- 1Department of Surgery, People’s Hospital of Deyang City, Deyang, China
- 2Department of Pathology, People’s Hospital of Deyang City, Deyang, China
There is mounting evidence on the implication of SLC6A8 in the initiation and progression of human cancers. However, a comprehensive understanding of the role of SLC6A8 in pan-cancer remains elusive yet. Bioinformatics analysis was performed to investigate the expression and mutation profiles of SLC6A8 in cancers, and the association of SLC6A8 expression with cancer patients’ survival and immune cell infiltration. In general, SLC6A8 is significantly upregulated across multiple cancers. SLC6A8 expression was inconsistently prognostic in different types of cancer, albeit associated with favorable survival in the vast majority of cancers. The receiver operating characteristic curves showed that SLC6A8 was relatively accurate in identifying possible cancer patients. The genetic alteration of SLC6A8, including mutation, amplification and deletion, was frequently present across various types of cancer. Mechanistically, SLC6A8 might be involved in tumorigenesis through “carbon metabolism” and “HIF-1 signaling pathway.” Besides, SLC6A8 expression had significant correlation with immune checkpoints genes and tumor-infiltrating immune cell abundances. The present study offers a novel insight into the roles of SLC6A8 in the oncogenesis and development of multiple common cancers.
Introduction
Despite the huge advances in medical resources and technology, the incidence and mortality of cancer are rapidly growing worldwide (Sung et al., 2021). Owing to the discovery and identification of star cancer-associated genes, targeted therapy and immunotherapy have transformed the treatment of cancer patients, leading to significantly prolonged survival and improving tolerance of cancer patients (Gotwals et al., 2017; Kennedy and Salama, 2020). For example, the implementation of first-generation tyrosine kinase inhibitors (TKIs) has significantly improved progression survival and overall survival as compared to traditional platinum-doublet chemotherapy in cancer patients harboring detectable sensitizing-EGFR mutations (Yoshioka et al., 2019). Nevertheless, the prognosis of most patients with advanced cancers remains poor due to the lack of available targetable driver mutations or emergence of therapy resistance (Gotwals et al., 2017; Camidge et al., 2019; Chai et al., 2022). Therefore, it is of urgent need to identify novel molecular targets that might play essential roles in the oncogenesis and malignant progression of cancers.
The solute carrier family 6 member 8 (SLC6A8) gene is located on chromosome X at the 28q locus and encodes creatine transporter protein which serves to transport creatine, a ubiquitous non-protein amino acid with pleiotropic effects (Bonilla et al., 2021), across cellular membranes (Joncquel-Chevalier Curt et al., 2015; Yıldız et al., 2020; Li et al., 2021). Initially, deletion or mutation of SLC6A8 was found to be associated with creatine transport deficiency (CTD) such as intellectual disability and verbal dyspraxia (Battini et al., 2007). Lately, it has been reported that SLC6A8 is implicated in the malignant progression of various tumor. For example, therapeutic targeting of SLC6A8 suppresses colon cancer progression by modulating creatine abundances (Kurth et al., 2021). SLC6A8 is required for the malignant progression of NSCLC via activating Notch signaling pathway (Feng et al., 2021). Moreover, deficiency in SLC6A8-mediated creatine transport is reported to impair immunogenic function (Di Biase et al., 2019). For instance, loss of SLC6A8 weakens TCR-mediated activation of mechanistic target of rapamycin complex 1 (mTORC1) signaling, resulting in hampered CD8+ T cell immunity (Samborska et al., 2022). Ablation of SLC6A8 reprogrammed macrophage polarization via regulating cytokine responses, contributing to altered immune responses in vivo (Ji et al., 2019). However, the oncogenic roles of SLC6A8 across different cancers remain unknown. Considering the involvement of SLC6A8 in multiple physical and pathological processes (Salomons et al., 2003), we sought to comprehensively explore the roles this gene plays in cancers through bioinformatics analysis.
Herein, we found that SLC6A8 expression was differentially expressed in 22 types of cancer while no significant difference was observed for SLC6A8 expression in the remaining 10 types of cancer. In addition, SLC6A8 was found to be a significant prognostic and diagnostic factor in various cancers. Moreover, SLC6A8 gene was often altered, with amplification, mutation and deletion being the dominant alteration types. Analysis of possible pathways involving SLC6A8 showed that SLC6A8 was mainly enriched in “Carbon metabolism,” “HIF-1 signaling pathway,” and “Glycolysis/Gluconeogenesis” pathways. Finally, the immunological roles of SLC6A8 in various cancers were evaluated.
Materials and Methods
Expression Analysis
The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) is a landmark cancer genomics program that sequenced and characterized the molecular landscape of tumors from 11,160 patients across 33 cancer types (Liu et al., 2018). Gene Expression Profiling Interactive Analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn) is a valuable and highly cited resource for gene expression analysis based on tumor and normal samples from the TCGA and the GTEx databases (Tang et al., 2019). The differential expression of SLC6A8 between tumor tissues and their normal counterparts were analyzed based on data from TCGA and GEPIA2 databases. p value <0.05 and |Log2FC| > 1 were set as the cutoffs of significantly differential expression of SLC6A8.
Survival Analysis and Receiver Operating Characteristic Analysis
PrognoScan (http://www.prognoscan.org/) is an online tool for evaluating the biological relationship between gene expression and patient prognosis based on publicly available cancer microarray datasets with clinical annotation (Mongre et al., 2019). Using the database, we conducted univariate cox regression analysis to evaluate the prognostic value of SLC6A8 in pan-cancer. p value <0.05 was considered to be statistically significant.
ROC curves were plotted to characterize the accuracy of SLC6A8 expression to discriminate between patients with or without cancer based on data from TCGA database.
Analysis of Mutation Landscape of SLC6A8
The cBio Cancer Genomics Portal (cbioportal, http://cbioportal.org) is an open-access resource for interactive exploration of multidimensional cancer genomics datasets including mutation data (Cerami et al., 2012). Via cbioportal database, the alteration types and sites of SLC6A8 in pan-cancer were identified based on the data of 10,967 samples from TCGA database. The filtering criteria of associations were set as following: Correlation ≥0.4, −Log10(p) ≥2, Max results = 200.
Protein-Protein Interaction Analysis and Pathway Enrichment Analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://string-db.org) aims to integrate all known and predicted associations between proteins (Szklarczyk et al., 2021). The interacting network of SLC6A8 protein with other associated proteins were visualized through STRING database. The cutoff value of interaction score was set as 0.15. The venn diagram was utilized to display intersecting members of STRING and GEPIA2.
For pathway enrichment analysis, both Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) enrichment analysis were conducted to visualize the cellular pathways that SLC6A8 was most significantly related to. The cutoff Q value and p value were set as 0.05 and 0.01, respectively.
Immunity-Related Analysis
Tumor Immune Estimation Resource (TIMER2, http://timer.cistrome.org/) web server provides comprehensive analysis and visualization of the functions of tumor infiltrating immune cells by integrating multiple algorithms that have been designed to estimate immune infiltration (Li et al., 2017). We analyzed immune infiltration in different cancer types based on the TCGA datasets through TIMER2 database. Besides, the associations between SLC6A8 expression and microsatellite instability (MSI), and tumor mutation burden (TMB) were assessed using SangerBox platform (http://sangerbox.com/Tool), an online platform for analysis of TCGA data. Pearson correlation coefficient was employed to analyze the correlation between SLC6A8 expression and other genes.
Results
Pan-Cancer Expression Profile of SLC6A8
To investigate SLC6A8 expression pattern in various human cancers, we analyzed the transcriptome data acquired from TCGA database. The results showed that SLC6A8 was significantly upregulated in 13 types of cancer including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), and uterine corpus endometrial carcinoma (UCEC). Of note, SLC6A8 expression in LUAD, LUSC, and CHOL was remarkably higher than in normal counterparts. Interestingly, 3 types of cancer, including colon adenocarcinoma (COAD), pheochromocytoma and paraganglioma (PCPG) and Rectum adenocarcinoma (READ), exhibited significantly lower SLC6A8 expression as compared to their respective normal tissues (Figure 1A). For the 11 types of cancer in which SLC6A8 expression in their corresponding normal tissues was lacking, the GTEx dataset in GEPIA2 website was used for comparison of SLC6A8 expression between tumor and normal tissue. According to the results, SLC6A8 expression was found to be significantly upregulated in skin cutaneous melanoma (SKCM), thymoma (THYM), and head and neck squamous cell carcinoma (HNSC), whereas the downregulation of SLC6A8 was found in testicular germ cell tumors (TGCT) and lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) (Figure 1B). Taken together, SLC6A8 was aberrantly expressed in diverse cancers.
