- 1Changzhi Medical College, Changzhi, China
- 2Department of Endocrinology and Metabolism, Guangxi Academy of Medical Sciences and the People’s Hospital of Guangxi Zhuang Autonomous Region, Nanning, China
- 3Department of Endocrinology and Metabolism, Heping Hospital Affiliated to Changzhi Medical College, Changzhi, China
- 4Department of Epidemiology, School of Public Health, Southern Medical University, Guangzhou, China
- 5Institute of Evidence-Based Medicine, Heping Hospital Affiliated to Changzhi Medical College, Changzhi, China
Background: Numerous studies reported the associations between endothelial nitric oxide synthase (eNOS) polymorphisms (4b/a VNTR (rs869109213), G894T (rs1799983) and T786C (rs2070744)) and type 2 diabetes mellitus (T2DM) risk. However, the conclusions were incongruent. Moreover, since no published meta-analyses were performed, a key issue regarding false-positive results needs to be addressed. Furthermore, four new articles have been published on these issues. Therefore, an updated meta-analysis was conducted to further explore these associations.
Objectives: To investigate the association between eNOS 4b/a, G894T and T786C polymorphisms and T2DM risk.
Methods: Studies were searched by using the PubMed, China National Knowledge Infrastructure (CNKI), Medline, Embase, International Statistical Institute (ISI) and the China Wanfang databases. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the associations using five genetic models. Furthermore, the false-positive report probability (FPRP), Bayesian false discovery probability (BFDP), and the Venice criteria were employed to assess the credibility of statistically significant associations.
Results: Overall, the eNOS 4b/a polymorphism was associated with a significantly decreased T2DM risk in Asians (bb vs. aa: OR = 0.44, 95% CI = 0.23–0.84; ab + bb vs. aa: OR = 0.45, 95% CI = 0.24–0.86; bb vs. aa + ab: OR = 0.73, 95% CI = 0.59–0.91; b vs. a: OR = 0.71, 95% CI = 0.57–0.88); the eNOS G894T polymorphism was associated with a significantly increased T2DM risk in Asians (GT vs. GG: OR = 1.52, 95% CI = 1.15–2.01; GT + TT vs. GG: OR = 1.52, 95% CI = 1.15–2.01; T vs. G: OR = 1.39, 95% CI = 1.09–1.76); the eNOS T786C polymorphism was associated with a significantly increased T2DM risk in Indian (TC vs. TT: OR = 1.93, 95% CI = 1.27–2.94; TC + CC vs. TT: OR = 2.06, 95%CI = 1.26–3.36; C vs. T: OR = 1.90, 95%CI = 1.17–3.08). However, when a sensitivity analysis was performed after excluding low quality and Hardy–Weinberg Disequilibrium (HWD) studies, no significant association was found for the eNOS G894T polymorphism. After credibility assessment, we identified “less-credible positive results” for the statistically significant associations in the current meta-analysis.
Conclusion: In conclusion, this article suggests that all substantial relationships between eNOS 4b/a, G894T, and T786C polymorphisms and T2DM risk are most likely due to false positive results rather than real connections or biological variables.
Introduction
Type 2 diabetes mellitus (T2DM), which is defined by chronic hyperglycemia caused by insulin resistance as well as multiple related micro-vascular and macro-vascular complications, is one of the most common chronic illnesses at home and abroad. Over the last 3 decades, the global prevalence of diabetes mellitus has more than quadrupled, making it one of the most serious global health issues (Moore et al., 2009). At the same time, it is reported that the incidence of T2DM is increasing at an alarming rate. Diabetes is anticipated to impact 702 million people by 2045, which means one in every eleven people will be affected, and huge amounts of money will be required globally to cure diabetes and manage its complications (https://diabetesatlas.org/en/). However, the pathogenesis of T2DM remains unclear and may be related to diet and exercise, obesity, geography, genetic susceptibility, environment, etc. Furthermore, there is abundant evidence that genetic predisposition plays a significant role in the etiology of T2DM (Ferland-McCollough et al., 2010). It has been reported that there is an important genetic predisposition for T2DM (Papazafiropoulou et al., 2017). At the same time, over 100 T2DM risk loci have been identified to date, although the molecular pathways of risk genes are unclear (Gaulton, 2017). In conclusion, genetic factors play an essential impact in the occurrence and development of T2DM.
Nitric oxide (NO) is a ubiquitous vasoactive substance, whose main function is to protect vascular endothelial cells from damage (Larsen et al., 2012). Endothelial dysfunction due to reduce in NO levels is an important mechanism for the development of T2DM. One of the essential enzymes in the process of NO generation is endothelial nitric oxide synthase (eNOS), which is encoded by the eNOS gene on chromosome 7q35-7q36 (Jamwal and Sharma, 2018). It has been observed that eNOS malfunction can cause a nitric oxide production problem, which can contribute to the development of characteristic T2DM aberrant metabolic phenotypes such as reduced glucose tolerance and insulin resistance (Li et al., 2002; Tsutsui et al., 2006). Therefore, eNOS polymorphisms may biologically be an ideal genetic marker for T2DM in biology.
Many eNOS gene polymorphisms have been identified in recent years, of which 4b/a, G894T, and T786C are the most investigated polymorphisms in T2DM (Wang et al., 1997; Veldman et al., 2002), although their associations remain controversial and equivocal. Several relevant meta-analyses have been performed to evaluate the correlations of T2DM with eNOS gene polymorphisms (Jia et al., 2013; Zhang et al., 2017; Dong et al., 2018), with conflicting results. And previously published meta-analyses did not evaluate the quality of the literature, nor did they evaluate the positive results to identify multiple comparisons. As a result, an updated meta-analysis was conducted to further investigate the possible association between eNOS genetic variants (4b/a, G894T, and T786C) and T2DM risk. This analysis included more papers and credible findings than previous meta-analyses (Jia et al., 2013; Zhang et al., 2017; Dong et al., 2018).
Materials and Methods
Search Strategy
The current study was performed according to the guidelines of the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) group (Moher et al., 2009). The literature was searched using PubMed, China National Knowledge Infrastructure (CNKI), Medline, Embase, ISI (International Statistical Institute) and the China Wanfang databases. The following search strategies were applied: (eNOS OR endothelial nitric oxide synthase OR nitric oxide synthase type III OR NOS3) AND (polymorphism OR variant OR mutation OR genotype OR allele) AND (diabetes OR mellitus OR diabetes mellitus OR DM). The literature search was updated to 15 March 2022. Furthermore, the reference lists of previously published meta-analyses were carefully reviewed to identify additional eligible studies (Jia et al., 2013; Zhang et al., 2017; Dong et al., 2018).
Selection Criteria
Inclusion criteria were as listed below: 1) case-control or cohort studies; 2) described the association between the eNOS 4b/a, G894T and T786C polymorphisms and risk of T2DM; 3) provided sufficient genotype data or the odds ratio (OR) with 95% confidence intervals (CI) in the selected literature. Exclusion criteria were as listed below: 1) duplicate genotype data; 2) studies with no available data; 3) meta-analyses of case reports, abstracts, reviews and letters.
Data Extraction and Quality Score Assessment
Two investigators independently extracted the data and cross-examined it, trying to resolve differences through discussion. If no consensus was reached after discussion, the third author would be invited to extract the data again for final review and confirmation. Moreover, the original authors could be contacted via e-mail if necessary. Races were divided into “Caucasians,” “Asians,” “Indians,” and “Africans.” “Mixed populations” was defined if race was not stated or the sample size of several races cannot be separated in original study.
The Newcastle-Ottawa Scale (NOS) (Wells et al., 2014) was applied by the two investigators to independently assess the quality of all included research. These scales are influenced by three factors: selection (four points), comparability (two points), and exposure (three points). Hardy–Weinberg Equilibrium (HWE) was employed to conduct a quality assessment on the basis of NOS (one point). The overall score varied from zero (worst) to ten (highest), with seven points or more as high quality.
Trial Sequential Analysis
Meta-analyses could increase the power and accuracy of evaluating intervention effects and are regarded as good evidence when available studies are used. However, misleading conclusions may be generated owing to random mistakes if the sample size is very small. Therefore, TSA was carried out to decrease random mistakes and predict the required information size (RIS) in this study (Brok et al., 2008; Thorlund et al., 2011). TSA was performed with the help of TSA 0.9 software (Copenhagen Trial Unit, Centre for Clinical Intervention Research, Copenhagen). The random effect model was used in this work. Alpha (type I error) and beta (type II error) were given as 0.05 and 0.2, respectively. The accruing information size (AIS) was used to determine information size, and the OR value was used to determine the combined effect amount. Based on O'Brien-Fleming-spending functions, a TSA employs trial sequential monitoring boundaries. In addition, the relative risk reduction (RRR) is set at 15% (Kulinskaya and Wood, 2014). If the cumulative Z-curve passes the monitoring border, the RIS line, or enters the futility region, strong evidence for our study may well be affirmed. Otherwise, additional research is required (Wetterslev et al., 2009).
Statistical Analysis
Potential associations between the eNOS genetic polymorphisms (4b/a VNTR, G894T and T786C) and T2DM risk were expressed by ORs and corresponding 95% CIs. Five genetic models were used for comparison: hybrid, homozygous, dominant, recessive and allele model. Chi-square-based Q-test and I2 value were employed in assessing the Heterogeneity. When P was less than 0.10 and/or I2 was greater than 50% (Li et al., 2005), the random-effects model (Mantel and Haenszel, 1959) was adopted because of the significant heterogeneity. On the contrary, the fixed-effects model (DerSimonian and Laird, 2015) was adopted. In addition, the source of heterogeneity was explored by meta-regression analysis. Subgroups were conducted by race, type of control, age and gender. Three methods were applied for sensitivity analyses: 1) excluded one study in turn; 2) eliminated low-quality and medium-quality or Hardy–Weinberg Disequilibrium (HWD) studies; 3) kept only high-quality and HWE studies. Furthermore, HWE was assessed using the Chi-square goodness-of-fit test. p > 0.05 was defined as HWE, otherwise as HWD in the control group. Begg’s funnel plot (Begg and Mazumdar, 1994) and Egger’s test (Egger et al., 1997) were applied to evaluate publication bias. If there were publication bias, the number of missing studies would be estimated and supplemented using a nonparametric “trim and fill” method (Dual and Tweedie, 2000). The false-positive report probability (FPRP) (Wacholder et al., 2004), Bayesian False Discovery Probability (BFDP) (Wakefield, 2007), and the Venice criteria (Ioannidis et al., 2008) were used to evaluate the credibility of statistically significant associations. Stata 12.0 software (STATA Corporation, College Station, TX) was applied to calculate all statistical analyses.
