
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 08 June 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.905581
This article is part of the Research TopicRecent Advances in the Molecular Genetics and Precision Medicine of Lung CarcinomaView all 17 articles
 Huihui Zhao1
Huihui Zhao1 Guojun Lu2*
Guojun Lu2*Background: Although previous studies reported that 26S proteasome non-ATPase regulatory subunit 2 (PSMD2) is involved in many human cancers. However, its clinical significance and function in lung adenocarcinoma remain unclear. Here, we examined the prognostic and immunological role of PSMD2 in lung adenocarcinoma.
Methods: The Cancer Genome Atlas (TCGA) was conducted to analyze PSMD2 expression and verified using UALCAN. PrognoScan and Kaplan-Meier curves were utilized to assess the effect of PSMD2 on survival. cBioPortal database was conducted to identify the mutation characteristics of PSMD2. Functional enrichment was performed to determine PSMD2-related function. Cancer Single-cell State Atlas (CancerSEA) was used to explore the cancer functional status of PSMD2 at single-cell resolution. PSMD2-related immune infiltration analysis was conducted. Tumor-Immune system interaction database (TISIDB) was performed to verify the correlation between PSMD2 expression and tumor-infiltrating lymphocytes (TILs).
Results: Both mRNA and protein expression of PSMD2 were significantly elevated in lung adenocarcinoma. High expression of PSMD2 was significantly correlated with high T stage (p = 0.014), lymph node metastases (p < 0.001), and TNM stage p = 0.005). Kaplan-Meier curves indicated that high expression of PSMD2 was correlated with poor overall survival (38.2 vs. 59.7 months, p < 0.001) and disease-specific survival (59.9 months vs. not available, p = 0.004). Multivariate analysis suggested that PSMD2 was an independent biomarker for poor overall survival (HR 1.471, 95%CI, 1.024–2.114, p = 0.037). PSMD2 had a high mutation frequency of 14% in lung adenocarcinoma. The genetic mutation of PSMD2 was also correlated with poor overall survival, disease-specific survival, and progression-free survival in lung adenocarcinoma. Functional enrichment suggested PSMD2 expression was involved in the cell cycle, RNA transport, and cellular senescence. CancerSEA analysis indicated PSMD2 expression was positively correlated with cell cycle, DNA damage, and DNA repair. Immune infiltration analysis suggested that PSMD2 expression was correlated with immune cell infiltration levels and abundance of TILs.
Conclusion: The upregulation of PSMD2 is significantly correlated with poor prognosis and immune infiltration levels in lung adenocarcinoma. Our findings suggest that PSMD2 is a potential biomarker for poor prognosis and immune therapeutic target in lung adenocarcinoma.
Lung cancer is still the most common cancer in China and the leading cause of cancer-related death in China and the United States of America (USA) (Lee et al., 2019; Siegel et al., 2022; Xia et al., 2022). Lung adenocarcinoma is the most common histological subtype of lung cancer and accounts for about 40% of diagnoses (Hua et al., 2020). The research to date indicates that targeted therapy and immune-checkpoint inhibitors plus chemotherapy have become standard therapy for lung adenocarcinoma and bring survival benefits (Hirsch et al., 2017; Gandhi et al., 2018; Mok et al., 2019). However, up to now, the 5-year survival rate of lung adenocarcinoma has remained a struggle at 16% (Huo et al., 2019). Furthermore, previous studies have reported that tumor-infiltrating lymphocytes (TILs) including tumor-associated macrophages and tumor-infiltrating neutrophils are correlated with the prognosis and sensitivity of chemotherapy and immunotherapy (Waniczek et al., 2017; Pan et al., 2019). As a result, it is urgent to explore the immune phenotype of lung adenocarcinoma and identify novel prognostic biomarkers and therapeutic strategies for lung adenocarcinoma.
The Proteasome 26S Subunit, Non-ATPase (PSMD) gene family, is composed of the PSMD1 to PSMD14. Previous studies have shown that PSMD family genes can play important role in the progress of circulation and tumorigenesis by regulating ubiquitinated protein breakdown. PSMD1-3 and PSMD7 were elevated in breast cancer and correlated with poor prognosis. They can promote cell proliferation and cell cycle progression prognosis in breast cancer cell lines (Oguro et al., 2015; Okumura et al., 2018; Fararjeh et al., 2019; Zhao et al., 2020). In esophageal cancer, upregulation of PSMD4 can reduce endoplasmic reticulum stress-induced cell apoptosis to promote tumor progression (Ma et al., 2019). It was also reported that PSMD9 was correlated with recurrence after radiotherapy in cervical cancer and can predict radiotherapy benefits in breast cancer (Langlands et al., 2014; Köster et al., 2020). A previous study has shown that PSMD2 was overexpressed in lung cancer and patients with higher expression of PSMD2 were correlated with poorer prognosis (Matsuyama et al., 2011). However, the number of lung cancer patients enrolled in this study was relatively small. Moreover, PSMD family genes are reported to correlate with immune infiltration profiles in breast cancer (Xuan et al., 2021). Up to now, there is no studies have designed to investigate the association between PSMD2 expression and immune cell infiltration in lung adenocarcinoma.
In this study, we first examined the expression of PSMD2 in lung adenocarcinoma in the TCGA and UALCAN databases. Then we conducted PrognoScan and Kaplan-Meier curves to assess the correlation between PSMD2 expression and prognosis. cBioPortal database was conducted to identify the mutation characteristics and prognostic significance of PSMD2. Furthermore, functional enrichment analysis, cancer functional status at single-cell resolution and immune cell infiltration analysis were also conducted. Our findings link the upregulation of PSMD2 with the poor prognosis and propose a therapeutic immunological target for lung adenocarcinoma.
TCGA (https://portal.gdc.cancer.gov/) is a landmark cancer genomics program (Tomczak et al., 2015). In the present study, the expression level of PSMD2 in different cancer types and related clinical data in lung adenocarcinoma were downloaded from TCGA for further analysis.
The UALCAN (http://ualcan.path.uab.edu/) is an online web resource to analyze cancer omics data (Chandrashekar et al., 2017). It can be used to perform protein expression analysis of the CPTAC dataset (Edwards et al., 2015). In this study, we input PSMD2 in the “scan by gene” module of UALCAN to explore the total protein expression between primary tumor and normal tissues with the CPTAC dataset of lung adenocarcinoma.
The HPA database (https://proteinatlas.org/) contains human protein expression profiles in tumor tissues and normal tissues (Uhlén et al., 2015; Uhlen et al., 2017). In the present study, HPA was conducted to confirm the protein expression of PSMD2 in lung adenocarcinoma.
PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) is an online database for prognosis analysis (Mizuno et al., 2009). In the present study, we input PSMD2 in the “Enter gene identifier” module of PrognoScan and validated the prognostic importance of PSMD2 in lung adenocarcinoma with two datasets (GSE31210, GSE13213).
The cBioPortal for Cancer Genomics (https://www.cbioportal.org/) is a Web resource to explore, visualize, and analyze multidimensional cancer genomics data (Cerami et al., 2012; Gao et al., 2013). In this study, we utilized the cBioPortal database to assess mutation data of PSMD2, capture its prognostic value in altered lung adenocarcinoma patients, and acquire co-expressed genes of PSMD2.
After acquiring co-expressed genes of PSMD2 from cBioPortal, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis with R package clusterProfiler to further quantify the functional annotations of these co-expressed genes (Yu et al., 2012). R package ggplot2 was adopted to visualize the results of GO and KEGG.
CancerSEA is an online database to comprehensively decode distinct functional states of cancer cells from different cancer types at single-cell resolution (Yuan et al., 2019). In this study, the relevant data of PSMD2 across different tumor functional states based on single-cell sequencing data were downloaded from CancerSEA and a heatmap was drawn. The t-SNE diagram was also downloaded from CancerSEA to describe the distribution of all individual cells.
The methods for analyzing immune infiltration were described as before (Li et al., 2021; Zhang et al., 2022). Tumor infiltration expression of 24 immune cell types was downloaded from published literature (Bindea et al., 2013) and quantified by the ssGSEA method with R package GSVA (Hänzelmann et al., 2013). Furthermore, Spearman correlation and Mann-Whitney U test were conducted to determine the correlation between low/high expression of PSMD2 and immune cell infiltration in lung adenocarcinoma.
TISIDB (http://cis.hku.hk/TISIDB/) is an integrated repository portal for tumor and immune system interaction (Ru et al., 2019). In the present study, TISIDB was employed to capture the relations between PSMD2 expression and the abundance of TILs in lung adenocarcinoma. The relations between PSMD2 expression and TILs were determined by Spearman’s test.
R (V 3.6.3, https://www.r-project.org/) was utilized for statistical analyses and R package ggplot2 was employed for visualization of expression differences. Paired t-test and Mann-Whitney U test were proposed to explore the expression differences between lung adenocarcinoma tissues and adjacent normal tissues. Kaplan-Meier curves and Cox regression were conducted with R package survminer and survival to assess the effect of PSMD2 on survival. p < 0.05 was considered statistically significant.
To assess the transcription level of PSMD2 in multiple tumors and normal samples, we conducted analyses on the TCGA database. As shown in Figure 1A, compared with normal tissue controls, the expression of PSMD2 was upregulated in 13 cancer types and only downregulated in Kidney Chromophobe. Looking at Figure 1B, the Mann-Whitney U test indicated that the PSMD2 mRNA expression level in lung adenocarcinoma (n = 535) was significantly upregulated relative to normal lung tissues (n = 59) (6.964 ± 0.698 vs. 6.510 ± 0.177, p < 0.001). As shown in Figure 1C, paired t-test analysis showed that PSMD2 mRNA expression level in lung adenocarcinoma (n = 57) were significantly upregulated relative to normal lung tissues (n = 57) (7.156 ± 0.662 vs. 6.512 ± 0.177, p < 0.001). Taken together, these results suggest that mRNA expression of PSMD2 is elevated in lung adenocarcinoma tissues.

FIGURE 1. Expression pattern of PSMD2 in Pan-cancer perspective and lung adenocarcinoma. (A) PSMD2 is increased in 13 cancer types and decreased only in Kidney Chromophobe from TCGA. (B,C) Both the Mann-Whitney U test and paired t-test indicate mRNA expression of PSMD2 is elevated in lung adenocarcinoma. (D) The protein expression of PSMD2 is upregulated in CPTAC. (E) The PSMD2 protein expression in normal tissue from HPA. (F) The PSMD2 protein expression in lung adenocarcinoma tissue from HPA. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, no significant).
To explore the total protein expression of PSMD2 between lung adenocarcinoma and normal tissues, we analyzed CPTAC with the UALCAN dataset. The result in Figure 1D also showed that the protein expression of PSMD2 in lung adenocarcinoma was significantly elevated than in normal tissues. Protein expression from HPA also suggested that PSMD2 in lung adenocarcinoma tissue was higher than that in normal lung tissue (Figures 1E,F). Overall, these results indicate that protein expression of PSMD2 is increased in lung adenocarcinoma tissues.
The basic clinicopathological factors of lung adenocarcinoma patients was listed in Table 1. To examine the clinical relevance of PSMD2 expression, we conducted the expression between different clinicopathological factors of lung adenocarcinoma patients with the Mann-Whitney U test. As listed in Figure 2, the high expression of PSMD2 was significantly correlated with T stage (T1 vs. T2-4, p = 0.014), N stage (N0 vs. N1-N3, p < 0.001), and TNM stage (Stage I-II vs. Stage III-IV, p = 0.005). However, PSMD2 expression had no significant correlation with other clinicopathological factors, such as M stage (p = 0.401), gender (p = 0.174), age (p = 0.884), smoking condition (p = 0.225), and anatomic subdivision (right vs. left, p = 0.999; peripheral vs. central, p = 0.149). On the whole, these results indicate that PSMD2 had a significant correlation with T stage, lymph node metastases, and high TNM stage.
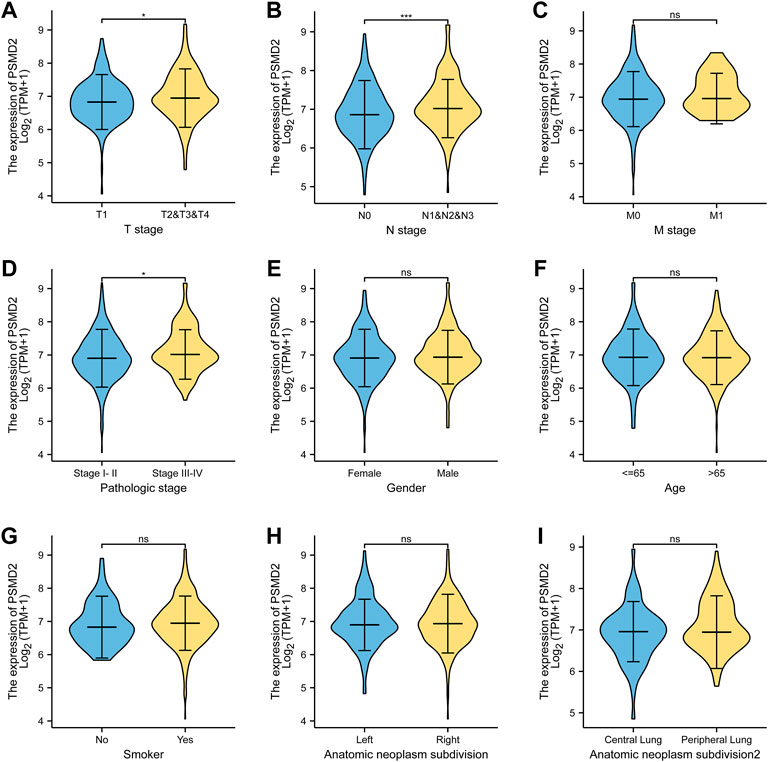
FIGURE 2. The clinical relevance of PSMD2 expression and clinicopathological factors in lung adenocarcinoma patients. PSMD2 mRNA expression was significantly increased with high T stage (A), N stage (B), and TNM stage (D). No observed differences in M stage (C), gender (E), age (F), smoke condition (G), and anatomic subdivision (right vs. left, central vs. peripheral) (H,I). (ns, no significance, ∗p < 0.05, ∗∗∗p < 0.001).
