- 1Clinical Medical College of Jining Medical University, Jining, China
- 2Department of Emergency, Jining No. 1 People’s Hospital, Jining, China
Several studies have reported that chromosome 9p21 is significantly associated with ischemic stroke (IS) risk, with the G allele associated with increased risk. However, controversial results have been reported in the literature. We systematically assessed the relationship between stroke and three 9p21 loci (rs2383206, rs2383207, and rs10757278) in this meta-analysis. First, we searched the PubMed and Embase databases for relevant studies. We then calculated odds ratios using the chi-squared test. The evaluation of experimental data was performed using bias tests and sensitivity analyses. We analyzed data from 16 studies involving 18,584 individuals of Chinese ancestry, including 14,033 cases and 14,656 controls. Our results indicated that chromosome 9p21 is significantly associated with IS (odds ratio: 1.15, 95% confidence interval: 1.1–1.20, p < 0.0001). Because the three single-nucleotide polymorphisms (rs2383206, rs2383207, and 10757278) have a linkage disequilibrium relationship, all three may increase the risk of IS.
Introduction
Stroke is a severe disease and is the leading cause of disability and death in China (Liu et al., 2011). It is an acute cerebrovascular disease that is characterized by focal loss of nerve function and high mortality and disability, and it currently poses a serious threat to human life and health (Li et al., 2021). Stroke is thought to be caused by environmental risk factors, multiple genes, and their interactions. To date, however, a large proportion of stroke risk remains unexplained (Ganesh et al., 2016). Genetic variation on chromosome 9p21 is widely believed to be linked to risk of coronary heart disease (McPherson et al., 2007; Samani et al., 2007), but it has a different role in stroke (Matarin et al., 2008; Gschwendtner et al., 2009). Previously, genome-wide association studies (GWAS) have analyzed genes associated with ischemic stroke (IS) (Söderholm et al., 2019). Single-nucleotide polymorphisms (SNPs) of rs2383206, rs2383207, and rs10757278 on chromosome 9p21 are linked to stroke. However, although several recent genetic studies have reported that chromosome 9p21 plays an important role in the mechanism of stroke, studies of different races and from different geographic locations have provided very different results. Therefore, an association between 9p21 polymorphisms and stroke has been established for individuals of European descent; the main aim of this meta-analysis was to study the relationship between three SNPs on chromosome 9p21 and stroke in the Chinese population.
Methods
Search Strategy
We searched the PubMed and Embase databases and selected all possible studies using the keywords “Stroke Chinese” and “rs2383206,” “rs2383207,” “rs10757278,” and “9p21.” The relevant literature was updated on 31 January 2022.
Selection Criteria
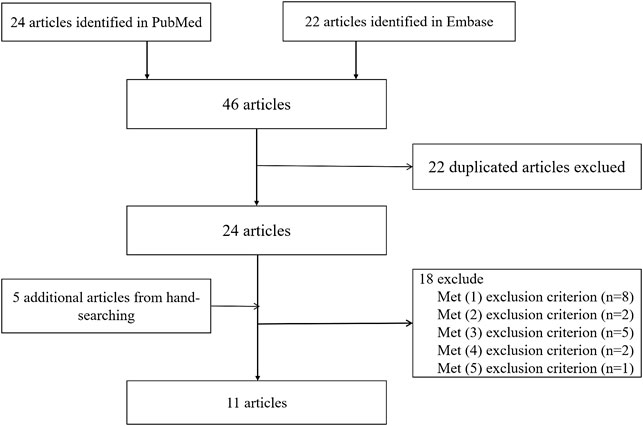
The following selection criteria were used: (1) an association between the proposed SNPs and stroke was evaluated using a case–control design; (2) an accurate genotype number was provided or could be calculated (Liu et al., 2013); (3) the odds ratio (OR) and 95% confidence interval (CI) were provided to measure the risk of disease; (4) the OR value and 95% CI were calculated by providing enough data; (5) the same diagnostic criteria were used for stroke. The exclusion criteria were (1) the research was presented as a poster presentation, summary, meta-analysis, conference summary or article, or case series analysis; (2) the study was not performed in a Chinese population; (3) the three SNPs were not used; (4) the study was not consistent with the research topic; and (5) the exact number of genotypes was not provided and could not be calculated and/or the OR and 95% CI were not provided and could not be calculated. Two authors (DW and XH) independently screened all studies by their title or abstract and then evaluated the full text. Any differences in opinion were resolved through discussion.
