21 connections, respectively. Of note, HSP90AA1 contributes to the estrogen signalling pathway, according to the KEGG pathway analysis (Table 2). In addition, KRAS protein, which plays a role in GnRH signalling, axon guidance, estrogen signalling and oxytocin signalling, interacted with 13 proteins in the network (Table 2 and Figure 7). PLCB1, a DE gene and DA protein, had nine connections in the network: GNAQ, GNAI2, PIP4K2A, GNAO1, SYNJ1, GNB2, PRKCG, CAMK2D, CAMK2A (Table 2 and Figure 7). These genes that were hubs in the network analysis and were associated with enriched pathways of biological relevance for puberty might be essential drivers of puberty in cattle.
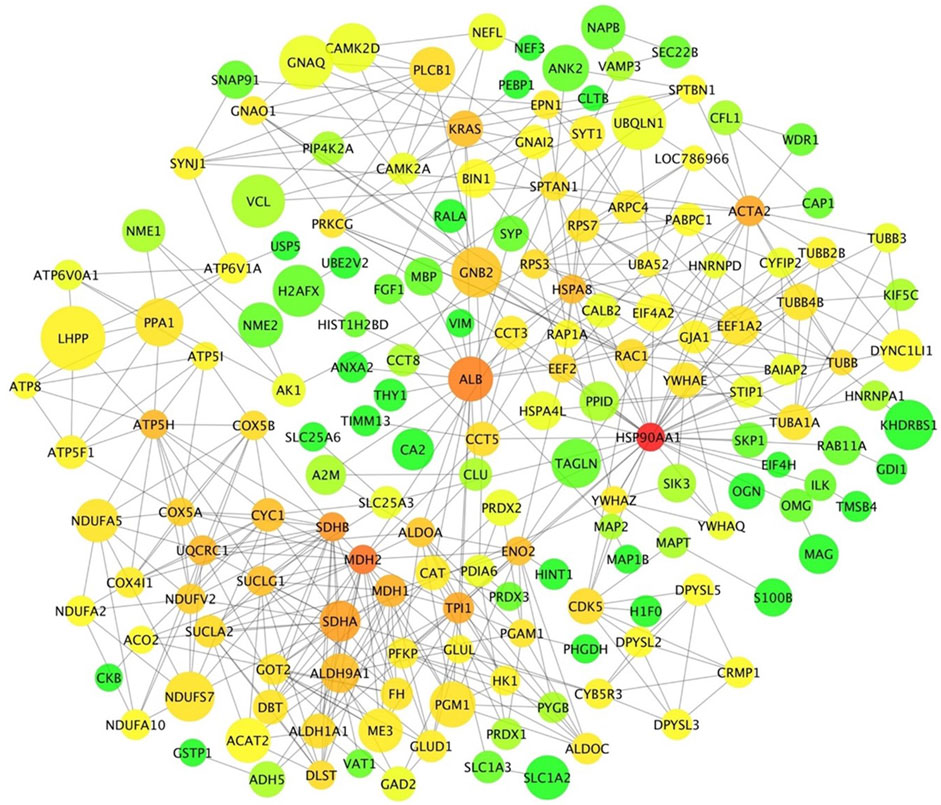
FIGURE 7. Protein-protein interaction network in the hypothalamic tissue. Each node represents a protein. Edges illustrate the interaction between proteins. Node size represents the level of differential abundance measured for each protein in absolute terms, where bigger nodes represent higher fold changes. Node colour range from green to red represent low to high connectivity in the network for each specific protein.
The interaction network from the pituitary DA proteins comprised 113 nodes with 254 edges (Figure 8). Among these nodes, ribosome-related proteins seem to form a dominant cluster in the network. The protein RPS5 had 17 connections within the network. A common DE gene and a DA protein, ENO1, was found in the network with 10 connections. This protein is involved in carbon metabolism, biosynthesis of amino acids and glycolysis/gluconeogenesis, as identified by the KEGG pathway analysis of DA proteins in the pituitary between pre- and post-pubertal Brahman heifers.
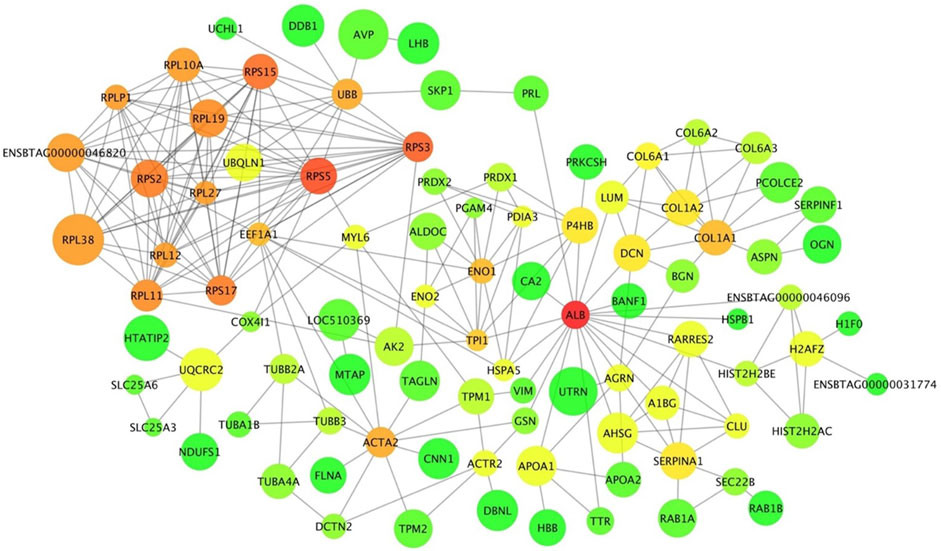
FIGURE 8. Protein-protein interaction network of differentially abundant proteins between pre- and postpubertal heifers in the pituitary gland. Each protein is a node and protein-protein interactions are illustrated as edges that link nodes. The size of node represents the increase or decrease in protein abundance post-puberty. Node colour represents the connectivity of a protein within the network; it is ranged from green to red for low to high connectivity.
