- 1Department of Pulmonary and Critical Care Medicine, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Research Unit of Respiratory Disease, Central South University, Changsha, China
- 3Hunan Diagnosis and Treatment Center of Respiratory Disease, Changsha, China
Background: The radial spoke head component 4A (RSPH4A) is involved in the assembly of radial spokes, which is essential for motile cilia function. Asthenoteratozoospermia in primary ciliary dyskinesia (PCD) related to RSPH4A variants has not been reported.
Materials and Methods: RSPH4A variants were identified and validated using whole-exome and Sanger sequencing in three unrelated Chinese families. High-speed video microscopy analysis (HSVA) was performed to measure the beating frequency and pattern of nasal cilia of the patients and healthy control. Papanicolaou staining and computer-aided sperm analysis were performed to analyze the morphology and motility of the sperm in patient 1. Immunofluorescence was adopted to confirm the structure deficiency of sperm and nasal cilia.
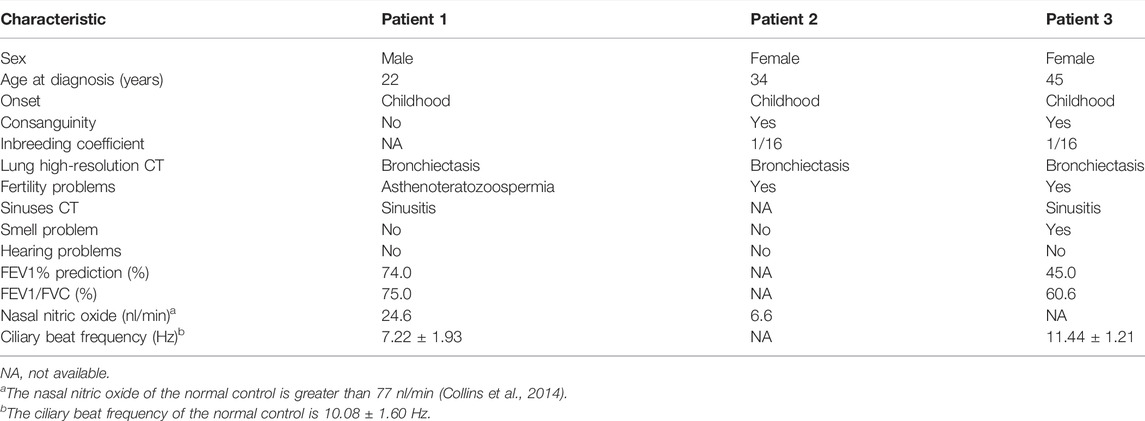
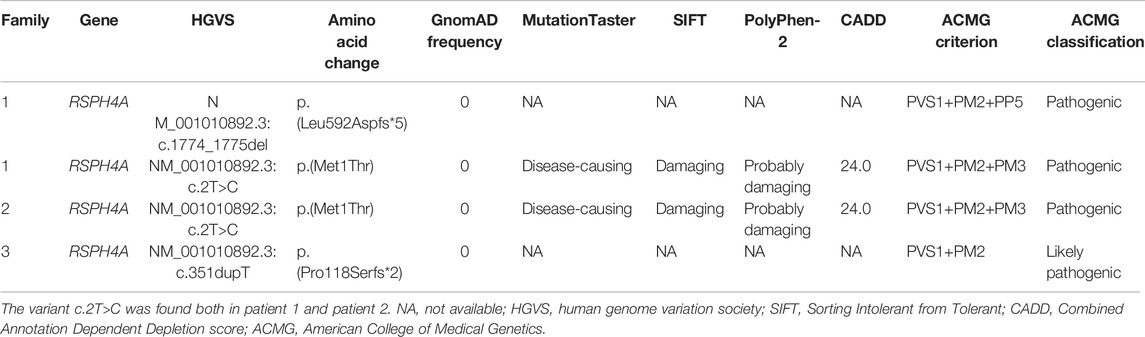
Results: Patient 1 from family 1 is a 22-year-old unmarried male presented with bronchiectasis. Semen analysis and sperm Papanicolaou staining confirmed asthenoteratozoospermia. Novel compound heterozygous RSPH4A variants c.2T>C, p.(Met1Thr) and c.1774_1775del, p.(Leu592Aspfs*5) were detected in this patient. Patients 2 and 3 are from two unrelated consanguineous families; they are both females and exhibited bronchiectasis and infertility. Two homozygous RSPH4A variants c.2T>C, p.(Met1Thr) and c.351dupT, p.(Pro118Serfs*2) were detected, respectively. HSVA showed that most of the cilia in patients 1 and 3 were with abnormal rotational movement. The absence of RSPH4A and RSPH1 in patient 1’s sperm and patient 3’s respiratory cilia was indicated by immunofluorescence. Patient 2 died of pulmonary infection and respiratory failure at the age of 35 during follow-up.
Conclusion: Dysfunctional sperm flagellum and motile cilia in the respiratory tract and the fallopian tube were found in patients with RSPH4A variants. Our study enriches the genetic spectrum and clinical phenotypes of RSPH4A variants in PCD, and c.2T>C, p.(Met1Thr) detected in our patients may be a hotspot RSPH4A variant in Chinese.
