- 1University of Chinese Academy of Sciences, Beijing, China
- 2Institute of Neuroscience, CAS Key Laboratory of Primate Neurobiology, State Key Laboratory of Neuroscience, Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China
- 3Shanghai Center for Brain Science and Brain-Inspired Intelligence Technology, Shanghai, China
Cloned mammals can be achieved through somatic cell nuclear transfer (SCNT), which involves reprogramming of differentiated somatic cells into a totipotent state. However, low cloning efficiency hampers its application severely. Cloned embryos have the same DNA as donor somatic cells. Therefore, incomplete epigenetic reprogramming accounts for low development of cloned embryos. In this review, we describe recent epigenetic barriers in SCNT embryos and strategies to correct these epigenetic defects and avoid the occurrence of abnormalities in cloned animals.
Introduction
Dolly, the first cloned mammal produced by SCNT, was born in 1996 (Wilmut et al., 1997). Since then, about 25 mammalian species have been cloned by SCNT (Wakayama et al., 1998; Lanza et al., 2000; McCreath et al., 2000; Onishi et al., 2000; Loi et al., 2001; Chesné et al., 2002; Forsberg et al., 2002; Keefer et al., 2002; Loi et al., 2002; Shin et al., 2002; Galli et al., 2003; Woods et al., 2003; Zhou et al., 2003; Gómez et al., 2004; Lee et al., 2005a; Lee et al., 2005b; Lagutina et al., 2005; Lan et al., 2006; Li et al., 2006; Berg et al., 2007; Green et al., 2007; Kim et al., 2007; Shi et al., 2007; Gómez et al., 2008; Folch et al., 2009; Hoshino et al., 2009; Wani et al., 2010; Yang et al., 2010; Hwang et al., 2013; Wani et al., 2017; Lu et al., 2018; Gavin et al., 2020), including our successful cloning of non-human primates (Liu et al., 2018b; Liu et al., 2019). Indeed, SCNT has great potential applications in many fields, such as agro-biotechnology, endangered species conservation, disease model production, and regenerative medicine (Kim et al., 2007; Rogers et al., 2008; Gómez et al., 2009; Hickey et al., 2011; Chung et al., 2014; Lu et al., 2014; Yamada et al., 2014; Chung et al., 2015; Lu et al., 2015). However, low efficiency hampers the application of SCNT and its extension. The standard SCNT efficiency is only 1%–5%, which results in a general failure of the technology to be applied in mammals extensively (Yang et al., 2007b; Loi et al., 2016; Matoba and Zhang, 2018). Somatic cells maintain some of their specific epigenetic memory, including DNA methylation, histone modification, chromosome configuration, and non-coding RNA expression, even though the oocyte has a powerful ability to reprogram (Dean et al., 2001; Mann et al., 2003; Santos et al., 2003; Yamagata et al., 2007; Zhang et al., 2009; Smith et al., 2012; Gao et al., 2018b; Matoba et al., 2018; Zhang et al., 2020). The efficiency of SCNT-based cloning in mammals remains at relatively or extremely low levels, and it appears to differ in a species-specific manner. To improve the SCNT efficiency, extensive efforts are needed to precisely identify the epigenetic factors and molecular mechanisms involved in the reprogramming of the nuclear donor cell’s (NDC’s) genome in somatic cell cloned embryos (Cao et al., 2020; Liu et al., 2020; Feng et al., 2022; Li et al., 2022). Thus far, the provenance and type of NDCs have been shown to influence the capability of donor genomic DNA to be epigenetically reprogrammed in the oocyte cytoplasm during SCNT (Samiec and Skrzyszowska, 2010, 2013; Sadeesh et al., 2016; Wiater et al., 2021b; Xu et al., 2021; Zhang et al., 2022). Moreover, the ability to reprogram the transcriptional activity of the donor cell genome in cloned embryos can be increased by extrinsic epigenetic modifiers (Sangalli et al., 2014; Sharma et al., 2018; Taweechaipaisankul et al., 2019; Skrzyszowska and Samiec, 2020; Wiater et al., 2021a). The approaches used to epigenomically modulate somatic cell nuclei in SCNT embryos have been thoroughly described in the review article by Samiec and Skrzyszowska (2018). Therefore, overcoming these epigenetic barriers and changing the somatic epigenome into an embryonic epigenome can allow for increased rates of full-term cloned embryo development.
Cloned mammals often have both placental and fetal abnormalities, which are derived from trophectoderm (TE) cells and the inner cell mass (ICM), respectively. For example, enlargement of the spongiotrophoblast (ST) layer, an irregular spongiotrophoblast–labyrinthine (LB) cell boundary, and proliferation of glycogen-rich cells in the placenta, as well as overweight conditions, respiratory disorders, and neonatal deaths have been observed in previous studies (Tanaka et al., 2001; Cibelli et al., 2002; Miglino et al., 2007; Palmieri et al., 2008; Matoba and Zhang, 2018). Fortunately, these abnormalities in cloned mammals cannot be transmitted to the offspring (Shimozawa et al., 2002; van der Berg et al., 2019; Shi et al., 2020). The two main barriers during SCNT embryo development are defective pre-implantation, which is the result of the failure of zygotic genome activation (ZGA) and embryonic arrest, and defective post-implantation development, which results in arrested gestation (Dean et al., 2001; Mann et al., 2003; Kishigami et al., 2006; Rybouchkin et al., 2006; Wang et al., 2007; Matoba et al., 2014; Gao et al., 2018b; Matoba et al., 2018; Yang et al., 2021). In this review, we describe and discuss the recent epigenetic barriers and strategies to improve pre-implantation and post-implantation development in cloned embryos and explore other strategies that can be implemented to overcome these barriers in mammals with a special focus on mice, owing to the many breakthroughs of SCNT in the mouse model (Figure 1).
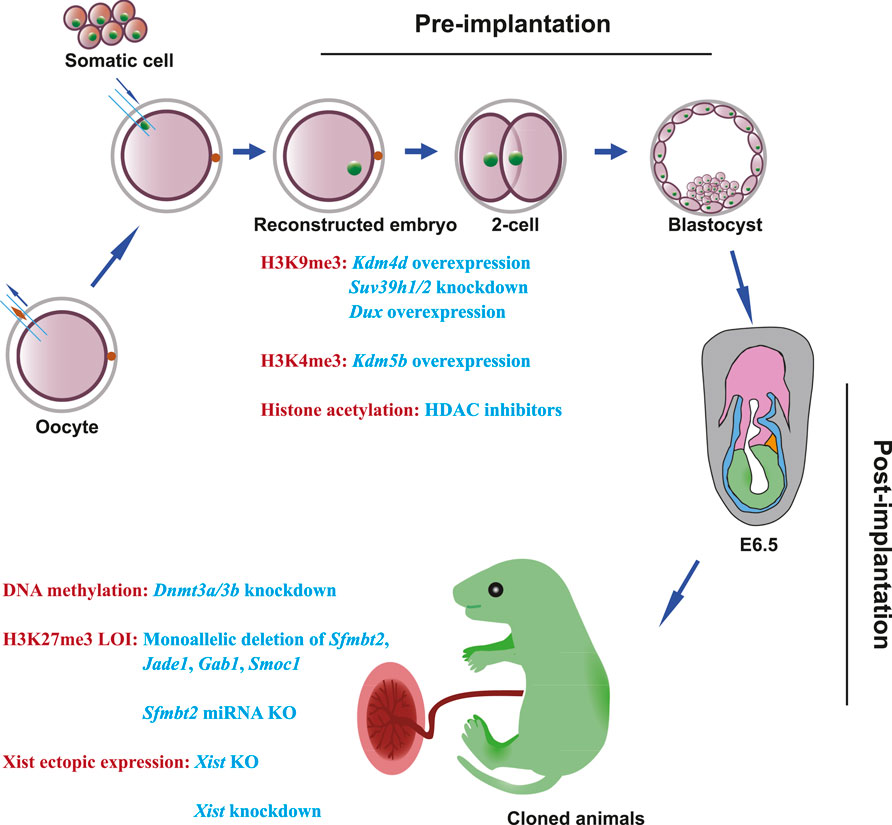
FIGURE 1. Epigenetic barriers and strategies to overcome developmental defects in SCNT embryos. During the development of SCNT embryos, epigenetic barriers can be classified into two categories: one is pre-implantation defects; the other is post-implantation defects. In pre-implantation of SCNT embryos, H3K9me3 deposition can be overcome by Kdm4 overexpression, Suv39h1/2 knockdown, and Dux transient overexpression. H3K4me3 deposition can be solved by Kdm5b injection. HDAC inhibitors can alleviate histone acetylation. In post-implantation of SCNT embryos, Dnmt3a/3b knockdown can improve post-implantation development. To overcome the loss of imprinting in H3K27me3 imprinted genes, generation of monoallelic deletions of Sfmbt2, Jade1, Gab1, and Smoc1, or deletion of Sfmbt2 miRNA can fix aberrant placentas and improve post-implantation development. Knockout or knockdown of Xist can solve the ectopic expression of Xist in cloned embryos.
Epigenetic defects during pre-implantation development in cloned embryos
Pre-implantation development in cloned embryos
Pre-implantation refers to the time during the zygote to blastocyst stages before implantation. There are two important events that occur during pre-implantation. One is the ZGA, and the other is the establishment of TE and ICM lineages (Braude et al., 1988; Mottla et al., 1995; Hamatani et al., 2004; Marikawa and Alarcón, 2009; Lee et al., 2014; Lu et al., 2016; Gao et al., 2018a; Schulz and Harrison, 2019). After fertilization, the genome is transcriptionally quiescent and is reprogrammed depending on maternal products. In mice, following fertilization, maternal RNA and protein are degraded, and the zygotic genome activates at the end of the 2-cell stage, wherein the maternal products are replaced by zygotic mRNA and protein. This process is known as ZGA (Lee et al., 2014; Jukam et al., 2017; Schulz and Harrison, 2019). Reconstructed cloned embryos also undergo ZGA since oocyte cytoplasm can initiate reprogramming when the somatic cell genome is introduced (Deng et al., 2020; Fu et al., 2021). Although this reprogramming can result in cloned animals successfully, the insufficient reprogramming by oocyte cytoplasm leads to low efficiency, and many SCNT embryos arrest at the ZGA stage (Matoba et al., 2014; Loi et al., 2016; Zhang et al., 2018c; Deng et al., 2018). The first two lineages emerge at the blastocyst stage: one is the ICM, which will give rise to the fetus, and the other is the TE, which will form the placenta. There are different epigenomes and transcriptomes between the fertilized blastocysts and SCNT blastocysts (Yang et al., 2007a; Zhao et al., 2010b; Marcho et al., 2015; Gao et al., 2018b). For example, many development-related genes are downregulated and affected by DNA methylation in SCNT blastocysts compared with the fertilized blastocysts (Gao et al., 2018b). Surprisingly, the transcriptomes are identical when comparing fertilized blastocysts to SCNT blastocysts after epigenetic modification, except that some H3K27me3-dependent imprinted genes are significantly upregulated in SCNT blastocysts (Matoba et al., 2018). These results indicate that additional epigenetic barriers remain in somatic cells, which prevent many SCNT embryos from going through ZGA and developing.
