
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 18 August 2022
Sec. Neurogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.905560
This article is part of the Research Topic Identifying the Key Pathogenic Factors of Neurological Disorders by Integrating Multi-omics Data View all 16 articles
A previous genome-wide association study (GWAS) has reported that variants rs2200733 and rs6843082 in the paired-like homeodomain transcription factor 2 (PITX2) gene may be one of the risk factors for ischemic stroke (IS) in European populations. However, more recently, studies in Asia have reported that rs2200733 and rs6843082 are only weakly or not associated with increased risk of IS. This difference may be caused by the sample size and genetic heterogeneity of rs2200733 and rs6843082 among different races. For this study, we selected eight articles with nine studies from the PubMed and Embase databases, including five articles from Asian and three articles from non-Asian, to evaluate the risk of IS caused by rs2200733 and rs6843082. Then, we investigated rs2200733 and rs6843082 single-nucleotide polymorphisms (SNPs) by analysis using allele, recessive, dominant, and additive models. We identified that rs2200733 and rs6843082 are weakly significantly associated with IS for the allele model (p = 0.8), recessive model (p = 0.8), dominant model (p = 0.49), and additive model (p = 0.76) in a pooled population. Next, we performed a subgroup analysis of the population, the result of which showed that rs2200733 and rs6843082 covey genetic risk for IS in a non-Asian population, but not in an Asian population. In conclusion, our analysis shows that the effect of PITX2 rs2200733 and rs6843082 SNPs on IS risk in Asia is inconsistent with the effect observed in European IS cohorts.
Ischemic stroke (IS) is the second leading cause of death worldwide and the main leading cause of intellectual disability in adults (Orellana-Urzua et al., 2020). The pathogenesis of IS has been studied using genome-wide association studies (GWAS), which provide a crucial direction for studying the genetic mechanism of IS (Chauhan and Debette, 2016; Liu et al., 2019b; Wei et al., 2019). In 2008, the PITX2 rs2200733 and rs10033464 variants were identified as significant contributors to IS in a European population (p = 2.18 × 10–10) (Gretarsdottir et al., 2008). However, a series of subsequent studies failed to replicate those results.
In 2009, Shi et al. analyzed 383 patients with atrial fibrillation (AF) versus (vs.) 851 patients without AF and 811 patients with IS vs. 688 patients without IS, all of Chinese. After analysis, rs2200733 was meaningfully correlated with AF (p = 4.1 × 10–12) but not IS in this Chinese population (Shi et al., 2009).
In 2012, Bertrand et al. analyzed 3548 patients with stroke vs. 5972 patients without stroke and then replicated their result in 5859 patients with stroke vs. 6281 patients without stroke, all of European ancestry. Their results showed that both rs2200733 and rs1906599 were associated with IS (OR = 1.32) (International Stroke Genetics Consortium et al., 2012). Their study again identified a significant association between rs2200733 and IS.
In 2022, Zhao et al. analyzed 476 patients with IS vs. 501 control individuals, all Chinese (Zhao et al., 2022). Their analysis found no meaningful association between rs6843082 and IS (p = 0.448).
In summary, previous studies have reported different results as to whether rs2200733 and rs6843082 increase susceptibility to IS. It is not clear whether the two SNPs (rs2200733 and rs6843082) are related to IS susceptibility. In this study, we further evaluate whether these two SNPs (rs2200733 and rs68430828) increase the risk of IS using nine studies from eight articles.
The relevant literature was searched in PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and Embase (https://www.embase.com/) databases. We filtered all relevant studies based on the keywords “Stroke,” “PITX2,” “rs2200733,” and “rs6843082.” The literature search was completed by 10 March 2022. In the following paragraph, we describe the criteria for inclusion.
The inclusion criteria for our meta-analysis were as follows: (1) the study used a case–control design, (2) the study evaluated whether the two SNPs (rs2200733 and rs6843082) are risk factors for IS, (3) the study provided a clear and definite number of genotypes or alleles or enough data to calculate these numbers, and (4) the study provided an explicit odds ratio (OR) and 95% confidence interval (CI) or sufficient data to calculate the OR and 95% CI. All studies that did not meet the inclusion criteria were eliminated.
For each study that met the inclusion criteria, we extracted the following information: (1) first author, (2) year of publication, (3) race of the study subjects, (4) number of cases and controls, and (5) quantity of rs2200733 and rs6843082 genotypes in cases and controls. The full results are shown in Table 1.
