- 1Department of Thoracic Surgery, Beilun District People’s Hospital, Ningbo, China
- 2Department of Hepatobiliary and Pancreatic Surgery, The People’s Hospital of jianyang City, Jianyang, China
- 3Department of Thoracic Surgery, Huanggang Central Hospital, Huanggang, China
- 4Department of Oncology, Huanggang Central Hospital, Huanggang, China
Background: Studies have reported that quercetin inhibits the growth and migration of lung adenocarcinoma (LUAD). This study aimed to explore the roles and mechanisms of quercetin target genes in the progression of LUAD.
Methods: The quercetin structure and potential target genes of quercetin were explored in the Traditional Chinese Medicine Systems Pharmacology and SwissTargetPrediction databases. The differentially expressed quercetin target genes were identified in The Cancer Genome Atlas (TCGA) database, and the clinical values of quercetin target genes were explored. Subsequently, a risk model was constructed via the Cox regression and survival analysis to evaluate the potential effects and possible mechanisms of quercetin target genes.
Results: The quercetin differential target genes involved in biological processes such as the oxidation-reduction process, cell proliferation, G2/M transition of the mitotic cell cycle, and were related to the lung cancer. NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 were related to the prognosis, and were independent factors influencing the prognosis of LUAD patients. The risk model was related to the gender, clinical stage, T stage, lymph node metastasis, and survival status of LUAD patients, and was independent risk factor associated with poor prognosis. In the high-risk group, the risk model involved signaling pathways such as cell cycle, DNA replication, spliceosome, and homologous recombination.
Conclusion: The quercetin potential target genes NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 were related to the diagnosis and prognosis of LUAD patients. A risk model based on 7 quercetin target genes could be used to assess the prognosis of patients with LUAD.
Introduction
Houttuynia cordata is the herbal medicine included in the Chinese pharmacopeia and is derived from the dry aerial part of the saururus chinensis plant family. At present, H. cordata has shown anti-tumor progression effects in cancer (Kim et al., 2017; Lou et al., 2019; Subhawa et al., 2020; Liu et al., 2021). For example, H. cordata enhances HIF-1A/FOXO3 signaling transduction, causing the up-regulation of MEF2A expression in HepG2 cells, inhibiting the expression of Bcl-2 family proteins (Bax, Bcl-2 and Bcl-Xl), promoting cell apoptosis, and inhibiting the growth of liver cancer cells transplanted into nude mice (Kim et al., 2017). H. cordata and 2-undecanone can significantly inhibit benzo(a)pyrene from inducing lung tumors without causing significant systemic toxicity in mice (Lou et al., 2019). In addition, H. cordata component quercetin features important biological values in the progression of lung cancer (Lee et al., 2015; Sonoki et al., 2015; Chuang et al., 2016; Klimaszewska-Wiśniewska et al., 2017; Dong et al., 2020). For example, quercetin reduces the viability of lung cancer cells, inhibits the expression of HSP70, and increases gemcitabine-induced cancer cell death. The combination therapy of quercetin and gemcitabine significantly down-regulates the expression of HSP70 (Lee et al., 2015). It has been reported that quercetin inhibits the proliferation, migration and invasion of non-small cell lung cancer (NSCLC) cells in vitro, and inhibits the growth of solid tumors in vivo. The Src expression and Fn14/NF-kappaB (NF-kB) signaling pathway are inhibited by the quercetin. Src overexpression in NSCLC cells leads to NSCLC cell proliferation and metastasis, and activating the Fn14/NF-kB signaling pathway inhibits the anti-NSCLC effect of quercetin (Dong et al., 2020). Therefore, the quercetin has the anti-tumor roles in lung cancer.
It has been confirmed that the quercetin has the anti-tumor properties in lung cancer and lung adenocarcinoma (LUAD) (Lee et al., 2015; Dong et al., 2020; Chuang et al., 2016; Klimaszewska-Wiśniewska et al., 2017; Sonoki et al., 2015). However, the mechanisms of quercetin in the development of LUAD has not yet been fully revealed. The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) database is a cancer database from the United States with reliable gene expression data and complete clinical information from a large sample of cancer patients (Guo Q et al., 2020; Liu et al., 2020). This study aimed to screen the hub target genes of quercetin in the progression of LUAD using the large data of TCGA database to elucidate the biological functions and mechanisms involved. A risk model of quercetin could then be constructed to assess the potential application as a prognostic tool for LUAD patients (Supplementary Figure S1), and evaluate the hub target genes as new therapeutic targets.
Materials and Methods
Screening of Quercetin Target Genes
The quercetin structure was obtained from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) Database (http://sm.nwsuaf.edu.cn/lsp/tcmsp.php), and the possible target genes of quercetin were predicted via the SwissTargetPrediction (http://www.swisstargetprediction.ch) database. Moreover, the quercetin structure from the pubchem (https://pubchem.ncbi.nlm.nih.gov/) database, and protein receptor structures from the PDB (http://www.rcsb.org/) database were downloaded to visualize molecular docking.
Identification of Quercetin Target Genes
The transcriptome data of gene expression and 522 clinical data of LUAD were downloaded from the TCGA database (Chen et al., 2019; Guo Q et al., 2020). The gene expression data included the 59 normal lung tissues and 535 LUAD tissues. The expression levels of quercetin target genes were identified via the Limma package, and the screening criteria was the logFC > 1 or < −1 and p < 0.05. The correlation between differentially expressed genes (DEGs) were analyzed using the correlation analysis. The roles of DEGs in LUAD were analyzed via the receiver operating characteristic (ROC) analysis. The area under the curve (AUC) and the p value were used as evaluation indicators.
Bioinformatics Analysis
The biological process (BP), cellular component (CC), molecular function (MF), signaling pathways and diseases of the quercetin related DEGs were explored in the DAVID database. We constructed a protein-protein interaction (PPI) network of the target genes of quercetin using the STRING (version: 11.5) database, and Cytoscape (version: 3.8.2) software was used for visualization.
Cox Regression Analysis
Univariate Cox regression analysis was used to analyze whether the DEGs of quercetin affected the prognosis of patients with LUAD, and the screening criteria was the p < 0.05. The independent factors affecting the prognosis of LUAD patients were screened using multivariate Cox regression analysis and akaike information standard (AIC), and the LUAD patients were divided into the high- and low-risk groups based on gene expression values. Subsequently, the clinical data were sorted out, and univariate Cox regression analysis was used to analyze the influence of clinicopathological characteristics of the high- and low-risk groups on the prognosis of LUAD patients.
Clinical Prognostic Value Analysis
The mortality of LUAD patients in the high- and low-risk groups was assessed by the Kaplan-Meier (K-M) survival analysis. The relationship between high- and low-risk and clinicopathological characteristics (age, gender, clinical stage, T stage, N stage, and M stage) of LUAD patients was evaluated via the correlation analysis. Furthermore, the roles of risk model related DEGs in the prognosis of LUAD patients were identified in the K-M plotter database and Gene Expression Profiling Interactive Analysis (GEPIA) database. p < 0.05 was considered statistically significant.
Consensus Clustering
The NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 expression data of 535 LUAD tissues were analyzed using the Consensus ClusterPlus package of R (version: 4.1.1), and principal component analysis (PCA) was carried out in cluster1 and cluster2 groups.
The Signaling Mechanism Involved in the Risk Model
The gene expression profiles of LUAD patients were divided into the high and low expression groups according to the median riskscore vlaue. The gene set enrichment analysis [GSEA (version: 4.1.0)] was used to assess the impact of potential mechanisms in the constructed risk model on the progression of LUAD. Each analysis performed 1,000 permutations of the genome. Gene sets with NOM p < 0.05 were considered significantly enriched.
Statistical Analysis
The data obtained from the TCGA database were processed using the Perl and R. The prognosis-related DEGs in LUAD were assessed using survival analysis, Cox regression analysis. Univariate Cox regression, multivariate Cox regression, and AIC method were used to construct the risk model for LUAD patients. p < 0.05 was considered the statistically significant.