H3K9me3 histone modification hampers proper zygotic genome activation
H3K9me3, a repressive histone modification, is associated with heterochromatin, and its deposition prevents transcription factor binding. Thus, H3K9me3 can silence genes and restrict developmental potency in early embryos (Becker et al., 2016). In 2014, Matoba et al. identified 222 genomic regions in cloned mouse embryos by comparative transcriptomic and epigenetic analysis. These genomic regions were termed reprogramming-resistant regions (RRRs) because they were aberrantly expressed in the mouse SCNT embryos at the late 2-cell stage when the major ZGA occurs. Furthermore, they found that the H3K9me3 modification was enriched in donor cell RRRs and served as a barrier to the initiation of ZGA and the activation of developmentally important genes in SCNT embryos (Matoba et al., 2014). Recently, Chen et al.(2020) found that the H3K9me3 barrier led to stronger topologically associating domains (TADs) and aberrant super-enhancer and promoter interactions in mouse SCNT embryos. Significantly, the H3K9me3 barrier was conserved in other mammalian SCNT embryos, including bovine, pig, human, monkey, and sheep (Cao et al., 2015; Chung et al., 2015; Huang et al., 2016; Liu et al., 2018a; Zhang et al., 2018a; Liu et al., 2018b; Zhang et al., 2018c; An et al., 2019). In mice, overcoming the somatic cell H3K9me3 barrier enables proper ZGA, and 88.6% of reconstructed embryos developed to the blastocyst stage, and 7.6% of transferred SCNT embryos developed to term (Matoba et al., 2014).
H3K4me3 histone modification may activate inappropriate genes after somatic cell nuclear transfer
H3K4me3 is often regarded as an active histone modification, which promotes the expression of genes during embryo development. In 2016, Liu et al. reported that injection of Kdm5b, an H3K4me3 demethylase, rescued 4-cell arrest and achieved high-quality blastocysts after SCNT in mice (Liu et al., 2016). This result suggests that H3K4me3 is an epigenetic barrier for SCNT-mediated reprogramming, and activating inappropriate genes with ON-memory from donor somatic cells can lead to developmental abnormalities or arrest (Liu et al., 2016; Hörmanseder et al., 2017). Aside from mice, the H3K4me3 barrier from donor somatic cells also exists in Xenopus, goats, and bovines and impedes the development of SCNT embryos (Hörmanseder et al., 2017; Zhang et al., 2018c; Deng et al., 2020; Zhou et al., 2020). However, based on transcriptome and epigenome analysis, H3K4me3 modification is not accounted for the failure of ZGA in pigs (Liu et al., 2021). This result indicates that H3K4me3 as an epigenetic barrier may not be conserved in all mammals.
Histone acetylation affects the development of somatic cell nuclear transfer embryos
Histone acetylation, which is the acetylation of lysines in histone tails, is associated with the active expression of genes. Acetylation of lysines can loosen chromatin and promote transcription (Clayton et al., 2006). Histone acetyltransferase (HAT) and histone deacetylase (HDAC) regulate the landscape of acetylation, and the sites of acetylation include highly conserved regions in histones H4 (K5, K8, K12, and K16) and H3 (K9, K14, K18, K23, and K27) (Kurdistani et al., 2004). Previous studies of reprogramming during SCNT demonstrated that H3K9ac, H3K14ac, and H4K16ac can be quickly deacetylated in donor somatic cells and acetylated after activation. However, H4K8ac and H4K12ac still maintain the acetylation pattern that resembles donor somatic cells (Wang et al., 2007). Moreover, in 2021, Yang et al. identified aberrant regions of H3K9ac in SCNT embryos that were associated with many important genes for embryonic development. Furthermore, these aberrant H3K9ac regions led to the failure of ZGA (Yang et al., 2021). Thus, H3K9ac may be an epigenetic barrier to proper ZGA. Notably, the barrier of histone acetylation in preimplantation of SCNT is conserved in large mammals because inhibitors of histone deacetylase (HDACi) can promote blastocyst development (Akagi et al., 2011; Hou et al., 2014; Wen et al., 2014; Min et al., 2016; Jin et al., 2017; Wang et al., 2018) but lead to no significant increase in birth rates (Sangalli et al., 2012; Hosseini et al., 2016; Loi et al., 2016; Ogura et al., 2021).
DNA methylation may act as a barrier to the development of somatic cell nuclear transfer embryos
DNA methylation is an abundant and vital epigenetic modification in development and is associated with gene silencing. 5-Methylcytosine (5mC) exists in the mammalian genome in 60%–80% of CpG sites (Smith and Meissner, 2013; Wu and Zhang, 2017). The DNA methyltransferase (DNMT) family is responsible for the generation of 5mC and is classified into two categories: de novo methyltransferase and maintenance methyltransferase. De novo methyltransferases (DNMT3A and DNMT3B during the development of embryos) are responsible for initial methylation (Okano et al., 1999; Wu and Zhang, 2017). DNMT1, which methylates hemi-methylation DNA through UHRF1, can maintain 5mC through cell divisions during development (Hermann et al., 2004; Bostick et al., 2007; Sharif et al., 2007; Wu and Zhang, 2017). Both de novo and maintenance methyltransferases are crucial because deficiencies of Dnmt3b or Dnmt1 result in fetuses that look normal before E9.5 but die at the later developmental stage in the mouse, and Dnmt3a-null mice die at 4 weeks of age (Li et al., 1992; Okano et al., 1999). It is crucial to regulate DNA methylation dynamically during development. There are two ways to mediate DNA demethylation. One is passive DNA demethylation, which occurs through dilution of 5mC following replication, and the other is active DNA demethylation by the TDG–TET pathway (Wu and Zhang, 2010; Wu and Zhang, 2014; Bochtler et al., 2017).
In mammalian pre-implantation development, DNA demethylation and re-methylation both occur and play a crucial role during this process. After fertilization, the oocyte and sperm lose their DNA methylation pattern except in imprinted regions, and the paternal genome is initially demethylated by the maternally stored Tet3 proteins (Greenberg and Bourc’his, 2019; Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011). The maternal genome also undergoes demethylation, but much less than the paternal genome (Shen et al., 2014). Then, passive demethylation takes place in two parental genomes through DNA replication-dependent dilution (Howell et al., 2001). At the blastocyst stage, DNA methylation is at its lowest and is followed by DNA re-methylation by DNMT3A and DNMT3B after implantation (Zhang et al., 2018b; Zhu et al., 2018).
DNA methylation erasure and establishment also take place in SCNT embryos. Like after fertilization in mice, the pseudopronucleus (PPN) in cloned embryos undergoes active demethylation through TET3 action stored in the oocyte (Wossidlo et al., 2010; Gu et al., 2011; Wossidlo et al., 2011; Matoba and Zhang, 2018). In the first division stage, the high methylation level of the SCNT embryo is still similar to that in the donor somatic cell, and reprogramming of DNA methylation is not complete. After several rounds of replication dilutes the DNA methylation, the global methylation patterns of SCNT blastocysts after epigenetic modification are similar to those of IVF blastocysts (Ma et al., 2014; Matoba et al., 2018). This result indicates that general DNA methylation is reprogrammed in SCNT embryos successfully. However, it has been shown that Magea and Xlr clusters of the X chromosomes still failed to be activated and maintain high levels of promoter DNA methylation in most of these genes, which suggests DNA methylation may act as a barrier to reprogramming (Matoba and Zhang, 2018). In 2018, Gao et al. found there were re-methylated regions enriched in LTR elements in mouse cloned embryos during the 2-cell and 4-cell stages, and these re-methylated regions resulted in improper expression of genes and retrotransposons that are important for ZGA (Gao et al., 2018b). In addition to mice, other mammals also have abnormal DNA methylation patterns during pre-implantation as revealed by 5mC antibody-based immunostaining, bisulfite sequencing, WGBS, and MeDIP-sequencing (Dean et al., 2001; Beaujean et al., 2004; Li et al., 2014; Zhang et al., 2016; Song et al., 2017; Wang et al., 2020b; Deng et al., 2020; Malpotra et al., 2022). From the aforementioned SCNT studies, overexpression of Kdm4d and Xist KO may also help SCNT embryos through the DNA methylation barrier. However, more evidence is needed to prove the DNA methylation barrier from donor somatic cells and improve cloning efficiency after modifying DNA methylation.
Epigenetic barriers after implantation in cloned embryos
Post-implantation development in cloned embryos
After the SCNT blastocyst implants into the uterus, TE cells undergo proliferation and form extraembryonic tissues including the placenta, and ICM cells develop into the embryo proper and yolk sac tissue (Arnold and Robertson, 2009; Rossant and Tam, 2009). During this process, not only the morphology of the embryo changes dramatically, but the epigenome of the post-implantation embryo also goes through reprogramming. For example, DNA methylation is re-established around implantation in mouse embryos. The loss of non-promoter (distal) H3K27me3 is seen after implantation. Moreover, H3K27me3 maternal imprinting was replaced with DNA methylation in extraembryonic tissues, and H3K27me3 maternal imprinting was missing after implantation (Wu and Zhang, 2010; Zheng et al., 2016; Chen et al., 2019; Xia and Xie, 2020).
DNA methylation may be an epigenetic barrier for somatic cell nuclear transfer post-implantation development
In post-implantation fertilized embryos, DNA hypomethylation in extraembryonic tissues can be sustained until birth, and the epiblast genome gains DNA methylation soon after implantation (Zhang et al., 2018b). However, due to a lack of critical data about DNA methylation in SCNT embryos post-implantation, we cannot compare the DNA methylation between fertilized and SCNT post-implantation embryos. Nevertheless, Gao et al. found the combination of DNA and histone modifier treatments not only enhanced the poor cloning efficiency but also alleviated abnormal SCNT placental development. It is worth mentioning that histone H3K9me3 modification could not rescue placental abnormalities. Thus, DNA methylation may be an epigenetic barrier for SCNT post-implantation (Gao et al., 2018b), and more evidence is needed to verify this conclusion.
Loss of H3K27me3-dependent imprinting in somatic cells impedes somatic cell nuclear transfer post-implantation development
It is well known that abnormal placentas are often seen in mammalian SCNT embryos. Recent studies showed that loss of H3K27me3-dependent imprinted genes leads to abnormal placentas (Wang et al., 2020a; Inoue et al., 2020; Xie et al., 2022). H3K27me3, similar to DNA methylation, is a repressive modification that can repress gene expression. In mice, H3K27me3 modifications from sperm are erased globally upon fertilization, and only non-promoter (distal) H3K27me3 modifications from the oocyte are maintained. Moreover, the erasure of promoter H3K27me3 upon fertilization was reestablished extensively in developmental genes in post-implantation embryos (Zheng et al., 2016).
