- 1Guangdong Provincial Key Laboratory of Genome Stability and Disease Prevention, Shenzhen University International Cancer Center, Department of Cell Biology and Genetics, School of Medicine, Department of Pharmacy, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital (Shenzhen Institute of Translational Medicine), Guangdong Key Laboratory for Biomedical Measurements and Ultrasound Imaging, Shenzhen University, Shenzhen, China
- 2Central Laboratory, Southern University of Science and Technology, Yantain Hospital, Shenzhen, China
N6-methyladenosine (m6A) is the most abundant internal chemical modification of eukaryotic mRNA and plays diverse roles in gene regulation. The m6A modification plays a significant role in numerous cancer types, including kidney, stomach, lung, bladder tumors, and melanoma, through varied mechanisms. As direct m6A readers, the YT521-B homology domain family proteins (YTHDFs) play a key role in tumor transcription, translation, protein synthesis, tumor stemness, epithelial–mesenchymal transition (EMT), immune escape, and chemotherapy resistance. An in-depth understanding of the molecular mechanism of YTHDFs is expected to provide new strategies for tumor treatment. In this review, we provide a systematic description of YTHDF protein structure and its function in tumor progression.
Introduction
N6-methyladenosine (m6A) is formed by methylation of adenosine at the N6 position. This plentiful posttranscriptional RNA modification extensively regulates multiple aspects of gene expression (He et al., 2019). It exists widely in eukaryotic mRNA and lncRNA and is involved in cell development, stem cell characteristic maintenance, tumor progression, sperm receiving ability, and T cell homeostasis (Dominissini et al., 2012; Maity et al., 2016; Cui et al., 2017; Ianniello et al., 2018). The dynamic m6A RNA modification is catalyzed by the methyltransferase complex (m6A “writer”), demethylases (m6A “eraser”), and m6A-binding proteins (m6A “readers”) (Helm et al., 2017; Meyer et al., 2017). M6A readers, including YT521-B homology (YTH)–containing domain family proteins, insulin-like growth factor binding protein 2, eukaryotic initiation factor, and heterogeneous nuclear ribonucleoproteins (Gilbert et al., 2016), recognize and bind RNA methylation modifications to mediate the translation and degradation of downstream RNA (Wu et al., 2018). Accumulating evidence supports the influence of the m6A modification on lung cancer, melanoma, gastric cancer, bladder cancer, and liver cancer through various mechanisms (Dai et al., 2018; Burbage et al., 2019; Chen X. et al., 2019; Li et al., 2019). In this review, we focus on the current status of our understanding of m6A readers. We describe the functions of YT521-B homologous domain family proteins (YTHDFs) in tumorigenesis and cancer progression. At last, we depict future research directions to fully characterize YTHDFs.
Biological characteristics of YTHDFs and m6A modification
As the most common m6A readers, YTH domain–containing proteins include the YTHDF and YTHDC subtypes, which specifically recognize m6A via an evolutionarily conserved aromatic cage in the YTH domain (Xu et al., 2014; Shi et al., 2017; Liao et al., 2018). Another two proteins, FMRP translational regulator 1, associated with embryonic development, and leucine-rich pentatricopeptide repeat containing, associated with human tumors, can also recognize m6A modification sites (Arguello et al., 2017; Edupuganti et al., 2017). As the main YTHDF proteins, YTHDF1, YTHDF2, and YTHDF3 were first discovered in the cytoplasm (Wang et al., 2014). YTHDFs contain two domains: a YTH domain at the C-terminal and the unstructured N-terminal of most remaining proteins, comprising a large and low-complexity domain that contains mainly Gln, Asn, and Pro residues (Stoilov et al., 2002). YTHDFs are prone to forming liquid–liquid phase separation and protein droplets because of the presence of this type of large low-complexity domain, especially those with RNA binding domains (Aguzzi et al., 2016; Ries et al., 2019). In fact, proteomic analysis revealed that each YTHDF exists in the form of droplet structures, involving stress particles and artificially created RNA particles (Kato et al., 2012; Jain et al., 2016). The m6A binding pocket of the YTHDF1 YTH domain is made up of two α helices (α1 and α2) and four β strands (β1, β2, β4, and β5). To be exact, m6A is properly matched in a pocket composed of Trp411, Trp465, and Trp470, parallel to the loop planes of Trp411 and Trp470 and perpendicular to the loop plane of Trp465 (Xu et al., 2015) (Figure 1A). The residues Lys416 and Arg527 of the YTHDF2 YTH domain play a critical role in binding RNA backbone and hydrophobic vesicles. M6A single nucleotide is located in the aromatic cage composed of three aromatic amino acids Trp432, Trp486, and Trp491, forming π–π interaction. These types of particular binding contribute to the specific recognition of m6A (Ma et al., 2019) (Figure 1B). YTHDF3 possesses 86% sequence identity with the YTH structural domain of both YTHDF1 and YTHDF2. Furthermore, X-ray structural analyses and molecular dynamics simulations demonstrated that all YTHDF YTH domains share essentially equal interactions with the m6A-modified RNA and have an analogous affinity (Figure 1C), suggesting the redundant characters of the three proteins in physiological functions (Li et al., 2020). Different binding proteins selectively recognize m6A-modified RNA and exert a crucial role in regulating target gene expression. YTHDF1 increases the transmission of mRNA transcription complex by binding to m6A at a site around the stop codon and coordinates translation initiation and protein synthesis (Wang et al., 2015). YTHDF2 was the first binding protein recognized to regulate mRNA stability and remodeling of RNA structure through m6A modification (Ivanova et al., 2017). YTHDF3 is usually coupled with YTHDF1 and YTHDF2, which control the attenuation or translation of methylated RNA (Li et al., 2017). Although many studies have provided evidence of the importance of the m6A recognition protein in multiple physiological and pathological processes, further studies are still needed to fully clarify its role in mRNA biology.