We used four common genetic models for this meta-analysis, including the allele model (A vs. G), recessive model (AA vs. AG+GG), dominant model (AA+AG vs. GG), and additive model (AA vs. GG). These results are helpful to evaluate the susceptibility to IS with the two SNPs (rs2200733 and rs6843082): A allele vs. G allele.
The HWE of the two SNPs (rs2200733 and rs6843082) in IS cases and the control group were analyzed using the Chi-square test. The relationship between the two SNPs (rs2200733 and rs6843082) and IS was analyzed using four gene models: allele model (A vs. G), recessive model (AA vs. AG+GG), dominant model (AA+AG vs. GG), and additive model (AA vs. GG). We performed all relevant Chi-square tests using the R program (http://www.r-project.org/).
First, we extracted the summary statistical information corresponding to the two SNPs (rs2200733 and rs6843082) in the above study. Then, Cochran’s Q test and I2 = [Q—(k—1)]/Q × 100% (Liu et al., 2017) were used to analyze the heterogeneity of the two SNPs (rs2200733 and rs6843082) among these datasets. The Q statistic approximately follows a χ2 distribution with k-1 degrees of freedom (k stands for the number of studies for analysis). When the P value from Cochran’s Q statistic <0.1 and the I2 value from Cochran’s Q statistic >50%, the data showed considerable heterogeneity (Hu et al., 2017; Liu et al., 2017).
In Cochran’s Q statistic, if p < 0.05 or I2 >50%, it indicated that there was heterogeneity between studies, and a random-effect model (DerSimonian–Laird) was used to calculate the pooled OR. If not, we used a fixed-effect model (Mantel–Haenszel). All statistical methods in the meta-analysis were applied by program R (http://www.r-project.org/).
In this analysis, we used funnel plots to assess the possible publication bias. When there was no publication bias, the plot of the funnel was symmetrically inverted. Otherwise, it was an asymmetric inverted funnel (Liu et al., 2014). The asymmetry of the funnel plot was evaluated by the Egger test. We performed all statistical tests using the R program (http://www.r-project.org/).
We retrieved 29 articles from PubMed and 44 articles from the Embase database. Finally, eight articles (Gretarsdottir et al., 2008; Shi et al., 2009; Bevan et al., 2012; Cao et al., 2013; Su et al., 2015; Wu et al., 2015; Ferreira et al., 2019; Zhao et al., 2022), including nine studies, were chosen for meta-analysis by excluding overlapping studies. A total of 55,829 participants were included in this meta-analysis: 39,231 cases in the case group (38,348 cases with rs2200733 and 1699 cases with rs6843082) and 16,598 cases in the control group (15,636 cases with rs2200733 and 1778 cases with rs6843082). The study by Su et al. analyzed the two SNPs based on 1632 participants, and so the total number of case and controls overlapped. The specifics are shown in Figure 1. The primary features of the studies we included are presented in Table 1.
The rs2200733 and rs6843082 SNPs were located within 10 kb on PITX2 gene (https://snipa.helmholtz-muenchen.de/snipa3/).
The study of Bevan et al. was excluded from the dominant, recessive, and additive models. For this analysis, we observed no remarkable heterogeneity in the pooled population when using the four genetic models (Table 2).
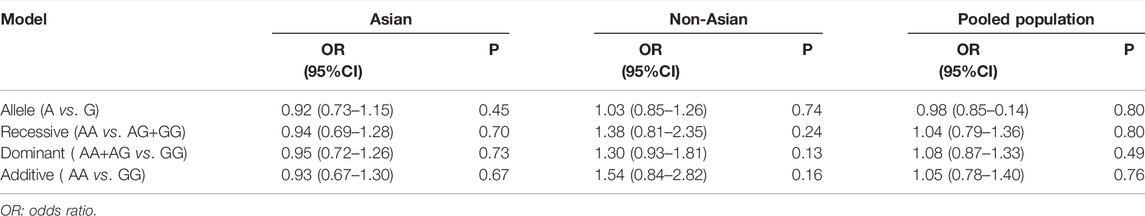
TABLE 2. Analysis of four genetic models’ association of rs2200733 and rs6843082 with ischemic stroke.