Okae et al. (2014) found the loss of imprinting of Gab1, Sfmbt2, and Slc38a4 in cloned mice, including in the brain and placenta through transcriptome-wide analyses. Furthermore, they found that Gab1, Sfmbt2, and Slc38a4 were maternally repressed independent of DNA methylation in oocytes for the establishment of imprinting (Okae et al., 2014). These results suggest there exists a DNA methylation-independent imprinting mechanism to account for the loss of imprinting of Gab1, Sfmbt2, and Slc38a4. In 2017, Inoue et al. used a low-input DNase I-sequencing (liDNase-seq) technique and found a DNA methylation-independent mechanism that prevents accessibility of certain maternal alleles in the zygotes. Moreover, they identified that oocyte-specific H3K27me3 was the DNA methylation-independent imprint. Interestingly, H3K27me3 imprinting was maintained during pre-implantation until the blastocyst stage. The ICM dilutes H3K27me3 imprinting and is completely lost in the epiblast at E6.5. However, at least Gab1, Phf17, Sfmbt2, Slc38a4, and Smoc1 maintain H3K27me3 imprinting until E9.5 in the mouse placenta (Inoue et al., 2017a). Matoba et al. (2018) found that H3K27me3 imprinted genes exhibited biallelic expression and were completely dysregulated in mouse SCNT blastocysts. Furthermore, they found the absence of H3K27me3 imprinting in donor cells resulted in the loss of imprinted gene expression in the mouse SCNT embryos. Given that H3K27me3 imprinted genes are important for the development of the placenta (Itoh et al., 2000; Miri et al., 2013; Inoue et al., 2017b), loss of H3K27me3 imprinting may act as a barrier for post-implantation development in SCNT.
Biallelic expression of Xist inhibits the development of somatic cell nuclear transfer embryos
In the female mammalian somatic cell, there are two X chromosomes (XX). One X chromosome and Y chromosome (XY) constitute sex chromosomes in males. To achieve X-linked dosage compensation between females and males, X-chromosome inactivation (XCI) has evolved (Sahakyan et al., 2018; Loda et al., 2022). In the XCI process, Xist, a non-coding RNA, is the trigger and master regulator of XCI (Penny et al., 1996; Engreitz et al., 2013; Loda and Heard, 2019). In mice, Xist first initiates at the paternally inherited X chromosome (Xp) and promotes imprinted XCI from the 4-cell to 8-cell stage in the female embryo. This type of XCI is called imprinted XCI. During pre-implantation, all cells maintain Xp imprinted XCI and a maternally inherited X chromosome (Xm) active state (Wang et al., 2016; Borensztein et al., 2017). Until the blastocyst stage, the trophectoderm still maintains imprinted XCI. However, the inner cell mass reactivates the paternal X chromosome and has two active X chromosomes. After implantation, random XCI, i.e., choosing Xp or Xm XCI at random, takes place in all cells of the embryo and is maintained in all descendent somatic cells throughout life (Lyon, 1962; Sahakyan et al., 2018).
In mouse cloned embryos, a previous study found that most X-linked genes are downregulated in SCNT pre-implantation embryos. Through immunofluorescent staining and RNA-FISH, they found ectopic expression of Xist in SCNT embryos, which caused two Xist puncta in female cells and a puncta in male cells; the result was both X chromosomes were inactivated (Inoue et al., 2010). Moreover, aberrant X chromosome inactivation patterns were reported in large animals (Xue et al., 2002; Jiang et al., 2008; Ruan et al., 2018). This result indicates that abnormal Xist expression is an epigenetic barrier for reprogramming in SCNT. It is worth mentioning that the XCI pattern is different among diverse mammals, and the abnormality of Xist expression may vary (Sahakyan et al., 2018; Patrat et al., 2020; Okamoto et al., 2021).
Strategies to overcome epigenetic barriers
Combination of various methods to achieve high cloning efficiency
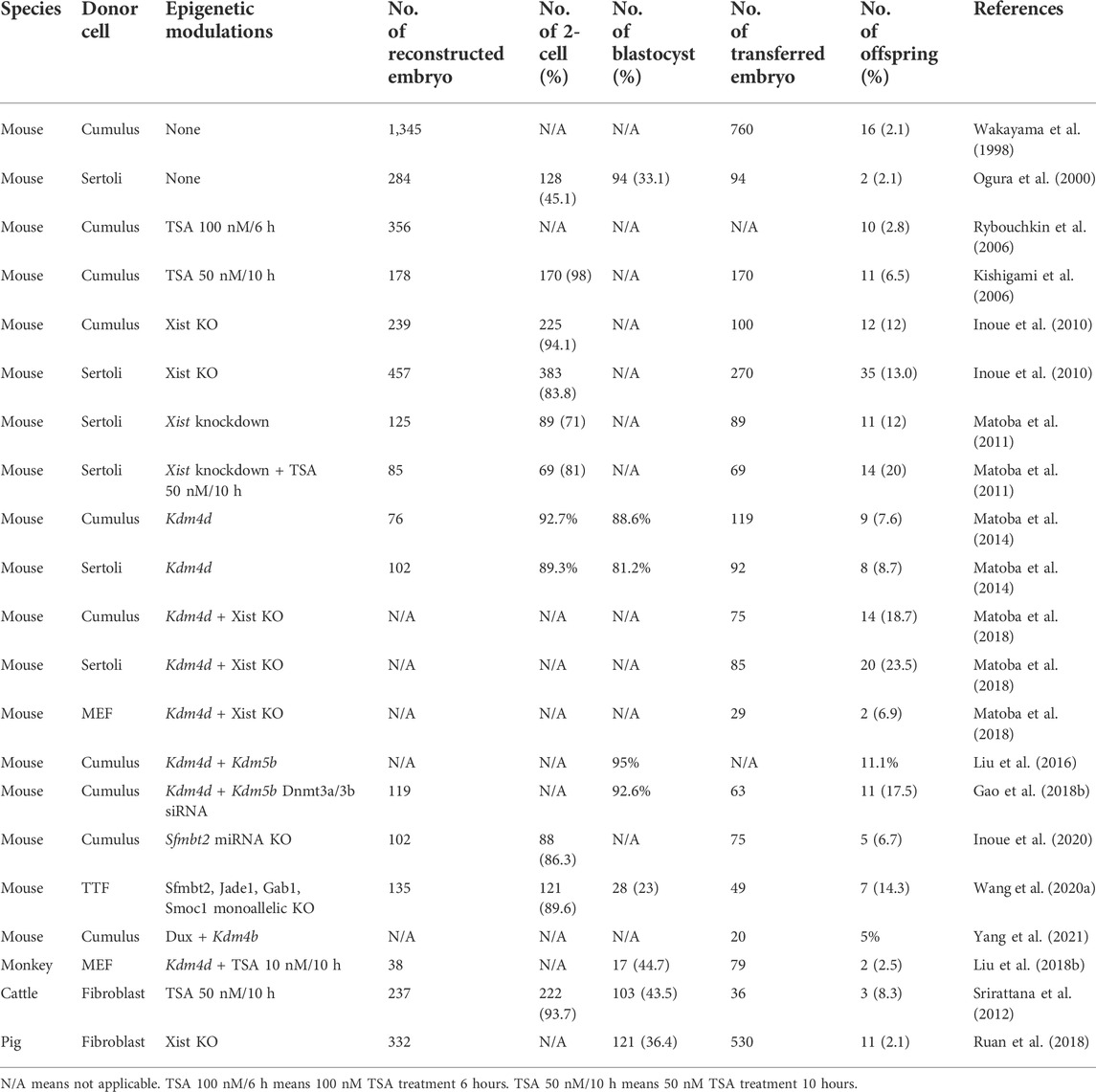
To achieve high cloning efficiency, strategies need to be applied to correct the aforementioned epigenetic defects. Owing to diverse epigenetic barriers that exist in cloned embryos, the combination of these strategies has been used in many studies, and we can identify which combination worked well or was redundant (Table 1).
Overcoming H3K9me3 barrier promotes zygotic genome activation and pre-implantation development
In mice, Matoba et al. showed that overexpression of Kdm4d, an H3K9me3 demethylase, or injection of siRNA targeting Suv39h1/2 H3K9me3 methyltransferases could repress the generation of H3K9me3 and improve the blastocyst rate to 87.5% in cumulus donors and the cloning efficiency up to 8.7% using Sertoli cells as the donor (Matoba et al., 2014). Liu et al. reported that overexpression of Kdm4b, another H3K9me3 demethylase, could also assist in overcoming the H3K9me3 barrier (Liu et al., 2016). The H3K9me3 barrier exists not only in mice but also in other large mammals. For example, KDM4D and KDM4E can rescue aberrant H3K9me3 deposition in bovine cloned embryos (Liu et al., 2018a). Injection of KDM4D or knockdown of SUV39H1/H2 can improve the development efficiency in pigs (Weng et al., 2020; Liu et al., 2021). Moreover, two cloned monkeys were achieved successfully by injection of KDM4D in reconstructed embryos (Liu et al., 2018b; Liu et al., 2019). In the derivation of human ntESCs, overexpression of KDM4A can improve the human SCNT blastocyst rate and ntESC derivation efficiency (Chung et al., 2015). Interestingly, Kdm4d only rescued pre-implantation in ZGA of embryos because enlargement of the placenta was not solved after Kdm4d injection in mice (Matoba et al., 2014). This result indicates that the H3K9me3 barrier impacts ZGA and pre-implantation development.
Removal of H3K4me3 barrier rescues 4-cell arrest in cloned embryos
In 2016, Liu et al. (2020) found that overexpression of Kdm5b, an H3K4me3 demethylase, rescued 4-cell arrest in mouse cloned embryos. Moreover, the combination of K4m4b and Kdm5b improved the SCNT blastocyst rate to over 95%, and 11.1% of reconstructed embryos developed to full term. Furthermore, in bovine cloned embryos, H3K4me3 in donor cells also was a barrier, and overexpression of Kdm5b improved the development of SCNT embryos.
Overcoming H3K27me3 imprinting barriers improves post-implantation development of cloned embryos
In 2020, Wang et al. (2020a) reported that the H3K27me3 imprinting barrier could be overcome through monoallelic imprinting gene deletions of Sfmbt2, Jade1, Gab1, and Smoc2 in fibroblasts, and these deletions increased fibroblast cloning efficiency to 14% compared with 0% in wild-type fibroblasts; these deletions also rescued the abnormal placental phenotype in SCNT. Moreover, monoallelic Sfmbt2, Jade1, and Gab1 triple deletions or monoallelic Sfmbt2 deletion also improved placental development and birth rate in SCNT. At the same time, Inoue et al. reported that correction of the expression of clustered miRNAs within the Sfmbt2 gene could rescue SCNT abnormal placentas, and the combination of Gab1 maternal deletion further alleviated abnormalities in SCNT placentas (Inoue et al., 2020). Recently, Xie et al. (2022) found that loss of Slc38a4 imprinting was a significant cause of mouse placenta hyperplasia in SCNT embryos at late gestation. Indeed, these studies demonstrated that the H3K27me3 imprinting barrier affects cloned placenta and post-implantation development of SCNT.
