- Department of Neuroscience of Disease, Brain Research Institute, Niigata University, Niigata, Japan
Animal models have been used to model human diseases, and among them, small fishes have been highlighted for their usefulness in various ways, such as the low cost of maintenance, ease of genetic modification, small size for easy handling, and strength in imaging studies due to their relative transparency. Recently, the use of turquoise killifish, Nothobranchius furzeri, which is known to exhibit various aging phenotypes in a short period, has attracted attention in research on aging and age-related diseases. However, when using animal models, it is important to keep their genetic background and interspecies differences in mind for translating them into human diseases. In this article, we obtained the gene symbols of protein-coding genes of turquoise killifish, medaka, zebrafish, and humans from NCBI datasets and extracted common shared genes among four species to explore the potential of interspecies translational research and to apply small fish models for human age-related disorders. Common shared protein-coding genes were analyzed with the Reactome Pathway Database to determine the coverage of these genes in each pathway in humans. We applied common shared genes to the Orphanet database to establish a list of human diseases that contain common shared genes among the four species. As examples, the senescence-related pathways and some pathways of human age-related diseases, such as Alzheimer’s disease, Parkinson’s disease, frontotemporal dementia, nonalcoholic fatty liver disease, progeria, hepatocellular carcinoma, and renal cell carcinoma, were extracted from the curated pathway and disease list to discuss the further utility of fish models for human age-related disorders.
Introduction
Zebrafish produce more than 100 eggs per oviposition, and medaka and turquoise killifish, Nothobranchius furzeri, also produce many but fewer eggs than zebrafish (Furutani-Seiki and Wittbrodt, 2004; Skinner and Watt, 2007; Polacik et al., 2016). Because small fish have a larger number of fertilized eggs than mice and because spawning and fertilization take place outside the parent’s body, it is very easy to microinject the desired gene-editing factor into the egg. The recent emergence of the CRISPR–Cas9 technique (Hwang et al., 2013; Ablain et al., 2015; Varshney et al., 2015) allows us to quickly knock out or edit specific genes in various organisms compared with zinc-finger nucleases (ZFNs) (Doyon et al., 2008; Meng et al., 2008) or transcription activator-like effectors (TALENs) (Huang et al., 2011; Sander et al., 2011). The CRISPR–Cas9 system also enables the knock-in of a specific DNA sequence mediated by homology-directed repair (HDR) or other mechanisms (Irion et al., 2014; Prykhozhij et al., 2018). These gene-editing techniques are applicable for zebrafish, medaka, and turquoise killifish. Because of the relatively hard chorion of turquoise killifish compared with zebrafish and medaka, it might be difficult to perform microinjection into the eggs of turquoise killifish. However, the methods of microinjection into the egg of turquoise killifish are being improved (Valenzano et al., 2011; Hartmann and Englert, 2012), and the use of genetic engineering in turquoise killifish has been expanding (Allard et al., 2013; Harel et al., 2016).
Turquoise killifish is a small fish species native to Africa, primarily Mozambique, that lives in ponds, swamps, and puddles (Poeschla and Valenzano, 2020). There is a short rainy season and a long dry season when adult fish cannot survive because the water dries up. Although adult fish cannot survive, turquoise killifish survive as a species through drought-resistant eggs laid in the soil, which can hatch during the future rainy season. In such a life cycle, turquoise killifish may not be subjected to a positive selection pressure to acquire various antiaging properties (Cui et al., 2019). Therefore, the lifespan of turquoise killifish is approximately four to six months, which is very short compared to zebrafish and medaka (Polacik et al., 2016). Around the age of three months, the turquoise killifish exhibits several signs of aging, such as organ atrophy, spine curvature, and increased levels of senescence-associated beta-galactosidase (Genade et al., 2005; Valenzano et al., 2006; Harel et al., 2015). Given their aging phenotypes, we examined the central nervous system of turquoise killifish and found that this fish showed age-dependent degeneration of dopaminergic and noradrenergic neurons, with gradually progressing alpha-synuclein pathology (Matsui et al., 2019). These pathological findings are similar to those of human Parkinson’s disease, and very interestingly, genetic depletion of alpha-synuclein with the CRISPR–Cas9 system mitigates neurodegeneration (Matsui et al., 2019). These findings suggest that alpha-synuclein can be a causative protein in the pathogenesis of Parkinson’s disease, and turquoise killifish could be a useful tool for unveiling the mechanisms of Parkinson’s disease and hopefully other age-related diseases.
To further utilize the potential of translational research of such fish models, it is important to know the genetic background of each small fish compared to those of humans and other small fishes. In this article, we analyzed the genetic backgrounds of turquoise killifish, medaka, zebrafish, and humans (Reichard et al., 2009; Kirchmaier et al., 2015; Valenzano et al., 2015; Delomas and Dabrowski, 2018; Poeschla and Valenzano, 2020) and explored the utility of small fish for translational research of human age-related disorders.