Results
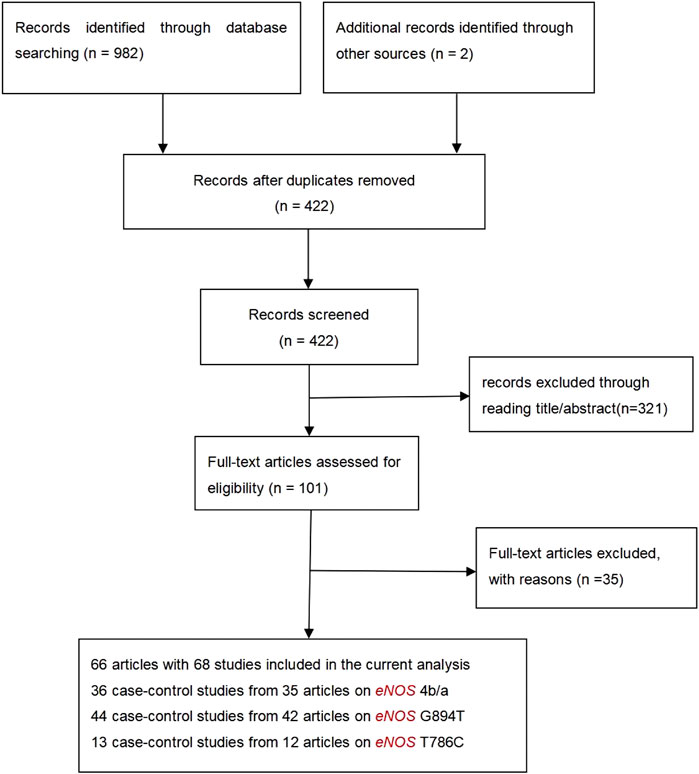
Study Characteristics
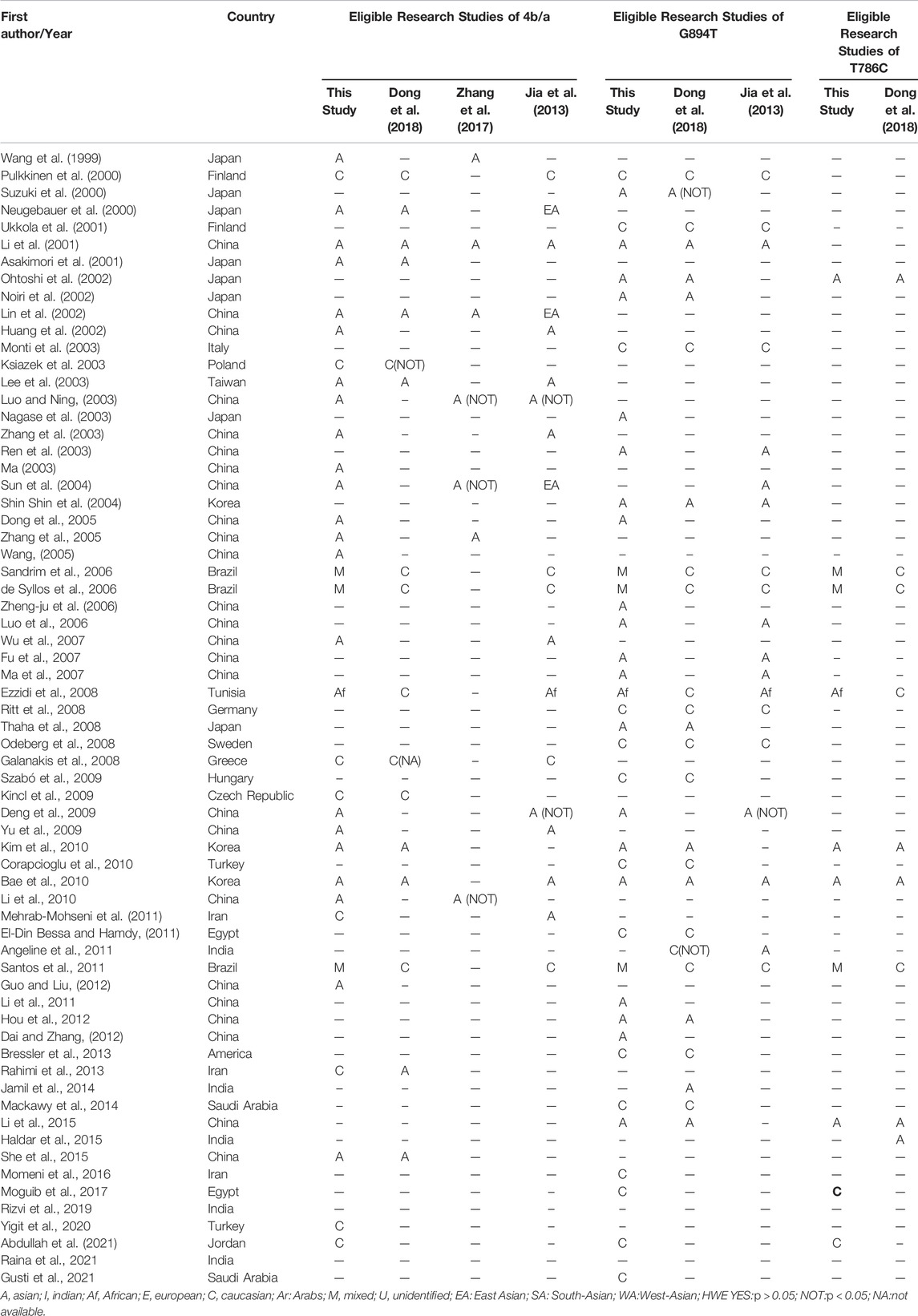
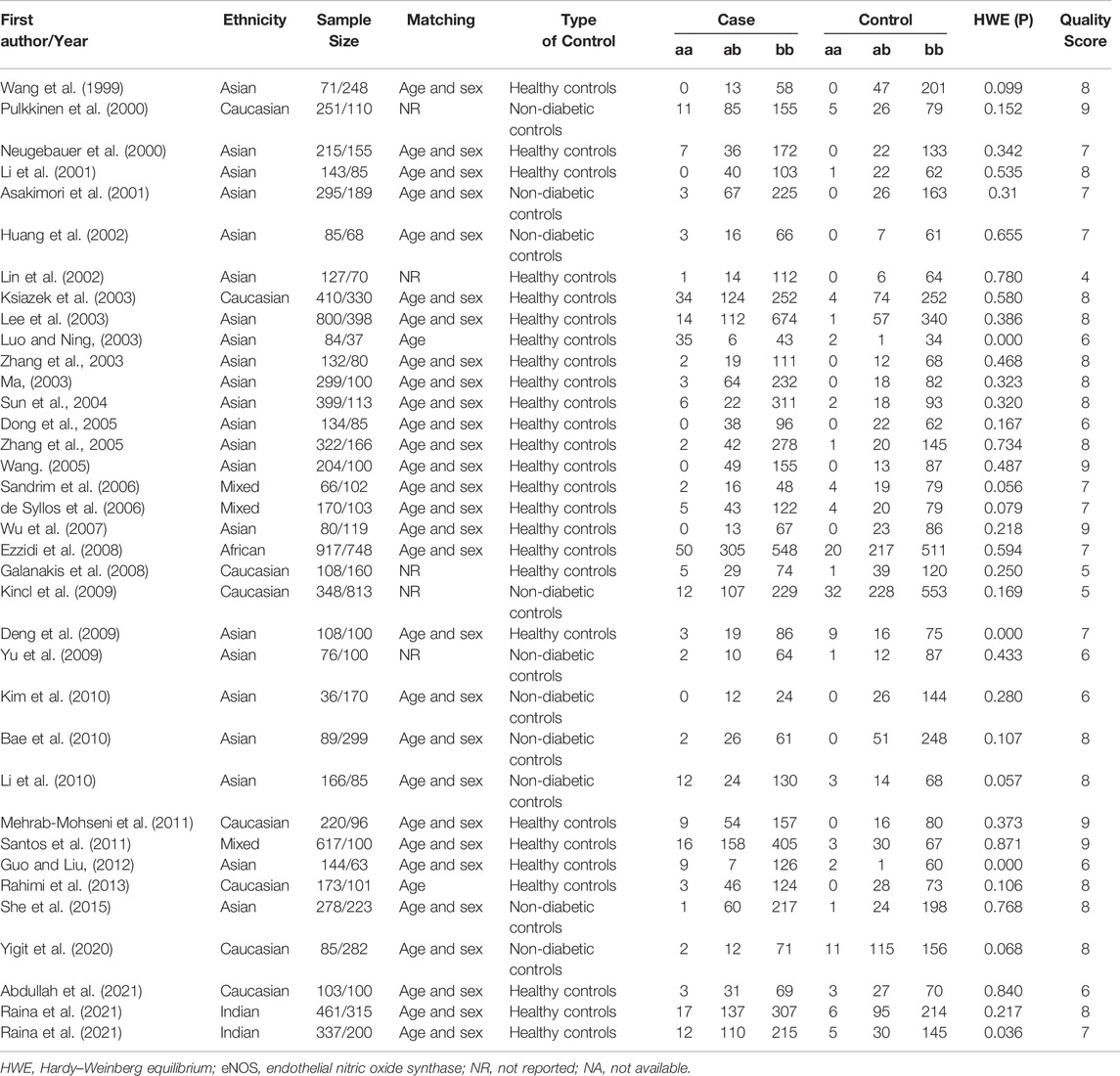
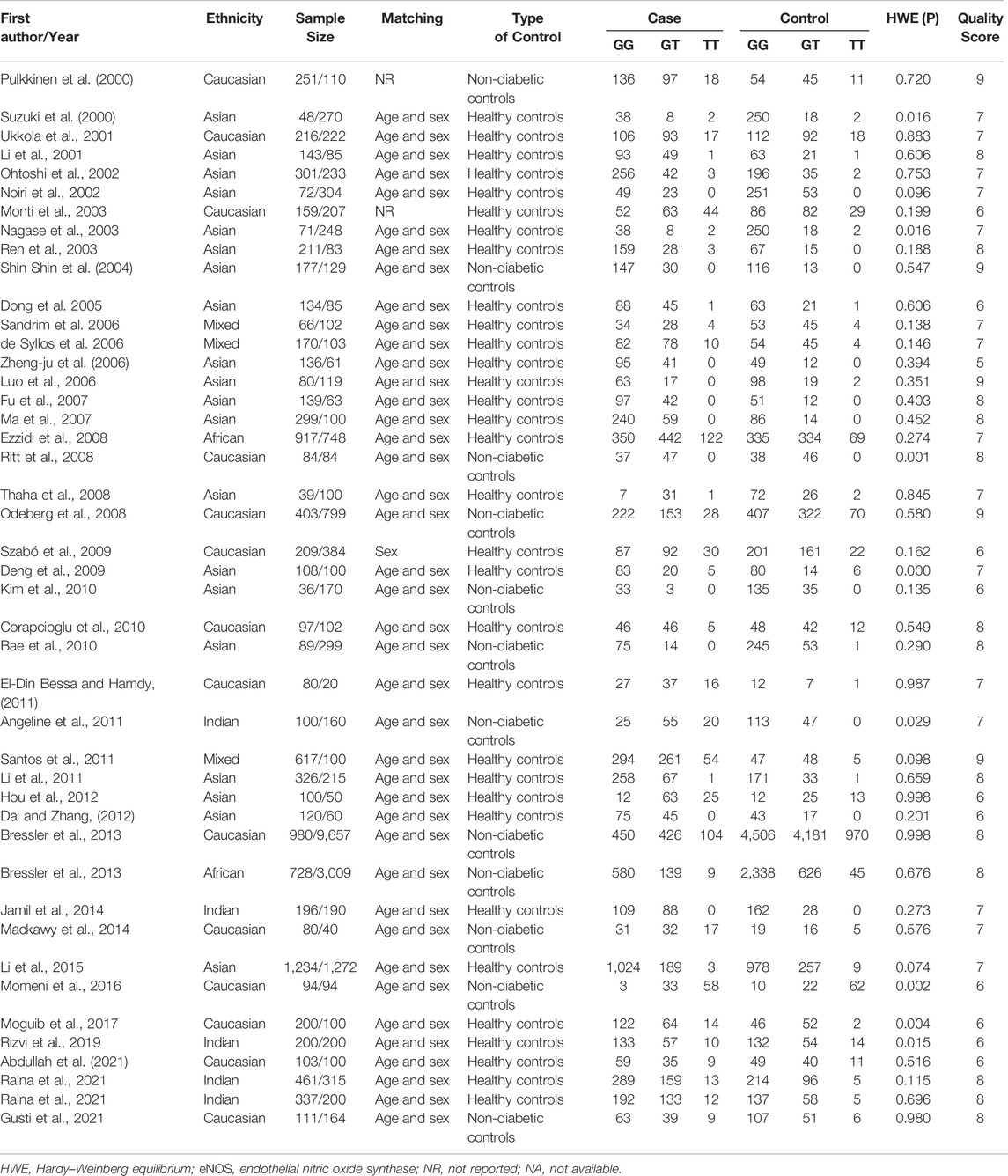
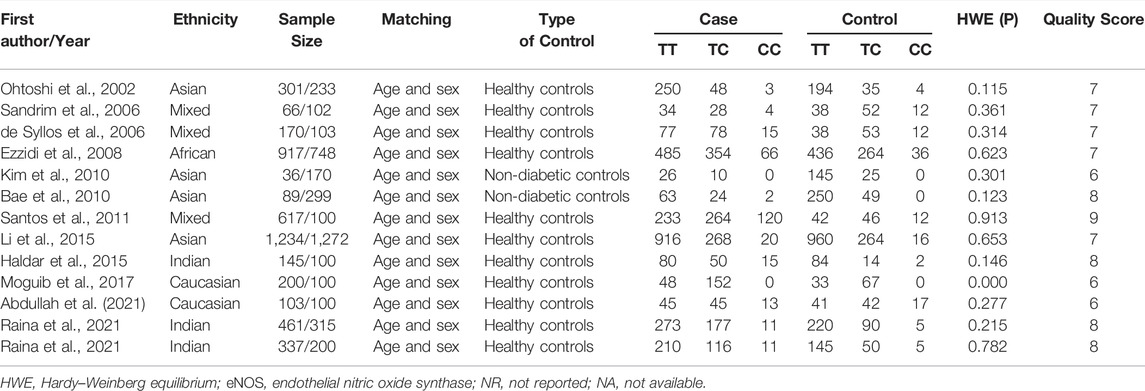
Initially, 984 articles were retrieved from PubMed, CNKI, Medline, Embase, ISI and the China Wan-fang databases. We excluded 321 papers by carefully evaluating titles and abstracts. Moreover, 16 were excluded due to duplication and invalidation of data, and 19 were excluded due to inadequate controls. Finally, 66 articles with 68 studies were eligible for inclusion in our meta-analysis (Table 1). The detailed investigation process is shown in Figure 1. A total of 68 studies (Figure 1) met our inclusion criteria (involving 15,988 T2DM cases and 25,452 controls), of which 36 studies reported the eNOS 4b/a (8,553 cases and 6,613 controls), 44 studies investigated the eNOS G894T (10,722 cases and 21,256 controls), and 13 studies reported the eNOS T786C (4,676 cases and 3,842 controls), as shown in Tables 2–4. Furthermore, twenty, thirty-seven, six, two and three studies were conducted to investigate Caucasians, Asians, Indians, Africans, and mixed groups, respectively. In addition, the eNOS 4b/a had 19 high-quality studies and 17 low-quality studies, the eNOS G894T had 18 high-quality studies