- 1Laboratory Medicine Center, Department of Clinical Laboratory, Zhejiang Provincial People’s Hospital (Affiliated People’s Hospital, Hangzhou Medical College), Hangzhou, China
- 2Department of Central Laboratory, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3School of Pharmacy, Hangzhou Medical College, Hangzhou, China
- 4School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, China
- 5Traditional Chinese Medicine Department, Zhejiang Provincial People’s Hospital (Affiliated People’s Hospital, Hangzhou Medical College), Hangzhou, China
Breast invasive carcinoma (BRCA) is a carcinoma with a fairly high incidence, and the therapeutic schedules are generally surgery and chemotherapy. However, chemotherapeutic drugs tend to produce serious toxic side effects, which lead to the cessation of treatment. Therefore, it is imperative to develop treatment strategies that are more effective and have fewer side effects at the genetic level. Centromeric protein W (CENPW) is an oncogene that plays an important part in nucleosome assembly. To date, no studies have reported the prognostic significance of CENPW in breast carcinoma. In this study, we verified that CENPW expression is up-regulated in breast carcinoma and positively associated with the level of immune cell infiltration. The clinicopathological characteristics further suggest that CENPW expression is correlated with a worse prognosis of breast carcinoma. Interestingly, the CENPW mutation contributes to the poor prognosis. Next, we discovered that the genes interacting with CENPW are mainly concentrated in the cell cycle pathway, and CENPW is co-expressed with CDCA7, which is also highly expressed in breast carcinoma and leads to a worse prognosis. Our subsequent studies verified that knockdown of CENPW significantly inhibits the proliferation and migration of breast carcinoma cells and promotes their apoptosis rate. Notably, inhibition of CEMPW sensitizes breast cancer cells to chemotherapeutic drugs that have been found to induce cell cycle arrest. In summary, these results provide extensive data and experimental evidence that CENPW can serve as a novel predictor of breast cancer and may act as a prospective therapeutic target.
Introduction
The International Agency for Research on Cancer (IARC) announced the latest global carcinoma burden data in 2020, showing that the new incidence of breast carcinoma in the world is up to 11.7%, which has overtaken lung carcinoma as the world’s largest most common cancer. Breast invasive carcinoma (BRCA) is the widest tumor type in women, with a poor prognosis and high recurrence rates (Lang et al., 2017). At present, paclitaxel combined with carboplatin has made great progress in the therapy of breast carcinoma, the long-term use of chemotherapeutic drugs produces obvious resistance and the therapeutic efficacy is still not ideal. (Fasching et al., 2021). In the general population, the pathogenesis of breast cancer is often directly related to alterations in gene expression (van den Broek et al., 2021). Therefore, finding the driver genes in tumorigenesis may open up new strategies for early breast cancer screening and the discovery of potential therapeutic targets, which will facilitate early detection and thereby reduce morbidity and mortality (Shen et al., 2021).
Centromeric protein W (CENPW), also known as cancer upregulated gene 2 (CUG2) protein, is overexpressed in multiple tumor tissues, including cervical, colon, liver, and lung cancers (Malilas et al., 2014). The CENPW is located at human chromosome 6q22.32 and is regulated by the upstream TSS region, which can transcribe a full-length mRNA of 600 bp (Lee et al., 2007). CENPW is a representative member of the Constitutive Centromere Associated Network (CCAN) family and is of crucial importance in the formation of centromeric nucleosomes (Klare et al., 2015). CENPW is encoded by CENPW and forms a DNA-binding heterodimer with CENPT (Prendergast et al., 2016), which is able to switch centromeric chromatin to a mitotic state (Prendergast et al., 2011). Previous studies have found that the regulation of CENPW can affect the migration of colon carcinoma cells and induce the epithelial-mesenchymal transition of lung carcinoma cells via TGF-β signaling (Malilas et al., 2013; Kaowinn et al., 2018). However, the role and value of CENPW in breast carcinoma remain unclear and require further study.
In the current study, several databases were used to evaluate the role of CENPW in breast carcinoma. We discovered that CENPW, which is highly expressed in breast carcinoma, is associated with a poor prognosis of breast carcinoma. We further found that CENPW mainly influences the cell cycle pathway, and the downregulation of CENPW expression could inhibit the migration and proliferation of breast cancer cells and promote apoptosis. Notably, we believe that CENPW could be a biomarker in the development process of breast carcinoma and propose that inhibition of CENPW may be a prospective strategy for inhibiting breast carcinoma development.
Materials and Methods
GEPIA Analysis Dataset
GEPIA is an interactive website server based on TCGA and GTEx databases, which provides us with customizable functional modules (http://gepia.cancer-pku.cn/). In this study, the CENPW expression was analyzed using GEPIA in different tumors and paracancerous tissues. The association between CENPW and the highly coexpressed gene (CDCA7) was evaluated according to the function of correlation analysis. In addition, the OS of CDCA7 gene was evaluated by the survival plots module.
Oncomine Database Analysis
The Oncomine Database is an openly available database for the analysis of CENPW expression levels in multiple tumor types. The difference of mRNA expression and co-expression of CENPW gene in breast carcinoma were studied by this database. The thresholds were set as two-fold change, top 10% gene rank and p-value = 1E-4.
UALCAN Cancer Database Analysis
The UALCAN Cancer Database comprehensively contains TCGA dataset, CPTAC dataset, and CBTTC dataset. The CENPW expression in BRCA was verified and the relationship between clinicopathological features and CENPW expression was identified by the UALCAN Cancer Database.
Bc-GenEx Miner Analysis
Bc-GenEx Miner is a full-featured online access database, which is specialized for breast cancer. According to this database, the CENPW expression in different clinical stages was mined, and the correlation between CENPW and the related genes was clustered.
Kaplan–Meier Survival Analysis
To evaluate the prognostic significance of a particular gene, the patient samples were divided into two groups on the basis of the median expression. The prognostic significance of CENPW in breast carcinoma was evaluated according to overall survival (OS), distant metastasis-free survival (DMFS), and relapse-free survival (RFS) by Kaplan–Meier plotter. Log-rank p-value and HRs with 95% confidence intervals were set.
cBioPortal Website Analysis
cBioPortal is a comprehensive and open website based on the TCGA database, which provides us with visual cancer gene mutation data (http://www.cbioportal.org/). The CENPW gene mutations were analyzed using seven breast cancer databases available on the cBioPortal website.
UCSC Xena Website Analysis
The UCSC Xena website contains multiple genomic databases for researching correlations between genomic or phenotypic variables (https://xenabrowser.net/). The TCGA database related to breast cancer provided by UCSC Xena was used for data mining to construct a heat map of CENPW and CDCA7.
GeneMANIA Website Analysis
The GeneMANIA site comprises a wealth of gene information, and can be used for functional analysis (http://www.genemania.org). Its prediction algorithm has high accuracy (Warde-Farley et al., 2010). Therefore, the gene clusters of mutual value with CENPW were predicted using this website.
Metascape Online Analysis
Metascape Online (https://metascape.org/gp/index.html) can be employed for function enrichment of CENPW-related gene sets and PPI network analysis (Zhou et al., 2019). And we used MCODE1 to further analyze densely connected regions. p-value < 0.05 was set as a cutoff value (Lu et al., 2021).
TIMER Website Analysis
The new version of TIMER 2.0 has seven main analysis systems, with gene and survival functions designed to assess the infiltration of multiple immune cells (https://cistrome.shinyapps.io/timer/) (Li et al., 2017). CENPW was selected to generate a scatter map through a gene module to observe the relevance between the expression of CENPW and the immune infiltration level in breast carcinoma.
Cell Culture
Human breast carcinoma cell lines (MDA-MB-231, BT-549) were purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). MDA-MB-231 cell was cultured in DMEM (Hyclone, Logan, UT, United States) with 10% FBS (Gibco, Grand Island, United States) at 37°C and 5% CO2. BT-549 cell was cultured in RPMI-1640 (Hyclone, Logan, UT, United States) with 10% FBS (Gibco, Grand Island, United States) at 37°C and 5% CO2.
Cell Transfection
Two pairs of siRNAs targeting CENPW, and negative control siRNA-NC (si-NC) were synthesized (RiboBio, Guangzhou, China). MDA-MB-231 and BT-549 cells were plated with appropriate cell density, and further transfected corresponding siRNA (si-NC, si1-CENPW and si2-CENPW) by Lipofectamine™ 3000 (Invitrogen, Carlsbad, CA, United States).