Methods
Finding the Common Shared Genes
Gene symbols of each species were extracted from protein-coding genes in the NCBI datasets (https://www.ncbi.nlm.nih.gov/datasets/; accessed on 18th January). The gene symbols were capitalized as normalization to extract the common shared genes by generating a Venn diagram in the exact match manner (https://bioinformatics.psb.ugent.be/webtools/Venn/).
Given another round of whole-genome duplication in teleosts, some genes did not correspond between humans and fishes in a 1:1 manner, and two orthologues could be present in the teleost fishes. To extract such duplicated genes in fishes, we first extracted fish genes that did not overlap with human genes in an exact match manner. Then, the gene symbols ending with A or B were picked up. After depleting the last letter, the genes with equal combinations of the remaining strings were considered to be a pair of duplicated genes. These extracted duplicated genes were examined to determine whether orthologues overlapped with human genes. This procedure was repeated through four species to find additional common shared genes.
Coverage of Common Shared Genes in Each Human Pathway
Common shared genes were applied to the Reactome pathway database (Jassal et al., 2020) to determine the coverage of genes in each human pathway.
Extraction of Orphanet Codes That Contain Common Shared Genes
Metascape (Zhou et al., 2019) was utilized by applying the Orphanet database (https://www.orpha.net/consor/cgi-bin/index.php). The list of ORPHAcodes related to each common shared gene was obtained, and the list was organized according to the list with the index of each ORPHAcode.
Results
Common Shared Genes Among Humans and three Fishes
Approximately 500 million years ago, vertebrates, including humans, experienced a whole-genome duplication in which the genome doubled twice in our ancestors (Ohno, 1970; Dehal and Boore, 2005). Another round of whole-genome duplication occurred in teleosts, including zebrafish, medaka, and turquoise killifish (Chiu et al., 2004; Hoegg et al., 2004; Jaillon et al., 2004). This is one of the most significant genetic differences between humans and fishes. Among these three fishes, sex chromosomes have not been identified in zebrafish. Zebrafish sex determinants remain unclear, but environmental factors are known to affect zebrafish sex determination (Baroiller and D'Cotta, 2001; Orn et al., 2003; Abozaid et al., 2012). Similar to humans, medaka or turquoise killifish sex is determined by XX/XY sex chromosomes (Schartl, 2004; Valenzano et al., 2009; Reichwald et al., 2015), which is another major genomic difference among the four species.
We attempted to extract the gene symbols of each species from public databases, such as the UniProt (UniProt, 2021), Ensembl genome browser (Howe et al., 2021), and NCBI datasets, and found that the NCBI datasets contained the most gene symbols of each species. Therefore, we extracted the gene symbols of each species from the NCBI datasets (https://www.ncbi.nlm.nih.gov/datasets/; accessed on 18 January) based on the protein-coding genes. The numbers of protein-coding genes were 19,671; 29,961; 22,140; and 22,207 for human, zebrafish, medaka, and turquoise killifish, respectively (Table 1; Supplementary Table S1). A Venn diagram was generated by exact matching with gene symbols normalized as strings (https://bioinformatics.psb.ugent.be/webtools/Venn/), and 8,726 genes were found to be common among the four species (Figure 1).
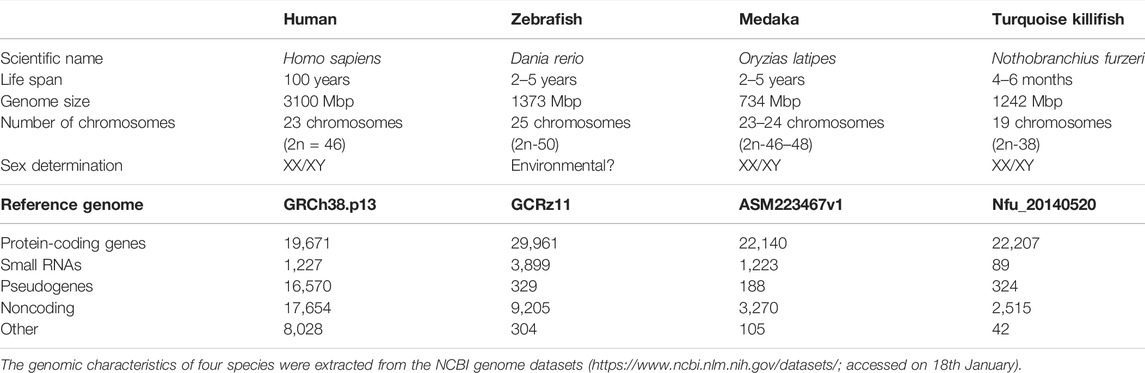
TABLE 1. Lifespans and genomic characteristics of humans, zebrafish, medaka, and turquoise killifish.