We computed the overall OR using a fixed-effect model in accordance with the outcomes of the heterogeneity test. The allele model tests showed that IS did not have a relationship with rs2200733 and rs6843082 in the Asian (p = 0.45), non-Asian (p = 0.74), and pooled populations (p = 0.80) (Table 2). Our results also showed that the two SNPs did not contribute to IS in Asian populations (OR = 0.92), but interestingly, the opposite results were seen in non-Asian populations, where both rs2200733 and rs6843082 were genetic risk factors for IS (OR = 1.03) (Figure 2).
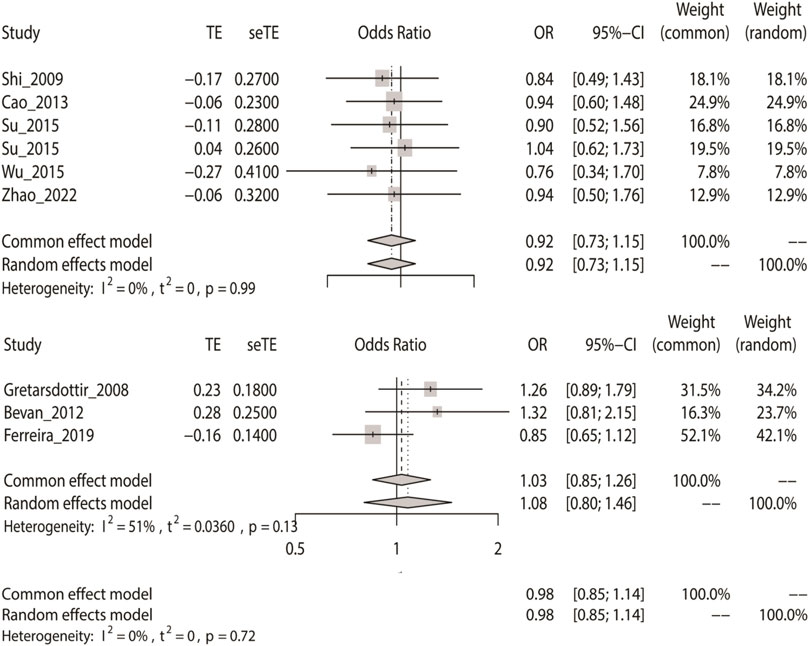
FIGURE 2. Fixed-effect meta-analysis of the allele model for rs2200733 and rs6843082 in the Asian, non-Asian, and pooled populations.
Similarly, we calculated the overall OR using a fixed-effect model based on the recessive model. The recessive model indicated that rs2200733 and rs6843082 and IS in the Asian (p = 0.70), non-Asian (p = 0.24), and pooled population (p = 0.80) (Table 2) were not closely related. The two SNPs were not associated with IS in Asian populations (OR = 0.94). Conversely, rs2200733 and rs6843082 could increase the incidence of IS disease in non-Asian populations (OR = 1.38) (Figure 3).
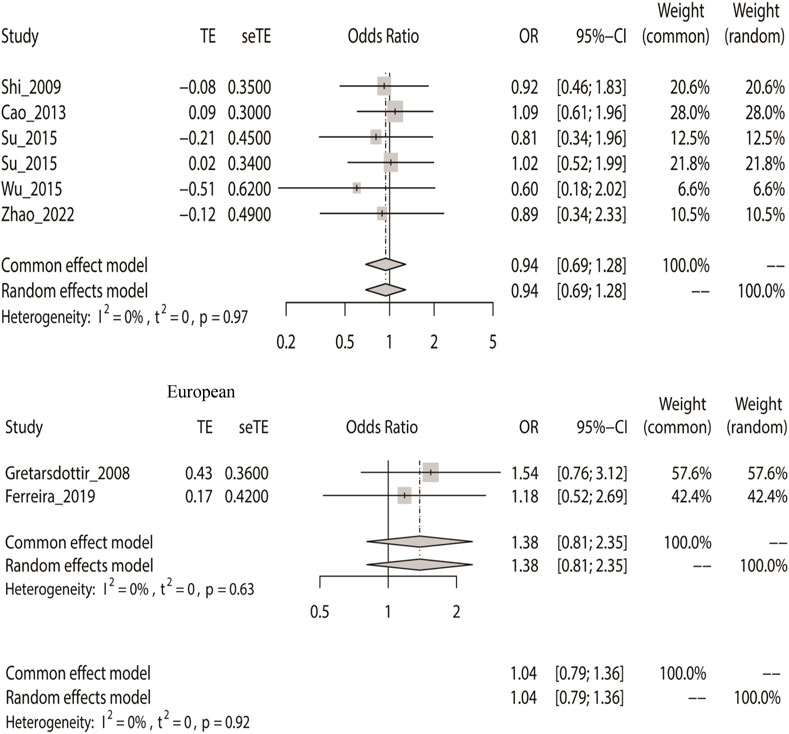
FIGURE 3. Fixed-effect meta-analysis of the recessive model for rs2200733 and rs6843082 in the Asian, non-Asian, and pooled populations.