Data Extraction
Trial data from each identified study were extracted separately by two investigators (DW and XH). Any differences were eliminated by discussing the data extraction for each study using standard data collection tables. The data and information that were extracted for inclusion in the analysis included the first author’s name, publication year, language, population, study type, sample size, numbers, and frequencies of rs2383206, rs2383207, and rs10757278 polymorphism genotypes in the cases and controls, ORs, and 95% CIs. All extracted data are presented in Tables 1, 2.
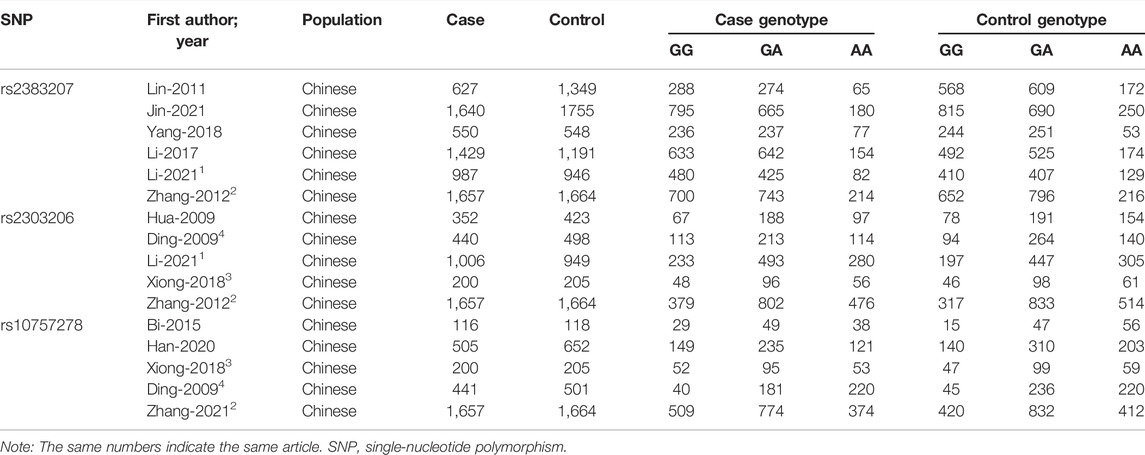
TABLE 1. Sixteen studies in 11 articles investigating the association between rs2383207, rs2383206, and rs10757278 and IS.
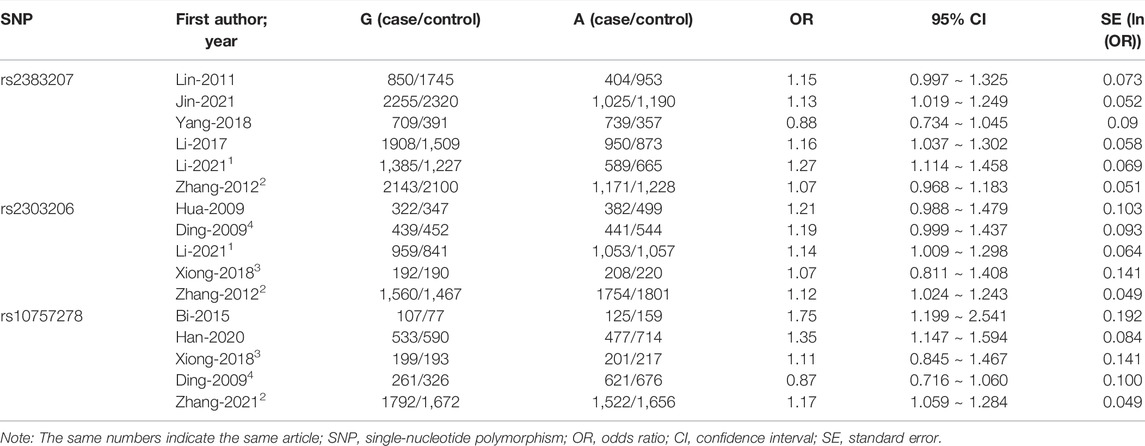
TABLE 2. Correlation analysis between different genetic patterns of rs2383207, rs2383206, and rs10757278 at 9p21 locus and IS susceptibility.