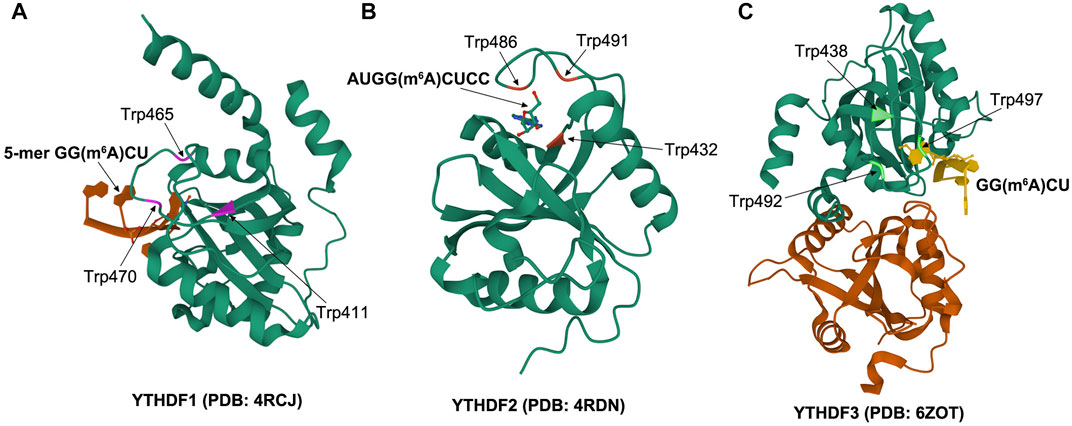
FIGURE 1. Schematic of crystal structure: (A) structure of YTHDF1 YTH domain in complex with 5-mer GG (m6A)CU (PDB id: 4RCJ), (B) structure of YTHDF2 YTH domain with AUGG (m6A)CUCC (PDB id: 4RDN), (C) structure of YTHDF3 YTH domain in complex with GG (m6A)CU (PDB id: 6ZOT).
YTHDFs and tumorigenesis
YTHDFs have been reported to elevate the growth and metastasis of various tumors, such as hepatic cell carcinoma (HCC), gastric cancer (GC), colorectal cancer (CRC), nonsmall cell lung cancer (NSCLC), and triple-negative breast cancer (TNBC) (Table 1). For example, YTHDF1 expression was dramatically upregulated in stage III and IV HCC patients compared with stage II disease, and the prognosis of patients with high YTHDF1 levels was worse (Zhao et al., 2018). Since the potential target genes regulated by YTHDF1 might be associated with amino acid degradation and lipid metabolism in the tumor cell cycle, abnormalities in these physiological functions could lead directly to the occurrence and progression of HCC. YTHDF2 also acts as an oncogene to promote the HCC lung metastasis and cancer stem cell (CSC) phenotype through increasing m6A methylation levels of OCT4 mRNA in the 5′-untranslated region (UTR) and OCT4 protein expression (Zhang et al., 2020). In addition, YTHDF2 attaches to m6A sites to boost mRNA degradation. When the m6A modification level in tumor suppressor gene SOCS2 mRNA was upregulated, the increased number of m6A binding sites in YTHDF2 ultimately promoted the degradation of SOCS2 mRNA, which was beneficial to HCC cell proliferation (Chen et al., 2018). These studies reveal that YTHDFs can accelerate HCC progression by promoting protein translation and regulating the stability of mRNA.
Huo et al. (2021) reported that YTHDF1 promoted upregulation of SPHK2 expression at the protein level via its specific structural domain, whereas SPHK2 coupled with KLF2 to induce phosphorylation modifications, thereby mediating KLF2 degradation and promoting the antiapoptotic function of GC cells. Compared with normal tissues, YTHDF2 was significantly upregulated in gastric cancer, Silencing of YTHDF2 suppressed gastric cancer cell multiplication, arresting the cell cycle in the G1/S phase and boosting cell apoptosis (Zhang et al., 2018).
In CRC, c-MYC initiated YTHDF1 expression, and high expression was associated with poor overall survival (OS) (Nishizawa et al., 2018). LncRNA-GAS5 was shown to directly bind to the WW domain of YAP to promote its phosphorylation and subsequently ubiquitin degradation, thus declining YTHDF3 transcription. YTHDF3 tended to selectively combine with GAS5 in methylated m6A site, thereby initiating its decay and the formation of a GAS5-YAP-YTHDF3 negative feedback loop to accelerate CRC development (Ni et al., 2019). This study suggests that targeting the lncRNA GAS5-YAP-YTHDF3 axis is a promising approach for CRC treatment.