Likewise, we calculated the overall OR using a fixed-effect model in accordance with the dominant model in the three groups. The dominant model showed that the two SNPs (rs2200733 and rs6843082) had no significant relationship with IS in Asian (p = 0.73), non-Asian (p = 0.13), and pooled (p = 0.49) populations (Table 2). The result of the subgroup analysis indicated that in the Asian population, the two SNPs were not genetic risk factors for IS (OR = 0.95); however, in the non-Asian population (OR = 1.30), the two SNPs were genetic risk factors for IS (OR = 1.08) (Figure 4).
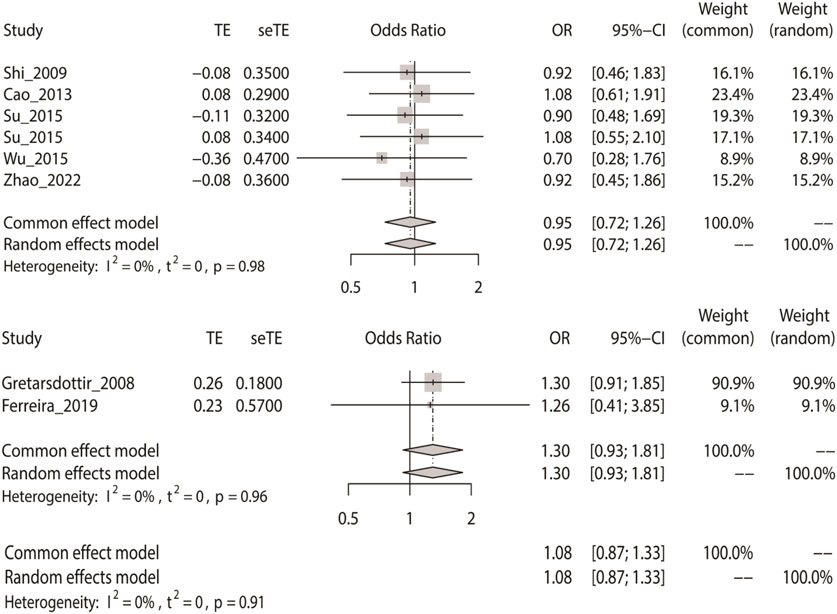
FIGURE 4. Fixed-effect meta-analysis of the dominant model for rs2200733 and rs6843082 in the Asian, non-Asian, and pooled populations.
Finally, we used the fixed-effect model to calculate the overall OR based on the additive model, where IS had no meaningful relationship with the two SNPs (rs2200733 and rs6843082) in Asian (p = 0.67), non-Asian (p = 0.16), and pooled populations (p = 0.76) (Table 2). The results were the same as the three previous genetic models; the two SNPs were not associated with IS in the Asian population (OR = 0.93); however, rs2200733 and rs6843082 were associated with an increased incidence of IS disease in the non-Asian population (OR = 1.54) (Figure 5).