To investigate the correlation between PSMD2 mRNA expression and prognosis of lung adenocarcinoma patients, Kaplan-Meier curves with R package survminer and survival were used. As can be seen in Figure 3A, the overall survival of lung adenocarcinoma patients with higher PSMD2 expression was significantly shorter than those with lower PSMD2 expression (38.2 vs. 59.7 months, p < 0.001). As shown in Figure 3B, the disease-specific survival of lung adenocarcinoma patients with higher PSMD2 expression was also significantly shorter than those with lower PSMD2 expression (59.9 months vs. not available, p = 0.004). However, as shown in Figure 3C, no significant difference was observed between higher and lower PSMD2 expression and progression-free interval (28.8 vs. 40.1 months, p = 0.089). Furthermore, we validated the prognostic importance of PSMD2 in PrognoScan. The result from two datasets in Figures 3D,E suggested that high PSMD2 expression was correlated with poor overall survival in lung adenocarcinoma. As a result, by associating PSMD2 and other clinical characteristics in Figures 4A,B, we established a nomogram and calibration plot for predicting the overall survival probability of lung adenocarcinoma patients.
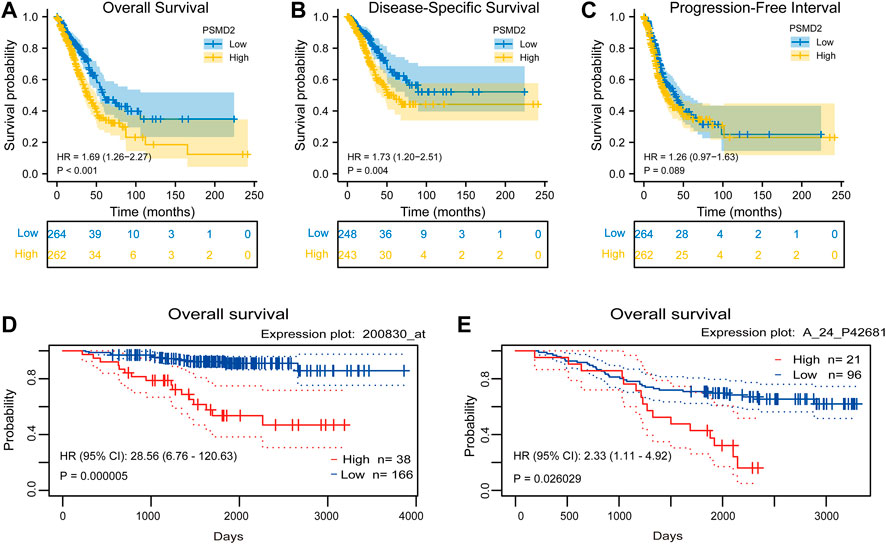
FIGURE 3. Correlation between PSMD2 expression and prognosis in lung adenocarcinoma. (A) Higher PSMD2 expression had a shorter overall survival. (B) Higher PSMD2 expression had a shorter disease-specific survival. (C) No significant difference was observed between PSMD2 expression and progression-free interval. (D,E) Two datasets (GSE31210, GSE13213) in PrognoScan suggested that high PSMD2 expression was correlated with poor overall survival in lung adenocarcinoma.
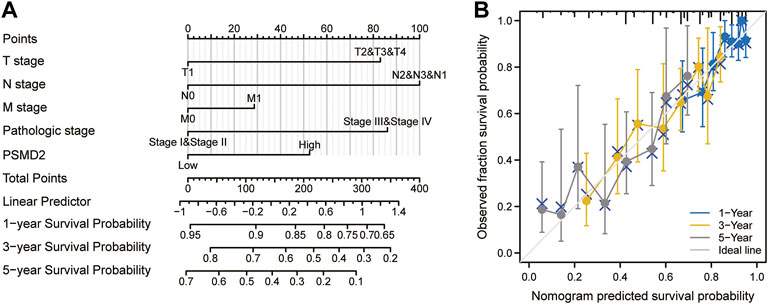
FIGURE 4. A nomogram and calibration plot for predicting the 1-, 3-, and 5-year overall survival probability of lung adenocarcinoma patients. (A) To predict the survival probability, first draw a vertical line from each clinical factor up to the points axis and get a point. Repeat until points for all clinical factors are decided. Sum all the points and then locate the total point on the total points axis. Draw a line straight down to the risk axis and finally obtain survival probability. (B) Calibration plot of the nomogram.
To further explore the prognostic role of PSMD2 for overall survival and disease-specific survival in lung adenocarcinoma patients, we performed Cox univariate and multivariate analyses. As shown in Figure 5A, univariate analysis showed that high expression of PSMD2 was significantly correlated with poor overall survival of lung adenocarcinoma patients (HR 1.694, 95%CI, 1.264–2.269, p < 0.001). Moreover, we conducted a multivariate analysis with the Cox proportional hazards model. The multivariate analysis in Figure 5B suggested that T stage (HR 1.767, 95%CI, 1.098–2.842, p = 0.019), N stage (HR 1.791, 95%CI, 1.181–2.716, p = 0.006), Pathologic stage (HR 1.693, 95%CI, 1.056–2.717, p = 0.029), and PSMD2 (HR 1.471, 95%CI, 1.024–2.114, p = 0.037) were independent factors for overall survival in lung adenocarcinoma patients. Furthermore, as shown in Supplementary Table S1, univariate analysis showed that high expression of PSMD2 was significantly correlated with poor disease-specific survival of lung adenocarcinoma patients (HR 1.734, 95%CI, 1.198–2.512, p = 0.004). However, the multivariate analysis suggested PSMD2 (HR 1.615, 95%CI, 0.998–2.614, p = 0.051) was not an independent factor for disease-specific survival in lung adenocarcinoma. Our data indicate that PSMD2 is an independent factor for overall survival in lung adenocarcinoma patients.