Statistical Analysis
We investigated the Hardy–Weinberg equilibrium of rs2383206, rs2383207, and rs10757278. We also investigated their association with stroke using the chi-squared test, which was performed using R (http://www.r-project.org/) (Liu et al., 2013). For the meta-analysis, we determined the heterogeneity among datasets using Cochran’s Q test and I2 = (Q – (k – 1))/Q × 100%. The Q statistic approximately follows a χ2 distribution, with k-1 degrees of freedom (k is the number of studies in the analysis) (Liu et al., 2017). When I2 was greater than 50% and the p-value was less than 0.1 (Higgins et al., 2021), the DerSimonian and Laird random-effects model was used as the pooling method; otherwise, the Mantel–Haenszel or inverse variance fixed-effects model was used as the pooling method, as appropriate. We also used funnel plots to assess potential publication bias. When there is no bias, funnel plots are symmetrical; conversely, when bias is present, funnel plots are asymmetrical (Liu et al., 2014).
Results
Characteristics of Included Studies
In this meta-analysis, 18,584 participants were included: 14,033 in the IS group (7,235 cases with rs2383207, 3,762 cases with rs2383206, and 3,036 cases with rs10757278) and 14,656 cases in the control group (7,653 cases with rs2383207, 3,807 cases with rs2383206, and 3,196 cases with rs10757278). Eleven articles were selected, comprising 16 studies, of which six investigated rs2383207 (Lin et al., 2011; Zhang et al., 2012; Li et al., 2017; Yang et al., 2018; Jin et al., 2021; Li et al., 2021), five investigated rs2383206 (Ding et al., 2009; Hu et al., 2009; Zhang et al., 2012; Xiong et al., 2018; Li et al., 2021), and five investigated rs10757278 (Ding et al., 2009; Zhang et al., 2012; Bi et al., 2015; Xiong et al., 2018; Han et al., 2020). The study identification and selection process is shown in detail in Figure 1.
Linkage Disequilibrium
The three SNPs—rs10757278, rs2383206, and rs2383207—are located within 10 kb of one another on chromosome 9p21 (https://snipa.helmholtz-muenchen.de/snipa3/).
Meta-Analysis Results of 9p21
There is a linkage disequilibrium relationship among the three SNPS (rs2303206, rs2383207, and rs10757278). Thus, we performed an analysis of the OR values of all studies involving rs2383206, rs2383207, and rs10757278 in which the G allele was a minor allele. Because I2 = 50%, a random-effects model was used to compare alleles (Figure 2). Chromosome 9p21 was significantly associated with IS risk, and the G allele was associated with increased IS risk (OR: 1.14, 95% CI: 1.08–1.19, p < 0.0001, Figure 2).
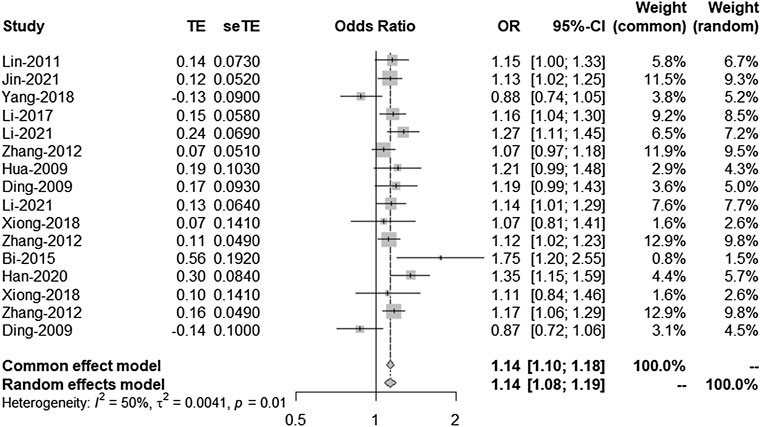
FIGURE 2. Random-effects meta-analysis of the association between the three single-nucleotide polymorphisms (SNPs; rs2383207, rs2383206, and rs10757278) and ischemic stroke (IS). CI, confidence interval; OR, odds ratios.
Publication Bias
The Harbord test was used to evaluate publication bias. The bias = 0.3228, p = 0.7661, indicating no publication bias in the studies of 9p21 (Figure 3).