Western Blot Analysis
The transfected cells were lysed with RIPA buffer (Sigma-Aldrich, United States) to extract protein solution. The protein concentration was gauged with BCA kit (Boxbio Science and vernight with primary antibodies, then removed for (HRP)-conjugated secondary antibody incubation. The blots were visualized by ECL-Plus kit (Thermo Scientific, United States).
Cell Proliferation Detection
Cell proliferation was detected by CCK-8 (Meilunbio, China). 5,000 transfected cells were inoculated into a 96-well plate (NEST Biotechnology). After culture for 0, 24, 48 and 72 h, 10 μL CCK-8 reagent was added into detecting well, and the optical density at 450 nm wavelength was detected.
Clone Formation Assay
Transfected cells (500 cells/well) were seeded into 6-well plates. A second transfection was performed 7 days after the first one to improve the transfection efficiency. After 14 days, the colonies were fixed with paraformaldehyde and stained with crystal violet for 20 min. Whereafter, the field of vision was photographed by a light microscope (Olympus, Japan).
Transwell Invasion Assay
The transfected cells were seeded into the upper chamber (Corning, United States), and the lower chamber was added with 500 μL 5% FBS complete medium. After 24 h of culture, the cells were fixed with paraformaldehyde solution and stained with crystal violet for 20 min. Finally, the invading cells were captured by a light microscope (Olympus, Japan).
Wound Healing Assay
The transfected cells were seeded into 6-well plates and grew to around 90% confluence. The monolayer cells were then scratched with the tip of a 10 μL pipette. After 24 h of culture, the wound healing area was captured by a light microscope (Nikon, Japan).
Flow Cytometry Analysis
The cell apoptosis was detected by flow cytometry using the Annexin V-FITC/PI apoptosis kit (MultiSciences, Hangzhou, China). The transfected cells (6×105 cells/well) were cultured in 6-well plates for 48 h and collected in 1×Binding buffer, then Annexin V-FITC (5 μL), and PI (10 μL) were added for 10 min incubation in the dark and subjected to the flow cytometry (Beckman coulter, CA, United States).
Statistical Analysis
All statistical computing was performed by GraphPad Prism 7.0. Results are expressed as mean ± SD. The discrepancies between two groups were accomplished by the student’s t-test. Statistical significance was established at *p < 0.05, **p < 0.01, compared to the corresponding control group.
Results
The Evaluation of CENPW Expression in Multiple Cancer Tissues
To clarify the molecular biological characteristics of CENPW in different tumors, we first investigated CENPW expression in cancers and adjoining normal tissues from various databases. The data from the Oncomine database verified that the expression of CENPW was enhanced in numerous tumors including brain, breast, colorectal, head and neck, lung carcinomas compared with normal samples, while CENPW expression was lower in leukemia than in normal samples (Figure 1A). GEPIA database was further employed to evaluate the expression differences of CENPW in 33 kinds of tumor types (Figure 1B). We found that the CENPW was highly expressed in many tumors such as in BLCA, BRCA, COAD, HNSC, LUAD and STAD. On the contrary, the CENPW expression was lower than that of normal controls in acute myeloid leukemia. Collectively, these results from different databases together revealed that CENPW is excessively upregulated in most tumors and may act as a potential indicator of cancer progression.
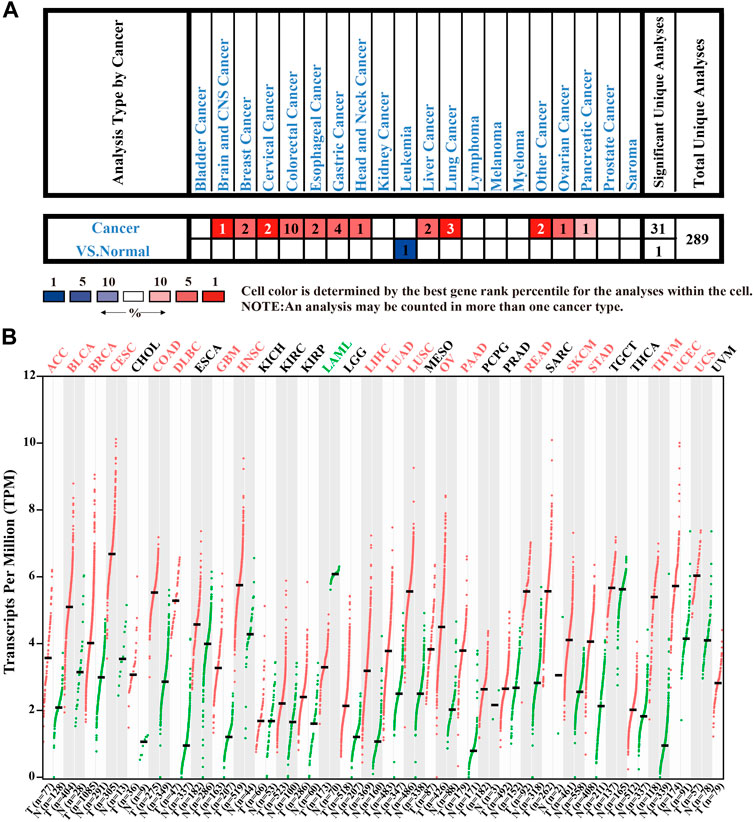
FIGURE 1. The expression levels of CENPW in different tissues. (A) The CENPW expression in multifarious tumor tissues and its corresponding normal tissues in the Oncomine database. (B) The CENPW expression in diverse types of carcinomas and normal tissues in GEPIA database.
CENPW Acts as an Oncogene in the Carcinogenesis and Progression of Breast Carcinoma
To better explore the potential value of CENPW in breast carcinoma patients, we further explored the expression and function of CENPW in breast carcinoma through several means. According to the Richardson Breast two dataset (Richardson et al., 2006), the CENPW expression in ductal breast cancer was distinctly higher than that in normal tissues (Figure 2A). In addition, the Curtis Breast dataset (Curtis et al., 2012) showed a 2.698-fold (p = 1.70E−09) increase in CENPW mRNA expression in medullary breast carcinoma (Figure 2B). Differences in mRNA expression of CENPW in breast carcinoma were also confirmed using the UALCAN database (Figure 2C). In addition, the UALCAN database was devoted to analyzing the relation between CENPW expression and different clinicopathological characteristics in BRCA patients. The data indicated that the CENPW expression was higher in female patients and was positively associated with patient age, nodal metastasis, and cancer stage (Figures 2D–G). Additionally, the relation between CENPW expression and triple-negative breast carcinoma classification was analyzed, and the expression level of CENPW was the highest in TNBC-BL2 (Figure 2H). The pairwise comparison p-values of the above results obtained by the UALCAN database are shown in Supplementary Table S1.