Discussions
High-throughput techniques such as transcriptomic and proteomic studies are potent approaches to understanding gene expression and regulation. Mowever, no single approach can stand alone if we are to understand the fundamental biology underlying complex traits. Analyses at multiple levels are needed to interpret complex biological systems. In this study, the correlation between protein abundance and mRNA expression in the hypothalamus and pituitary gland was insignificant. To date, reports on the correlation between mRNA and proteins that use high-throughput approaches are scarce. Yeast, bacteria and human cancer data reports found limited correlations between mRNA and protein levels (Anderson and Seilhamer, 1997; Gygi et al., 1999; Chen et al., 2002; Griffin et al., 2002; Ghaemmaghami et al., 2003; Greenbaum et al., 2003; Washburn et al., 2003; Tian et al., 2004; Nie et al., 2006; Jayapal et al., 2008; de Sousa Abreu et al., 2009; Haider and Pal, 2013; Bai et al., 2015; Carvalhais et al., 2015; Payne, 2015; Edfors et al., 2016). A weak correlation of 0.05 was reported for 17 DE genes and corresponding protein levels analysed in the bovine mammary gland (Dai et al., 2017). The weak correlation between mRNA and protein expression levels may result from methodological constraints and biological factors, such as translational regulation and protein in vivo half-lives (Maier et al., 2009; Vogel and Marcotte, 2012; Haider and Pal, 2013). Additional data on factors affecting protein levels and in-depth studies, can help address this conundrum.
Hypothalamus transcriptome and proteome analyses in pre- and post-pubertal Brahman heifers revealed eight DE genes that are also DA proteins. Among these DA proteins, tubulin beta 2B class IIb (TUBB2B) is involved in neuronal migration and axonal guidance that are prominent brain remodeling mechanisms during pubertal development (Bond et al., 1984; Romaniello et al., 2015). A study in female rats noted that the neurotrophic effects of estrogen on the central nervous system at the onset of puberty were partly mediated by the increase of tubulin beta class II (Rogers et al., 1993). Further, the significant increase of TUBB2B and TUBB2A mRNA expression in low progesterone endometrium was reported in heifers (Forde et al., 2012). Our study revealed a decreased abundance of TUBB2B post-puberty, suggesting the correlation between TUBB2B and steroid hormones. High expression of TUBB2B in pre-pubertal Brahman heifers is proposed to play a part in remodeling the brain and how it responds the feedback of ovarian hormones. Changes to the central nervous system are intrincical to pubertal development.
Another protein among the eight that were DA that were also DE genes in the hypothalamus is spectrin alpha nonerythrocytic 1 (SPTAN1). The SPTAN1 gene is known to play a vital role in brain development and epileptic encephalophathy (Wang et al., 2018). This protein was identified as an upregulated gene in the mammary epithelial cells of pre-pubertal female mice compared to post-pubertal mice (Pal et al., 2017). These results in mice seem contrasting to ours, as SPTAN1 was downregulated in post-pubertal cattle. Species or tissue differences and the definition for puberty (and therefore timing of sampling) could explain such discrepancies. Nonetheless, SPTAN1 is emerging as a candidate for future research targeting puberty machanisnsm in mammals.
Perhaps the Phospholipase C beta 1 (PLCB1) was the DA protein, which is also a DE gene, with more evidence of its role in puberty. PLCB1 plays a crucial role in reproductive physiology. Genome-wide association studies in pigs reported the contribution of PLCB1 to growth at puberty onset and in the gonadotropin signalling pathway (Meng et al., 2017). Further, PLCB1 disruption resulted in infertile mice with pleiotropic reproductive defects (Filis et al., 2013). PLCB1 is a crucial factor modulating GPCR signalling, which controls reproductive physiology in mice (Jiang et al., 1997; Kim et al., 1997; Xie et al., 1999; Bohm et al., 2002; Ballester et al., 2004; Filis et al., 2013). Our results serve as evidence that PLCB1 may have a role in puberty onset in Bos indicus cattle: it interacts in the predicted network with other genes (GNAQ, GNAI2, PIP4K2A, GNAO1, SYNJ1, GNB2, PRKCG, CAMK2D, CAMK2A) related to GnRH signalling and other hormonal and neuronal signalling pathways known to be involved in the feedback mechanisms driving the activation of HPO axis. More importantly, PLCB1 was one of the upregulated DA proteins at post-puberty involving in GnRH signalling, estrogen signalling, calcium signalling and oxytocin signalling. In hypothalamic neurons, the pulsatile release of GnRH is depended on voltage-gated calcium influx (Krsmanović et al., 1992). Therefore, it is logical to propose that the increase of GnRH release at the hypothalamus of Brahman heifers may be initiated via oxytocin signalling, via PLCB1.
Pituitary transcriptome and proteome analyses revealed five differentially expressed genes: IGFBP2, TAGLN, CHGB, ENO1 and HIST2H2AC, significantly less abundant post-puberty. Among these, IGFBP2 was the DA and DE gene with mounting evidence of its roles in puberty. IGFBP2 is a predominant insulin growth factor binding protein synthesised and secreted in the anterior pituitary during the pre-ovulatory and early luteal phase in beef cattle (Funston et al., 1995; Roberts et al., 2001). In the anterior pituitary, the expression of IGFBP2 fluctuated with changes in the estrous cycle that were associated with serum progesterone (Funston et al., 1995). In addition, estrogen signalling increased IGFBP2 expression in the anterior pituitary of ewes, cattle, pigs and rats (Michels et al., 1993; Clapper et al., 1998; Roberts et al., 2001; Rempel and Clapper, 2002). Roberts et al. (2001) reported that IGFBPs levels in the anterior pituitary decreased from pre-ovulatory to early luteal development in beef cattle. Consistent with Robert et al. (2001) observations, the mRNA expression of IGFBP2 was decreased in the transcriptome study (Nguyen et al., 2017) and the current study. In short, we observed a lower abundancy of IGFBP2 in the bovine pituitary gland at the luteal phase post-puberty. The release of IGFBP2 in the pituitary gland was stimulated by gonadotropin-releasing hormone, as demonstrated by Robert et al. (2001). Also, polymorphisms in IGFBP2 were associated with age at puberty in Brahman cattle (Fortes et al., 2013b).
