
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 08 September 2022
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.933148
This article is part of the Research Topic Genome-Wide Association Studies of COVID-19 Among Diverse Human Populations View all 12 articles
Objectives: To explore the connection of hypertension and severe COVID-19 outcomes.
Methods: A total of 68 observational studies recording mortality and/or general severity of COVID-19 were pooled for meta-analyses of the relationship of severe COVID-19 outcomes with hypertension as well as systolic and diastolic blood pressure. Genome-wide cross-trait meta-analysis (GWCTM) was performed to explore the genes linking between hypertension and COVID-19 severity.
Results: The results of meta-analysis with the random effect model indicated that pooled risk ratios of hypertension on mortality and severity of COVID-19 were 1.80 [95% confidence interval (CI) 1.54–2.1] and 1.78 (95% confidence interval 1.56–2.04), respectively, although the apparent heterogeneity of the included studies was detected. In subgroup analysis, cohorts of severe and mild patients of COVID-19 assessed in Europe had a significant pooled weighted mean difference of 6.61 mmHg (95% CI 3.66–9.55) with no heterogeneity found (p = 0.26). The genes in the shared signature of hypertension and the COVID-19 severity were mostly expressed in lungs. Analysis of molecular networks commonly affected both by hypertension and by severe COVID-19 highlighted CCR1/CCR5 and IL10RB signaling, as well as Th1 and Th2 activation pathways, and also a potential for a shared regulation with multiple sclerosis.
Conclusion: Hypertension is significantly associated with the severe course of COVID-19. Genetic variants within inflammation- and immunity-related genes may affect their expression in lungs and confer liability to both elevated blood pressure and to severe COVID-19.
The coronavirus disease 2019 (COVID-19) pandemic, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has created a public health crisis worldwide. This disease is associated with a wide spectrum of clinical manifestations ranging from mild to life-threatening, with a possibility of adverse outcomes. Across 2 years, the prevention and therapy went through a few rounds of optimization, which decreased the global fatality rate for COVID-19 to 1.22% (COVID-19. who.int/). Further improvements require comprehensive understanding of COVID-19 pathogenesis through elucidating both the viral and the host determinants of the severe course of COVID-19.
Hypertension is a common disease defined as the systolic blood pressure (BP) readings on two different days being over 140 mmHg, and/or the diastolic BP readings ≥90 mmHg (Organization, 2019). While hypertension is repeatedly reported as one of the predictors of adverse SARS-CoV-2 infection outcomes in various cohorts (Manohar, 2021; Stanetic et al., 2021; Kumaran et al., 2022), the connection of this multifactorial condition and severe COVID-19 remains not very well characterized, especially when compared to that of obesity or diabetes (Madjid et al., 2020; Schiffrin, 2020). Relative lack of systemic efforts in clinical dissection of interactions between hypertension and COVID-19 is surprising, especially in light of an active involvement of ACE2, a receptor for SARS-CoV-2, in the hypertension control (Verano-Braga et al., 2020). On the other hand, an importance of hypertension as a factor contributing to COVID-19 outcomes is underlined by notions that adequate control of BP is one of the prerequisites for alleviating COVID-19-associated organ damage (Zheng et al., 2020; Hessami et al., 2021; Wu et al., 2021).
The genome-wide association study (GWAS) is a powerful tool to identify genetic variants contributing to complex phenotypic traits, which are influenced by many factors at once. Under the assumption that many gene variants may be associated with multiple traits, cross-phenotype (CP) associations analyses were made possible recently by introducing the CPASSOC package that uses summary-level data from GWAS to analyze multiple phenotypes for each SNP by accounting for their correlations among traits and among cohorts (Park et al., 2016; Li and Zhu, 2017). We mined multiple existing GWASs that have reported various outcomes related to COVID-19, including hospitalization, severe respiratory problems, and even death in an attempt to extract a list of genes possibly contributing to hypertension and the severity of COVID-19. We analyzed the functions of these genes and extracted additional insights into common pathophysiology of these two diseases.
An extensive search was performed within the databases of PubMed, COVID-19 Portfolio, Embase, Scopus, and China National Knowledge Infrastructure (CNKI) by using the keywords of (COVID-19 OR SARS-CoV-2 OR coronavirus) AND (severity OR clinical outcome) AND (hypertension OR blood pressure). All relevant sources from 30 December 2019 to 20 June 2021 were retrieved without language restrictions. Two authors (CH and YS) independently reviewed the collected literature works. Furthermore, the reference lists of all relevant studies were also manually checked for additional entries.