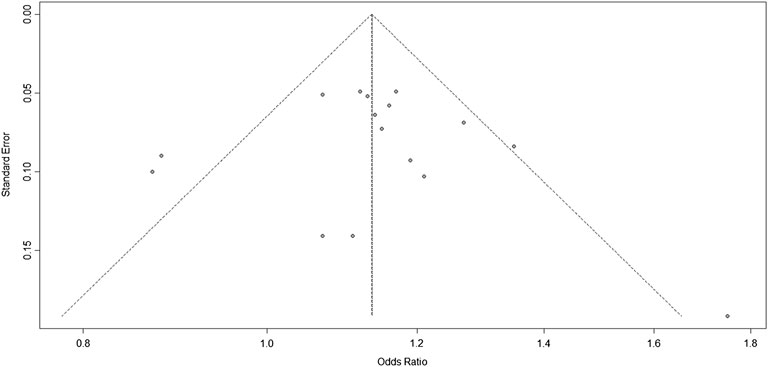
FIGURE 3. Funnel plots corresponding to the random-effects meta-analysis of all studies of the three single-nucleotide polymorphisms (SNPs; rs2383207, rs2383206, and rs10757278) and ischemic stroke (IS).
Sensitivity Analysis
Because the I2 is > 50% in this meta-analysis, a random-effects model was used. To assess the impact of each individual study on the pooled effect estimate, we performed a sensitivity analysis by removing one study at a time. The pooled estimate I2 = 49.8%; thus, no single study significantly affected the results of each single-locus sensitivity analysis.
Second and Third Analyses
According to the results of the bias test and sensitivity analysis, it was found that the studies by Yang et al. (rs2383207) (Yang et al., 2018) and Ding et al. (rs10757278) (Ding et al., 2009) had roughly the same weight and were outside the funnel plot. We decided to remove the two studies and re-analyze the results. After removing two studies, we used R program to re-analyze the remaining studies. In the second analysis, chromosome 9p21 remained significantly associated with IS risk, and the G allele was associated with increased IS risk (OR: 1.16, 95% CI: 1.12–1.20, p < 0.0001, Figure 4). The results of this second analysis further confirmed that the two removed studies had little influence on the initial results. Moreover, there was homogeneity between the studies (I2 = 5%, p = 0.40), and the two experiments were outliers. A second bias test revealed that bias = 1.4217, p = 0.0696 (Figure 5). Sensitivity tests for the individual studies were again performed to ensure that no single study significantly affected the results of each single-locus sensitivity analysis.
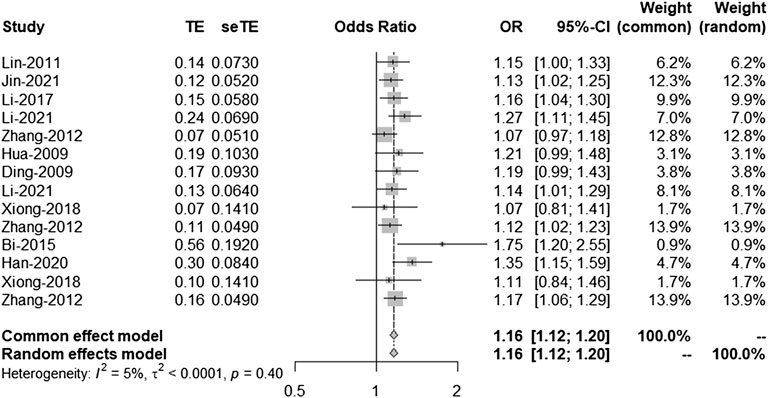
FIGURE 4. Fixed effects meta-analysis of the association between the three single-nucleotide polymorphisms (SNPs; rs2383207, rs2383206, and rs10757278) and ischemic stroke (IS) in the second analysis. CI, confidence interval; OR, odds ratios.
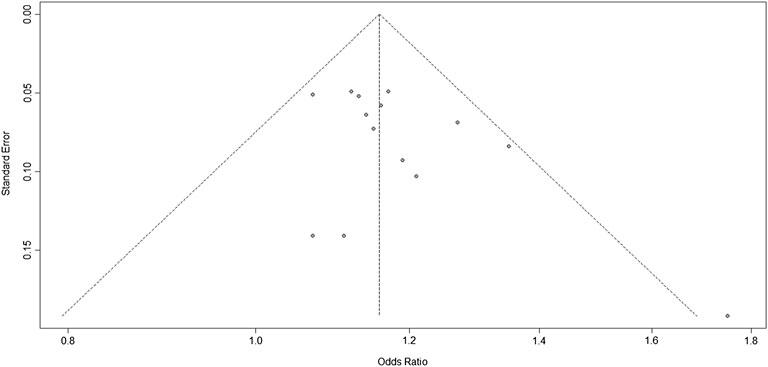
FIGURE 5. Funnel plots corresponding to the fixed-effects meta-analysis of the three single-nucleotide polymorphisms (SNPs; rs2383207, rs2383206, and rs10757278) and ischemic stroke (IS) in the second analysis.