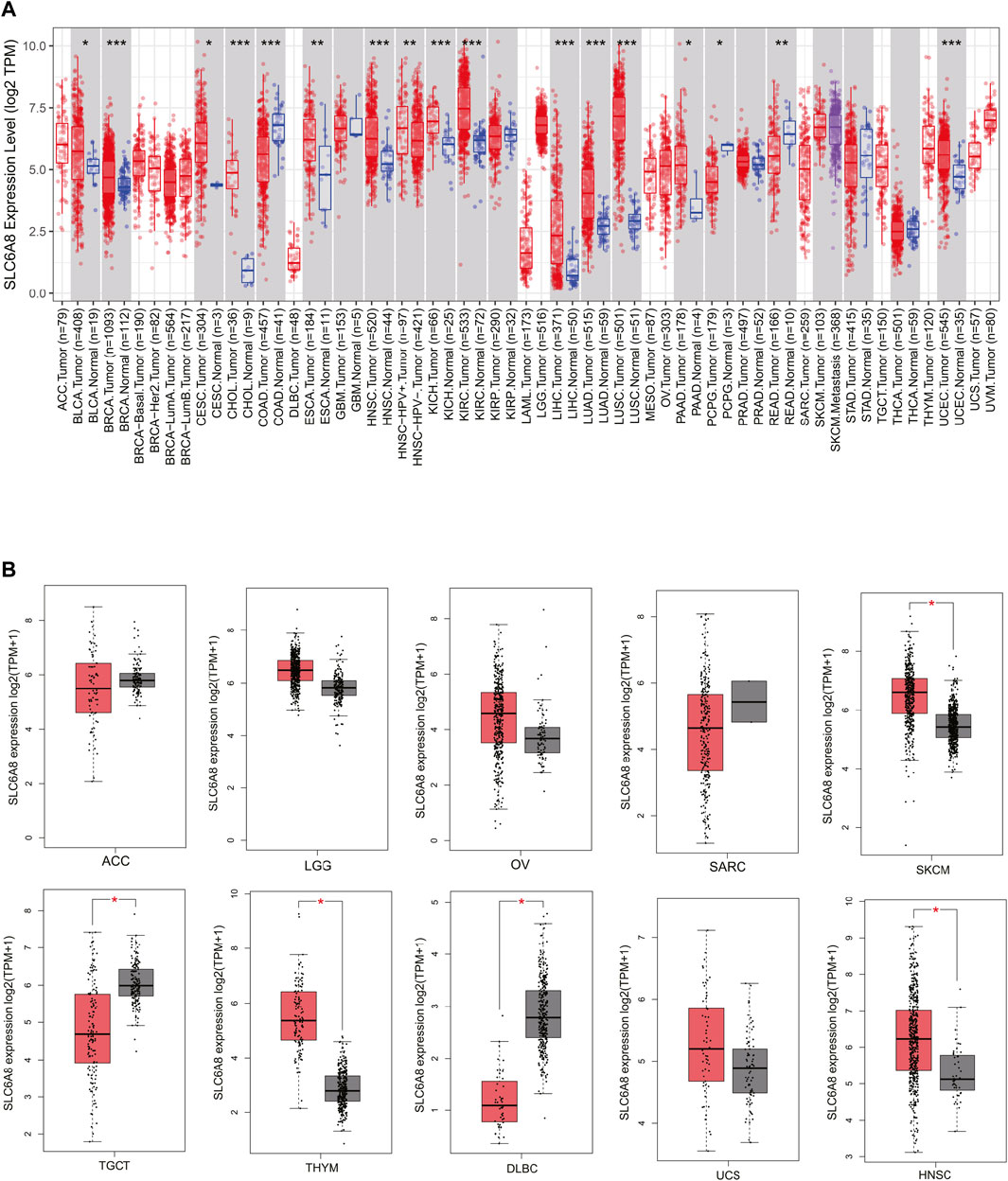
FIGURE 1. SLC6A8 expression profiles in cancers. (A) Pan-cancer differential-expression data for SLC6A8 between tumor tissues and paired normal tissues based on TCGA data. (B) Analysis of SLC6A8 expression levels in tumor tissues versus corresponding normal tissues via GEPIA2 platform. *p < 0.05, **p < 0.01, ***p < 0.001.
Pan-Cancer Analysis of the Prognostic and Predictive Value of SLC6A8
In order to assess the relationship between SLC6A8 expression and patients’ prognoses in 33 types of cancer, univariate cox regression analysis was performed based on data from TCGA. SLC6A8 expression was significantly associated with the overall survival (OS) of BRCA, LGG, mesothelioma (MESO), and prostate adenocarcinoma (PRAD), disease free interval (DFI) of BRCA and PRAD, disease specific survival (DSS) of LGG, LUAD, MESO, PRAD, and uveal Melanoma (UVM), progression free interval (PFI) in BRCA, MESO, and PRAD (Figures 2A–D). Kaplan–Meier survival curves of pan-cancer patients showed an adverse pan-cancer prognostic association of SLC6A8 expression (Figures 2E,F). Specifically, SLC6A8 expression was negatively associated with overall survival in LGG, LUAD, MESO, BRCA, and PRAD while positively associated with OS in UVM (Figure 3A). Consistently, SLC6A8 expression was negatively associated with the disease-free survival in LGG, MESO and, unexpectedly, SKCM (Figure 3B). Furthermore, we investigated the association between SLC6A8 expression and the prognosis of cancer patients through bioinformatics analysis of the GEO dataset via the Prognoscan database. Notably, upregulation of SLC6A8 was found to be inversely related to the overall survival in cancer of lung, breast and skin, relapse free survival (RFS) in cancer of the lung and breast, and distant metastasis-free survival (DMFS) in breast cancer. However, SLC6A8 expression was favorably related to the OS in blood, colorectal and ovarian cancer (Figure 3C).
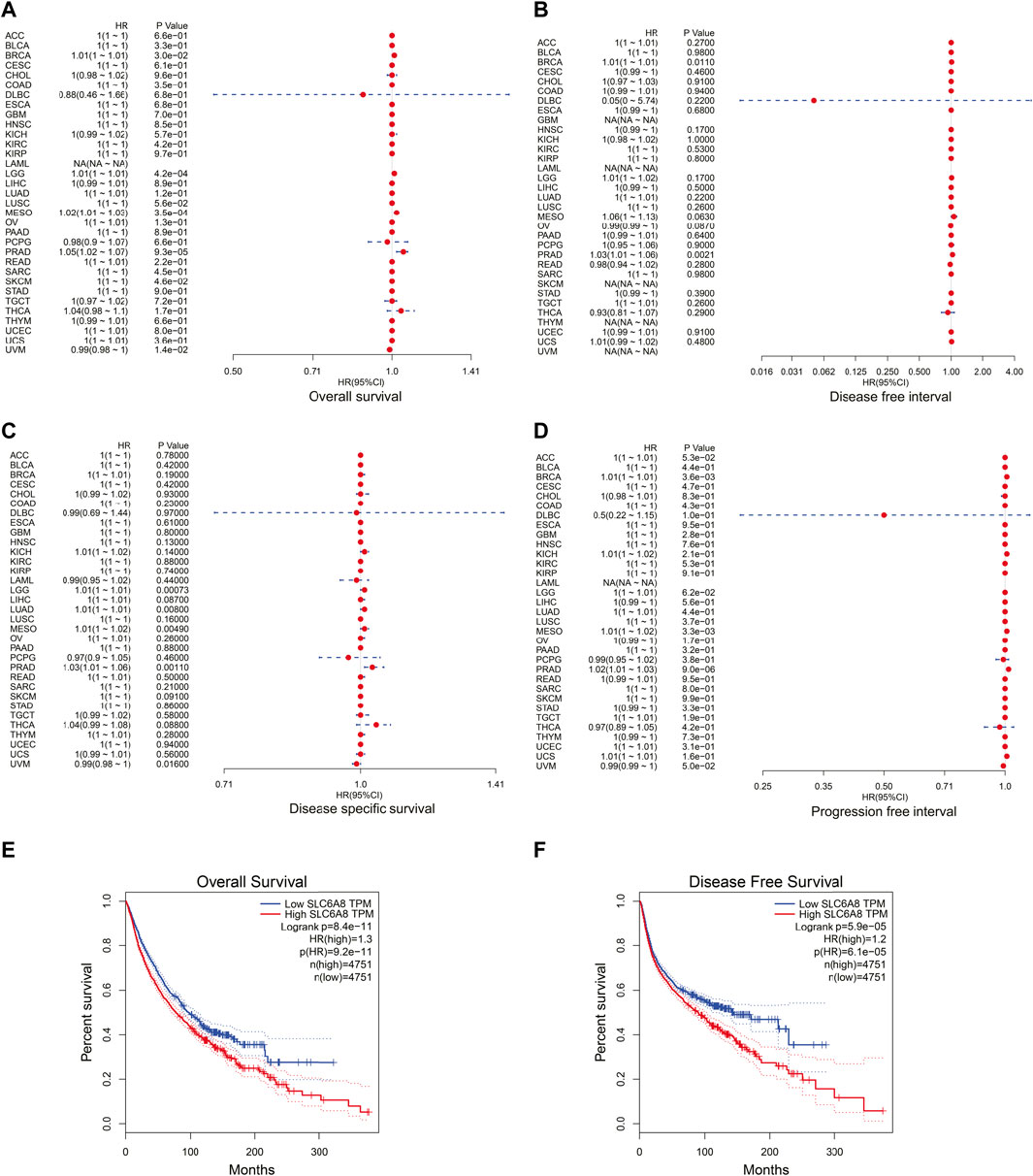
FIGURE 2. Correlation between SLC6A8 expression and cancer patients’ survival. (A–D) Forest plots of hazard ratios of SLC6A8 in different cancers regarding overall survival (A), disease-free interval (B), disease specific survival (C), and progression free interval (D). (E) and (F) Kaplan-Meier survival curves of overall survival (E) and disease-free survival (F) according to SLC6A8 expression in pan-cancer according in GEPIA2. *p < 0.05, **p < 0.01, ***p < 0.001.