Eligible studies were considered when they met the following criteria: 1. there was properly established COVID-19 diagnosis; 2. complete data for survivor/non-survivor or severe COVID-19 infection/mild; 3. complete data for hypertension prevalence or BP measurement; and 4. complete information for subject characteristics. Here, COVID-19 diagnosis was based on SAR-CoV-2 reverse transcription-polymerase chain reaction (RT-PCR) testing of nasopharyngeal (NP) swab, throat swab, or other types of respiratory sampling, and the severe COVID-19 infection was detected in patients requiring oxygen support, or admitted to intensive care, or reported as dead. In case of any degree of non-clarity in the data, the corresponding authors were contacted for full information. The exclusion criteria were as follows: 1. duplicated studies; 2. meta-analyses, reviews, case reports, and nonhuman studies; 3. containing COVID-19 patients with pneumonia or other lung diseases; 4. containing pediatric patients; and 5. the quality assessment scores of studies were below 7. The quality score of each study was less than 7 based on the principles of AHRQ (Rostom, 2004) and QUADAS (JB, 2003), which was assessed based on five items (Supplementary Table S1). The third party (LC and YL) took part in discussion to solve the disagreement of the evaluation result.
The following information was extracted from each study: first author’s name, publication year, period of patients’ admission, study type, country, sample size, age, gender, hypertension prevalence, systolic blood pressure, diastolic blood pressure, cases of non-survivors or severe COVID-19 infection, and controls of non-survivor of mild COVID-19 infection were recorded into a standardized information sheet.
The cross-phenotype association (CPASSOC) approach (Zhu et al., 2015) was employed to identify genetic variants shared between COVID-19 and hypertension. CPASSOC allows for the presence of heterogeneous effects across traits and provides SHet statistics and p-values weighted by a sample size. In two-step CPASSOC analysis, the correlation matrix was built with SNPs whose summary statistics Z-scores were greater than 1.96 or less than −1.96 and which had a linkage disequilibrium (LD) pattern from 1,000 Genomes Project phase 3, and then SHom and SHet tests were performed. SHom is more powerful when heterogeneity is not present, while SHet allows for trait heterogeneity. Significant levels of less than 5 × 10–8 were used as cut-off values. Here, λmeta statistics were calculated to test the possibility of sample overlap through measuring concordance of effect sizes (Chen et al., 2016). Under the null hypothesis, λmeta = 1 means that the pair of cohorts are completely independent, while λmeta <1 indicates samples overlap between cohorts.
The FUMA (v1.3.5 d) (https://fuma.ctglab.nl/), a platform for annotation, visualization, and interpretation of GWAS results, was utilized for functional mapping of the genes found by GWCTM (Watanabe et al., 2017), with the summary statistics obtained from GWCTM treated as input. In the beginning, positional gene mapping of SNP2GENE function within FUMA was carried out. Then, the tissue specificity of these genes was explored with GENE2FUNC within FUMA and GTEx v8 54 tissue-types data. Finally, the eQTL gene mapping was carried out within the aforementioned identified specific tissue of the GTEx v8 dataset.
IPA (Ingenuity Pathway Analysis) software (Ingenuity Systems; Qiagen China Co., Ltd.) was employed to perform core enrichment analysis in human lung tissue with genes identified for COVID-19 severity and its adverse outcomes, respectively. The Gene Ontology (GO) enrichment analysis was performed as previously described (Cai et al., 2018).
For studies reporting the interquartile range, the standard deviation values were obtained as described in the Cochrane Handbook for Systematic Reviews (Cumpston et al., 2019). The weighted mean differences (WMDs) were calculated with 95% confidence intervals (CIs) (Cai et al., 2015). The heterogeneity was evaluated using the Q-test and I2 statistic (Cai et al., 2013). I2 > 50% or p < 0.1 were considered significantly heterogeneous. A fixed-effects model was used when the result showed no significant heterogeneity; otherwise, a random-effects model was applied. The publication bias was evaluated by Egger’s regression test (Lei et al., 2005). All the analyses were conducted in accordance with the Cochrane Handbook for Systematic Reviews (Version 5.0) or R software, with two-sided p < 0.05 set for statistical significance.
The process of literature selection is shown in Supplementary Figure S1. Through electronic-database searching and manual examination of the reference lists, we collected a total of 4,284 records. After three rounds of filtering, a total of 68 studies were retained for further statistics analysis, among which there were 21 studies containing 32,015 COVID-19 patients with the outcome of survivor/non-survivor, and 47 studies including 230,941 COVID-19 patients with the outcome of clinical severity or mildness. The main characteristics of studies included in the current meta-analysis are summarized in Table 1.