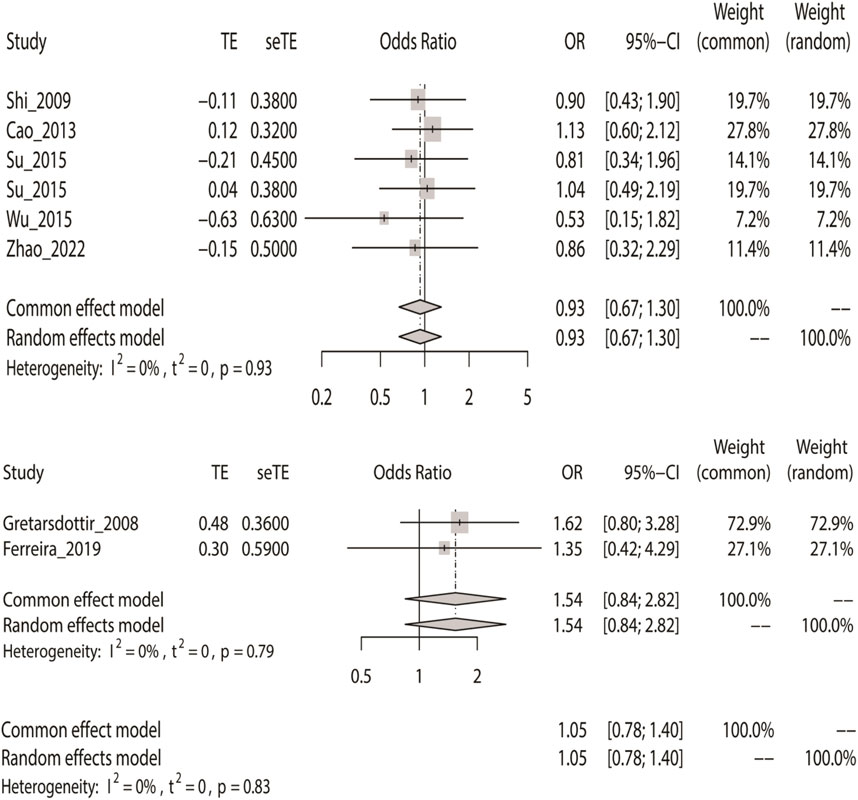
FIGURE 5. Fixed-effect meta-analysis of the additive model for rs2200733 and rs6843082 in the Asian, non-Asian, and pooled populations.
The funnel plot and Egger’s test were applied to assess the existence of the potential publication bias in the four genetic models. There was no bias in the four plots, which were symmetrical inverted funnels. For the allele, recessive, dominant, and additive models, p = 0.943, 0.133, 0.053, and 0.204, respectively (Figure 6).
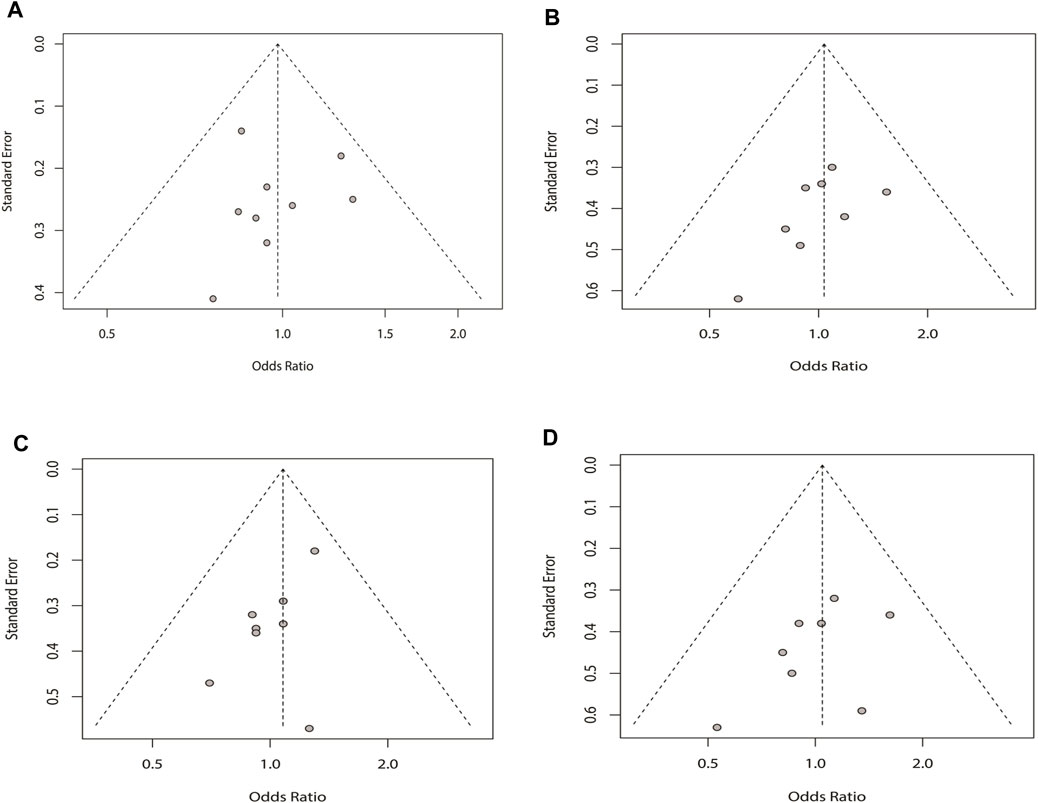
FIGURE 6. Sensitivity analysis of the four genetic models for rs2200733 and rs6843082 in the pooled population: (A) allele model for the two SNPs in the pooled population, (B) recessive model for the two SNPs in the pooled population, (C) dominant model for the two SNPs in the pooled population, and (D) additive model for the two SNPs in the pooled population.
