
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 17 August 2022
Sec. RNA
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.929477
This article is part of the Research Topic Role of Non-coding RNAs, Metabolites and Extracellular Vesicles in Disease Regulation and Health View all 16 articles
 Mustafa Hitit1*
Mustafa Hitit1* Mehmet Kose2
Mehmet Kose2 Mehmet Salih Kaya3
Mehmet Salih Kaya3 Mesut Kırbas4
Mesut Kırbas4 Sukru Dursun5
Sukru Dursun5 Ilyas Alak6
Ilyas Alak6 Mehmet Osman Atli7*
Mehmet Osman Atli7*MicroRNA (miRNA) plays an important role in the control of gene expression and is implied in many biological functions, including embryo implantation and development. The aim was to assess plasma miRNA profiles during the peri-implantation and ascertain potential candidate miRNA markers for early pregnancy diagnosis in ovine plasma. The plasma samples were obtained from a total of 24 ewes on days 12 (pre-implantation; P12, n = 4), 16 (implantation; P16, n = 4) and 22 (post-implantation; P22, n = 4) after mating, and on their corresponding days of 12 (Pre-C; C12, n = 4), 16 (Imp-C; C16, n = 4) and 22 (Post-C; C22, n = 4) of the estrous cycle. The miRNA profiles in plasma were assessed by microarray technology. We detected the presence of 60 ovine-specific miRNAs in plasma samples. Of these miRNAs, 22 demonstrated a differential expression pattern, especially between the estrous cycle and early pregnancy, and targeted 521 genes. Two miRNAs (oar-miR-218a and oar-miR-1185-3p) were confirmed using RT-qPCR in the ovine plasma samples. Protein-protein interaction (PPI) network of target genes established six functional modules, of which modules 1 and 3 were enriched in the common GO terms, such as inflammatory response, defense response, and regulation of immune response. In contrast, module 2 was enriched in the developmental process involved in reproduction, embryo development, embryonic morphogenesis, and regulation of the developmental process. The results indicate that miRNAs profiles of plasma seemed to be modulated during the peri-implantation stage of pregnancy in ewes. Circulating miRNAs could be promising candidates for diagnosis in early ovine pregnancy.
In order to have a successful pregnancy, the uterine environment and a viable embryo must work together during the peri-implantation stage of pregnancy (Kose et al., 2016c; Kaya et al., 2017). Many molecules controlled at the gene expression level are implicated in the regulation of implantation (Kose et al., 2016a; Alak et al., 2020), but their particular regulatory mechanisms remain still unclear. Previous studies indicated that changes in microRNA (miRNA) expression play a functional role in embryo implantation in many species (Song et al., 2015; Gebremedhn et al., 2018). Apart from roles in expressed tissues of interest, particularly in the endometrium (Kiyma et al., 2015; Yan et al., 2021), miRNAs also could be released into the extracellular environment by cells, facilitating cell-cell interactions and providing valuable information related to the specific conditions (Bidarimath et al., 2021; Shi et al., 2021).
miRNAs, non-coding RNA molecules with 21–24 nucleotides, post-transcriptionally modulate gene expression and thus are implied in biological processes (Bartel, 2004; Hitit et al., 2015; WB, 2016). An understanding of miRNAs expressions and their functions in the female reproductive tract has been recently developed. Accordingly, miRNAs are functional at different stages of the reproduction process in the female (oocyte growth, maturation, embryo development, implantation, and placentation) (Guzeloglu et al., 2013; Hitit et al., 2013; Kaczmarek et al., 2020). miRNA expression profiles were shown to be different during the peri-implantation period and even differed between implantation and non-implantation regions in mice (Chakrabarty et al., 2007; Liu et al., 2015). Moreover, gene expression profiles concerning embryo implantation in ovine endometrium revealed pregnancy-associated miRNAs, of which many were differentially expressed in the endometrium on day 13 of pregnancy (Kiyma et al., 2015). Also, we determined differentially expressed miRNA profiles in the endometrium during the early pregnancy on days 12, 16, and 22 (Kose et al., 2022).
Placenta or embryo-derived miRNA molecules pass into body fluids, such as plasma, serum, and milk, within the extracellular vesicle (Tan et al., 2020). Therefore, their resistance with the stable structure to adverse effects such as freezing–thawing and high temperature is encouraging for detecting early pregnancy and monitoring the maintenance of pregnancy (De Bem et al., 2017). More specifically, in livestock animals, plasma samples from cattle and goats provide unique miRNA profiles between pregnant and non-pregnant groups, which circulating miRNAs were shown to be potential markers of early pregnancy stages and endometrial receptivity in the field of reproductive biology (Ioannidis and Donadeu, 2016; Zang et al., 2021; Zhao et al., 2021). El-Shorafa and Sharif (2016) determined that healthy fertile women exhibit different expression profiles of some miRNAs in maternal plasma than women who experienced recurrent pregnancy loss of unknown cause (El-Shorafa and Sharif, 2016). Similarly, Kotlabova et al. (2011) reported that seven miRNAs of placental origin were identified in the maternal circulation throughout pregnancy.
Considering the literature indicated above, we hypothesized that early pregnancy causes a change in plasma miRNA profiles, and this may be a candidate marker for early pregnancy diagnosis in sheep. The current study, therefore, aimed to ascertain the circulating miRNA profile in the plasma during peri-implantation, which is one of the most crucial stages for the establishment and maintenance of pregnancy in the ewe, and identify candidate miRNA markers for early pregnancy diagnosis.
All experimental steps were confirmed by the Bahri Dagdas International Agricultural Research Institute Ethical Research Committee (Number: 29/01/2016-49-7). Twenty-four multiparous ewes (n = 24) were used; assigned into cyclic (C, n = 12) and pregnant (P, n = 12) groups, randomly. Animal diets for 3- to 5-year-olds were adjusted to satisfy the NRC (2007) nutritional requirements. All other supplementals were given ad libitum during the study.
Before the experiment, cycles of the ewes were synchronized with two cloprostenol (a synthetic analog of prostaglandin F2alpha; PGF2α, 125 mcg) injections 11 days apart. Immediately after the second injection, estrus was checked three times a day using teaser rams. Estrus of ewes was obtained through teaser ram at 8 h intervals for 5 days after the second injection, and the ewes that showed estrus were recorded. Then, these ewes were followed to accomplish their entire cycle and noted for new natural estrus using teaser rams. In this new estrus, the pregnant ewes mated (day 0) two times, 12 h alone, using fertility-proven rams. The estrus day in the cyclic group was accepted as day zero (day 0). Ewes were scheduled for slaughter on days of 12 (pre-implantation, n = 4; P12), 16 (implantation, n = 4; P16) or 22 (post-implantation, n = 4; P22) of gestation following mating, and on their corresponding days of 12 (n = 4, C12), 16 (n = 4, C16) or 22 (n = 4, C22) of the estrous cycle. To provide a similar effect of progesterone and to observe the only effect of pregnancy or embryo on plasma miRNA expression, cyclic ewes were exposed to a natural progesterone implant via intravaginal on day 13 of the cycle for days 16 and 22 groups, and progesterone implants were kept until the ewes were slaughtered. The presence of only one embryonic trophoblast was observed for 12, 16, and 22 days of pregnancy in the uterine lumen (Spencer et al., 2004; Bazer et al., 2012).
We collected blood samples from ewes in tubes containing EDTA for plasma isolation just before the ewes were slaughtered. We centrifuged collected blood samples at 1,600×g for 13 min, and plasma was extracted and kept at − 80°C. Plasma was thawed at 20°C in the dry bath. Then, total cell-free RNA was extracted from 250 μl of plasma through miRCURY RNA Isolation Kit—Biofluids (Exiqon #300112 Vedbaek- Denmark) according to the manufacturer’s protocol. Plasma samples underwent on-column DNase to get rid of DNA contamination using the manufacturer’s protocols. Consequently, we eluted RNA samples using 40 μl RNase-free water. Tubes containing the miRNA were kept at −80°C until the miRNA array analysis.