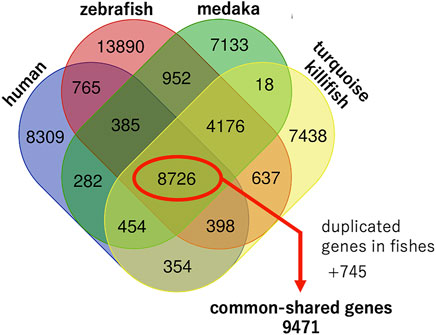
FIGURE 1. Genomic characteristics of humans, zebrafish, medaka, and turquoise killifish. Venn diagram generated by an exact match with gene symbols normalized as strings. In addition to 8,726 common shared genes, duplicated orthologues in fishes were extracted and an additional 745 common shared genes were found. In total, 9,471 common shared genes were found among the four species.
As mentioned previously, teleost fishes experienced an additional round of whole-genome duplication compared to humans, and so, some genes did not correspond one-to-one between humans and fish. In such cases, two orthologues could be present in the teleost fishes. We extracted such duplicated orthologues from fishes and found an additional 745 common shared genes among four species out of a total of 9,471 genes (Supplementary Table S2). A study comparing short- and long-lived turquoise killifish strains showed that long-lived killifish acquired specific SNPs in several aging genes (grn, tnfb, pdgfr, brca1, tp53, bp, ercc6, ghr, irs4, foxo4, myc, egr1, med1, ncor1, polg, gsr, mgat5, tert, hsf1, and hspa9) under positive selection (Valenzano et al., 2015). Among these 19 genes, 16 genes were included in the common shared genes except for brca1, irs4, and tnfb.
Human Pathways and Diseases With Common-Shared Genes
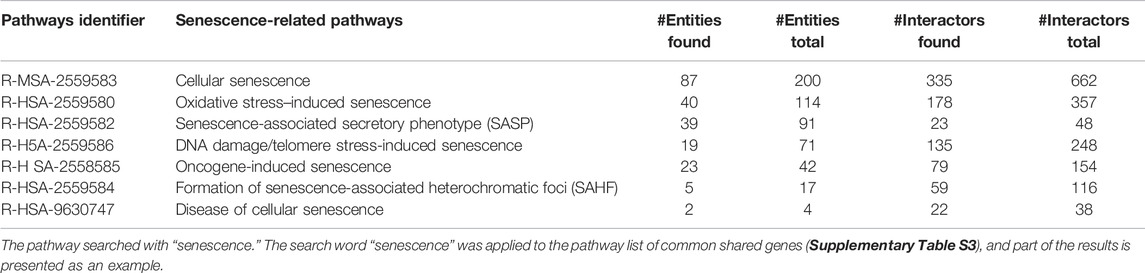
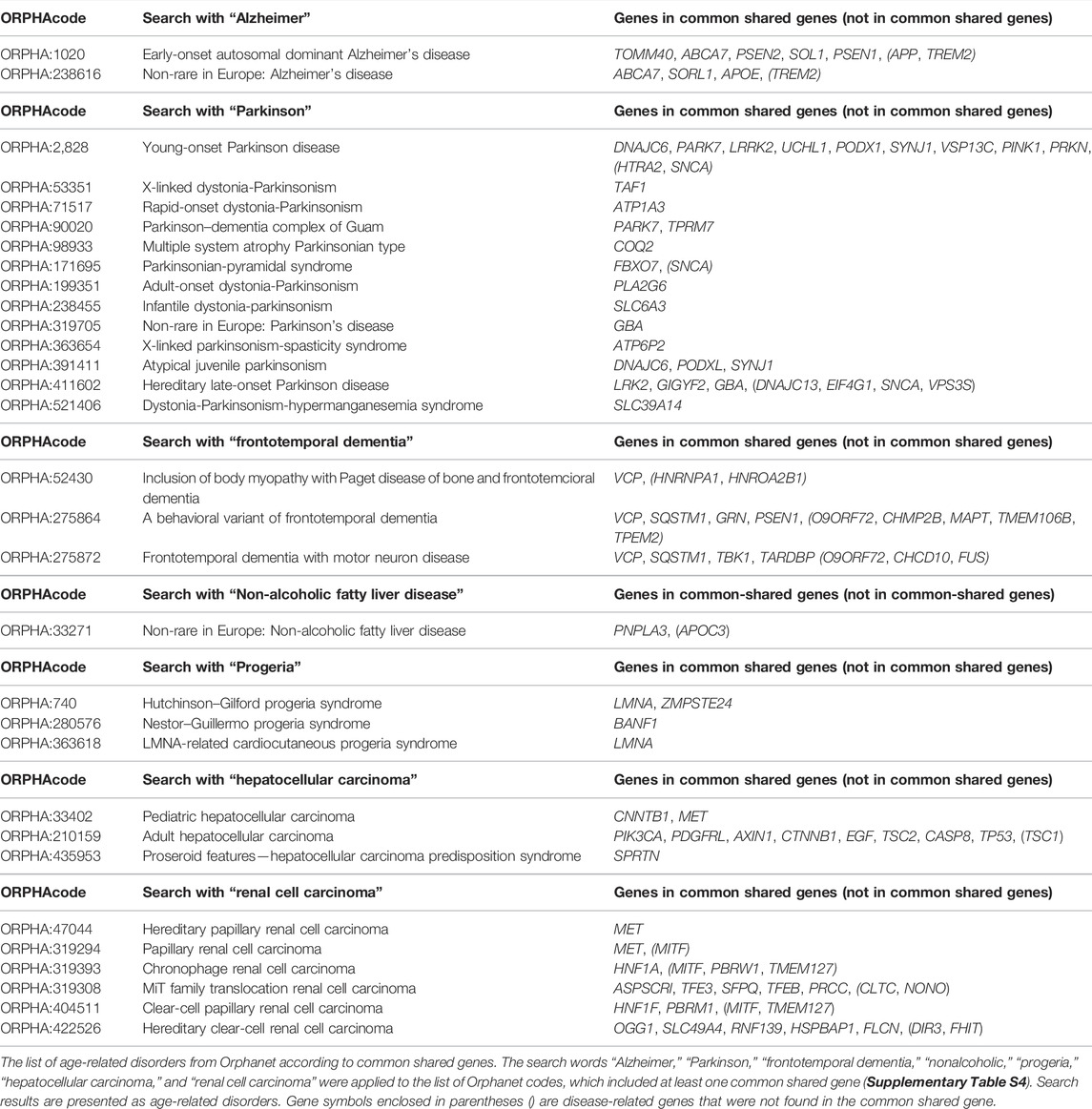
Then, the 9,471 common shared genes were applied to the Reactome Pathway Database (Jassal et al., 2020), a web tool for human pathway analysis, to determine the proportion and number of common shared genes in known human biological pathways. A total of 2,405 pathways were found to be associated with at least one of the common shared genes. Given the list of 2,405 pathways, we extracted some of the senescence-related pathways and noticed that common shared genes covered approximately 30–50% of the genes in the human senescence-related pathways (Table 2). Complete gene lists in each pathway are available in Supplementary Table S3. Next, we applied the 9,471 common shared genes to the Orphanet database (https://www.orpha.net/consor/cgi-bin/index.php) (Weinreich et al., 2008) to establish the human disease list, which included at least one gene from the common shared genes. From this application, 2,677 Orphacodes were extracted. We extracted several human age-related disorders, such as Alzheimer’s disease, Parkinson’s disease, frontotemporal dementia, nonalcoholic fatty liver disease, and progeria, from our Orphanet disease list containing common shared genes. We also extracted hepatocellular carcinoma and renal cell carcinoma because turquoise killifish are known to develop hepatocellular carcinoma and renal cell carcinoma at high rates despite their short lifespan (Di Cicco et al., 2011) (Table 3; Supplementary Table S4).