Results
Screening and Identification of Quercetin Differentially Expressed Target Genes
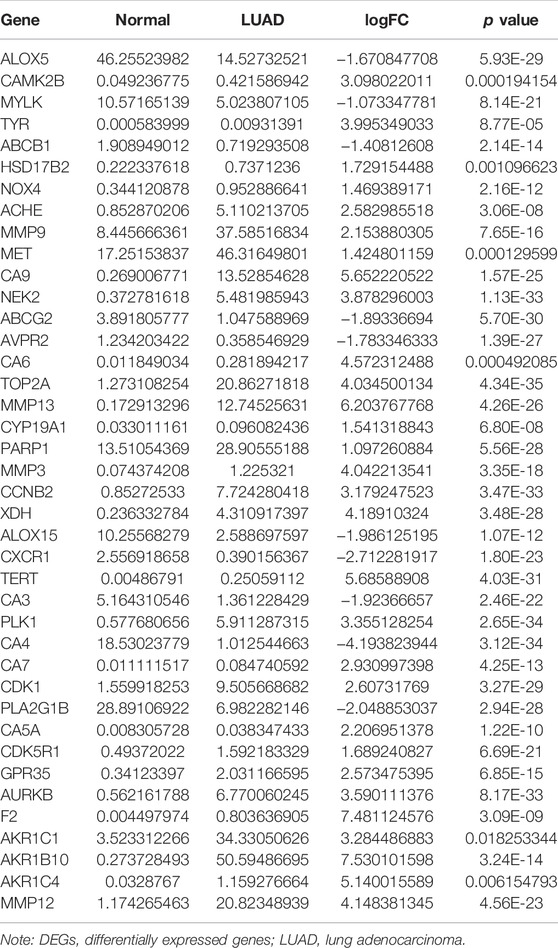
The structure of quercetin was obtained from the TCMSP database (Figure 1A). Figure 1B shows the distribution of quercetin target genes in the SwissTargetPrediction database (Supplementary Table S1). In the TCGA database, compared with normal lung tissues, there were the 79 DEGs in LUAD tissues (Figure 1C; Supplementary Table S2). Among them, 40 DEGs exhibited a fold change greater than 1 (Supplementary Figure S2; Table 1).
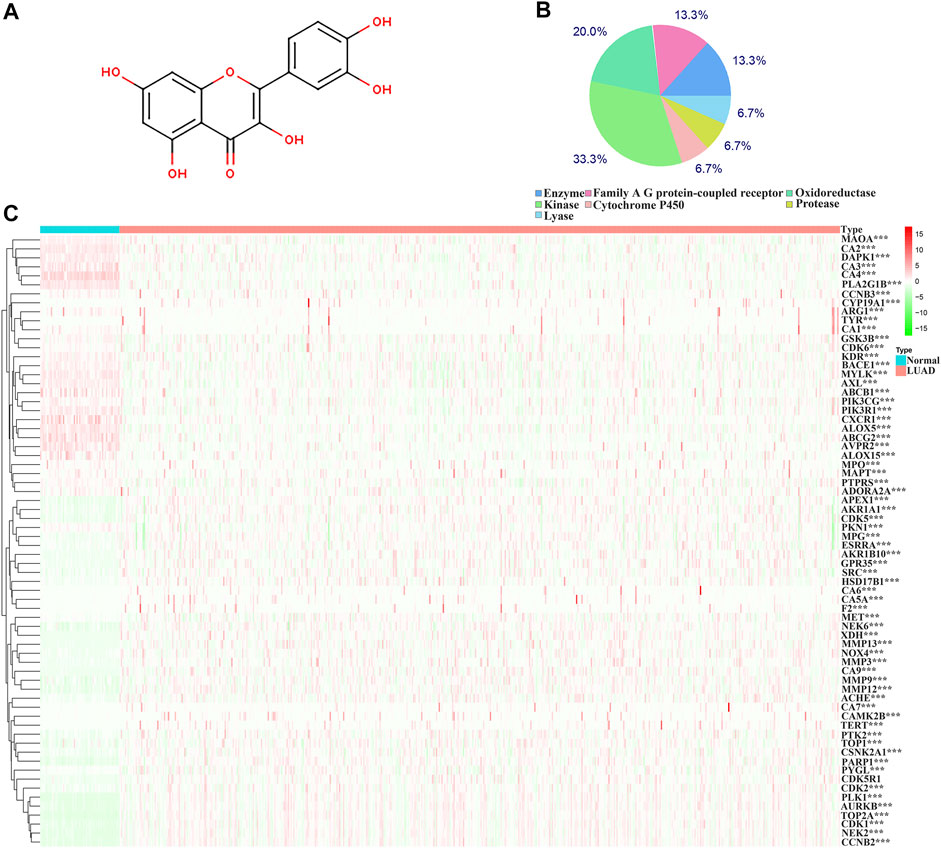
FIGURE 1. Structure and target genes of quercetin. (A) Structure; (B) The distribution ratio of quercetin target genes; (C) Quercetin-related differentially expressed genes in TCGA LUAD tissues. Note: TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma.
The Biological Functions Involved in the Differential Expression of Quercetin Target Genes
The biological functions involved in 40 DEGs were explored using gene ontology (GO) in the DAVID database. The GO (BP) results showed that the DEGs were involved in the oxidation-reduction process, cell proliferation, G2/M transition of the mitotic cell cycle, positive regulation of reactive oxygen species metabolic process, mitotic nuclear division, and others. The GO (CC) results indicated that the DEGs were involved in the cytosol, spindle microtubule, proteinaceous extracellular matrix, and others. The GO (MF) results showed that the DEGs were involved in protein kinase activity, cyclin-dependent protein serine/threonine kinase activity, protein serine/threonine kinase activity, oxidoreductase activity, and others (Figures 2A–C; Table 2). Furthermore, KEGG results demonstrated that the DEGs were involved in nitrogen metabolism, steroid hormone biosynthesis, oocyte meiosis, ovarian steroidogenesis and arachidonic acid metabolism signaling mechanisms (Figure 2D). In addition, the DEGs of quercetin were associated with lung cancer, breast cancer, chronic obstructive pulmonary disease, bladder cancer, esophageal adenocarcinoma, liver carcinoma, colorectal cancer, and other cancers (Figure 2E).
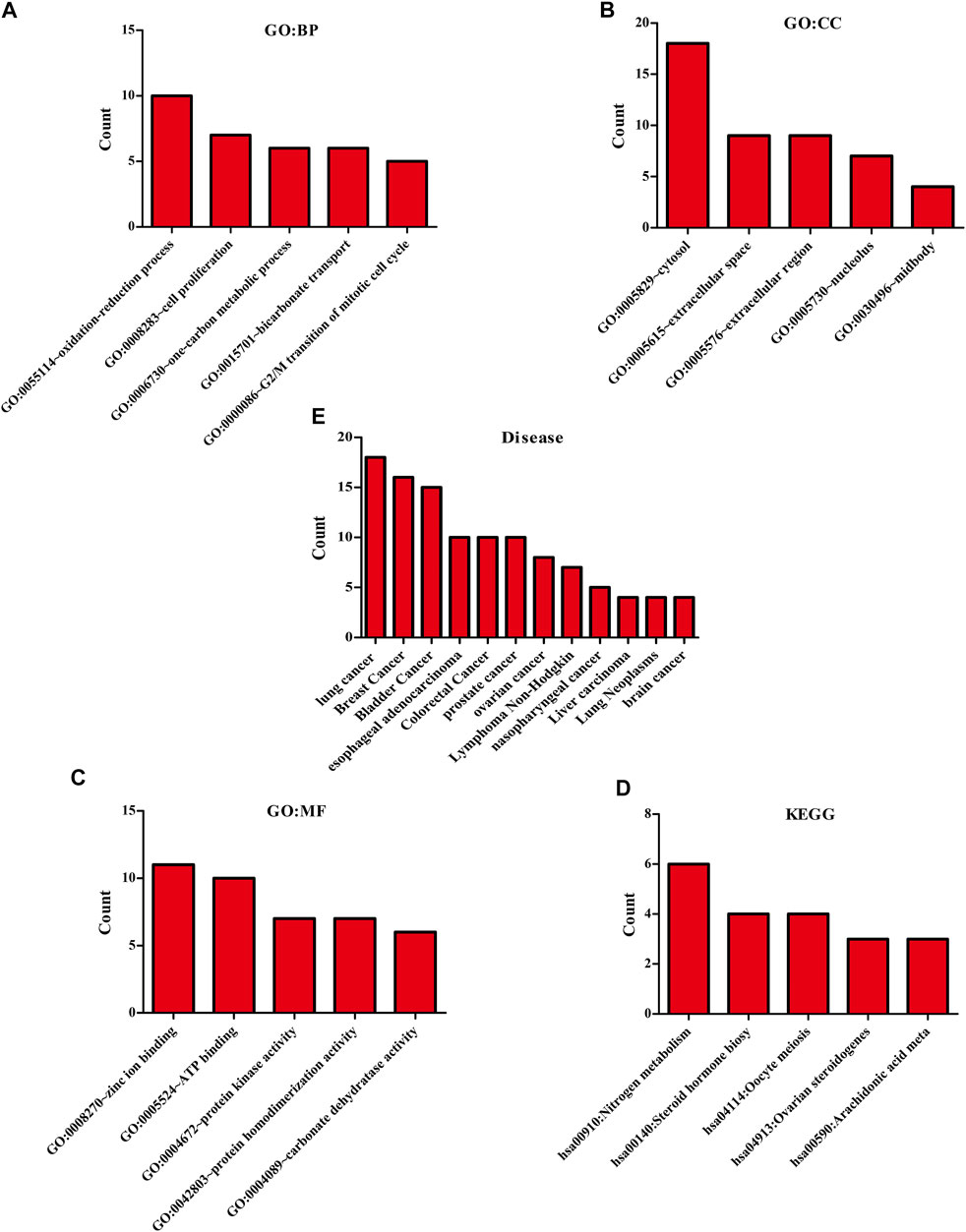
FIGURE 2. The biological functions, mechanisms and diseases of 40 quercetin-related differentially expressed target genes were explored in the DAVID database.