In a large-scale genome and transcriptome sequencing, Shi et al. (2019) found that YTHDF1 was amplified in NSCLC from Tibetan domestic mammals. Knockdown of YTHDF1 inhibited NSCLC cell growth and the formation of xenograft tumor by adjusting the translational efficiency of CDK2, CDK4, and cyclin D1 and also restrained de novo lung adenocarcinoma progression. It is of interest that patients with high expression of YTHDF1 in high altitudes have a better clinical prognosis. This is because the knockdown of YTHDF1 activates the Keap1-Nrf2-AKR1C1 axis, which increases the sensitivity of NSCLC patients to cisplatin chemotherapy. The above results indicate that the specific recognition and regulation of YTHDF1 on m6A-modified mRNA may be associated with time and cell environment, which contributes to different functions of YTHDF1 in various tumors. Moreover, YTHDF2 was reported to be upregulated in lung cancer and shown to bind straightly to the m6A modification site in the 3′-UTR of the 6-phosphogluconate dehydrogenase (6-PGD) mRNA. This event promoted the translation of 6-PGD mRNA and enhanced the pentose phosphate pathway flux to facilitate the growth of lung cancer cells (Sheng et al., 2020).
It was found that m6A abundantly existed in ITGA6 transcripts, and methyltransferase-like 3 (METTL3) promoted YTHDF1/YTHDF3 binding to the ITGA6 mRNA 3′-UTR, thereby enhancing ITGA6 mRNA translation and changing the adhesion ability of bladder cancer cells (Laudato et al., 2017; Jin H. et al., 2019). In TNBC patients, YTHDF3 expression was correlated with poorer disease-free survival (DFS) and OS. YTHDF3 improved ZEB1 mRNA stability in an m6A-dependent manner and positively regulated cell migration, invasion, and EMT in TNBC cells (Lin et al., 2022). The above evidence demonstrates that YTHDF3 can not only function alone but also couple with METTL3 to elevate the translation of methylated RNA, playing a vital role in tumor progression.
It is worth noting that some studies have shown that YTHDF1 inhibits tumor progression. Knockdown of YTHDF1 could improve the crosspresentation ability of dendritic cells in mouse melanoma, and when combined with a PD-L1 checkpoint inhibitor, the tumor was almost completely controlled (Han et al., 2019). Another study showed that m6A modification dramatically inhibited ocular melanoma cell growth, and its low expression suggested a poor prognosis. YTHDF1 functions as a tumor suppressor of ocular melanoma by recognizing m6A-modified RNA and accelerating the translation of histidine trinucleotide binding protein 2 (HINT2) (Jia et al., 2019). Hence, as m6A readers, YTHDFs regulate tumor-specific oncogene expression and biological function via different mechanisms. Thus, targeting YTHDFs is implicated as a new strategy for cancer treatment.
YTHDFs and tumor immune responses
T cell immune responses play important roles in tumor immunotherapy. Although neoantigens in cancer patients are abundant, thorough removal of tumors is hindered by the failure to produce lasting antitumor immune responses (Yarchoan et al., 2017). Han et al. (2019) reported that YTHDF1 regulated neoantigen-specific immunity via m6A modification. YTHDF1 depletion in classical dendritic cells not only increased the mutual expression of tumor antigens but also enhanced CD8+ T cells crosspriming. The transcripts of lysosomal proteases were labeled by m6A and recognized by YTHDF1, thus promoting the translation of lysosomal cathepsin in dendritic cells and significantly inhibiting crosspresentation by wild-type dendritic cells. This study also showed that T cells in YTHDF1-deficient mice produced high levels of IFN-γ, indicating that YTHDF1 knockout in host cells promotes T cell activation at an early stage. In addition, the therapeutic effect of a PD-L1 immune checkpoint inhibitor was heightened in YTHDF1−/− mice, with almost complete inhibition of tumor growth, suggesting the potential importance of YTHDF1 as a therapeutic target for antitumor immune therapy.
Lin et al. (2020) found that YTHDF2 expression was positively associated with the expression of multiple immune checkpoint biomarkers, including CTLA-4, TIM-3, and PD-1, as well as isocitrate dehydrogenase 1, and tumor-associated macrophage factors in lower-grade glioma. Further experiments are needed to confirm this bioinformatic conclusion. In addition, high YTHDF2 transcription was associated with a higher survival rate and increased tumor-infiltrating lymphocytes in clear cell renal cell carcinoma and NSCLC patients, suggesting the antitumor function of YTHDF2 (Su et al., 2021; Tsuchiya et al., 2021). It is of interest that there are two aspects to the regulatory role of YTHDF2 in tumor immunity. Wang et al. (2020) reported that YTHDF2 stabilized STAT1 and IRF1 mRNA in tumors deficient in METTL3 or METTL4, thus promoting the signal transduction of the IFN-γ-STAT1-IRF1 axis and enhancing the response of immunotherapy-resistant colorectal cancer to PD-1 treatment. Methylated YTHDF2 decreased the stability of mRNA transcribed from the melanoma-promoting genes PD-1, CXCR4, and SOX10, to inhibit the growth of melanoma (Yang et al., 2019). A natural killer (NK) cell is a type of lymphocyte that mediates antitumor immune response. Ma et al. (2021) showed that YTHDF2 was upregulated in NK cells to maintain the steady state and terminal maturation of NK cells after activation of the immune system. In a mechanistic manner, YTHDF2 mediated the release of IFN-γ, granzyme B, and perforin in NK cells by forming a STAT5-YTHDF2 positive feedback loop, which finally promoted TARDBP degradation and enhanced NK proliferation and survival. However, YTHDF2 was found to inhibit innate immunity by binding with m6A-modified circZKSCAN1 in mammalian cells, to inhibit RIG-I recognition and k63-PolyUb and promote tumor immune escape (Chen Y. G. et al., 2019). Therefore, YTHDF2 not only enhances immunity but also promotes tumor immune escape, which are functions that may depend on differences in tumor types and the mechanisms of tumorigenesis.