Previous GWAS studies have shown that rs2200733 and rs6843082 SNPs in PITX2 are associated with genetic susceptibility to IS in European populations (Gretarsdottir et al., 2008). Subsequently, however, our results indicated that rs2200733 and rs6843082 conveyed no increased risk of IS. Overall, most studies have shown that the rs2200733 SNP in PITX2 is associated with European IS, but five studies conducted in Chinese populations all concluded that the rs2200733 SNP was not associated with the IS risk. Meanwhile, three studies analyzed the association between rs2200733 and AF in the Chinese population, and the results suggested that the expression of rs2200733 had a potential genetic risk for AF, but not IS (Shi et al., 2009; Cao et al., 2013; Su et al., 2015).
In accordance with the analysis of the two SNPs (rs2200733 and rs6843082) in a pooled population, we can conclude that the G allele has low importance for the risk of IS. In the subgroup analysis, the results showed that the two SNPs had no correlation with the risk of IS in an Asian population, but the results in a non-Asian population showed a significant relevance with the risk of IS. These results suggest that the specific gene expression of that population and/or disease could be affected by genetic variation (Liu et al., 2019a). Therefore, two possibilities may lead to different associations between the two SNPs and human IS gene expression. The first factor is the racial difference, such as the genetic difference between Asian and non-Asian populations. For example, Gretarsdottir et al. indicated that rs2200733 has a strong association with IS (Gretarsdottir et al., 2008), but Cao et al. showed that rs2200733 has no association with any type of stroke (Cao et al., 2013). The second probability is that the disease condition has an effect on gene expression (Tammen et al., 2013; Maruthai et al., 2022). Haplotype association analysis by Su et al. showed that rs6843082 was significantly correlated with serum total cholesterol (TC) in IS patients in the additive and dominant models (Su et al., 2015). Moreover, for female individuals, the result of the recessive model also showed that rs2200733 was associated with high levels of TC, increasing the risk of IS (Su et al., 2015). The two SNPs (rs2200733 and rs6843082) are closely related to the PITX2 gene on chromosome 4q25, which is associated with cardiac morphogenesis, particularly the differential identity of the left atrium from the right atrium and the growth of myocardial sleeves of pulmonary veins (Logan et al., 1998; Mommersteeg et al., 2007; Tessari et al., 2008). Therefore, the PITX2 gene may be expressed during the development of the circulatory system and play a role in IS-related risk factors. Further studies are required to determine whether these two SNPs are risk factors for IS and provide a new direction for the treatment of IS.
Numerous studies on the relationship between PITX2 and IS have produced conflicting results. So far, there is still no final and unanimous conclusion. Our analysis includes samples from multiple researchers; therefore, the results of our study may be more reliable than those of single studies. However, some potential limitations in our meta-analysis should be acknowledged. First, there was a small sample of GWAS and candidate gene studies, which may influence the pooled estimated value. Second, environmental factors, such as smoking and alcohol use, may affect the risk of IS, but some studies did not consider these risk factors. Third, different populations have different genetic susceptibilities both to the two SNPs (rs2200733 and rs6843082) and to IS, which can make the primary cause difficult to distinguish. IS is a disease that is effected by the interactions of multiple environmental and genetic factors (Dichgans, 2007; Cai et al., 2020), and so the influence of genes and environment on the pathogenesis of IS needs to be more deeply investigated.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Written informed consent was not obtained from the individual(s) for the publication of no identifiable images or data included in this article.
MY and QW participated in the design of this study. XH, DW, and YX conducted the literature search. DW, XH, YX, MY, and QW retrieved and selected the articles. DW and XH conducted data extraction. DW and XH performed the statistical analysis of the data. DW and XH wrote the manuscript draft. MY and QW supervised the study. All authors contributed to the article. All authors read and approved the final manuscript.