Chromogranin B (CHGB), another DA protein that was also a DE gene from the pituitary studies, belongs to a chromogranin protein family, which has been noted to affect the secretion and storage of FSH and LH at different periods of estrous cycle in sheep (Crawford and McNeilly, 2002). A study of granin-gonadotropin interactions in sheep observed a decrease of CHGB mRNA level after the pre-ovulatory LH surge (Crawford and McNeilly, 2002). In agreement with the sheep study, mRNA expression of CHGB decreased at post-puberty in the pituitary transcriptome of Brahman heifers (Nguyen at el., 2017). Additionally, the neuropeptide CHGB was selected as one of the fertility-related neuropeptides in the pituitary gland neuropeptidome of pre- and post-pubertal Brangus heifers (DeAtley et al., 2018). The neuropeptidome study of Brangus heifers also reported higher levels of CHGB neuropeptide products in pre-pubertal pituitary tissue (DeAtley et al., 2018). This study observed a lower abundance of CHGB protein after puberty in Brahman heifers. Although measuring neuropeptides is different from measuring protein levels, both studies point to the importance of CHGB in the pituitary glands during pubertal development in cattle.
The DA protein α-enolase (ENO1) is one of the three isoforms of glycolytic enzymes responsible for catalysing the conversion of 2-phosphoglycerate to phosphoenolpyruvate (Pancholi, 2001). A study in the ovaries of pre-laying and laying geese suggested that ENO1 may regulate reproductive function in female geese (Ji et al., 2014; Kang et al., 2014). The overexpression of ENO1 in granular cells induced the mRNA expression of FSH and LH receptors (Ji et al., 2014). Activation of these receptors is necessary for the hormonal functioning of pituitary glycoprotein hormones (LH and FSH). Pituitary FSH and LH concentrations decreased at puberty in heifers (Desjardins and Hafs, 1968). Our current and transcriptome studies (Fortes et al., 2016; Nguyen et al., 2017) found lower mRNA and protein expression of ENO1 post-puberty. On the contrary, single-cell RNA profiling found ENO1 to be down-regulated in a pre-puberty gene network of mouse mammary glands (Pal et al., 2017). It is apparent that ENO1 impacts female puberty, but its precise function needs to be considered in terms of tissue and species-specific.
The DA protein coded by the DE gene HIST2H2AC in the pituitary, a member of the histone 2A family, was mapped to a genetic locus reported linking puberty timing and pubertal height growth in humans (Cousminer et al., 2013). A genetic correlation between age at puberty and high height in Brahman heifers was also reported (Vargas et al., 1998). Differential expression of HIST2H2AC at mRNA and proteins level between pre- and post-pubertal heifers proposes it has a role in pubertal development. This candidate gene could be mined for mutations that could be tested for their effects on height and puberty in cattle.
The activation of HPO axis culminates with the pulsatile release of GnRH from the hypothalamus. GnRH triggers the synthesis and secretion of LH and FSH in the pituitary gland, which stimulates the production of gonadal hormones leading to ovulation (Bliss et al., 2010). Both positive and negative feedback at several levels regulate HPO function. One positive feedback on the activation of GnRH secretion is commenced by GnRH receptor (GnRHR) - G-protein αq subunit (Gαq/11), which induces increased intracellular calcium (Khadra and Li, 2006). In agreement with this positive feedback, our proteomic study in the hypothalamus revealed the up-regulation of the calcium signalling pathway (FDR = 0.05) and GnRH signalling pathway (FDR = 0.02). G protein subunit alpha q (GNAQ), phospholipase C beta 1 (PLCB1), calcium/calmodulin-dependent protein kinase II alpha (CAMK2A), and calcium/calmodulin-dependent protein kinase II delta (CAMK2D) showed increased abundance at post-puberty in Brahman heifers and were associated with the upregulated pathways. As a result, enrichment analysis of DA proteins from the hypothalamus confirmed the involvement of these proteins on the onset of puberty via multiple pathways.
Among enriched GO terms in the pituitary libraries, six of these 11 GO terms, including regulation of multicellular organismal process, extracellular region part, extracellular matrix, proteinaceous extracellular matrix, calcium ion binding and extracellular matrix structural constituent were also found as GO terms involved in the onset of puberty in goat and rats (Gao et al., 2018). These findings and the referenced literature suggest the involvement of these GO terms in the regulation of puberty in goats, rats and Brahman heifers. It seems that many of the mechanisms of female puberty are conserved across mammals.
The interaction network in the hypothalamus revealed three DA proteins as highly connected hubs: HSP90AA1, MDH2 and ALB. The protein HSP90AA1 was involved in estrogen signalling, and it interacted with other proteins of the PPAR and oxytocin pathways (ILK, EEF2 and PRKCG). Recently, a transcriptomic study in pre-versus post-pubertal mammary epithelial cells noted HSP90AA1 as a DE gene (Pal et al., 2017). Even though there is little evidence of a relationship between MDH2 and female reproduction, MDH2 was characterised as a protein marker for male fertility (Kwon et al., 2015a; Kwon et al., 2015b). A study in Egyptian boys suggested ALB status involved in each puberty stage (Cole et al., 1982). The increase of albumin excretion rate in the urinary tract was significantly associated with puberty in non-diabetic children and adolescents (Bangstad et al., 1993). Bovine serum albumin–estrogen compounds impacted GnRH1 neuronal activity (Temple and Wray, 2005). The inferred network predicted interactions between ALB and proteins involved in estrogen (HSPA8) and oxytocin (EEF2) signalling.