Histone deacetylase inhibitor improves pre-implantation development of cloned embryos
In 2006, two unrelated groups found trichostatin A (TSA), a histone deacetylase inhibitor, led to a more than 5-fold increase in birth rate and establishment of ntESCs successfully in mice (Kishigami et al., 2006; Rybouchkin et al., 2006). Since then, HDAC inhibitors have proven to increase in vitro development of SCNT embryos in other mammals, including sheep (Wen et al., 2014), cattle (Akagi et al., 2011; Min et al., 2016), pigs (Hou et al., 2014; Jin et al., 2017; Wang et al., 2018), gaurs (Srirattana et al., 2012), and monkeys (Liu et al., 2018b; Liu et al., 2019). However, the increase in birth rate has not been seen in these large animals aside from a study reported in pigs after scriptaid treatment (Zhao et al., 2010a; Ogura et al., 2021). Furthermore, the mechanism involved in the HDAC inhibitor’s improvement of cloning efficiency remains largely unknown. Recently, by using ULI-NChIP-seq (ultra-low-input native ChIP-seq), Yang et al. generated a high-resolution H3K9ac landscape of fertilized and SCNT embryos from the 1-cell to morula stage in mice and identified aberrantly acetylated regions (AARs) in SCNT embryos. Surprisingly, they found that TSA treatment was able to fix AARs without H3K9me3 occupancy and improve cloning efficiency. They also identified DUX, a crucial transcription factor that can correct aberrant H3K9ac signals in most AARs regardless of RRRs. Appropriate expression of exogenous Dux promotes Dux target genes and 2-cell expressed genes in mouse embryos. Consequently, it improves SCNT efficiency. Thus, in this study, TSA treatment worked in the aberrant H3K9ac regions of SCNT and rescued the efficiency of Kdm4b, and TSA largely relied on DUX (Yang et al., 2021). Prior to this result, Yang et al. (2020) found transient expression of Dux was able to facilitate reprogramming in cloned embryos.
Fixing aberrant DNA methylation improves post-implantation development of somatic cell nuclear transfer embryos
In 2018, Gao et al. (2018b) found DNA re-methylation in pre-implantation SCNT embryos, and the knockdown of Dnmt3a and Dnmt3b improved the development of SCNT embryos, resulting in 5.33% of transferred embryos developing to term. More surprisingly, the increased weight of the SCNT placenta was alleviated after the knockdown of Dnmt3a and Dnmt3b. Through co-injection of Kdm4b+5b mRNA and Dnmt3a+3b siRNA, an impressive 17.2% of transferred embryos developed to term.
Heterozygous knockout or knockdown of Xist improves cloning efficiency
In 2010, Inoue et al. (2010) reported that heterozygous knockout of Xist, which is overexpressed in cloned embryos, can improve the efficiency of SCNT by about 9-fold. Similarly, injection of Xist siRNA can improve female cloning efficiency by 10-fold compared to controls (Matoba et al., 2011). Moreover, the combination of Xist KO and Kdm4d overexpression can increase the development to full term of transferred SCNT embryos by 23% (Matoba et al., 2018).
Modification of culture media improves the development of somatic cell nuclear transfer embryos
SCNT embryos are more sensitive to culture media than in vivo or in vitro fertilized embryos (Heindryckx et al., 2001; Chung et al., 2002; Dai et al., 2009; Cordova et al., 2017). Thus, much effort has been dedicated to improving the culture media for the development of SCNT embryos. In the mouse culture systems, KSOM, M16, and CZB are common mediums for fertilized embryos but cannot improve the development of SCNT using the canonical SCNT technology (Dai et al., 2009). Dai et al. (2009) reported that D media, a sequential culture method that involves M16 or CZB without EDTA and glutamine, used during early SCNT embryo development followed by transfer of the SCNT embryos to KSOM at the late 2-cell stage could significantly improve SCNT embryo development. Furthermore, 62.3% of SCNT embryos in D media culture can reach the blastocyst stage. Moreover, they found that deprivation of EDTA and glutamine from the D media overcame the arrest of the 2-cell stage in the development of SCNT, but the detailed mechanism responsible remains unknown. In 2002, Chung et al. (2002) found that supplementation of glucose in the CZB medium can improve early development in SCNT embryos but not in fertilized embryos. This result indicated that the metabolism and the requirements may differ between SCNT and fertilized embryos. Moreover, supplementation of melatonin or vitamin C, which is a free radical scavenger, in SCNT media can improve the blastocyst rate (Salehi et al., 2014; Mallol et al., 2015; Cordova et al., 2017). During the development of SCNT embryos, the metabolism and needs are diverse at different stages. Hence, transferring the SCNT embryos into the oviduct at the 2-cell stage is preferable (Cordova et al., 2017). The two-step cultured medium is also suitable for other mammals in the development of SCNT, including cattle, goats, sheep, and rabbits (Wang et al., 2011; Kwong et al., 2012; Hosseini et al., 2015; Sugimoto et al., 2015).
Conclusion
Somatic cell nuclear transfer technology was developed more than 60 years ago, and the first cloned mammal, Dolly, was generated over 25 years ago. Many breakthroughs have been made, especially recently. Following the development of low-input sequencing technology, we can analyze epigenetic changes in SCNT embryos and explore the mechanisms of SCNT reprogramming.
In this review, we describe the recently discovered epigenetic barriers and discuss the strategies to overcome these barriers. From the aforementioned studies, most barriers in SCNT embryos, including pre-and post-implantation barriers, could be overcome using multiple methods. Moreover, a combination of different and non-redundant methods has the potential to achieve high cloning efficiency.
Author contributions
QS designed the manuscript. QS and YL wrote the manuscript. All authors have read and approved publication of this manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31825018, 82021001, and 2021ZD020900 to QS), the Biological Resource Program of CAS (KFJ-BRP-005), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB32060100 to QS), and Lingang Lab (LG202106-01-01 and LG202106-02-01 to QS.).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akagi, S., Matsukawa, K., Mizutani, E., Fukunari, K., Kaneda, M., Watanabe, S., et al. (2011). Treatment with a histone deacetylase inhibitor after nuclear transfer improves the preimplantation development of cloned bovine embryos. J. Reprod. Dev. 57, 120–126. doi:10.1262/jrd.10-058a
An, Q., Peng, W., Cheng, Y., Lu, Z., Zhou, C., Zhang, Y., et al. (2019). Melatonin supplementation during in vitro maturation of oocyte enhances subsequent development of bovine cloned embryos. J. Cell. Physiol. 234, 17370–17381. doi:10.1002/jcp.28357
Arnold, S. J., and Robertson, E. J. (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103. doi:10.1038/nrm2618
Beaujean, N., Taylor, J., Gardner, J., Wilmut, I., Meehan, R., and Young, L. (2004). Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol. Reprod. 71, 185–193. doi:10.1095/biolreprod.103.026559
Becker, J. S., Nicetto, D., and Zaret, K. S. (2016). H3K9me3-Dependent heterochromatin: Barrier to cell fate changes. Trends Genet. 32, 29–41. doi:10.1016/j.tig.2015.11.001
Berg, D. K., Li, C., Asher, G., Wells, D. N., and Oback, B. (2007). Red deer cloned from antler stem cells and their differentiated progeny. Biol. Reprod. 77, 384–394. doi:10.1095/biolreprod.106.058172
Bochtler, M., Kolano, A., and Xu, G. L. (2017). DNA demethylation pathways: Additional players and regulators. Bioessays 39, 1–13. doi:10.1002/bies.201600178
Borensztein, M., Syx, L., Ancelin, K., Diabangouaya, P., Picard, C., Liu, T., et al. (2017). Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat. Struct. Mol. Biol. 24, 226–233. doi:10.1038/nsmb.3365
Bostick, M., Kim, J. K., Estève, P. O., Clark, A., Pradhan, S., and Jacobsen, S. E. (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. doi:10.1126/science.1147939
Braude, P., Bolton, V., and Moore, S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459–461. doi:10.1038/332459a0
Cao, P., Li, H., Zuo, Y., and Nashun, B. (2020). Characterization of DNA methylation patterns and mining of epigenetic markers during genomic reprogramming in SCNT embryos. Front. Cell Dev. Biol. 8, 570107. doi:10.3389/fcell.2020.570107
Cao, Z., Li, Y., Chen, Z., Wang, H., Zhang, M., Zhou, N., et al. (2015). Genome-wide dynamic profiling of histone methylation during nuclear transfer-mediated porcine somatic cell reprogramming. PLoS One 10, e0144897. doi:10.1371/journal.pone.0144897