Furthermore, some studies have shown that YTHDF3 contributes to the formation of the tumor microenvironment and could be used as an immune-related marker. High YTHDF3 expression indicates poor survival with a diversity of lymphocyte infiltration in breast cancer, as well as esophageal squamous cell carcinoma and head and neck squamous cell carcinoma (Zhang et al., 2021; Zhao et al., 2021; Pu et al., 2022). The above research shows that YTHDF3 may be a potential therapeutic target related to tumor-infiltrating immune cells.
YTHDFs and EMT
EMT is the key process of tumor cell metastasis, with the deficit of E-cadherin, which is considered to be the most basic event in this process. As a key transcription factor in this process, Snail reduces the expression of E-cadherin, which promotes tumor recurrence, metastasis, and drug resistance (Jang et al., 2019). YTHDF1 promotes cancer progression by regulating EMT. Lin et al. (2019) found that Snail was methylated in the CDS region and that YTHDF1 was significantly enriched in the CDS region of Snail mRNA leading to increased TGF-β-induced snail expression in HeLa cells. In addition, YTHDF1 silencing significantly increased the expression levels of Claudin-1 and zonula occludens protein 1, whereas expression of matrix metalloproteinase-9, matrix metalloproteinase-2, Vimentin, and N-cadherin was inhibited. These findings indicated that YTHDF1-mediated EMT is responsible for promoting the migration and invasion of HCC cells (Luo et al., 2021).
Recent studies have confirmed the involvement of YTHDF2 in tumor EMT and its role in regulating tumor cell migration and invasion. The METTL3/YTHDF2 m6A axis regulates miR-1915-3p expression by inhibiting the activity of the transcription factor KLF4, thereby targeting SET and inhibiting NSCLC cell migration, invasion, and EMT (Pan et al., 2021). Chen et al. (2017b) reported that YTHDF2 orchestrated two cellular processes in pancreatic cancer cells. YTHDF2 promoted pancreatic cancer cell proliferation and inhibited migration and invasion, a phenomenon termed “migration-proliferation dichotomy,” and also repressed EMT, probably by negatively regulating the YAP signaling pathway (Supplementary Figure S1). Therefore, YTHDF2 should be seriously considered as a therapeutic target for pancreatic cancer. Other studies showed that YTHDF3 promotes tumor migration and invasion by regulating EMT. Knockdown of YTHDF3 increased E-cadherin expression and reduced the expression of N-cadherin and Vimentin, which finally inhibited the migration and invasion of TNBC cells (Lin et al., 2022). Wang M. et al. (2020) reported that YTHDF3 improved ZEB1 mRNA stability in an m6A-dependent manner, maintained ZEB1 mRNA stability, and finally, promoted the translation and expression of ZEB1.
YTHDFs and drug resistance in tumors
Abnormal AKR1C1 expression induces the development of drug resistance in chemotherapy of cancers including NSCLC, breast cancer, and ovarian cancer (Chen et al., 2017a). YTHDF1 knockout was associated with decreased Keap1 expression, whereas the expression of Nrf2 and its downstream factor AKR1C1 was increased. Furthermore, YTHDF1 knockout reduced the translation efficiency of m6A-modified Keap1 transcripts, resulting in activation of the antioxidant oxygen radical scavenging system (Nrf2-AKR1C1) and resistance of NSCLC to cisplatin chemotherapy, leading to a worse clinical prognosis. This study suggested that the Keap1-Nrf2-AKR1C1 axis was the downstream regulatory target of YTHDF1, emphasizing the important character of YTHDF1 in hypoxia adjustment and NSCLC progression (Shi et al., 2019). In addition, YTHDF1 was recruited to the m6A modification site of TRIM29, promoting TRIM29 translation in cisplatin-resistant ovarian cancer cells. Furthermore, YTHDF1 knockdown inhibited cisplatin-resistant ovarian cancer cells with CSC-like characteristics (Hao et al., 2021), suggesting that YTHDF1 is a promising cancer marker to trace the recurrence of ovarian cancer.