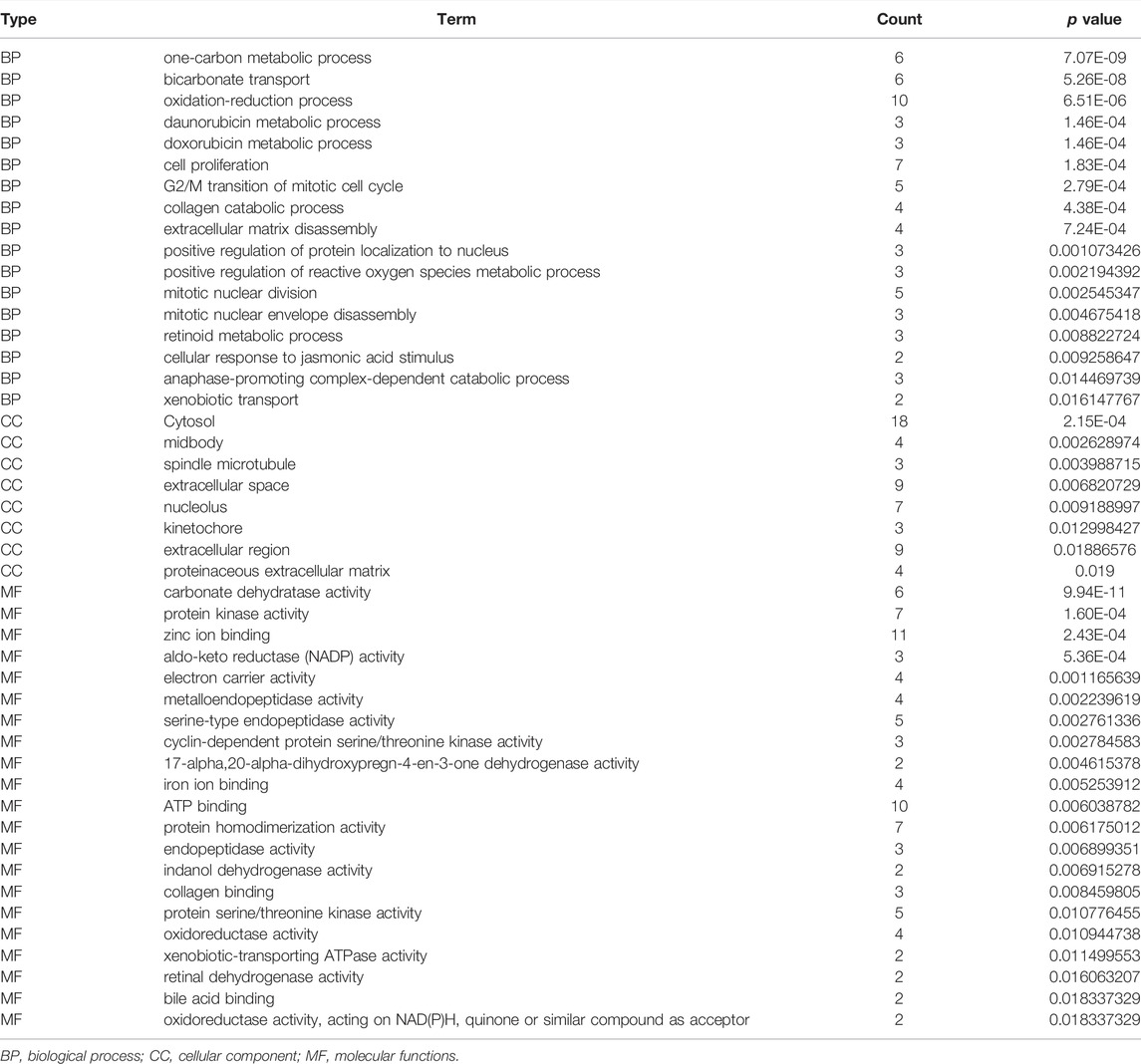
TABLE 2. The biological functions of 40 differentially expressed target genes were explored in the DAVID database.
Correlation Analysis of the Differentially Expressed Target Genes
Correlation analysis revealed a partial correlation between the DEGs expression levels in 535 LUAD tissues (Supplementary Figure S3). The degree of color was positively correlated with the degree of correlation. Furthermore, the PPI network constructed from the String database revealed the protein interaction relationship between quercetin differentially expressed target genes (Supplementary Figure S4).
Evaluating the Diagnostic Value of Quercetin Differentially Expressed Target Genes in Lung Adenocarcinoma
The diagnostic values of the DEGs in LUAD were evaluated via the ROC analysis. The results shows that the AUC of ALOX5 was 0.9426, CAMK2B was 0.6477, MYLK was 0.8709, ABCB1 was 0.8209, HSD17B2 was 0.6294, NOX4 was 0.7748, ACHE was 0.7195, MMP9 was 0.8195, MET was 0.6517, CA9 was 0.9139, NEK2 was 0.9794, ABCG2 was 0.9508, AVPR2 was 0.9313, CA6 was 0.6369, TOP2A was 0.9899, MMP13 was 0.9188, CYP19A1 was 0.7139, PARP1 was 0.9346, MMP3 was 0.8448, CCNB2 was 0.9757, XDH was 0.9363, ALOX15 was 0.7823, CXCR1 was 0.8957, TERT was 0.9597, CA3 was 0.8853, PLK1 was 0.9841, CA4 was 0.9835, CA7 was 0.7873, CDK1 was 0.9593, PLA2G1B was 0.9373, CA5A was 0.7564, CDK5R1 was 0.8686, GPR35 was 0.806, AURKB was 0.9691, F2 was 0.7314, AKR1C1 was 0.5859, AKR1B10 was 0.7992, AKR1C4 was 0.6086, and MMP12 was 0.8884 (Figure 3).