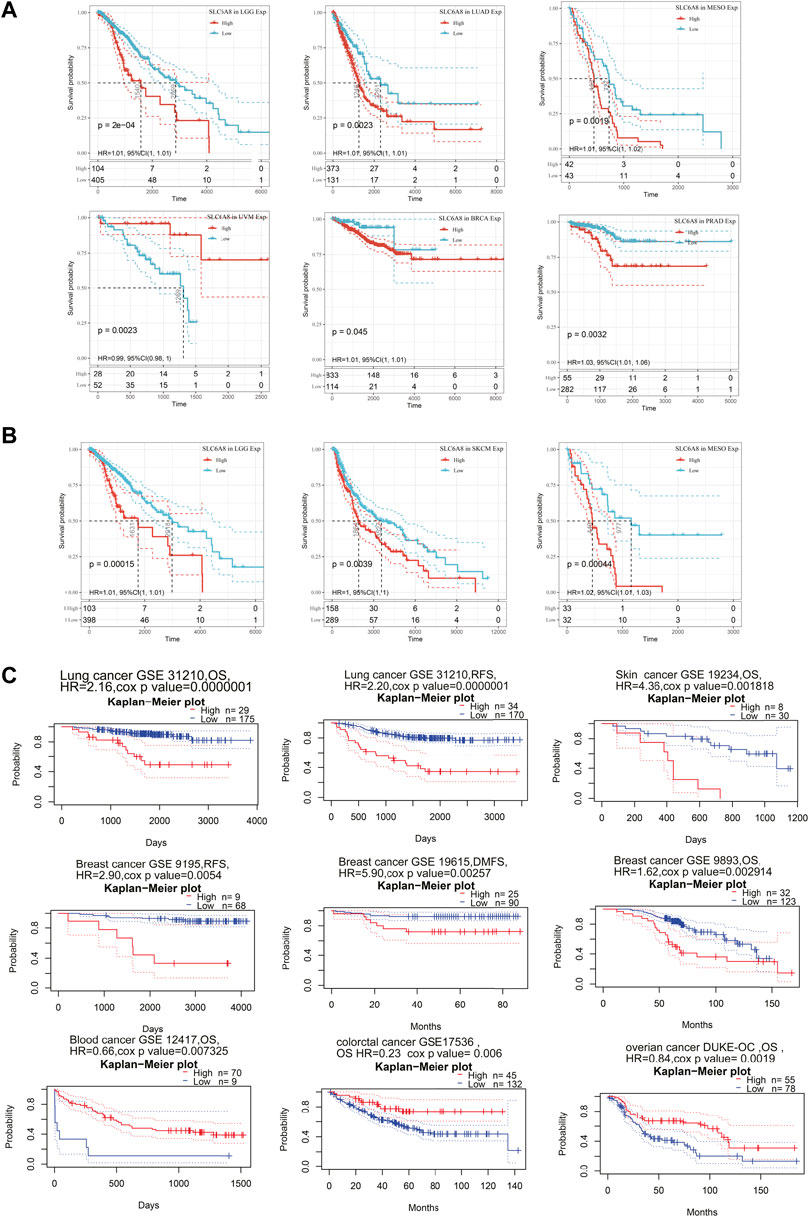
FIGURE 3. Survival analysis based on SLC6A8 expression in cancers. (A) and (B) Survival curves with significance of overall survival (A) and disease-free survival (B) based on SLC6A8 expression downloaded from TCGA. (C) Kaplan–Meier plots according to prognoscan database. *p < 0.05, **p < 0.01, ***p < 0.001.
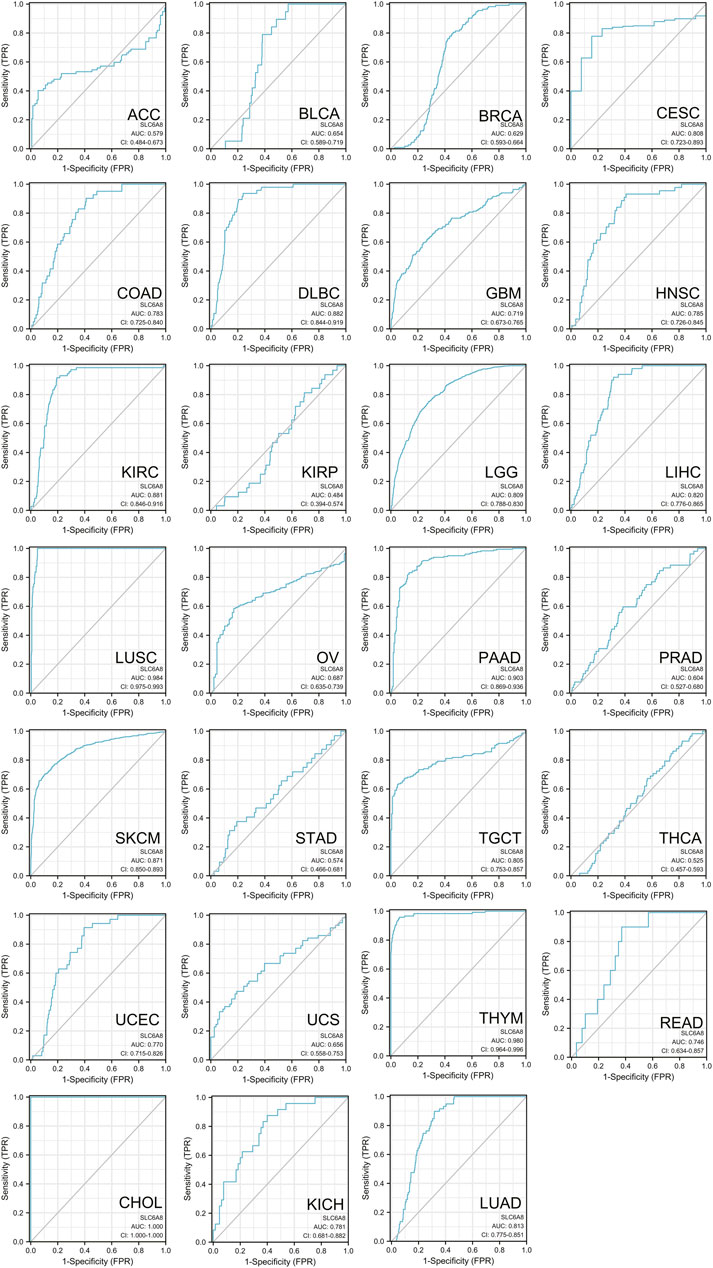
Besides, ROC curve was used to evaluate the clinical prediction accuracy of SLC6A8 in 27 cancer types. Remarkably, SLC6A8 was showed to be able to discriminate cancer patients from those without cancer in 26 of 27 types of cancer (AUC >0.5) (Figure 4). Specifically, the AUC value of three fourths of cancer exceeded 0.7, meaning that SLC6A8 was relatively accurate in identifying possible cancer patients. Taken together, SLC6A8 might be a potential prognostic and predictive factor in various cancers.