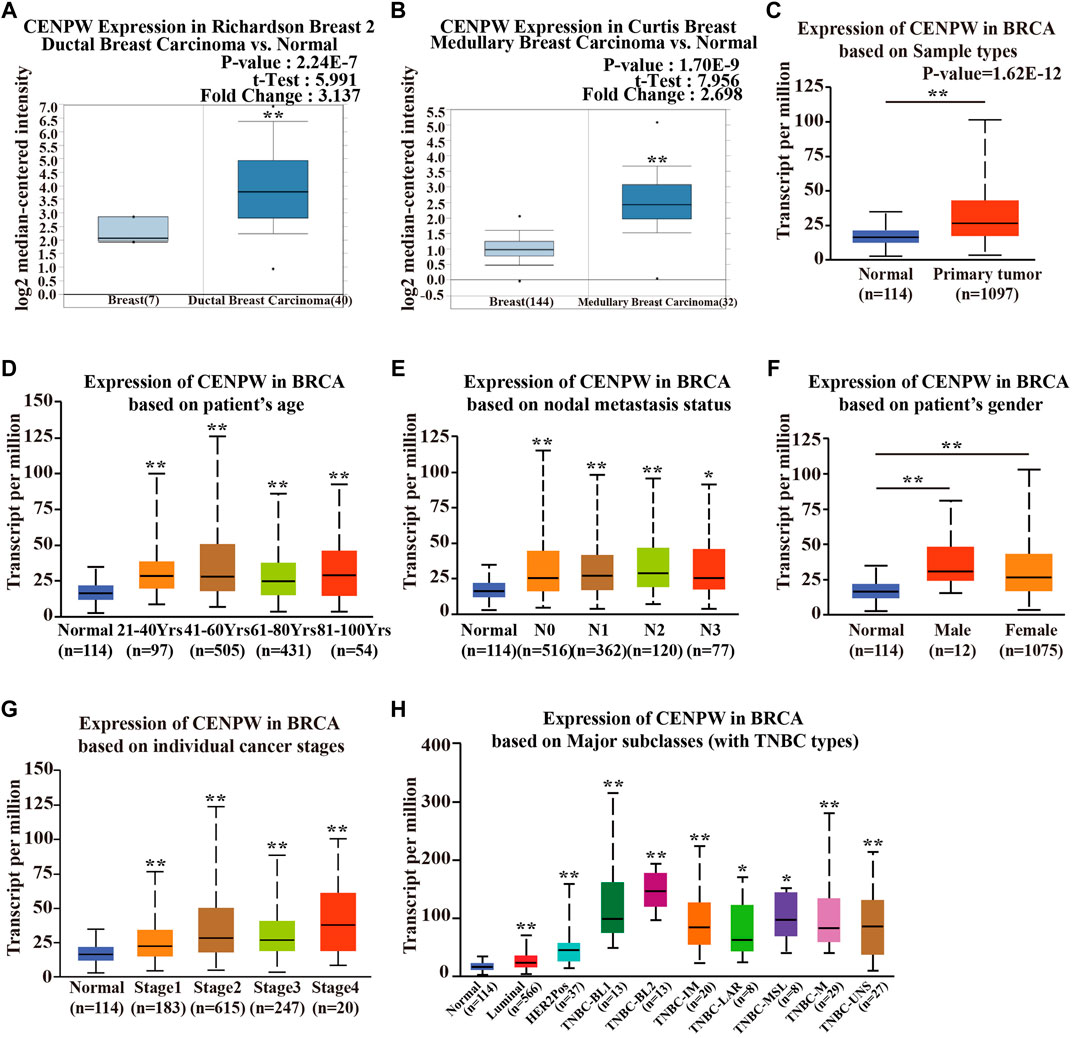
FIGURE 2. The CENPW expression in multiple subtypes of breast carcinoma. Expression levels of CENPW in (A) ductal breast carcinoma and (B) medullary breast carcinoma. (C) The CENPW expression in multiple subtypes of BRCA compared to normal samples derived UALCAN cancer database. Box plots of CENPW mRNA levels in the UALCAN cancer database based on (D) patient’s age, (E) gender, (F) nodal metastasis status, (G) individual cancer stages and (H) major subclasses (with TNBC types) of breast cancer patients were shown. *p < 0.05, **p < 0.01 compared with control group.
We also evaluated the expression of CENPW with different clinicopathological features in breast carcinoma patients by the bc-GenExMiner online tool. The CENPW expression was higher in the less than 51-year-old group, and higher CENPW expression was correlated with a more advanced Scarff–Bloom–Richardson (SBR) grade (Figures 3A,B). CENPW levels in breast carcinoma patients with positive node status (N) were higher than those in patients with negative node status (Figure 3C). Progesterone receptor (PR) and estrogen receptor (ER) status were negatively correlated with CENPW expression (Figures 3D–F). In contrast, human epidermal growth factor receptor-2 (HER-2) status was positively correlated with CENPW expression (Figure 3G). In addition, CENPW increased in base-like subtypes compared to non-base-like subtypes (Figure 3H). Simultaneously, CENPW was strongly elevated in triple-negative breast cancer (TNBC) patients (Figure 3I). Detailed data on the clinicopathological features are presented in Supplementary Table S2.
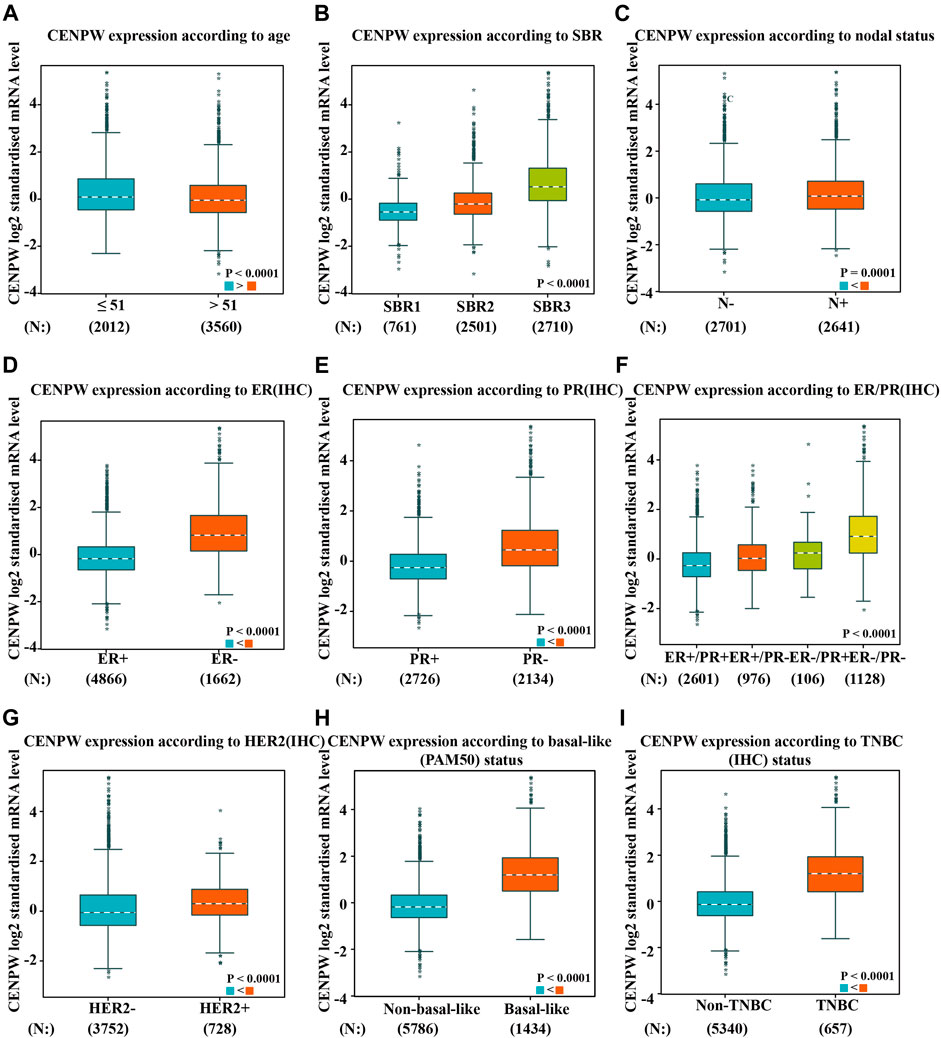
FIGURE 3. The correlation between CENPW expression and various clinicopathological parameters. Data are displayed for (A) age, (B) SBR, (C) nodal status, (D) ER, (E) PR, (F) ER/PR, (G) HER-2, (H) basal-like status and (I) triple-negative status.
The High Level of CENPW is Correlated With Poor Prognosis in Breast Carcinoma Patients
We further explored the relationship between CENPW expression and breast carcinoma prognosis. Results of the Kaplan-Meier plotter showed that higher expressions of CENPW were linked to poorer OS (Figure 4A). Similarly, DMFS and RFS were worse in breast cancer patients with increased CENPW expression (Figures 4B,C). To reconfirm the function of CENPW in breast carcinoma prognosis, we illustrated the effect of CENPW on OS, DMFS, and DFS by bc-GenExMiner database. Breast cancer patients with upregulated CENPW presented with worse OS, DMFS, and DFS (Figures 4D–F), which were similar to the results of the Kaplan-Meier plotter. In summary, these data showed that CENPW expression is associated with poor prognosis in breast cancer.