Gao et al. (2018) performed KEGG analysis of long non-coding RNA targets in the hypothalamus of pubertal rats and reported significant enrichment for the ribosome pathway. In this current study, the ribosome pathway was also enriched, and the proteins in this pathway were less abundant post-puberty in the pituitary gland. Further, these ribosomal proteins formed a dominant cluster in the pituitary interaction network. A study in pubertal Brangus heifers observed high expression of the ribosomal protein L39 gene in the pituitary gland (Cánovas et al., 2014). Further, ribosomal DA proteins in the interaction network, such as RPS5, RPS3 and RPLP1, were also listed in the networks constructed from endometrial gene expression of low and high fertility heifers (Killeen et al., 2014). Recently, miR-503-3p was proposed as a new repressor of the initiation of puberty in female mice (Tong et al., 2018). The stable overexpression of miR-503-3p in the GT1-7 cell line can influence ribosome biogenesis pathways and result in down-regulation of puberty-related genes (Tong et al., 2018). The ribosomal protein RPL22 inhibited the expression of Lin28B, a puberty-related gene (Ong et al., 2009; Rao et al., 2012). Another ribosomal protein, namely RPS7, also regulated the PI3K and MAPK signalling pathways that are involved in puberty (Bliss et al., 2010; Acosta-Martínez, 2011; Wang et al., 2013; Nelson et al., 2017; Pal et al., 2017). From the pituitary protein-protein interaction network, there were seven ribosomal proteins that interacted with UBB protein. Disruption in the UBB gene in both male and female mice resulted in infertile animals (Ryu et al., 2008). These collective evidences suggested proteins in the ribosomal pathway are relevant for pubertal development in mammals. Further work could target the DA ribosomal proteins and their targets in the pituitary gland to investigate the molecular mechanisms of puberty in Bos indicus.
Conclusion
In summary, The study confirmed the poor global correlation between mRNAs and proteins, suggesting the need for multiple omics analyses for interpreting complex biological systems. A total of 275 and 186 DA proteins, between pre-pubertal and post-pubertal Brahman heifers in the hypothalamus and pituitary gland were identified, respectively. These DA proteins may regulate puberty onset directly or indirectly. DA proteins in the hypothalamus were mainly associated with metabolic pathways, energy metabolism, and nervous system development. In agreement with previous findings of complex feedback effects on GnRH release, the KEGG pathway analysis indicated that these DA proteins are involved in pathways that convey both inhibitory and excitatory inputs for hypothalamic neurons. In the pituitary, the abundance of proteins involved in protein digestion and absorption, focal adhesion, and ECM-receptor interaction were significantly increased post-puberty. The decreased abundance of ribosomal proteins post-puberty can be interpreted in the context of these genes’ inhibitory input to puberty-related genes. Proteins related to energy production and amino acid biosynthesis may have a crucial role in the neuroendocrine regulation of the pubertal process, meriting further investigation.
Data Availability Statement
The proteomic datasets for the hypothalamus and pituitary gland have been submitted into Proteome Xchange Consortium via PRIDE partner repository with identifier PXD009620 and PXD009619, respectively.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of The University of Queensland ethics board (Animal Ethics Approval Number QAAFI/279/12).
Author Contributions
LN designed the study, performed all experiments and statistical analyses, prepared figures and wrote the manuscript. LL contributed to Figure 2. MF designed the study and collected samples. All authors commented on the manuscript, read and approved the final manuscript.
Funding
This work was supported by the start-up grant to Stephen Moore from The University of Queensland, which covered the initial animal trial.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the contributions to the field experiment and sample collection of Gry Boe-Hansen, Laercio R. Porto-Neto, Lisa Kidd, and Joao Paulo A. do Rego. The authors would also like to thank Amanda Nouwens for running LC-ESI-/MS-MS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.935433/full#supplementary-material
References
Acosta-Martínez, M. (2011). PI3K: An Attractive Candidate for the Central Integration of Metabolism and Reproduction. Front. Endocrin. 2, 110. doi:10.3389/fendo.2011.00110
Anderson, L., and Seilhamer, J. (1997). A Comparison of Selected mRNA and Protein Abundances in Human Liver. Electrophoresis 18 (3-4), 533–537. doi:10.1002/elps.1150180333
Bai, Y., Wang, S., Zhong, H., Yang, Q., Zhang, F., Zhuang, Z., et al. (2015). Integrative Analyses Reveal Transcriptome-Proteome Correlation in Biological Pathways and Secondary Metabolism Clusters in A. flavus in Response to Temperature. Sci. Rep. 5, 14582. doi:10.1038/srep14582