The profile of miRNAs from ovine plasma samples was investigated using the Affymetrix Microarray system with the GeneChip miRNA 4.0 Array (Affymetrix, United States) that is arranged to retrieve mature miRNA sequences in miRBase (20.0) (http://mirbase.org/ftp.shtml). Mature miRNA sequences of one hundred fifty from sheep are demonstrated in miRBase (20.0). A total of one 1) microgram of RNA was labeled with a FlashTagTM Biotin HSR RNA Labeling Kit (Affymetrix, United States). Following RNA labeling, through a GeneChip Hybridization Control Kit (Affymetrix, United States), microarray chips were hybridized with agitation at 60 rpm for 15 h. Then, the chip arrays were washed and subsequently stained through a Fluidics Station 450 (Affymetrix, Santa Clara, California, United States) with AGCC Fluidics Control Software. Fluorescence was detected from the array chip with an Affymetrix® GeneChip Scanner 3000.
The signal of probes was generated as cell intensity files (*CEL files) computed using Affymetrix GeneChip Command Console software and analyzed in Transcriptome Analysis Console software. The intensity data of each chip was processed through the robust multi-array average (RMA) and identified above background (DABG) normalization with a default analysis setting of Affymetrix. Probe set summarization was performed through Median Polish. Probe values were generated as log2 transformed. Comparison between the cyclic and the pregnant samples was accomplished through fold-change with an independent t-test. One-way ANOVA was used to reveal statistically significant genes at the significance level of p ≤ 0.05. To demonstrate biologically relevant gene expression changes of each of cyclic and pregnant conditions, the standard approach was employed through a p-value (p ≤ 0.05) as the primary criterion followed by fold change (−1.5 ≥ FC ≥ 1.5) as the secondary criterion to select differentially expressed genes. Upon first analysis, criteria were relaxed to (−1.25 ≥ FC ≥ 1.25; p < 0.05) to identify all the family members of miRNAs of interest in all the ovine plasma samples.
The target gene prediction of miRNAs in plasma samples was accomplished using the miRNAconsTarget online tool from sRNAtoolbox (http://bioinfo5.ugr.es/srnatoolbox), providing consensus target prediction. The given input data are developed on independent prediction from animal-based tools. TargetSpy, PITA (energy score < −15), and miRanda (pairing score >150 and an energy score < −15), making a total of three prediction algorithms. The common target genes predicted by all three tools were considered a potential miRNA target.
The functional network association among target genes was established using the database of STRING (version 11.5, http://string-db.org), and eventually visualized in Cytoscape (version 3.9.0). The PPI network of target genes was transferred and subsequently assessed in Cytoscape. The functional modules were determined and shown by Molecular Complex Detection (MCODE), a plugin in Cytoscape for identifying intensively connected nodes in a current network. The module was set as follows: k-score = 2, cut-off degree = 2, max depth size = 100, MCODE score >5, and node score cut-off = 0.2 (Lou et al., 2019). The nodes included in the main modules are demonstrated as densely connected genes that show significant biological functions. CytoHubb CytoScape plugin was used to reveal important nodes by integrating topological calculations such as Maximal clique centrality (MCC), Maximum neighborhood component (MNC), Degree, Edge percolated component (EPC), and (EcCentricity) EC (Chin et al., 2014). The overlapping genes were ranked using the aforementioned five algorithms.
The KEGG pathway and GO enrichment for the predicted target genes from the modules were analyzed using Cytoscape software with the ClueGO V2.5.7 plug-in (Bindea et al., 2009). The ClueGO plug-in generates functionally grouped GO annotation networks for many target genes. The GO categories were assigned to molecular function (MF), cellular component (CC), and biological process (BP). Two-sided hypergeometric tests set the p-value to 0.05, and multiple test corrections were performed using Bonferroni step-down adjustment. The threshold of the kappa score was adjusted to 0.7.
Forward primer, universal reverse primer, and Stem-Loop primer sequences of two miRNAs to be confirmed by RT-qPCR (Supplementary Material S1). For the confirmation with RT-qPCR, firstly, a reverse transcription reaction was prepared using the First-Strand cDNA (1 μg RNA to cDNA) Synthesis Kit for RT-qPCR (USB, Cat no: 75780). Reverse transcriptase reaction conditions were as follows: 32 min at 16°C, 60 min at 44°C, 10 min at 95°C, and 5 min at 4°C. Following cDNA synthesis by reverse transcriptase reaction, RT-qPCR analyses of the samples were performed using VeriQuest Fast SYBR Green RT-qPCR Master Mix (USB, Cat no: 75690). RT-qPCR reaction was used as follows: Polymerase for 8 min at 95°C, then 45 cycles of denaturation for 25 s at 94°C, annealing for 42 s at 58°C, and extension for 50 s at 70°C. Log transformation of the data was performed according to the previously mentioned 2−ΔCt method (Livak and Schmittgen, 2001). Statistical analysis of values normalized to reference genes was calculated by Relative Expression Software Tool (REST 2009) (Pfaffl et al., 2002).
Microarray results in plasma samples revealed a total of 183 miRNAs in all species between the estrous cycle and early pregnancy, while 60 were identified in ovine plasma samples (Supplementary Material S2). However, when respectively compared, there were 6, 12, and 4 statistically significant plasma-specific miRNAs between C12 vs. P12, C16 vs. P16, and C22 vs. P22, respectively. Among these, one was shared between C12 vs. P12, C16 vs. P16, C16 vs. P16 and C22 vs. P22, whereas 5, 10, and 3 miRNAs were unique to C12 vs. P12, C16 vs. P16, and C22 vs. P22, respectively (Figure 1).
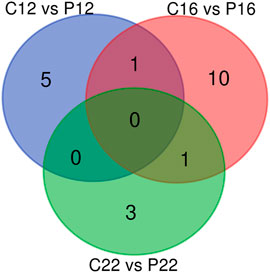
FIGURE 1. miRNA profiles in estrus cycle and pregnancy in ovine plasma. The Venn diagrams represent miRNA profiles between C12 and P12, C16 and P16, and C22 and P22.
A total of 22 differentially expressed ovine plasma miRNAs were determined (p < 0.05, difference of miRNAs greater than 1.25-fold change) when cyclic ovine plasma samples compared to pregnant ovine plasma samples. While five miRNAs (oar-miR-23b, oar-let-7i, oar-miR-19b, oar-miR-21, oar-miR-487b-3p were upregulated between C12 and P12, one of them (oar-miR-329a-5p) was downregulated (Figure 2A). Between C16 and P16, 8 miRNAs (oar-miR-29a, oar-miR-299-3p, oar-miR-30d, oar-miR-379-5p, oar-miR-152, oar-miR-323b, oar-miR-329a-5p, and oar-miR-654-5p) were upregulated while 4 miRNAs (oar-let-7b, oar-miR-218a, oar-miR-487a-5p, and oar-miR-758-3p) were downregulated (Figure 2B). In C22 vs. P22, miRNAs (oar-miR-29b, oar-miR-1185-3p, oar-miR-487a-5p, and oar-miR-543-3p) were downregulated (Figure 2C). The list of the 22 significant miRNAs from cyclic and pregnant groups with their fold change is depicted in Table 1. Furthermore, the detailed information on the differentially expressed ovine plasma miRNAs is demonstrated in Supplementary Material S2.
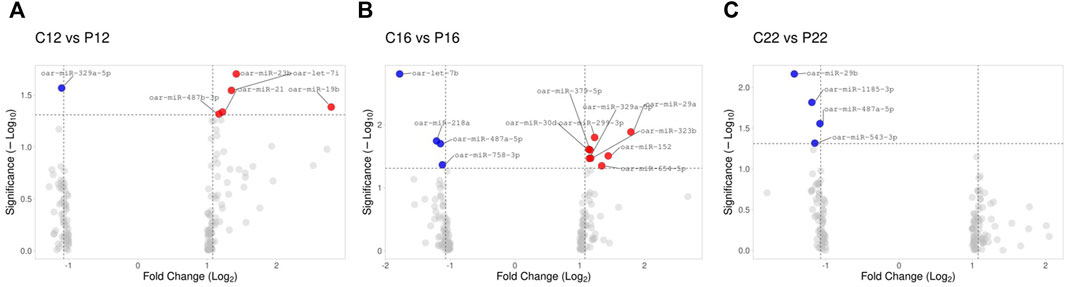
FIGURE 2. Illustration of volcano plot for differentially expressed miRNAs in estrus cycle and pregnancy in ovine plasma. The −log10 is set versus the log2 (fold change: cycle: pregnancy). (A) C12 vs. P12, (B) C16 vs. P16, and (C) C22 vs. P22. Color represents the fold change, red: up and blue: down.