Chen, M., Zhu, Q., Li, C., Kou, X., Zhao, Y., Li, Y., et al. (2020). Chromatin architecture reorganization in murine somatic cell nuclear transfer embryos. Nat. Commun. 11, 1813. doi:10.1038/s41467-020-15607-z
Chen, Z., Yin, Q., Inoue, A., Zhang, C., and Zhang, Y. (2019). Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci. Adv. 5, eaay7246. doi:10.1126/sciadv.aay7246
Chesné, P., Adenot, P. G., Viglietta, C., Baratte, M., Boulanger, L., and Renard, J. P. (2002). Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol. 20, 366–369. doi:10.1038/nbt0402-366
Chung, Y. G., Eum, J. H., Lee, J. E., Shim, S. H., Sepilian, V., Hong, S. W., et al. (2014). Human somatic cell nuclear transfer using adult cells. Cell Stem Cell 14, 777–780. doi:10.1016/j.stem.2014.03.015
Chung, Y. G., Mann, M. R., Bartolomei, M. S., and Latham, K. E. (2002). Nuclear-cytoplasmic "tug of war" during cloning: effects of somatic cell nuclei on culture medium preferences of preimplantation cloned mouse embryos. Biol. Reprod. 66, 1178–1184. doi:10.1095/biolreprod66.4.1178
Chung, Y. G., Matoba, S., Liu, Y., Eum, J. H., Lu, F., Jiang, W., et al. (2015). Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 17, 758–766. doi:10.1016/j.stem.2015.10.001
Cibelli, J. B., Campbell, K. H., Seidel, G. E., West, M. D., and Lanza, R. P. (2002). The health profile of cloned animals. Nat. Biotechnol. 20, 13–14. doi:10.1038/nbt0102-13
Clayton, A. L., Hazzalin, C. A., and Mahadevan, L. C. (2006). Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell 23, 289–296. doi:10.1016/j.molcel.2006.06.017
Cordova, A., King, W. A., and Mastromonaco, G. F. (2017). Choosing a culture medium for SCNT and iSCNT reconstructed embryos: from domestic to wildlife species. J. Anim. Sci. Technol. 59, 24. doi:10.1186/s40781-017-0149-1
Dai, X., Hao, J., and Zhou, Q. (2009). A modified culture method significantly improves the development of mouse somatic cell nuclear transfer embryos. Reproduction 138, 301–308. doi:10.1530/REP-09-0069
Dean, W., Santos, F., Stojkovic, M., Zakhartchenko, V., Walter, J., Wolf, E., et al. (2001). Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. U. S. A. 98, 13734–13738. doi:10.1073/pnas.241522698
Deng, M., Liu, Z., Chen, B., Wan, Y., Yang, H., Zhang, Y., et al. (2020). Aberrant DNA and histone methylation during zygotic genome activation in goat cloned embryos. Theriogenology 148, 27–36. doi:10.1016/j.theriogenology.2020.02.036
Deng, M., Liu, Z., Ren, C., Zhang, G., Pang, J., Zhang, Y., et al. (2018). Long noncoding RNAs exchange during zygotic genome activation in goat. Biol. Reprod. 99, 707–717. doi:10.1093/biolre/ioy118
Engreitz, J. M., Pandya-Jones, A., McDonel, P., Shishkin, A., Sirokman, K., Surka, C., et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973. doi:10.1126/science.1237973
Feng, T., Qi, X., Zou, H., Ma, S., Yu, D., Gao, F., et al. (2022). TSA activates pluripotency factors in porcine recloned embryos. Genes (Basel) 13, 649. doi:10.3390/genes13040649
Folch, J., Cocero, M. J., Chesné, P., Alabart, J. L., Domínguez, V., Cognié, Y., et al. (2009). First birth of an animal from an extinct subspecies (Capra pyrenaica pyrenaica) by cloning. Theriogenology 71, 1026–1034. doi:10.1016/j.theriogenology.2008.11.005
Forsberg, E. J., Strelchenko, N. S., Augenstein, M. L., Betthauser, J. M., Childs, L. A., Eilertsen, K. J., et al. (2002). Production of cloned cattle from in vitro systems. Biol. Reprod. 67, 327–333. doi:10.1095/biolreprod67.1.327
Fu, B., Ma, H., and Liu, D. (2021). Functions and regulation of endogenous retrovirus elements during zygotic genome activation: Implications for improving somatic cell nuclear transfer efficiency. Biomolecules 11. doi:10.3390/biom11060829
Galli, C., Lagutina, I., Crotti, G., Colleoni, S., Turini, P., Ponderato, N., et al. (2003). Pregnancy: a cloned horse born to its dam twin. Nature 424, 635. doi:10.1038/424635a
Gao, L., Wu, K., Liu, Z., Yao, X., Yuan, S., Tao, W., et al. (2018a). Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173, 248–259. e215. doi:10.1016/j.cell.2018.02.028
Gao, R., Wang, C., Gao, Y., Xiu, W., Chen, J., Kou, X., et al. (2018b). Inhibition of aberrant DNA Re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell 23, 426–435. e425. doi:10.1016/j.stem.2018.07.017
Gavin, W., Buzzell, N., Blash, S., Chen, L., Hawkins, N., Miner, K., et al. (2020). Generation of goats by nuclear transfer: a retrospective analysis of a commercial operation (1998-2010). Transgenic Res. 29, 443–459. doi:10.1007/s11248-020-00207-w
Gómez, M. C., Pope, C. E., Giraldo, A., Lyons, L. A., Harris, R. F., King, A. L., et al. (2004). Birth of African Wildcat cloned kittens born from domestic cats. Cloning Stem Cells 6, 247–258. doi:10.1089/clo.2004.6.247
Gómez, M. C., Pope, C. E., Kutner, R. H., Ricks, D. M., Lyons, L. A., Ruhe, M., et al. (2008). Nuclear transfer of sand cat cells into enucleated domestic cat oocytes is affected by cryopreservation of donor cells. Cloning Stem Cells 10, 469–483. doi:10.1089/clo.2008.0021
Gómez, M. C., Pope, C. E., Ricks, D. M., Lyons, J., Dumas, C., and Dresser, B. L. (2009). Cloning endangered felids using heterospecific donor oocytes and interspecies embryo transfer. Reprod. Fertil. Dev. 21, 76–82. doi:10.1071/rd08222
Green, A. L., Wells, D. N., and Oback, B. (2007). Cattle cloned from increasingly differentiated muscle cells. Biol. Reprod. 77, 395–406. doi:10.1095/biolreprod.106.058164
Greenberg, M. V. C., and Bourc'his, D. (2019). The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607. doi:10.1038/s41580-019-0159-6
Gu, T. P., Guo, F., Yang, H., Wu, H. P., Xu, G. F., Liu, W., et al. (2011). The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610. doi:10.1038/nature10443
Hamatani, T., Carter, M. G., Sharov, A. A., and Ko, M. S. (2004). Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117–131. doi:10.1016/s1534-5807(03)00373-3
Heindryckx, B., Rybouchkin, A., Van Der Elst, J., and Dhont, M. (2001). Effect of culture media on in vitro development of cloned mouse embryos. Cloning 3, 41–50. doi:10.1089/15204550152475545
Hermann, A., Goyal, R., and Jeltsch, A. (2004). The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279, 48350–48359. doi:10.1074/jbc.M403427200
Hickey, R. D., Lillegard, J. B., Fisher, J. E., McKenzie, T. J., Hofherr, S. E., Finegold, M. J., et al. (2011). Efficient production of Fah-null heterozygote pigs by chimeric adeno-associated virus-mediated gene knockout and somatic cell nuclear transfer. Hepatology 54, 1351–1359. doi:10.1002/hep.24490
Hörmanseder, E., Simeone, A., Allen, G. E., Bradshaw, C. R., Figlmüller, M., Gurdon, J., et al. (2017). H3K4 methylation-dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer embryos. Cell Stem Cell 21, 135–143. e136. doi:10.1016/j.stem.2017.03.003
Hoshino, Y., Hayashi, N., Taniguchi, S., Kobayashi, N., Sakai, K., Otani, T., et al. (2009). Resurrection of a bull by cloning from organs frozen without cryoprotectant in a -80 degrees c freezer for a decade. PLoS One 4, e4142. doi:10.1371/journal.pone.0004142
Hosseini, S. M., Dufort, I., Nieminen, J., Moulavi, F., Ghanaei, H. R., Hajian, M., et al. (2016). Epigenetic modification with trichostatin A does not correct specific errors of somatic cell nuclear transfer at the transcriptomic level; highlighting the non-random nature of oocyte-mediated reprogramming errors. BMC Genomics 17, 16. doi:10.1186/s12864-015-2264-z
Hosseini, S. M., Hajian, M., Ostadhosseini, S., Forouzanfar, M., Abedi, P., Jafarpour, F., et al. (2015). Contrasting effects of G1.2/G2.2 and SOF1/SOF2 embryo culture media on pre- and post-implantation development of non-transgenic and transgenic cloned goat embryos. Reprod. Biomed. Online 31, 372–383. doi:10.1016/j.rbmo.2015.06.008
Hou, L., Ma, F., Yang, J., Riaz, H., Wang, Y., Wu, W., et al. (2014). Effects of histone deacetylase inhibitor oxamflatin on in vitro porcine somatic cell nuclear transfer embryos. Cell. Reprogr. 16, 253–265. doi:10.1089/cell.2013.0058
Howell, C. Y., Bestor, T. H., Ding, F., Latham, K. E., Mertineit, C., Trasler, J. M., et al. (2001). Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838. doi:10.1016/s0092-8674(01)00280-x
Huang, J., Zhang, H., Yao, J., Qin, G., Wang, F., Wang, X., et al. (2016). BIX-01294 increases pig cloning efficiency by improving epigenetic reprogramming of somatic cell nuclei. Reproduction 151, 39–49. doi:10.1530/REP-15-0460