and 26 low-quality studies, and the eNOS T786C had five high-quality studies and eight low-quality studies. Moreover, the complete features, scores, HWE and the genotype frequencies of the selected literature were shown in Tables 2–4. Furthermore, Table 5 showed the results of the detailed quality scores for the included articles according to the NOS.
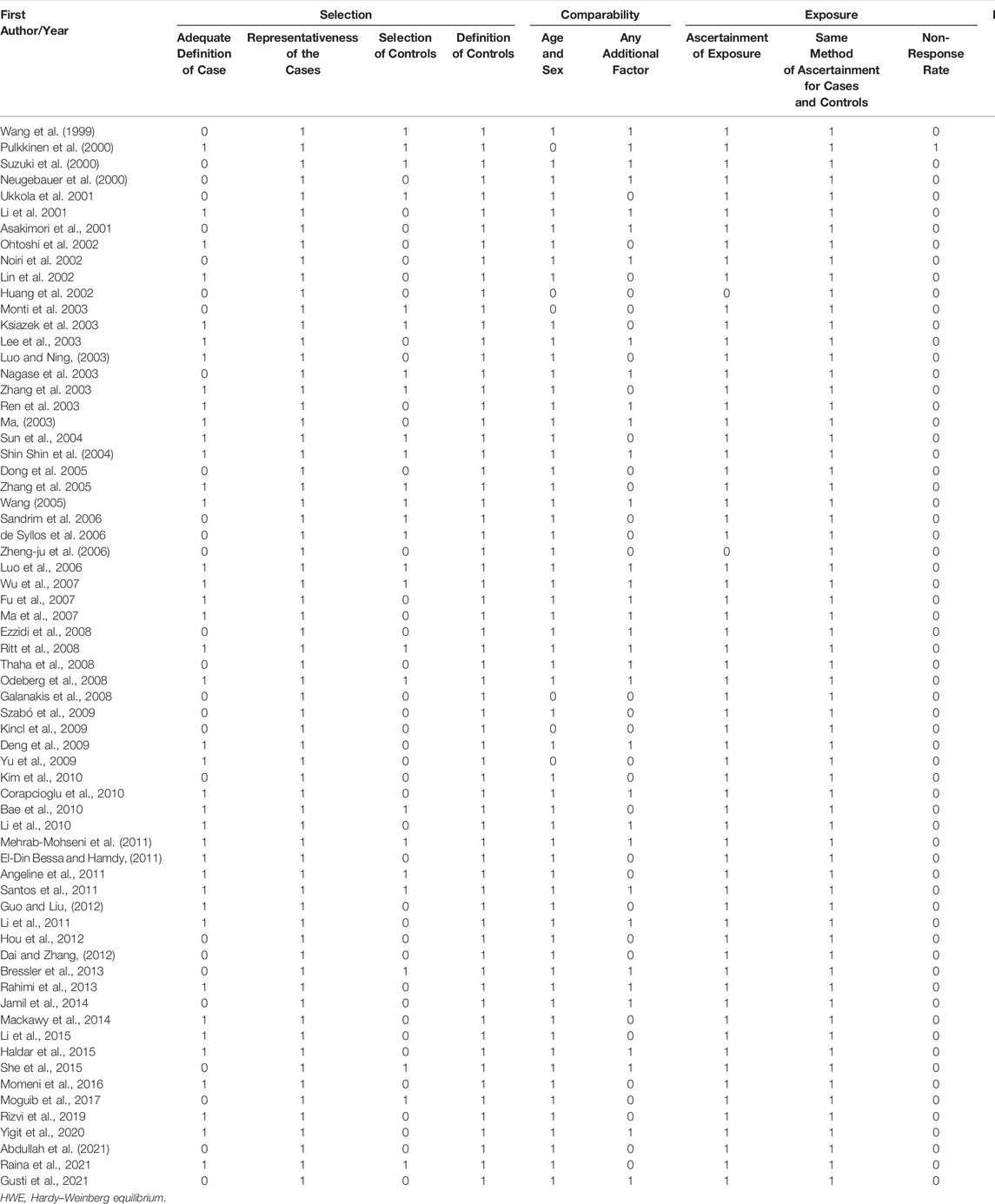
TABLE 5. Quality assessment of included studies based on the Newcastle-Ottawa Scale for assessing the quality of case control studies.
Quantitative Synthes
In the total analysis, the eNOS 4b/4a was associated with a substantially lower T2DM risk (ab vs. aa: OR = 0.71, 95% CI = 0.52–0.96; bb vs. aa: OR = 0.55, 95% CI = 0.38–0.79; ab + bb vs. aa: OR = 0.58, 95% CI = 0.40–0.82; bb vs. aa + ab: OR = 0.77, 95% CI = 0.66–0.89; b vs. a: OR = 0.76, 95% CI = 0.65–0.87, Table 6; Figure 2). In the following ethnic subgroup analysis, we discovered a significant association between eNOS 4b/4a polymorphism and T2DM susceptibility in the Asian population (bb vs. aa: OR = 0.44, 95% CI = 0.23–0.84; ab + bb vs. aa: OR = 0.45, 95% CI = 0.24–0.86; bb vs. aa + ab: OR = 0.73, 95% CI = 0.59–0.91; b vs. a: OR = 0.71, 95% CI = 0.57–0.88, Table 6; Figure 2). Also, similar association was also found in the healthy control and matched studies (Table 6).