We know from Figure 4 that the included studies were homogeneous, and the sensitivity analysis of each study also indicated that no single experiment significantly affected the experimental results. Therefore, based on the forest map and funnel plot, we also removed the study by Bi et al. (Bi et al., 2015), located outside the funnel plot, in the third analysis. In this third analysis, an increased risk of IS was associated with the G allele (OR: 1.16, 95% CI: 1.11–1.20, p < 0.0001, Figure 6). Further analysis confirmed that the homogeneity between studies was more significant after removing the study by Bi-2015 (I2 = 0%, p = 0.71, Figure 7), and there was a more significant correlation between chromosome 9p21 and IS risk. Thus, the removal of the study by Bi et al. (2015) further verified our original conclusions. The experimental results indicate that chromosome 9p21 is significantly associated with IS risk, and an increased risk of IS is associated with the G allele.

FIGURE 6. Fixed-effects meta-analysis of the associated between the three single-nucleotide polymorphisms (SNPs; rs2383207, rs2383206, and rs10757278) and ischemic stroke (IS) in the third analysis. CI, confidence interval; OR, odds ratios.
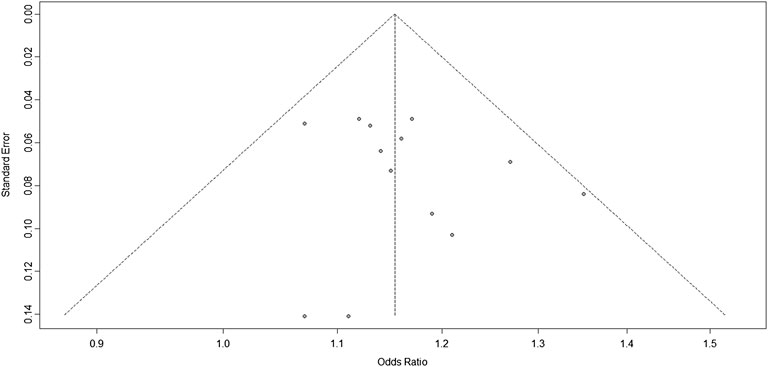
FIGURE 7. Funnel plots corresponding to the fixed-effects meta-analysis of the three single-nucleotide polymorphisms (SNPs; rs2383207, rs2383206, and rs10757278) and ischemic stroke (IS) in the third analysis.
Discussion
Stroke is currently the main cause of death in China; it has high morbidity, mortality, and disability rates (Kim et al., 2015). Stroke can be clinically divided into two types: IS and hemorrhagic stroke. Among the stroke subtypes, hemorrhagic stroke accounts for 20–40% of strokes in Chinese population, while in most Western populations, the majority of strokes (80–90%) are cerebral infarctions (Reed, 1990). Furthermore, IS accounts for approximately 87% of all stroke types, and IS a multifactorial disease that is influenced by both genetic and environmental factors (Wang et al., 2021). Chromosome 921 was originally reported to be associated with coronary heart disease (Matarin et al., 2008). There are some similarities between the etiologies and mechanisms of coronary heart disease and stroke, and 9p21 variants are associated with both diseases (Matarin et al., 2008). However, when investigating the association between 9p21 and IS, the conclusions drawn by researchers in China and in the rest of the world have been inconsistent. Stroke is influenced by many factors, including genetic, environmental, and vascular risk factors. The main method of studying susceptibility sites and genes in complex diseases is GWAS, based on SNPs (McPherson et al., 2007).