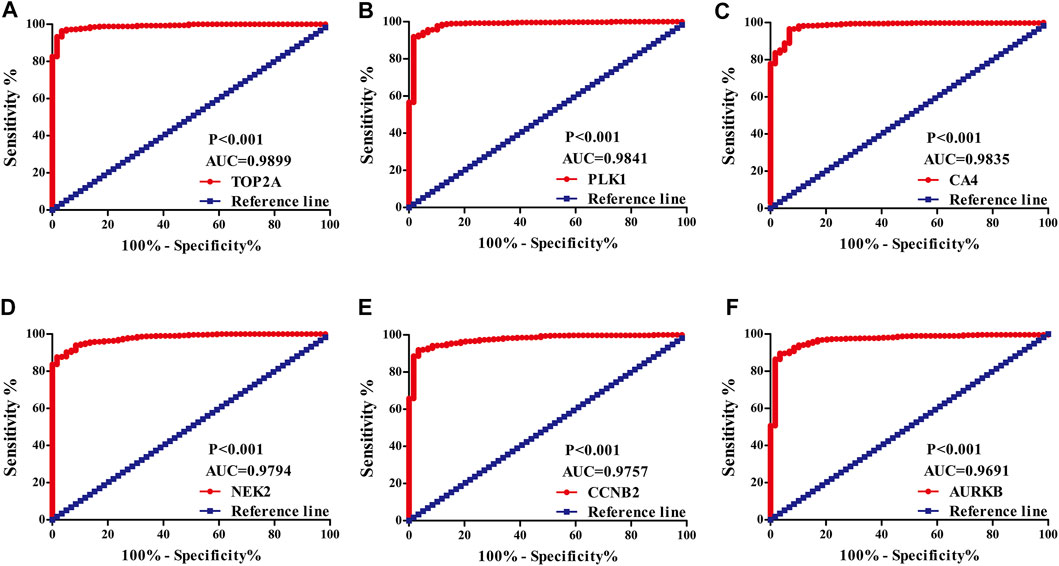
FIGURE 3. The diagnostic value of quercetin-related differentially expressed target genes in LUAD via the ROC analysis. Note: ROC, Receiver operating characteristic; LUAD, lung adenocarcinoma.
Screening and Constructing a Risk Model for Lung Adenocarcinoma Patients
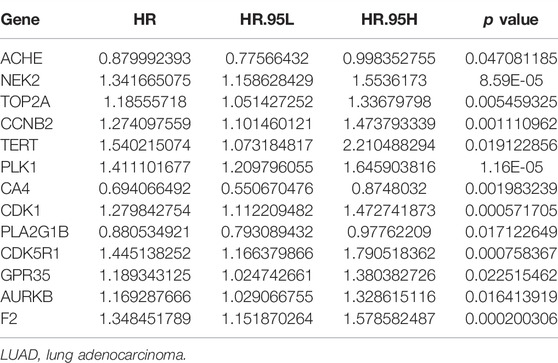
Univariate Cox regression analysis demonstrated that ACHE, NEK2, TOP2A, CCNB2, TERT, PLK1, CA4, CDK1, PLA2G1B, CDK5R1, GPR35, AURKB, and F2 were independent factors influencing the prognosis of LUAD patients (Figure 4A; Table 3). On this basis, multivariate Cox regression analysis revealed that NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 were independent factors affecting the prognosis of LUAD patients (Table 4), which were used to construct a risk model. Furthermore, K-M survival analysis confirmed that the prognosis of LUAD patients in the high-risk group was worse (Figure 4B), with the AUC being 0.677 (Figure 4C). The risk factors were further evaluated by correlation analysis and were found to be related to the gender, clinical stage, T stage, lymph node metastasis, and survival status of LUAD patients (Figure 4D). Univariate and multivariate Cox regression analysis showed that the high riskscore was a risk factor for poor prognosis in LUAD patients (Figures 4E,F; Table 5). Quercetin structure and risk factor structures were visualized via molecular docking (Figure 5).
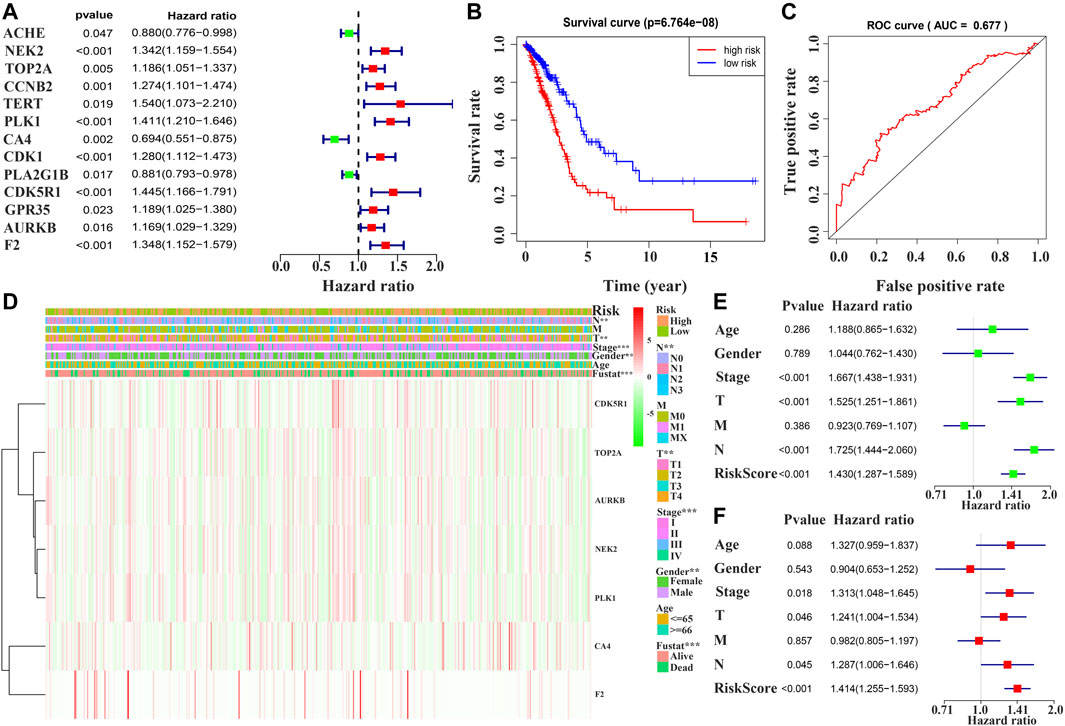
FIGURE 4. Prognostic value of quercetin-related differentially expressed target genes in patients with LUAD via the COX and survival analysis. Note: LUAD, lung adenocarcinoma.
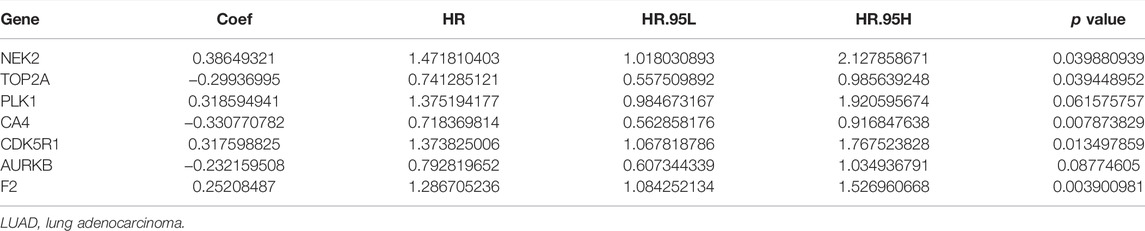
TABLE 4. Risk model factors of LUAD patients were shown via the multivariate Cox regression analysis.
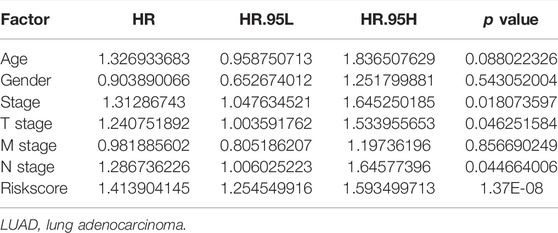
TABLE 5. Prognosis-related clinicopathological features of LUAD patients were shown via the multivariate Cox regression analysis.