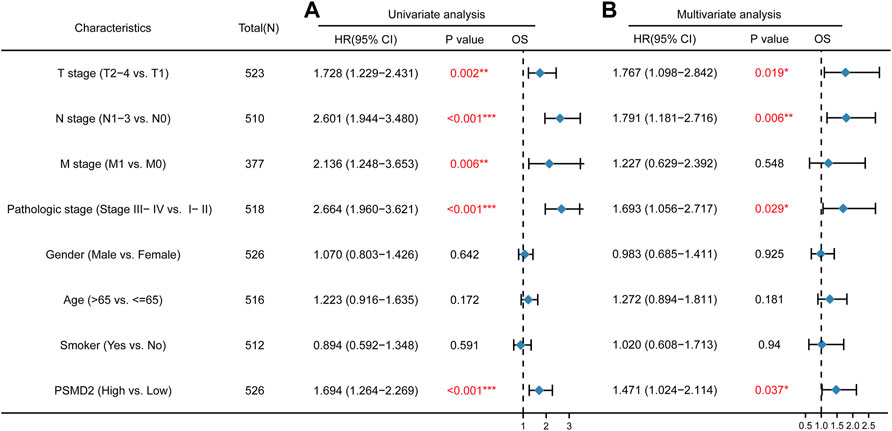
FIGURE 5. Cox regression analyses and forest plots of prognostic factors for overall survival. (A) Univariate Cox analysis and forest plot results of PSMD2 for OS. (B) Multivariate Cox analysis and forest plot indicated PSMD2 was an independent prognostic biomarker for OS in lung adenocarcinoma. (Red colors mean significant results. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
To identify the mutation characteristics of PSMD2 and its prognostic value in altered lung adenocarcinoma patients, we utilized an analysis on the cBioPortal database. It can be seen from Figure 6A that PSMD2 had a high mutation frequency of 14% in lung adenocarcinoma (TCGA, PanCancer Atlas). The main alteration frequency of PSMD2 was mRNA upregulation and amplification (Figure 6B). Moreover, compared with the unaltered group, the result of Kaplan-Meier curves found that the altered group was associated with poor prognosis in overall survival (Figure 6C, 28.57 vs. 52.60 months, p = 1.737e-4), disease-specific survival (Figure 6D, 36.66 vs. 88.14 months, p = 2.559e-4), and progression-free survival (Figure 6E, 19.00 vs. 39.52 months, p = 3.377e-4). Overall, these results suggest that genetic mutation of PSMD2 was correlated with poor prognosis in lung adenocarcinoma.
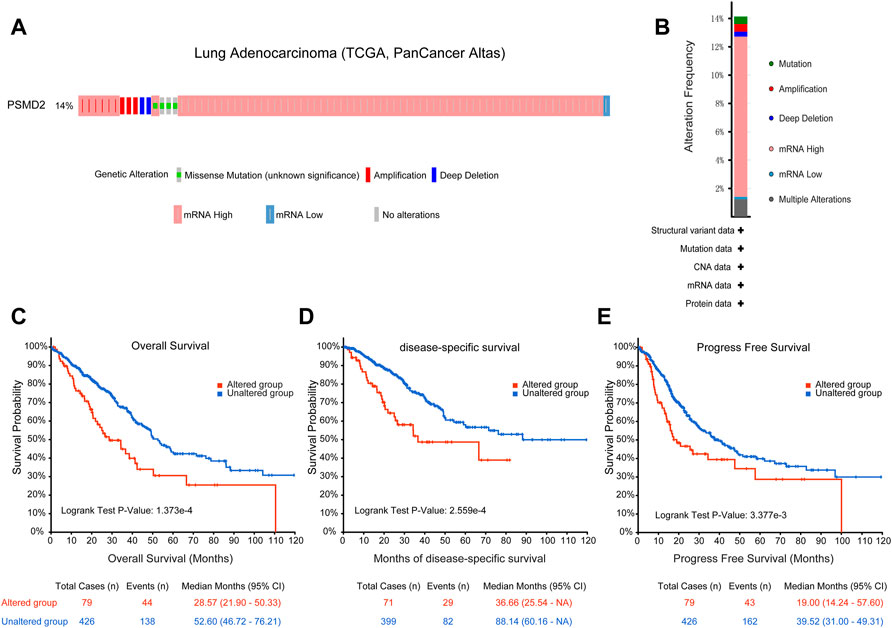
FIGURE 6. Genomic mutation of PSMD2 in lung adenocarcinoma. (A) OncoPrint presented the different mutation types and proportions of PSMD2. (B) Cancer types summary showed the alteration frequency. (C–E) Genomic mutation of PSMD2 was correlated with poor overall survival, disease-specific survival, and progression-free survival.
To perform GO and KEGG analysis, we first downloaded co-expressed genes of PSMD2 from cBioPortal. As shown in Supplementary Table S2, with |cor Spearman| > 0.5 and p < 0.05, PSMD2 had 139 co-expressed genes, including 137 positive and two negative co-expressed genes. R package clusterProfiler was utilized for GO and KEGG analysis. As shown in Figure 7A, GO analysis revealed that these co-expressed genes of PSMD2 were involved in the biological progress of organelle fission, chromosome segregation, and regulation of cell cycle phase transition. They acted as structural constituents in the condensed chromosome, spindle, and chromosomal region (Figure 7B), and played an important part in the molecular function of ATPase activity, tubulin binding, and DNA helicase activity (Figure 7C). KEGG pathway analysis in Figure 7D indicated enrichment function in cell cycle, RNA transport, and cellular senescence.
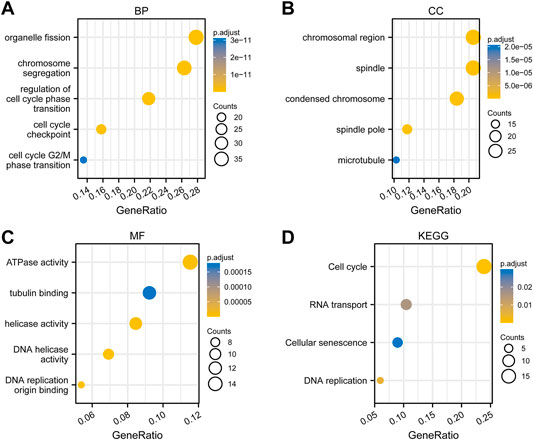
FIGURE 7. Functional enrichment analysis of co-expressed genes of PSMD2. (A) Biological progress of organelle fission, chromosome segregation, and regulation of cell cycle phase transition. (B) Structural constituents in the condensed chromosome, spindle, and chromosomal region. (C) The molecular function of ATPase activity, tubulin binding, and DNA helicase activity. (D) KEGG pathway analysis indicated enrichment function in the cell cycle, RNA transport, and cellular senescence.