Ballester, M., Molist, J., Lopez-Bejar, M., Sánchez, A., Santaló, J., M. Folch, J., et al. (2004). Disruption of the Mouse Phospholipase C-Β1 Gene in a β-lactoglobulin Transgenic Line Affects hourbility, Growth, and Fertility in Mice. Gene 341, 279–289. doi:10.1016/j.gene.2004.07.007
Bangstad, H.-J., Dahl-Jørgensen, K., Kjaexsgaard, P., Mevold, K., and Hanssen, K. (19931992). Urinary Albumin Excretion Rate and Puberty in Non-diabetic Children and Adolescents. Acta Paediatr. (Oslo, Nor. 82 (10), 857–862. doi:10.1111/j.1651-2227.1993.tb17628.x
Bliss, S. P., Navratil, A. M., Xie, J., and Roberson, M. S. (2010). GnRH Signaling, the Gonadotrope and Endocrine Control of Fertility. Front. Neuroendocrinol. 31 (3), 322–340. doi:10.1016/j.yfrne.2010.04.002
Bohm, D., Schwegler, H., Kotthaus, L., Nayernia, K., Rickmann, M., Kohler, M., et al. (2002). Disruption of PLC-$beta;1-Mediated Signal Transduction in Mutant Mice Causes Age-dependent Hippocampal Mossy Fiber Sprouting and Neurodegeneration. Mol. Cell. Neurosci. 21 (4), 584–601. doi:10.1006/mcne.2002.1199
Bond, J. F., Robinson, G. S., and Farmer, S. R. (1984). Differential Expression of Two Neural Cell-specific Beta-Tubulin mRNAs during Rat Brain Development. Mol. Cell. Biol. 4 (7), 1313–1319. doi:10.1128/mcb.4.7.1313-1319.1984
Cánovas, A., Reverter, A., DeAtley, K. L., Ashley, R. L., Colgrave, M. L., Fortes, M. R. S., et al. (2014). Multi-Tissue Omics Analyses Reveal Molecular Regulatory Networks for Puberty in Composite Beef Cattle. PloS one 9 (7), e102551. doi:10.1371/journal.pone.0102551
Carvalhais, V., França, A., Pier, G. B., Vilanova, M., Cerca, N., and Vitorino, R. (2015). Comparative Proteomic and Transcriptomic Profile of Staphylococcus Epidermidis Biofilms Grown in Glucose-Enriched Medium. Talanta 132, 705–712. doi:10.1016/j.talanta.2014.10.012
Chen, G., Gharib, T. G., Huang, C.-C., Taylor, J. M. G., Misek, D. E., Kardia, S. L. R., et al. (2002). Discordant Protein and mRNA Expression in Lung Adenocarcinomas. Mol. Cell. Proteomics 1 (4), 304–313. doi:10.1074/mcp.M200008-MCP200
Chenoweth, P. (1994). Aspects of Reproduction in Female Bos indicus Cattle: a Review. Aust. Vet. J. 71 (12), 422–426. doi:10.1111/j.1751-0813.1994.tb00961.x
Clapper, J. A., Snyder, J. L., Roberts, A. J., Hamernik, D. L., and Moss, G. E. (1998). Estradiol Increases Relative Amounts of Insulin-like Growth Factor Binding Protein (IGFBP)-3 in Serum and Expression of IGFBP-2 in Anterior Pituitaries of Ewes1. Biol. Reprod. 59 (1), 124–130. doi:10.1095/biolreprod59.1.124
Cole, T. J., Salem, S. I., Hafez, A. S., Galal, O. M., and Massoud, A. (1982). Plasma Albumin, Parasitic Infection and Pubertal Development in Egyptian Boys. Trans. R. Soc. Trop. Med. Hyg. 76 (1), 17–20. doi:10.1016/0035-9203(82)90007-4
Cousminer, D. L., Berry, D. J., Timpson, N. J., Ang, W., Thiering, E., Byrne, E. M., et al. (2013). Genome-wide Association and Longitudinal Analyses Reveal Genetic Loci Linking Pubertal Height Growth, Pubertal Timing and Childhood Adiposity. Hum. Mol. Genet. 22 (13), 2735–2747. doi:10.1093/hmg/ddt104
Crawford, J., and McNeilly, A. (2002). Co-localisation of Gonadotrophins and Granins in Gonadotrophs at Different Stages of the Oestrous Cycle in Sheep. J. Endocrinol. 174 (2), 179–194. doi:10.1677/joe.0.1740179
Dai, W., Chen, Q., Wang, Q., White, R. R., Liu, J., and Liu, H. (2017). Complementary Transcriptomic and Proteomic Analyses Reveal Regulatory Mechanisms of Milk Protein Production in Dairy Cows Consuming Different Forages. Sci. Rep. 7, 44234. doi:10.1038/srep44234
Day, M. L., and Anderson, L. H. (1998). Current Concepts on the Control of Puberty in Cattle. J. Animal Sci. 76 (Suppl. l_3), 1–15. doi:10.2527/1998.76suppl_31x
Day, M. L., Imakawa, K., Garcia-Winder, M., Zalesky, D. D., Schanbacher, B. D., Kittok, R. J., et al. (1984). Endocrine Mechanisms of Puberty in Heifers: Estradiol Negative Feedback Regulation of Luteinizing Hormone Secretion 1. Biol. Reprod. 31 (2), 332–341. doi:10.1095/biolreprod31.2.332
Day, M. L., Imakawa, K., Wolfe, P. L., Kittok, R. J., and Kinder, J. E. (1987). Endocrine Mechanisms of Puberty in Heifers. Role of Hypothalamo-Pituitary Estradiol Receptors in the Negative Feedback of Estradiol on Luteinizing Hormone Secretion1. Biol. Reprod. 37 (5), 1054–1065. doi:10.1095/biolreprod37.5.1054
de Sousa Abreu, R., Penalva, L. O., Marcotte, E. M., and Vogel, C. (2009). Global Signatures of Protein and mRNA Expression Levels. Mol. Biosyst. 5 (12), 1512–1526. doi:10.1039/b908315d
DeAtley, K. L., Colgrave, M. L., Cánovas, A., Wijffels, G., Ashley, R. L., Silver, G. A., et al. (2018). Neuropeptidome of the Hypothalamus and Pituitary Gland of Indicine × Taurine Heifers: Evidence of Differential Neuropeptide Processing in the Pituitary Gland before and after Puberty. J. Proteome Res. 17 (5), 1852–1865. doi:10.1021/acs.jproteome.7b00875
Desjardins, C., and Hafs, H. D. (1968). Levels of Pituitary FSH and LH in Heifers from Birth through Puberty. J. animal Sci. 27 (2), 472–477. doi:10.2527/jas1968.272472x
Edfors, F., Danielsson, F., Hallström, B. M., Käll, L., Lundberg, E., Pontén, F., et al. (2016). Gene‐specific Correlation of RNA and Protein Levels in Human Cells and Tissues. Mol. Syst. Biol. 12 (10), 883. doi:10.15252/msb.20167144