TABLE 1. The list of the differentially expressed miRNAs between cyclic days (C12, C16, and C22) and pregnant days (P12, P16, and P22).
The target genes of the twenty-two miRNAs were predicted using the miRNAconsTarget online tool from sRNAtoolbox based on animal-based prediction with three algorithms (TargetSpy, PITA, and miRanda). Six differentially expressed miRNAs between C12 vs. P12 targeted 156 genes. Twelve miRNAs targeted 257 target genes between C16 and P16, whereas there were 108 in C22 and P22 by four differentially expressed miRNAs (Supplementary Material S3). Overlapping target genes from cyclic and pregnant were excluded, and then 326 were submitted to STRING online database. A PPI network including 328 nodes and 1,936 edges was directed employing the Cytoscape software (Supplementary Material S4).
Among the miRNAs, oar-miR-1185-3p and oar-miR-218a were detected to be regulated in ovine plasma samples. The expression of oar-miR-1185-3p mRNA was similar between C12 and P12. It was found to be greater in P16 than in C16, while it did not change between C22 and P22 (Figure 3A). oar-miR-218a mRNA was found to be lower in pregnancy groups than in cyclic groups (C12 vs. P12, C16 vs. P16, and C22 vs. P22) identified (Figure 3B).
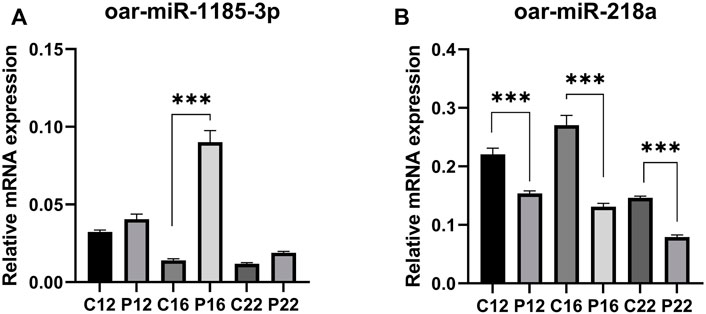
FIGURE 3. The relative abundance of selected miRNAs quantified by RT-qPCR were statistically significant. Expression of (A) oar-miR-218a and (B) oar-miR-1185-3p, between C12 vs P12, C16 vs P16, and C22 vs P22. Data are shown as relative abundance ± SEM, p < 0.05; (Estrous cyclic day 12: C12, Pregnant day 12: P12, Estrous cyclic day 16:C16, Pregnant day 16: P16, Estrous cyclic day 22:C22, Pregnant day 22: P22, indicates the group).
Six clusters were attained from the PPI network following module analysis through the MCODE plugin of Cytoscape, and we sorted out three modules among all as hub modules constructed from MCODE scores (Figures 4A–C). Module 1 consisted of 19 nodes and 151 edges and had the highest MCODE score (16.778) of all modules (Figure 4A). Module 2 has 30 nodes and 131edges (Figure 4B) Supplementary Material S5, whereas module 3 contained 21 nodes and 84 edges (Figure 4C). All five classifications of methods within the CytoHubba plugin were accepted, and the highest ranked genes of each method (top 10) were shown (Table 2). Module 1 was enriched in (GO:0006468) protein phosphorylation, (GO:00069540) inflammatory response, (GO:0006952) defense response, (GO:0042127) regulation of cell population proliferation, (GO:0010033) response to organic substance, (GO:0009893) positive regulation of metabolic process, (GO:0002684) positive regulation of immune system process, (GO:2000026) regulation of multicellular organismal development, (GO:0009966) regulation of signal transduction (Figure 5). Module 2 was involved in (GO:0009790) embryo development, (GO:0003006) developmental process involved in reproduction, (GO:0048598) embryonic morphogenesis, (GO:0050793) regulation of developmental process, (GO:0035295) tube development, (GO:0060429) epithelium development, (GO:0061138) morphogenesis of a branching epithelium, (GO:0009891) positive regulation of the biosynthetic process (Figure 6). Although module 3 had common GO terms with module 1, it contained (GO:0006952) defense response, (GO:0050776) regulation of immune response, (GO:0045087) innate immune response, (GO:0010033) response to organic substance, (GO:0006954) inflammatory response, (GO:0051240) positive regulation of the multicellular organismal process (Figure 7) (Supplementary Material S6).
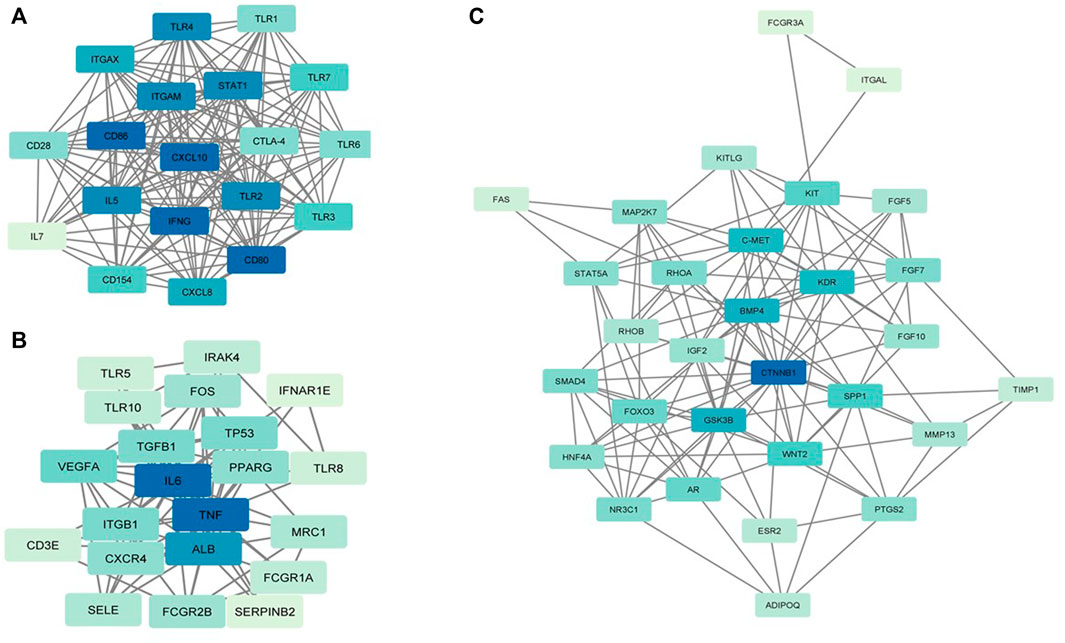
FIGURE 4. MCODE clustering of the top three clusters. (A) Module 1. (B) Module 2. (C) Module 3. In the modules, the nodes are colored in a continuous manner compliant with their |log2FC| values.