Introduction
Primary ciliary dyskinesia (PCD) is an autosomal recessive inherited disease, characterized by motile ciliary dysfunction (Mirra et al., 2017). The clinical manifestations of primary ciliary dyskinesia are varied including chronic upper and lower respiratory tract disease, left-right laterality defects, and infertility (Goutaki et al., 2016). Until now, more than 50 PCD-related pathogenic genes have been identified (Wallmeier et al., 2020).
Each radial spoke dimer contains at least 23 proteins and plays roles in controlling the activity of the dynein arm (Yang et al., 2006). As dynein serves as a molecular motor and the radial spoke complex functions as a mechanochemical senor underpinning the flagellar motility, the disruption of these axonemal components can therefore compromise the motility-related signal transmission and cause asthenospermia (Zhang et al., 2021). The radial spoke head component 4A (RSPH4A) is involved in the assembly of radial spokes, which is essential for motile cilia function. The genes belong to the radial spoke complex which is related to PCD including DNAJB13, RSPH1, RSPH9, RSPH3, and RSPH4A (Castleman et al., 2009; Wallmeier et al., 2020). Variants in RSPH1, RSPH3, and RSPH9 genes have been reported to cause male infertility in humans (Reish et al., 2010; Knowles et al., 2014; Jeanson et al., 2015). However, the phenotype of the spermatozoa in patients with RSPH4A variants has not been reported.
In this study, we identified three novel RSPH4A variants in patients. We first demonstrated asthenoteratozoospermia was related to RSPH4A variants and reported two infertile female patients caused by RSPH4A variants.
Case Presentation
Three patients with RSPH4A variants were presented in our study. Patient 1 was a 22-year-old male from a non-consanguineous family I (Figure 1A). Patient 2 was a 34-year-old female from a consanguineous family II. Patient 3 was a 45-year-old female from a consanguineous family III. They all presented over 10 years of history of chronic cough, daily nasal congestion, and yellowish purulent (Table 1). High-resolution computed tomography (HRCT) showed bronchiectasis in all patients (Figure 1B). The nasal nitric oxide level was markedly reduced both in patient 1 (24.6 nl/min) and patient 2 (6.6 nl/min). Patient 1 was unmarried, but the semen analysis showed asthenoteratozoospermia. Both patients 2 and 3 exhibited infertility. Because of the failure to achieve pregnancy, their husbands had completed the fertility examination, and the semen test results were both reported to be normal. Patient 2 died of pulmonary infection and respiratory failure at 35 years of age during follow-up. Two sisters of patient 3 died when they were 1 year old due to repeated pulmonary infection and respiratory failure. One of the older brothers of patient 3 also presented over 10 years of history of chronic cough, daily nasal congestion, and yellowish purulent and died at 37 years of age for the same reason. All family members had no situs inversus reported.
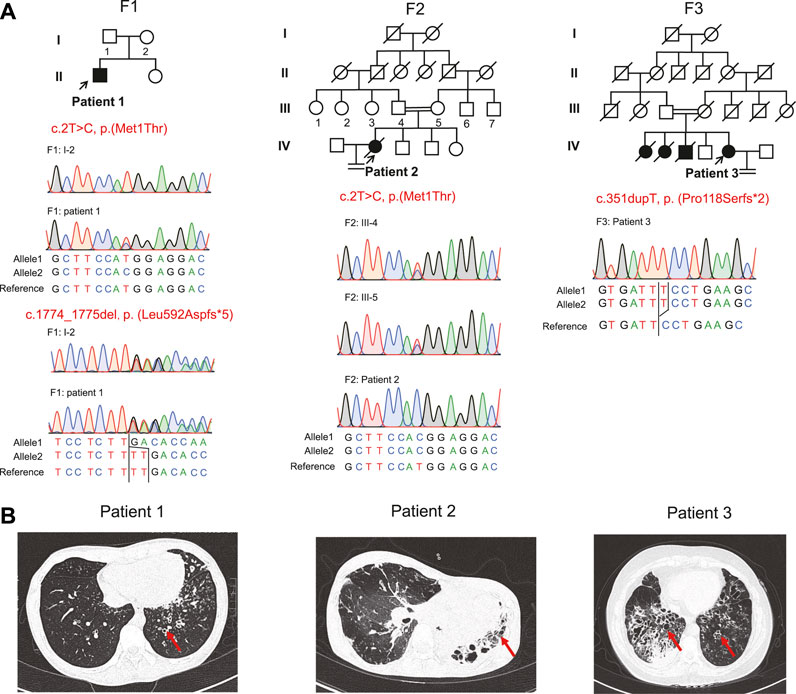
FIGURE 1. Pedigrees, variants, and clinical features of these patients. (A) Pedigrees of three families (F1, F2, and F3) with RSPH4A variants and the segregation analysis by Sanger sequencing. The filled symbol represents the affected member, the empty symbol represents unaffected members, and the slashed symbols represent deceased members. Squares, males; circles, females. The patients (patient 1, patient 2, and patient 3) included in this study are indicated by a black arrow. (B) Chest high-resolution computed tomography showed bronchiectasis (red arrows) in these patients.
Materials and Methods
Ethical Compliance
This study was approved by the Review Board of the Second Xiangya Hospital of Central South University in China. Written informed consent was obtained from all participants.