Filis, P., Kind, P. C., and Spears, N. (2013). Implantation Failure in Mice with a Disruption in Phospholipase C Beta 1 Gene: Lack of Embryonic Attachment, Aberrant Steroid Hormone Signalling and Defective Endocannabinoid Metabolism. Mol. Hum. Reprod. 19 (5), 290–301. doi:10.1093/molehr/gas067
Forde, N., Mehta, J. P., Minten, M., Crowe, M. A., Roche, J. F., Spencer, T. E., et al. (2012). Effects of Low Progesterone on the Endometrial Transcriptome in Cattle1. Biol. Reprod. 87 (5), 124. doi:10.1095/biolreprod.112.103424
Fortes, M. R. S., Kemper, K., Sasazaki, S., Reverter, A., Pryce, J. E., Barendse, W., et al. (2013a). Evidence for Pleiotropism and Recent Selection in thePLAG1region in Australian Beef Cattle. Anim. Genet. 44 (6), 636–647. doi:10.1111/age.12075
Fortes, M. R. S., Li, Y., Collis, E., Zhang, Y., and Hawken, R. J. (2013b). The IGF1 Pathway Genes and Their Association with Age of Puberty in Cattle. Anim. Genet. 44 (1), 91–95. doi:10.1111/j.1365-2052.2012.02367.x
Fortes, M. R. S., Nguyen, L. T., Weller, M. M. D. C. A., Cánovas, A., Islas-Trejo, A., Porto-Neto, L. R., et al. (2016). Transcriptome Analyses Identify Five Transcription Factors Differentially Expressed in the Hypothalamus of Post- versus Prepubertal Brahman Heifers1. J. animal Sci. 94 (9), 3693–3702. doi:10.2527/jas.2016-0471
Funston, R. N., Moss, G. E., and Roberts, A. J. (1995). Insulin-like Growth Factor-I (IGF-I) and IGF-Binding Proteins in Bovine Sera and Pituitaries at Different Stages of the Estrous Cycle. Endocrinology 136 (1), 62–68. doi:10.1210/endo.136.1.7530196
Gao, X., Ye, J., Yang, C., Luo, L., Liu, Y., Ding, J., et al. (2018). RNA-seq Analysis of lncRNA-Controlled Developmental Gene Expression during Puberty in Goat & Rat. BMC Genet. 19, 19. doi:10.1186/s12863-018-0608-9
Gasser, C. L., Grum, D. E., Mussard, M. L., Fluharty, F. L., Kinder, J. E., and Day, M. L. (2006). Induction of Precocious Puberty in Heifers I: Enhanced Secretion of Luteinizing Hormone1. J. Anim. Sci. 84 (8), 2035–2041. doi:10.2527/jas.2005-636
Ghaemmaghami, S., Huh, W.-K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., et al. (2003). Global Analysis of Protein Expression in Yeast. Nature 425 (6959), 737–741. doi:10.1038/nature02046
Greenbaum, D., Colangelo, C., Williams, K., and Gerstein, M. (2003). Comparing Protein Abundance and mRNA Expression Levels on a Genomic Scale. Genome Biol. 4 (9), 117. journal article). doi:10.1186/gb-2003-4-9-117
Griffin, T. J., Gygi, S. P., Ideker, T., Rist, B., Eng, J., Hood, L., et al. (2002). Complementary Profiling of Gene Expression at the Transcriptome and Proteome Levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1 (4), 323–333. doi:10.1074/mcp.m200001-mcp200
Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. (1999). Correlation between Protein and mRNA Abundance in Yeast. Mol. Cell. Biol. 19 (3), 1720–1730. doi:10.1128/mcb.19.3.1720
Haider, S., and Pal, R. (2013). Integrated Analysis of Transcriptomic and Proteomic Data. Cg 14 (2), 91–110. doi:10.2174/1389202911314020003
Jayapal, K. P., Philp, R. J., Kok, Y.-J., Yap, M. G. S., Sherman, D. H., Griffin, T. J., et al. (2008). Uncovering Genes with Divergent mRNA-Protein Dynamics in Streptomyces Coelicolor. PloS one 3 (5), e2097. doi:10.1371/journal.pone.0002097
Ji, H., Yang, H., Wang, J., Guo, J., Hu, Z., Zhang, H., et al. (2014). Effect of Adenovirus-Mediated Up-Regulation of α-enolase Gene Products on Follicle-Stimulating Hormone Receptor mRNA and Luteinizing Hormone Receptor mRNA of Granular Cells from Goose F1 Follicles. Res. Veterinary Sci. 96 (3), 526–532. doi:10.1016/j.rvsc.2014.02.010
Jiang, H., Kuang, Y., Wu, Y., Xie, W., Simon, M. I., and Wu, D. (1997). Roles of Phospholipase C β2 in Chemoattractant-Elicited Responses. Proc. Natl. Acad. Sci. U.S.A. 94 (15), 7971–7975. doi:10.1073/pnas.94.15.7971
Johnston, D. J., Barwick, S. A., Fordyce, G., Holroyd, R. G., Williams, P. J., Corbet, N. J., et al. (2013). Genetics of Early and Lifetime Annual Reproductive Performance in Cows of Two Tropical Beef Genotypes in Northern Australia. Animal Prod. Sci. 54 (1), 1–15.
Kang, B., Jiang, D. M., Bai, L., He, H., and Ma, R. (2014). Molecular Characterisation and Expression Profiling of the ENO1 Gene in the Ovarian Follicle of the Sichuan White Goose. Mol. Biol. Rep. 41 (4), 1927–1935. doi:10.1007/s11033-014-3039-3
Khadra, A., and Li, Y.-X. (2006). A Model for the Pulsatile Secretion of Gonadotropin-Releasing Hormone from Synchronized Hypothalamic Neurons. Biophysical J. 91 (1), 74–83. doi:10.1529/biophysj.105.080630
Killeen, A. P., Morris, D. G., Kenny, D. A., Mullen, M. P., Diskin, M. G., and Waters, S. M. (2014). Global Gene Expression in Endometrium of High and Low Fertility Heifers during the Mid-luteal Phase of the Estrous Cycle. BMC Genomics 15, 234. doi:10.1186/1471-2164-15-234
Kim, D., Jun, K. S., Lee, S. B., Kang, N.-G., Min, D. S., Kim, Y.-H., et al. (1997). Phospholipase C Isozymes Selectively Couple to Specific Neurotransmitter Receptors. Nature 389 (6648), 290–293. doi:10.1038/38508
Krsmanović, L., Stojilković, S., Merelli, F., Dufour, S. M., Virmani, M. A., and Catt, K. J. (1992). Calcium Signaling and Episodic Secretion of Gonadotropin-Releasing Hormone in Hypothalamic Neurons. Proc. Natl. Acad. Sci. 89 (18), 8462–8466.