Matthew Traylor et al. found that chromosome 9p21 and histone deacetylase were associated with stroke in individuals of European ancestry (Traylor et al., 2012). Furthermore, Akinyemi et al. reported that rs2383207 increases IS incidence in indigenous West African men (Akinyemi et al., 2017). Previously, GWAS was also used to demonstrate that the antisense non-coding RNA in the INK4 locus (ANRIL) variants rs2383207 and rs1333049 increases the risk of IS and coronary heart disease in Caucasian populations (Dichgans et al., 2014; Dehghan et al., 2016). Notably, studies investigating the genetic associations of chromosome 9p21 variants have mainly been performed among Caucasian populations, and relatively few studies have been carried out in Han Chinese populations. Although Chen et al. (2019) studied chromosome 9p21 variants in Chinese populations, they concluded that mutations in rs2383207 may reduce the risk of IS but reported no definite correlation between rs10757278 and IS (Chen et al., 2019). In the present study, we once again focused on the relationship between stroke and chromosome 9p21.
In this meta-analysis, 18,584 participants were included; the IS and control groups contained 14,033 and 14,656 individuals, respectively. The three investigated SNPs have a linkage disequilibrium relationship, and we arrived at the same conclusions through unified analysis. All three SNPs were associated with IS risk. However, there was heterogeneity between the experimental results and studies; thus, bias detection and sensitivity analyses were carried out. Figure 3 suggested that the research may have been biased; therefore, to remove any possible bias, we performed another set of analyses. These further analyses had similar results that were more significant than those of the original analysis, further confirming that our analysis was correct.
In conclusion, our results indicate that rs2383206, rs2383207, and rs10757278 are significantly associated with IS risk and the G allele is associated with an increased risk of IS. Because the three SNPs in the present study have linkage disequilibrium and are in similar positions on chromosome 9p21, a unified analysis was performed. Environmental factors such as smoking and alcohol use may also be associated with IS risk, but not all studies considered these risk factors. Therefore, the influence of genes and the environment on IS pathogenesis needs to be further studied.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
DW, XH, CC, and QW participated in the design of this study. XH and DW conducted the literature search. DW, XH, CC, and QW retrieved and selected the articles. DW and XH conducted the data extraction. DW and XH performed the statistical analysis of the data. XH, DW, and CC wrote the manuscript draft. QW supervised the study. All authors contributed to the article and read and approved the final manuscript.
Funding
This study was supported by the Shandong Medicine and Health Science Technology Development Program (Grant No. 2018WS470), the Shandong Traditional Chinese Medicine Science and Technology Development Program (Grant No. 2019-0746), the Natural Science Foundation of Shandong Province (Grant No. ZR2021MH133), and the Jining Key Research and Development Project (Grant No. 2020YXNS035). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are deeply grateful to all the participants in this study. We also thank Bronwen Gardner, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
References
Akinyemi, R., Arnett, D. K., Tiwari, H. K., Ovbiagele, B., Sarfo, F., Srinivasasainagendra, V., et al. (2017). Interleukin-6 (IL-6) rs1800796 and Cyclin Dependent Kinase Inhibitor (CDKN2A/CDKN2B) rs2383207 are Associated with Ischemic Stroke in Indigenous West African Men. J. Neurological Sci. 379, 229–235. doi:10.1016/j.jns.2017.05.046