GWAS summary datasets for hypertension and COVID-19 are listed in Table 2. Briefly, we utilized one GWAS dataset on hypertension with the largest sample size as found in the MR-base database (Elsworth, 2020) and three GWAS datasets on COVID-19 of varying severity, including recorded cases of death, confirmed very severe respiratory COVID-19, and hospitalized COVID-19.
In mortality meta-analysis of 16,548 survivors and 3,297 non-survivors, the random-effects model was optimal. Using this model, hypertension had an estimated pooled RR of 1.80 (95% CI 1.54–2.1) for COVID-19-related death, with heterogeneity being detected (I2 = 91%, p < 0.001). With univariable meta-regression model and 1,000 permutations in the permutation test, both age and gender displayed significant influence on the pooled RR, p = 0.001 and 0.015, respectively. Taking separately, age and gender regression models explained 66.76 and 26.29% of heterogeneity, respectively. Interestingly, in each sequentially increasing age bracket, the RR of hypertension was decreasing by 0.03 (95% CI −0.043 to −0.019).
In meta-analysis of 19,011 severe and 211,930 mild cases of COVID-19, the random-effects model detected hypertension-related pooled RR of 1.78 (95% CI 1.56–2.04) for severe course of COVID-19, also with evidence of heterogeneity (I2 = 97.2%, p < 0.001). With univariable meta-regression model and 1,000 permutations in the permutation test, age, but not gender had significant influence on the pooled RR, with p = 0.009 and 0.25, respectively. Taking separately, age and gender regression models explained 18.00 and 2.92% of heterogeneity, respectively. Similar to that observed in mortality analysis, in each sequentially increasing age bracket, the RR of hypertension was decreasing by 0.02 (95% CI −0.036 to −0.007).
For both types of analyses, mortality, and severity, the likelihood ratio tests pointed at better fits provided by the univariable meta-regression models than multivariable meta-regression models. For the mortality study, the sensitivity analysis confirmed that omitting any one study had no effect on the pooled RR (Supplementary Figure S2A), with no risk of publication bias (p > 0.05). For the severity study, Schȍnfeld’s study performed differently than others, and its omission affected RR (Supplementary Figure S2B), with risk of publication bias detected (p = 0.03).
For systolic blood pressure (SBP), a total of 14 studies, which profiled cohorts with either severe or mild course of COVID-19, were analyzed. The pooled weighted mean difference between these two cohorts was significant at 3.30 mmHg (95% CI 0.20–6.40), with the random-effects model pointing at evident heterogeneity (I2 = 86%, p < 0.0001, Figure 1A). In further subgroup analysis of three European studies, there was significant pooled weighted mean difference of 6.61 mmHg between severe and mild patients (95% CI 3.66–9.55), with no apparent heterogeneity (I2 = 27%, p = 0.26, Figure 1B). When a total of nine mortality studies were analyzed, the pooled weighted mean difference between survivors and non-survivors was at -1.54 mmHg (95% CI -4.33 to 1.24), with moderate heterogeneity (I2 = 58%, p = 0.01).
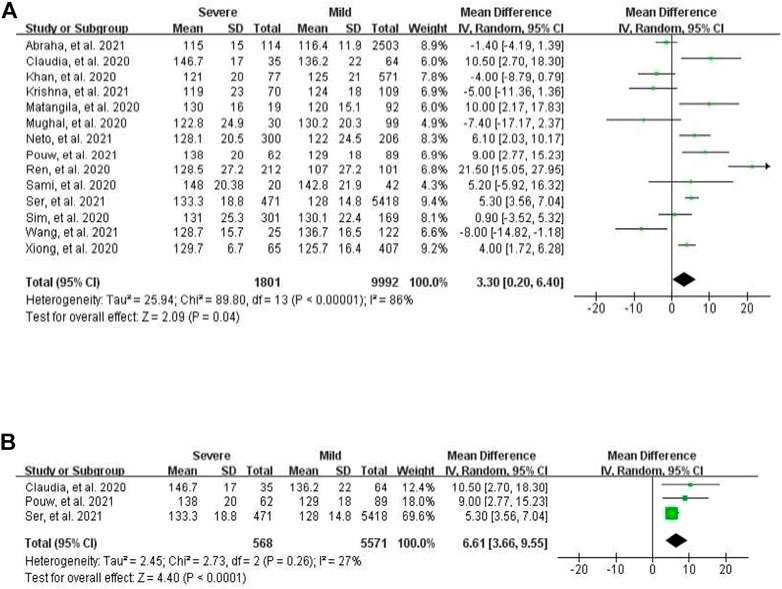
FIGURE 1. Forest plot for the meta-analysis of mean difference of SBP between severe and mild cases. (A) In all populations; (B) in the European population.