Pan-Cancer Analysis of the Mutation Landscape of SLC6A8
To investigate the genetic alteration of SLC6A8 across different types of cancer, bioinformatics analysis was performed based on the data of 10,967 samples from TCGA database via cBioPortal database. As shown in Figure 5A, “amplification” represented the primary mutation type for the majority of cancer types. And the highest alteration frequency (>10%) occurred in DLBC, also with “amplification” as the dominant type. PRAD showed the lowest alteration frequency of <1%. Notably, Mutation was the dominant type of genetic alteration in UCEC. Moreover, among the 72 mutations that are distributed between amino acids 52 and 581, X260 splice (from SKCM, STAD, and BRCA samples) was found to be the site with the highest mutation frequency (Figure 5B). The details of all SLC6A8 mutation profiles were summarized in Supplementary Table S1. Besides mutation, deletion and amplification, we further investigated the association between SLC6A8 and other genomic signatures, including gene expression, somatic copy number, somatic mutation, DNA methylation microRNA expression and protein level-RPPA. As illustrated by the circos plots, SLC6A8 was associated with other genomic signatures in ACC, BLCA, BRCA, CRC, ESCA&STAD, GBM, HNSC, LIHC, LUAD, LUSC, PRAD, and SKCM (Figure 5C). Thus, SLC6A8 is a potential factor contributing to modulation of the mutational landscape in various cancers.
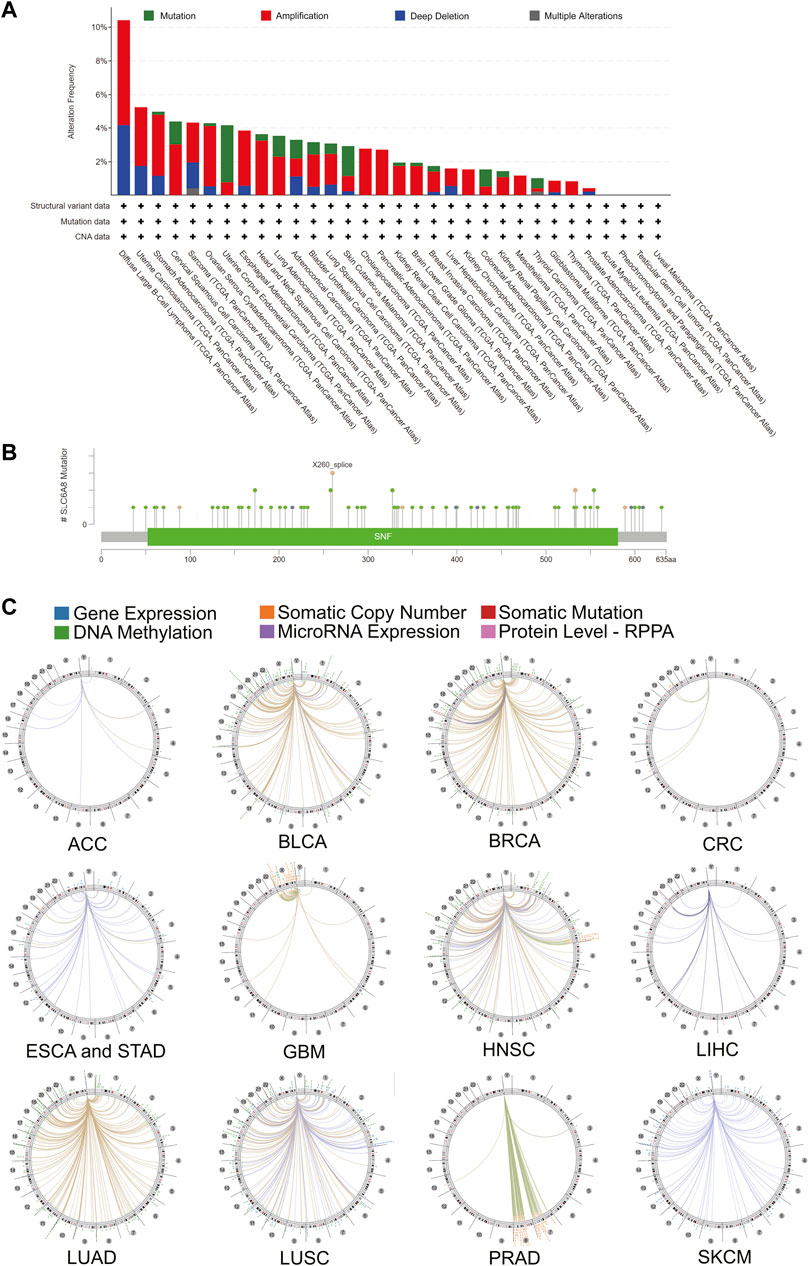
FIGURE 5. Mutational landscape of SLC6A8 and its association with genome in cancers. (A) SLC6A8 gene alteration frequency across cancers. (B) Mutation sites of SLC6A8 in pan-cancer. (C) The correlation between SLC6A8 and other signatures of genome based on TCGA data.
Pan-Cancer Analysis of the Biological Functions of SLC6A8
To explore the mechanisms underlying the oncogenic role of SLC6A8, pathway enrichment analyses were conducted following identification of genes and proteins that were associated with SLC6A8. As shown in Figure 6A, a total of 49 SLC6A8-interacting proteins were identified. Moreover, we analyzed the top 100 genes that were associated with SLC6A8 expression based on tumor data from GEPIA2. The expression of SLC6A8 exhibited the strongest positive correlation with that of ADM, GPI, NDRG1, PGK1, and PNCK (Figure 6B), and their correlation with SLC6A8 expression in distinct cancer types were illustrated by the heatmap, respectively (Figure 6C). Of note, interaction analysis of STRING-based interacting proteins and GEPIA-based correlated genes of SLC6A8 identified 2 common members (e.g., PNK1 and SGK1) (Figure 6D).
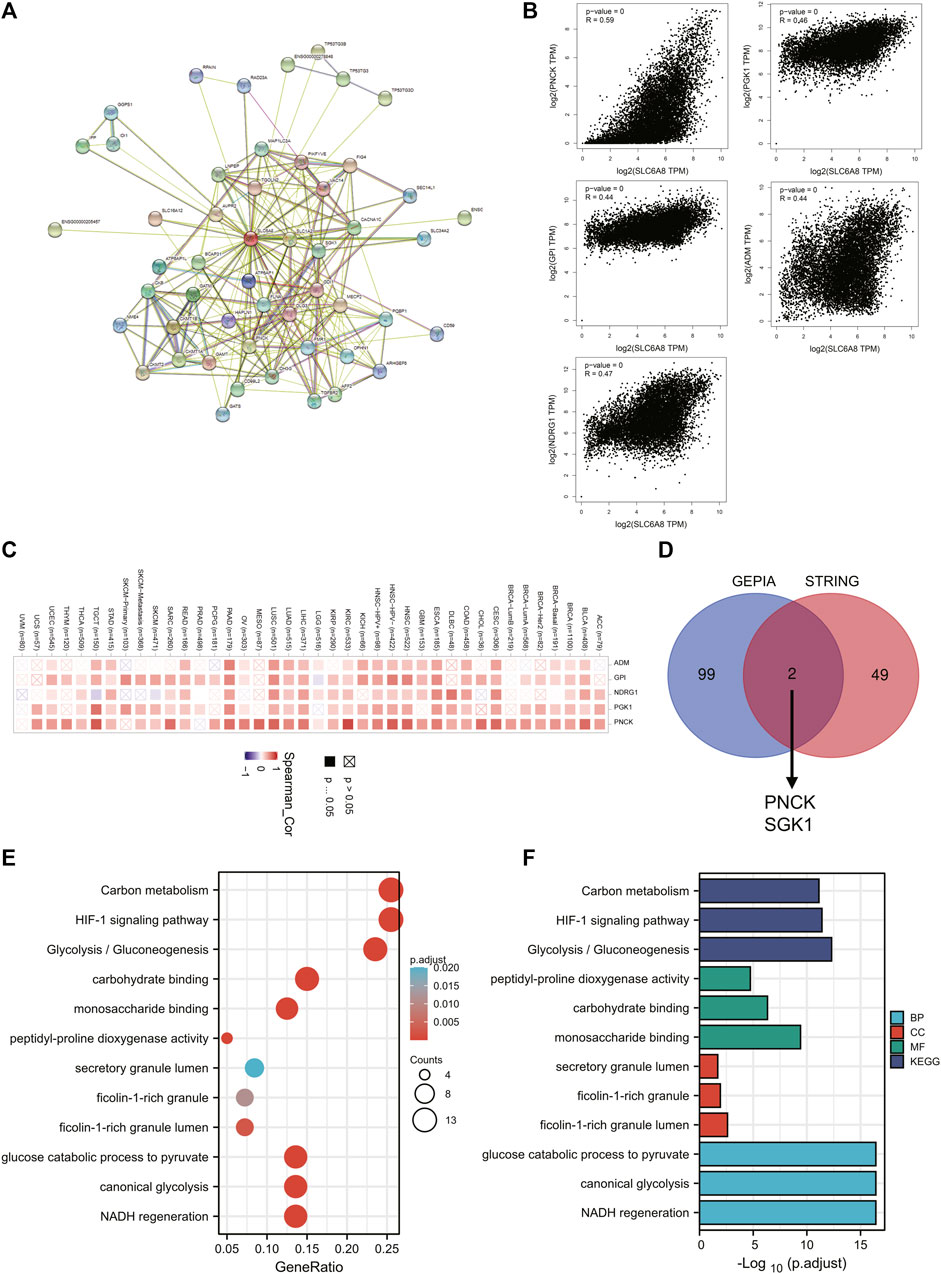
FIGURE 6. Analysis of pathways enriched for SLC6A8 in cancers. (A) Protein-protein interaction network of SLC6A8 was determined via STRING database. (B) Pearson correlation analysis of SLC6A8 expression and its top 5 related genes according to GEPIA2. (C) The expressions of the 5 genes that are most related to SLC6A8 expression in detailed cancer types were illustrated by the heatmap. (D) The venn diagram showing the common members of GEPIA2-based SLC6A8-associated genes and STRING-based SLC6A8-interacting proteins. (E) and (F) KEGG pathway (E) and GO (F) enrichment analysis based on SLC6A8-related genes (proteins).