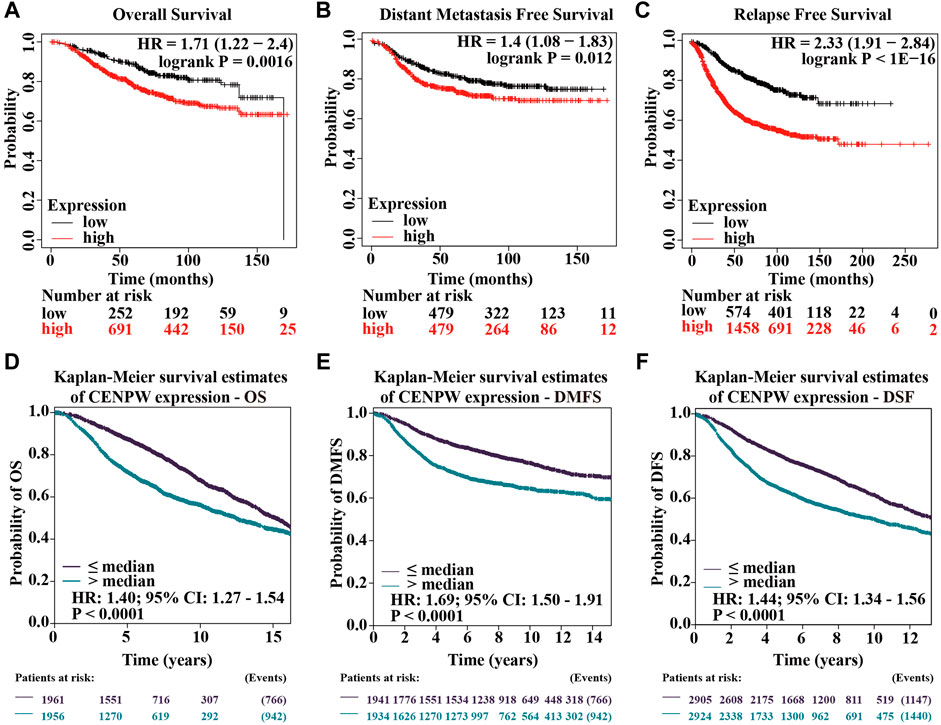
FIGURE 4. Survival data assessing the prognostic value of CENPW. Kaplan-Meier survival analysis of (A) OS, (B) DMFS and (C) RFS in breast cancer. And survival analysis of (D) OS, (E) DMFS and (F) DFS analyzed by bc-GenExMiner database in patients with breast cancer.
The Analysis of Gene Mutation and Interaction of CENPW in Breast Carcinoma Patients
To further understand the influence of CENPW gene mutations on breast carcinoma patients, we used the cBioPortal database and detected mutations in seven breast cancer databases. It was found that in 6,801 breast cancer patients, 2% had mutations in the CENPW gene (Figure 5A), and alterations with amplification were more common (Figure 5B). Furthermore, we predicted the 3D structure of CENPW and identified the mutation site using the cBioPortal online tool (Figure 5C). Next, we evaluated the effect of mutation on the prognosis of breast carcinoma patients and found that the high mutation group was obviously linked to shorter DFS and PFS in breast carcinoma patients (Figures 5D,E). In addition, the UALCAN database was used to assess the methylation levels of the CENPW promoters in BRCA (Figure 5F). The data indicated that the methylation level of the CENPW promoter in patients with BRCA was dramatically reduced, which may give rise to an increase in CENPW expression. We also revealed the relation between CENPW expression and TP53 mutations and found that CENPW was highly expressed in TP53 mutant groups (Figure 5G).
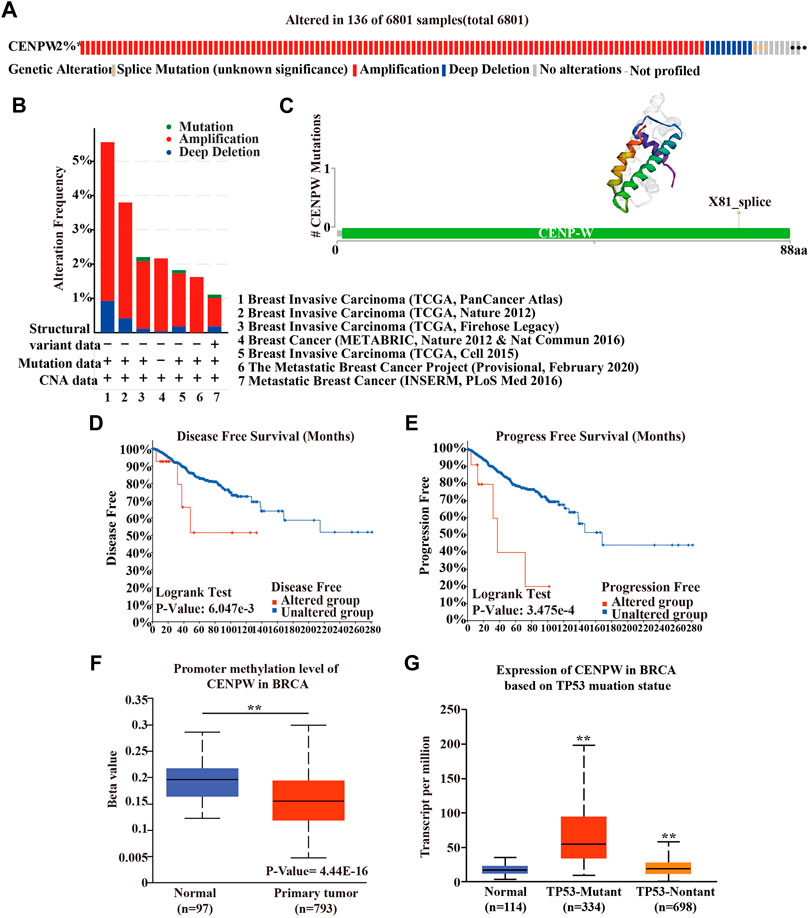
FIGURE 5. Gene mutation analyses of CENPW. (A) The mutation rate of CENPW in 6,801 samples. (B) Summary of alterations in various expressed CENPW in the cBioPortal database. (C) The mutation site and 3D structure of CENPW. Survival analysis of (D) DFS and (E) PFS of the mutated CENPW gene. (F) The promoter methylation level of CENPW and (G) the mRNA expression of CENPW based on TP53 mutation statue in BRCA by UALCAN cancer database. *p < 0.05, **p < 0.01 compared with control group.
The Co-expression of CENPW and CDCA7 in Breast Carcinoma
To further explore the mechanism of CENPW in BRCA, we employed the Oncomine database to construct the co-expression network of CENPW (Figure 6A). The co-expression profile showed that CENPW had the highest association coefficient with CDCA7 among 129 breast carcinoma samples in the LU Breast dataset (Lu et al., 2008). We also confirmed the correlation between CENPW and CDCA7 through the GEPIA website and bc-GenExMiner database and obtained results consistent with those of Oncomine (Figures 6B,C). Furthermore, CENPW and CDCA7 expression heat maps were derived using the UCSC Xena website. The CENPW expression was positively linked to the transcription level of CDCA7 in a 50-gene qPCR assay (PAM50) for breast carcinoma subtypes (Figure 6D). These data indicated that CENPW might be associated with the CDCA7 signaling pathway in breast cancer. Then the UALCAN database was performed to verify differences in CDCA7 expression. The results revealed that CDCA7 was obviously upregulated in BRCA (Figure 6E). According to the outcome of the GEPIA database survival analysis, CDCA7 upregulation was correlated with poorer OS in breast carcinoma patients (Figure 6F), which was consistent with the effect of CENPW on breast cancer.