DNA Extraction, Whole-Exome Sequencing, and Variant Calling
We collected blood samples from the patients and their family members, which were obtained with informed consent. According to the manufacturer’s instructions, we used the QIAamp DNA Blood Mini Kit (250) (Cat #51106; QIAGEN, Valencia, CA, United States) to extract genomic DNA. As previously described, whole-exome capture and high-throughput sequencing were performed (Guo et al., 2017). Briefly, the genomic DNA of the patient was captured with the Agilent SureSelect Human All Exon V6 Kit (Agilent, CA, United States) and sequenced on Illumina Hiseq 4000 (Illumina Inc., San Diego, United States). After quality control, the sequencing reads were aligned to the NCBI human reference genome (GRCh37/hg19) by the bwa v7.1.0-r789 (RRID: SCR_010910) (Li and Durbin, 2010). GATK4.0.3.0 (RRID: SCR_001876) was used to call variants, and the variants were annotated with ANNOVAR (RRID: SCR_012821) (Wang et al., 2010; Van der Auwera et al., 2013).
Variant Filtering and In Silico Analysis
We filtered single-nucleotide variants (SNVs) and short insertions and deletions (InDels) as follows: 1) we ruled out high-frequency variants (minor allele frequency >0.01) in the 1000 Genomes Project data set (RRID: SCR_008801), NHLBI Exome Sequencing Project Exome Variant Server (RRID: SCR_012761), Genome Aggregation Database (all datasets and East Asian population datasets of gnomAD v2.1.1 RRID: SCR_014964 genome database), and the inhouse database of Novogene. 2) Noncoding and intronic variants were filtered. 3) Synonymous missense variants were excluded. 4) Homozygous or compound heterozygous variants were retained 5) Disease-related variants were identified by a PCD or PCD-candidate gene list collected from the literature (Lei et al., 2022).
Sanger Sequencing
We adopted Sanger sequencing to validate the variants in the patients. An online tool (PrimerQuest, IDT, https://sg.idtdna.com/PrimerQuest) was used to design primers. The primer sequences used for Sanger sequencing are listed in Supplementary Table S1. PCR products were sequenced by using the ABI PRISM 3730 DNA Analyzer (RIDD: SCR_021899) using the BigDye Terminator v3.1 Cycle Sequencing Kit (Cat #4337454; Thermo Scientific).
Semen Analysis and Immunostaining of the Sperm and Cilia
Semen sample was collected via masturbation from patient 1 after 5 days of sexual abstinence. To obtain the sperm smears, we used a saline solution to wash, centrifuge, and smear spermatozoa from patient 1 and the fertile control individual and fixed in 4% paraformaldehyde. We performed semen evaluation with semen parameters, according to the World Health Organization guideline (Cooper et al., 2010). On the basis of Papanicolaou staining, morphological abnormalities of the flagella were classified as absent, short, bent, coiled, or irregular width (Ben Khelifa et al., 2014). The percentages of morphologically normal and abnormal spermatozoa were assessed according to the WHO guidelines (Sánchez-Álvarez et al., 2010). Immunostaining of the slides was performed, as described previously (Dai et al., 2019). Sperm smears were incubated with primary antibodies: RSPH4A (Cat# HPA031196, RRID: AB_10601612; 1:25; Sigma-Aldrich), RSPH1 (Cat# HPA017382, RRID: AB_1858392; 1:25; Sigma-Aldrich), and anti-acetylated tubulin monoclonal antibody (Cat# T7451, RRID: AB_609894; 1:500; Sigma-Aldrich) for 2 h at 37°C. The binding of the antibodies was detected by incubating with Alexa Fluor 488 anti-mouse IgG (Cat# A-21121, RRID: AB_2535764; 1:400; Molecular Probes) and Alexa Fluor 555 anti-rabbit IgG (Cat# A-31572, RRID: AB_162543; 1:500; Molecular Probes) for 1 h at 37°C. Subsequently, the sperm smears were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 5 min. Finally, they were viewed using an Olympus BX53 microscope (Olympus, Tokyo, Japan) and a scientific complementary metal-oxide semiconductor (sCMOS) camera (Prime BSI; Teledyne Photometrics Inc., United States). Nasal brushing samples from patient 3 were suspended in Gibco Medium 199 (12350039; Gibco). We used an anti-acetylated tubulin monoclonal antibody (Cat# T7451, RRID: AB_609894; 1:500; Sigma-Aldrich) to mark the ciliary axoneme, RSPH4A (Cat# HPA031196, RRID: AB_10601612; 1:100; Sigma-Aldrich) and RSPH1 (Cat# HPA017382, RRID: AB_1858392; 1:100; Sigma-Aldrich) to label the radial spoke in the ciliary axoneme and DAPI to label the nuclei. Immunofluorescence was employed to profile the ciliary structure of patients with RSPH4A variants.
High-Speed Microscopy Analysis
Nasal brush biopsy samples from patients 1 and 3 were suspended in Gibco Medium 199 (12350039 Gibco). An Upright Olympus BX53 microscope (Olympus, Tokyo, Japan) with a ×40 objective lens was used to image strips of the ciliated epithelium. We recorded videos by using a scientific complementary metal-oxide semiconductor (sCMOS) camera (Prime BSI; Teledyne Photometrics Inc., United States) at a rate of 500 frames per second (fps) at room temperature and reviewed at 50 fps to perform an analysis of the ciliary beat pattern, as described previously (Lei et al., 2022). Six separate ciliated epithelium strips (five sideway edges and one from above) with mucus-free regions from the subject were analyzed. The ciliary beat frequency was calculated using the validated automated open-source software CiliarMove (Sampaio et al., 2021).