For diastolic blood pressure (DBP), a total of 13 studies, which profiled cohorts of patients with either severe or milder form of COVID-19, were analyzed. The pooled mean difference between mild and severe groups was −0.44 mmHg and non-significant (95% CI −1.52 to 0.64), with moderate heterogeneity (I2 = 45%, p = 0.04). In four studies presenting DBP values of deceased sufferers and survivors, the pooled weighted mean difference was 2.95 mmHg and non-significant (95 percent CI −3.28–9.18), with evidence of heterogeneity (I2 = 91%, p < 0.0001).
In the analyzed GWAS dataset, λmeta values were 1.06 ± 0.01, 1.04 ± 0.01, and 1.10 ± 0.01 between hypertension and three adverse COVID-19 outcomes, i.e., death, very severe respiratory problems, and hospitalization. CPASSOC-driven genome-wide cross-trait meta-analysis was performed to search for a common variant contributing both to hypertension and to adverse COVID-19 outcomes, including death, very severe respiratory problems, and hospitalization.
The summary statistics from the aforementioned GWCTM results were further explored by GENE2FUNC implemented in the platform of FUMA. A total of 149 genes were identified as shared between hypertension and COVID-19 death by positional gene mapping of SNP2GENE function (Supplementary Figure S3A); a total of 222 genes were identified as shared between hypertension and very severe, respiratory confirmed COVID-19 by positional gene mapping of SNP2GENE function (Supplementary Figure S3B); and a total of 187 genes were identified as shared between hypertension and the outcomes of the requirement of oxygen support and hospitalization (Supplementary Figure S3C). In both shared gene lists, the top positions were occupied by CLCN6, MTHFR, C10orf107, FES, and FURIN. Gene expression analysis of shared gene sets highlighted the sigmoid colon as the most relevant to the association of COVID-19 death (Figure 2A), suggesting that the disruption of gut homeostasis in the course of COVID-19 (Varshney et al., 2021) may contribute to COVID-19 mortality disproportionally.
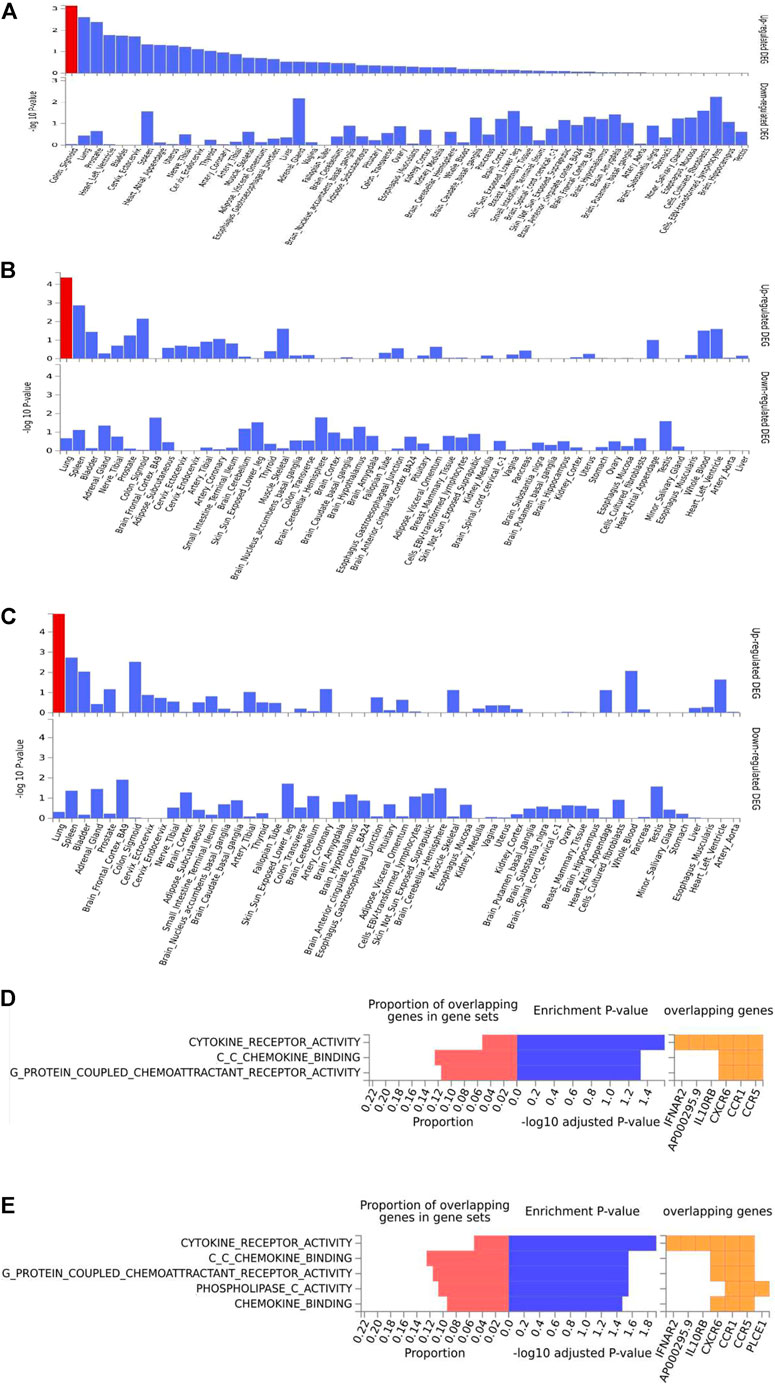
FIGURE 2. Enrichment analysis of shared genes between hypertension and adverse outcomes of COVID-19, i.e., death, very severe respiration, and hospitalization. (A–C) Tissue enrichment of shared genes between hypertension and COVID-19 death, very severe respiration, and hospitalization, respectively. (D) and (E) GO function enrichment of shared genes between hypertension and COVID-19 very severe respiration and hospitalization, respectively.
On the other hand, a similarly executed analysis pointed at the lung tissue as the most relevant association of hypertension with other severe COVID-19 outcomes (Figures 2B,C). After integrating GWAS results with lung eQTL data from GTEx (version 8), a total of 67 protein-coding genes were pinpointed as shared between hypertension and the severe respiratory involvement in COVID-19 (Supplementary Table S2) and a total of 57 genes were shared between hypertension and the hospitalization due to COVID-19 (Supplementary Table S3). A majority of these genes were involved in immune function, with cytokine receptor pathways, chemokine binding, and the signaling by G-protein-coupled chemoattractant receptors being particularly highlighted (Figures 2C,D).