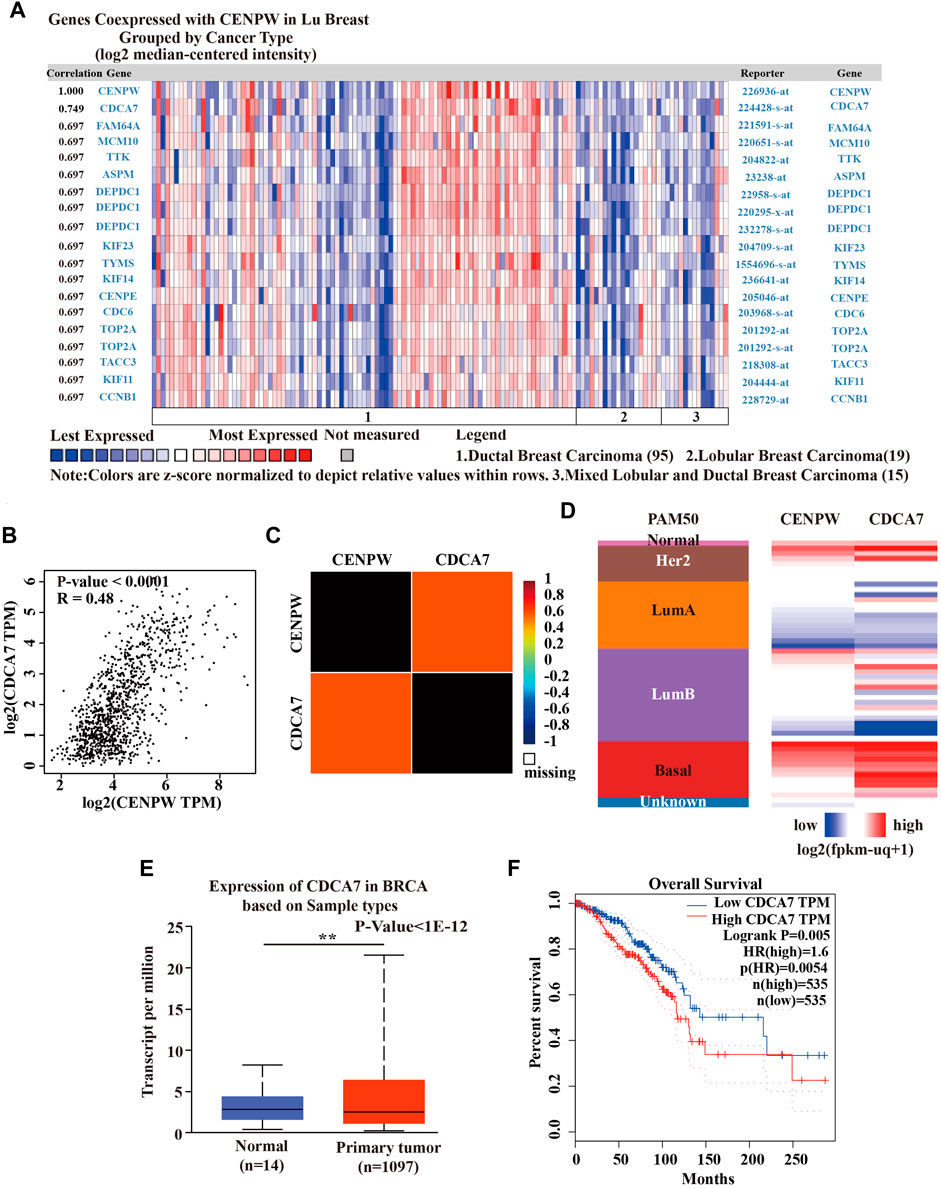
FIGURE 6. Coexpression analysis of CENPW. (A) Coexpression profile of CENPW derived from the Oncomine database. (B) The relation between CENPW and CDCA7 expression in breast carcinoma in GEPIA website and (C) the bc-GenExMiner database. (D) Heat map of CENPW and CDCA7 expression across PAM50 subtypes by UCSC Xena website. (E) The CDCA7 expression of breast cancer in Ualcan database. (F) The OS status according to the CDCA7 expression in GEPIA database. *p < 0.05, **p < 0.01 between the compared groups.
The Enrichment and Interaction Analysis of CENPW-Related Genes
Twenty genes (including CENPT, CENPH, CENPA, FOXO3, and CKS2) interacting with CENPW were excavated using the GeneMANIA website (Figure 7A). To explore the signaling pathway associated with CENPW, we used Metascape Online for functional analysis and discovered that the genes interacting with CENPW were primarily enriched in cell cycle, cell division, and chromosome maintenance (Figure 7B). These data inspired us to explore the relationship between CENPW and the cell cycle pathway. Next, the PPI network based on Metascape Online and the MCODE1 plug-in, further helped to identify these important modules of genes interacting with CENPW (Figures 7C,D). To further verify the co-expression of CENPW and CDCA7, we also conducted the above study on CDCA7. The functional analysis of twenty genes interacting with CDCA7 showed that they were also mainly enriched in cell cycle signaling pathway (Supplementary Figure S1). This was consistent with the main biological effects of CENPW on breast cancer cells.
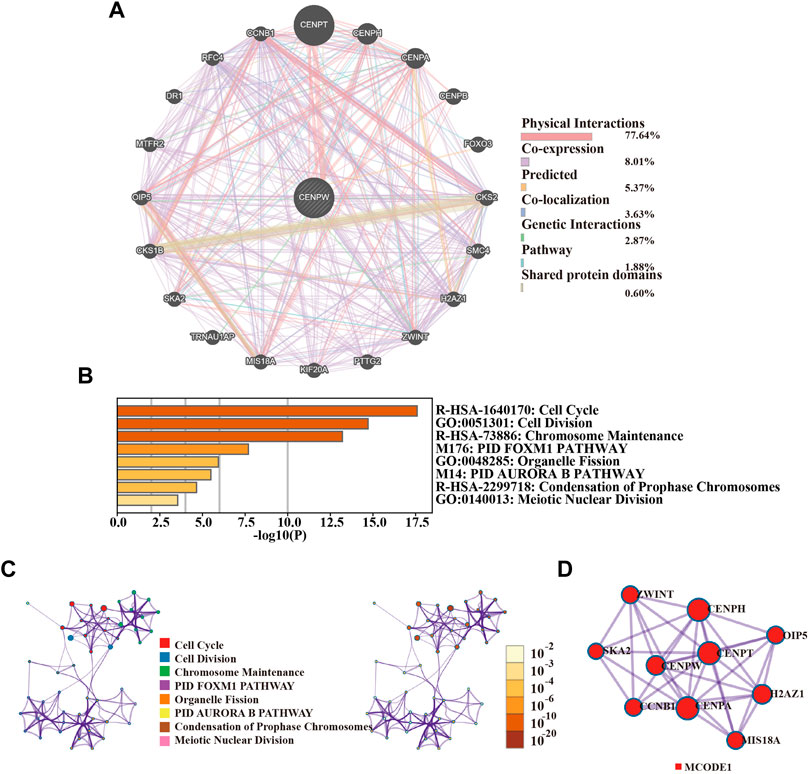
FIGURE 7. The enrichment and interaction of CENPW related genes. (A) PPI network of CENPW from the Metascape database. (B) Functional enrichment and signal path of the overlapping genes. (C) The GO and KEGG analysis suggesting the distribution and relation of the diverse functions according to Metascape Online database. (D) PPI network and MCODE showed associated genes interacting with CENPW.
The Correlation Between CENPW Expression and Immune Cell Infiltration
The relation between CENPW expression and the level of immune cell infiltration in BRCA was further investigated by TIMER2.0 database. Specifically, we investigated the association between CENPW expression and specific immune cell subsets. The data showed that CENPW expression was positively associated with most infiltrated immune cells in BRCA but negatively associated with the immune invasion levels of macrophages (Figure 8). There were noticeable discrepancies between CENPW expression and immune invasion levels of B cells, neutrophils, dendritic cells and macrophages. In addition, we analyzed the immune process of CDCA7 in the breast cancer microenvironment, and the expression level of CDCA7 was negatively correlated with macrophages, positively correlated with immune invasion of B cells, neutrophil, dendritic cell (Supplementary Figure S2), which is consistent with the results of CENPW.
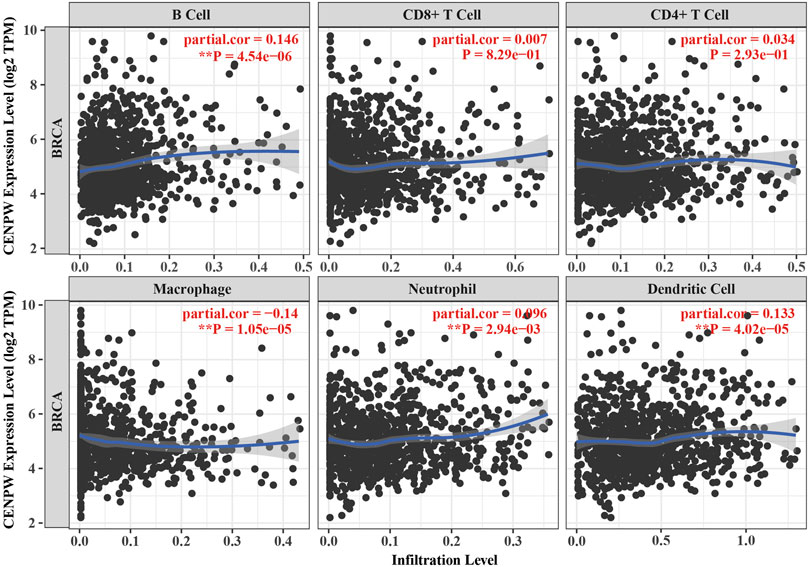
FIGURE 8. Immune infiltration data from the TIMER2.0 database. The relation between CENPW expression and the immune infiltration level in BRCA.