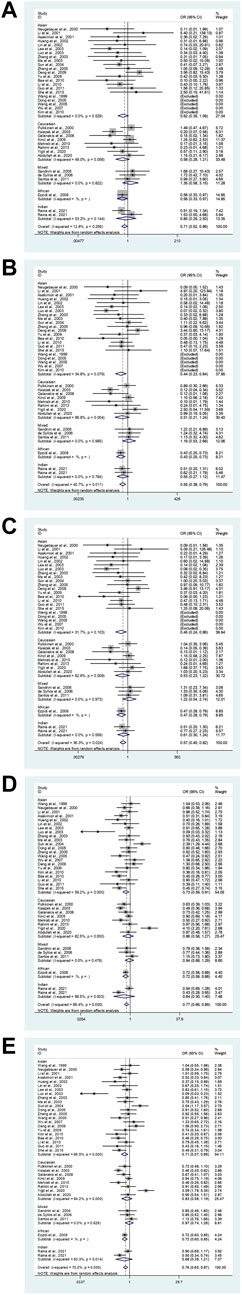
FIGURE 2. The forest plots of all selected studies on the association between eNOS 4b/a polymorphism and the risk of T2DM in different races [(A): hybrid model; (B) homozygous model; (C) dominant model; (D) recessive model; (E) allele model].
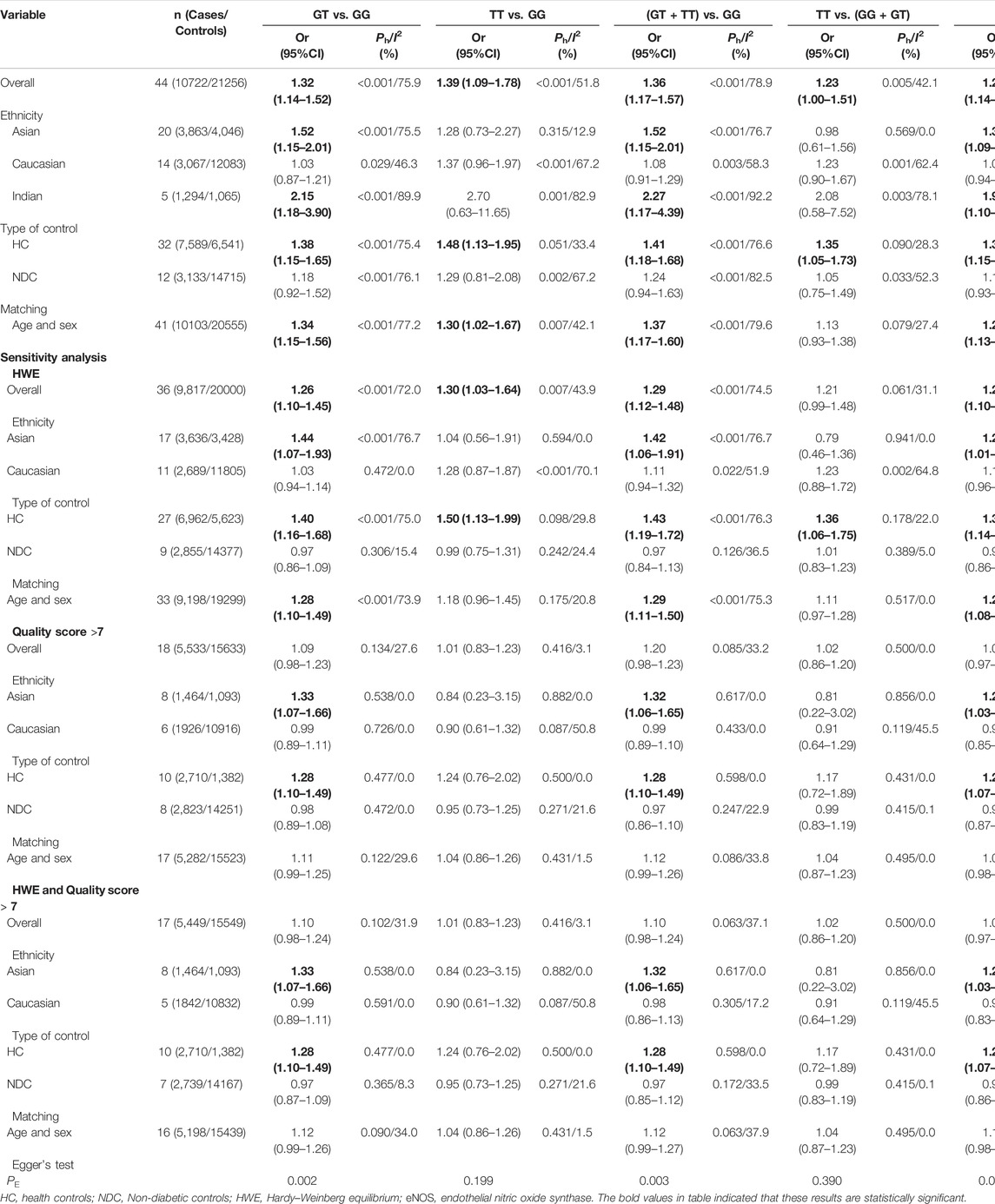
Overall, a substantial association was found between the eNOS G894T polymorphism and an increased risk of T2DM (GT vs. GG: OR = 1.32, 95% CI = 1.14–1.52; TT vs. GG: OR = 1.39, 95% CI = 1.09–1.78; GT + TT vs. GG: OR = 1.36, 95% CI = 1.17–1.57; TT vs. GG + GT: OR = 1.23, 95% CI = 1.00–1.51; T vs. G: OR = 1.29, 95% CI = 1.14–1.45, Table 7; Figure 3). Moreover, a significantly increased risk of T2DM was also found in Asians (GT vs. GG: OR = 1.52, 95% CI = 1.15–2.01; GT + TT vs. GG: OR = 1.52, 95% CI = 1.15–2.01; T vs. G: OR = 1.39, 95% CI = 1.09–1.45) and Indians (GT vs. GG: OR = 2.15, 95% CI = 1.18–3.90; GT + TT vs. GG: OR = 2.27, 95% CI = 1.17–4.39; T vs. G: OR = 1.97, 95% CI = 1.10–3.55, Table 7; Figure 3). Furthermore, similar results were also observed in the healthy control and matched analyses (Table 7).
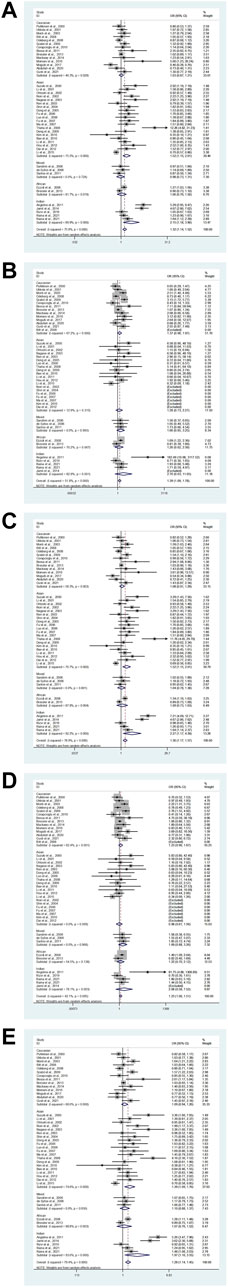
FIGURE 3. The forest plots of all selected studies on the association between eNOS G894T polymorphism and the risk of T2DM in different races [(A): hybrid model; (B) homozygous model; (C) dominant model; (D) recessive model; (E) allele model].
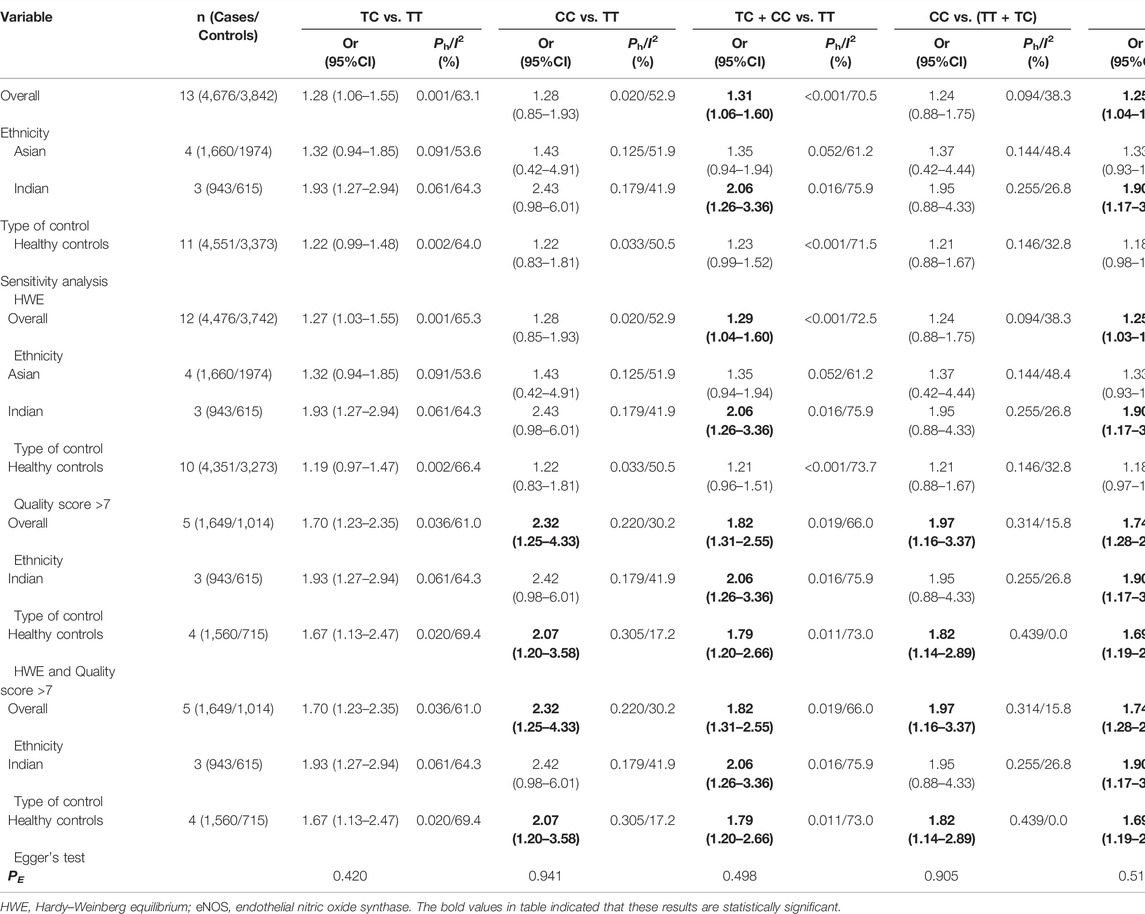
Our study exposed an overall powerful association between eNOS T786C and T2DM susceptibility (TC vs. TT: OR = 1.28, 95%CI = 1.06–1.55; TC + CC vs. TT: OR = 1.31, 95%CI = 1.06–1.60; C vs. T: OR = 1.25, 95%CI = 1.04–1.49, Table 8; Figure 4). At the same time, subgroup studies revealed that Indians had a significantly increased risk of T2DM (TC vs. TT: OR = 1.93, 95% CI = 1.27–2.94; TC + CC vs. TT: OR = 2.06, 95%CI = 1.26–3.36; C vs. T: OR = 1.90, 95%CI = 1.17–3.08, Table 8; Figure 4). Moreover, no signification association was observed in the healthy population according to type of control (Table 8).