This study was supported by the Natural Science Foundation of Shandong Province (Grant No. ZR2021MH133), the Shandong Medicine and Health Science Technology Development Program (Grant No.2018WS470), the Shandong Traditional Chinese Medicine Science and Technology Development Program (Grant No.2019-0746), the Jining Key Research and Development Project (Grant No.2020YXNS035), and the National Natural Science Foundation of China (Grant No. 81871855).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Catherine Perfect, MA (Cantab), from Liwen Bianji (Edanz) (www.liwenbianji.cn), for editing the English text of a draft of this manuscript.
International Stroke Genetics Consortium Bellenguez, C., Bevan, S., Gschwendtner, A., Spencer, C. C. A., Burgess, A. I., Pirinen, M., et al. (2012). Genome-Wide Association Study Identifies a Variant in HDAC9 Associated with Large Vessel Ischemic Stroke. Nat. Genet. 44, 328–333. doi:10.1038/ng.1081
Bevan, S., Bertrand, S., Bellenguez, C., Bevan, S., Gschwendtner, A., Spencer, C. C., et al. (2012). Genome-Wide Association Study Identifies a Variant in HDAC9 Associated with Large Vessel Ischemic Stroke. Nat. Genet. 44, 328–333. doi:10.1038/ng.1081
Cai, H., Cai, B., Liu, Z., Wu, W., Chen, D., Fang, L., et al. (2020). Genetic Correlations and Causal Inferences in Ischemic Stroke. J. Neurol. Sci. 267, 1980–1990. doi:10.1007/s00415-020-09786-4
Cao, Y.-y., Ma, F., Wang, Y., Wang, D. W., and Ding, H. (2013). Rs2200733 and Rs10033464 on Chromosome 4q25 Confer Risk of Cardioembolic Stroke: An Updated Meta-Analysis. Mol. Biol. Rep. 40, 5977–5985. doi:10.1007/s11033-013-2707-z
Chauhan, G., and Debette, S. (2016). Genetic Risk Factors for Ischemic and Hemorrhagic Stroke. Curr. Cardiol. Rep. 18, 124. doi:10.1007/s11886-016-0804-z
Dichgans, M. (2007). Genetics of Ischaemic Stroke. Lancet Neurol. 6, 149–161. doi:10.1016/s1474-4422(07)70028-5
Ferreira, L. E., Secolin, R., Lopes-Cendes, I., Cabral, N. L., and França, P. H. C. D. (2019). Association and Interaction of Genetic Variants with Occurrence of Ischemic Stroke Among Brazilian Patients. Gene 695, 84–91. doi:10.1016/j.gene.2019.01.041
Gretarsdottir, S., Thorleifsson, G., Manolescu, A., Styrkarsdottir, U., Helgadottir, A., Gschwendtner, A., et al. (2008). Risk Variants for Atrial Fibrillation on Chromosome 4q25 Associate with Ischemic Stroke. Ann. Neurol. 64, 402–409. doi:10.1002/ana.21480
Hu, Y., Zheng, L., Cheng, L., Zhang, Y., Bai, W., Zhou, W., et al. (2017). GAB2 Rs2373115 Variant Contributes to Alzheimer's Disease Risk Specifically in European Population. J. Neurol. Sci. 375, 18–22. doi:10.1016/j.jns.2017.01.030
Liu, G., Hu, Y., and Jiang, Q. (2019a). Population Difference and Disease Status Affect the Association between Genetic Variants and Gene Expression. Gastroenterology 157, 894–896. doi:10.1053/j.gastro.2019.01.278
Liu, G., Wang, H., Liu, J., Li, J., Li, H., Ma, G., et al. (2014). The CLU Gene Rs11136000 Variant is Significantly Associated with Alzheimer's Disease in Caucasian and Asian Populations. Neuromol Med. 16, 52–60. doi:10.1007/s12017-013-8250-1
Liu, G., Xu, Y., Jiang, Y., Zhang, L., Feng, R., and Jiang, Q. (2017). PICALM Rs3851179 Variant Confers Susceptibility to Alzheimer's Disease in Chinese Population. Mol. Neurobiol. 54, 3131–3136. doi:10.1007/s12035-016-9886-2