Knockdown of CENPW Inhibits the Proliferation of Breast Carcinoma Cells
The above bioinformatics data proved that under-expression of CENPW was correlated with better survival in breast cancer patients. To explore the role of CENPW in breast cancer cells, we transfected MDA-MB-231 and BT-549 cells with two specific siRNA sequences to induce knockdown of CENPW expression. Growth curves suggested that CENPW knockdown suppressed the proliferation of these two breast carcinoma cell lines (*p < 0.05, Figures 9A,B).
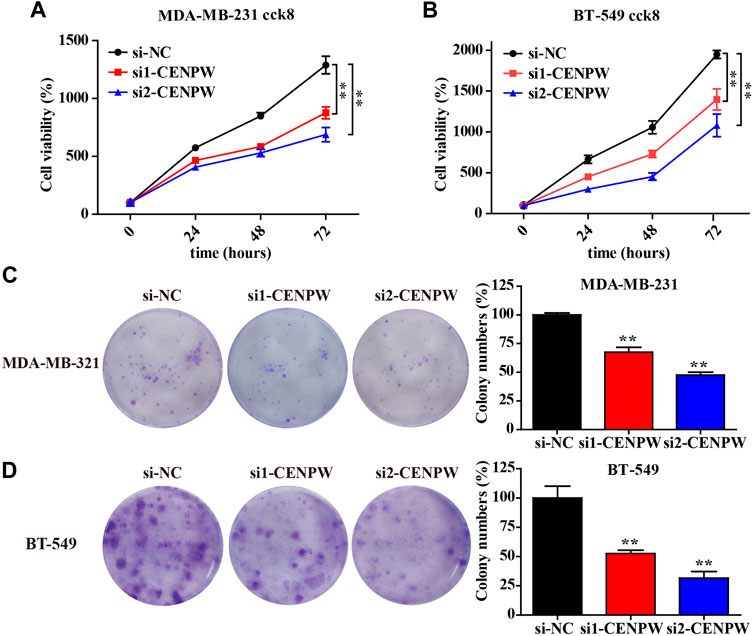
FIGURE 9. Knockdown of CENPW inhibits breast cancer cell proliferation. (A,B) Cell proliferation capability of control and CENPW knockdown cells. (C,D) The colony formation of breast cancer cells with or without CENPW knockdown. Quantitative histograms of colony formation are shown on the right. *p < 0.05, **p < 0.01 compared with si-NC group.
In addition, we examined the effect of CENPW knockdown on long-term cell viability using a clonal formation survival assay in MDA-MB-231 and BT-549 cells (Figures 9C,D). And then after 14 days of culture, the clonal number of the two breast carcinoma cell treated with si1-CENPW and si2-CENPW decreased compared to the control cells.
Knockdown of CENPW Inhibits Migration and Invasion of Breast Cancer Cells
The most concerning aspect of cancer is its ability to migrate and invade. Therefore, the invasion and migration of MDA-MB-231 and BT-549 cells were evaluated using adherent cell scratch migration and transwell invasion assays. Transwell analysis indicated that knockdown of CENPW reduced the number of invading cells (**p < 0.01, Figures 10A,B). In addition, as indicated by the scratch test in Figures 10C,D, the wound healing rates of MDA-MB-231 and BT-549 cells after CENPW silencing were significantly reduced (**p < 0.01). Moreover, these results together revealed that knockdown of CENPW can reduce the migration and invasion ability of breast carcinoma cells.
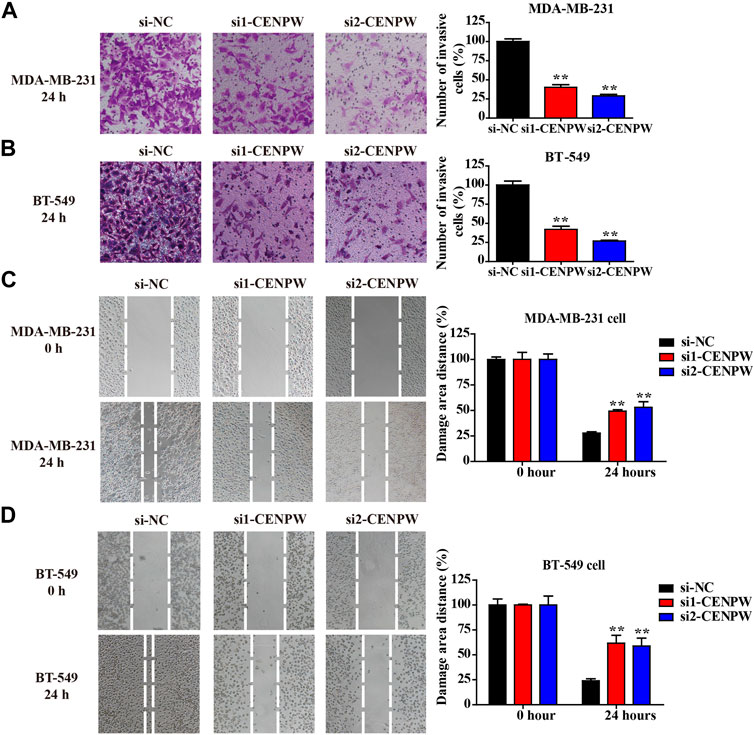
FIGURE 10. Knockdown of CENPW suppresses breast carcinoma cell invasion and migration. (A,B) The invasive ability was evaluated by transwell assay. (C,D) The migration capacity of control and CENPW knockdown cells was detected by wound healing assay. The statistical column diagrams are displayed on the right. *p < 0.05, **p < 0.01 compared with si-NC group.
Knockdown of CENPW Blocks the Cell Cycle Process and Increases the Probability of DNA Damage and Apoptosis in Breast Carcinoma Cells
To figure out the causes of these phenomena, we discovered the protein expression levels of the related pathways. The western blot results indicated that si1-CENPW and si2-CENPW could induce the degradation of CDK6 and Cyclin-D1, suggesting that inhibition of CENPW could block the cell cycle process, which was consistent with the data of GO and KEGG analysis. Furthermore, the significantly decreased protein expression levels of Wee1 and CHK1 indicate that knockdown of CENPW may cause DNA damage, which may be closely related to the participation of CENPW in the nucleosome assembly process. Meanwhile, we discovered that the expression of the apoptosis-related gene BCL2 decreased after CENPW knockdown (Figure 11A). Further apoptosis assay detected by flow cytometry reflected that knockdown of CENPW increased the proportion of apoptotic cells (Figure 11B). Notably, we found that breast cancer cells with reduced CENPW expression were more sensitive to chemotherapeutic drugs that have been found to induce cell cycle arrest. Improved tumor cell killing effect of cisplatin and doxorubicin was found in the CENPW inhibited breast carcinoma cells (Figures 11C,D).
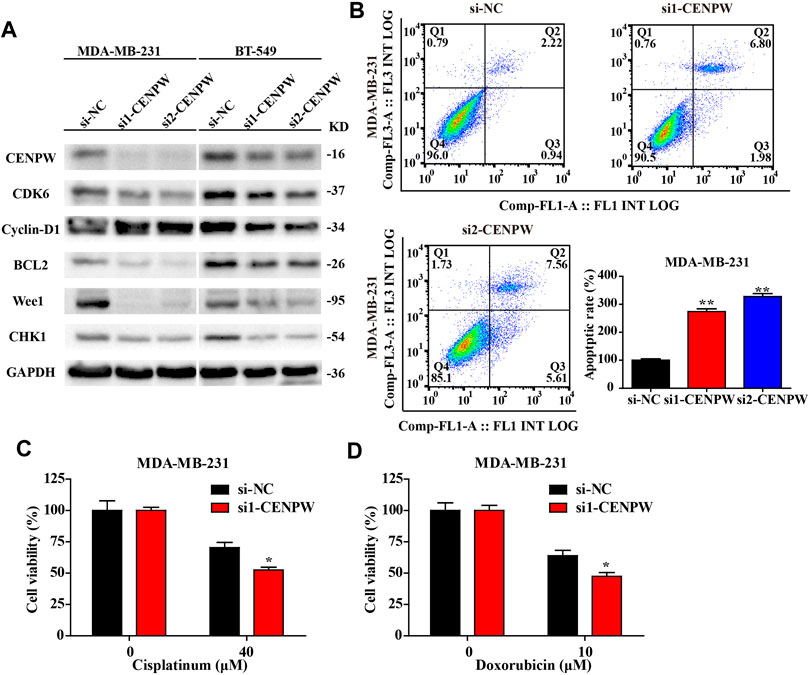
FIGURE 11. Knockdown of CENPW decreased the expression level of related pathway proteins and increased the apoptosis rate of breast carcinoma cells. (A) Western blot assay of control and CENPW knockdown cells. (B) The apoptosis rate was assessed by flow cytometry. (C,D) The cytotoxicity of cisplatin and doxorubicn in the CENPW inhibited breast carcinoma cells. *p < 0.05, **p < 0.01 compared with si-NC group.