Studies have shown that temozolomide (TMZ) resistance was acquired in recurrent glioblastoma multiforme (GBM) tumors after radiotherapy and chemotherapy. After 60 Gy radiation, YTHDF2 synthesis in GBM cells was significantly upregulated (Pak et al., 2021), suggesting that YTHDF2 plays an important role in TMZ resistance to recurrent glioblastoma. Huang et al. (2019) showed that TUSC7, a strong inhibitory lnc-RNA, was expressed at low levels in NSCLC tissues and inhibited the nondivision stem cells by obstructing Notch signaling. YTHDF2 promoted the renewal of the stem cell–like population in erlotinib-resistant cells by inhibiting TUSC7 and promoted the characteristics of tumor resistance (Li et al., 2021). Furthermore, YTHDF1/3 increased the drug resistance of tumors by promoting the translation of drug resistance–related proteins. Jin et al. (2019) found that METTL3 improved the m6A modification levels of YAP and further accelerated YAP translation by engaging YTHDF1/3 and the eEIF3a translation initiation complex, thereby inducing the NSCLC drug resistance and metastasis. Therefore, there is strong evidence that YTHDFs play an important regulatory role in the responses of tumors to chemotherapy and represent a promising therapeutic target for drug-resistant cancer.
Conclusion and perspective
Based on the current research, YTHDFs mainly recognize m6A-modified target genes, increase translation through triggering translational initiation and elongation, and then control mRNA stability and target gene expression. YTHDFs affect tumorigenesis, metastasis, tumor immunity, EMT, and chemoresistance by regulating target gene expression. Many studies have shown the various functions of YTHDFs and further revealed the crosstalk and competition between different YTHDFs. Here, we summarized the current status of the structure and biological characteristics of m6A readers—YTHDFs—in human cancers. However, a full and comprehensive understanding of YTHDFs remains a distant prospect for several reasons. First, YTHDFs exert either oncogenic or tumor-suppressive effects that may depend on the type of regulated targets or interactome in specific cancer species. Second, the biological significance of liquid–liquid separation induced by the low-complexity domain of YTHDFs in tumors is still unknown. Third, we need to dissect the interactions among YTHDFs as well as those between YTHDFs and erasers and readers in tumorigenesis, metastasis, and immunity of human cancers. Fourth, we need to determine how the expression and activity of YTHDFs are regulated in cancer and whether the recognition of m6A by YTHDFs is RNA sequence specific. At last, it is unclear whether YTHDFs can regulate the occurrence and development of cancer by recognizing m6A-modified noncoding RNA, such as microRNA or lncRNA. The functions of YTHDF mutants and isoforms in tumor molecular diagnosis also represent an important topic for future research.
Several groups have reported that targeting m6A-modifying enzymes and regulatory proteins, such as FTO (Su et al., 2020), METTL3 (Yankova et al., 2021), and ALKBH5 (Fang et al., 2022), is beneficial to the treatment of malignant leukemia and solid tumors. We also found a covalent small molecule drug, which could activate the tumor immune microenvironment and play an antihepatoma role by targeting the m6A recognition protein IGF2BP1 (Liu et al., 2022). Thus, the targeted regulation of YTHDFs by small chemical molecules may be a promising therapeutic strategy for human cancer. Overall, more basic and clinical studies are required to fully elucidate the mechanism underlying the functions of YTHDFs, which will provide new hope for cancer diagnosis and targeted treatment.
Author contributions
DZ and LZ contributed equally to the design and coordination of the study. HY, CYC, and QL collected the data with help from the CLC, and HY wrote the manuscript. All authors reviewed and edited the manuscript.
Funding
This study was supported by grants from the Natural Science Foundation of Guangdong Province (2021A1515011154, 2019A1515010210, 2021A1515011046, 2021A1515010996), Regional Joint Fund of Guangdong Province (2019B1515120080), the Shenzhen Municipal Government of China (JCYJ20210324093408024, JCYJ20180507182427559), and Shenzhen Key Medical Discipline Construction Fund (No. SZXK060). The authors would like to thank Jessica Tamanini for editing the manuscript prior to submission.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.934223/full#supplementary-material
References
Aguzzi, A., and Altmeyer, M. (2016). Phase separation: Linking cellular compartmentalization to disease. Trends Cell. Biol. 26 (7), 547–558. doi:10.1016/j.tcb.2016.03.004