To capture the expression of PSMD2 at single-cell resolution and its correlation with cancer functional states, we conducted an analysis on CancerSEA. The results listed in Figure 8A suggested that PSMD2 expression was significantly positively correlated with angiogenesis and DNA damage, while negatively correlated with proliferation in acute lymphoblastic leukemia (ALL). PSMD2 expression was positively correlated with cell cycle, DNA damage, and DNA repair, while negatively correlated with quiescence in lung adenocarcinoma (Figure 8B). The t-SNE diagram in Figure 8C described the PSMD2 expression profile in single cells of lung adenocarcinoma.
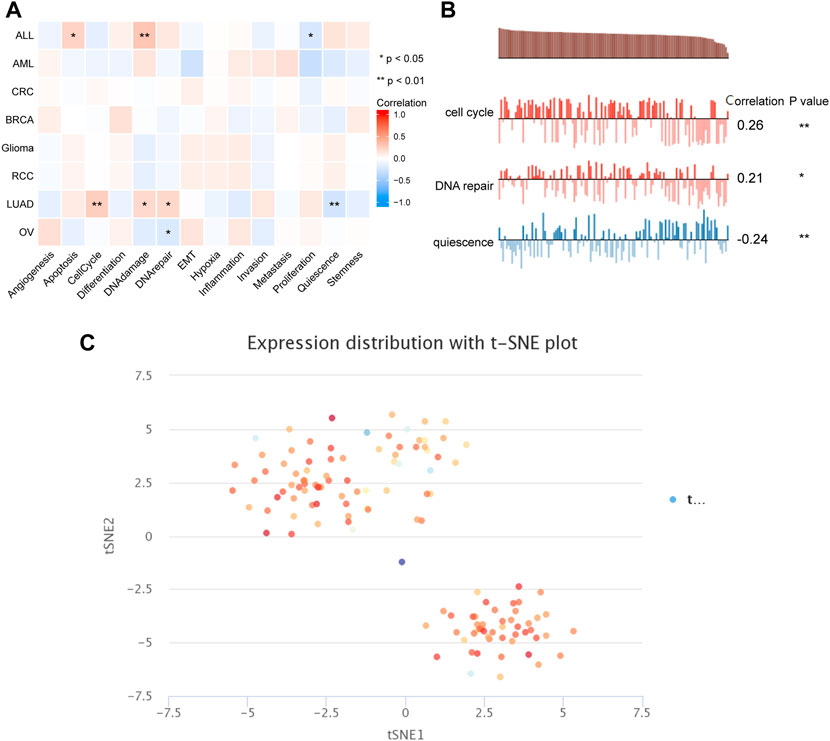
FIGURE 8. The correlation between PSMD2 expression and cancer functional states. (A) A heatmap indicated the relationship between PSMD2 expression and cancer functional states from CancerSEA. (B) PSMD2 expression was correlated with cell cycle, DNA repair, and quiescence in lung adenocarcinoma. (C) The t-SNE diagram described the PSMD2 expression profile in single cells of lung adenocarcinoma.
To analyze the correlation between the expression of PSMD2 and immune cell infiltration, we conducted ssGSEA method with R package GSVA. The results of Spearman correlation analysis were listed in Figures 9A–G and Table 2. It was apparent that PSMD2 expression was negatively correlated with the immune cell infiltration levels of CD8 T cells (r = −0.238, p < 0.001), B cells (r = −0.225, p < 0.001), pDC (r = −0.194, p < 0.001), mast cells (r = −0.191, p < 0.001), TFH (r = −0.190, p < 0.001), Tcells (r = −0.160, p < 0.001), Th17 cells (r = −0.143, p < 0.001), NK CD56bright cells (r = −0.128, p < 0.001), cytotoxic cells (r = −0.122, p < 0.001), and positively correlated with the immune cell infiltration levels of Th2 cells (r = 0.438, p < 0.001), Tgd (r = 0.146, p < 0.001), aDC (r = 0.142, p < 0.001), NK CD56dim cells (r = 0.121, p < 0.001), and NK cells (r = 0.142, p < 0.001). On the whole, these results suggest that PSMD2 may regulate the level of tumor-infiltrating immune cells to affect lung adenocarcinoma progression.
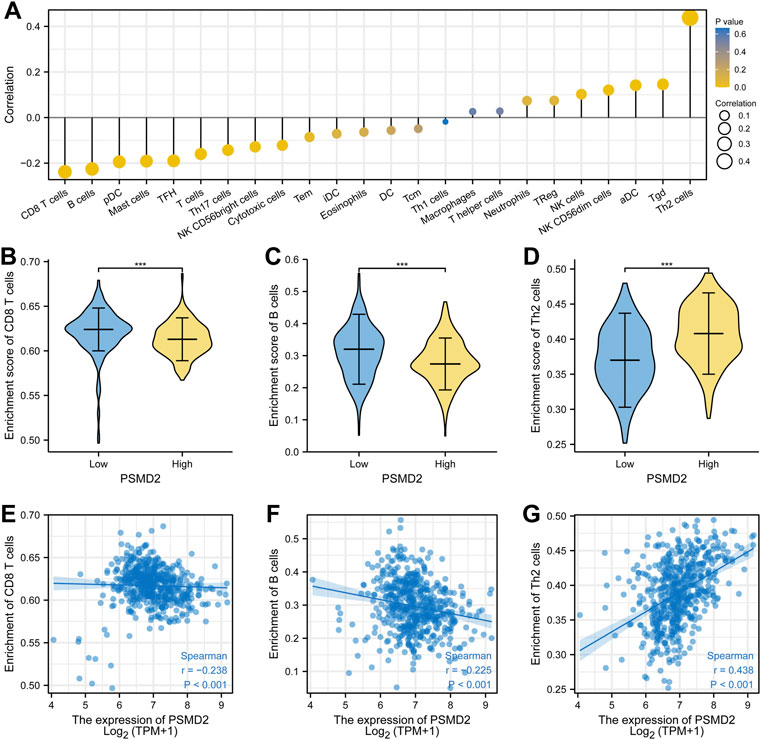
FIGURE 9. Spearman correlation between PSMD2 expression and immune cell infiltration. (A) The correlation between PSMD2 expression and the relative abundances of 24 immune cells. (B–D) Mann-Whitney U test indicated there were differences in immune cells (CD8 T cells, B cells, Th2 cells) between high/low expression of PSMD2. (E–G) Scatter diagrams for correlation between immune cells (CD8 T cells, B cells, Th2 cells) and PSMD2 expression.