Hwang, I., Jeong, Y. W., Kim, J. J., Lee, H. J., Kang, M., Park, K. B., et al. (2013). Successful cloning of coyotes through interspecies somatic cell nuclear transfer using domestic dog oocytes. Reprod. Fertil. Dev. 25, 1142–1148. doi:10.1071/RD12256
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017a). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424. doi:10.1038/nature23262
Inoue, K., Hirose, M., Inoue, H., Hatanaka, Y., Honda, A., Hasegawa, A., et al. (2017b). The rodent-specific MicroRNA cluster within the Sfmbt2 gene is imprinted and essential for placental development. Cell Rep. 19, 949–956. doi:10.1016/j.celrep.2017.04.018
Inoue, K., Kohda, T., Sugimoto, M., Sado, T., Ogonuki, N., Matoba, S., et al. (2010). Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330, 496–499. doi:10.1126/science.1194174
Inoue, K., Ogonuki, N., Kamimura, S., Inoue, H., Matoba, S., Hirose, M., et al. (2020). Loss of H3K27me3 imprinting in the Sfmbt2 miRNA cluster causes enlargement of cloned mouse placentas. Nat. Commun. 11, 2150. doi:10.1038/s41467-020-16044-8
Iqbal, K., Jin, S. G., Pfeifer, G. P., and Szabó, P. E. (2011). Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 108, 3642–3647. doi:10.1073/pnas.1014033108
Itoh, M., Yoshida, Y., Nishida, K., Narimatsu, M., Hibi, M., and Hirano, T. (2000). Role of Gab1 in heart, placenta, and skin development and growth factor- and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol. Cell. Biol. 20, 3695–3704. doi:10.1128/mcb.20.10.3695-3704.2000
Jiang, L., Lai, L., Samuel, M., Prather, R. S., Yang, X., and Tian, X. C. (2008). Expression of X-linked genes in deceased neonates and surviving cloned female piglets. Mol. Reprod. Dev. 75, 265–273. doi:10.1002/mrd.20758
Jin, J. X., Lee, S., Taweechaipaisankul, A., Kim, G. A., and Lee, B. C. (2017). The HDAC inhibitor LAQ824 enhances epigenetic reprogramming and in vitro development of porcine SCNT embryos. Cell. Physiol. biochem. 41, 1255–1266. doi:10.1159/000464389
Jukam, D., Shariati, S. A. M., and Skotheim, J. M. (2017). Zygotic genome activation in vertebrates. Dev. Cell 42, 316–332. doi:10.1016/j.devcel.2017.07.026
Keefer, C. L., Keyston, R., Lazaris, A., Bhatia, B., Begin, I., Bilodeau, A. S., et al. (2002). Production of cloned goats after nuclear transfer using adult somatic cells. Biol. Reprod. 66, 199–203. doi:10.1095/biolreprod66.1.199
Kim, M. K., Jang, G., Oh, H. J., Yuda, F., Kim, H. J., Hwang, W. S., et al. (2007). Endangered wolves cloned from adult somatic cells. Cloning Stem Cells 9, 130–137. doi:10.1089/clo.2006.0034
Kishigami, S., Mizutani, E., Ohta, H., Hikichi, T., Thuan, N. V., Wakayama, S., et al. (2006). Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340, 183–189. doi:10.1016/j.bbrc.2005.11.164
Kurdistani, S. K., Tavazoie, S., and Grunstein, M. (2004). Mapping global histone acetylation patterns to gene expression. Cell 117, 721–733. doi:10.1016/j.cell.2004.05.023
Kwong, P. J., Abdullah, R. B., and Wan Khadijah, W. E. (2012). Increasing glucose in KSOMaa basal medium on culture Day 2 improves in vitro development of cloned caprine blastocysts produced via intraspecies and interspecies somatic cell nuclear transfer. Theriogenology 78, 921–929. doi:10.1016/j.theriogenology.2012.04.009
Lagutina, I., Lazzari, G., Duchi, R., Colleoni, S., Ponderato, N., Turini, P., et al. (2005). Somatic cell nuclear transfer in horses: effect of oocyte morphology, embryo reconstruction method and donor cell type. Reproduction 130, 559–567. doi:10.1530/rep.1.00772
Lan, G. C., Chang, Z. L., Luo, M. J., Jiang, Y. L., Han, D., Wu, Y. G., et al. (2006). Production of cloned goats by nuclear transfer of cumulus cells and long-term cultured fetal fibroblast cells into abattoir-derived oocytes. Mol. Reprod. Dev. 73, 834–840. doi:10.1002/mrd.20443
Lanza, R. P., Cibelli, J. B., Diaz, F., Moraes, C. T., Farin, P. W., Farin, C. E., et al. (2000). Cloning of an endangered species (Bos gaurus) using interspecies nuclear transfer. Cloning 2, 79–90. doi:10.1089/152045500436104
Lee, B. C., Kim, M. K., Jang, G., Oh, H. J., Yuda, F., Kim, H. J., et al. (2005a). Dogs cloned from adult somatic cells. Nature 436, 641. doi:10.1038/436641a
Lee, G. S., Kim, H. S., Hyun, S. H., Lee, S. H., Jeon, H. Y., Nam, D. H., et al. (2005b). Production of transgenic cloned piglets from genetically transformed fetal fibroblasts selected by green fluorescent protein. Theriogenology 63, 973–991. doi:10.1016/j.theriogenology.2004.04.017
Lee, M. T., Bonneau, A. R., and Giraldez, A. J. (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613. doi:10.1146/annurev-cellbio-100913-013027
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi:10.1016/0092-8674(92)90611-f
Li, G., Jia, Q., Zhao, J., Li, X., Yu, M., Samuel, M. S., et al. (2014). Dysregulation of genome-wide gene expression and DNA methylation in abnormal cloned piglets. BMC Genomics 15, 811. doi:10.1186/1471-2164-15-811
Li, Z., Ruan, Z., Zhao, X., Qin, X., Zhang, J., Feng, Y., et al. (2022). RNAi-mediated knockdown of Xist improves development of the female buffalo (Bubalus bubalis) nuclear transfer embryos. Theriogenology 187, 27–33. doi:10.1016/j.theriogenology.2022.04.020
Li, Z., Sun, X., Chen, J., Liu, X., Wisely, S. M., Zhou, Q., et al. (2006). Cloned ferrets produced by somatic cell nuclear transfer. Dev. Biol. 293, 439–448. doi:10.1016/j.ydbio.2006.02.016
Liu, W., Liu, X., Wang, C., Gao, Y., Gao, R., Kou, X., et al. (2016). Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2, 16010. doi:10.1038/celldisc.2016.10
Liu, X., Chen, L., Wang, T., Zhou, J., Li, Z., Bu, G., et al. (2021). TDG is a pig-specific epigenetic regulator with insensitivity to H3K9 and H3K27 demethylation in nuclear transfer embryos. Stem Cell Rep. 16, 2674–2689. doi:10.1016/j.stemcr.2021.09.012
Liu, X., Wang, Y., Gao, Y., Su, J., Zhang, J., Xing, X., et al. (2018a). H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development 145. doi:10.1242/dev.158261
Liu, Z., Cai, Y., Liao, Z., Xu, Y., Wang, Y., Wang, Z., et al. (2019). Cloning of a gene-edited macaque monkey by somatic cell nuclear transfer. Natl. Sci. Rev. 6, 101–108. doi:10.1093/nsr/nwz003
Liu, Z., Cai, Y., Wang, Y., Nie, Y., Zhang, C., Xu, Y., et al. (2018b). Cloning of macaque monkeys by somatic cell nuclear transfer. Cell 172, 881–887. e887. doi:10.1016/j.cell.2018.01.020
Liu, Z., Xiang, G., Xu, K., Che, J., Xu, C., Li, K., et al. (2020). Transcriptome analyses reveal differential transcriptional profiles in early- and late-dividing porcine somatic cell nuclear transfer embryos. Genes 11, 1499. doi:10.3390/genes11121499
Loda, A., Collombet, S., and Heard, E. (2022). Gene regulation in time and space during X-chromosome inactivation. Nat. Rev. Mol. Cell Biol. 23, 231–249. doi:10.1038/s41580-021-00438-7
Loda, A., and Heard, E. (2019). Xist RNA in action: Past, present, and future. PLoS Genet. 15, e1008333. doi:10.1371/journal.pgen.1008333
Loi, P., Clinton, M., Barboni, B., Fulka, J., Cappai, P., Feil, R., et al. (2002). Nuclei of nonviable ovine somatic cells develop into lambs after nuclear transplantation. Biol. Reprod. 67, 126–132. doi:10.1095/biolreprod67.1.126
Loi, P., Iuso, D., Czernik, M., and Ogura, A. (2016). A new, dynamic era for somatic cell nuclear transfer? Trends Biotechnol. 34, 791–797. doi:10.1016/j.tibtech.2016.03.008
Loi, P., Ptak, G., Barboni, B., Fulka, J., Cappai, P., and Clinton, M. (2001). Genetic rescue of an endangered mammal by cross-species nuclear transfer using post-mortem somatic cells. Nat. Biotechnol. 19, 962–964. doi:10.1038/nbt1001-962
Lu, D., Li, Q., Wu, Z., Shang, S., Liu, S., Wen, X., et al. (2014). High-level recombinant human lysozyme expressed in milk of transgenic pigs can inhibit the growth of Escherichia coli in the duodenum and influence intestinal morphology of sucking pigs. PLoS One 9, e89130. doi:10.1371/journal.pone.0089130
Lu, D., Liu, S., Shang, S., Wu, F., Wen, X., Li, Z., et al. (2015). Production of transgenic-cloned pigs expressing large quantities of recombinant human lysozyme in milk. PLoS One 10, e0123551. doi:10.1371/journal.pone.0123551
Lu, F., Liu, Y., Inoue, A., Suzuki, T., Zhao, K., and Zhang, Y. (2016). Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165, 1375–1388. doi:10.1016/j.cell.2016.05.050
Lu, F., Luo, C., Li, N., Liu, Q., Wei, Y., Deng, H., et al. (2018). Efficient generation of transgenic buffalos (Bubalus bubalis) by nuclear transfer of fetal fibroblasts expressing enhanced green fluorescent protein. Sci. Rep. 8, 6967. doi:10.1038/s41598-018-25120-5
Lyon, M. F. (1962). Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 14, 135–148.