Results
Identification of the RSPH4A Variants
Novel compound heterozygous variants c.2T>C, p.(Met1Thr) and c.1774_1775del, p.(Leu592Aspfs*5), a homozygous stat loss variant c.2T>C, p.(Met1Thr), and a novel frame-shift variant c.351dupT, p.(Pro118Serfs*2) in RSPH4A (NM_001010892.3) were identified in Proband 1, 2, and 3 through whole-exome sequencing. We did not identify these variants in the 1000 Genomes Project data set and NHLBI Exome Sequencing Project Exome Variant Server, gnomAD v2.1.1 database, and the Human Gene Mutation Database. Sanger sequencing validated the variants in this patient (Figure 1A). These variants were classified into pathogenic or likely pathogenic (Table 2), according to the ACMG guidelines.
Motile Cilia Analysis and Immunofluorescence of Nasal Epithelial Cilia
Rotational movement of the cilia was observed in patients 1 and 3 (Figure 2A and Supplementary Videos S1 and S2), while planar beating was observed in the healthy control (Supplementary Video S3). High-speed video microscopy analysis of nasal brush biopsy samples indicated that the nasal epithelial ciliary beat frequency of patients 1 and 3 with RSPH4A variants was not significantly different from the healthy control (Table1). Immunofluorescence of the nasal epithelial ciliary in patient 3 with RSPH4A variants showed both RSPH4A and RSPH1 were absent (Figure 2B).
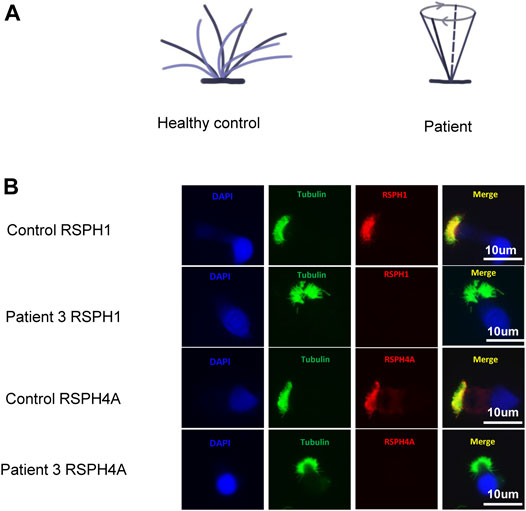
FIGURE 2. (A) Ciliary beating patterns of healthy cilia and our patients with RSPH4A variants. (B) Immunofluorescence of nasal ciliated cells revealed the absence of RSPH4A and RSPH1 in patient 3 (red). An anti-acetylated tubulin monoclonal antibody was used to mark the ciliary axoneme (green), and DAPI was used to label the nuclei (blue).
Sperm Morphology and Immunofluorescence Analysis
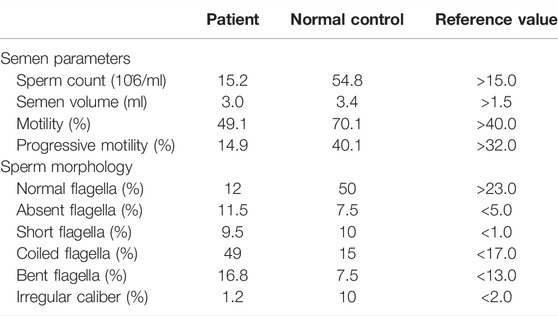
The abnormalities of sperm morphology were revealed by Papanicolaou staining. Although there were also some spermatozoa with seemingly normal morphology, Papanicolaou staining showed morphological abnormalities in patient 1’s sperm, including absent, bent, coiled, and short flagella, compared with the control individual (Figure 3A). The absent, bent, coiled, and short flagella, respectively, accounted for 11.5, 16.8, 49, and 9.5% in patient 1’s sperm (n = 200; Table 3). We conducted semen analysis according to the WHO guidelines (Table 3) and concluded that the patient had asthenoteratospermia given the abnormal morphology of the sperm (Auger et al., 2016). To confirm abnormalities in the axonemal or peri-axonemal structures of the sperm flagella of patient 1, we performed immunostaining. In patient 1’s spermatozoa, the staining signal of RSPH4A was undetectable compared with the normal control (Figure 3B). Moreover, we also could not detect the staining signal of RSPH1 in the sperm of patient 1 (Figure 3B).
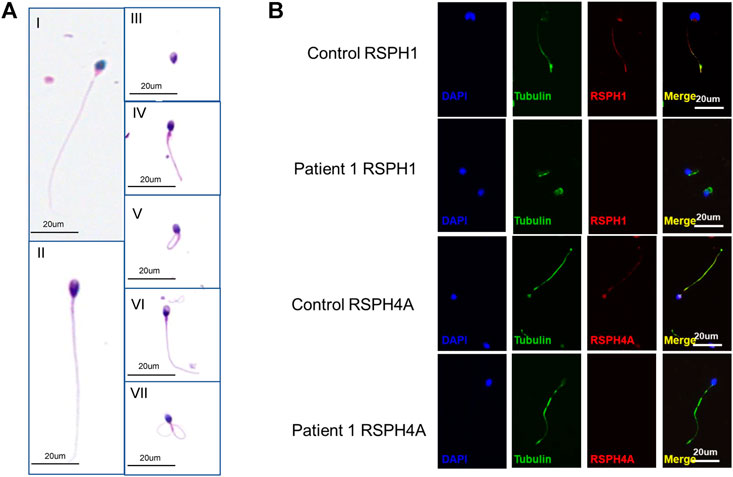
FIGURE 3. Phenotypic features of the RSPH4A mutant sperm and RSPH4A immunostaining in human spermatozoa. (A) Papanicolaou staining of spermatozoa from patient 1 and a healthy control. (A-I) Normal morphology of spermatozoa from a healthy control male. (A-II) Observed normal length of the sperm from patient 1 with RSPH4A variants. In addition, multiple malformations can be observed, including (A-III) absent, (A-IV) short, (A-V) coiled, (A-VI) bent, and (A-VII) irregular caliber flagella. (B) RSPH4A and RSPH1 are expressed in the whole length of the sperm flagella in the normal control but absent in patient 1.