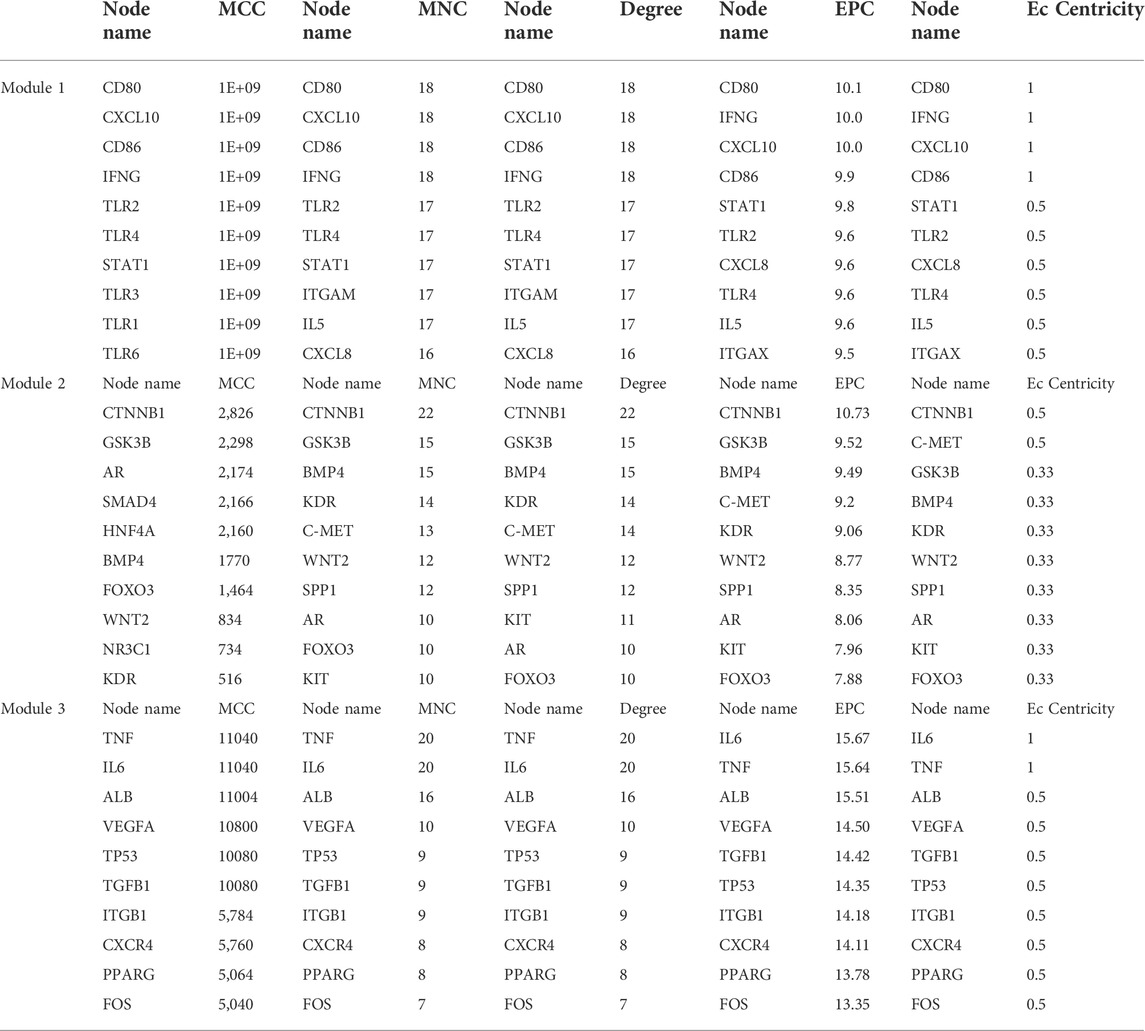
TABLE 2. Hub target genes of differentially expressed miRNAs from the modules. Overlapping hub protein symbols in the top 10, ranked methods were highlighted as bold. MCC: maximal clique centrality; MNC: maximum neighborhood component; Degree: node degree; EPC: edge percolated component; EC: EcCentricity.
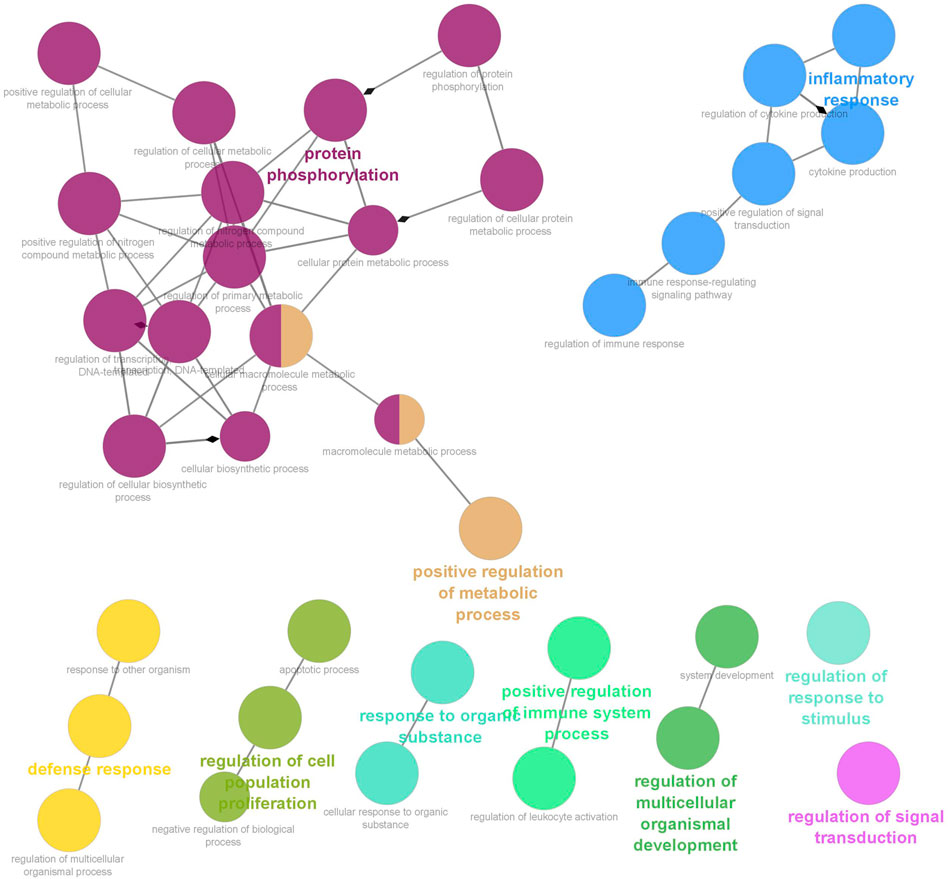
FIGURE 5. Functional interaction of network analysis of the target genes from module 1 between estrus cycle and pregnancy. GO terms are shown as nodes, the color depth of node shows distinct proportions of target genes. Nodes located in the same cluster are nominated as the same node color and size of nodes demonstrates the number of mapped genes in each GO term. Relationship between terms is shown by edges. Functionally associated groups partly overlap and are randomly colored.
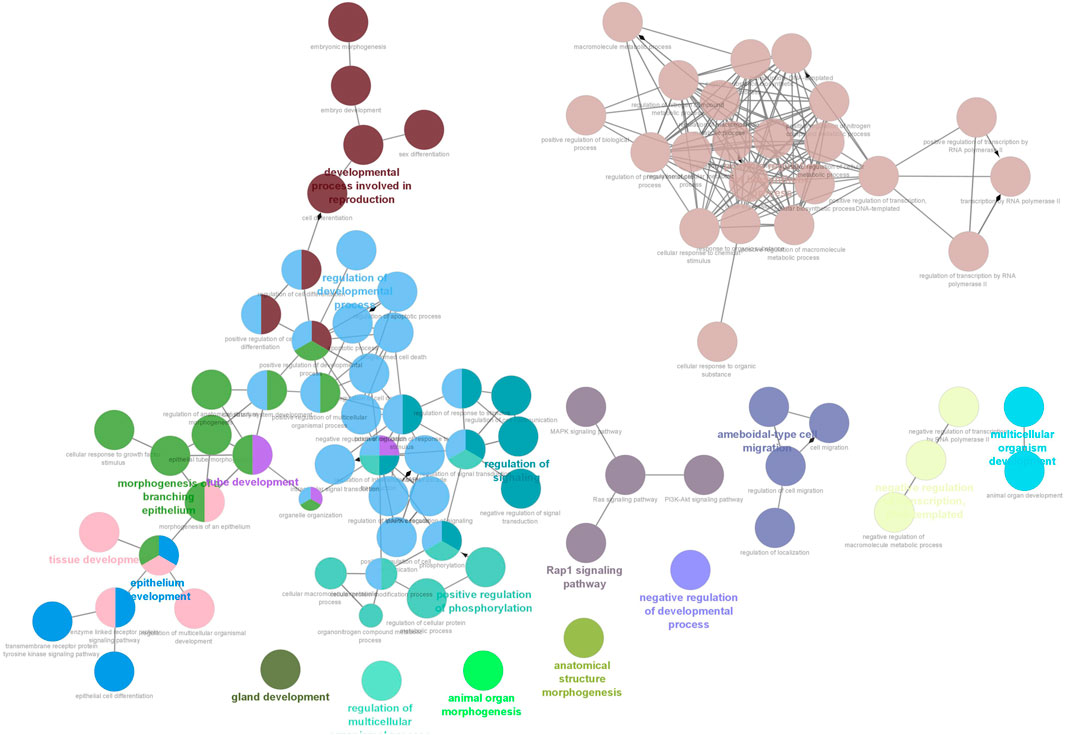
FIGURE 6. Functional interaction of network analysis of the target genes from module 2 between estrus cycle and pregnancy. GO terms are shown as nodes, the color depth of node shows distinct proportions of target genes. Nodes located in the same cluster are nominated as the same node color and size of nodes demonstrates the number of mapped genes in each GO term. Relationship between terms is shown by edges. Functionally associated groups partly overlap and are randomly colored.