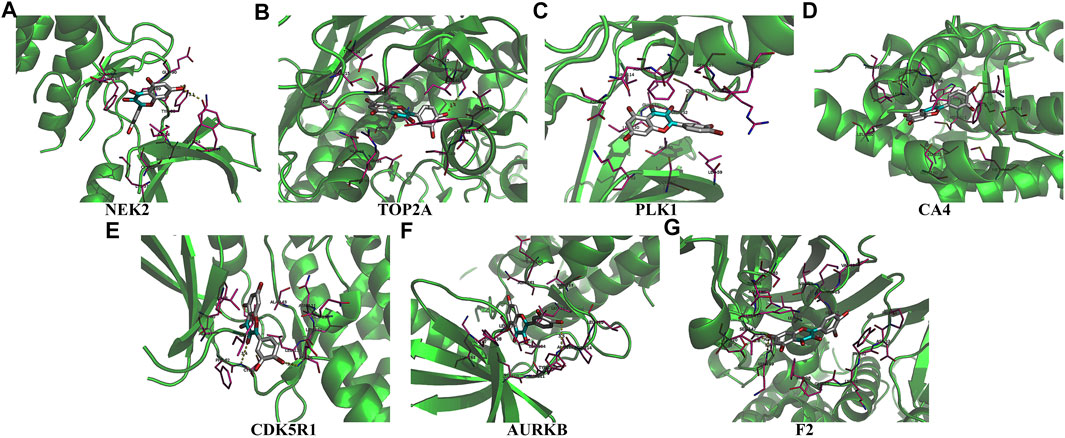
FIGURE 5. The molecular docking between quercetin and risk model genes. (A) NEK2; (B) TOP2A; (C) PLK1; (D) CA4; (E) CDK5R1; (F) AURKB; (G) F2.
The Value of Identifying Model Factors in the Prognosis of Lung Adenocarcinoma Patients
In the GEPIA database, the expression levels of NEK2, TOP2A, PLK1, AURKB, F2, and CA4 were confirmed to be related to the OS of LUAD patients (Figures 6A–F). Moreover, the expression levels of NEK2, PLK1, CA4, and AURKB were correlated with the disease-free survival (DFS) of LUAD patients (Figures 6G–J). In the K-M Plotter database, the expression levels of NEK2, TOP2A, PLK1, RP17, CDK5R1, STK12, and F2 were correlated with the OS in LUAD patients (Figures 7A–I). Furthermore, the expression levels of NEK2, TOP2A, PLK1, RP17, CDK5R1, and STK12 were correlated with the progression-free survival (PFS) in LUAD patients (Supplementary Figures S5A–I).
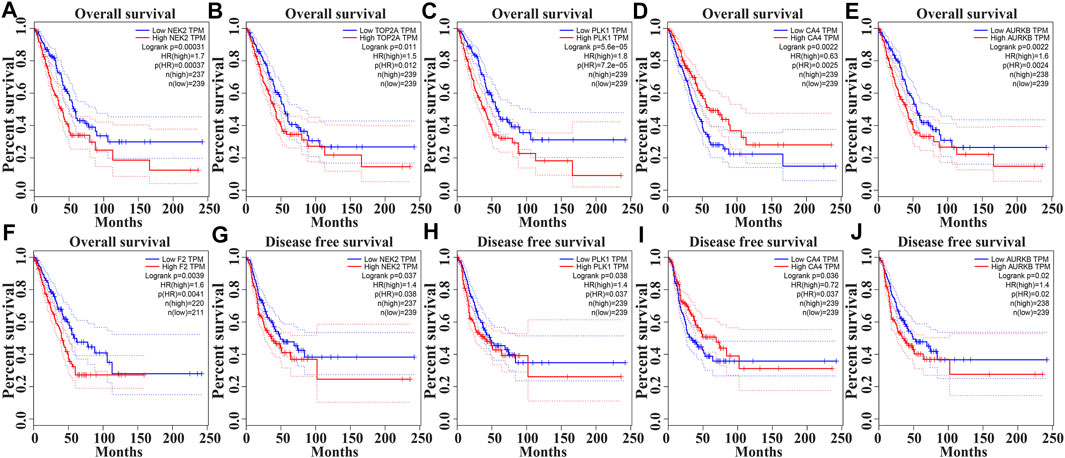
FIGURE 6. The risk model factors were correlated with the prognosis of LUAD patients in the GEPIA database. Note: LUAD, lung adenocarcinoma; GEPIA, gene expression profiling interactive analysis.
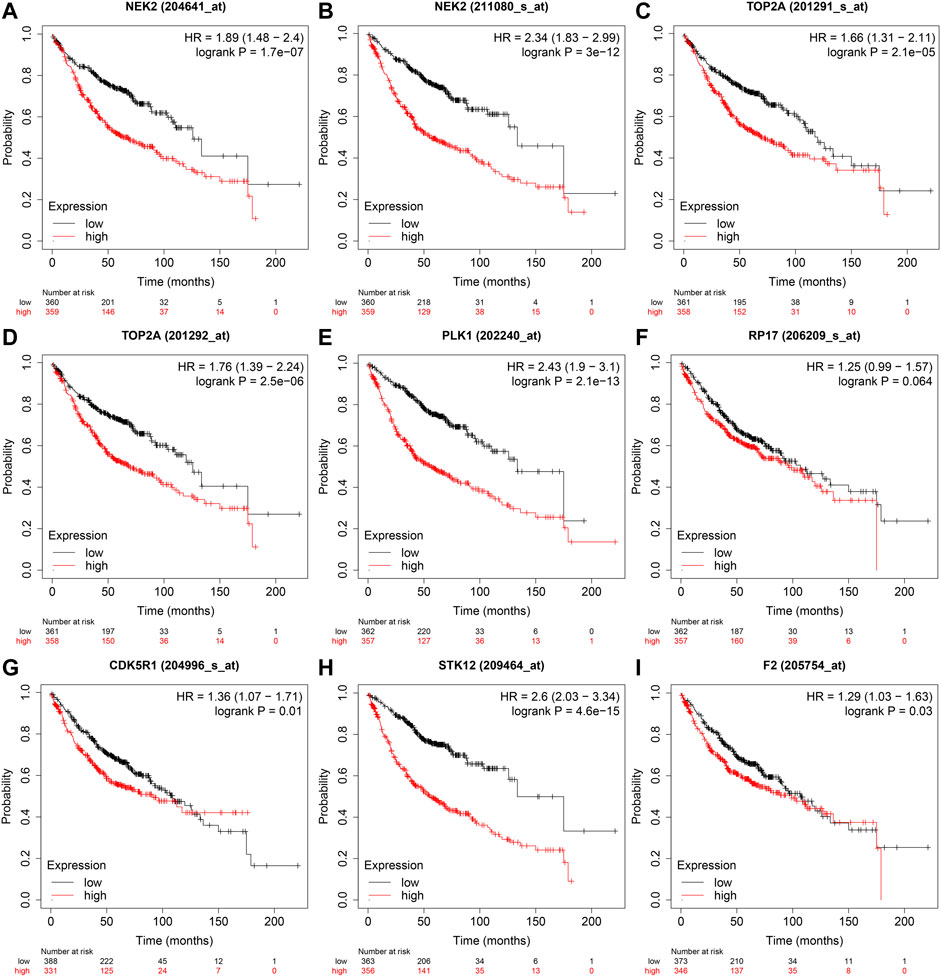
FIGURE 7. The risk model factors were correlated with the OS of LUAD patients in the Kaplan-Meier Plotter database. Note: OS, overall survival; LUAD, lung adenocarcinoma.
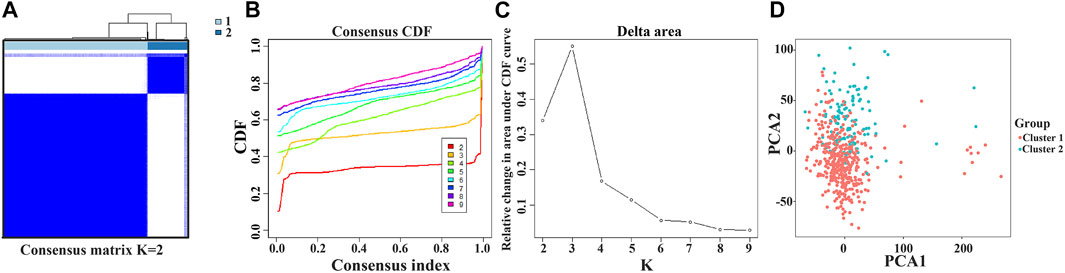
Consensus Clustering of the Prognostic Factors Identified two Clusters of Lung Adenocarcinoma With Different Outcomes
The NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 genes were analyzed via cluster analysis. As the clustering evolves from k = 2 to 9, k = 2 might be the most appropriate choice for minimal interference (Figures 8A–C). Therefore, k = 2 was used for consensus cluster analysis, defined as the cluster1 and cluster2 groups. PCA analysis was performed in 535 LUAD tissues from the TCGA database, and the results revealed that the cluster1 and cluster2 groups were significantly different (Figure 8D).