Arguello, A. E., DeLiberto, A. N., and Kleiner, R. E. (2017). RNA chemical proteomics reveals the N6-methyladenosine (m6A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 139 (48), 17249–17252. doi:10.1021/jacs.7b09213
Burbage, M., Gros, M., and Amigorena, S. (2019). Translate less, prime better, to improve anti-tumor responses. Nat. Immunol. 20 (5), 518–520. doi:10.1038/s41590-019-0371-8
Chen, J., Solomides, C., Simpkins, F., and Simpkins, H. (2017a). The role of Nrf2 and ATF2 in resistance to platinum-based chemotherapy. Cancer Chemother. Pharmacol. 79 (2), 369–380. doi:10.1007/s00280-016-3225-1
Chen, J., Sun, Y., Xu, X., Wang, D., He, J., Zhou, H., et al. (2017b). YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell. Cycle 16 (23), 2259–2271. doi:10.1080/15384101.2017.1380125
Chen, M., Wei, L., Law, C. T., Tsang, F. H., Shen, J., Cheng, C. L., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67 (6), 2254–2270. doi:10.1002/hep.29683
Chen, X., Zhang, J., and Zhu, J. (2019). The role of m6A RNA methylation in human cancer. Mol. Cancer 18 (1), 103. doi:10.1186/s12943-019-1033-z
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell. 76 (1), 96–109. doi:10.1016/j.molcel.2019.07.016
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). M6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell. Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Dai, D., Wang, H., Zhu, L., Jin, H., and Wang, X. (2018). N6-methyladenosine links RNA metabolism to cancer progression. Cell. Death Dis. 9 (2), 124. doi:10.1038/s41419-017-0129-x
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Edupuganti, R. R., Geiger, S., Lindeboom, R., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24 (10), 870–878. doi:10.1038/nsmb.3462
Fang, R., Chen, X., Zhang, S., Shi, H., Ye, Y., Shi, H., et al. (2021). EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 12 (1), 177. doi:10.1038/s41467-020-20379-7
Fang, Z., Mu, B., Liu, Y., Guo, N., Xiong, L., Guo, Y., et al. (2022). Discovery of a potent, selective and cell active inhibitor of m6A demethylase ALKBH5. Eur. J. Med. Chem. 238, 114446. doi:10.1016/j.ejmech.2022.114446
Gilbert, W. V., Bell, T. A., and Schaening, C. (2016). Messenger RNA modifications: Form, distribution, and function. Science 352 (6292), 1408–1412. doi:10.1126/science.aad8711
Han, D., Liu, J., Chen, C., Dong, L., Liu, Y., Chang, R., et al. (2019). Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 566 (7743), 270–274. doi:10.1038/s41586-019-0916-x
Hao, L., Wang, J., Liu, B., Yan, J., Li, C., Jiang, J., et al. (2021). M6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim. Biophys. Acta. Mol. Cell. Res. 1868 (1), 118878. doi:10.1016/j.bbamcr.2020.118878
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18 (1), 176. doi:10.1186/s12943-019-1109-9
Helm, M., and Motorin, Y. (2017). Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 18 (5), 275–291. doi:10.1038/nrg.2016.169
Huang, G., Wang, M., Li, X., Wu, J., Chen, S., Du, N., et al. (2019). TUSC7 suppression of Notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cells. Life Sci. 232, 116630. doi:10.1016/j.lfs.2019.116630
Huo, F. C., Zhu, Z. M., Zhu, W. T., Du, Q. Y., Liang, J., and Mou, J. (2021). METTL3-mediated m6A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene 40 (16), 2968–2981. doi:10.1038/s41388-021-01753-1
Ianniello, Z., and Fatica, A. (2018). N6-Methyladenosine role in acute myeloid leukaemia. Int. J. Mol. Sci. 19 (8), 2345. doi:10.3390/ijms19082345
Ivanova, I., Much, C., Di Giacomo, M., Azzi, C., Morgan, M., Moreira, P. N., et al. (2017). The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell. 67 (6), 1059–1067. doi:10.1016/j.molcel.2017.08.003
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016). ATPase-Modulated stress granules contain a diverse proteome and substructure. Cell. 164 (3), 487–498. doi:10.1016/j.cell.2015.12.038
Jang, D., Kwon, H., Choi, M., Lee, J., and Pak, Y. (2019). Sumoylation of Flotillin-1 promotes EMT in metastatic prostate cancer by suppressing Snail degradation. Oncogene 38 (17), 3248–3260. doi:10.1038/s41388-018-0641-1
Jia, R., Chai, P., Wang, S., Sun, B., Xu, Y., Yang, Y., et al. (2019). M6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol. Cancer 18 (1), 161. doi:10.1186/s12943-019-1088-x
Jin, D., Guo, J., Wu, Y., Du, J., Yang, L., Wang, X., et al. (2019). M6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 12 (1), 135. doi:10.1186/s13045-019-0830-6
Jin, H., Ying, X., Que, B., Wang, X., Chao, Y., Zhang, H., et al. (2019). N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine 47, 195–207. doi:10.1016/j.ebiom.2019.07.068
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149 (4), 753–767. doi:10.1016/j.cell.2012.04.017