Summary of Clinical and Functional Phenotypes of Patients With RSPH4A Variants
After a comprehensive literature search, 45 patients with RSPH4A biallelic pathogenic variants were identified (Ziętkiewicz et al., 2012; Daniels et al., 2013; Casey et al., 2015; Frommer et al., 2015; Emiralioğlu et al., 2020; De Jesús-Rojas et al., 2021) (Figure 4, Supplementary Table S2). About half of the 45 patients with RSPH4A variants are Hispanic (21/45), and 15 patients are from consanguineous families. There are 31 females (31/45) and 13 adult patients (≥18 years old, 13/45). Situs inversus is not present in all patients. Only one female patient reported infertility. The nasal nitric oxide production rate was markedly reduced in all 15 patients. A stiff and rotatory beating pattern of HSVA was presented in 11 patients with RSPH4A deficiency. Central apparatus abnormalities or normal ciliary ultrastructure were shown in the TEM analysis of respiratory cilia.
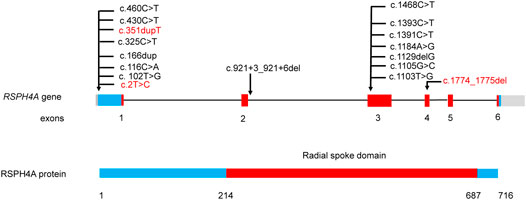
FIGURE 4. RSPH4A gene and protein structure and the reported disease-causing variants of RSPH4A. Domains and their relative exons were filled using the same colors. Variants in our study were highlighted using red.
Discussion
This study describes one male and two females with PCD that carried biallelic disease-causing variants in the RSPH4A gene. Our cases enrich the variant spectrum and clinical phenotypes of RSPH4A variants in PCD.
According to our summary, no situs inversus was reported in patients with RSPH4A variants, and the nasal nitric oxide level was low. This is consistent with the clinical manifestations of the three patients in our study. In the rsph4a knock-out zebrafish model, there is no situs inversus, which may be related to the “9+0” structure of embryonic nodes without radial spokes, while other motile cilia are of the “9+2” structure (Han et al., 2018). Among the spoke head-related genes, variants of RSPH4A lead to the most severe phenotype on the cilia ultrastructure in the PCD patients (Daniels et al., 2013; Knowles et al., 2014). The absence of RSPH4A results in the deficient axonemal assembly of radial spoke head components RSPH1 and RSPH9 (Frommer et al., 2015). Therefore, it is probable that patients with RSPH4A variants presented the most serious clinical manifestations among the patients with radial spoke family deficiency. As previously reported, a male patient with the RSPH4A variant died at the age of 27 because of respiratory failure caused by pulmonary infection (Bian et al., 2021). In this study, patient 2 also died at the age of 35 because of respiratory failure.
The nasal cilia HSVA of patients with RSPH4A variants presented a reduced amplitude and rotatory beating pattern. The HSVA of motile cilia in the airway of the Rsph4a knock-out mouse presented rotational movement, which may be caused by the deficiency of radial spokes (Shinohara et al., 2015). The HSVA of our patients 1 and 3 showed rotational movement, and the nasal epithelial ciliary beat frequency was in the normal range. Among previously reported patients, the transmission electron microscope examination of respiratory cilia presented with central apparatus abnormalities and a normal structure. However, because of the COVID-19 epidemic situation, these patients are not able to finish the transmission electron microscope examination. According to European Respiratory Society guidelines (Lucas et al., 2017), we confirmed the diagnosis of PCD by the detection of biallelic variants or a low nasal nitric oxide level combined with the characteristic HSVA presentation of the nasal cilia.
One splicing variant, seven missense variants, two frame-shift variants, and two nonsense variants have been reported (Supplementary Table S2). The most reported variant was c.921+3_921+6del from the Hispanic family, which has been reported in 17 patients. There were two reported RSPH4A variants from the Chinese family including c.730+1G>A and c.1454C>G. In our study, three novel variants were identified. Two patients from two unrelated families carried the same variant c.2T>C, which may be the hotspot variant in Chinese patients with RSPH4A variants.
Infertility is defined as the failure to achieve pregnancy after 12 months of regular unprotected sexual intercourse (Carson and Kallen, 2021). PCD is a rare inherited disease in which genetic variants impair motile cilia function (Afzelius, 1976). Female and male infertility can be caused by defects in the cilia in the fallopian tubes or in sperm flagella, respectively (Wallmeier et al., 2020). The central apparatus and radial spokes play roles in mechano-chemical sensors to control motility in 9+2 cilia and flagella (Smith and Yang, 2004). The RPSH4A gene belongs to the radial spoke family. Therefore, it is persuasive to assume that male patients with RSPH4A variants present infertility such as RSPH1, RSPH3, and RSPH9 (Castleman et al., 2009; Reish et al., 2010; Knowles et al., 2014).