Discussion
At present, breast cancer has become the largest number of diagnoses and the highest incidence in women, and it has serious adverse effects on health (Joko-Fru et al., 2021). Therefore, it is crucial to investigate the pathogenesis, diagnosis, treatment, and prognosis of breast carcinoma (Liang et al., 2020). In the study, we explored and revealed that abnormal CENPW expression is closely linked to breast tumors through various databases and in vitro cell experiments. Ziliang Zhou, et al. (Zhou et al., 2020) reported that the CENPW expression level in liver carcinoma was obviously higher than normal liver tissues. Similarly, we discovered that CENPW expression was up-regulated in ductal breast carcinoma and medullary breast carcinoma using two different online databases. According to the clinicopathological features of breast carcinoma, compared with normal tissue, the HER-2 status, SBR grade, basal-like status, nodular status, and triple-negative status were positively associated with CENPW levels in breast carcinoma samples. In contrast, age, ER, and PR were negatively correlated with CENPW expression. These results indicated that CENPW expression was linked to poor prognosis of breast carcinoma, which was also confirmed by subsequent data from the bc-GenexMiner database. Therefore, based on the above series of data analyses, we believe that CENPW may be a promising indicator for screening and may be used as a new prognostic factor for breast carcinoma.
CENPW is a crucial member of the CCAN family, which plays a critical role in nucleosome assembly and is related to the deregulation of centromere proteins and carcinogenesis (Hori et al., 2008). Our findings showed that breast carcinoma patients with high CENPW expression had a worse prognosis than those with low CENPW expression. Notably, through gene mutation analysis in the cBioPortal database, we identified that mutations in CENPW were associated with poor prognosis. Interestingly, a highly correlated gene, cell division cycle-associated protein 7 (CDCA7) (Ye et al., 2018), was discovered by CENPW co-expression analysis. Cai et al. (Cai et al., 2021) reported that downregulation of CDCA7 not only inhibits cell proliferation, but also intercepts the cell cycle in ovarian carcinoma. In the current study, functional enrichment analysis indicated that genes interacting with CENPW were mainly enriched in the cell cycle. These data indicated that CENPW and CDCA7 may play a joint role in the cell cycle pathway; however, further direct evidence is required to confirm this. In addition, studies on CDCA7 have found that its high expression can help to predict poor prognosis and tumor progression in colorectal carcinoma and kidney carcinoma (Li et al., 2020; Liu et al., 2021). Further investigation reveals that CDCA7 expression is also up-regulated in breast carcinoma and leads to poor prognosis.
Notably, CENPW principally affects the cell cycle process of tumor cells by regulating nucleosome assembly (Sridhar et al., 2021). Therefore, we used the GeneMANIA website and Metascape online tools to analyze the gene enrichment and pathway enrichment of CENPW, and the data suggested that CENPW has a vital influence on the cell cycle pathway. In addition, Zhou et al. (Zhou et al., 2021) reported that knockdown of CENPW restrains HCC progression by inactivating E2F signaling. However, the significance of its downregulation in breast carcinoma prognosis has not yet been studied. To illustrate the specific molecular function of the CENPW gene in breast cancer, we constructed CNEPW gene silenced cells and found that CENPW suppression could inhibit the proliferation and migration of MDA-MB-231 and BT-549 cells, hindering the cell cycle signaling pathway, thereby causing apoptosis of breast tumor cells. Previous studies have confirmed that CENPW accelerates cell transformation and stemness through nuclear NPM1 protein and TGF-β signaling (Kaowinn et al., 2019a; Yawut et al., 2020). Moreover, Kaowinn et al. (Kaowinn et al., 2019b) found that CENPW elevated the expression of YAP1, resulting in the increase of the EMT in lung cancer cells. Interestingly, we found that breast cancer cells with CENPW inhibited were more sensitive to chemotherapeutic drugs that have been found to induce cell cycle arrest. These effects may be resulting from the inhibition of TGF-β signaling pathway, but required further exploration.
These cellular results were consistent with the above bioinformatics analysis and fully illustrated the fact that CNEPW may be a crucial oncogene and accelerant for the progression of breast carcinoma, which could help in the development of new breast carcinoma treatment strategies. However, there are still some limitations in our current study. A gene regulates multiple metabolic pathways, but its specific mechanism of action cannot be clearly explained by a single signaling pathway. Therefore, further experimental studies are required to better understand its functions. Further clinical trials are indispensable for elucidating the value of CENPW in the treatment of breast carcinoma.
Conclusion
This study presents evidence that high CENPW expression in breast cancer generates poor prognosis and can therefore be used as a novel biomarker for breast carcinoma. In particular, mutations in the CENPW gene also led to a poor prognosis. Therefore, knockdown CENPW expression can suppress the proliferation and migration of breast cancer cells and induce apoptosis. Our results provide bioinformatics data and experimental evidence that CENPW can be used as a unique biomarker for breast carcinoma, and as a hopeful therapeutic option for the development of related inhibitors to treat breast cancer at the genetic level.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
FZ, XT, and YW conceived and designed the experiments. LW, HW, and CY performed the experiments. YW, GL, YY, and YG analyzed the data. YL and LW wrote the paper. FZ, JD, and YW revised and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by Zhejiang Provincial Natural Science Foundation of China (Grant Nos. LGF21H010008, LGF20H080005, LGF22H160038, LGF22H160027); major projects of Hangzhou health science and technology (Z20220024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.900111/full#supplementary-material
References
Cai, C., Peng, X., and Zhang, Y. (2021). Downregulation of Cell Division Cycle-Associated Protein 7 (CDCA7) Suppresses Cell Proliferation, Arrests Cell Cycle of Ovarian Cancer, and Restrains Angiogenesis by Modulating Enhancer of Zeste Homolog 2 (EZH2) Expression. Bioengineered 12 (1), 7007–7019. doi:10.1080/21655979.2021.1965441
Curtis, C., Shah, S. P., Shah, S. P., Chin, S.-F., Turashvili, G., Rueda, O. M., et al. (2012). The Genomic and Transcriptomic Architecture of 2,000 Breast Tumours Reveals Novel Subgroups. Nature 486 (7403), 346–352. doi:10.1038/nature10983
Fasching, P. A., Link, T., Hauke, J., Seither, F., Jackisch, C., Klare, P., et al. (2021). Neoadjuvant Paclitaxel/olaparib in Comparison to Paclitaxel/carboplatinum in Patients with HER2-Negative Breast Cancer and Homologous Recombination Deficiency (GeparOLA Study). Ann. Oncol. 32 (1), 49–57. doi:10.1016/j.annonc.2020.10.471
Hori, T., Amano, M., Suzuki, A., Backer, C. B., Welburn, J. P., Dong, Y., et al. (2008). CCAN Makes Multiple Contacts with Centromeric DNA to Provide Distinct Pathways to the Outer Kinetochore. Cell 135 (6), 1039–1052. doi:10.1016/j.cell.2008.10.019