Laudato, S., Patil, N., Abba, M. L., Leupold, J. H., Benner, A., Gaiser, T., et al. (2017). P53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int. J. Cancer 141 (9), 1879–1890. doi:10.1002/ijc.30854
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell. Res. 27 (3), 444–447. doi:10.1038/cr.2017.10
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019). The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 512 (3), 479–485. doi:10.1016/j.bbrc.2019.03.093
Li, K., Peng, Z., Gao, S., Wang, Q., Wang, R., Li, X., et al. (2021). M6A associated TSUC7 inhibition contributed to Erlotinib resistance in lung adenocarcinoma through a notch signaling activation dependent way. J. Exp. Clin. Cancer Res. 40 (1), 325. doi:10.1186/s13046-021-02137-9
Li, Y., Bedi, R. K., Moroz-Omori, E. V., and Caflisch, A. (2020). Structural and dynamic insights into redundant function of YTHDF proteins. J. Chem. Inf. Model. 60 (12), 5932–5935. doi:10.1021/acs.jcim.0c01029
Liao, S., Sun, H., and Xu, C. (2018). YTH domain: A family of N(6)-methyladenosine (m(6)A) readers. Genomics Proteomics Bioinforma. 16 (2), 99–107. doi:10.1016/j.gpb.2018.04.002
Lin, X., Chai, G., Wu, Y., Li, J., Chen, F., Liu, J., et al. (2019). RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 10 (1), 2065. doi:10.1038/s41467-019-09865-9
Lin, X., Wang, Z., Yang, G., Wen, G., and Zhang, H. (2020). YTHDF2 correlates with tumor immune infiltrates in lower-grade glioma. Aging 12 (18), 18476–18500. doi:10.18632/aging.103812
Lin, Y., Jin, X., Nie, Q., Chen, M., Guo, W., Chen, L., et al. (2022). YTHDF3 facilitates triple-negative breast cancer progression and metastasis by stabilizing ZEB1 mRNA in an m6A-dependent manner. Ann. Transl. Med. 10 (2), 83. doi:10.21037/atm-21-6857
Liu, Y., Guo, Q., Yang, H., Zhang, X. W., Feng, N., Wang, J. K., et al. (2022). Allosteric regulation of IGF2BP1 as a novel strategy for the activation of tumor immune microenvironment. ACS Cent. Sci. doi:10.1021/acscentsci.2c00107
Luo, X., Cao, M., Gao, F., and He, X. (2021). YTHDF1 promotes hepatocellular carcinoma progression via activating PI3K/AKT/mTOR signaling pathway and inducing epithelial-mesenchymal transition. Exp. Hematol. Oncol. 10 (1), 35. doi:10.1186/s40164-021-00227-0
Ma, C., Liao, S., and Zhu, Z. (2019). Crystal structure of human YTHDC2 YTH domain. Biochem. Biophys. Res. Commun. 518 (4), 678–684. doi:10.1016/j.bbrc.2019.08.107
Ma, S., Yan, J., Barr, T., Zhang, J., Chen, Z., Wang, L., et al. (2021). The RNA m6A reader YTHDF2 controls NK cell anti-tumor and antiviral immunity. J. Exp. Med. 218 (8), e20210279. doi:10.1084/jem.20210279
Maity, A., and Das, B. (2016). N6‐methyladenosine modification in mRNA: Machinery, function and implications for health and diseases. Febs. J. 283 (9), 1607–1630. doi:10.1111/febs.13614
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell. Dev. Biol. 33 (1), 319–342. doi:10.1146/annurev-cellbio-100616-060758
Ni, W., Yao, S., Zhou, Y., Liu, Y., Huang, P., Zhou, A., et al. (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol. Cancer 18 (1), 143. doi:10.1186/s12943-019-1079-y
Nishizawa, Y., Konno, M., Asai, A., Koseki, J., Kawamoto, K., Miyoshi, N., et al. (2018). Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget 9 (7), 7476–7486. doi:10.18632/oncotarget.23554
Pak, O., Zaitsev, S., Shevchenko, V., Sharma, A., Sharma, H. S., and Bryukhovetskiy, I. (2021). Effectiveness of bortezomib and temozolomide for eradication of recurrent human glioblastoma cells, resistant to radiation. Prog. Brain Res. 266, 195–209. doi:10.1016/bs.pbr.2021.06.010
Pan, H., Pan, Z., Guo, F., Meng, F., Zu, L., Fan, Y., et al. (2021). MicroRNA-1915-3p inhibits cell migration and invasion by targeting SET in non-small-cell lung cancer. BMC Cancer 21 (1), 1218. doi:10.1186/s12885-021-08961-8
Pi, J., Wang, W., Ji, M., Wang, X., Wei, X., Jin, J., et al. (2021). YTHDF1 promotes gastric carcinogenesis by controlling translation of FZD7. Cancer Res. 81 (10), 2651–2665. doi:10.1158/0008-5472.CAN-20-0066
Pu, Y., Lu, X., Yang, X., Yang, Y., Wang, D., Li, M., et al. (2022). Estimating the prognosis of esophageal squamous cell carcinoma based on the Cancer Genome Atlas (TCGA) of m6A methylation-associated genes. J. Gastrointest. Oncol. 13 (1), 1–12. doi:10.21037/jgo-21-686
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). M6A enhances the phase separation potential of mRNA. Nature 571 (7765), 424–428. doi:10.1038/s41586-019-1374-1
Sheng, H., Li, Z., Su, S., Sun, W., Zhang, X., Li, L., et al. (2020). YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis 41 (5), 541–550. doi:10.1093/carcin/bgz152
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell. Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Shi, Y., Fan, S., Wu, M., Zuo, Z., Li, X., Jiang, L., et al. (2019). YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 10 (1), 4892. doi:10.1038/s41467-019-12801-6