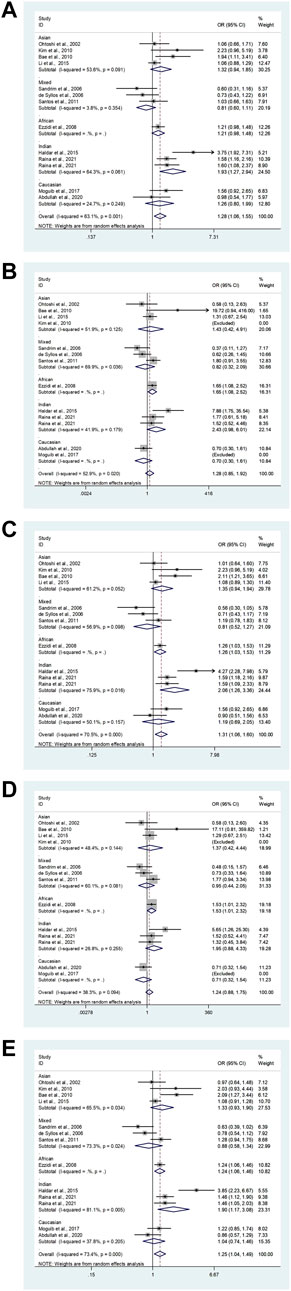
FIGURE 4. The forest plots of all selected studies on the association between eNOS t786C polymorphism and the risk of T2DM in different races [(A): hybrid model; (B) homozygous model; (C) dominant model; (D) recessive model; (E) allele model].
Heterogeneity and Sensitivity Analyses
Several possible causes of variation were discovered, including ethnicity, gender, sample size, age, quality score, type of controls and HWE. Therefore, a meta-regression analysis was used to identify causes of heterogeneity. For the eNOS G894T, no covariate was found as a possible cause of between-study variation. A meta-regression analysis revealed that HWE (ab vs. aa: p = 0.045) was the source of heterogeneity between the eNOS 4b/a polymorphism and the risk of T2DM. At the same time, the quality score (TC vs. TT: p = 0.045; TC + CC vs. TT: p = 0.042; C vs. T: p = 0.041) and HWE (CC vs. TT: p = 0.029; CC vs. TC + TT: p = 0.041) were the sources of heterogeneity between the eNOS T786C polymorphism and the risk of T2DM.
Three methods were employed for sensitivity analyses in this meta-analysis. Firstly, results did not alter when a single study was removed each time. Second, when HWD studies were omitted, Asians were found to have a significantly lower risk of eNOS 4b/a polymorphism and T2DM in the overall analysis (Table 6). For the eNOS G894T polymorphism, significantly increased T2DM risk was only observed in Asians and healthy population when we retained high-quality and HWE studies in the control group (Table 7). For the eNOS T786C polymorphism, a significant association was also discovered in the healthy population when we only included high-quality and HWE studies in the control group (Table 8).
Publication Bias
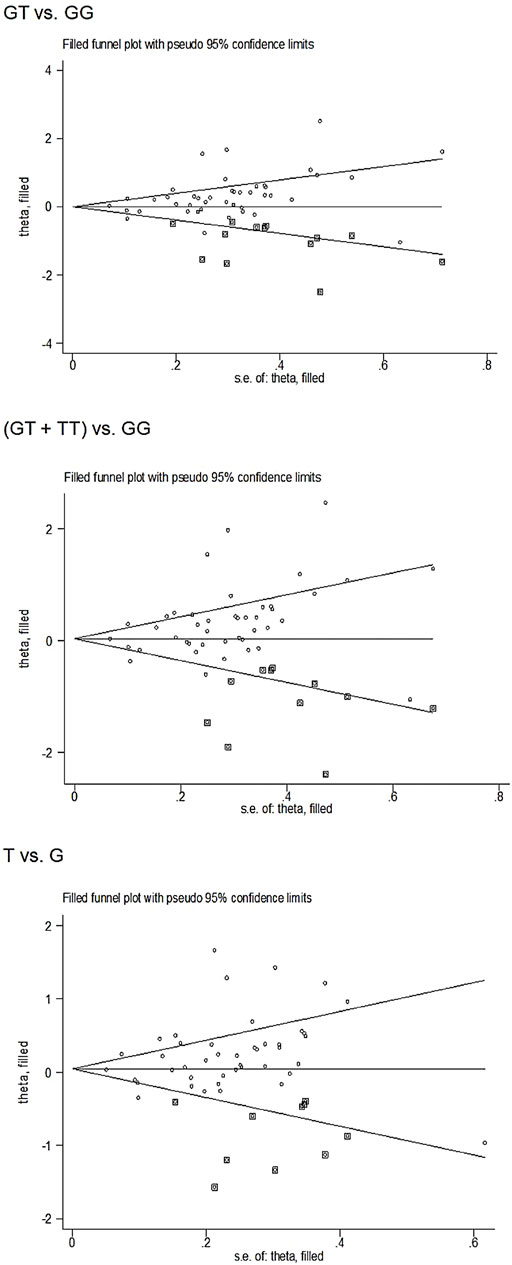
Begg’s funnel plot and Egger’s test revealed only publication bias between the eNOS G894T polymorphism and T2DM risk (GT vs. GG: p = 0.002; GT + TT vs. GG: p = 0.003; T against G: p = 0.014, Table 7). Then, publication bias was adjusted using the nonparametric “trim and fill” method. And we need to add 13, 11, and 10 articles in the future for GT vs. GG, GT + TT vs. GG, and T vs. G models, respectively (Figure 5). In the overall analysis, the findings for GT vs. GG, GT + TT vs. GG, and T vs. G models did not change (data not shown), demonstrating that more research cannot alter the merger outcomes.
TSA Results
The TSA of the dominant model for the eNOS 4b/a and T786C polymorphisms revealed that the cumulative z-curve passed both the RIS line and the TSA threshold, indicating that no more evidence was required to confirm the conclusion. However, multiple comparisons and other confounding factors, we believe, can still increase the occurrence of false positive errors, so credibility analysis is still required for the eNOS 4b/a and T786C polymorphisms. The cumulative Z-curve of the dominant model for the eNOS G894T polymorphism did not surpass the TSA threshold, and the total number of cases and controls was smaller than the RIS, according to the TSA. Therefore, more trials were still required to confirm the association between eNOS G894T polymorphism and T2DM risk. Figure 6 displays the above results.
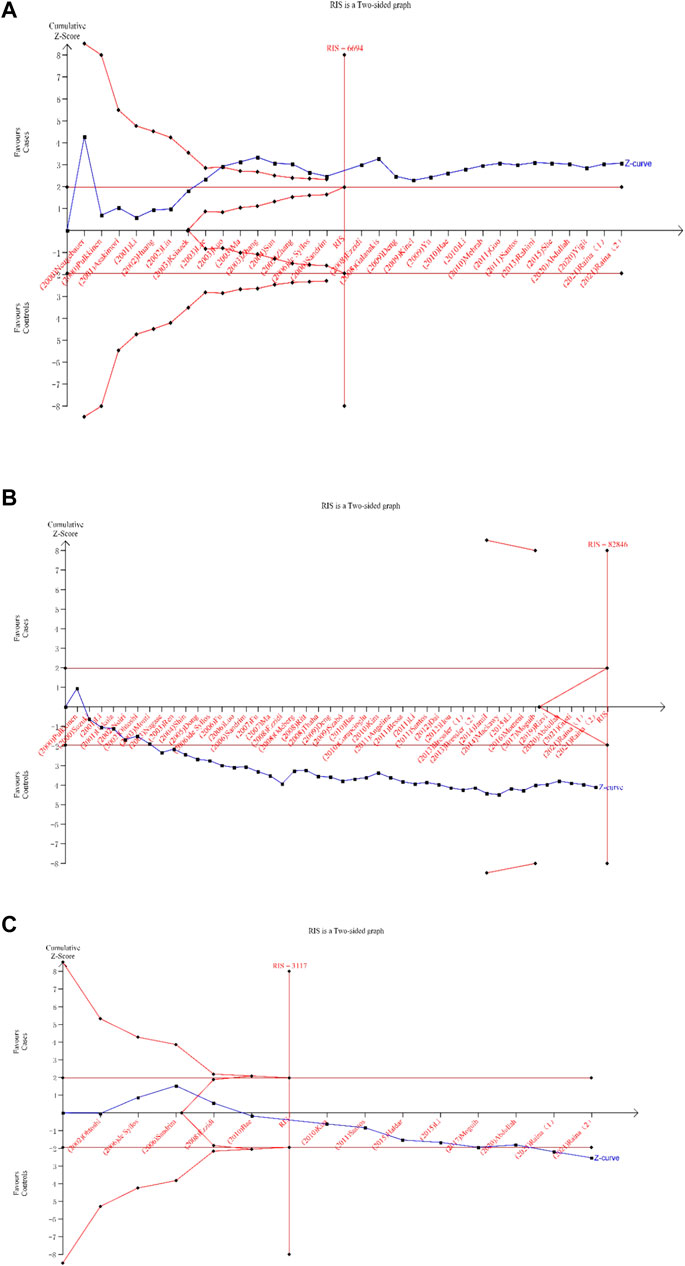
FIGURE 6. Trial sequential analysis for the eNOS polymorphisms under dominant gene model [(A): (bb + ab) vs. aa; (B) (TT + GT) vs. GG; (C) (CC + TC) vsTT)].
Credibility of the Identified Genetic Associations
The credibility of this meta-analysis was assessed using the FPRP, BFDP, and Venice criteria. Associations meeting the following criteria were regarded to be of high credibility [29]: 1) statistically significant associations were observed in at least two of the genetic models; 2) FPRP <0.2 and BFDP <0.8; 3) I2 < 50%; and 4) statistical power >80%. All other major findings were viewed as “less credible results”. All statistically significant associations were deemed “less credible” in this study.
Discussion
T2DM is a polygenic genetic disease, which is also greatly influenced by environmental factors. And it is the outcome of the combined action of numerous genes and environmental factors. Several studies have shown that diabetes is the most important risk factor for mortality and disability caused by cardiovascular and cerebrovascular illnesses, according to several research. However, the molecular mechanism of the genetics of T2DM has not been elucidated. Much significant evidence indicates that the eNOS polymorphisms have been considered as potential genetic factors for T2DM. Numerous eNOS polymorphisms have been reported, and their relationship with various disorders has been studied, including coronary artery disease, myocardial infarction, coronary spasm, hypertension, end-stage renal disease (ESRD), and T2DM. Previous research has focused on three eNOS polymorphisms: the intronic 427-bp repeat (4b/a) in the promoter region; the G894T (Glu298Asp) missense mutation in exon seven and the T786C single nucleotide polymorphism. T786C inhibits eNOS transcription, G894T inhibits eNOS activity, and 4b/a inhibits plasma NO concentrations, which may be a reflection of eNOS activity. Many researchers have sought to investigate the potential relationship between eNOS polymorphisms and T2DM risk. Regrettably, no credible evidence is available, which might be attributable to a variety of factors such as small sample numbers, ethnic and geographical disparities. As a result, meta-analysis is an effective method to conquer these flaws.