The analysis of top canonical pathways identified three common sets of shared genes, namely, pathogenesis of multiple sclerosis, IL-10 signaling, and Th1 and Th2 activation (Table 3). Furthermore, these three sets of genes were enriched within the sets describing some other relevant pathophysiological conditions, including “infectious diseases,” “organismal injury and abnormalities,” “cellular function of altering cell morphology,” and “connective tissue development.”
Blood pressure is commonly recorded as two numbers: one for systolic BP representing the pressure in blood vessels when the heart contracts or beats, and the other for diastolic BP representing the pressure in the vessels when the heart rests between beats. Hypertension, i.e., elevated blood pressure, is a chronic and serious medical condition that significantly increases the risks of disease of heart, brain, kidney, and other organs. Since hypertension co-occurring with COVID-19 is reported as a risk factor for severe clinical outcomes (Hessami et al., 2021), understanding the association between these two conditions and their mechanistic underpinnings remains a priority. Due to confounding factors, observational studies with a limited sample size may not produce robust results. Thus, we explored the relationship between blood pressure and two types of COVID-19 outcomes, i.e., mortality and severe course of the disease, in a meta-analysis of the existing literature. Despite apparent heterogeneity of the studies included, the random-effects model of the meta-analysis suggested that both for mortality and for severity of COVID-19, their associations with hypertension were significant and substantial, with RR of 1.80 and 1.78, respectively. In mortality meta-analysis, age and gender explained the major share of heterogeneity, while in the meta-analysis of severe COVID-19, the heterogeneity was majorly due to other confounding factors rather than age and gender, thus suggesting that some other parameters should be recorded for studies aiming at exploring relationships of elevated BP with outcomes of COVID-19. In the subgroup meta-analysis of the European populations using the random-effects model, relationships between the severe course of COVID-19 and SBP were significant, with a pooled weighted mean difference of 6.61 mmHg between COVID-19 cohorts with the severe and mild course of illness and no heterogeneity. These findings were in contrast to the analysis of the entire dataset, which detected a significant pooled weighted mean difference of 3.30 mmHg between COVID-19 cohorts with the severe and mild course of illness, with apparent heterogeneity. These observations suggest that elevated BP is significantly associated with the COVID-19 severity, in presence of complex, yet-to-be identified confounding factors.