According to a previous report, three male patients with RSPH4A variants were likely to be fertile (Vanaken, 2017). But there was no detailed clinical information, variant descriptions, and phenotype of motile cilia of those patients, and it is still unclear about the fertility status of the male patients with RSPH4A variants. Patient 1 in this study presented asthenozoospermia which is one of the main factors contributing to male infertility (Shahrokhi et al., 2020). Male infertility caused by the RSPH1 or RSPH9 variant has been clarified (Reish et al., 2010; Knowles et al., 2014). The absence of RSPH4A due to variants in RSPH4A results in deficient axonemal assembly of radial spoke head components RSPH1 and RSPH9 (Frommer et al., 2015). Therefore, we have reason to assume that RSPH4A variants cause male infertility. Moreover, in a previous report, the sperm of a 21-year-old male with the RSPH4A variant was immotile (Bian et al., 2021). According to the semen analysis and sperm morphology, it is probable that the RSPH4A variants cause male infertility.
As previously reported, two female patients with RSPH4A variants presented infertility, which is consistent with the two female patients in our study. In a previous study, in the Rsph4a knock-out mice, the oviduct cilia showed two types of abnormal motion patterns: anti-clockwise rotation and beating with small amplitude, which may cause the failure of oocyte pickup (Yuan et al., 2021) and led to female mouse infertility (Yoke et al., 2020). The reason that female patients with RSPH4A variants presented infertility is probably because of the change in the beating pattern of the oviduct cilia. However, because of the difficulty to obtain the clinical sample, we cannot confirm the beating pattern of oviduct cilia in our patients.
In conclusion, our study reported three novel RSPH4A variants and first demonstrated asthenoteratozoospermia related to RSPH4A variants, which revealed the relationship between RSPH4A variants and male infertility. Moreover, we also described the detailed information of two infertile female patients with RSPH4A variants. Our study enriches the genetic spectrum and clinical phenotypes of RSPH4A variants in PCD and provides more evidence for future genetic counseling and gene-targeted therapy for this disease.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Review Board of the Second Xiangya Hospital of Central South University in China. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
LW: data collection, data analysis, basic experiments, and writing the manuscript; RW: data collection, data analysis, and writing the manuscript; DY: data collection and data analysis; CYL: basic experiments; YX: basic experiments; YL: basic experiments; TG: basic experiments; HL and CL: conceived and designed the experiments.
Funding
This study was supported by the National Natural Science Foundation of China (82070003 to HL), the Hunan Province Health Commission Scientific Research Project (B2019159 to HL), the Science and Technology Program of Changsha, China (kq1901120 to HL), the Xiangya Clinical Big Data System Construction Project in Pulmonary Inflammatory Disease of Central South University, and the National Key Clinical Specialty Construction Projects of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the patients and their families who participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.922287/full#supplementary-material
References
Afzelius, B. A. (1976). A Human Syndrome Caused by Immotile Cilia. Science 193 (4250), 317–319. doi:10.1126/science.1084576
Auger, J., Jouannet, P., and Eustache, F. (2016). Another Look at Human Sperm Morphology. Hum. Reprod. 31 (1), 10–23. doi:10.1093/humrep/dev251
Ben Khelifa, M., Coutton, C., Zouari, R., Karaouzène, T., Rendu, J., Bidart, M., et al. (2014). Mutations in DNAH1, Which Encodes an Inner Arm Heavy Chain Dynein, Lead to Male Infertility from Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 94 (1), 95–104. doi:10.1016/j.ajhg.2013.11.017
Bian, C., Zhao, X., Liu, Y., Chen, M., Zheng, S., Tian, X., et al. (2021). Case Report of Neurofibromatosis Type 1 Combined with Primary Ciliary Dyskinesia. Front. Med. 15 (6), 933–937. doi:10.1007/s11684-021-0860-7
Carson, S. A., and Kallen, A. N. (2021). Diagnosis and Management of Infertility. JAMA 326 (1), 65–76. doi:10.1001/jama.2021.4788
Casey, J. P., McGettigan, P. A., Healy, F., Hogg, C., Reynolds, A., Kennedy, B. N., et al. (2015). Unexpected Genetic Heterogeneity for Primary Ciliary Dyskinesia in the Irish Traveller Population. Eur. J. Hum. Genet. 23 (2), 210–217. doi:10.1038/ejhg.2014.79
Castleman, V. H., Romio, L., Chodhari, R., Hirst, R. A., de Castro, S. C. P., Parker, K. A., et al. (2009). Mutations in Radial Spoke Head Protein Genes RSPH9 and RSPH4A Cause Primary Ciliary Dyskinesia with Central-Microtubular-Pair Abnormalities. Am. J. Hum. Genet. 84 (2), 197–209. doi:10.1016/j.ajhg.2009.01.011