Bi, J., Yang, L., Liu, D., Wu, J., Tong, X., Cen, S., et al. (2015). Sequence Variants on Chromosome 9p21 are Associated with Ischemic Stroke and the Lipids Level in Chinese Han Population. J. Stroke Cerebrovasc. Dis. 24, 894–900. doi:10.1016/j.jstrokecerebrovasdis.2014.12.020
Chen, J.-X., Liu, J., Hu, F., Bi, Y., Li, M., and Zhao, L. (2019). Genetic Variants on Chromosome 9p21 Confer Risks of Cerebral Infarction in the Chinese Population: A Meta-Analysis. Int. J. Immunopathol. Pharmacol. 33, 2058738419847852. doi:10.1177/2058738419847852
Dehghan, A., Bis, J. C., White, C. C., Smith, A. V., Morrison, A. C., Cupples, L. A., et al. (2016). Genome-Wide Association Study for Incident Myocardial Infarction and Coronary Heart Disease in Prospective Cohort Studies: The CHARGE Consortium. PLoS One 11, e0144997. doi:10.1371/journal.pone.0144997
Dichgans, M., Malik, R., König, I. R., Rosand, J., Clarke, R., Gretarsdottir, S., et al. (2014). Shared Genetic Susceptibility to Ischemic Stroke and Coronary Artery Disease: a Genome-Wide Analysis of Common Variants. Stroke 45, 24–36. doi:10.1161/strokeaha.113.002707
Ding, H., Xu, Y., Wang, X., Wang, Q., Zhang, L., Tu, Y., et al. (2009). 9p21 is a Shared Susceptibility Locus Strongly for Coronary Artery Disease and Weakly for Ischemic Stroke in Chinese Han Population. Circ. Cardiovasc Genet. 2, 338–346. doi:10.1161/circgenetics.108.810226
Ganesh, C., Corey, A., Audrey, C., Myriam, F., Azadeh, R., Joshua, B., et al. (2016). Identification of Additional Risk Loci for Stroke and Small Vessel Disease: A Meta-Analysis of Genome-Wide Association Studies. Lancet Neurol. 15, 695–707. doi:10.1016/s1474-4422(16)00102-2
Gschwendtner, A., Bevan, S., Cole, J. W., Plourde, A., Matarin, M., Ross-Adams, H., et al. (2009). Sequence Variants on Chromosome 9p21.3 Confer Risk for Atherosclerotic Stroke. Ann. Neurol. 65, 531–539. doi:10.1002/ana.21590
Han, X., Wang, C., Tang, D., Shi, Y., and Gao, M. (2020). Association of Genetic Polymorphisms in Chromosome 9p21 with Risk of Ischemic Stroke. Cytokine 127, 154921. doi:10.1016/j.cyto.2019.154921
Higgins, J., Thomas, J., Chandler, J., Cumpston, M., Li, T., Page, M., et al. (2021). Cochran Handbook for Systematic Reviews of Interventions. version 6.2. Available at: http://www.trainingcochraneorg/handbook (Accessed January 2, 2022).
Hu, W.-l., Li, S.-j., Liu, D.-t., Wang, Y., Niu, S.-q., Yang, X.-c., et al. (2009). Genetic Variants on Chromosome 9p21 and Ischemic Stroke in Chinese. Brain Res. Bull. 79, 431–435. doi:10.1016/j.brainresbull.2009.04.001
Jin, M., Zhang, H., Zhang, Q., Li, X., Wu, N., Hu, Y., et al. (2021). Association of Single-Nucleotide Polymorphism on Chromosome 9 and Ischemic Stroke in Heilongjiang Province in China. Int. J. Clin. Exp. Pathol. 14, 726–733.
Kim, A. S., Cahill, E., and Cheng, N. T. (2015). Global Stroke Belt: Geographic Variation in Stroke Burden Worldwide. Stroke 46, 3564–3570. doi:10.1161/strokeaha.115.008226
Li, R., Zhang, X., Yin, W., Wang, Y., and Liu, Y. (2021). Common Genetic Variants on Chromosome 9p21 Confers Risk of Ischemic Stroke: A Large-Scale Genetic Association Study. Cell Mol. Biol. (Noisy-le-grand) 67, 132–137. doi:10.14715/cmb/2021.67.2.20
Li, S., Xu, Y. M., Zheng, H., Randell, E., Wang, H. Z., Cui, J., et al. (2017). A Reduced Interval of Chromosome 9p21 Locus Is Associated with Ischemic Stroke in Chinese Northern Han Population. Int. J. Genet. Genomics 5, 14–18. doi:10.11648/j.ijgg.20170501.12
Lin, H.-F., Tsai, P.-C., Liao, Y.-C., Lin, T.-H., Tai, C.-T., Juo, S.-H. H., et al. (2011). Chromosome 9p21 Genetic Variants Are Associated with Myocardial Infarction but Not with Ischemic Stroke in a Taiwanese Population. J. Investig. Med. 59, 926–930. doi:10.2310/JIM.0b013e318214ea49
Liu, G., Wang, H., Liu, J., Li, J., Li, H., Ma, G., et al. (2014). The CLU Gene rs11136000 Variant Is Significantly Associated with Alzheimer's Disease in Caucasian and Asian Populations. Neuromol Med. 16, 52–60. doi:10.1007/s12017-013-8250-1