Based on these lists obtained previously, several age-related disease genes were explored. APOE is known to be associated with Alzheimer’s disease and is located in the lifespan-related loci of the turquoise killifish (Kirschner et al., 2012). APOE was also reported as an aging marker of the short-lived fish N. guentheri (Wang et al., 2014). PARK7 is one of the causative genes of familial Parkinsonism, and polymorphisms in PARK7 were reported to be related to the lifespan of turquoise killifish (Genade and Wilcox, 2021). GRN, one of the causative genes of frontotemporal dementia, is known to be regulated downstream of PARK7 in the context of neuroprotection (Genade and Wilcox, 2021). A common biological pathway linking PINK–PARKIN–PARK7 was reported in the pathogenesis of Parkinson’s disease (van der Merwe et al., 2015), and these three genes were observed among the common shared genes. A search on Orphanet for “Parkinson” seems to cover the common shared genes well, except for hereditary late-onset Parkinson’s disease (ORPHA:411602). Referring to the gene list of turquoise killifish, dnaj13, eif4g1, and vsp35 were found, but the orthologous genes corresponding to human SNCA and human HTRA2 were not found. Although snca was not included in the gene list of turquoise killifish constructed with NCBI datasets, snca is known to be expressed in turquoise killifish (Matsui et al., 2019) and was found in turquoise killifish in the UniProt database (UniProt, 2021). Thus, turquoise killifish possesses most of the Parkinson’s disease-related genes, which could be applied to translational research on Parkinson’s disease. Compared to age-related diseases, common shared genes only covered 30–50% of the genes in senescence-related pathways. This difference in gene coverage may be due to the relatively small number of genes in common or to genomic differences that occur during evolution. In interpreting the effects of aging on the pathogenesis of age-related diseases using fish as a model, it will be necessary to keep in mind that the effects may be species dependent.
Discussion
In this article, we explored the genetic background and common shared genes among humans and three fishes by using the available gene symbol data from the NCBI database (https://www.ncbi.nlm.nih.gov/datasets/). We applied common shared genes to the Reactome pathway database (Jassal et al., 2020) to determine the coverage of genes in human pathways. To determine the involvement of commonly shared genes in human diseases, we utilized the Orphanet database (Weinreich et al., 2008). These genetic background data between humans and fishes are important and worth considering before translating the previous findings of fish models in the context of human disease studies, including aging studies. Additionally, these data allow us to design an experiment that applies a fish model including turquoise killifish in aging studies. Although this kind of knowledge-based approach allows us to observe the genomic landscape from a broad perspective in an interspecies manner, several caveats and limitations exist.