Kwon, W.-S., Rahman, M. S., Lee, J.-S., Yoon, S.-J., Park, Y.-J., and Pang, M.-G. (2015b). Discovery of Predictive Biomarkers for Litter Size in Boar Spermatozoa*. Mol. Cell. Proteomics 14 (5), 1230–1240. doi:10.1074/mcp.M114.045369
Kwon, W.-S., Rahman, M. S., Ryu, D.-Y., Park, Y.-J., and Pang, M.-G. (2015a). Increased Male Fertility Using Fertility-Related Biomarkers. Sci. Rep. 5, 15654. doi:10.1038/srep15654
Lau, L. Y., Nguyen, L. T., Reverter, A., Moore, S. S., Lynn, A., McBride‐Kelly, L., et al. (2020). Gene Regulation Could Be Attributed to TCF3 and Other Key Transcription Factors in the Muscle of Pubertal Heifers. Vet. Med. Sci. 6 (4), 695–710. doi:10.1002/vms3.278
Lesmeister, J. L., Burfening, P. J., and Blackwell, R. L. (1973). Date of First Calving in Beef Cows and Subsequent Calf Production. J. Animal Sci. 36 (1), 1–6. doi:10.2527/jas1973.3611
Maier, T., Güell, M., and Serrano, L. (2009). Correlation of mRNA and Protein in Complex Biological Samples. FEBS Lett. 583 (24), 3966–3973. doi:10.1016/j.febslet.2009.10.036
Meng, Q., Wang, K., Liu, X., Zhou, H., Xu, L., Wang, Z., et al. (2017). Identification of Growth Trait Related Genes in a Yorkshire Purebred Pig Population by Genome Wide Association Studies. Asian-Australas J. Anim. Sci. 30 (4), 462–469. doi:10.5713/ajas.16.0548
Michels, K. M., Lee, W. H., Seltzer, A., Saavedra, J. M., and Bondy, C. A. (1993). Up-regulation of Pituitary [125I]insulin-like Growth Factor-I (IGF-I) Binding and IGF Binding Protein-2 and IGF-I Gene Expression by Estrogen. Endocrinology 132 (1), 23–29. doi:10.1210/endo.132.1.7678216
Nelson, V. L. B., Negrón, A. L., Reid, I., Thomas, J. A., Yang, L., Lin, R. Z., et al. (2017). Loss of PI3K P110αin the Adipose Tissue Results in Infertility and Delayed Puberty Onset in Male Mice. BioMed Res. Int. 2017, 1–13. doi:10.1155/2017/3756089
Nguyen, L. T., Reverter, A., Cánovas, A., Venus, B., Anderson, S. T., Islas-Trejo, A., et al. (2018a). STAT6, PBX2, and PBRM1 Emerge as Predicted Regulators of 452 Differentially Expressed Genes Associated with Puberty in Brahman Heifers. Front. Genet. 9 (87). Original Research). doi:10.3389/fgene.2018.00087
Nguyen, L. T., Reverter, A., Cánovas, A., Venus, B., Islas-Trejo, A., Porto-Neto, L. R., et al. (2017). Global Differential Gene Expression in the Pituitary Gland and the Ovaries of Pre- and Postpubertal Brahman Heifers. J. animal Sci. 95 (2), 599–615. doi:10.2527/jas.2016.092110.2527/jas2016.0921
Nguyen, L. T., Zacchi, L. F., Schulz, B. L., Moore, S. S., and Fortes, M. R. S. (2018b). Adipose Tissue Proteomic Analyses to Study Puberty in Brahman Heifers. J. animal Sci. 96, 2392–2398. doi:10.1093/jas/sky128
Nguyen, L. T., Zacchi, L. F., Schulz, B. L., Moore, S. S., and Fortes, M. R. S. (2018c). Liver Proteomics to Study the Onset of Puberty in Brahman Heifers Proceedings of the World Congress on Genetics Applied to Livestock Production No. 11. Auckland, New Zealand, 256.
Nie, L., Wu, G., and Zhang, W. (2006). Correlation between mRNA and Protein Abundance in Desulfovibrio Vulgaris: A Multiple Regression to Identify Sources of Variations. Biochem. Biophysical Res. Commun. 339 (2), 603–610. doi:10.1016/j.bbrc.2005.11.055
Ojeda, S. R., Lomniczi, A., Mastronardi, C., Heger, S., Roth, C., Parent, A.-S., et al. (2006). Minireview: the Neuroendocrine Regulation of Puberty: Is the Time Ripe for a Systems Biology Approach? Endocrinology 147 (3), 1166–1174. doi:10.1210/en.2005-1136
Ong, K. K., Elks, C. E., Li, S., Zhao, J. H., Luan, J. a., Andersen, L. B., et al. (2009). Genetic Variation in LIN28B Is Associated with the Timing of Puberty. Nat. Genet. 41, 729–733. doi:10.1038/ng.382
Pal, B., Chen, Y., Vaillant, F., Jamieson, P., Gordon, L., Rios, A. C., et al. (2017). Construction of Developmental Lineage Relationships in the Mouse Mammary Gland by Single-Cell RNA Profiling. Nat. Commun. 8 (1), 1627. doi:10.1038/s41467-017-01560-x
Pancholi, V. (2001). Multifunctional α-enolase: its Role in Diseases. CMLS, Cell. Mol. Life Sci. 58 (7), 902–920. doi:10.1007/pl00000910
Payne, S. H. (2015). The Utility of Protein and mRNA Correlation. Trends Biochem. Sci. 40 (1), 1–3. doi:10.1016/j.tibs.2014.10.010
Rao, S., Lee, S.-Y., Gutierrez, A., Perrigoue, J., Thapa, R. J., Tu, Z., et al. (2012). Inactivation of Ribosomal Protein L22 Promotes Transformation by Induction of the Stemness Factor, Lin28B. Blood 120 (18), 3764–3773. doi:10.1182/blood-2012-03-415349
Rempel, L. A., and Clapper, J. A. (2002). Administration of Estradiol-17β Increases Anterior Pituitary IGF-I and Relative Amounts of Serum and Anterior Pituitary IGF-Binding Proteins in Barrows. J. animal Sci. 80 (1), 214–224. doi:10.2527/2002.801214x