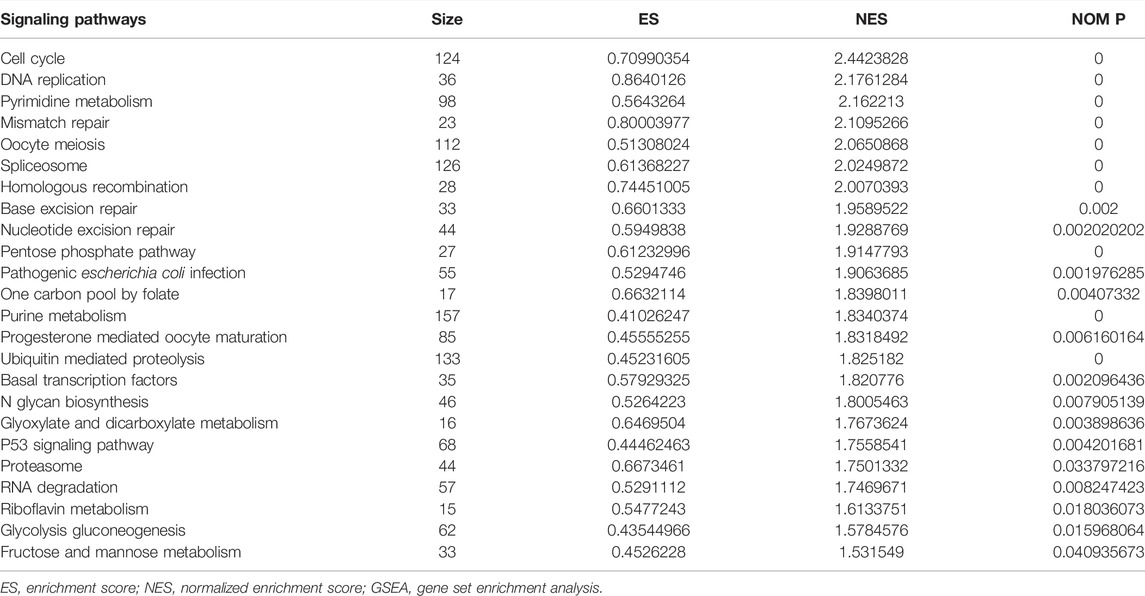
Possible Mechanisms of Lung Adenocarcinoma Progression in the High-Risk Model
In the high-risk group, the risk model involved the cell cycle, DNA replication, oocyte meiosis, spliceosome, homologous recombination, base excision repair, ubiquitin-mediated proteolysis, basal transcription factors, and other signaling pathways (Figure 9; Table 6).
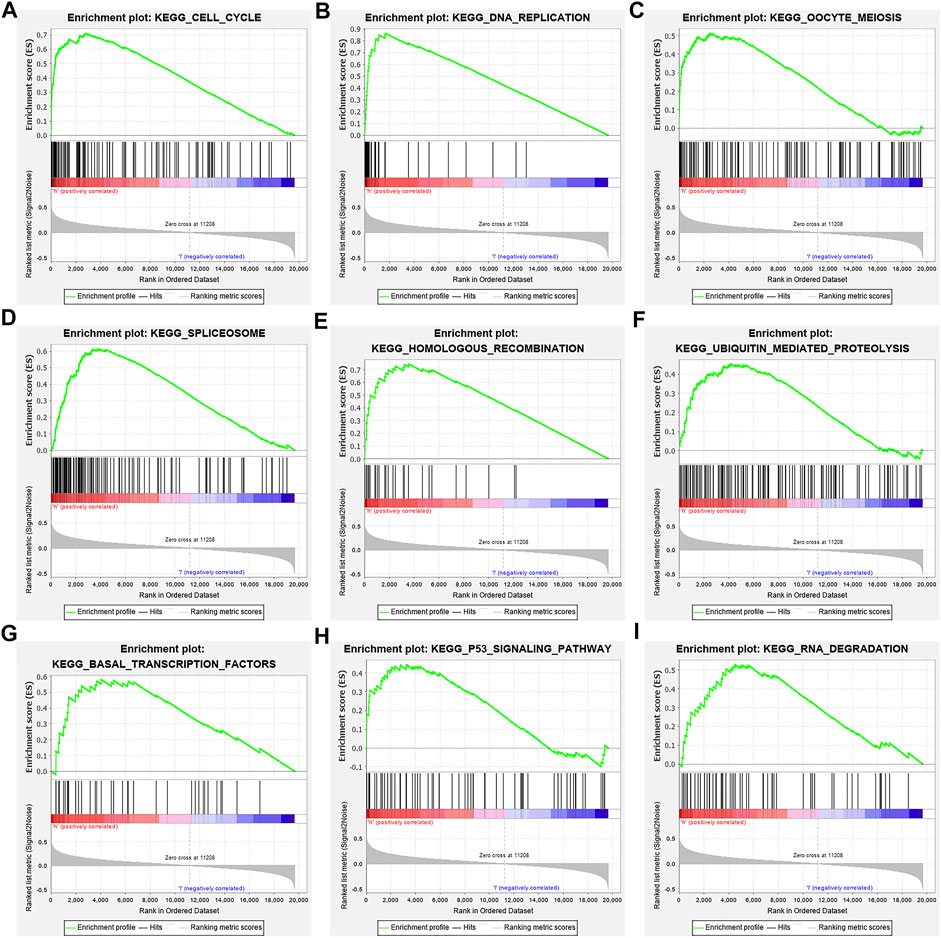
FIGURE 9. Signaling pathways involved in the risk model via the GSEA. Note: GSEA, gene set enrichment analysis.
Discussion
Quercetin has been shown to exert a beneficial effect in cancer treatment, and could delay the progression of tumors (Zheng et al., 2012; Lee et al., 2015; Sonoki et al., 2015; Reyes-Farias and Carrasco-Pozo, 2019; Zhao et al., 2019; Li et al., 2020). For example, quercetin can inhibit the expression of Bcl-2 and promote the Bax expression to increase the apoptosis rate of A549 cells (Zheng et al., 2012). Claudin-2 is highly expressed in LUAD tissues and cells. Knockout of claudin-2 reduces the proliferation and migration of LUAD cells. Quercetin reduces the expression of claudin-2, and promotes the expression of miR-16 in LUAD A549 cells. However, miR-16 inhibitors can rescue the decreased expression of claudin-2 induced by quercetin (Sonoki et al., 2015). This demonstrates that quercetin has an anti-tumor progression effect in LUAD. At present, many studies have reported the mechanism of quercetin in the progress of LUAD. However, the roles of quercetin in the progress of LUAD has not yet been fully revealed. Therefore, this study uses biological methods to find quercetin target genes, and the risk model was built by screening valuable target genes to provide a new direction and vision for quercetin treatment in LUAD.