First, in the available database, even among protein-coding genes, fairly many genes are still waiting to be annotated in many species. As observed in turquoise killifish, snca was not obtained from NCBI datasets. A blastp search (Altschul et al., 1990; States and Gish, 1994; Boratyn et al., 2012) of the amino acid sequence of human SNCA based on the turquoise killifish protein dataset yielded the answer “PREDICTED: alpha-synuclein-like” (Supplementary Table S5). This might be the reason why the Snca protein of turquoise killifish has not been annotated and could not be found in the list of gene symbols in the NCBI dataset. There are many gene symbols beginning with LOC, a notation that indicates that a published symbol is not available for this gene, and orthologues have not yet been determined (https://www.ncbi.nlm.nih.gov/books/NBK3840/#genefaq.Conventions). The numbers of gene symbols beginning with LOC are 266; 4,473; 6,924; and 7,332 for human, zebrafish, medaka, and turquoise killifish, respectively. This caveat of a lack of annotation should be kept in mind when attempting to explore genomic information.
Second, pseudogenes, miRNAs, noncoding RNAs, and others would be worth investigating (Esteller, 2011; Cheetham et al., 2020; Statello et al., 2021); we did not explore such RNAs in an interspecies manner in this study. It is often difficult to obtain sufficient insight by comparing such genes in an interspecies manner. Thus, some meaningful information may be overlooked.
Third, whole-genome alignment is suitable for covering the entire genomes of multiple species. However, whole-genome comparison studies among multiple species require long computation times, complex algorithms, and expensive computational resources and are difficult for researchers not familiar with bioinformatics to repeat in a timely manner (Armstrong et al., 2019). If the target gene has already been determined, blastp (Altschul et al., 1990; States and Gish, 1994; Boratyn et al., 2012) is an effective way to evaluate coverage, e-values, and percent identity by using human sequence data as a reference. It should be noted that a gene may have multiple amino acid sequences, and a single amino acid sequence may be annotated to multiple genes in the protein dataset of the reference organism. For example, medaka and turquoise killifish Tp53 amino acid sequences showed higher homology to human TP63 and human TP73 (TP53 family proteins) (Belyi et al., 2010) than to human TP53 (Supplementary Table S6).
Finally, in the disease-related genes, we only focused on Orphanet (Weinreich et al., 2008); genes potentially involved in disease modification were not included. Such regulatory genes would be better to be included when we consider applying an animal model for specific diseases. In addition, there are many variants of uncertain significance (VUS) (Elliott, 2020; Sullivan, 2021) in human genes, and their evaluation is also rapidly progressing (Mahecha et al., 2022; Postel et al., 2022). Thus, the identification of disease-related genes will be increasing, and their significance will be determined in the future.
Given the current situation, timely updates with recent genomics data would be ideal to translate and interpret the data and phenomena with model animals. A goal should be to establish a platform where anyone can easily compare genomes between species, even if they cannot write the code for analysis.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/datasets/.
Author Contributions
ED analyzed data, conducted bioinformatic analysis, and co-wrote the manuscript. HM supervised the project, analyzed results, and co-wrote the manuscript. All authors have approved the final version of the manuscript.
Funding
This work was supported by grants from AMED (Grant Numbers JP22gm6110028 (HM)), JSPS KAKENHI (Grant Numbers JP 22484842, JP 18955907 (HM)), and JST (Moonshot R&D) (Grant Number JPMJMS 2024) (HM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge Ms. Shinano Kobayashi and Ms. Noriko Matsui for participating in helpful discussions and providing continuous support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.928597/full#supplementary-material