Ma, H., Morey, R., O'Neil, R. C., He, Y., Daughtry, B., Schultz, M. D., et al. (2014). Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183. doi:10.1038/nature13551
Mallol, A., Santaló, J., and Ibáñez, E. (2015). Improved development of somatic cell cloned mouse embryos by vitamin C and latrunculin A. PLoS One 10, e0120033. doi:10.1371/journal.pone.0120033
Malpotra, S., Goel, P., Shyam, S., Singh, M. K., and Palta, P. (2022). Global DNA methylation profiles of buffalo (Bubalus bubalis) preimplantation embryos produced by handmade cloning and in vitro fertilization. Sci. Rep. 12, 5161. doi:10.1038/s41598-022-09207-8
Mann, M. R., Chung, Y. G., Nolen, L. D., Verona, R. I., Latham, K. E., and Bartolomei, M. S. (2003). Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69, 902–914. doi:10.1095/biolreprod.103.017293
Marcho, C., Cui, W., and Mager, J. (2015). Epigenetic dynamics during preimplantation development. Reproduction 150, R109–R120. doi:10.1530/REP-15-0180
Marikawa, Y., and Alarcón, V. B. (2009). Establishment of trophectoderm and inner cell mass lineages in the mouse embryo. Mol. Reprod. Dev. 76, 1019–1032. doi:10.1002/mrd.21057
Matoba, S., Inoue, K., Kohda, T., Sugimoto, M., Mizutani, E., Ogonuki, N., et al. (2011). RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. U. S. A. 108, 20621–20626. doi:10.1073/pnas.1112664108
Matoba, S., Liu, Y., Lu, F., Iwabuchi, K. A., Shen, L., Inoue, A., et al. (2014). Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895. doi:10.1016/j.cell.2014.09.055
Matoba, S., Wang, H., Jiang, L., Lu, F., Iwabuchi, K. A., Wu, X., et al. (2018). Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos Disrupts post-implantation development. Cell Stem Cell 23, 343–354. e345. doi:10.1016/j.stem.2018.06.008
Matoba, S., and Zhang, Y. (2018). Somatic cell nuclear transfer reprogramming: Mechanisms and applications. Cell Stem Cell 23, 471–485. doi:10.1016/j.stem.2018.06.018
McCreath, K. J., Howcroft, J., Campbell, K. H., Colman, A., Schnieke, A. E., and Kind, A. J. (2000). Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature 405, 1066–1069. doi:10.1038/35016604
Miglino, M. A., Pereira, F. T., Visintin, J. A., Garcia, J. M., Meirelles, F. V., Rumpf, R., et al. (2007). Placentation in cloned cattle: Structure and microvascular architecture. Theriogenology 68, 604–617. doi:10.1016/j.theriogenology.2007.04.060
Min, B., Cho, S., Park, J. S., Jeon, K., and Kang, Y. K. (2016). The HIST1 locus escapes reprogramming in cloned bovine embryos. G3 (Bethesda) 6, 1365–1371. doi:10.1534/g3.115.026666
Miri, K., Latham, K., Panning, B., Zhong, Z., Andersen, A., and Varmuza, S. (2013). The imprinted polycomb group gene Sfmbt2 is required for trophoblast maintenance and placenta development. Development 140, 4480–4489. doi:10.1242/dev.096511
Mottla, G. L., Adelman, M. R., Hall, J. L., Gindoff, P. R., Stillman, R. J., and Johnson, K. E. (1995). Lineage tracing demonstrates that blastomeres of early cleavage-stage human pre-embryos contribute to both trophectoderm and inner cell mass. Hum. Reprod. 10, 384–391. doi:10.1093/oxfordjournals.humrep.a135949
Ogura, A., Inoue, K., Ogonuki, N., Noguchi, A., Takano, K., Nagano, R., et al. (2000). Production of male cloned mice from fresh, cultured, and cryopreserved immature Sertoli cells. Biol. Reprod. 62, 1579–1584. doi:10.1095/biolreprod62.6.1579
Ogura, A., Matoba, S., and Inoue, K. (2021). 25th ANNIVERSARY OF CLONING BY SOMATIC-CELL NUCLEAR TRANSFER: Epigenetic abnormalities associated with somatic cell nuclear transfer. Reproduction 162, F45–f58. doi:10.1530/REP-21-0013
Okae, H., Matoba, S., Nagashima, T., Mizutani, E., Inoue, K., Ogonuki, N., et al. (2014). RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum. Mol. Genet. 23, 992–1001. doi:10.1093/hmg/ddt495
Okamoto, I., Nakamura, T., Sasaki, K., Yabuta, Y., Iwatani, C., Tsuchiya, H., et al. (2021). The X chromosome dosage compensation program during the development of cynomolgus monkeys. Science 374, eabd8887. doi:10.1126/science.abd8887
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi:10.1016/s0092-8674(00)81656-6
Onishi, A., Iwamoto, M., Akita, T., Mikawa, S., Takeda, K., Awata, T., et al. (2000). Pig cloning by microinjection of fetal fibroblast nuclei. Science 289, 1188–1190. doi:10.1126/science.289.5482.1188
Palmieri, C., Loi, P., Ptak, G., and Della Salda, L. (2008). Review paper: a review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep, and mice. Vet. Pathol. 45, 865–880. doi:10.1354/vp.45-6-865
Patrat, C., Ouimette, J. F., and Rougeulle, C. (2020). X chromosome inactivation in human development. Development 147. doi:10.1242/dev.183095
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S., and Brockdorff, N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. doi:10.1038/379131a0
Rogers, C. S., Hao, Y., Rokhlina, T., Samuel, M., Stoltz, D. A., Li, Y., et al. (2008). Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J. Clin. Invest. 118, 1571–1577. doi:10.1172/JCI34773
Rossant, J., and Tam, P. P. (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713. doi:10.1242/dev.017178
Ruan, D., Peng, J., Wang, X., Ouyang, Z., Zou, Q., Yang, Y., et al. (2018). XIST derepression in active X chromosome Hinders pig somatic cell nuclear transfer. Stem Cell Rep. 10, 494–508. doi:10.1016/j.stemcr.2017.12.015
Rybouchkin, A., Kato, Y., and Tsunoda, Y. (2006). Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol. Reprod. 74, 1083–1089. doi:10.1095/biolreprod.105.047456
Sadeesh, E., Shah, F., and Yadav, P. S. (2016). Differential developmental competence and gene expression patterns in buffalo (Bubalus bubalis) nuclear transfer embryos reconstructed with fetal fibroblasts and amnion mesenchymal stem cells. Cytotechnology 68, 1827–1848. doi:10.1007/s10616-015-9936-z
Sahakyan, A., Yang, Y., and Plath, K. (2018). The role of xist in X-chromosome dosage compensation. Trends Cell Biol. 28, 999–1013. doi:10.1016/j.tcb.2018.05.005
Salehi, M., Kato, Y., and Tsunoda, Y. (2014). Effect of melatonin treatment on developmental potential of somatic cell nuclear-transferred mouse oocytes in vitro. Zygote 22, 213–217. doi:10.1017/S0967199413000336
Samiec, M., and Skrzyszowska, M. (2013). Assessment of in vitro Developmental Capacity of Porcine Nuclear-Transferred Embryos Reconstituted with Cumulus Oophorus Cells Undergoing Vital Diagnostics for Apoptosis Detection/Ocena zdolności rozwojowych in vitro klonalnych zarodków świni rekonstytuowanych z jąder komórek wzgórka jajonośnego poddawanych przyżyciowej diagnostyce w kierunku wykrywania apoptozy. Ann. Animal Sci. 13, 513–529. doi:10.2478/aoas-2013-0035
Samiec, M., and Skrzyszowska, M. (2018). Intrinsic and extrinsic molecular determinants or modulators for epigenetic remodeling and reprogramming of somatic cell-derived genome in mammalian nuclear-transferred oocytes and resultant embryos. Pol. J. Vet. Sci. 21, 217–227. doi:10.24425/119040
Samiec, M., and Skrzyszowska, M. (2010). Preimplantation developmental capability of cloned pig embryos derived from different types of nuclear donor somatic cells. Ann. Anim. Sci. 10, 385–398.
Sangalli, J. R., Chiaratti, M. R., De Bem, T. H., de Araújo, R. R., Bressan, F. F., Sampaio, R. V., et al. (2014). Development to term of cloned cattle derived from donor cells treated with valproic acid. PLoS One 9, e101022. doi:10.1371/journal.pone.0101022
Sangalli, J. R., De Bem, T. H., Perecin, F., Chiaratti, M. R., Oliveira Lde, J., de Araújo, R. R., et al. (2012). Treatment of nuclear-donor cells or cloned zygotes with chromatin-modifying agents increases histone acetylation but does not improve full-term development of cloned cattle. Cell. Reprogr. 14, 235–247. doi:10.1089/cell.2011.0079
Santos, F., Zakhartchenko, V., Stojkovic, M., Peters, A., Jenuwein, T., Wolf, E., et al. (2003). Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr. Biol. 13, 1116–1121. doi:10.1016/s0960-9822(03)00419-6
Schulz, K. N., and Harrison, M. M. (2019). Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234. doi:10.1038/s41576-018-0087-x
Sharif, J., Muto, M., Takebayashi, S., Suetake, I., Iwamatsu, A., Endo, T. A., et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. doi:10.1038/nature06397
Sharma, P., Yadav, A. S., Selokar, N. L., Kumar, D., Dhaka, S. S., and Yadav, P. S. (2018). Epigenetic status of buffalo fibroblasts treated with sodium butyrate a chromatin remodeling agent. Tissue Cell 50, 51–58. doi:10.1016/j.tice.2017.12.006
Shen, L., Inoue, A., He, J., Liu, Y., Lu, F., and Zhang, Y. (2014). Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–471. doi:10.1016/j.stem.2014.09.002
Shi, D., Lu, F., Wei, Y., Cui, K., Yang, S., Wei, J., et al. (2007). Buffalos (Bubalus bubalis) cloned by nuclear transfer of somatic cells. Biol. Reprod. 77, 285–291. doi:10.1095/biolreprod.107.060210
Shi, J., Tan, B., Luo, L., Li, Z., Hong, L., Yang, J., et al. (2020). Assessment of the growth and reproductive performance of cloned pietrain boars. Anim. (Basel) 10, 2053. doi:10.3390/ani10112053
Shimozawa, N., Ono, Y., Kimoto, S., Hioki, K., Araki, Y., Shinkai, Y., et al. (2002). Abnormalities in cloned mice are not transmitted to the progeny. Genesis 34, 203–207. doi:10.1002/gene.10143
Shin, T., Kraemer, D., Pryor, J., Liu, L., Rugila, J., Howe, L., et al. (2002). A cat cloned by nuclear transplantation. Nature 415, 859. doi:10.1038/nature723
Skrzyszowska, M., and Samiec, M. (2020). Enhancement of in vitro developmental outcome of cloned goat embryos after epigenetic modulation of somatic cell-inherited nuclear genome with trichostatin A. Ann. Animal Sci. 20, 97–108. doi:10.2478/aoas-2019-0063
Smith, L. C., Suzuki, J., Goff, A. K., Filion, F., Therrien, J., Murphy, B. D., et al. (2012). Developmental and epigenetic anomalies in cloned cattle. Reprod. Domest. Anim. 47 (4), 107–114. doi:10.1111/j.1439-0531.2012.02063.x
Smith, Z. D., and Meissner, A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220. doi:10.1038/nrg3354
Song, X., Liu, Z., He, H., Wang, J., Li, H., Li, J., et al. (2017). Dnmt1s in donor cells is a barrier to SCNT-mediated DNA methylation reprogramming in pigs. Oncotarget 8, 34980–34991. doi:10.18632/oncotarget.16507
Srirattana, K., Imsoonthornruksa, S., Laowtammathron, C., Sangmalee, A., Tunwattana, W., Thongprapai, T., et al. (2012). Full-term development of gaur-bovine interspecies somatic cell nuclear transfer embryos: effect of trichostatin A treatment. Cell. Reprogr. 14, 248–257. doi:10.1089/cell.2011.0099
Sugimoto, H., Kida, Y., Oh, N., Kitada, K., Matsumoto, K., Saeki, K., et al. (2015). Production of somatic cell nuclear transfer embryos using in vitro-grown and in vitro-matured oocytes in rabbits. Zygote 23, 494–500. doi:10.1017/S0967199414000082