To further confirm the relations between PSMD2 expression and TILs, we performed an analysis on TISIDB. As shown in Figure 10A, we found that PSMD2 expression was negatively correlated with abundance of eosinophil cells (r = −0.39, p < 2.2e-16), mast cells (r = −0.267, p = 7.49e-10), activated B cells (r = −0.262, p = 1.79e-09), Th17 (r = −0.216, p = 7.23e-07), immature B cells (r = −0.204, p = 3.03e-06), tem CT8 T cells (r = −0.191, p = 1.28e-05), pDC (r = −0.169, p = 1.19e-04), macrophage cells (r = −0.163, p = 2.09e-04), and neutrophil cells (r = −0.152, p = 5.34e-04), NK cells (r = −0.136, p = 1.89e-03), TFH (r = −0.117, p = 7.59e-03), Th1 (r = −0.106, p = 0.016). PSMD2 expression was negatively correlated with abundance of activated CD4 cells (r = 0.284, p = 6.14e-11), CD56dim cells (r = 0.107, p = 1.46e-02). The results were similar to Figure 9. Scatter diagrams for correlation between PSMD2 expression and abundance of eosinophil cells, mast cells, activated CD4 cells and CD56dim cells were listed in Figures 10B–E. Taken together, these results indicate that PSMD2 expression is correlated with most TILs in lung adenocarcinoma, further suggesting that PSMD2 may play an important role in the lung adenocarcinoma microenvironment.

FIGURE 10. The relations between PSMD2 expression and abundance of TILs in TISIDB. (A) Relations between PSMD2 expression and 28 types of TILs across human cancers. (B–E) Scatter diagrams for correlation between PSMD2 expression and abundance of eosinophil cells, mast cells, activated CD4 cells, and CD56dim cells.
In this study, we reported that the expression level of PSMD2 is significantly elevated in lung adenocarcinoma and its upregulation is a reliable predictor of high T stage, lymph node metastases, and high TNM stage. In light of the PrognoScan database, Kaplan-Meier curves, and multivariate Cox analysis, our results confirmed that high expression of PSMD2 is correlated with poor prognosis and PSMD2 is an independent prognostic biomarker for overall survival of lung adenocarcinoma patients. Our results further indicated that the genetic mutation of PSMD2 was also correlated with poor overall survival, disease-specific survival, and progression-free survival in lung adenocarcinoma. The genetic mutation of PSMD2 was also correlated with poor overall survival, disease-specific survival, and progression-free survival in lung adenocarcinoma. Moreover, immune infiltration analysis suggested that PSMD2 expression had a significant correlation with the level of tumor-infiltrating immune cells, further suggesting a specific role for PSMD2 in the immunological interactions in lung adenocarcinoma.
As a member of The PSMD gene family, PSMD2 has been characterized as an important non-ATPase regulatory subunit of the 19S proteasome (Li et al., 2019). Previous studies elucidated that proteasome can induce ubiquitination and degradation of protein, which in turn play an important role in the development of cell proliferation, apoptosis, and cell cycle (Chesnel et al., 2006). Functioning as a component of the proteasome, PSMD2 was reported to play a vital role in tumor progression. In colorectal cancer, PSMD2 can facilitate the degradation of diverse Ras-related GTPase and then inhibit cell proliferation and affect the expression of cell-cycle protein via blocking NF-kappaB signaling (Ying et al., 2022). In breast cancer, a paper from Li et al. reported that PSMD2 can interact with p21 and p27, mediate the degradation of their ubiquitin-proteasome, and then promote cell proliferation and cell cycle progression in breast cancer (Oguro et al., 2015). In hepatocellular carcinoma, PSMD2 can modulate cellular lipid metabolism to regulate HepG2 cell proliferation via p38-JNK and AKT signaling (Tan et al., 2019). In the present study, our results showed that the mRNA and protein expression of PSMD2 is elevated in lung adenocarcinoma tissues. Moreover, we analyzed the correlation between PSMD2 expression and the clinicopathological factors of lung adenocarcinoma patients. The current study suggests that high expression of PSMD2 is significantly correlated with high T stage, N stage, and TNM stage. Given our results, it is likely that PSMD2 is involved in tumorigenesis and metastasis of lung adenocarcinoma. Furthermore, both functional enrichment analysis and CancerSEA results indicate PSMD2 expression is correlated with cell cycle, further suggesting that PSMD2 can regulate cell cycle to promote cancer progression. However, this should be tested in other experiments.
Previous studies demonstrated that upregulation of PSMD2 is correlated with poor prognosis in many cancers. A paper from established that patients with high PSMD2 expression have poor overall survival and progression-free survival in bladder urothelial carcinoma Salah Fararjeh et al. (2021). Based on the result of univariate and multivariate analysis, PSMD2 has been identified as an independent prognostic biomarker for overall survival in bladder urothelial carcinoma (Salah Fararjeh et al., 2021). In breast cancer, reported that upregulation of PSMD2 is associated with shorter overall survival and distant-metastasis-free survival, further suggesting that PSMD2 could act as a factor for an unfavorable prognosis in breast cancer Li et al. (2018). Our findings on the prognostic value of PSMD2 in lung adenocarcinoma are consistent with these reports. In this study, our results indicate that lung adenocarcinoma patients with high PSMD2 expression have poor overall survival and progression-free survival. Univariate and multivariate analysis found that PSMD2 is an independent prognostic factor for overall survival in lung adenocarcinoma patients. Furthermore, mutation characteristics of PSMD2 from the cBioPortal database also suggest that the altered group is associated with poor prognosis in overall survival, disease-specific survival, and progression-free survival in lung adenocarcinoma. Based on our data, we conclude that PSMD2 is a potential biomarker for poor prognosis in lung adenocarcinoma.