Overall, the eNOS 4b/a was connected with a substantially lower the risk of T2DM in Asians; the eNOS G894T was connected with a significantly higher risk of T2DM in Asians, however, it had no significant effect on the risk of T2DM in Caucasians. the eNOS T786C was connected with a significantly higher risk of T2DM in Indians. However, after omitting low-quality and HWD studies, we observed that eNOS 4b/a polymorphism substantially lowered T2DM risk in the entire population while eNOS T786C polymorphism considerably raised T2DM risk in the whole population. However, after omitting low-quality and HWD studies, we observed that eNOS 4b/a polymorphism substantially lowered T2DM risk in the entire population while eNOS T786C polymorphism considerably raised T2DM risk in the whole population. The current study used many subgroups and distinct genetic models, which resulted in multiple comparisons, so the pooled p value must be corrected. FPRP has been described as a proper method for assessing the likelihood of significant outcomes in molecular epidemiology investigations using multiple hypothesis testing. Furthermore, Wakefield suggested a more accurate Bayesian metric of false detection in genetic epidemiology investigations in 2007. Many factors may lead to errors and biases, such as genotyping errors and phenotypic misclassification, of which statistical power was a significant factor. A substantial amount of evidence (statistical power>80%) can achieve a higher degree of statistical significance or reduce the false-discovery rate. As a result, in this study, we used the FPRP test, BFDP test and the Venice criteria to evaluate false discovery. All the statistically significant connections were less-credible in the current meta-analysis after assessing credibility. Our meta-analysis has also revealed heterogeneity. According to the results of the meta-regression study, the quality score and HWE were the sources of heterogeneity. Furthermore, bias and mistakes were widespread in several HWD studies with low quality and small sample size, making the conclusion of these original studies untrustworthy, particularly in molecular epidemiology studies. And small sample studies with positive results may be easier to accept since they are likely to produce false-positive results because their research is less rigorous and frequently of poor quality. The asymmetry of the funnel plot was created by a study of low-quality small samples. Therefore, we added high-quality and HWE to evaluate sensitivity analyses in control studies.
We hypothesized that the single and combined effects of the eNOS 4b/a, G894T, and T786C polymorphisms were linked with T2DM risk in all races based on the biochemical features outlined for these genes. Nevertheless, when we applied the FPRP, BFDP test, and Venice criteria to assess the credibility of this meta-analysis, all statistically significant relationships were declared “less credible” (greater heterogeneity, FPRP >0.2, BRDP >0.8, and lower statistic power). Therefore, these results indicated that much larger sample size was needed to study the potential gene-gene interactions.
A total of three previously published meta-analyses investigated the relationship between the eNOS 4b/a, G894T, and T786C polymorphisms and the risk of T2DM. There was a clear mismatch in the categorization of ethnic groupings between the previous related meta-analyses and the current meta-analysis. Furthermore, the sample size in this study was substantially greater. A total of 66 articles were included in this study, of which 36 articles reported the eNOS 4b/a (8,553 cases and 6,613 controls), 44 articles investigated the eNOS G894T (10,722 cases and 21,256 controls), and 13 articles reported the eNOS T786C (4,676 cases and 3,842 controls). Five genetic models were compared separately in this study. However, Dong et al., Zhang et al. and Jia et al. applied four genetic models. In addition, when we used the FPRP, BFDP test, and Venice criteria to assess the credibility of the previous meta-analyses, all statistically significant relationships were deemed “less credible.” As a result, their findings may be unreliable.
Compared with the previous meta-analysis, the new meta-analysis had several advantages: 1) credibility was investigated using FPRP, BFDP test and Venice criteria; 2) the quality of the eligible research was evaluated; 3) The sample size was larger and the data collected was more detailed than the previous meta-analyses; 4) more subgroup analyses were performed according to the type of control, matching and quality score; 5) TSA was carried out to decrease random mistakes. However, there are still some potential limitations in the current meta-analysis. First, the current meta-analysis included only published research, although positive outcomes are known to be published more frequently than negative ones. Second, T2DM is a complex multi-genetic disorder, and the link between an individual SNP and T2DM risk is relatively weak. However, we did not retrieve the corresponding data on the combined impacts of gene-gene and gene-environment. Third, the relationship between the eNOS polymorphisms and the risk of T2DM complications has not been investigated. Therefore, the current meta-analysis has a large sample size and a sufficiently large subgroup to help confirm our findings.
To summarize, our study reveals that all substantial relationships between the eNOS 4b/a, G894T, and T786C polymorphisms and T2DM risk are most likely due to false-positive results rather than real connections or biological variables. larger-scale epidemiological studies on this topic should be conducted in the future to confirm or disprove our findings.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
DW: research design and performance, data collection, data analysis, paper writing. LL: data collection. CZ: data recheck. WL: methodology. FW and XH: research design and paper review.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AIS, accruing information size; BFDP, Bayesian false discovery probability; CIs, confidence intervals; CNKI, China National Knowledge Infrastructure; eNOS, endothelial nitric oxide synthase; FPRP, false-positive report probability; HWD, Hardy–Weinberg Disequilibrium; HWE, Hardy–Weinberg equilibrium; ISI; International Statistical Institute; NOS, Newcastle-Ottawa Scale; NO, Nitric oxide; ORs, odds ratios; RIS, required information size; RRR, relative risk reduction; T2DM, type 2 diabetes mellitus; TSA, Trial sequential analysis.
References
Abdullah, S., Jarrar, Y., Alhawari, H., Abed, E., and Zihlif, M. (2021). The Influence of Endothelial Nitric Oxide Synthase (eNOS) Genetic Polymorphisms on Cholesterol Blood Levels Among Type 2 Diabetic Patients on Atorvastatin Therapy. Endocr. Metab. Immune Disord. Drug Targets 21 (2), 352–359. doi:10.2174/1871530320666200621174858
Angeline, T., Krithiga, H. R., Isabel, W., Asirvatham, A. J., and Poornima, A. (2011). Endothelial Nitric Oxide Synthase Gene Polymorphism (G894T) and Diabetes Mellitus (Type II) Among South Indians. Oxid. Med. Cell. Longev. 2011, 462607. doi:10.1155/2011/462607
Asakimori, Y., Yorioka, N., Yamamoto, I., Okumoto, S., Doi, S., Hirai, T., et al. (2001). Endothelial Nitric Oxide Synthase Intron 4 Polymorphism Influences the Progression of Renal Disease. Nephron 89 (2), 219–223. doi:10.1159/000046071
Bae, J., Kim, I. J., Hong, S. H., Sung, J. H., Lim, S. W., Cha, D. H., et al. (2010). Association of Endothelial Nitric Oxide Synthase Polymorphisms with Coronary Artery Disease in Korean Individuals with or without Diabetes Mellitus. Exp. Ther. Med. 1 (4), 719–724. doi:10.3892/etm_00000111
Begg, C. B., and Mazumdar, M. (1994). Operating Characteristics of a Rank Correlation Test for Publication Bias. Biometrics 50, 1088–1101. doi:10.2307/2533446
Bressler, J., Pankow, J. S., Coresh, J., and Boerwinkle, E. (2013). Interaction between the NOS3 Gene and Obesity as a Determinant of Risk of Type 2 Diabetes: the Atherosclerosis Risk in Communities Study. PLoS One 8 (11), e79466. doi:10.1371/journal.pone.0079466
Brok, J., Thorlund, K., Gluud, C., and Wetterslev, J. (2008). Trial Sequential Analysis Reveals Insufficient Information Size and Potentially False Positive Results in Many Meta-Analyses. J. Clin. Epidemiol. 61, 763–769. doi:10.1016/j.jclinepi.2007.10.007
Corapcioglu, D., Sahin, M., Emral, R., Celebi, Z. K., Sener, O., and Gedik, V. T. (2010). Association of the G894T Polymorphism of the Endothelial Nitric Oxide Synthase Gene with Diabetic Foot Syndrome Foot Ulcer, Diabetic Complications, and Comorbid Vascular Diseases: a Turkish Case-Control Study. Genet. Test. Mol. Biomarkers 14 (4), 483–488. doi:10.1089/gtmb.2010.0023
Dai, H. S., and Zhang, Y. (2012). An Association Study of MTHFR and eNOS Genes Polymorphism with Diabetic Nephropathy. Chin. J. Trauma Disabil. Med. 20 (6), 4–6.
de Syllos, R. W., Sandrim, V. C., Lisboa, H. R., Tres, G. S., and Tanus-Santos, J. E. (2006). Endothelial Nitric Oxide Synthase Genotype and Haplotype Are Not Associated with Diabetic Retinopathy in Diabetes Type 2 Patients. Nitric Oxide 15 (4), 417–422. doi:10.1016/j.niox.2006.02.002
Deng, W. Q., Wei, J., Wu, Y. N., Wang, F. H., Song, D. Y., Jiang, Y. Z., et al. (2009). Association between Polymorphism of Endothe-Lial Nitric Oxide Synthase Gene and Type 2 Diabetic Patients with Erectile Dysfunction. Lin. Chuang Hui Cui 24, 481–485 (In Chinese).
DerSimonian, R., and Laird, N. (2015). Meta-analysis in Clinical Trials Revisited. Contemp. Clin. Trials 45, 139–145. doi:10.1016/j.cct.2015.09.002
Dong, J., Ping, Y., Wang, Y., and Zhang, Y. (2018). The Roles of Endothelial Nitric Oxide Synthase Gene Polymorphisms in Diabetes Mellitus and its Associated Vascular Complications: a Systematic Review and Meta-Analysis. Endocrine 62 (2), 412–422. doi:10.1007/s12020-018-1683-4
Dong, Y. H., Qu, S. P., Lu, W. S., Ding, M., Dong, C., Jiang, H. W., et al. (2005). Gene Polymorphism in Chromosome 7q35 and Susceptibility to Diabetic Nephropathy. Chin. J. Endocrinol. Metab. (1), 51–54.