In further analysis of the gene sets shared between hypertension and severe outcomes of COVID-19, the mainly affected tissue compartment was in the lungs. Thus, the relevant genes were identified in the lungs by integrating GWCTM results with lung eQTL. In the pathway enrichment analysis, the only pathway directly related to the regulation of BP was one involved in cardiac hypertrophy, with its fifth place among top shared pathways connecting hypertension with COVID-19 severity. Instead, predominant involvement of the signaling related to immune function was highlighted, with an emphasis on CCR1 and CCR5, the members of the beta chemokine receptor family, as well as IL10RB, an accessory chain essential for the active interleukin 10 (IL10) receptor complex.
Notably, both CCR1 and CCR5 serve as receptors for the same set of cytokine/chemokine ligands, including macrophage inflammatory protein 1 alpha (MIP-1 alpha), monocyte chemoattractant protein 3 (MCP-3), myeloid progenitor inhibitory factor-1 (MPIF-1), and regulated on activation normal T expressed and secreted protein (RANTES). The latter is well-known as a biomarker of vascular dysfunction in the pulmonary interface and a major driver of hypertension (Nosalski and Guzik, 2017; Funk-Hilsdorf et al., 2022). Its receptor CCR5 is expressed at the surface of T cells and macrophages and serves as a co-receptor for the cell entrance for macrophage-tropic viruses. One single-cell RNA sequencing study of immune-epithelial interactions within the lung tissue (Chua et al., 2020) indicated that during the infection of SARS-CoV-2, the activated resident macrophages in general and inducible ligands for CCR1 and CCR5 in particular, contribute to inflammatory tissue damage, lung injury, and respiratory failure. In addition to CCR1 and CCR5, our study highlighted involvement of IL10RB, whose co-expression with IL10RA is required for IL10-induced signal transduction. IL10 has pleiotropic effects on immunoregulation and inflammation of mucosal tissues. Notably, mucosal integrity and immunity are indispensable for the prevention of symptomatic COVID-19 (Arrieta et al., 2009; Cortes et al., 2022). In a recent genome-wide study, IL10RB was identified as the top key regulator of COVID-19 host susceptibility, with higher IL10RB expression in patient blood being associated with worse COVID-19 outcomes (Voloudakis et al., 2021). Furthermore, in multiple rodent models, the recombinant IL10 can exert direct antihypertensive action by increasing the production of nitric oxide (NO), a kind of potent vasodilator (Lima et al., 2016; Gillis et al., 2020). Incidentally, the same sets of molecular networks have been reported to be involved in the pathogenesis of multiple sclerosis (Szczucinski and Losy, 2007; Porro et al., 2020), one of the diseases highlighted by our ingenuity-driven analysis of canonical pathways.
The strengths of this study include its multi-pronged design, which included two different definitions of severe COVID-19 outcomes as well as three types of hypertension-related parameters. We should also stress that all or a majority of study participants were of European ancestry, thus reducing the potential population heterogeneity. Several limitations should be acknowledged, one is the heterogeneity of our findings. Observed levels of heterogeneity indicate inherent complexity of relationships between hypertension and severe COVID-19. In part, heterogeneity may be explained by differing age and gender structure of studied populations. In addition, severity of COVID-19 may be affected by a large number of other confounders such as BMI, smoking, history of medications, and presence of comorbidities (Cai and He, 2019; Cai and He, 2021). We have also detected a moderate publication bias, possibly explained by a lack of interest in publishing largely negative association trends. A further meta-analysis of larger sets of independent publications is warranted.
In conclusion, our results suggest that elevated blood pressure is significantly associated with the COVID-19 severity. The genetic liabilities to elevated blood pressure and to severe COVID-19 intertwine, and among them, the immune-regulating receptors CCR1/CCR5 and IL10RB signaling pathway are highlighted in the common effective tissue of the lung.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Bioethics Committee of Bio-X Institutes of Shanghai Jiaotong University. The Ethics Committee waived the requirement of written informed consent for participation.
LC, LH and AB designed the study and prepared the manuscript, CH and YS collected the primary data for the meta-analysis, LC and YL performed the primary analyses, LC, YL, LH and AB contributed to the interpretations of the findings and the critical revision of the manuscript.