Collins, S. A., Gove, K., Walker, W., and Lucas, J. S. A. (2014). Nasal Nitric Oxide Screening for Primary Ciliary Dyskinesia: Systematic Review and Meta-Analysis. Eur. Respir. J. 44 (6), 1589–1599. doi:10.1183/09031936.00088614
Cooper, T. G., Noonan, E., von Eckardstein, S., Auger, J., Baker, H. W. G., Behre, H. M., et al. (2010). World Health Organization Reference Values for Human Semen Characteristics*‡. Hum. Reprod. update 16 (3), 231–245. doi:10.1093/humupd/dmp048
Dai, C., Hu, L., Gong, F., Tan, Y., Cai, S., Zhang, S., et al. (2019). ZP2 Pathogenic Variants Cause In Vitro Fertilization Failure and Female Infertility. Genet. Med. 21 (2), 431–440. doi:10.1038/s41436-018-0064-y
Daniels, M. L. A., Leigh, M. W., Davis, S. D., Armstrong, M. C., Carson, J. L., Hazucha, M., et al. (2013). Founder Mutation inRSPH4AIdentified in Patients of Hispanic Descent with Primary Ciliary Dyskinesia. Hum. Mutat. 34 (10), 1352–1356. doi:10.1002/humu.22371
De Jesús-Rojas, W., Reyes-De Jesús, D., and Mosquera, R. A. (2021). Primary Ciliary Dyskinesia Diagnostic Challenges: Understanding the Clinical Phenotype of the Puerto Rican Founder Mutation. Diagn. Base 11 (2). 281. doi:10.3390/diagnostics11020281
Emiralioğlu, N., Taşkıran, E. Z., Koşukcu, C., Bilgiç, E., Atilla, P., Kaya, B., et al. (2020). Genotype and Phenotype Evaluation of Patients with Primary Ciliary Dyskinesia: First Results from Turkey. Pediatr. Pulmonol. 55 (2), 383–393. doi:10.1002/ppul.24583
Frommer, A., Hjeij, R., Loges, N. T., Edelbusch, C., Jahnke, C., Raidt, J., et al. (2015). Immunofluorescence Analysis and Diagnosis of Primary Ciliary Dyskinesia with Radial Spoke Defects. Am. J. Respir. Cell Mol. Biol. 53 (4), 563–573. doi:10.1165/rcmb.2014-0483oc
Goutaki, M., Meier, A. B., Halbeisen, F. S., Lucas, J. S., Dell, S. D., Maurer, E., et al. (2016). Clinical Manifestations in Primary Ciliary Dyskinesia: Systematic Review and Meta-Analysis. Eur. Respir. J. 48 (4), 1081–1095. doi:10.1183/13993003.00736-2016
Guo, T., Tan, Z.-P., Chen, H.-M., Zheng, D.-y., liu, L., Huang, X.-G., et al. (2017). An Effective Combination of Whole-Exome Sequencing and Runs of Homozygosity for the Diagnosis of Primary Ciliary Dyskinesia in Consanguineous Families. Sci. Rep. 7 (1), 7905. doi:10.1038/s41598-017-08510-z
Han, X., Xie, H., Wang, Y., and Zhao, C. (2018). Radial Spoke Proteins Regulate Otolith Formation during Early Zebrafish Development. FASEB J. 32 (7), 3984–3992. doi:10.1096/fj.201701359r
Jeanson, L., Copin, B., Papon, J.-F., Dastot-Le Moal, F., Duquesnoy, P., Montantin, G., et al. (2015). RSPH3 Mutations Cause Primary Ciliary Dyskinesia with Central-Complex Defects and a Near Absence of Radial Spokes. Am. J. Hum. Genet. 97 (1), 153–162. doi:10.1016/j.ajhg.2015.05.004
Knowles, M. R., Ostrowski, L. E., Leigh, M. W., Sears, P. R., Davis, S. D., Wolf, W. E., et al. (2014). Mutations inRSPH1Cause Primary Ciliary Dyskinesia with a Unique Clinical and Ciliary Phenotype. Am. J. Respir. Crit. Care Med. 189 (6), 707–717. doi:10.1164/rccm.201311-2047oc
Lei, C., Yang, D., Wang, R., Ding, S., Wang, L., Guo, T., et al. (2022). DRC1 Deficiency Caused Primary Ciliary Dyskinesia and MMAF in a Chinese Patient. J. Hum. Genet. 67 (4), 197–201. doi:10.1038/s10038-021-00985-z
Li, H., and Durbin, R. (2010). Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinforma. Oxf. Engl. 26 (5), 589–595. doi:10.1093/bioinformatics/btp698
Lucas, J. S., Barbato, A., Collins, S. A., Goutaki, M., Behan, L., Caudri, D., et al. (2017). European Respiratory Society Guidelines for the Diagnosis of Primary Ciliary Dyskinesia. Eur. Respir. J. 49 (1). 1601090. doi:10.1183/13993003.01090-2016
Mirra, V., Werner, C., and Santamaria, F. (2017). Primary Ciliary Dyskinesia: An Update on Clinical Aspects, Genetics, Diagnosis, and Future Treatment Strategies. Front. Pediatr. 5, 135. doi:10.3389/fped.2017.00135
Reish, O., Slatkin, M., Chapman-Shimshoni, D., Elizur, A., Chioza, B., Castleman, V., et al. (2010). Founder Mutation(s) in theRSPH9Gene Leading to Primary Ciliary Dyskinesia in Two Inbred Bedouin Families. Ann. Hum. Genet. 74 (2), 117–125. doi:10.1111/j.1469-1809.2009.00559.x
Sampaio, P., da Silva, M. F., Vale, I., Roxo-Rosa, M., Pinto, A., Constant, C., et al. (2021). CiliarMove: New Software for Evaluating Ciliary Beat Frequency Helps Find Novel Mutations by a Portuguese Multidisciplinary Team on Primary Ciliary Dyskinesia. ERJ Open Res. 7 (1). 792. doi:10.1183/23120541.00792-2020
Sánchez-Álvarez, J., Cano-Corres, R., and Fuentes-Arderiu, X. (2010). A Complement for the WHO Laboratory Manual for the Examination and Processing of Human Semen (First Edition. EJIFCC 23 (3), 103–106.