To investigate the effects of SLC6A8 on cellular pathways and functions, we conducted KEGG and GO enrichment analysis based on SLC6A8-associated genes in tumor. KEGG analysis indicated that “Carbon metabolism,” “HIF-1 signaling pathway” and “Glycolysis/Gluconeogenesis” were three most enriched pathways (Figure 6E). Additionally, GO molecular function analysis revealed that SLC6A8 was associated to “peptidyl-proline dioxygenase activity,” “carbohydrate binding” and “monosaccharide binding” (Figure 6F). Thereof, these results suggest that SLC6A8 might influence tumor progression via reshaping cellular metabolism.
Pan-Cancer Analysis of the Relationship Between Immune Landscape and SLC6A8 Expression
To explore the relationship between tumor immune microenvironment and SLC6A8, we applied R software package to evaluate the stromal, immune, and estimate scores (Petitprez et al., 2020). SLC6A8 expression was negatively correlated with stromal, immune, and estimate scores in the majority of cancers analyzed (Figures 7A–C). Notably, the negative correlation between SLC6A8 and all three scores was the strongest in LUSC (Figure 8A). Next, we analyzed the relationship between SLC6A8 expression and scores of tumor-infiltrating immune cells based on data from TIMER database, and found that the most negatively related tumors were LUSC, SKCM and TGCT (Figure 8B). In addition, the correlation of SLC6A8 expression with various tumor-infiltrating immune cells was further explored in 30 types of cancer (Figure 9). SLC6A8 expression was found to be negatively associated with the majority of tumor-infiltrating immune cells in most of the cancers such as BRCA, UCS, LUSC, SKCM, KIRP, and KICH. Collectively, those results indicate that SLC6A8 might be involved in the regulation of tumor immune microenvironment.
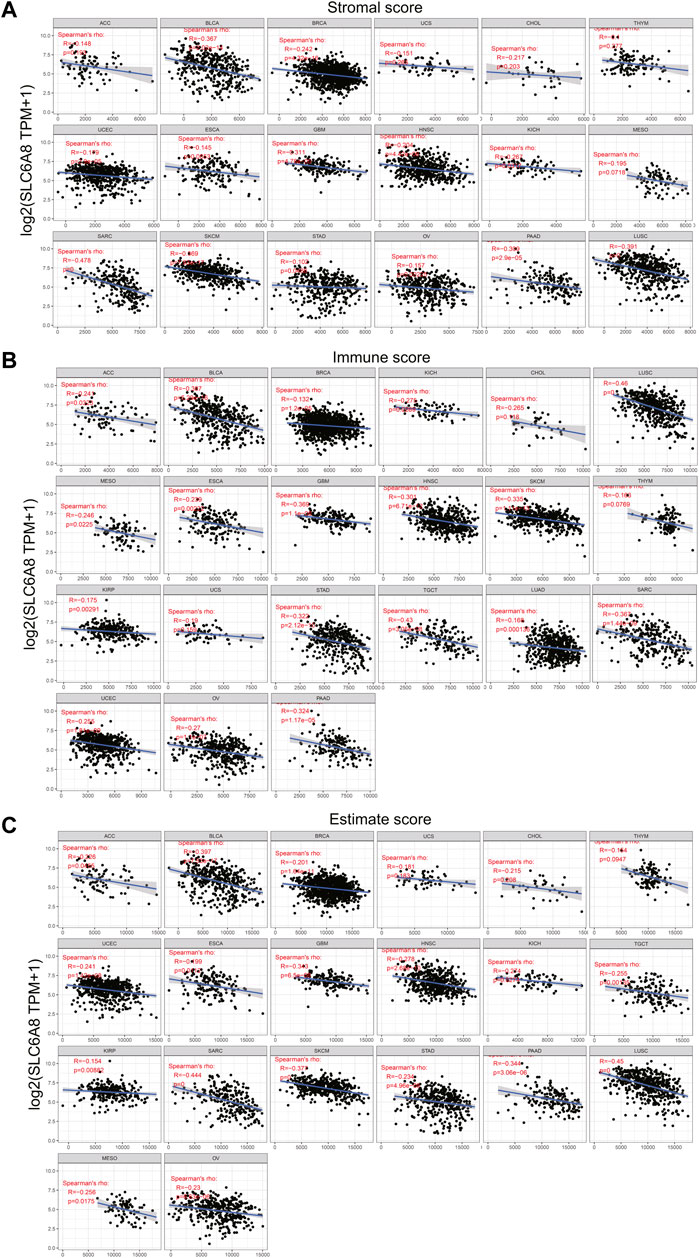
FIGURE 7. SLC6A8 expression was associated with immune conditions. (A)–(C) SLC6A8 expression was negatively related with immune score (A), stromal score (B) and estimate score (C) in various cancers.
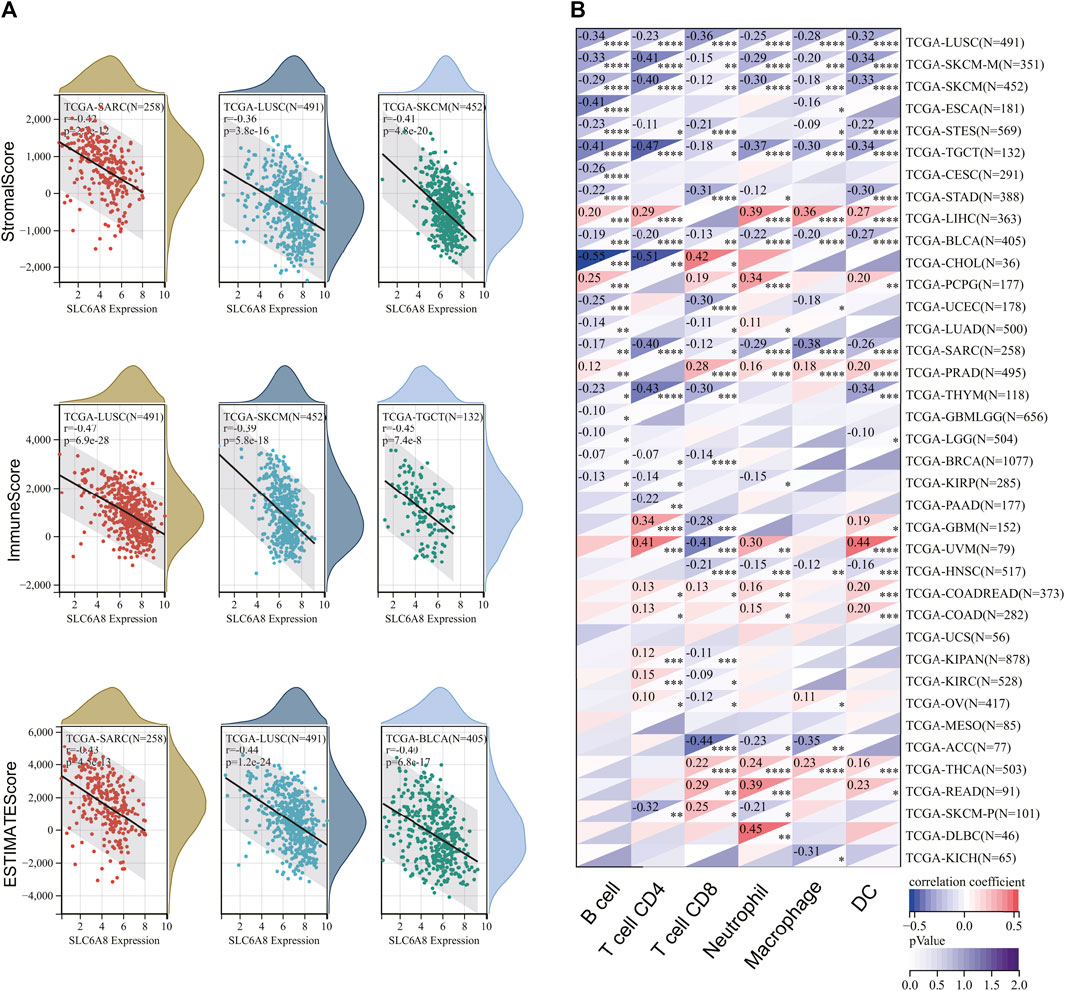
FIGURE 8. The correlation between SLC6A8 and immune status in different cancers. (A) The negative correlation between SLC6A8 and stromal, immune, and estimate scores is the strongest in LUSC. (B) Analysis of associations between SLC6A8 expression and scores of tumor-infiltrating immune cells in TIMER database.