Liu, G., Zhang, H., Liu, B., and Ji, X. (2019b). Rs2293871 Regulates HTRA1 Expression and Affects Cerebral Small Vessel Stroke and Alzheimer's Disease. Brain 142, e61. doi:10.1093/brain/awz305
Logan, M., Pagán-Westphal, S. M., Smith, D. M., Paganessi, L., and Tabin, C. J. (1998). The Transcription Factor Pitx2 Mediates Situs-Specific Morphogenesis in Response to Left-Right Asymmetric Signals. Cell 94, 307–317. doi:10.1016/s0092-8674(00)81474-9
Maruthai, K., Sankar, S., and Subramanian, M. (2022). Methylation Status of VDR Gene and its Association with Vitamin D Status and VDR Gene Expression in Pediatric Tuberculosis Disease. Immunol. Investig. 51, 73–87. doi:10.1080/08820139.2020.1810702
Mommersteeg, M. T. M., Brown, N. A., Prall, O. W. J., de Gier-de Vries, C., Harvey, R. P., Moorman, A. F. M., et al. (2007). Pitx2c and Nkx2-5 are Required for the Formation and Identity of the Pulmonary Myocardium. Circulation Res. 101, 902–909. doi:10.1161/CIRCRESAHA.107.161182
Orellana-Urzúa, S., Rojas, I., Líbano, L., and Rodrigo, R. (2020). Pathophysiology of Ischemic Stroke: Role of Oxidative Stress. Curr. Pharm. Des. 26, 4246–4260. doi:10.2174/1381612826666200708133912
Shi, L., Li, C., Wang, C., Xia, Y., Wu, G., Wang, F., et al. (2009). Assessment of Association of Rs2200733 on Chromosome 4q25 with Atrial Fibrillation and Ischemic Stroke in a Chinese Han Population. Hum. Genet. 126, 843–849. doi:10.1007/s00439-009-0737-3
Su, L., Shen, T., Xie, J., Yan, Y., Chen, Z., Wu, Y., et al. (2015). Association of GWAS-Supported Variants Rs2200733 and Rs6843082 on Chromosome 4q25 with Ischemic Stroke in the Southern Chinese Han Population. J. Mol. Neurosci. 56, 585–592. doi:10.1007/s12031-015-0520-y
Tammen, S. A., Friso, S., and Choi, S.-W. (2013). Epigenetics: The Link between Nature and Nurture. Mol. Aspects Med. 34, 753–764. doi:10.1016/j.mam.2012.07.018
Tessari, A., Pietrobon, M., Notte, A., Cifelli, G., Gage, P. J., Schneider, M. D., et al. (2008). Myocardial Pitx2 Differentially Regulates the Left Atrial Identity and Ventricular Asymmetric Remodeling Programs. Circulation Res. 102, 813–822. doi:10.1161/CIRCRESAHA.107.163188
Wei, C.-J., Cui, P., Li, H., Lang, W.-J., Liu, G.-Y., and Ma, X.-F. (2019). Shared Genes between Alzheimer's Disease and Ischemic Stroke. CNS Neurosci. Ther. 25, 855–864. doi:10.1111/cns.13117
Wu, Q., Wu, H., Gegentana, G., Huo, W., Suyalatu, S., Wu, N., et al. (2015). “Identification of the Susceptibility Gene Loci Associated with Ischemic Stroke in a Mongolian Population in China,” in Computational Molecular Biology MCCMB 2015 16, 50.
Keywords: ischemic stroke, genome-wide association study, rs2200733, rs6843082, population
Citation: Wang D, Hu X, Yang X, Yang M and Wu Q (2022) Variants rs2200733 and rs6843082 Show Different Associations in Asian and Non-Asian Populations With Ischemic Stroke. Front. Genet. 13:905560. doi: 10.3389/fgene.2022.905560
Received: 27 March 2022; Accepted: 30 May 2022;
Published: 18 August 2022.
Edited by:
Guiyou Liu, Tianjin Institute of Industrial Biotechnology (CAS), ChinaReviewed by:
Hui Liu, Capital Medical University, ChinaCopyright © 2022 Wang, Hu, Yang, Yang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingfeng Yang, bWZ5YW5nQDE2My5jb20=; Qingjian Wu, d3F3MTEwQDE2My5jb20=
†ORCID: Mingfeng Yang, orcid.org/0000-0002-6857-4743; Qingjian Wu, orcid.org/0000-0002-8746-8743
‡These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.