References
Ablain, J., Durand, E. M., Yang, S., Zhou, Y., and Zon, L. I. (2015). A CRISPR/Cas9 Vector System for Tissue-specific Gene Disruption in Zebrafish. Dev. Cell. 32 (6), 756–764. doi:10.1016/j.devcel.2015.01.032
Abozaid, H., Wessels, S., and Hörstgen-Schwark, G. (2012). Elevated Temperature Applied during Gonadal Transformation Leads to Male Bias in Zebrafish (Danio rerio). Sex. Dev. 6 (4), 201–209. doi:10.1159/000336297
Allard, J. B., Kamei, H., and Duan, C. (2013). Inducible Transgenic Expression in the Short-Lived fishNothobranchius Furzeri. J. Fish. Biol. 82 (5), 1733–1738. doi:10.1111/jfb.12099
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215 (3), 403–410. doi:10.1016/S0022-2836(05)80360-2
Armstrong, J., Fiddes, I. T., Diekhans, M., and Paten, B. (2019). Whole-Genome Alignment and Comparative Annotation. Annu. Rev. Anim. Biosci. 7, 41–64. doi:10.1146/annurev-animal-020518-115005
Baroiller, J. F., and D'Cotta, H. (2001). Environment and Sex Determination in Farmed Fish. Comp. Biochem. Physiology Part C Toxicol. Pharmacol. 130 (4), 399–409. doi:10.1016/s1532-0456(01)00267-8
Belyi, V. A., Ak, P., Markert, E., Wang, H., Hu, W., Puzio-Kuter, A., et al. (2010). The Origins and Evolution of the P53 Family of Genes. Cold Spring Harb. Perspect. Biol. 2 (6), a001198. doi:10.1101/cshperspect.a001198
Boratyn, G. M., Schäffer, A. A., Agarwala, R., Altschul, S. F., Lipman, D. J., and Madden, T. L. (2012). Domain Enhanced Lookup Time Accelerated BLAST. Biol. Direct 7, 12. doi:10.1186/1745-6150-7-12
Cheetham, S. W., Faulkner, G. J., and Dinger, M. E. (2020). Overcoming Challenges and Dogmas to Understand the Functions of Pseudogenes. Nat. Rev. Genet. 21 (3), 191–201. doi:10.1038/s41576-019-0196-1
Chiu, C.-h., Dewar, K., Wagner, G. P., Takahashi, K., Ruddle, F., Ledje, C., et al. (2004). Bichir HoxA Cluster Sequence Reveals Surprising Trends in Ray-Finned Fish Genomic Evolution. Genome Res. 14 (1), 11–17. doi:10.1101/gr.1712904
Cui, R., Medeiros, T., Willemsen, D., Iasi, L. N. M., Collier, G. E., Graef, M., et al. (2019). Relaxed Selection Limits Lifespan by Increasing Mutation Load. Cell. 178 (2), 385–399. doi:10.1016/j.cell.2019.06.004
Dehal, P., and Boore, J. L. (2005). Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 3 (10), e314. doi:10.1371/journal.pbio.0030314
Delomas, T. A., and Dabrowski, K. (2018). Larval Rearing of Zebrafish at Suboptimal Temperatures. J. Therm. Biol. 74, 170–173. doi:10.1016/j.jtherbio.2018.03.017
Di Cicco, E., Tozzini, E. T., Rossi, G., and Cellerino, A. (2011). The Short-Lived Annual Fish Nothobranchius Furzeri Shows a Typical Teleost Aging Process Reinforced by High Incidence of Age-dependent Neoplasias. Exp. Gerontol. 46 (4), 249–256. doi:10.1016/j.exger.2010.10.011
Doyon, Y., McCammon, J. M., Miller, J. C., Faraji, F., Ngo, C., Katibah, G. E., et al. (2008). Heritable Targeted Gene Disruption in Zebrafish Using Designed Zinc-Finger Nucleases. Nat. Biotechnol. 26 (6), 702–708. doi:10.1038/nbt1409
Elliott, A. M. (2020). Genetic Counseling and Genome Sequencing in Pediatric Rare Disease. Cold Spring Harb. Perspect. Med. 10 (3), a036632. doi:10.1101/cshperspect.a036632
Esteller, M. (2011). Non-coding RNAs in Human Disease. Nat. Rev. Genet. 12 (12), 861–874. doi:10.1038/nrg3074
Furutani-Seiki, M., and Wittbrodt, J. (2004). Medaka and Zebrafish, an Evolutionary Twin Study. Mech. Dev. 121 (7-8), 629–637. doi:10.1016/j.mod.2004.05.010
Genade, T., Benedetti, M., Terzibasi, E., Roncaglia, P., Valenzano, D. R., Cattaneo, A., et al. (2005). Annual Fishes of the Genus Nothobranchius as a Model System for Aging Research. Aging Cell. 4 (5), 223–233. doi:10.1111/j.1474-9726.2005.00165.x
Genade, T., and Wilcox, D. A. (2021). Nothobranchius Fish: An Untapped Resource for Studying Aging-Related Neurodegeneration. Cas 14 (2), 78–93. doi:10.2174/1874609814666210202091301
Harel, I., Benayoun, B. A., Machado, B., Singh, P. P., Hu, C.-K., Pech, M. F., et al. (2015). A Platform for Rapid Exploration of Aging and Diseases in a Naturally Short-Lived Vertebrate. Cell. 160 (5), 1013–1026. doi:10.1016/j.cell.2015.01.038