Shahrokhi, S. Z., Salehi, P., Alyasin, A., Taghiyar, S., and Deemeh, M. R. (2020). Asthenozoospermia: Cellular and Molecular Contributing Factors and Treatment Strategies. Andrologia 52 (2), e13463. doi:10.1111/and.13463
Shinohara, K., Chen, D., Nishida, T., Misaki, K., Yonemura, S., and Hamada, H. (2015). Absence of Radial Spokes in Mouse Node Cilia Is Required for Rotational Movement but Confers Ultrastructural Instability as a Trade-Off. Dev. Cell 35 (2), 236–246. doi:10.1016/j.devcel.2015.10.001
Smith, E. F., and Yang, P. (2004). The Radial Spokes and Central Apparatus: Mechano-Chemical Transducers that Regulate Flagellar Motility. Cell Motil. Cytoskelet. 57 (1), 8–17. doi:10.1002/cm.10155
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From FastQ Data to High Confidence Variant Calls: the Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinforma. 43, 11–33. doi:10.1002/0471250953.bi1110s43
Vanaken, G. J. (2017). "Infertility in an Adult Cohort with Primary Ciliary Dyskinesia: Phenotype-Gene association." Gert Jan Vanaken, Laurence Bassinet, Mieke Boon, Rahma Mani, Isabelle Honoré, Jean-Francois Papon, Harry Cuppens, Martine Jaspers, Natalie Lorent, André Coste, Estelle Escudier, Serge Amselem, Bernard Maitre, Marie Legendre and Sophie Christin-Maitre. Eur Respir J 2017; 50: 1700314. Eur. Respir. J. 50 (5). 1750314. doi:10.1183/13993003.50314-2017
Wallmeier, J., Nielsen, K. G., Kuehni, C. E., Lucas, J. S., Leigh, M. W., Zariwala, M. A., et al. (2020). Motile Ciliopathies. Nat. Rev. Dis. Prim. 6 (1), 77. doi:10.1038/s41572-020-0209-6
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 38 (16), e164. doi:10.1093/nar/gkq603
Yang, P., Diener, D. R., Yang, C., Kohno, T., Pazour, G. J., Dienes, J. M., et al. (2006). Radial Spoke Proteins of Chlamydomonas Flagella. J. Cell Sci. 119 (Pt 6), 1165–1174. doi:10.1242/jcs.02811
Yoke, H., Ueno, H., Narita, A., Sakai, T., Horiuchi, K., Shingyoji, C., et al. (2020). Rsph4a Is Essential for the Triplet Radial Spoke Head Assembly of the Mouse Motile Cilia. PLoS Genet. 16 (3), e1008664. doi:10.1371/journal.pgen.1008664
Yuan, S., Wang, Z., Peng, H., Ward, S. M., Hennig, G. W., Zheng, H., et al. (2021). Oviductal Motile Cilia Are Essential for Oocyte Pickup but Dispensable for Sperm and Embryo Transport. Proc. Natl. Acad. Sci. U. S. A. 118 (22). e2102940118. doi:10.1073/pnas.2102940118
Zhang, X., Sun, J., Lu, Y., Zhang, J., Shimada, K., Noda, T., et al. (2021). LRRC23 Is a Conserved Component of the Radial Spoke that Is Necessary for Sperm Motility and Male Fertility in Mice. J. Cell Sci. 134 (20). jcs259381. doi:10.1242/jcs.259381
Keywords: primary ciliary dyskinesia, RSPH4A, infertility, asthenoteratozoospermia, MMAF, bronchiectasis
Citation: Wang L, Wang R, Yang D, Lu C, Xu Y, Liu Y, Guo T, Lei C and Luo H (2022) Novel RSPH4A Variants Associated With Primary Ciliary Dyskinesia–Related Infertility in Three Chinese Families. Front. Genet. 13:922287. doi: 10.3389/fgene.2022.922287
Received: 17 April 2022; Accepted: 02 May 2022;
Published: 22 June 2022.
Edited by:
Steven LC Pei, Yale University, United StatesReviewed by:
Martin Breuss, University of Colorado, United StatesLance Lee, Sanford Research, United States
Copyright © 2022 Wang, Wang, Yang, Lu, Xu, Liu, Guo, Lei and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Lei, bGVpY2hlbmdAY3N1LmVkdS5jbg==; Hong Luo, bHVvaG9uZ2h1eGlAY3N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Lin Wang
Lin Wang Rongchun Wang1,2,3†
Rongchun Wang1,2,3† Danhui Yang
Danhui Yang Chenyang Lu
Chenyang Lu Yingjie Xu
Yingjie Xu Ying Liu
Ying Liu Ting Guo
Ting Guo Cheng Lei
Cheng Lei Hong Luo
Hong Luo