Many studies have described that tumor-infiltrating immune cells are the representative cellular components and play an important part in host antitumor immune responses (Oguro et al., 2015). There is also some evidence that infiltrating immune cells is closely associated with the efficacy of immunotherapy (Zhang et al., 2020). The findings indicate that compared with high immune cell infiltration, patients with low immune cell infiltration levels may appear poor outcomes to conventional therapy in some solid tumors (Tobin et al., 2019). A paper from reported that the expression of PSMD member genes including PSMD2 is related to markers of six tumor-infiltrating immune cell types in breast cancer Xuan et al. (2021). In this study, we analyzed the correlation between the expression of PSMD2 and immune cell infiltration. By ssGSEA method, we found that PSMD2 expression was negatively correlated with the immune cell infiltration levels of CD8 T cells, B cells, pDC, mast cells, TFH, T cells, Th17 cells, NK CD56bright cells, and cytotoxic cells. Moreover, PSMD2 expression is positively correlated with the immune cell infiltration levels of Th2 cells, Tgd, aDC, NK CD56dim cells, and NK cells. To further confirm the relations between PSMD2 expression and TILs, we performed an analysis on TISIDB. The results from TISIDB were similar to the ssGSEA method. Our findings suggest that PSMD2 expression is correlated with immune cell infiltration and raise the possibility that PSMD2 can be a potential immunotherapy target in lung adenocarcinoma. However, this hypothesis should be tested with further research.
In conclusion, our findings showed that PSMD2 expression is significantly elevated in lung adenocarcinoma and its upregulation is a reliable predictor of high T stage, lymph node metastases, and high TNM stage. Our results confirmed that high expression of PSMD2 is correlated with poor prognosis and PSMD2 is an independent prognostic biomarker for lung adenocarcinoma patients. Moreover, PSMD2 expression had a significant correlation with the level of tumor-infiltrating immune cells.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
GL designed the study and contributed analysis tools. HZ performed data analysis and wrote the manuscript. All authors reviewed the manuscript.
This study was funded by Wu JiePing Medical Foundation (320.6750.19059).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.905581/full#supplementary-material
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 39 (4), 782–795. doi:10.1016/j.immuni.2013.10.003
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2 (5), 401–404. doi:10.1158/2159-8290.CD-12-0095
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19 (8), 649–658. doi:10.1016/j.neo.2017.05.002
Chesnel, F., Bazile, F., Pascal, A., and Kubiak, J. Z. (2006). Cyclin B Dissociation from CDK1 Precedes its Degradation upon MPF Inactivation in Mitotic Extracts of Xenopus laevis Embryos. Cell. Cycle 5 (15), 1687–1698. doi:10.4161/cc.5.15.3123
Edwards, N. J., Oberti, M., Thangudu, R. R., Cai, S., McGarvey, P. B., Jacob, S., et al. (2015). The CPTAC Data Portal: A Resource for Cancer Proteomics Research. J. Proteome Res. 14 (6), 2707–2713. doi:10.1021/pr501254j
Fararjeh, A. S., Chen, L. C., Ho, Y. S., Cheng, T. C., Liu, Y. R., Chang, H. L., et al. (2019). Proteasome 26S Subunit, Non-ATPase 3 (PSMD3) Regulates Breast Cancer by Stabilizing HER2 from Degradation. Cancers 11 (4), 527. doi:10.3390/cancers11040527
Gandhi, L., Rodríguez-Abreu, D., Gadgeel, S., Esteban, E., Felip, E., De Angelis, F., et al. (2018). Pembrolizumab Plus Chemotherapy in Metastatic Non-small-cell Lung Cancer. N. Engl. J. Med. 378 (22), 2078–2092. doi:10.1056/NEJMoa1801005
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 6 (269), pl1. doi:10.1126/scisignal.2004088
Hänzelmann, S., Castelo, R., and Guinney, J. (2013). GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinforma. 14, 7. doi:10.1186/1471-2105-14-7
Hirsch, F. R., Scagliotti, G. V., Mulshine, J. L., Kwon, R., Curran, W. J., Wu, Y.-L., et al. (2017). Lung Cancer: Current Therapies and New Targeted Treatments. Lancet 389 (10066), 299–311. doi:10.1016/S0140-6736(16)30958-8
Hua, X., Zhao, W., Pesatori, A. C., Consonni, D., Caporaso, N. E., Zhang, T., et al. (2020). Genetic and Epigenetic Intratumor Heterogeneity Impacts Prognosis of Lung Adenocarcinoma. Nat. Commun. 11 (1), 2459. doi:10.1038/s41467-020-16295-5
Huo, J., Xu, Y., Sheu, T., Volk, R. J., and Shih, Y.-C. T. (2019). Complication Rates and Downstream Medical Costs Associated with Invasive Diagnostic Procedures for Lung Abnormalities in the Community Setting. JAMA Intern Med. 179 (3), 324–332. doi:10.1001/jamainternmed
Köster, F., Sauer, L., Hoellen, F., Ribbat-Idel, J., Bräutigam, K., Rody, A., et al. (2020). PSMD9 Expression Correlates with Recurrence after Radiotherapy in Patients with Cervical Cancer. Oncol. Lett. 20 (1), 581–588. doi:10.3892/ol.2020.11622
Langlands, F. E., Dodwell, D., Hanby, A. M., Horgan, K., Millican-Slater, R. A., Speirs, V., et al. (2014). PSMD9 Expression Predicts Radiotherapy Response in Breast Cancer. Mol. Cancer 13, 73. doi:10.1186/1476-4598-13-73
Lee, J. J.-K., Park, S., Park, H., Kim, S., Lee, J., Lee, J., et al. (2019). Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell. 177 (7), 1842–1857. doi:10.1016/j.cell.2019.05.013
Li, C., Hu, J., Hu, X., Zhao, C., Mo, M., Zu, X., et al. (2021). LncRNA SNHG9 Is a Prognostic Biomarker and Correlated with Immune Infiltrates in Prostate Cancer. Transl. Androl. Urol. 10 (1), 215–226. doi:10.21037/tau-20-1134
Li, P., Li, H., Zhao, Y., Guo, Q., Yu, Y., Zhu, S., et al. (2019). Asporin Promotes Cell Proliferation via Interacting with PSMD2 in Gastric Cancer. Front. Biosci. 24 (6), 1178–1189. doi:10.2741/4774