This study was supported by the grants from the Natural Science Foundation of Shanghai (No. 19ZR1427700), and cooperation foundation for Sanya Maternal and Child Health Hospital.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.933148/full#supplementary-material
Arrieta, M. C., Madsen, K., Doyle, J., and Meddings, J. (2009). Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut 58 (1), 41–48. doi:10.1136/gut.2008.150888
Cai, L., Chen, T., Yang, J., Zhou, K., Yan, X., Chen, W., et al. (2015). Serum trace element differences between Schizophrenia patients and controls in the han chinese population. Sci. Rep. 5, 15013. doi:10.1038/srep15013
Cai, L., Deng, S. L., Liang, L., Pan, H., Zhou, J., Wang, M. Y., et al. (2013). Identification of genetic associations of SP110/MYBBP1A/RELA with pulmonary tuberculosis in the chinese han population. Hum. Genet. 132 (3), 265–273. doi:10.1007/s00439-012-1244-5
Cai, L., and He, L. (2019). Placebo effects and the molecular biological components involved. Gen. Psychiatr. 32 (5), e100089. doi:10.1136/gpsych-2019-100089
Cai, L., and He, L. (2021). The novel coronavirus and humans: who can dominate who? J. BioX. Res. 4 (2), 45. doi:10.1097/JBR.0000000000000098
Cai, L., Huang, T., Su, J., Zhang, X., Chen, W., Zhang, F., et al. (2018). Implications of newly identified brain eQTL genes and their interactors in schizophrenia. Mol. Ther. Nucleic Acids 12, 433–442. doi:10.1016/j.omtn.2018.05.026
Chen, G. B., Lee, S. H., Zhu, Z. X., Benyamin, B., and Robinson, M. R. (2016). EigenGWAS: finding loci under selection through genome-wide association studies of eigenvectors in structured populations. Hered. (Edinb) 117 (1), 51–61. doi:10.1038/hdy.2016.25
Chua, R. L., Lukassen, S., Trump, S., Hennig, B. P., Wendisch, D., Pott, F., et al. (2020). COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38 (8), 970–979. doi:10.1038/s41587-020-0602-4
Cortes, G. M., Marcialis, M. A., Bardanzellu, F., Corrias, A., Fanos, V., and Mussap, M. (2022). Inflammatory bowel disease and COVID-19: how microbiomics and metabolomics depict two sides of the same coin. Front. Microbiol. 13, 856165. doi:10.3389/fmicb.2022.856165
Cumpston, M., Li, T., Page, M. J., Chandler, J., Welch, V. A., Higgins, J. P., et al. (2019). Updated guidance for trusted systematic reviews: a new edition of the cochrane handbook for systematic reviews of interventions. Cochrane Database Syst. Rev. 10, ED000142. doi:10.1002/14651858.ED000142
Elsworth, B. (2020). The MRC IEU OpenGWAS data infrastructure. BioRxiv. doi:10.1002/14651858.ED000142
Funk-Hilsdorf, T. C., Behrens, F., Grune, J., and Simmons, S. (2022). Dysregulated immunity in pulmonary hypertension: From companion to composer. Front. Physiol. 13, 819145. doi:10.3389/fphys.2022.819145
Gillis, E. E., Musall, J. B., Baban, B., and Sullivan, J. C. (2020). IL-10 treatment decreases blood pressure in male, but not female, spontaneously hypertensive rats. Am. J. Physiol. Ren. Physiol. 319 (3), F359-F365. doi:10.1152/ajprenal.00206.2020
Hessami, A., Shamshirian, A., Heydari, K., Pourali, F., Alizadeh-Navaei, R., Moosazadeh, M., et al. (2021). Cardiovascular diseases burden in COVID-19: systematic review and meta-analysis. Am. J. Emerg. Med. 46, 382–391. doi:10.1016/j.ajem.2020.10.022
Jb, W. P. R. A. R. (2003). Bossuyt PM kleijnen J the development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med. Res. Methodol. 3, 25.