Liu, G., Xu, Y., Jiang, Y., Zhang, L., Feng, R., and Jiang, Q. (2017). PICALM rs3851179 Variant Confers Susceptibility to Alzheimer's Disease in Chinese Population. Mol. Neurobiol. 54, 3131–3136. doi:10.1007/s12035-016-9886-2
Liu, G., Zhang, S., Cai, Z., Ma, G., Zhang, L., Jiang, Y., et al. (2013). PICALM Gene rs3851179 Polymorphism Contributes to Alzheimer's Disease in an Asian Population. Neuromol Med. 15, 384–388. doi:10.1007/s12017-013-8225-2
Liu, L., Wang, D., Wong, K. S. L., and Wang, Y. (2011). Stroke and Stroke Care in China: Huge Burden, Significant Workload, and a National Priority. Stroke 42, 3651–3654. doi:10.1161/strokeaha.111.635755
Matarin, M., Brown, W. M., Singleton, A., Hardy, J. A., and Meschia, J. F. (2008). Whole Genome Analyses Suggest Ischemic Stroke and Heart Disease Share an Association with Polymorphisms on Chromosome 9p21. Stroke 39, 1586–1589. doi:10.1161/strokeaha.107.502963
McPherson, R., Pertsemlidis, A., Kavaslar, N., Stewart, A., Roberts, R., Cox, D. R., et al. (2007). A Common Allele on Chromosome 9 Associated with Coronary Heart Disease. Science 316, 1488–1491. doi:10.1126/science.1142447
Reed, D. M. (1990). The Paradox of High Risk of Stroke in Population with Low Risk of Coronary Heart Disease. Am. J. Epidemiol. 131, 579–588. doi:10.1093/oxfordjournals.aje.a115542
Samani, N. J., Erdmann, J., Hall, A. S., Hengstenberg, C., Mangino, M., Mayer, B., et al. (2007). Genomewide Association Analysis of Coronary Artery Disease. N. Engl. J. Med. 357, 443–453. doi:10.1056/NEJMoa072366
Söderholm, M., Pedersen, A., Lorentzen, E., Stanne, T. M., Bevan, S., Olsson, M., et al. (2019). Genome-Wide Association Meta-Analysis of Functional Outcome after Ischemic Stroke. Neurology 92, e1271–e1283. doi:10.1212/wnl.0000000000007138
Traylor, M., Farrall, M., Holliday, E. G., Sudlow, C., Hopewell, J. C., Cheng, Y.-C., et al. (2012). Genetic Risk Factors for Ischaemic Stroke and its Subtypes (The METASTROKE Collaboration): A Meta-Analysis of Genome-Wide Association Studies. Lancet Neurol. 11, 951–962. doi:10.1016/s1474-4422(12)70234-x
Wang, Q., Zhao, J., Chang, H., Liu, X., and Zhu, R. (2021). Association between lncRNA ANRIL Genetic Variants with the Susceptibility to Ischemic Stroke: From a Case-Control Study to Meta-Analysis. Med. Baltim. 100, e25113. doi:10.1097/md.0000000000025113
Xiong, L., Liu, W., Gao, L., Mu, Q., Liu, X., Feng, Y., et al. (2018). The ANRIL Genetic Variants and Their Interactions with Environmental Risk Factors on Atherothrombotic Stroke in a Han Chinese Population. J. Stroke Cerebrovasc. Dis. 27, 2336–2347. doi:10.1016/j.jstrokecerebrovasdis.2018.04.020
Yang, J., Gu, L., Guo, X., Huang, J., Chen, Z., Huang, G., et al. (2018). LncRNA ANRIL Expression and ANRIL Gene Polymorphisms Contribute to the Risk of Ischemic Stroke in the Chinese Han Population. Cell Mol. Neurobiol. 38, 1253–1269. doi:10.1007/s10571-018-0593-6
Keywords: ischemic stroke, chromosome 9p21, rs2383206, rs2383207, rs10757278, Chinese
Citation: Hu X, Wang D, Cui C and Wu Q (2022) Association of Single-Nucleotide Polymorphisms of rs2383206, rs2383207, and rs10757278 With Stroke Risk in the Chinese Population: A Meta-analysis. Front. Genet. 13:905619. doi: 10.3389/fgene.2022.905619
Received: 27 March 2022; Accepted: 13 May 2022;
Published: 28 June 2022.
Edited by:
Guiyou Liu, Tianjin Institute of Industrial Biotechnology (CAS), ChinaReviewed by:
Yiyan Li, University of Chinese Academy of Sciences, ChinaXingli Xu, Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, China
Copyright © 2022 Hu, Wang, Cui and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingjian Wu, d3F3MTEwQDE2My5jb20=
†ORCID: Qingjian Wu, orcid.org/0000-0002-8746-8743
‡These authors have contributed equally to this work and share first authorship
 Xuemei Hu1,2‡
Xuemei Hu1,2‡ Qingjian Wu
Qingjian Wu