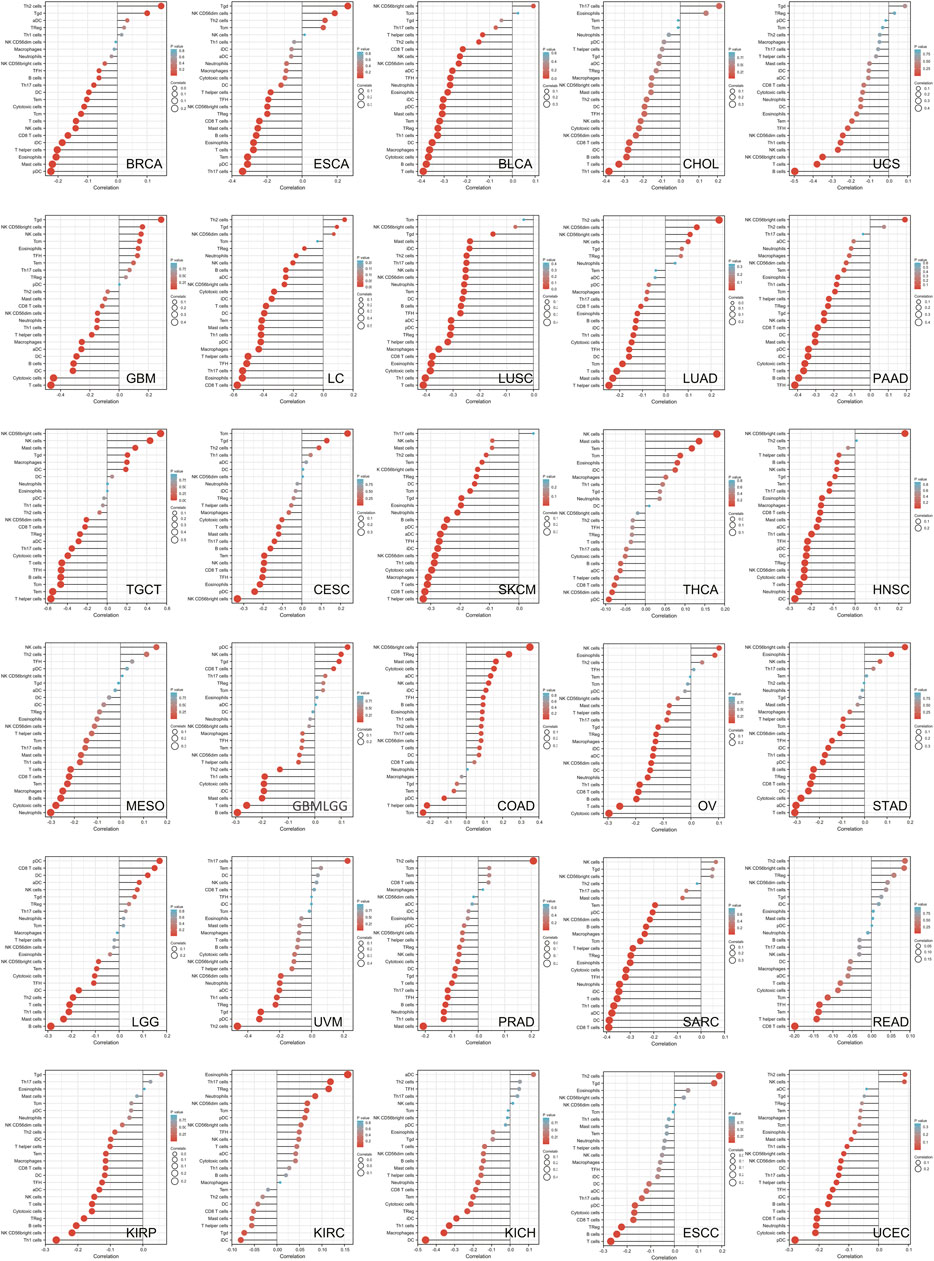
FIGURE 9. The stick charts showing the relationship between SLC6A8 expression and levels of diverse tumor-infiltrating immune cells in various cancers.
Pan-Cancer Analysis of the Association Between SLC6A8 Expression and Immunotherapy
To evaluate the association between SLC6A8 expression and immunotherapy, we analyzed the correlation between SLC6A8 expression and some representative immune checkpoints genes (Dijkstra et al., 2016). As displayed in Figure 10A, the expression of SLC6A8 was positively correlated with that of immune checkpoints genes in LAML, PCGC, and LIHC while negatively correlated with them in BLCA, SKCM and LUSC. Notably, CD276 expression was remarkably associated with SLC6A8 in 11 of 32 types of cancer (Figure 10A). MutL homologue 1 (MLH1), mutS homologue 2 (MSH2), mutS 6 (MSH6), postmeiotic segregation increased 2 (PMS2), and epithelial cell adhesion molecule (EPCAM) are the key components of the DNA mismatch repair (MMR) system, deficiency in which accounts for a significant proportion of human cancers by inducing instability in genome (McGrail et al., 2020). As illustrated by the heatmap, SLC6A8 expression had significantly positive correlation with these MMR genes in 8 types of cancer, including BRCA, HNSC, LIHC, LUSC, OV, PRAD, STAD, and UCEC (Figure 10B). MSI and TMB are both considered to be valuable predictive biomarkers of patients’ response to immune checkpoints inhibitors (ICIs) (Shum et al., 2022). SLC6A8 expression was significantly higher in MSI-high tumor tissues than MSI-low ones in KICH, LUSC, STAD and TGCT (Figure 10C). Additionally, SLC6A8 expression was also positively associated with TMB in BRCA, HNSC, KIRC, LAML, LUAD, MESO, PAAD, PRAD, SKCM, STAD, THCA, THYM, and UCEC (Figure 10D). Nevertheless, all statistically significant coefficients of SLC6A8 with MSI/TMB were below 0.4, indicating that SLC6A8 was a weak biomarker to predict response to immunotherapy.
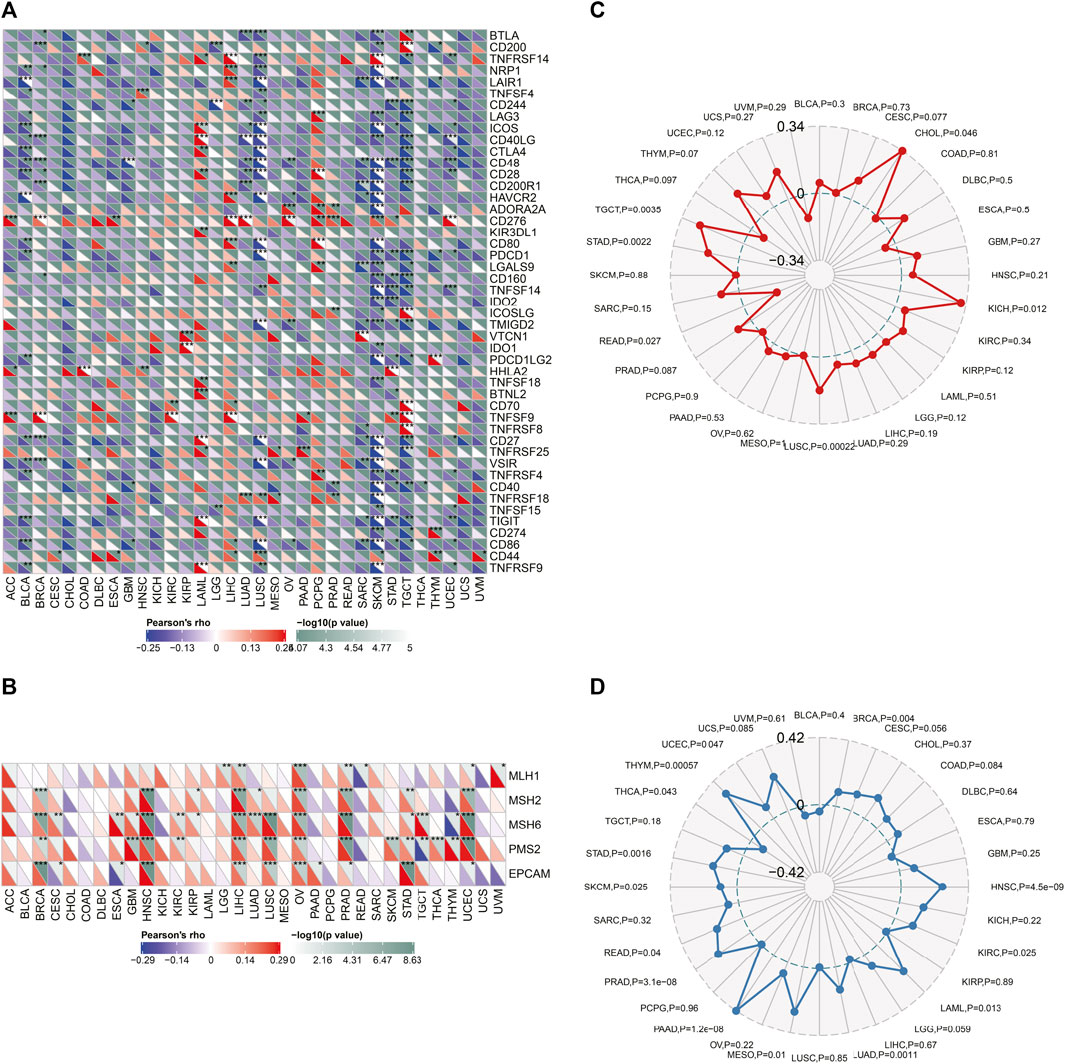
FIGURE 10. Relationship between SLC6A8 and response to immunotherapy. (A) Correlations between expression of SLC6A8 and representative immune checkpoint genes in different cancers. (B) Correlations of SLC6A8 and 5 component genes of MMR system across cancers. (C) and (D) Correlations of SLC6A8 with MSI (C) and TMB (D). *p < 0.05, **p < 0.01, ***p < 0.001.