Harel, I., Valenzano, D. R., and Brunet, A. (2016). Efficient Genome Engineering Approaches for the Short-Lived African Turquoise Killifish. Nat. Protoc. 11 (10), 2010–2028. doi:10.1038/nprot.2016.103
Hartmann, N., and Englert, C. (2012). A Microinjection Protocol for the Generation of Transgenic Killifish (Species: Nothobranchius Furzeri). Dev. Dyn. 241 (6), 1133–1141. doi:10.1002/dvdy.23789
Hoegg, S., Brinkmann, H., Taylor, J. S., and Meyer, A. (2004). Phylogenetic Timing of the Fish-specific Genome Duplication Correlates with the Diversification of Teleost Fish. J. Mol. Evol. 59 (2), 190–203. doi:10.1007/s00239-004-2613-z
Howe, K. L., Achuthan, P., Allen, J., Allen, J., Alvarez-Jarreta, J., Amode, M. R., et al. (2021). Ensembl 2021. Nucleic Acids Res. 49 (D1), D884–D891. doi:10.1093/nar/gkaa942
Huang, P., Xiao, A., Zhou, M., Zhu, Z., Lin, S., and Zhang, B. (2011). Heritable Gene Targeting in Zebrafish Using Customized TALENs. Nat. Biotechnol. 29 (8), 699–700. doi:10.1038/nbt.1939
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient Genome Editing in Zebrafish Using a CRISPR-Cas System. Nat. Biotechnol. 31 (3), 227–229. doi:10.1038/nbt.2501
Irion, U., Krauss, J., and Nüsslein-Volhard, C. (2014). Precise and Efficient Genome Editing in Zebrafish Using the CRISPR/Cas9 System. Development 141 (24), 4827–4830. doi:10.1242/dev.115584
Jaillon, O., Aury, J.-M., Brunet, F., Petit, J.-L., Stange-Thomann, N., Mauceli, E., et al. (2004). Genome Duplication in the Teleost Fish Tetraodon nigroviridis Reveals the Early Vertebrate Proto-Karyotype. Nature 431 (7011), 946–957. doi:10.1038/nature03025
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., et al. (2020). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 48 (D1), D498–D503. doi:10.1093/nar/gkz1031
Kirchmaier, S., Naruse, K., Wittbrodt, J., and Loosli, F. (2015). The Genomic and Genetic Toolbox of the Teleost Medaka (Oryzias latipes). Genetics 199 (4), 905–918. doi:10.1534/genetics.114.173849
Kirschner, J., Weber, D., Neuschl, C., Franke, A., Böttger, M., Zielke, L., et al. (2012). Mapping of Quantitative Trait Loci Controlling Lifespan in the Short‐lived fishNothobranchius Furzeri- a New Vertebrate Model for Age Research. Aging Cell. 11 (2), 252–261. doi:10.1111/j.1474-9726.2011.00780.x
Mahecha, D., Nuñez, H., Lattig, M. C., and Duitama, J. (2022). Machine Learning Models for Accurate Prioritization of Variants of Uncertain Significance. Hum. Mutat. 43 (4), 449–460. doi:10.1002/humu.24339
Matsui, H., Kenmochi, N., and Namikawa, K. (2019). Age- and α-Synuclein-Dependent Degeneration of Dopamine and Noradrenaline Neurons in the Annual Killifish Nothobranchius Furzeri. Cell. Rep. 26 (7), 1727–1733. e1726. doi:10.1016/j.celrep.2019.01.015
Meng, X., Noyes, M. B., Zhu, L. J., Lawson, N. D., and Wolfe, S. A. (2008). Targeted Gene Inactivation in Zebrafish Using Engineered Zinc-Finger Nucleases. Nat. Biotechnol. 26 (6), 695–701. doi:10.1038/nbt1398
Örn, S., Holbech, H., Madsen, T. H., Norrgren, L., and Petersen, G. I. (2003). Gonad Development and Vitellogenin Production in Zebrafish (Danio rerio) Exposed to Ethinylestradiol and Methyltestosterone. Aquat. Toxicol. 65 (4), 397–411. doi:10.1016/s0166-445x(03)00177-2
Poeschla, M., and Valenzano, D. R. (2020). The Turquoise Killifish: a Genetically Tractable Model for the Study of Aging. J. Exp. Biol. 223 (Pt Suppl. 1). doi:10.1242/jeb.209296
Polačik, M., Blažek, R., and Reichard, M. (2016). Laboratory Breeding of the Short-Lived Annual Killifish Nothobranchius Furzeri. Nat. Protoc. 11 (8), 1396–1413. doi:10.1038/nprot.2016.080
Postel, M. D., Culver, J. O., Ricker, C., and Craig, D. W. (2022). Transcriptome Analysis Provides Critical Answers to the "variants of Uncertain Significance" Conundrum. Hum. Mutat. doi:10.1002/humu.24394
Prykhozhij, S. V., Fuller, C., Steele, S. L., Veinotte, C. J., Razaghi, B., Robitaille, J. M., et al. (2018). Optimized Knock-In of Point Mutations in Zebrafish Using CRISPR/Cas9. Nucleic Acids Res. 46 (17), e102. doi:10.1093/nar/gky512