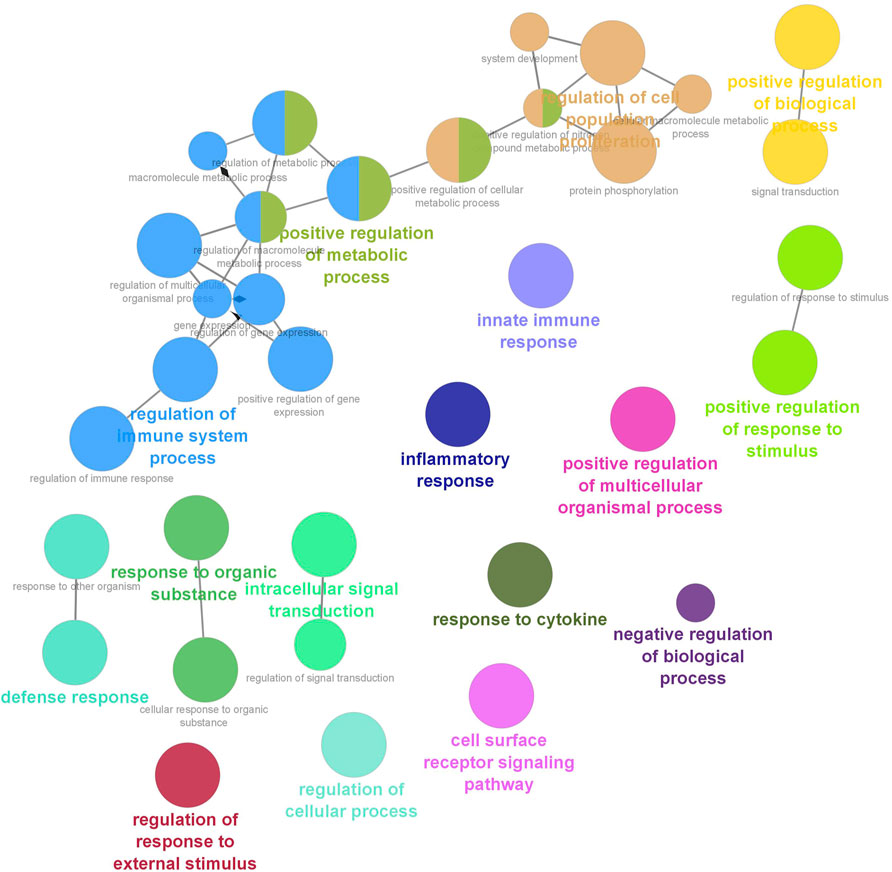
FIGURE 7. Functional interaction of network analysis of the target genes from module 3 between estrus cycle and pregnancy. GO terms are shown as nodes, the color depth of node shows distinct proportions of target genes. Nodes located in the same cluster are nominated as the same node color and size of nodes demonstrates the number of mapped genes in each GO term. Relationship between terms is shown by edges. Functionally associated groups partly overlap and are randomly colored.
In this current study, we carried out profiling of plasma miRNA expression to determine potential miRNAs involved in the critical stages of peri-implantation. The circulating miRNA expression profile was evaluated on days 12, 16, and 22 during pregnancy after mating and on their corresponding days of 12, 16, and 22 of the estrous cycle. We already know that the estrous cycle in ewes is 17 days and declining progesterone levels start on day 16 after mating. Therefore, in the C16 and C22 groups, an external supply of progesterone was added. As a result, we were able to compare the plasma miRNA profiles of cyclic and pregnant sheep without being affected by the fluctuating levels of progesterone. We may therefore hypothesize that whatever changes we detected in the levels of miRNA expression in ovine plasma in pregnant ewes were a direct result of the conceptus given that progesterone levels were similar across the cyclic and pregnant groups.
Circulating plasma miRNAs have been detected in several studies that indicated a significant number of miRNAs to reveal the importance of transcriptomics in reproductive biology (Hitit and Kurar, 2017; Zong et al., 2021). As such, many miRNAs originating from villous trophoblasts circulate through maternal blood within or linked with extracellular vesicles (Miura et al., 2010; Cameron et al., 2011; Kotlabova et al., 2011). It has been identified that 374 human placenta-specific miRNAs, of which 10 with the highest global expression were also reflected in plasma (Smith et al., 2021). In another exploratory study, 208 plasma-specific miRNAs were identified in cattle; of these miRNAs, sixteen were statistically different between non-pregnant and pregnant plasma samples (Ioannidis and Donadeu, 2016). However, this is the first study elucidating the identification of circulating miRNA profiles in ovine plasma during the estrous cycle and pregnancy. In the current study, we discovered a total of 60 ovine circulating miRNAs between cyclic and pregnant plasma, indicating remarkable outcomes for sheep reproductive biology. Among these, 22 were differentially expressed in microarray results upon pregnancy groups were compared with cyclic (Table 1). The results may indicate that circulating plasma miRNAs may function as potential biomarkers in the regulation of pregnancy during the estrous cycle and early pregnancy (Ioannidis and Donadeu, 2017; Lim et al., 2021).
Our study is noteworthy that the target prediction of selected miRNAs uncovered a great many GO terms that ovine circulating miRNAs in plasma might regulate biological and molecular control of gene expression during early pregnancy. Accordingly, we particularly ascertained critical GO terms that may be involved in early pregnancy, such as (GO:0006468) protein phosphorylation, (GO:00069540) inflammatory response, (GO:0006952) defense response, (GO:0042127) regulation of cell population proliferation, (GO:0009893 positive regulation of metabolic process, (GO:0010033) response to organic substance, (GO:0002684) positive regulation of immune system process, (GO:2000026) regulation of multicellular organismal development, (GO:0009966) regulation of signal transduction. This may illuminate that circulating miRNAs clarify molecular events during pregnancy recognition and embryo implantation because extracellular vesicles derived from the serum, trophectoderm, and uterine epithelia in pregnant ewes are implicated in intercellular communication, and microRNA-mediated pathways are involved in the key pathway in the pregnancy (Brooks et al., 2016; Ioannidis and Donadeu, 2017; Sun et al., 2021; Ono et al., 2022).
The potential of circulating miRNAs has been demonstrated, in many animals, to be identified in maternal circulation directly following initiation of implantation (Sun et al., 2021). Based upon this knowledge, for instance, elevated expression levels of circulating miRNAs (miR-23b) are shown to be expressed within either pregnant endometria (Krawczynski et al., 2015) or serum (Reliszko et al., 2017) on a gestational day 16 of pregnant pigs. In sheep serum, expression of miR-23b, miR-30d, and miR-379 was found to be at greater levels in exosomes obtained from the umbilical vein on gestational day of 90 (Cleys et al., 2014). However, in line with the expression of miR-23b in pregnancy, our study detected higher expression levels of oar-miR-23b as early as day 12 of pregnancy compared to C12. In addition, our unpublished data showed that oar-miR-23b tended to be higher in P16 than C16 in the endometrium. We also showed that oar-miR-30d and oar-miR-379-5p were higher in P16 than in C16. Because the capability to detect miRNAs for endometrium has enabled us to reveal pregnancy-related markers (Kose et al., 2022), it is intriguing to monitor common miRNAs for conceptuses and maternal bloodstream. As such, broadly conserved miRNAs (Let-7b) have been detected at different stages of pregnancy in trophoblast (Liu et al., 2015) and serum (Ioannidis and Donadeu, 2017). We demonstrated that let-7i and let-7b were differentially expressed between C12 vs. P12 and C16 vs. P16. Likewise, as identified higher expression of miR-29a in cattle maternal blood on day 39 of pregnancy (Sánchez et al., 2021), conserved miR-29 family members, oar-miR-29a and oar-miR-29b, were found to be expressed in C16 vs. P16 and C22 vs. P22 in ovine plasma, respectively.