Disscussion
The SLC6A8 gene is located on the long arm of chromosome X and contains 14 exons. By acting as a plasma membrane transporter, SLC6A8 protein enables cells to import extracellular creatine or its precursor, guanidinoacetate, into cells against steep concentrations, thus maintaining the intracellular creatine levels (Jomura et al., 2022). Recently, SLC6A8 was reported to be involved in the malignant progression of several cancers, including NSCLC, CRC, and HCC via regulation of key signaling pathways. Our previous works have demonstrated that SLC6A8 could promote the survival of triple-negative breast cancer cells by maintaining intracellular creatine levels (Li et al., 2021). Nevertheless, analysis of the role of SLC6A8 in pan-cancer remains unexplored. Here, we conducted a pan-cancer bioinformatics analysis of the expression and mutation profile, and clinical significance of SLC6A8 and its potential effect on malignancy.
In this work, we sought to thoroughly investigate the function of SLC6A8 in various cancers by integrating complementary bioinformatics methods and public databases. The results of gene expression analysis showed that SLC6A8 was upregulated in most types of the cancers while downregulated in a few types of cancers as compared to their corresponding normal counterparts (Figures 1A,B). According to the results of the survival analysis, SLC6A8 expression predicted poor overall survival and disease-free survival of cancer patients (Figures 2E,F). Intriguingly, SLC6A8 expression had positive correlation with the overall survival of patients diagnosed with blood, colorectal and ovarian cancer (Figure 3C). ROC curve analysis results indicated that SLC6A8 expression was able to discriminate cancer patients with relatively good accuracy (AUC>0.5) (Figure 4). Our findings unprecedentedly revealed the potential prognostic and diagnostic value of SLC6A8 in pan-cancer.
Cancer immunotherapies have been proved to induce durable responses in multiple malignancies, and thus have transformed the treatment landscapes for a subset of cancers: NSCLC, GC, and BLCA, to name a few (Borcoman et al., 2019). One of the vital strategies of immunotherapy is to overcome cancer immune evasion by modulating the functionality of immune cells (Marin-Acevedo et al., 2021). For instance, GBM stem cells (GSCs) escape immune clearance via increasing recruitment of tumor-associated macrophages (Gangoso et al., 2021). Accumulating studies have illustrated that SLC6A8 was intricately implicated in cell immunity by regulating immune cells including T cells and macrophages (Ji et al., 2019; Peng et al., 2021). Herein, we found that SLC6A8 expression was negatively correlated with the stromal, immune, and estimate score in the majority of cancers (Figures 7, 8). Besides, SLC6A8 had negative correlation with the levels of various tumor-infiltrating immune cells in pan-cancer (Figure 9). These results collectively indicated that SLC6A8 might be associated with enhanced immunosuppressive tumor microenvironment. Notably, ICIs have revolutionized the paradigm in cancer immunotherapy (Nakajima and Nakatsura, 2021). Mechanistically, ICIs could competitively interact with immune checkpoint proteins to relieve cancer-mediated inhibition of T-cell function (Liu et al., 2020). Our study firstly uncovered the association between SLC6A8 and several common biomarkers of response to ICIs (e.g., levels of checkpoint genes, MSI, and TMB) in multiple cancers (Figure 10). These results suggest that SLC6A8 expression is a potential biomarker of patient prognosis and response to immunotherapy.
In spite of the fact that numerous public databases have been integrated to clarify the role of SLC6A8 in pan-cancer, several limitations to the present work should be pointed out. First, simple combination of sequencing and microarray data across different databases for analysis of tumor information might bias the data and reduce the reliability of the analysis due to a lack of analysis of data covering cellular-level information. Second, all the conclusions in this study were solely based on the results of bioinformatics analysis. Further in vitro/vivo experiments are necessarily needed to validate the results. Our next step is to confirm and elucidate the role of SLC6A8 across different cancers experimentally. Finally, according to the results of the study, SLC6A8 exhibited conflicting trends in expression profiles, patient prognosis, and tumor immunology in different types of cancer, which greatly complicated our understanding of the oncogenic role of SLC6A8. Therefore, more comprehensive and coherent scientific research on this gene should be conducted.
Conclusion
To summarize, our study uncovered the expression profile and clinical significance of SLC6A8 in pan-cancer through bioinformatics analysis for the first time. The aberrant expression of SLC6A8 predicts poor survival and distinguishes cancer patients from those without cancer in numerous cancers. SLC6A8 gene is often altered, with amplification being the most common type of alteration. Mechanistically, the results of KEGG and GO analyses showed that SLC6A8 was significantly enriched in metabolism-associated pathways such as “Carbon metabolism” and “Glycolysis/Gluconeogenesis.” Additionally, SLC6A8 expression is related to immune infiltration levels, expression of immune checkpoint genes, TMB, and MSI, suggesting the potential of SLC6A8 for prediction of the efficacy of ICIs.