Tanaka, S., Oda, M., Toyoshima, Y., Wakayama, T., Tanaka, M., Yoshida, N., et al. (2001). Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol. Reprod. 65, 1813–1821. doi:10.1095/biolreprod65.6.1813
Taweechaipaisankul, A., Kim, G. A., Jin, J. X., Lee, S., Qasim, M., Kim, E. H., et al. (2019). Enhancement of epigenetic reprogramming status of porcine cloned embryos with zebularine, a DNA methyltransferase inhibitor. Mol. Reprod. Dev. 86, 1013–1022. doi:10.1002/mrd.23178
van der Berg, J. P., Kleter, G. A., and Kok, E. J. (2019). Regulation and safety considerations of somatic cell nuclear transfer-cloned farm animals and their offspring used for food production. Theriogenology 135, 85–93. doi:10.1016/j.theriogenology.2019.06.001
Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R., and Yanagimachi, R. (1998). Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374. doi:10.1038/28615
Wang, F., Kou, Z., Zhang, Y., and Gao, S. (2007). Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol. Reprod. 77, 1007–1016. doi:10.1095/biolreprod.107.063149
Wang, F., Shin, J., Shea, J. M., Yu, J., Bošković, A., Byron, M., et al. (2016). Regulation of X-linked gene expression during early mouse development by Rlim. Elife 5, e19127. doi:10.7554/eLife.19127
Wang, H., Cui, W., Meng, C., Zhang, J., Li, Y., Qian, Y., et al. (2018). MC1568 enhances histone acetylation during oocyte meiosis and improves development of somatic cell nuclear transfer embryos in pig. Cell. Reprogr. 20, 55–65. doi:10.1089/cell.2017.0023
Wang, L. Y., Li, Z. K., Wang, L. B., Liu, C., Sun, X. H., Feng, G. H., et al. (2020a). Overcoming intrinsic H3K27me3 imprinting barriers improves post-implantation development after somatic cell nuclear transfer. Cell Stem Cell 27, 315–325. e315. doi:10.1016/j.stem.2020.05.014
Wang, M., Feng, S., Ma, G., Miao, Y., Zuo, B., Ruan, J., et al. (2020b). Whole-genome methylation analysis reveals epigenetic variation in cloned and donor pigs. Front. Genet. 11, 23. doi:10.3389/fgene.2020.00023
Wang, Y. S., Tang, S., An, Z. X., Li, W. Z., Liu, J., Quan, F. S., et al. (2011). Effect of mSOF and G1.1/G2.2 media on the developmental competence of SCNT-derived bovine embryos. Reprod. Domest. Anim. 46, 404–409. doi:10.1111/j.1439-0531.2010.01679.x
Wani, N. A., Vettical, B. S., and Hong, S. B. (2017). First cloned bactrian camel (Camelus bactrianus) calf produced by interspecies somatic cell nuclear transfer: A step towards preserving the critically endangered wild bactrian camels. PLoS One 12, e0177800. doi:10.1371/journal.pone.0177800
Wani, N. A., Wernery, U., Hassan, F. A., Wernery, R., and Skidmore, J. A. (2010). Production of the first cloned camel by somatic cell nuclear transfer. Biol. Reprod. 82, 373–379. doi:10.1095/biolreprod.109.081083
Wen, B. Q., Li, J., Li, J. J., Tian, S. J., Sun, S. C., Qi, X., et al. (2014). The histone deacetylase inhibitor Scriptaid improves in vitro developmental competence of ovine somatic cell nuclear transferred embryos. Theriogenology 81, 332–339. doi:10.1016/j.theriogenology.2013.09.032
Weng, X. G., Cai, M. M., Zhang, Y. T., Liu, Y., Liu, C., and Liu, Z. H. (2020). Improvement in the in vitro development of cloned pig embryos after kdm4a overexpression and an H3K9me3 methyltransferase inhibitor treatment. Theriogenology 146, 162–170. doi:10.1016/j.theriogenology.2019.11.027
Wiater, J., Samiec, M., Skrzyszowska, M., and Lipiński, D. (2021a). Trichostatin A-assisted epigenomic modulation affects the expression profiles of not only recombinant human α1, 2-fucosyltransferase and α-galactosidase A enzymes but also Galα1→3Gal epitopes in porcine Bi-transgenic adult cutaneous fibroblast cells. Int. J. Mol. Sci. 22, 1386. doi:10.3390/ijms22031386
Wiater, J., Samiec, M., Wartalski, K., Smorąg, Z., Jura, J., Słomski, R., et al. (2021b). Characterization of mono- and Bi-transgenic pig-derived epidermal keratinocytes expressing human FUT2 and GLA genes-in vitro studies. Int. J. Mol. Sci. 22, 9683. doi:10.3390/ijms22189683
Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J., and Campbell, K. H. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813. doi:10.1038/385810a0
Woods, G. L., White, K. L., Vanderwall, D. K., Li, G. P., Aston, K. I., Bunch, T. D., et al. (2003). A mule cloned from fetal cells by nuclear transfer. Science 301, 1063. doi:10.1126/science.1086743
Wossidlo, M., Arand, J., Sebastiano, V., Lepikhov, K., Boiani, M., Reinhardt, R., et al. (2010). Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 29, 1877–1888. doi:10.1038/emboj.2010.80
Wossidlo, M., Nakamura, T., Lepikhov, K., Marques, C. J., Zakhartchenko, V., Boiani, M., et al. (2011). 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241. doi:10.1038/ncomms1240
Wu, H., and Zhang, Y. (2014). Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. doi:10.1016/j.cell.2013.12.019
Wu, S. C., and Zhang, Y. (2010). Active DNA demethylation: many roads lead to rome. Nat. Rev. Mol. Cell Biol. 11, 607–620. doi:10.1038/nrm2950
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534. doi:10.1038/nrg.2017.33
Xia, W., and Xie, W. (2020). Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Rep. 15, 1158–1175. doi:10.1016/j.stemcr.2020.09.005
Xie, Z., Zhang, W., and Zhang, Y. (2022). Loss of Slc38a4 imprinting is a major cause of mouse placenta hyperplasia in somatic cell nuclear transferred embryos at late gestation. Cell Rep. 38, 110407. doi:10.1016/j.celrep.2022.110407
Xu, L., Song, S. H., Idrees, M., Mesalam, A., Joo, M. D., Sidrat, T., et al. (2021). Effects of donor cell types on the development of bovine embryos using cytoplasm injection cloning technology. Int. J. Mol. Sci. 22, 5841. doi:10.3390/ijms22115841
Xue, F., Tian, X. C., Du, F., Kubota, C., Taneja, M., Dinnyes, A., et al. (2002). Aberrant patterns of X chromosome inactivation in bovine clones. Nat. Genet. 31, 216–220. doi:10.1038/ng900
Yamada, M., Johannesson, B., Sagi, I., Burnett, L. C., Kort, D. H., Prosser, R. W., et al. (2014). Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature 510, 533–536. doi:10.1038/nature13287
Yamagata, K., Yamazaki, T., Miki, H., Ogonuki, N., Inoue, K., Ogura, A., et al. (2007). Centromeric DNA hypomethylation as an epigenetic signature discriminates between germ and somatic cell lineages. Dev. Biol. 312, 419–426. doi:10.1016/j.ydbio.2007.09.041
Yang, B. Z., Yang, C. Y., Li, R. C., Qin, G. S., Zhang, X. F., Pang, C. Y., et al. (2010). An inter-subspecies cloned buffalo (Bubalus bubalis) obtained by transferring of cryopreserved embryos via somatic cell nuclear transfer. Reprod. Domest. Anim. 45, e21–25. doi:10.1111/j.1439-0531.2009.01510.x
Yang, G., Zhang, L., Liu, W., Qiao, Z., Shen, S., Zhu, Q., et al. (2021). Dux-mediated corrections of aberrant H3K9ac during 2-cell genome activation optimize efficiency of somatic cell nuclear transfer. Cell Stem Cell 28, 150–163.e5. e155. doi:10.1016/j.stem.2020.09.006
Yang, J., Yang, S., Beaujean, N., Niu, Y., He, X., Xie, Y., et al. (2007a). Epigenetic marks in cloned rhesus monkey embryos: comparison with counterparts produced in vitro. Biol. Reprod. 76, 36–42. doi:10.1095/biolreprod.106.051383
Yang, L., Liu, X., Song, L., Di, A., Su, G., Bai, C., et al. (2020). Transient Dux expression facilitates nuclear transfer and induced pluripotent stem cell reprogramming. EMBO Rep. 21, e50054. doi:10.15252/embr.202050054
Yang, X., Smith, S. L., Tian, X. C., Lewin, H. A., Renard, J. P., and Wakayama, T. (2007b). Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 39, 295–302. doi:10.1038/ng1973
Zhang, K., Wu, D. Y., Zheng, H., Wang, Y., Sun, Q. R., Liu, X., et al. (2020). Analysis of genome architecture during SCNT reveals a role of cohesin in impeding minor ZGA. Mol. Cell 79, 234–250. e239. doi:10.1016/j.molcel.2020.06.001
Zhang, M., Wang, F., Kou, Z., Zhang, Y., and Gao, S. (2009). Defective chromatin structure in somatic cell cloned mouse embryos. J. Biol. Chem. 284, 24981–24987. doi:10.1074/jbc.M109.011973
Zhang, S., Chen, X., Wang, F., An, X., Tang, B., Zhang, X., et al. (2016). Aberrant DNA methylation reprogramming in bovine SCNT preimplantation embryos. Sci. Rep. 6, 30345. doi:10.1038/srep30345
Zhang, Y. T., Yao, W., Chai, M. J., Liu, W. J., Liu, Y., Liu, Z. H., et al. (2022). Evaluation of porcine urine-derived cells as nuclei donor for somatic cell nuclear transfer. J. Vet. Sci. 23, e40. doi:10.4142/jvs.21297
Zhang, Y., Wang, Q., Liu, K., Gao, E., Guan, H., and Hou, J. (2018a). Treatment of donor cells with recombinant KDM4D protein improves preimplantation development of cloned ovine embryos. Cytotechnology 70, 1469–1477. doi:10.1007/s10616-018-0224-6
Zhang, Y., Xiang, Y., Yin, Q., Du, Z., Peng, X., Wang, Q., et al. (2018b). Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 50, 96–105. doi:10.1038/s41588-017-0003-x
Zhang, Z., Zhai, Y., Ma, X., Zhang, S., An, X., Yu, H., et al. (2018c). Down-Regulation of H3K4me3 by MM-102 facilitates epigenetic reprogramming of porcine somatic cell nuclear transfer embryos. Cell. Physiol. biochem. 45, 1529–1540. doi:10.1159/000487579
Zhao, J., Hao, Y., Ross, J. W., Spate, L. D., Walters, E. M., Samuel, M. S., et al. (2010a). Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell. Reprogr. 12, 75–83. doi:10.1089/cell.2009.0038
Zhao, J., Whyte, J., and Prather, R. S. (2010b). Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 341, 13–21. doi:10.1007/s00441-010-1000-x
Zheng, H., Huang, B., Zhang, B., Xiang, Y., Du, Z., Xu, Q., et al. (2016). Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079. doi:10.1016/j.molcel.2016.08.032
Zhou, C., Zhang, J., Zhang, M., Wang, D., Ma, Y., Wang, Y., et al. (2020). Transcriptional memory inherited from donor cells is a developmental defect of bovine cloned embryos. FASEB J. 34, 1637–1651. doi:10.1096/fj.201900578RR
Zhou, Q., Renard, J. P., Le Friec, G., Brochard, V., Beaujean, N., Cherifi, Y., et al. (2003). Generation of fertile cloned rats by regulating oocyte activation. Science 302, 1179. doi:10.1126/science.1088313
Keywords: somatic cell nuclear transfer, cloning efficiency, epigenetic barriers, pre-implantation, post-implantation
Citation: Li Y and Sun Q (2022) Epigenetic manipulation to improve mouse SCNT embryonic development. Front. Genet. 13:932867. doi: 10.3389/fgene.2022.932867
Received: 30 April 2022; Accepted: 29 July 2022;
Published: 30 August 2022.
Edited by:
Sadie L. Marjani, Central Connecticut State University, United StatesReviewed by:
Maria Skrzyszowska, National Research Institute of Animal Production, PolandQingran Kong, Wenzhou Medical University, China
Copyright © 2022 Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Sun, cXN1bkBpb24uYWMuY24=
 Yamei Li
Yamei Li Qiang Sun
Qiang Sun