A growing body of studies has shown that regulating gene expression is a potentially effective method to delay the progress of LUAD (Sekimoto et al., 2017; Guo W et al., 2020; Hu et al., 2020; Kou et al., 2020; Huang et al., 2021). Hu et al. reported that miR-486-5p was downregulated in cancer patient tissues and serum exosomes, inhibiting the proliferation, and metastasis of LUAD cells. In contrast, up-regulating NEK2 expression eliminates the inhibitory effect of miR-486-5p overexpression on the progress of LUAD (Hu et al., 2020). TOP2A is highly expressed in LUAD tissues. TOP2A is an independent prognostic factor for LUAD patients (Guo W et al., 2020; Kou et al., 2020). Interfering with the expression of TOP2A in A549 and GLC82 cells has been reported to inhibit the proliferation, migration and invasion of LUAD cells and reduce the expression levels of CCNB1 and CCNB2 (Kou et al., 2020). In our study, ACHE, NEK2, TOP2A, CCNB2, TERT, PLK1, CA4, CDK1, PLA2G1B, CDK5R1, GPR35, AURKB, and F2 were independent factors affecting the prognosis of LUAD patients via the univariate Cox regression analysis. Furthermore, NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 were independent factors affecting the prognosis of LUAD patients via the multivariate Cox regression analysis. K-M survival analysis demonstrated that the LUAD patients in the high-risk group had the poorer prognosis. Correlation analysis showed that the riskscore was related to the gender, clinical stage, T stage, lymph node metastasis, and survival status of LUAD patients. Consequently, univariate and multivariate Cox regression analysis revealed that the high riskscore was a risk factor for poor prognosis in LUAD patients. It has been reported in previous literature that NEK2, TOP2A, PLK1, CA4, and AURKB can play oncogenic or tumor suppressing roles in the progression of lung cancer (Bertran-Alamillo et al., 2019; Chen et al., 2020; Du et al., 2020; Gong et al., 2020; Shin et al., 2020). For example, TOP2A expression level was elevated in LUAD tissues and cells. TOP2A overexpression is associated with the prognosis in LUAD patients. The proliferation ability of A549 cells decreased and the apoptosis rate increased when TOP2A was inhibited in LUAD cells. Decreased TOP2A expression level could regulate LUAD growth by activating the ERK/JNK/p-P38/CHOP signaling pathway (Du et al., 2020). PLK1 expression level is the predictor of poor prognosis in patients with metastatic NSCLC. Active PLK1 promotes cancer metastasis by upregulating TGF-β signaling and amplifies metastatic properties by forming a positive feedback loop (Shin et al., 2020). It preliminarily showed that the quercetin target genes have crucial biological functions in LUAD progress.
Studies have found that NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 have important regulatory roles in cell cycle, DNA replication, homologous recombination, and others (Chabalier-Taste et al., 2016; Fan et al., 2019; Zeng et al., 2019; Wang et al., 2020; Ha et al., 2022). The expression level of NEK2 in human gastric cancer (GC) tissues is significantly upregulated. The expression level of ERK in human tissues is correlated with the expression of NEK2, and their overexpression might predict a poor prognosis in GC patients. Inhibiting NEK2 expression in GC cells can attenuate the ERK and c-JUN phosphorylation, reducing the cyclin D1 transcription. NEK2 can rescue the GC cell viability, proliferation and cell cycle progression (Fan et al., 2019). TOP2A was overexpressed in bladder urothelial carcinoma (BLCA) samples compared with normal epithelial tissue. Univariate Cox regression analysis showed that high expression of TOP2A was a factor affecting cancer-specific survival, PFS and recurrence-free survival of BLCA patients. Inhibiting the expression of TOP2A in BLCA cells could inhibit cell proliferation, migration and invasion, and promote anti-apoptosis (Zeng et al., 2019). Furthermore, the results of GO and KEGG showed that the DEGs of quercetin are involved in cell proliferation, G2/M transition of the mitotic cell cycle, positive regulation of the reactive oxygen species metabolic process, mitotic nuclear division, oocyte meiosis, etc. GSEA results demonstrated that the risk model involved signaling pathways such as cell cycle, DNA replication, oocyte meiosis, spliceosome, homologous recombination, base excision repair, ubiquitin-mediated proteolysis, and basal transcription factors. However, further research is required to confirm those results in the future.
This bioinformatics results revealed the potential role of hub quercetin target genes in the progression of LUAD and constructed a risk model to provide potential therapeutic targets for quercetin treatment of LUAD patients. However, our research results remain to be confirmed by basic research in the future. The prognostic value of the cellular level model was evaluated to explore the impact of quercetin on hub targets, as well as the expression of hub target genes in LUAD. Our results preliminarily indicated that the quercetin target genes NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2 were abnormally expressed in LUAD tissues, and their expression levels were related to the diagnosis and prognosis of LUAD patients. The constructed risk model is expected to effectively evaluate the prognosis of LUAD patients. In conclusion, the quercetin potential target genes NEK2, TOP2A, PLK1, CA4, CDK5R1, AURKB, and F2, were abnormally expressed in LUAD tissues, and their expression levels were related to the diagnosis and prognosis of LUAD patients. The constructed risk model is expected to effectively evaluate the prognosis of LUAD patients.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
This study didn’t require ethical approval/written informed consent was not required.
Author Contributions
DL and QG designed themes, formulated research plans, and guided the implementation of the research. Y-QZ drafted the manuscript and processed the data. KL provided financial support, manuscript revision and language editing, and QG was responsible for revised guidance. The final manuscript was read and approved by all the authors.
Funding
Our study was funded by the General project of Scientific Research Fund of Beilun District People’s Hospital (No.2022YJJJYB08).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.890079/full#supplementary-material
References
Bertran-Alamillo, J., Cattan, V., Schoumacher, M., Codony-Servat, J., Giménez-Capitán, A., Cantero, F., et al. (2019). AURKB as a Target in Non-small Cell Lung Cancer with Acquired Resistance to Anti-EGFR Therapy. Nat. Commun. 10 (1), 1812. doi:10.1038/s41467-019-09734-5
Chabalier-Taste, C., Brichese, L., Racca, C., Canitrot, Y., Calsou, P., and Larminat, F. (2016). Polo-like Kinase 1 Mediates BRCA1 Phosphorylation and Recruitment at DNA Double-Strand Breaks. Oncotarget 7 (3), 2269–2283. doi:10.18632/oncotarget.6825
Chen, C., Peng, S., Li, P., Ma, L., and Gan, X. (2020). High Expression of NEK2 Promotes Lung Cancer Progression and Drug Resistance and Is Regulated by Mutant EGFR. Mol. Cel Biochem 475, 15–25. doi:10.1007/s11010-020-03854-z
Chen, D., Song, Y., Zhang, F., Wang, X., Zhu, E., Zhang, X., et al. (2019). Genome-Wide Analysis of Lung Adenocarcinoma Identifies Novel Prognostic Factors and a Prognostic Score. Front. Genet. 10, 493. doi:10.3389/fgene.2019.00493