Dual, S., and Tweedie, R. (2000). A Nonparametric “Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 95, 89–98. doi:10.1080/01621459.2000.10473905
Egger, M., Smith, D. G., Schneider, M., and Minder, C. (1997). Bias in Meta-Analysis Detected by a Simple, Graphical Test. Br. Med. J. 315, 629–634. doi:10.1136/bmj.315.7109.629
El-Din Bessa, S. S., and Hamdy, S. M. (2011). Impact of Nitric Oxide Synthase Glu298Asp Polymorphism on the Development of End-Stage Renal Disease in Type 2 Diabetic Egyptian Patients. Ren. Fail 33 (9), 878–884. doi:10.3109/0886022X.2011.605978
Ezzidi, I., Mtiraoui, N., Mohamed, M. B., Mahjoub, T., Kacem, M., and Almawi, W. Y. (2008). Association of Endothelial Nitric Oxide Synthase Glu298Asp, 4b/a, and -786T>C Gene Variants with Diabetic Nephropathy. J. Diabetes Complicat. 22 (5), 331–338. doi:10.1016/j.jdiacomp.2007.11.011
Ferland-McCollough, D., Ozanne, S. E., Siddle, K., Willis, A. E., and Bushell, M. (2010). The Involvement of microRNAs in Type 2 Diabetes. Biochem. Soc. Trans. 38, 1565–1570. doi:10.1042/bst0381565
Fu, Z. J., Li, C. G., Wang, Z. C., and Yan, S. L. (2007). Coexistence of Aldose Reduction Gene and Endothelial Nitric Oxide Synthase Polymorphisms Associates with Diabetic Nephropathy. J. Clin. Rehabil. Tissue Eng. Res. 11 (34), 6893–6896.
Galanakis, E., Kofteridis, D., Stratigi, K., Petraki, E., Vazgiourakis, V., Fragouli, E., et al. (2008). Intron 4 A/b Polymorphism of the Endothelial Nitric Oxide Synthase Gene Is Associated with Both Type 1 and Type 2 Diabetes in a Genetically Homogeneous Population. Hum. Immunol. 69 (4-5), 279–283. doi:10.1016/j.humimm.2008.03.001
Gaulton, K. J. (2017). Mechanisms of Type 2 Diabetes Risk Loci. Curr. Diab Rep. 17 (9), 72. doi:10.1007/s11892-017-0908-x
Guo, X. J., and Liu, S. J. (2012). The Association between Endothelial Nitric Oxide Synthase Gene Polymorphism and Type 2 Diabetic Nephropathy. China Healthc. Innov. 6 (19), 3–5.
Gusti, A. M. T., Qusti, S. Y., Bahijri, S. M., Toraih, E. A., Bokhari, S., Attallah, S. M., et al. (2021). Glutathione S-Transferase (GSTT1rs17856199) and Nitric Oxide Synthase (NOS2 Rs2297518) Genotype Combination as Potential Oxidative Stress-Related Molecular Markers for Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 14, 1385–1403. doi:10.2147/DMSO.S300525
Haldar, S. R., Chakrabarty, A., Chowdhury, S., Haldar, A., Sengupta, S., and Bhattacharyya, M. (2015). Oxidative Stress-Related Genes in Type 2 Diabetes: Association Analysis and Their Clinical Impact. Biochem. Genet. 53 (4-6), 93–119. doi:10.1007/s10528-015-9675-z
Hou, H., Zhang, F., Zhao, M., Cao, G., Huang, H., and Huang, P. (2012). Relationship between Endothelial Nitric Oxide Synthase Glu298Asp Gene Polymorphisms and the Chronic Periodontitis with Type 2 Diabetes Mellitus. Hua Xi Kou Qiang Yi Xue Za Zhi 30 (6), 628–631.
Huang, H. S., Lin, L. X., and Chen, M. (2002). Association of Poly-Morphism of Endothelial Nitric Oxide Synthase Gene with Essential Hypertension and Type 2 Diabetes Mellitus. Zhong Hua Nei Fen Mi Dai Xie Za Zhi 18.
Ioannidis, J. P., Boffetta, P., Little, J., O'Brien, T. R., Uitterlinden, A. G., Vineis, P., et al. (2008). Assessment of Cumulative Evidence on Genetic Associations: Interim Guidelines. Int. J. Epidemiol. 37, 120–132. doi:10.1093/ije/dym159
Jamil, K., Kandula, V., Kandula, R., Asimuddin, M., Joshi, S., and Yerra, S. K. (2014). Polymorphism of CYP3A4 2 and eNOS Genes in the Diabetic Patients with Hyperlipidemia Undergoing Statin Treatment. Mol. Biol. Rep. 41 (10), 6719–6727. doi:10.1007/s11033-014-3557-z
Jamwal, S., and Sharma, S. (2018). Vascular Endothelium Dysfunction: a Conservative Target in Metabolic Disorders. Inflamm. Res. 67 (5), 391–405. doi:10.1007/s00011-018-1129-8
Jia, Z., Zhang, X., Kang, S., and Wu, Y. (2013). Association of Endothelial Nitric Oxide Synthase Gene Polymorphisms with Type 2 Diabetes Mellitus: a Meta-Analysis. Endocr. J. 60 (7), 893–901. doi:10.1507/endocrj.ej12-0463
Kim, O. J., Kim, U. K., Oh, S. H., Cho, Y. W., Oh, K. I., Oh, D., et al. (2010). Association of Endothelial Nitric Oxide Synthase Polymorphisms and Haplotypes with Ischemic Stroke in Korean Individuals with or without Diabetes Mellitus. Mol. Med. Rep. 3 (3), 509–513. doi:10.3892/mmr_00000289
Kincl, V., Vasků, A., Meluzín, J., Panovský, R., Seménka, J., and Groch, L. (2009). Association of the eNOS 4a/b and -786T/C Polymormphisms with Coronary Artery Disease, Obesity and Diabetes Mellitus. Folia Biol. (Praha) 55 (5), 187–191.
Ksiazek, P., Wojewoda, P., Muc, K., and Buraczynska, M. (2003). Endothelial Nitric Oxide Synthase Gene Intron 4 Polymorphism in Type 2 Diabetes Mellitus. Mol. Diagn 7 (2), 119–123. doi:10.1007/BF03260027
Kulinskaya, E., and Wood, J. (2014). Trial Sequential Methods for Meta-Analysis. Res. Synth. Methods 5, 212–220. doi:10.1002/jrsm.1104
Larsen, T., Mose, F. H., Bech, J. N., and Pedersen, E. B. (2012). Effect of Nitric Oxide Inhibition on Blood Pressure and Renal Sodium Handling: a Dose-Response Study in Healthy Man. Clin. Exp. Hypertens. 34, 567–574. doi:10.3109/10641963.2012.681727
Lee, Y. J., Chang, D. M., and Tsai, J. C. (2003). Association of a 27-bp Repeat Polymorphism in Intron 4 of Endothelial Constitutive Nitric Oxide Synthase Gene with Serum Uric Acid Levels in Chinese Subjects with Type 2 Diabetes. Metabolism 52 (11), 1448–1453. doi:10.1016/s0026-0495(03)00258-0
Li, C., Dong, Y., and Lü, W. (2001). The Association between Polymorphism of Endothelial Nitric Oxide Synthase Gene and Diabetic Nephropathy. Zhonghua Nei Ke Za Zhi 40 (11), 729–732.
Li, H. M., Miao, H., Lu, Y. B., and Cheng, J. L. (2005). Association between the Polymorphism of Human Vitamin D Receptor Gene and the Susceptibility of Diabetic Nephropathy in Chinese Han Population. Chin. J. Clin. Rehabil. 9, 1–4.
Li, H., Wallerath, T., Münzel, T., and Förstermann, U. (2002). Regulation of Endothelial-type NO Synthase Expression in Pathophysiology and in Response to Drugs. Nitric Oxide 7 (3), 149–164. doi:10.1016/s1089-8603(02)00111-8
Li, J. Y., Tao, F., Wu, X. X., Tan, Y. Z., He, L., and Lu, H. (2015). Polymorphic Variations in Manganese Superoxide Dismutase (MnSOD) and Endothelial Nitric Oxide Synthase (eNOS) Genes Contribute to the Development of Type 2 Diabetes Mellitus in the Chinese Han Population. Genet. Mol. Res. 14 (4), 12993–13002. doi:10.4238/2015.October.21.20
Li, M. W., Li, X. X., Zhao, M. W., and Ji, L. N. (2010). Association of Genetic Polymorphism of Nitric Oxide Synthase and Diabetic Retinopathy (Chinese). Chin. J. Ocul. Fundus Dis. 26, 135–138.
Li, X. N., Yu, M. X., Wu, X. Y., Shui, H., and Xiao, J. S. (2011). Association of Endothelial Nitric Oxide Synthase Gene Glu298Asp Polymorphism with Diabetic Kidney Disease. J. Clin. Nephrol. 11 (8), 351–353.
Lin, S., Qu, H., and Qiu, M. (2002). Allele A in Intron 4 of ecNOS Gene Will Not Increase the Risk of Diabetic Nephropathy in Type 2 Diabetes of Chinese Population. Nephron 91 (4), 768. doi:10.1159/000065048
Luo, H., and Ning, Y. Y. (2003). Relationship between Gene Polymorphism of Endothelial Nitricoxide Synthase and Early Intervention of Patients with Diabetic Nephropathy. Chin. J. Clin. Rehabil. 7 (12), 1766–1767.
Luo, R., Fan, J. Y., Zhang, P., Liu, W., Gao, Z. H., and Zhu, T. H. (2006). A Study on Relationship between the Endothelial Consti-Tutive Nitric Oxide Synthase Gene Polymorphism and the Coronary Disease in Patients with Type 2 Diabetes. J. Chin. Phys. (1), 5–7.
Ma, J. R. (2003). Study on the Relationship between the Endothelial Constitutive Nitric Oxide Synthase (ecNOS) Gene Polymorphism and the Development of Diabetic Vascular Complication in Type 2 Diabetic Patients. Tianjin: Tianjin Medical University.
Ma, J. R., Yu, D. M., and Liu, D. (2007). The Relationship between the G894T Mutation of the Endothelial-Constitutive Nitric Oxide Synthase (ecNOS) and Diabetic Microangiopa-Thies. Zhong Guo Tang Niao Bing Za Zhi 15, 471–473.
Mackawy, A. M., Khan, A. A., and Badawy Mel-, S. (2014). Association of the Endothelial Nitric Oxide Synthase Gene G894T Polymorphism with the Risk of Diabetic Nephropathy in Qassim Region, Saudi Arabia-A Pilot Study. Meta Gene 2, 392–402. doi:10.1016/j.mgene.2014.04.009
Mantel, N., and Haenszel, W. (1959). Statistical Aspects of the Analysis of Data from Retrospective Studies of Disease. J. Natl. Cancer Inst. 22, 719–748.