Reichard, M., Polačik, M., and Sedláček, O. (2009). Distribution, Colour Polymorphism and Habitat Use of the African killifishNothobranchius Furzeri, the Vertebrate with the Shortest Life Span. J. Fish. Biol. 74 (1), 198–212. doi:10.1111/j.1095-8649.2008.02129.x
Reichwald, K., Petzold, A., Koch, P., Downie, B. R., Hartmann, N., Pietsch, S., et al. (2015). Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell. 163 (6), 1527–1538. doi:10.1016/j.cell.2015.10.071
Sander, J. D., Cade, L., Khayter, C., Reyon, D., Peterson, R. T., Joung, J. K., et al. (2011). Targeted Gene Disruption in Somatic Zebrafish Cells Using Engineered TALENs. Nat. Biotechnol. 29 (8), 697–698. doi:10.1038/nbt.1934
Schartl, M. (2004). A Comparative View on Sex Determination in Medaka. Mech. Dev. 121 (7-8), 639–645. doi:10.1016/j.mod.2004.03.001
Skinner, A. M. J., and Watt, P. J. (2007). Strategic Egg Allocation in the Zebra Fish, Danio rerio. Behav. Ecol. 18 (5), 905–909. doi:10.1093/beheco/arm059
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell. Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
States, D. J., and Gish, W. (1994). QGB: Combined Use of Sequence Similarity and Codon Bias for Coding Region Identification. J. Comput. Biol. 1 (1), 39–50. doi:10.1089/cmb.1994.1.39
Sullivan, K. E. (2021). The Scary World of Variants of Uncertain Significance (VUS): A Hitchhiker's Guide to Interpretation. J. Allergy Clin. Immunol. 147 (2), 492–494. doi:10.1016/j.jaci.2020.06.011
UniProt, C. (2021). UniProt: the Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 49 (D1), D480–D489. doi:10.1093/nar/gkaa1100
Valenzano, D. R., Benayoun, B. A., Singh, P. P., Zhang, E., Etter, P. D., Hu, C.-K., et al. (2015). The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell. 163 (6), 1539–1554. doi:10.1016/j.cell.2015.11.008
Valenzano, D. R., Kirschner, J., Kamber, R. A., Zhang, E., Weber, D., Cellerino, A., et al. (2009). Mapping Loci Associated with Tail Color and Sex Determination in the Short-Lived Fish Nothobranchius Furzeri. Genetics 183 (4), 1385–1395. doi:10.1534/genetics.109.108670
Valenzano, D. R., Sharp, S., and Brunet, A. (2011). Transposon-Mediated Transgenesis in the Short-Lived African KillifishNothobranchius Furzeri, a Vertebrate Model for Aging. G3 (Bethesda) 1 (7), 531–538. doi:10.1534/g3.111.001271
Valenzano, D. R., Terzibasi, E., Cattaneo, A., Domenici, L., and Cellerino, A. (2006). Temperature Affects Longevity and Age-Related Locomotor and Cognitive Decay in the Short-Lived Fish Nothobranchius Furzeri. Aging Cell. 5 (3), 275–278. doi:10.1111/j.1474-9726.2006.00212.x
van der Merwe, C., Jalali Sefid Dashti, Z., Christoffels, A., Loos, B., and Bardien, S. (2015). Evidence for a Common Biological Pathway Linking Three Parkinson's Disease-Causing genes:parkin,PINK1andDJ-1. Eur. J. Neurosci. 41 (9), 1113–1125. doi:10.1111/ejn.12872
Varshney, G. K., Pei, W., LaFave, M. C., Idol, J., Xu, L., Gallardo, V., et al. (2015). High-throughput Gene Targeting and Phenotyping in Zebrafish Using CRISPR/Cas9. Genome Res. 25 (7), 1030–1042. doi:10.1101/gr.186379.114
Wang, X., Shang, X., Luan, J., and Zhang, S. (2014). Identification, Expression and Function of Apolipoprotein E in Annual Fish Nothobranchius Guentheri: Implication for an Aging Marker. Biogerontology 15 (3), 233–243. doi:10.1007/s10522-014-9493-4
Weinreich, S. S., Mangon, R., Sikkens, J. J., Teeuw, M. E., and Cornel, M. C. (2008). Orphanet: a European Database for Rare Diseases. Ned. Tijdschr. Geneeskd. 152 (9), 518–519.
Keywords: small fishes, zebrafish, medaka, turquoise killifish, age-related disorders, genomes
Citation: Dohi E and Matsui H (2022) The Utility of Small Fishes for the Genetic Study of Human Age-Related Disorders. Front. Genet. 13:928597. doi: 10.3389/fgene.2022.928597
Received: 26 April 2022; Accepted: 08 June 2022;
Published: 15 July 2022.
Edited by:
Mariana Härter Remião, Federal University of Pelotas, BrazilReviewed by:
Tyrone Genade, East Tennessee State University, United StatesFrederico Schmitt Kremer, Federal University of Pelotas, Brazil
Copyright © 2022 Dohi and Matsui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hideaki Matsui, aGlkZTA3MjlAYnJpLm5paWdhdGEtdS5hYy5qcA==
 Eisuke Dohi
Eisuke Dohi Hideaki Matsui
Hideaki Matsui