Li, Y., Huang, J., Zeng, B., Yang, D., Sun, J., Yin, X., et al. (2018). PSMD2 Regulates Breast Cancer Cell Proliferation and Cell Cycle Progression by Modulating P21 and P27 Proteasomal Degradation. Cancer Lett. 430, 109–122. doi:10.1016/j.canlet.2018.05.018
Ma, A. G., Yu, L. M., Zhao, H., Qin, C. W., Tian, X. Y., and Wang, Q. (2019). PSMD4 Regulates the Malignancy of Esophageal Cancer Cells by Suppressing Endoplasmic Reticulum Stress. Kaohsiung J. Med. Sci. 35 (10), 591–597. doi:10.1002/kjm2.12093
Matsuyama, Y., Suzuki, M., Arima, C., Huang, Q. M., Tomida, S., Takeuchi, T., et al. (2011). Proteasomal Non-catalytic Subunit PSMD2 as a Potential Therapeutic Target in Association with Various Clinicopathologic Features in Lung Adenocarcinomas. Mol. Carcinog. 50 (4), 301–309. doi:10.1002/mc.20632
Mizuno, H., Kitada, K., Nakai, K., and Sarai, A. (2009). PrognoScan: a New Database for Meta-Analysis of the Prognostic Value of Genes. BMC Med. Genomics 2, 18. doi:10.1186/1755-8794-2-18
Mok, T. S. K., Wu, Y. L., Kudaba, I., Kowalski, D. M., Cho, B. C., Turna, H. Z., et al. (2019). Pembrolizumab versus Chemotherapy for Previously Untreated, PD-L1-Expressing, Locally Advanced or Metastatic Non-small-cell Lung Cancer (KEYNOTE-042): a Randomised, Open-Label, Controlled, Phase 3 Trial. Lancet 393 (10183), 1819–1830. doi:10.1016/S0140-6736(18)32409-7
Oguro, S., Ino, Y., Shimada, K., Hatanaka, Y., Matsuno, Y., Esaki, M., et al. (2015). Clinical Significance of Tumor‐infiltrating Immune Cells Focusing on BTLA and Cbl‐b in Patients with Gallbladder Cancer. Cancer Sci. 106 (12), 1750–1760. doi:10.1111/cas.12825
Okumura, T., Ikeda, K., Ujihira, T., Okamoto, K., Horie-Inoue, K., Takeda, S., et al. (2018). Proteasome 26S Subunit PSMD1 Regulates Breast Cancer Cell Growth through P53 Protein Degradation. J. Biochem. 163 (1), 19–29. doi:10.1093/jb/mvx053
Pan, J.-h., Zhou, H., Cooper, L., Huang, J.-l., Zhu, S.-b., Zhao, X.-x., et al. (2019). LAYN Is a Prognostic Biomarker and Correlated with Immune Infiltrates in Gastric and Colon Cancers. Front. Immunol. 10, 6. doi:10.3389/fimmu.2019.00006
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). TISIDB: an Integrated Repository Portal for Tumor-Immune System Interactions. Bioinformatics 35 (20), 4200–4202. doi:10.1093/bioinformatics/btz210
Salah Fararjeh, A., Al-Khader, A., Al-Saleem, M., and Abu Qauod, R. (2021). The Prognostic Significance of Proteasome 26S Subunit, Non-ATPase (PSMD) Genes for Bladder Urothelial Carcinoma Patients. Cancer Inf. 20, 117693512110676. doi:10.1177/11769351211067692
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer Statistics, 2022. CA A Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Tan, Y., Jin, Y., Wu, X., and Ren, Z. (2019). PSMD1 and PSMD2 Regulate HepG2 Cell Proliferation and Apoptosis via Modulating Cellular Lipid Droplet Metabolism. BMC Mol. Biol. 20 (1), 24. doi:10.1186/s12867-019-0141-z
Tobin, J. W. D., Keane, C., Gunawardana, J., Mollee, P., Birch, S., Hoang, T., et al. (2019). Progression of Disease within 24 Months in Follicular Lymphoma Is Associated with Reduced Intratumoral Immune Infiltration. J. Clin. Oncol. 37 (34), 3300–3309. doi:10.1200/JCO.18.02365
Tomczak, K., Czerwińska, P., and Wiznerowicz, M. (2015). The Cancer Genome Atlas (TCGA): an Immeasurable Source of Knowledge. Contemp. Oncol. 1A (1A), 68–77. doi:10.5114/wo.2014.47136
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based Map of the Human Proteome. Science 347 (6220), 1260419. doi:10.1126/science.1260419
Uhlen, M., Zhang, C., Lee, S., Sjöstedt, E., Fagerberg, L., Bidkhori, G., et al. (2017). A Pathology Atlas of the Human Cancer Transcriptome. Science 357 (6352), eaan2507. doi:10.1126/science.aan2507
Waniczek, D., Lorenc, Z., Śnietura, M., Wesecki, M., Kopec, A., and Muc-Wierzgoń, M. (2017). Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch. Immunol. Ther. Exp. 65 (5), 445–454. doi:10.1007/s00005-017-0463-9
Xia, C., Dong, X., Li, H., Cao, M., Sun, D., He, S., et al. (2022). Cancer Statistics in China and United States, 2022: Profiles, Trends, and Determinants. Chin. Med. J. Engl. 135 (5), 584–590. doi:10.1097/CM9.0000000000002108
Xuan, D. T. M., Wu, C.-C., Kao, T.-J., Ta, H. D. K., Anuraga, G., Andriani, V., et al. (2021). Prognostic and Immune Infiltration Signatures of Proteasome 26S Subunit, Non-ATPase (PSMD) Family Genes in Breast Cancer Patients. Aging 13 (22), 24882–24913. doi:10.18632/aging.203722
Ying, K., Wang, C., Liu, S., Kuang, Y., Tao, Q., and Hu, X. (2022). Diverse Ras-Related GTPase DIRAS2, Downregulated by PSMD2 in a Proteasome-Mediated Way, Inhibits Colorectal Cancer Proliferation by Blocking NF-κB Signaling. Int. J. Biol. Sci. 18 (3), 1039–1050. doi:10.7150/ijbs.68312
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Yuan, H., Yan, M., Zhang, G., Liu, W., Deng, C., Liao, G., et al. (2019). CancerSEA: a Cancer Single-Cell State Atlas. Nucleic Acids Researc 47 (D1), D900–D908. doi:10.1093/nar/gky939
Zhang, S., Yang, H., Xiang, X., Liu, L., Huang, H., and Tang, G. (2022). THBS2 Is Closely Related to the Poor Prognosis and Immune Cell Infiltration of Gastric Cancer. Front. Genet. 13, 803460. doi:10.3389/fgene.2022.803460
Zhang, W., Shen, Y., Huang, H., Pan, S., Jiang, J., Chen, W., et al. (2020). A Rosetta Stone for Breast Cancer: Prognostic Value and Dynamic Regulation of Neutrophil in Tumor Microenvironment. Front. Immunol. 11, 1779. doi:10.3389/fimmu.2020.01779
Keywords: PSMD2, lung adenocarcinoma, prognosis, biomarker, immune infiltration
Citation: Zhao H and Lu G (2022) Prognostic Implication and Immunological Role of PSMD2 in Lung Adenocarcinoma. Front. Genet. 13:905581. doi: 10.3389/fgene.2022.905581
Received: 27 March 2022; Accepted: 23 May 2022;
Published: 08 June 2022.
Edited by:
Wei Zhao, City University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Jin Bai, Xuzhou Medical University, ChinaCopyright © 2022 Zhao and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojun Lu, Z3VvanVubHVAbmptdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.