Roberts, A. J., Funston, R. N., and Moss, G. E. (2001). Insulin-like Growth Factor Binding Proteins in the Bovine Anterior Pituitary. Endo 14 (3), 399–406. doi:10.1385/endo:14:3:399
Rogers, L. C., de Boer, I., Junier, M.-P., and Ojeda, S. R. (1993). Estradiol Increases Neural-specific Class II-β-Tubulin mRNA Levels in the Developing Female Hypothalamus by Regulating mRNA Stability. Mol. Cell. Neurosci. 4 (5), 424–431. doi:10.1006/mcne.1993.1053
Romaniello, R., Arrigoni, F., Bassi, M. T., and Borgatti, R. (2015). Mutations in α- and β-tubulin Encoding Genes: Implications in Brain Malformations. Brain Dev. 37 (3), 273–280. doi:10.1016/j.braindev.2014.06.002
Ryu, K.-Y., Garza, J. C., Lu, X.-Y., Barsh, G. S., and Kopito, R. R. (2008). Hypothalamic Neurodegeneration and Adult-Onset Obesity in Mice Lacking the Ubb Polyubiquitin Gene. Proc. Natl. Acad. Sci. U.S.A. 105 (10), 4016–4021. doi:10.1073/pnas.0800096105
Schillo, K. K., Hall, J. B., and Hileman, S. M. (1992). Effects of Nutrition and Season on the Onset of Puberty in the Beef Heifer. J. Anim. Sci. 70 (12), 3994–4005. doi:10.2527/1992.70123994x
Snelling, W. M., Allan, M. F., Keele, J. W., Kuehn, L. A., McDaneld, T., Smith, T. P. L., et al. (2010). Genome-wide Association Study of Growth in Crossbred Beef Cattle12. J. animal Sci. 88 (3), 837–848. doi:10.2527/jas.2009-2257
Tahir, M. S., Nguyen, L. T., Schulz, B. L., Boe-Hansen, G. A., Thomas, M. G., Moore, S. S., et al. (2019). Proteomics Recapitulates Ovarian Proteins Relevant to Puberty and Fertility in Brahman Heifers (Bos indicus L.). Genes. 10 (11), 923. doi:10.3390/genes10110923
Tan, N. Y., Bailey, U.-M., Jamaluddin, M. F., Mahmud, S. H. B., Raman, S. C., and Schulz, B. L. (2014). Sequence-based Protein Stabilization in the Absence of Glycosylation. Nat. Commun. 5, 3099. (Article). doi:10.1038/ncomms4099
Temple, J. L., and Wray, S. (2005). Bovine Serum Albumin-Estrogen Compounds Differentially Alter Gonadotropin-Releasing Hormone-1 Neuronal Activity. Endocrinology 146 (2), 558–563. doi:10.1210/en.2004-1117
Tian, Q., Stepaniants, S. B., Mao, M., Weng, L., Feetham, M. C., Doyle, M. J., et al. (2004). Integrated Genomic and Proteomic Analyses of Gene Expression in Mammalian Cells. Mol. Cell. Proteomics 3 (10), 960–969. doi:10.1074/mcp.M400055-MCP200
Tong, L., Wang, M., Zhou, Y., Li, Y., Chen, L., Xu, F., et al. (2018). MiR-505-3p, a New Repressor of Puberty Onset in Female Mice. bioRxiv. doi:10.1101/271718
Vargas, C. A., Elzo, M. A., Chase, C. C., Chenoweth, P. J., and Olson, T. A. (1998). Estimation of Genetic Parameters for Scrotal Circumference, Age at Puberty in Heifers, and Hip Height in Brahman Cattle. J. animal Sci. 76 (10), 2536–2541. doi:10.2527/1998.76102536x
Vogel, C., and Marcotte, E. M. (2012). Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 13 (4), 227–232. doi:10.1038/nrg3185
Wang, Y., Ji, T., Nelson, A. D., Glanowska, K., Murphy, G. G., Jenkins, P. M., et al. (2018). Critical Roles of αII Spectrin in Brain Development and Epileptic Encephalopathy. J. Clin. Investig. 128 (2), 760–773. doi:10.1172/JCI95743
Wang, Z., Hou, J., Lu, L., Qi, Z., Sun, J., Gao, W., et al. (2013). Small Ribosomal Protein Subunit S7 Suppresses Ovarian Tumorigenesis through Regulation of the PI3K/AKT and MAPK Pathways. PloS one 8 (11), e79117. doi:10.1371/journal.pone.0079117
Washburn, M. P., Koller, A., Oshiro, G., Ulaszek, R. R., Plouffe, D., Deciu, C., et al. (2003). Protein Pathway and Complex Clustering of Correlated mRNA and Protein Expression Analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 100 (6), 3107–3112. doi:10.1073/pnas.0634629100
Xie, W., Samoriski, G. M., McLaughlin, J. P., Romoser, V. A., Smrcka, A., Hinkle, P. M., et al. (1999). Genetic Alteration of Phospholipase C β3 Expression Modulates Behavioral and Cellular Responses to μ Opioids. Proc. Natl. Acad. Sci. U.S.A. 96 (18), 10385–10390. doi:10.1073/pnas.96.18.10385
Keywords: puberty, hypothalamus, pituitary gland, brahman heifers, proteomics
Citation: Nguyen LT, Lau LY and Fortes MRS (2022) Proteomic Analysis of Hypothalamus and Pituitary Gland in Pre and Postpubertal Brahman Heifers. Front. Genet. 13:935433. doi: 10.3389/fgene.2022.935433
Received: 03 May 2022; Accepted: 25 May 2022;
Published: 14 June 2022.
Edited by:
Ana Fabrícia Braga Magalhães, Universidade Federal dos Vales do Jequitinhonha e Mucuri (UFVJM), BrazilReviewed by:
Kate Keogh, Teagasc Grange Animal and Bioscience Research Department (ABRD), IrelandGuillermo Giovambattista, CONICET Institute of Veterinary Genetics (IGEVET), Argentina
Copyright © 2022 Nguyen, Lau and Fortes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Loan To Nguyen, dC5uZ3V5ZW4zQHVxLmVkdS5hdQ==
 Loan To Nguyen
Loan To Nguyen Li Yieng Lau
Li Yieng Lau Marina Rufino Salinas Fortes
Marina Rufino Salinas Fortes