Stoilov, P., Rafalska, I., and Stamm, S. (2002). Yth: A new domain in nuclear proteins. Trends biochem. Sci. 27 (10), 495–497. doi:10.1016/s0968-0004(02)02189-8
Su, G., Liu, T., Han, X., Sun, H., Che, W., Hu, K., et al. (2021). YTHDF2 is a potential biomarker and associated with immune infiltration in kidney renal clear cell carcinoma. Front. Pharmacol. 12, 709548. doi:10.3389/fphar.2021.709548
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 38 (1), 79–96. e11. doi:10.1016/j.ccell.2020.04.017
Tsuchiya, K., Yoshimura, K., Inoue, Y., Iwashita, Y., Yamada, H., Kawase, A., et al. (2021). YTHDF1 and YTHDF2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non–small-cell lung cancer. OncoImmunology 10 (1), 1962656. doi:10.1080/2162402X.2021.1962656
Wang, L., Hui, H., Agrawal, K., Kang, Y., Li, N., Tang, R., et al. (2020). M6a RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 39 (20), e104514. doi:10.15252/embj.2020104514
Wang, M., Yang, Y., Yang, J., Yang, J., and Han, S. (2020). Circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m6A-YTHDF3-Zeb1. Life Sci. 257, 118082. doi:10.1016/j.lfs.2020.118082
Wang, S., Gao, S., Zeng, Y., Zhu, L., Mo, Y., Wong, C., et al. (2022). N6-Methyladenosine reader YTHDF1 promotes ARHGEF2 translation and RhoA signaling in colorectal cancer. Gastroenterology 162 (4), 1183–1196. doi:10.1053/j.gastro.2021.12.269
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wu, X., Sang, L., and Gong, Y. (2018). N6-methyladenine RNA modification and cancers. Am. J. Cancer Res. 8 (10), 1957–1966.
Xie, H., Li, J., Ying, Y., Yan, H., Jin, K., Ma, X., et al. (2020). METTL3/YTHDF2 m6A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J. Cell. Mol. Med. 24 (7), 4092–4104. doi:10.1111/jcmm.15063
Xu, C., Liu, K., Ahmed, H., Loppnau, P., Schapira, M., and Min, J. (2015). Structural basis for the discriminative recognition of N6-Methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290 (41), 24902–24913. doi:10.1074/jbc.M115.680389
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10 (11), 927–929. doi:10.1038/nchembio.1654
Xu, C., Yuan, B., He, T., Ding, B., and Li, S. (2020). Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J. Cell. Mol. Med. 24 (13), 7538–7549. doi:10.1111/jcmm.15382
Yang, S., Wei, J., Cui, Y., Park, G., Shah, P., Deng, Y., et al. (2019). M6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10 (1), 2782. doi:10.1038/s41467-019-10669-0
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593 (7860), 597–601. doi:10.1038/s41586-021-03536-w
Yarchoan, M., Johnson, B. A., Lutz, E. R., Laheru, D. A., and Jaffee, E. M. (2017). Targeting neoantigens to augment anti-tumour immunity. Nat. Rev. Cancer 17 (4), 209–222. doi:10.1038/nrc.2016.154
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020). YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39 (23), 4507–4518. doi:10.1038/s41388-020-1303-7
Zhang, J., Pi, J., Liu, Y., Yu, J., and Feng, T. (2018). Knockdown of YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) inhibits proliferation and promotes apoptosis in MGC-803 gastric cancer cells. Cell. Mol. Immunol. 33 (12), 1628–1634.
Zhang, Y., Li, L., Ye, Z., Zhang, L., Yao, N., and Gai, L. (2021). Identification of m6A methyltransferase-related genes predicts prognosis and immune infiltrates in head and neck squamous cell carcinoma. Ann. Transl. Med. 9 (20), 1554. doi:10.21037/atm-21-4712
Zhao, G., An, J., Pu, Q., Geng, W., Song, H., Zhao, Q., et al. (2021). Gene signatures and Cancer-Immune phenotypes based on m6A regulators in breast cancer. Front. Oncol. 11, 756412. doi:10.3389/fonc.2021.756412
Zhao, X., Chen, Y., Mao, Q., Jiang, X., Jiang, W., Chen, J., et al. (2018). Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 21 (4), 859–868. doi:10.3233/CBM-170791
Keywords: M6A, YTHDFs, tumor, immune escape, EMT, chemotherapy resistance
Citation: Yang H, Chiang C, Luo Q, Chen C, Huang J, Zhu L and Zheng D (2022) YT521-B homology domain family proteins as N6-methyladenosine readers in tumors. Front. Genet. 13:934223. doi: 10.3389/fgene.2022.934223
Received: 02 May 2022; Accepted: 11 July 2022;
Published: 09 August 2022.
Edited by:
Yan-Ming Xu, Shantou University, ChinaReviewed by:
Xuyu Gu, Southeast University, ChinaCopyright © 2022 Yang, Chiang, Luo, Chen, Huang, Zhu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lizhi Zhu, bHp6aHU4NkBwa3UuZWR1LmNu; Duo Zheng, ZHpoZW5nQHN6dS5lZHUuY24=
†These authors have contributed equally to this work
 Heng Yang1†
Heng Yang1† Chengyao Chiang
Chengyao Chiang Chunlan Chen
Chunlan Chen Junrong Huang
Junrong Huang Lizhi Zhu
Lizhi Zhu Duo Zheng
Duo Zheng