Kumaran, M., Pham, T. M., Wang, K., Usman, H., Norris, C. M., MacDonald, J., et al. (2022). Predicting the risk factors associated with severe outcomes among COVID-19 patients-decision tree modeling approach. Front. Public Health 10, 838514. doi:10.3389/fpubh.2022.838514
Lei, C., Dongqing, Z., Yeqing, S., Oaks, M. K., Lishan, C., Jianzhong, J., et al. (2005). Association of the CTLA-4 gene with rheumatoid arthritis in chinese han population. Eur. J. Hum. Genet. 13 (7), 823–828. doi:10.1038/sj.ejhg.5201423
Li, X., and Zhu, X. (2017). Cross-phenotype association analysis using summary statistics from GWAS. Methods Mol. Biol. 1666, 455–467. doi:10.1007/978-1-4939-7274-6_22
Lima, V. V., Zemse, S. M., Chiao, C. W., Bomfim, G. F., Tostes, R. C., Clinton Webb, R., et al. (2016). Interleukin-10 limits increased blood pressure and vascular RhoA/Rho-kinase signaling in angiotensin II-infused mice. Life Sci. 145, 137–143. doi:10.1016/j.lfs.2015.12.009
Manohar, J., Abedian, S., Martini, R., Kulm, S., Salvatore, M., Ho, K., et al. (2021). Social and clinical determinants of COVID-19 outcomes: modeling real-world data from a pandemic epicenter. medRxiv. doi:10.1101/2021.04.06.21254728
Madjid, M., Safavi-Naeini, P., Solomon, S. D., and Vardeny, O. (2020). Potential effects of coronaviruses on the cardiovascular system: a review. JAMA Cardiol. 5 (7), 831–840. doi:10.1001/jamacardio.2020.1286
Nosalski, R., and Guzik, T. J. (2017). Perivascular adipose tissue inflammation in vascular disease. Br. J. Pharmacol. 174 (20), 3496–3513. doi:10.1111/bph.13705
Organization, W. H. (2019). Hypertension. Fact sheets. WHO, 16. Dostęp. Dostępny w internecie Available at: https://www.who.int/news-room/fact--sheets/detail/hypertension.
Park, H., Li, X., Song, Y. E., He, K. Y., and Zhu, X. (2016). Multivariate analysis of anthropometric traits using summary statistics of genome-wide association studies from GIANT consortium. PLoS One 11 (10), e0163912. doi:10.1371/journal.pone.0163912
Porro, C., Cianciulli, A., and Panaro, M. A. (2020). The regulatory role of IL-10 in neurodegenerative diseases. Biomolecules 10 (7), E1017. doi:10.3390/biom10071017
Rostom, A. (2004). Celiac disease. Appendix D. quality assessment forms. Rockville (MD): Agency for Healthcare Research and Quality (US).
Stanetic, K., Stanetic, B., Petrovic, V., Markovic, B., Kevic, V., Todorovic, N., et al. (2021). The influence of different risk factors on COVID-19 outcomes in adult patients - an observational-descriptive study. Acta Med. Acad. 50 (2), 308–316. doi:10.5644/ama2006-124.348
Szczucinski, A., and Losy, J. (2007). Chemokines and chemokine receptors in multiple sclerosis. potential targets for new therapies. Acta Neurol. Scand. 115 (3), 137–146. doi:10.1111/j.1600-0404.2006.00749.x
Varshney, R., Bansal, N., Khanduri, A., Gupta, J., and Gupta, R. (2021). Colonic gangrene: a sequela of coronavirus disease 2019. Cureus 13 (4), e14687. doi:10.7759/cureus.14687
Verano-Braga, T., Martins, A. L. V., Motta-Santos, D., Campagnole-Santos, M. J., and Santos, R. A. S. (2020). ACE2 in the renin-angiotensin system. Clin. Sci. 134 (23), 3063–3078. doi:10.1042/CS20200478
Voloudakis, G., Hoffmann, G., Venkatesh, S., Lee, K. M., Dobrindt, K., Vicari, J. M., et al. (2021). IL10RB as a key regulator of COVID-19 host susceptibility and severity. medRxiv. doi:10.1101/2021.05.31.21254851
Watanabe, K., Taskesen, E., van Bochoven, A., and Posthuma, D. (2017). Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8 (1), 1826. doi:10.1038/s41467-017-01261-5
Wu, C., Qu, G., Wang, L., Cao, S., Xia, D., Wang, B., et al. (2021). Clinical characteristics and inflammatory immune responses in COVID-19 patients with hypertension: a retrospective study. Front. Pharmacol. 12, 721769. doi:10.3389/fphar.2021.721769
Zheng, Z., Peng, F., Xu, B., Zhao, J., Liu, H., Peng, J., et al. (2020). Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J. Infect. 81 (2), e16–e25. doi:10.1016/j.jinf.2020.04.021
Keywords: COVID-19, hypertension, DBP, SBP, meta-analysis, multiple sclerosis
Citation: Cai L, He C, Liu Y, Sun Y, He L and Baranova A (2022) Inflammation and immunity connect hypertension with adverse COVID-19 outcomes. Front. Genet. 13:933148. doi: 10.3389/fgene.2022.933148
Received: 30 April 2022; Accepted: 08 July 2022;
Published: 08 September 2022.
Edited by:
Zhongshan Cheng, St. Jude Children’s Research Hospital, United StatesCopyright © 2022 Cai, He, Liu, Sun, He and Baranova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ancha Baranova, YWJhcmFub3ZAZ211LmVkdQ==; Lei Cai, bGNhaUBzanR1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.