Joko-Fru, W. Y., Griesel, M., Mezger, N. C. S., Hämmerl, L., Seraphin, T. P., Feuchtner, J., et al. (2021). Breast Cancer Diagnostics, Therapy, and Outcomes in Sub-saharan Africa: A Population-Based Registry Study. J. Natl. Compr. Canc Netw. doi:10.6004/jnccn.2021.7011
Kaowinn, S., Kaewpiboon, C., Koh, S. S., Krämer, O. H., and Chung, Y. H. (2018). STAT1-HDAC4 S-ignaling I-nduces E-pithelial-mesenchymal T-ransition and S-phere F-ormation of C-ancer C-ells O-verexpressing the O-ncogene, CUG2. Oncol. Rep. 40 (5), 2619–2627. doi:10.3892/or.2018.6701
Kaowinn, S., Seo, E. J., Heo, W., Bae, J.-H., Park, E.-J., Lee, S., et al. (2019). Cancer Upregulated Gene 2 (CUG2), a Novel Oncogene, Promotes Stemness-like Properties via the NPM1-TGF-β Signaling axis. Biochem. Biophysical Res. Commun. 514 (4), 1278–1284. doi:10.1016/j.bbrc.2019.05.091
Kaowinn, S., Yawut, N., Koh, S. S., and Chung, Y.-H. (2019). Cancer Upregulated Gene (CUG)2 Elevates YAP1 Expression, Leading to Enhancement of Epithelial-Mesenchymal Transition in Human Lung Cancer Cells. Biochem. Biophysical Res. Commun. 511 (1), 122–128. doi:10.1016/j.bbrc.2019.02.036
Klare, K., Weir, J. R., Basilico, F., Zimniak, T., Massimiliano, L., Ludwigs, N., et al. (2015). CENP-C Is a Blueprint for Constitutive Centromere-Associated Network Assembly within Human Kinetochores. J. Cell Biol. 210 (1), 11–22. doi:10.1083/jcb.201412028
Lang, G.-T., Shi, J.-X., Hu, X., Zhang, C.-H., Shan, L., Song, C.-G., et al. (2017). The Spectrum of BRCA Mutations and Characteristics of BRCA-Associated Breast Cancers in China: Screening of 2,991 Patients and 1,043 Controls by Next-Generation Sequencing. Int. J. Cancer 141 (1), 129–142. doi:10.1002/ijc.30692
Lee, S., Gang, J., Jeon, S. B., Choo, S. H., Lee, B., Kim, Y.-G., et al. (2007). Molecular Cloning and Functional Analysis of a Novel Oncogene, Cancer-Upregulated Gene 2 (CUG2). Biochem. Biophysical Res. Commun. 360 (3), 633–639. doi:10.1016/j.bbrc.2007.06.102
Li, S., Huang, J., Qin, M., Zhang, J., and Liao, C. (2020). High Expression of CDCA7 Predicts Tumor Progression and Poor Prognosis in Human Colorectal Cancer. Mol. Med. Rep. 22 (1), 57–66. doi:10.3892/mmr.2020.11089
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 77 (21), e108–e110. doi:10.1158/0008-5472.can-17-0307
Liang, Y., Zhang, H., Song, X., and Yang, Q. (2020). Metastatic Heterogeneity of Breast Cancer: Molecular Mechanism and Potential Therapeutic Targets. Seminars Cancer Biol. 60, 14–27. doi:10.1016/j.semcancer.2019.08.012
Liu, S., Wang, Y., Miao, C., Xing, Q., and Wang, Z. (2021). High Expression of CDCA7 Predicts Poor Prognosis for Clear Cell Renal Cell Carcinoma and Explores its Associations with Immunity. Cancer Cell Int. 21 (1), 140. doi:10.1186/s12935-021-01834-x
Lu, T., Xu, R., Li, Q., Zhao, J.-y., Peng, B., Zhang, H., et al. (2021). Systematic Profiling of Ferroptosis Gene Signatures Predicts Prognostic Factors in Esophageal Squamous Cell Carcinoma. Mol. Ther. - Oncolytics 21, 134–143. doi:10.1016/j.omto.2021.02.011
Lu, X., Lu, X., Wang, Z. C., Iglehart, J. D., Zhang, X., and Richardson, A. L. (2008). Predicting Features of Breast Cancer with Gene Expression Patterns. Breast Cancer Res. Treat. 108 (2), 191–201. doi:10.1007/s10549-007-9596-6
Malilas, W., Koh, S. S., Kim, S., Srisuttee, R., Cho, I.-R., Moon, J., et al. (2013). Cancer Upregulated Gene 2, a Novel Oncogene, Enhances Migration and Drug Resistance of Colon Cancer Cells via STAT1 Activation. Int. J. Oncol. 43 (4), 1111–1116. doi:10.3892/ijo.2013.2049
Malilas, W., Koh, S. S., Lee, S., Srisuttee, R., Cho, I.-R., Moon, J., et al. (2014). Suppression of Autophagic Genes Sensitizes CUG2-Overexpressing A549 Human Lung Cancer Cells to Oncolytic Vesicular Stomatitis Virus-Induced Apoptosis. Int. J. Oncol. 44 (4), 1177–1184. doi:10.3892/ijo.2014.2264
Prendergast, L., van Vuuren, C., Kaczmarczyk, A., Doering, V., Hellwig, D., Quinn, N., et al. (2011). Premitotic Assembly of Human CENPs -T and -W Switches Centromeric Chromatin to a Mitotic State. PLoS Biol. 9 (6), e1001082. doi:10.1371/journal.pbio.1001082
Prendergast, L., Müller, S., Liu, Y., Huang, H., Dingli, F., Loew, D., et al. (2016). The CENP-T/-W Complex Is a Binding Partner of the Histone Chaperone FACT. Genes Dev. 30 (11), 1313–1326. doi:10.1101/gad.275073.115
Richardson, A. L., Wang, Z. C., De Nicolo, A., Lu, X., Brown, M., Miron, A., et al. (2006). X Chromosomal Abnormalities in Basal-like Human Breast Cancer. Cancer Cell 9 (2), 121–132. doi:10.1016/j.ccr.2006.01.013
Shen, S. J., Xu, Y. L., Zhou, Y. D., Ren, G. S., Jiang, J., Jiang, H. C., et al. (2021). A Comparative Study of Breast Cancer Mass Screening and Opportunistic Screening in Chinese Women. Zhonghua Wai Ke Za Zhi 59 (2), 109–115. doi:10.3760/cma.j.cn112139-20201015-00753
Sridhar, S., Hori, T., Nakagawa, R., Fukagawa, T., and Sanyal, K. (2021). Bridgin Connects the Outer Kinetochore to Centromeric Chromatin. Nat. Commun. 12 (1), 146. doi:10.1038/s41467-020-20161-9
van den Broek, J. J., Schechter, C. B., van Ravesteyn, N. T., Janssens, A. C. J. W., Wolfson, M. C., Trentham-Dietz, A., et al. (2021). Personalizing Breast Cancer Screening Based on Polygenic Risk and Family History. J. Natl. Cancer Inst. 113 (4), 434–442. doi:10.1093/jnci/djaa127
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 38 (Web Server issue), W214–W220. doi:10.1093/nar/gkq537
Yawut, N., Kaewpiboon, C., Budluang, P., Cho, I.-R., Kaowinn, S., Koh, S. S., et al. (2020). Overexpression of Cancer Upregulated Gene 2 (CUG2) Decreases Spry2 through C-Cbl, Leading to Activation of EGFR and β-Catenin Signaling. Cmar Vol. 12, 10243–10250. doi:10.2147/cmar.s271109
Ye, L., Li, F., Song, Y., Yu, D., Xiong, Z., Li, Y., et al. (2018). Overexpression of CDCA7 Predicts Poor Prognosis and Induces EZH2˗mediated Progression of Triple˗negative Breast Cancer. Int. J. Cancer 143 (10), 2602–2613. doi:10.1002/ijc.31766
Zhou, Y., Chai, H., Guo, L., Dai, Z., Lai, J., Duan, J., et al. (2021). Knockdown of CENPW Inhibits Hepatocellular Carcinoma Progression by Inactivating E2F Signaling. Technol. Cancer Res. Treat. 20, 15330338211007253. doi:10.1177/15330338211007253
Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A. H., Tanaseichuk, O., et al. (2019). Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 10 (1), 1523. doi:10.1038/s41467-019-09234-6
Keywords: CENPW, poor prognosis, biomarker, breast cancer, therapeutic strategy
Citation: Wang L, Wang H, Yang C, Wu Y, Lei G, Yu Y, Gao Y, Du J, Tong X, Zhou F, Li Y and Wang Y (2022) Investigating CENPW as a Novel Biomarker Correlated With the Development and Poor Prognosis of Breast Carcinoma. Front. Genet. 13:900111. doi: 10.3389/fgene.2022.900111
Received: 20 March 2022; Accepted: 24 May 2022;
Published: 17 June 2022.
Edited by:
Myvizhi Esai Selvan, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Atrayee Bhattacharya, Dana–Farber Cancer Institute, United StatesLin Shan, Capital Medical University, China
Copyright © 2022 Wang, Wang, Yang, Wu, Lei, Yu, Gao, Du, Tong, Zhou, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feifei Zhou, NTUxMDAyNUB6anUuZWR1LmNu; Yanchun Li, bHljbWVkQDE2My5jb20=; Ying Wang, bmFuY3l3YW5neWluZ0AxNjMuY29t
†These authors have contributed equally to this work
 Luyang Wang
Luyang Wang Hairui Wang
Hairui Wang Chen Yang
Chen Yang Yunyi Wu1
Yunyi Wu1 Jing Du
Jing Du Feifei Zhou
Feifei Zhou Yanchun Li
Yanchun Li Ying Wang
Ying Wang