The gene targets of differentially expressed miRNA are underrepresented in common categories (inflammatory response (GO:0006954), defense response (GO:0006952), and regulation of immune response (GO:0050776)) relating to immune responses in functional interaction of network analysis of module 1 and 3, meaning that the pregnancy-stimulated changes require both activation and suppression of immune functions through circulating miRNAs (Bidarimath et al., 2014) Among them, CD80 and CD86, costimulatory molecules, involved in signaling in T cell activation (Gool et al., 1996) and upregulated during early pregnancy in the cattle endometrium (Kamat et al., 2016). We showed that CD80 and CD86 was modulated by oar-miR-329a-5p, oar-miR-23b, oar-miR-654-5p, oar-miR-29b, and oar-miR-29a. In similar lines to our results, the miR-29 family has been implied in the regulation of the immune system (Liston et al., 2012) and detected as a potential biomarker for adverse pregnancy outcomes (Deng et al., 2020; Sørensen et al., 2021). As determined in our study, miR-19b and miR-29a were potential candidates for pregnancy detection in cows (Ono et al., 2022). miR-19b was also detected in ovine extracellular vesicles of cyclic and pregnant uterine luminal fluid on day 14 (Burns et al., 2014). In our unpublished data, as in ovine plasma, we found that miR-19b and miR-29a were differentially expressed in C16 vs. P16 and C22 vs. P22 in the ovine endometrium. In module 3, we illuminated that differentially expressed miRNAs targeted a couple of hub genes, which are interleukins, in other words, proinflammatory cytokines (TNF and IL6). oar-miR-19b between C12 and P12 targeted TNF, which plays an indispensable role in very early pregnancy. On the other hand, miRNAs may directly regulate the expression of interferon tau (IFNT)-stimulated genes (ISGs) in peripheral blood cells as they are secreted into the uterine cavity and partially enter the blood circulation directly. As such, CXCL10, classical type I IFN-stimulated genes, was among the hub gene in module 1 and targeted by oar-let-7b and oar-let-7i. In addition, we detected that miRNAs targeted toll-like receptors (TLRs) that are part of the innate immune system because they have roles in the stimulation of the acquired immune system (Akira and Takeda, 2004; Atli et al., 2018). TLR2 and TLR4 in module 1 enriched in (GO:0045087) innate immune response, targeted by oar-miR-30d, oar-miR-758-3p, and oar-miR-654-5p.
Module 2 showed that targets (ADIPOQ, BMP4, CTNNB1, FGF10, HNF4A, IGF2, SMAD4, WNT2) were related to functional GOs that may be involved in early pregnancy because it was enriched in (GO:0009790) embryo development and (GO:0048598) embryonic morphogenesis. CTNNB1 was one of hub genes targeted by oar-miR-218a, oar-miR-543-3p, oar-miR-1185-3p, and oar-miR-19b. Wnt/β-catenin signaling pathway has an important function in early embryogenesis (Liu et al., 2016), that β-catenin-regulated adhesion is essential for successful preimplantation of embryo development in mice (Messerschmidt et al., 2016). In line with gene expression, the Wnt/β-catenin signaling pathway requires strict control of miRNA regulation in embryo development (Stepicheva et al., 2015; Caronia-Brown et al., 2016). As discussed above, the development process occurring during embryonic development requires branching morphogenesis of organs and tissues. Our study revealed that target genes were enriched in (GO:0061138) morphogenesis of a branching epithelium and (GO:0060429) epithelium development. Of these, BMP4 was one of the early developmental genes expressed in the epithelium (Warburton et al., 2005; Geng et al., 2011) and reported post-transcriptionally regulated by miRNAs (Spruce et al., 2010; Battista et al., 2013). Accordingly, we showed that WNT2 and BMP4, other hub genes, were targeted by the same miRNAs (oar-miR-30d, oar-miR-29a). These results were corroborative with those circulating miRNAs that may play a functional role in early pregnancy to modulate embryonic development.
Although the inconsistency in the expression of oar-miR-1185-3p between microarray and RT-PCR emerged, miRNA expression of oar-miR-218a is consistent with microarray results between C16 and P16. This is consistent with reports that oar-miR-218a has been recently reported to be an early pregnancy marker of extracellular vesicles in ovine serum (Sun et al., 2021). Also, serum and plasma identification of miR-218 expression was associated with pregnancy disorder (Lu et al., 2017; Akgör et al., 2020). The limitations of this study were that due to the lack of compatible miRNA expression probes with ewe, we were unable to confirm more miRNA markers using RT-PCR.
In our study, using microarray and qPCR profiling in ovine, we discovered for the first time changes in the levels of miRNAs in plasma during early pregnancy. Differently expressed miRNA profiles in the ovine plasma during the estrus cycle and early pregnancy indicate that miRNAs may function as promising candidates for diagnosis in early ovine pregnancy. More specifically, predicted target genes in modules showed different functional GO such as (GO:0003006) developmental process involved in reproduction, (GO:0009790) embryo development, and (GO:0002684) regulation of immune system process that circulating miRNAs may enable the identification of networks to reveal the molecular basis of early events during pregnancy in sheep.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: EMBL-EBI ArrayExpress, E-MTAB-11709.
The animal study was reviewed and approved by the Bahri Dagdas International Agricultural Research Institute Ethical Research Committee (Number: 29/01/2016−49-7).
MH, MKo, and MA designed the study. MKo, MKi, MSK, SD, MA, and MH collected the data. MKo, MH, and MA analyzed the data. MKo, MKi, MSK, IA, MH, SD, and MA wrote the manuscript. All authors approved the final version of the manuscript and accepted the submission.
The study was financially funded by Scientific and Technological Research Council of Turkey (TUBITAK) projects (214O643) to MK. This study was partially submitted to the Proceedings of the 22nd Annual Conference of the European Society for Domestic Animal Reproduction (ESDAR), Cordoba, Spain, 27–29 September 2018.