Chuang, C.-H., Yeh, C.-L., Yeh, S.-L., Lin, E.-S., Wang, L.-Y., and Wang, Y.-H. (2016). Quercetin Metabolites Inhibit MMP-2 Expression in A549 Lung Cancer Cells by PPAR-γ Associated Mechanisms. J. Nutr. Biochem. 33 (undefined), 45–53. doi:10.1016/j.jnutbio.2016.03.011
Dong, Y., Yang, J., Yang, L., and Li, P. (2020). Quercetin Inhibits the Proliferation and Metastasis of Human Non-small Cell Lung Cancer Cell Line: The Key Role of Src-Mediated Fibroblast Growth Factor-Inducible 14 (Fn14)/Nuclear Factor Kappa B (NF-Κb) Pathway. Med. Sci. Monit. 26, e920537. doi:10.12659/MSM.920537
Du, X., Xue, Z., Lv, J., and Wang, H. (2020). Expression of the Topoisomerase II Alpha (TOP2A) Gene in Lung Adenocarcinoma Cells and the Association with Patient Outcomes. Med. Sci. Monit. 26, e929120. doi:10.12659/MSM.929120
Fan, W.-D., Chen, T., and Liu, P.-J. (2019). NIMA Related Kinase 2 Promotes Gastric Cancer Cell Proliferation via ERK/MAPK Signaling. Wjg 25 (23), 2898–2910. doi:10.3748/wjg.v25.i23.2898
Gong, F. L., Wang, L., Yu, L. G., Dang, Y. F., Jiang, X. N., Zhao, L., et al. (2020). DHPAC, a Novel Microtubule Depolymerizing Agent, Suppresses Angiogenesis and Vasculogenic Mimicry Formation of Human Non‐small Cell Lung Cancer. J. Cel Biochem 121, 4756–4771. doi:10.1002/jcb.29690
Guo, Q., Ke, X.-x., Liu, Z., Gao, W.-L., Fang, S.-X., Chen, C., et al. (2020). Evaluation of the Prognostic Value of STEAP1 in Lung Adenocarcinoma and Insights into its Potential Molecular Pathways via Bioinformatic Analysis. Front. Genet. 11, 242. doi:10.3389/fgene.2020.00242
Guo, W., Sun, S., Guo, L., Song, P., Xue, X., Zhang, H., et al. (2020). Elevated TOP2A and UBE2C Expressions Correlate with Poor Prognosis in Patients with Surgically Resected Lung Adenocarcinoma: a Study Based on Immunohistochemical Analysis and Bioinformatics. J. Cancer Res. Clin. Oncol. 146 (4), 821–841. doi:10.1007/s00432-020-03147-4
Ha Kim, Y., Yeol Choi, J., Jeong, Y., Wolgemuth, D. J., and Rhee, K. (2002). Nek2 Localizes to Multiple Sites in Mitotic Cells, Suggesting its Involvement in Multiple Cellular Functions during the Cell Cycle. Biochem. Biophysical Res. Commun. 290 (2), 730–736. doi:10.1006/bbrc.2001.6212
Hu, H., Xu, H., Lu, F., Zhang, J., Xu, L., Xu, S., et al. (2020). Exosome-Derived miR-486-5p Regulates Cell Cycle, Proliferation and Metastasis in Lung Adenocarcinoma via Targeting NEK2. Front. Bioeng. Biotechnol. 8 (undefined), 259. doi:10.3389/fbioe.2020.00259
Huang, J., Zhang, Q., Shen, J., Chen, X., and Ma, S. (2021). Multi-omics Analysis Identifies Potential Mechanisms of AURKB in Mediating Poor Outcome of Lung Adenocarcinoma. Aging 13 (4), 5946–5966. doi:10.18632/aging.202517
Kim, J. M., Hwang, I.-H., Jang, I.-S., Kim, M., Bang, I. S., Park, S. J., et al. (2017). Houttuynia Cordata Thunb Promotes Activation of HIF-1a-FOXO3 and MEF2A Pathways to Induce Apoptosis in Human HepG2 Hepatocellular Carcinoma Cells. Integr. Cancer Ther. 16 (3), 360–372. doi:10.1177/1534735416670987
Klimaszewska-Wiśniewska, A., Hałas-Wiśniewska, M., Izdebska, M., Gagat, M., Grzanka, A., and Grzanka, D. (2017). Antiproliferative and Antimetastatic Action of Quercetin on A549 Non-small Cell Lung Cancer Cells through its Effect on the Cytoskeleton. Acta Histochem. 119 (2), 99–112. doi:10.1016/j.acthis.2016.11.003
Kou, F., Sun, H., Wu, L., Li, B., Zhang, B., Wang, X., et al. (2020). TOP2A Promotes Lung Adenocarcinoma Cells' Malignant Progression and Predicts Poor Prognosis in Lung Adenocarcinoma. J. Cancer 11 (9), 2496–2508. doi:10.7150/jca.41415
Lee, S. H., Lee, E. J., Min, K. H., Hur, G. Y., Lee, S. H., Lee, S. Y., et al. (2015). Quercetin Enhances Chemosensitivity to Gemcitabine in Lung Cancer Cells by Inhibiting Heat Shock Protein 70 Expression. Clin. Lung Cancer 16 (6), e235–e243. doi:10.1016/j.cllc.2015.05.006
Li, Y., Wang, Z., Jin, J., Zhu, S.-X., He, G.-Q., Li, S.-H., et al. (2020). Quercetin Pretreatment Enhances the Radiosensitivity of colon Cancer Cells by Targeting Notch-1 Pathway. Biochem. Biophysical Res. Commun. 523 (4), 947–953. doi:10.1016/j.bbrc.2020.01.048
Liu, J., Hou, S., Wang, J., Chai, Z., Hong, X., Zhao, T., et al. (2020). A Prognostic 14-Gene Expression Signature for Lung Adenocarcinoma: A Study Based on TCGA Data Mining. Oxidative Med. Cell Longevity 2020 (undefined), 1–11. doi:10.1155/2020/8847226
Liu, J., Zhu, X., Yang, D., Li, R., and Jiang, J. (2021). Effect of Heat Treatment on the Anticancer Activity of Houttuynia Cordata Thunb Aerial Stem Extract in Human Gastric Cancer SGC-7901 Cells. Nutr. Cancer 73 (1), 160–168. doi:10.1080/01635581.2020.1737153
Lou, Y., Guo, Z., Zhu, Y., Kong, M., Zhang, R., Lu, L., et al. (2019). Houttuynia Cordata Thunb. And its Bioactive Compound 2-undecanone Significantly Suppress Benzo(a)pyrene-Induced Lung Tumorigenesis by Activating the Nrf2-HO-1/nqo-1 Signaling Pathway. J. Exp. Clin. Cancer Res. 38 (1), 242. doi:10.1186/s13046-019-1255-3
Reyes-Farias, M., and Carrasco-Pozo, C. (2019). The Anti-cancer Effect of Quercetin: Molecular Implications in Cancer Metabolism. Ijms 20 (13), 3177. undefined. doi:10.3390/ijms20133177
Sekimoto, N., Suzuki, Y., and Sugano, S. (2017). Decreased KPNB1 Expression Is Induced by PLK1 Inhibition and Leads to Apoptosis in Lung Adenocarcinoma. J. Cancer 8 (19), 4125–4140. doi:10.7150/jca.21802
Shin, S.-B., Jang, H.-R., Xu, R., Won, J.-Y., and Yim, H. (2020). Active PLK1-Driven Metastasis Is Amplified by TGF-β Signaling that Forms a Positive Feedback Loop in Non-small Cell Lung Cancer. Oncogene 39 (4), 767–785. doi:10.1038/s41388-019-1023-z
Sonoki, H., Sato, T., Endo, S., Matsunaga, T., Yamaguchi, M., Yamazaki, Y., et al. (2015). Quercetin Decreases Claudin-2 Expression Mediated by Up-Regulation of microRNA miR-16 in Lung Adenocarcinoma A549 Cells. Nutrients 7 (6), 4578–4592. doi:10.3390/nu7064578
Subhawa, S., Chewonarin, T., and Banjerdpongchai, R. (2020). The Effects of Houttuynia Cordata Thunb and Piper Ribesioides Wall Extracts on Breast Carcinoma Cell Proliferation, Migration, Invasion and Apoptosis. Molecules 25 (5), 1196. undefined. doi:10.3390/molecules25051196
Wang, B., Jiang, H., Wan, X., Wang, Y., Zheng, X., Li, P., et al. (2020). Carbonic Anhydrase IV Inhibits Cell Proliferation in Gastric Cancer by Regulating the Cell Cycle. Oncol. Lett. 20 (4), 4. doi:10.3892/ol.2020.11865
Zeng, S., Liu, A., Dai, L., Yu, X., Zhang, Z., Xiong, Q., et al. (2019). Yet alPrognostic Value of TOP2A in Bladder Urothelial Carcinoma and Potential Molecular Mechanisms. BMC Cancer 19 (1), 604. doi:10.1186/s12885-019-5814-y
Zhao, J., Fang, Z., Zha, Z., Sun, Q., Wang, H., Sun, M., et al. (2019). Quercetin Inhibits Cell Viability, Migration and Invasion by Regulating miR-16/HOXA10 axis in Oral Cancer. Eur. J. Pharmacol. 847 (undefined), 11–18. doi:10.1016/j.ejphar.2019.01.006
Keywords: LUAD, risk model, quercetin, prognosis, GSEA
Citation: Zhang Y-Q, Li K, Guo Q and Li D (2022) A New Risk Model Based on 7 Quercetin-Related Target Genes for Predicting the Prognosis of Patients With Lung Adenocarcinoma. Front. Genet. 13:890079. doi: 10.3389/fgene.2022.890079
Received: 05 March 2022; Accepted: 19 April 2022;
Published: 13 May 2022.
Edited by:
Heriberto Prado-Garcia, Instituto Nacional de Enfermedades Respiratorias-México (INER), MexicoReviewed by:
Nahid Eskandari, Isfahan University of Medical Sciences, IranLidan Sun, Jiaxing University, China
Copyright © 2022 Zhang, Li, Guo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Guo, guoqianglidan@163.com; Dan Li, 804180423@qq.com
†These authors have contributed equally to this work
 Yun-Qiang Zhang1†
Yun-Qiang Zhang1†