Data Availatbility Statenent
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
QL conceived and supervised the study. XY analyzed the data and drew the figures. QL and XY wrote and edited the manuscript. Both authors read and approved the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.916439/full#supplementary-material
References
Battini, R., Chilosi, A., Mei, D., Casarano, M., Alessandrì, M. G., Leuzzi, V., et al. (2007). Mental Retardation and Verbal Dyspraxia in a New Patient with De Novo Creatine Transporter (SLC6A8) Mutation. Am. J. Med. Genet. 143A, 1771–1774. doi:10.1002/ajmg.a.31827
Bonilla, D., Kreider, R., Stout, J., Forero, D., Kerksick, C., Roberts, M., et al. (2021). Metabolic Basis of Creatine in Health and Disease: A Bioinformatics-Assisted Review. Nutrients 13, 1238. doi:10.3390/nu13041238
Borcoman, E., Kanjanapan, Y., Champiat, S., Kato, S., Servois, V., Kurzrock, R., et al. (2019). Novel Patterns of Response under Immunotherapy. Ann. Oncol. 30, 385–396. doi:10.1093/annonc/mdz003
Camidge, D. R., Doebele, R. C., and Kerr, K. M. (2019). Comparing and Contrasting Predictive Biomarkers for Immunotherapy and Targeted Therapy of NSCLC. Nat. Rev. Clin. Oncol. 16, 341–355. doi:10.1038/s41571-019-0173-9
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio Cancer Genomics Portal: an Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2, 401–404. doi:10.1158/2159-8290.cd-12-0095
Chai, A. W. Y., Yee, P. S., and Cheong, S. C. (2022). Rational Combinations of Targeted Therapy and Immune Checkpoint Inhibitors in Head and Neck Cancers. Front. Oncol. 12, 837835. doi:10.3389/fonc.2022.837835
Di Biase, S., Ma, X., Wang, X., Yu, J., Wang, Y.-C., Smith, D. J., et al. (2019). Creatine Uptake Regulates CD8 T Cell Antitumor Immunity. J. Exp. Med. 216, 2869–2882. doi:10.1084/jem.20182044
Dijkstra, K. K., Voabil, P., Schumacher, T. N., and Voest, E. E. (2016). Genomics- and Transcriptomics-Based Patient Selection for Cancer Treatment with Immune Checkpoint Inhibitors: A Review. JAMA Oncol. 2, 1490–1495. doi:10.1001/jamaoncol.2016.2214
Feng, Y., Guo, X., and Tang, H. (2021). SLC6A8 Is Involved in the Progression of Non-small Cell Lung Cancer through the Notch Signaling Pathway. Ann. Transl. Med. 9, 264. doi:10.21037/atm-20-5984
Gangoso, E., Southgate, B., Bradley, L., Rus, S., Galvez-Cancino, F., Mcgivern, N., et al. (2021). Glioblastomas Acquire Myeloid-Affiliated Transcriptional Programs via Epigenetic Immunoediting to Elicit Immune Evasion. Cell 184, 2454–2470. doi:10.1016/j.cell.2021.03.023
Gotwals, P., Cameron, S., Cipolletta, D., Cremasco, V., Crystal, A., Hewes, B., et al. (2017). Prospects for Combining Targeted and Conventional Cancer Therapy with Immunotherapy. Nat. Rev. Cancer 17, 286–301. doi:10.1038/nrc.2017.17
Ji, L., Zhao, X., Zhang, B., Kang, L., Song, W., Zhao, B., et al. (2019). Slc6a8-Mediated Creatine Uptake and Accumulation Reprogram Macrophage Polarization via Regulating Cytokine Responses. Immunity 51, 272–284. doi:10.1016/j.immuni.2019.06.007
Jomura, R., Akanuma, S.-i., Tachikawa, M., and Hosoya, K.-i. (2022). SLC6A and SLC16A Family of Transporters: Contribution to Transport of Creatine and Creatine Precursors in Creatine Biosynthesis and Distribution. Biochim. Biophys. Acta Biomembr. 1864, 183840. doi:10.1016/j.bbamem.2021.183840
Joncquel-Chevalier Curt, M., Voicu, P.-M., Fontaine, M., Dessein, A.-F., Porchet, N., Mention-Mulliez, K., et al. (2015). Creatine Biosynthesis and Transport in Health and Disease. Biochimie 119, 146–165. doi:10.1016/j.biochi.2015.10.022
Kennedy, L. B., and Salama, A. K. S. (2020). A Review of Cancer Immunotherapy Toxicity. CA Cancer J. Clin. 70, 86–104. doi:10.3322/caac.21596
Kurth, I., Yamaguchi, N., Andreu-Agullo, C., Tian, H. S., Sridhar, S., Takeda, S., et al. (2021). Therapeutic Targeting of SLC6A8 Creatine Transporter Suppresses Colon Cancer Progression and Modulates Human Creatine Levels. Sci. Adv. 7, eabi7511. doi:10.1126/sciadv.abi7511
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 77, e108–e110. doi:10.1158/0008-5472.can-17-0307
Li, Q., Liu, M., Sun, Y., Jin, T., Zhu, P., Wan, X., et al. (2021a). SLC6A8-mediated Intracellular Creatine Accumulation Enhances Hypoxic Breast Cancer Cell Survival via Ameliorating Oxidative Stress. J. Exp. Clin. Cancer Res. 40, 168. doi:10.1186/s13046-021-01933-7
Li, S., Bianconi, S., Van Der Veen, J. W., Dang Do, A., Stolinski, J., Cecil, K. M., et al. (2021b). Oxidative Phosphorylation in Creatine Transporter Deficiency. NMR Biomed. 34, e4419. doi:10.1002/nbm.4419
Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., Lazar, A. J., Cherniack, A. D., et al. (2018). An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–e11. doi:10.1016/j.cell.2018.02.052
Liu, X., Bao, X., Hu, M., Chang, H., Jiao, M., Cheng, J., et al. (2020). Inhibition of PCSK9 Potentiates Immune Checkpoint Therapy for Cancer. Nature 588, 693–698. doi:10.1038/s41586-020-2911-7
Marin-Acevedo, J. A., Kimbrough, E. O., and Lou, Y. (2021). Next Generation of Immune Checkpoint Inhibitors and beyond. J. Hematol. Oncol. 14, 45. doi:10.1186/s13045-021-01056-8
McGrail, D. J., Garnett, J., Yin, J., Dai, H., Shih, D. J. H., Lam, T. N. A., et al. (2020). Proteome Instability is a Therapeutic Vulnerability in Mismatch Repair-Deficient Cancer. Cancer cell 37, 371–386. doi:10.1016/j.ccell.2020.01.011
Mongre, R. K., Jung, S., Mishra, C. B., Lee, B. S., Kumari, S., and Lee, M. S. (2019). Prognostic and Clinicopathological Significance of SERTAD1 in Various Types of Cancer Risk: A Systematic Review and Retrospective Analysis. Cancers (Basel) 11, 337. doi:10.3390/cancers11030337
Nakajima, H., and Nakatsura, T. (2021). Towards the Era of Immune Checkpoint Inhibitors and Personalized Cancer Immunotherapy. Immunol. Med. 44, 10–15. doi:10.1080/25785826.2020.1785654
Peng, M., Ren, J., Jing, Y., Jiang, X., Xiao, Q., Huang, J., et al. (2021). Tumour-derived Small Extracellular Vesicles Suppress CD8+ T Cell Immune Function by Inhibiting SLC6A8-Mediated Creatine Import in NPM1-Mutated Acute Myeloid Leukaemia. J. Extracell. Vesicles 10, e12168. doi:10.1002/jev2.12168
Petitprez, F., Levy, S., Sun, C.-M., Meylan, M., Linhard, C., Becht, E., et al. (2020). The Murine Microenvironment Cell Population Counter Method to Estimate Abundance of Tissue-Infiltrating Immune and Stromal Cell Populations in Murine Samples Using Gene Expression. Genome Med. 12, 86. doi:10.1186/s13073-020-00783-w
Salomons, G. S., Van Dooren, S. J. M., Verhoeven, N. M., Marsden, D., Schwartz, C., Cecil, K. M., et al. (2003). X-linked Creatine Transporter Defect: An Overview. J. Inherit. Metab. Dis. 26, 309–318. doi:10.1023/a:1024405821638
Samborska, B., Roy, D. G., Rahbani, J. F., Hussain, M. F., Ma, E. H., Jones, R. G., et al. (2022). Creatine Transport and Creatine Kinase Activity is Required for CD8+ T Cell Immunity. Cell Rep. 38, 110446. doi:10.1016/j.celrep.2022.110446
Shum, B., Larkin, J., and Turajlic, S. (2022). Predictive Biomarkers for Response to Immune Checkpoint Inhibition. Seminars Cancer Biol. 79, 4–17. doi:10.1016/j.semcancer.2021.03.036
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/measurement Sets. Nucleic acids Res. 49, D605–D612. doi:10.1093/nar/gkaa1074
Tang, Z., Kang, B., Li, C., Chen, T., and Zhang, Z. (2019). GEPIA2: an Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 47, W556–W560. doi:10.1093/nar/gkz430
Yıldız, Y., Göçmen, R., Yaramış, A., Coşkun, T., and Haliloğlu, G. (2020). Creatine Transporter Deficiency Presenting as Autism Spectrum Disorder. Pediatrics 146, e20193460. doi:10.1542/peds.2019-3460
Yoshioka, H., Shimokawa, M., Seto, T., Morita, S., Yatabe, Y., Okamoto, I., et al. (2019). Final Overall Survival Results of WJTOG3405, a Randomized Phase III Trial Comparing Gefitinib versus Cisplatin with Docetaxel as the First-Line Treatment for Patients with Stage IIIB/IV or Postoperative Recurrent EGFR Mutation-Positive Non-small-cell Lung Cancer. Ann. Oncol. 30, 1978–1984. doi:10.1093/annonc/mdz399
Keywords: SLC6A8, pan-cancer, prognosis, immunology, bioinformatics analysis
Citation: Yang X and Li Q (2022) Pan-Cancer Analysis of the Oncogenic and Immunological Role of Solute Carrier Family 6 Member 8 (SLC6A8). Front. Genet. 13:916439. doi: 10.3389/fgene.2022.916439
Received: 09 April 2022; Accepted: 31 May 2022;
Published: 17 August 2022.
Edited by:
Hauke Thomsen, ProtaGene CGT GmbH, GermanyReviewed by:
Liudmila V. Spirina, Russian Academy of Sciences, RussiaOlivier Braissant, Centre Hospitalier Universitaire Vaudois (CHUV), Switzerland
Copyright © 2022 Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiao Li, bGlxaWFvMTk5NzUxOEAxNjMuY29t
 Xin Yang
Xin Yang Qiao Li
Qiao Li