The authors thank Dr. Aydin Guzeloglu (Department of Biomedical Sciences, College of Veterinary Medicine) for the support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.929477/full#supplementary-material
Akgör, U., Ayaz, L., and Çayan, F. (2020). Expression levels of maternal plasma microRNAs in preeclamptic pregnancies, J. Obstet. Gynaecol. 41, 910. doi:10.1080/01443615.2020.1820465
Akira, S., and Takeda, K. (2004). Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511. doi:10.1038/NRI1391
Alak, I., Hitit, M., Kose, M., Kaya, M. S., Ucar, E. H., Atli, Z., et al. (2020). Relative abundance and localization of interferon-stimulated gene 15 mRNA transcript in intra- and extra-uterine tissues during the early stages of pregnancy in sheep. Anim. Reprod. Sci. 216, 106347. doi:10.1016/J.ANIREPROSCI.2020.106347
Atli, M. O., Kose, M., Hitit, M., Kaya, M. S., and Bozkaya, F. (2018). Expression patterns of Toll-like receptors in the ovine corpus luteum during the early pregnancy and prostaglandin F2α-induced luteolysis. Theriogenology 111, 25–33. doi:10.1016/J.THERIOGENOLOGY.2018.01.010
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi:10.1016/S0092-8674(04)00045-5
Battista, M., Musto, A., Navarra, A., Minopoli, G., Russo, T., and Parisi, S. (2013). miR-125b regulates the early steps of ESC differentiation through Dies1 in a TGF-independent manner. Int. J. Mol. Sci. 14, 13482–13496. doi:10.3390/ijms140713482
Bazer, F. W., Song, G., Kim, J., Dunlap, K. A., Satterfield, M. C., Johnson, G. A., et al. (2012). Uterine biology in pigs and sheep. J. Anim. Sci. Biotechnol. 313, 23–21. doi:10.1186/2049-1891-3-23
Bidarimath, M., Khalaj, K., Wessels, J. M., and Tayade, C. (2014). MicroRNAs, immune cells and pregnancy. Cell. Mol. Immunol. 11, 538–547. doi:10.1038/CMI.2014.45
Bidarimath, M., Lingegowda, H., Miller, J. E., Koti, M., and Tayade, C. (2021). Insights into extracellular vesicle/exosome and miRNA mediated Bi-directional communication during porcine pregnancy. Front. Vet. Sci. 8, 654064. doi:10.3389/FVETS.2021.654064
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi:10.1093/bioinformatics/btp101
Brooks, K., Burns, G. W., Moraes, J. G. N., and Spencer, T. E. (2016). Analysis of the uterine epithelial and conceptus transcriptome and luminal fluid proteome during the peri-implantation period of pregnancy in sheep. Biol. Reprod. 95, 88–17. doi:10.1095/BIOLREPROD.116.141945
Burns, G., Brooks, K., Wildung, M., Navakanitworakul, R., Christenson, L. K., and Spencer, T. E. (2014). Extracellular vesicles in luminal fluid of the ovine uterus. PLoS One 9, e90913. doi:10.1371/JOURNAL.PONE.0090913
Cameron, A., da Silveira, J. C., Bouma, G., and Bruemmer, J. E. (2011). Evaluation of exosomes containing miRNA as an indicator of pregnancy status in the mare. J. Equine Vet. Sci. 31, 314–315. doi:10.1016/J.JEVS.2011.03.148
Caronia-Brown, G., Anderegg, A., and Awatramani, R. (2016). Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 11, 9–15. doi:10.1186/s13064-016-0065-y
Chakrabarty, A., Tranguch, S., Daikoku, T., Jensen, K., Furneaux, H., and Dey, S. K. (2007). MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. U. S. A. 104, 15144–15149. doi:10.1073/PNAS.0705917104
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11. doi:10.1186/1752-0509-8-S4-S11
Cleys, E. R., Halleran, J. L., Mcwhorter, E., Hergenreder, J., Enriquez, V. A., da Silveira, J. C., et al. (2014). Identification of microRNAs in exosomes isolated from serum and umbilical cord blood, as well as placentomes of gestational day 90 pregnant sheep. Mol. Reprod. Dev. 81, 983–993. doi:10.1002/MRD.22420
De Bem, T. H. C., Da Silveira, J. C., Sampaio, R. V., Sangalli, J. R., Oliveira, M. L. F., Ferreira, R. M., et al. (2017). Low levels of exosomal-miRNAs in maternal blood are associated with early pregnancy loss in cloned cattle. Sci. Rep. 71 7, 14319–14411. doi:10.1038/s41598-017-14616-1
Deng, L., Huang, Y., Li, L., Chen, H., and Su, J. (2020). Serum miR-29a/b expression in gestational diabetes mellitus and itsinfluence on prognosis evaluation. J. Int. Med. Res. 48, 300060520954763. doi:10.1177/0300060520954763
El-Shorafa, H. M., and Sharif, F. A. (2016). Dysregulation of micro-RNA contributes to the risk of unexplained recurrent pregnancy loss. Int. J. Reprod. Contracept. Obstet. Gynecol. 2. doi:10.5455/2320-1770.ijrcog20130914
Gebremedhn, S., Salilew-Wondim, D., Hoelker, M., Held-Hoelker, E., Neuhoff, C., Tholen, E., et al. (2018). Exploring maternal serum microRNAs during early pregnancy in cattle. Theriogenology 121, 196–203. doi:10.1016/J.THERIOGENOLOGY.2018.08.020
Geng, Y., Dong, Y., Yu, M., Zhang, L., Yan, X., Sun, J., et al. (2011). Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc. Natl. Acad. Sci. U. S. A. 108, 7058–7063. doi:10.1073/PNAS.1007293108
Gool, S. W., Vandenberghe, P., Boer, M. de, and Ceuppens, J. L. (1996). CD80, CD86 and CD40 provide accessory signals in a multiple-step T-cell activation model. Immunol. Rev. 153, 47–83. doi:10.1111/J.1600-065X.1996.TB00920.X
Guzeloglu, A., Atli, M., Kurar, E., Kayis, S., Semacan, A., Aslan, S., et al. (2013). Preliminary expression analysis of global microRNA and biogenesis pathway in equine endometrium during the estrous cycle and early pregnancy: OC 8.1. Reprod. Domest. Anim. 48.
Hitit, M., Atli, M., Kurar, E., Kose, M., Guzeloglu, A., Kaya, M., et al. (2013). Investigation of microRNA biogenesis at mRNA level in bovine corpus luteum and endometrium during the oestrous cycle: P93. Reprod. Domest. Anim. 48.
Hitit, M., Kurar, E., and Güzeloğlu, A. (2015). MikroRNA biyogenezi. Atatürk üniversitesi vet. Bilim. Derg. 10. doi:10.17094/avbd.35776
Hitit, M., and Kurar, E. (2017). Putative role of micro - RNA s i n female reproductive tract. Open Access J. Vet. Sci. Res. 2, 1–5. doi:10.23880/OAJVSR-16000131
Ioannidis, J., and Donadeu, F. X. (2017). Changes in circulating microRNA levels can be identified as early as day 8 of pregnancy in cattle. PLoS One 12, e0174892. doi:10.1371/JOURNAL.PONE.0174892
Ioannidis, J., and Donadeu, F. X. (2016). Circulating miRNA signatures of early pregnancy in cattle. BMC Genomics 17, 184. doi:10.1186/S12864-016-2529-1
Kaczmarek, M. M., Najmula, J., Guzewska, M. M., and Przygrodzka, E. (2020). MiRNAs in the peri-implantation period: contribution to embryo-maternal communication in pigs. Int. J. Mol. Sci. 21, E2229. doi:10.3390/ijms21062229
Kamat, M. M., Vasudevan, S., Maalouf, S. A., Townson, D. H., Pate, J. L., and Ott, T. L. (2016). Changes in myeloid lineage cells in the uterus and peripheral blood of dairy heifers during early pregnancy. Biol. Reprod. 95, 68–12. doi:10.1095/biolreprod.116.141069
Kaya, M. S., Kose, M., Guzeloglu, A., Kıyma, Z., and Atli, M. O. (2017). Early pregnancy-related changes in toll-like receptors expression in ovine trophoblasts and peripheral blood leukocytes. Theriogenology 93, 40–45. doi:10.1016/J.THERIOGENOLOGY.2017.01.031
Kiyma, Z., Hitit, M., Ozel, C., Sen, G., Kose, M., Guzeloglu, A., et al. (2015). Preliminary investigation of microRNAs in ovine endometrium: P70. Reprod. Domest. Anim. 50. doi:10.1111/rda.12581
Köse, M., Kaya, M. S., and Atli, M. O. (2016c). Effect of exogenous progesterone and gonadotropin-releasing hormone application on maintenance of pregnancy in early pregnant ewes after prostaglandin F2 alpha injection. EurasianJVetSci. 32, 26–29. doi:10.15312/EurasianJVetSci.2016115446
Kose, M., Kaya, M. S., Aydilek, N., Kucukaslan, I., Bayril, T., Bademkiran, S., et al. (2016a). Expression profile of interferon tau–stimulated genes in ovine peripheral blood leukocytes during embryonic death. Theriogenology 85, 1161–1166. doi:10.1016/J.THERIOGENOLOGY.2015.11.032
Kose, M., Hitit, M., Kaya, M. S., Kirbas, M., Dursun, S., Alak, I., et al. (2022). Expression pattern of microRNAs in ovine endometrium during the peri-implantation. Theriogenology. doi:10.1016/j.theriogenology.2022.07.015
Kotlabova, K., Doucha, J., and Hromadnikova, I. (2011). Placental-specific microRNA in maternal circulation--identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 89, 185–191. doi:10.1016/J.JRI.2011.02.006
Krawczynski, K., Bauersachs, S., Reliszko, Z. P., Graf, A., and Kaczmarek, M. M. (2015). Expression of microRNAs and isomiRs in the porcine endometrium: Implications for gene regulation at the maternal-conceptus interface. BMC Genomics 16, 906–919. doi:10.1186/s12864-015-2172-2
Lim, H.-J., Kim, H. J., Lee, J. H., Lim, D. H., Son, J. K., Kim, E.-T., et al. (2021). Identification of plasma miRNA biomarkers for pregnancy detection in dairy cattle. J. Anim. Reprod. Biotechnol. 36, 35–44. doi:10.12750/JARB.36.1.35
Liston, A., Papadopoulou, A. S., Danso-Abeam, D., and Dooley, J. (2012). MicroRNA-29 in the adaptive immune system: Setting the threshold. Cell. Mol. Life Sci. 69, 3533–3541. doi:10.1007/S00018-012-1124-0
Liu, R., Wang, M., Su, L., Li, X., Zhao, S., and Yu, M. (2015). The expression pattern of micrornas and the associated pathways involved in the development of porcine placental folds that contribute to the expansion of the exchange surface area. Biol. Reprod. 93, 62. doi:10.1095/biolreprod.114.126540
Liu, Z., Guo, J., Wang, Y., Weng, Z., Huang, B., Yu, M. K., et al. (2016). CFTR- β -catenin interaction regulates mouse embryonic stem cell differentiation and embryonic development. Cell Death Differ. 241 24, 98–110. doi:10.1038/cdd.2016.118
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lou, W., Liu, J., Ding, B., Chen, D., Xu, L., Ding, J., et al. (2019). Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC 11 medical and health sciences 1112 oncology and carcinogenesis 06 biological Sciences 0604 Genetics. J. Transl. Med. 17, 1–14. doi:10.1186/S12967-018-1761-7/FIGURES/10
Lu, Q., Yan, Q., Xu, F., Li, Y., Zhao, W., Wu, C., et al. (2017). MicroRNA-873 is a potential serum biomarker for the detection of ectopic pregnancy. Cell. Physiol. biochem. 41, 2513–2522. doi:10.1159/000475946
Messerschmidt, D., De Vries, W. N., Lorthongpanich, C., Balu, S., Solter, D., and Knowles, B. B. (2016). β-catenin-mediated adhesion is required for successful preimplantation mouse embryo development. Development 143, 1993–1999. doi:10.1242/DEV.133439
Miura, K., Miura, S., Yamasaki, K., Higashijima, A., Kinoshita, A., Yoshiura, K. I., et al. (2010). Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 56, 1767–1771. doi:10.1373/CLINCHEM.2010.147660
Ono, K., Okamoto, S., Ninomiya, C., Toji, N., Kanazawa, T., Ishiguro-Oonuma, T., et al. (2022). Analysis of circulating microRNA during early gestation in Japanese black cattle. Domest. Anim. Endocrinol. 79, 106706. doi:10.1016/J.DOMANIEND.2021.106706
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. doi:10.1093/NAR/30.9.E36
Reliszko, Z. P., Gajewski, Z., and Kaczmarek, M. M. (2017). Signs of embryo-maternal communication: miRNAs in the maternal serum of pregnant pigs. Reproduction 154, 217–228. doi:10.1530/REP-17-0224
Sánchez, J. M., Gómez-Redondo, I., Browne, J. A., Planells, B., Gutiérrez-Adán, A., and Lonergan, P. (2021). MicroRNAs in amniotic fluid and maternal blood plasma associated with sex determination and early gonad differentiation in cattle. Biol. Reprod. 105, 345–358. doi:10.1093/BIOLRE/IOAB079
Shi, S., Tan, Q., Liang, J., Cao, D., Wang, S., Liang, J., et al. (2021). Placental trophoblast cell-derived exosomal microRNA-1290 promotes the interaction between endometrium and embryo by targeting LHX6, Mol. Ther. - Nucleic Acids 26, 760–772. doi:10.1016/J.OMTN.2021.09.009/ATTACHMENT/3DEC40E6-E6C4-4605-A186-3F0015C9DD55/MMC1
Smith, M. D., Pillman, K., Jankovic-Karasoulos, T., McAninch, D., Wan, Q., Bogias, K. J., et al. (2021). Large-scale transcriptome-wide profiling of microRNAs in human placenta and maternal plasma at early to mid gestation. RNA Biol. 18, 507–520. doi:10.1080/15476286.2021.1963105/SUPPL_FILE/KRNB_A_1963105_SM3313
Song, Y., An, X., Zhang, L., Fu, M., Peng, J., Han, P., et al. (2015). Identification and profiling of microRNAs in goat endometrium during embryo implantation. PLoS One 10, e0122202. doi:10.1371/JOURNAL.PONE.0122202
Sørensen, A. E., van Poppel, M. N. M., Desoye, G., Damm, P., Simmons, D., Jensen, D. M., et al. (2021). The predictive value of miR-16, -29a and -134 for early identification of gestational diabetes: a nested analysis of the dali cohort. Cells 10, 170. doi:10.3390/CELLS10010170
Spencer, T. E., Johnson, G. A., Bazer, F. W., and Burghardt, R. C. (2004). Implantation mechanisms: insights from the sheep. Reproduction 128, 657–668. doi:10.1530/REP.1.00398
Spruce, T., Pernaute, B., Di-Gregorio, A., Cobb, B. S., Merkenschlager, M., Manzanares, M., et al. (2010). An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev. Cell 19, 207–219. doi:10.1016/J.DEVCEL.2010.07.014
Stepicheva, N., Nigam, P. A., Siddam, A. D., Fu Peng, C., and Song, J. L. (2015). microRNAs regulate β-catenin of the Wnt signaling pathway in early sea urchin development. Dev. Biol. 402, 127–141. doi:10.1016/j.ydbio.2015.01.008
Sun, Y., Xu, M., Gao, R., Xie, S., Sun, X., He, J., et al. (2021). Identification of differentially expressed miRNAs in serum extracellular vesicles (EVs) of Kazakh sheep at early pregnancy. Reprod. Domest. Anim. 56, 713–724. doi:10.1111/RDA.13910
Tan, Q., Shi, S., Liang, J., Zhang, X., Cao, D., and Wang, Z. (2020). MicroRNAs in small extracellular vesicles indicate successful embryo implantation during early pregnancy. Cells. doi:10.3390/CELLS90306459
Warburton, D., Bellusci, S., De Langhe, S., Del Moral, P. M., Fleury, V., Mailleux, A., et al. (2005). Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr. Res. 577 57, 26R-37R–37R. doi:10.1203/01.PDR.0000159570.01327.ED
Wb, N. (2016). Non-coding RNAs in uterine development, function and disease. Adv. Exp. Med. Biol. 886, 171–189. doi:10.1007/978-94-017-7417-8_9
Yan, C., Lv, H., Peng, Z., Yang, D., Shen, P., Yu, J., et al. (2021). Analysis of miRNA expression changes in bovine endometrial stromal cells treated with lipopolysaccharide. Theriogenology 167, 85–93. doi:10.1016/J.THERIOGENOLOGY.2021.03.012
Zang, X., Zhou, C., Wang, W., Gan, J., Li, Y., Liu, D., et al. (2021). Differential MicroRNA expression involved in endometrial receptivity of goats. Biomolecules 202111, 472. doi:10.3390/BIOM11030472
Zhao, L., Haili, Y., Rong, Z., Haoyu, T., Cheng, W., et al. (2021). Screening and identification of microRNAs from plasma-derived extracellular vesicles (EVs) of dazu black goat (Capra hircus) in early pregnant stages. Gene 790, 145706. doi:10.1016/J.GENE.2021.145706
Keywords: ovine, plasma, microRNA, circulating, early pregnancy, expression
Citation: Hitit M, Kose M, Kaya MS, Kırbas M, Dursun S, Alak I and Atli MO (2022) Circulating miRNAs in maternal plasma as potential biomarkers of early pregnancy in sheep. Front. Genet. 13:929477. doi: 10.3389/fgene.2022.929477
Received: 26 April 2022; Accepted: 18 July 2022;
Published: 17 August 2022.
Edited by:
Naseer A. Kutchy, Children’s National Hospital, United StatesCopyright © 2022 Hitit, Kose, Kaya, Kırbas, Dursun, Alak and Atli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mustafa Hitit, dmV0ZHJtdXN0YWZhaGl0aXRAZ21haWwuY29t; Mehmet Osman Atli, bW9hdGxpQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.