Mehrab-Mohseni, M., Tabatabaei-Malazy, O., Hasani-Ranjbar, S., Amiri, P., Kouroshnia, A., Bazzaz, J. T., et al. (2011). Endothelial Nitric Oxide Synthase VNTR (Intron 4 A/b) Polymorphism Association with Type 2 Diabetes and its Chronic Complications. Diabetes Res. Clin. Pract. 91 (3), 348–352. doi:10.1016/j.diabres.2010.12.030
Moguib, O., Raslan, H. M., Abdel Rasheed, I., Effat, L., Mohamed, N., El Serougy, S., et al. (2017). Endothelial Nitric Oxide Synthase Gene (T786C and G894T) Polymorphisms in Egyptian Patients with Type 2 Diabetes. J. Genet. Eng. Biotechnol. 15 (2), 431–436. doi:10.1016/j.jgeb.2017.05.001
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G. PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA Statement. J. Clin. Epidemiol. 62 (10), 1006–1012. doi:10.1016/j.jclinepi.2009.06.005
Momeni, A., Chaleshtori, M. H., Saadatmand, S., and Kheiri, S. (2016). Correlation of Endothelial Nitric Oxide Synthase Gene Polymorphism (GG, TT and GT Genotype) with Proteinuria and Retinopathy in Type 2 Diabetic Patients. J. Clin. Diagn Res. 10 (2), OC32–OC35. doi:10.7860/JCDR/2016/14975.7291
Monti, L. D., Barlassina, C., Citterio, L., Galluccio, E., Berzuini, C., Setola, E., et al. (2003). Endothelial Nitric Oxide Synthase Polymorphisms Are Associated with Type 2 Diabetes and the Insulin Resistance Syndrome. Diabetes 52 (5), 1270–1275. doi:10.2337/diabetes.52.5.1270
Nagase, S., Suzuki, H., Wang, Y., Kikuchi, S., Hirayama, A., Ueda, A., et al. (2003). Association of ecNOS Gene Polymorphisms with End Stage Renal Diseases. Mol. Cell. Biochem. 244 (1-2), 113–118. doi:10.1007/978-1-4615-0247-0_16
Neugebauer, S., Baba, T., and Watanabe, T. (2000). Association of the Nitric Oxide Synthase Gene Polymorphism with an Increased Risk for Progression to Diabetic Nephropathy in Type 2 Diabetes. Diabetes 49 (3), 500–503. doi:10.2337/diabetes.49.3.500
Noiri, E., Satoh, H., Taguchi, J., Brodsky, S. V., Nakao, A., Ogawa, Y., et al. (2002). Association of eNOS Glu298Asp Polymorphism with End-Stage Renal Disease. Hypertension 40 (4), 535–540. doi:10.1161/01.hyp.0000033974.57407.82
Odeberg, J., Larsson, C. A., Råstam, L., and Lindblad, U. (2008). The Asp298 Allele of Endothelial Nitric Oxide Synthase Is a Risk Factor for Myocardial Infarction Among Patients with Type 2 Diabetes Mellitus. BMC Cardiovasc Disord. 8, 36. doi:10.1186/1471-2261-8-36
Ohtoshi, K., Yamasaki, Y., Gorogawa, S., Hayaishi-Okano, R., Node, K., Matsuhisa, M., et al. (2002). Association of (-)786t-C Mutation of Endothelial Nitric Oxide Synthase Gene with Insulin Resistance. Diabetologia 45 (11), 1594–1601. doi:10.1007/s00125-002-0922-6
Papazafiropoulou, A., Papanas, N., Melidonis, A., and Maltezos, E. (2016). Family History of Type 2 Diabetes: Does Having a Diabetic Parent Increase the Risk? Cdr 13 (1), 19–25. doi:10.2174/1573399812666151022143502
Pulkkinen, A., Viitanen, L., Kareinen, A., Lehto, S., Vauhkonen, I., and Laakso, M. (2000). Intron 4 Polymorphism of the Endothelial Nitric Oxide Synthase Gene Is Associated with Elevated Blood Pressure in Type 2 Diabetic Patients with Coronary Heart Disease. J. Mol. Med. Berl. 78 (7), 372–379. doi:10.1007/s001090000124
Rahimi, Z., Rahimi, Z., Shahvaisi-Zadeh, F., Sadeghei, S., Vessal, M., and Yavari, N. (2013). eNOS 4a/b Polymorphism and its Interaction with eNOS G894T Variants in Type 2 Diabetes Mellitus: Modifying the Risk of Diabetic Nephropathy. Dis. Markers 34 (6), 437–443. doi:10.3233/DMA-13098810.1155/2013/512107
Raina, P., Sikka, R., Gupta, H., Matharoo, K., Bali, S. K., Singh, V., et al. (2021). Association of eNOS and MCP-1 Genetic Variants with Type 2 Diabetes and Diabetic Nephropathy Susceptibility: A Case-Control and Meta-Analysis Study. Biochem. Genet. 59, 966–996. doi:10.1007/s10528-021-10041-2
Ren, T., Xiang, S. S., and Liu, L. (2003). An Association of Dia-Betic Neuropathy with Polymorphisms of eNOS, PON1,RAGE and ALR2 Genes. Shang Hai Yi Yao 26, 24–27.
Ritt, M., Ott, C., Delles, C., Schneider, M. P., and Schmieder, R. E. (2008). Impact of the Endothelial Nitric Oxide Synthase Gene G894T Polymorphism on Renal Endothelial Function in Patients with Type 2 Diabetes. Pharmacogenet Genomics 18 (8), 699–707. doi:10.1097/FPC.0b013e32830500b1
Rizvi, S., Raza, S. T., Rahman, Q., Eba, A., Zaidi, Z. H., and Mahdi, F. (2019). Association of Endothelial Nitric Oxide Synthase (eNOS) and Norepinephrine Transporter (NET) Genes Polymorphism with Type 2 Diabetes Mellitus. Mol. Biol. Rep. 46 (5), 5433–5441. doi:10.1007/s11033-019-04998-y
Sandrim, V. C., de Syllos, R. W., Lisboa, H. R., Tres, G. S., and Tanus-Santos, J. E. (2006). Endothelial Nitric Oxide Synthase Haplotypes Affect the Susceptibility to Hypertension in Patients with Type 2 Diabetes Mellitus. Atherosclerosis 189 (1), 241–246. doi:10.1016/j.atherosclerosis.2005.12.011
Santos, K. G., Crispim, D., Canani, L. H., Ferrugem, P. T., Gross, J. L., and Roisenberg, I. (2011). Association of eNOS Gene Polymorphisms with Renal Disease in Caucasians with Type 2 Diabetes. Diabetes Res. Clin. Pract. 91 (3), 353–362. doi:10.1016/j.diabres.2010.12.029
She, C., Yang, X., Gu, H., Deng, Y., Xu, J., Ma, K., et al. (2015). The Association of Variable Number of Tandem Repeats Polymorphism in the Endothelial Nitric Oxide Synthase Gene and Diabetic Retinopathy. Zhonghua Yan Ke Za Zhi 51 (5), 338–343.
Shin Shin, Y., Baek, S. H., Chang, K. Y., Park, C. W., Yang, C. W., Jin, D. C., et al. (2004). Relations between eNOS Glu298Asp Polymorphism and Progression of Diabetic Nephropathy. Diabetes Res. Clin. Pract. 65 (3), 257–265. doi:10.1016/j.diabres.2004.01.010
Simmons, J., Gregory, J. M., Kumah-Crystal, Y. A., and Simmons, J. H. (2009). Mitigating Micro- and Macro-Vascular Complications of Diabetes Beginning in Adolescence. Vhrm 5, 1015–1031. doi:10.2147/vhrm.s4891
Sun, H-Y., Yang, M., Liu, S., Wang, C., Zhang, Q., and Chen, M. (2004). Study on the Correlation of the Polymorphisms of Endothelial Nitric Oxide Synthase Gene with Type 2 Diabetes Mellitus and Diabetic Nephropathy. Chin. J. Prev. Contr Chron. Non-Commun Dis. 12 (3), 101–103.
Suzuki, H., Nagase, S., Kikuchi, S., Wang, Y., and Koyama, A. (2000). Association of a Missense Glu298Asp Mutation of the Endothelial Nitric Oxide Synthase Gene with End Stage Renal Disease. Clin. Chem. 46 (11), 1858–1860. doi:10.1093/clinchem/46.11.1858
Szabó, G. V., Kunstár, A., and Acsády, G. (2009). Methylentetrahydrofolate Reductase and Nitric Oxide Synthase Polymorphism in Patients with Atherosclerosis and Diabetes. Pathol. Oncol. Res. 15 (4), 631–637. doi:10.1007/s12253-009-9163-z
Thaha, M., Pranawa, Yogiantoro, M., Sutjipto, , Sunarjo, , Tanimoto, M., et al. (2008). Association of Endothelial Nitric Oxide Synthase Glu298Asp Polymorphism with End-Stage Renal Disease. Clin. Nephrol. 70 (2), 144–154. doi:10.5414/cnp70144
Thorlund, K., Wetterslev, J., Brok, J., Imberger, G., and Gluud, G. (2011). User Manual for Trial Sequential Analysis (TSA). Copenhagen, Denmark: Copenhagen Trial Unit, Centre for Clinical Intervention Research.
Tsutsui, M., Shimokawa, H., Morishita, T., Nakashima, Y., and Yanagihara, N. (2006). Development of Genetically Engineered Mice Lacking All Three Nitric Oxide Synthases. J. Pharmacol. Sci. 102 (2), 147–154. doi:10.1254/jphs.cpj06015x
Ukkola, O., Erkkilä, P. H., Savolainen, M. J., and Kesäniemi, Y. A. (2001). Lack of Association between Polymorphisms of Catalase, Copper-Zinc Superoxide Dismutase (SOD), Extracellular SOD and Endothelial Nitric Oxide Synthase Genes and Macroangiopathy in Patients with Type 2 Diabetes Mellitus. J. Intern Med. 249 (5), 451–459. doi:10.1046/j.1365-2796.2001.00828.x
Veldman, B. A., Spiering, W., Doevendans, P. A., Vervoort, G., Kroon, A. A., de Leeuw, P. W., et al. (2002). The Glu298Asp Polymorphism of the NOS 3 Gene as a Determinant of the Baseline Production of Nitric Oxide. J. Hypertens. 20 (10), 2023–2027. doi:10.1097/00004872-200210000-00022
Wacholder, S., Chanock, S., Garcia-Closas, M., El Ghormli, L., and Rothman, N. (2004). Assessing the Probability that a Positive Report Is False: an Approach for Molecular Epidemiology Studies. J. Natl. Cancer Inst. 96, 434–442. doi:10.1093/jnci/djh075
Wakefield, J. (2007). A Bayesian Measure of the Probability of False Discovery in Genetic Epidemiology Studies. Am. J. Hum. Genet. 81, 208–227. doi:10.1086/519024
Wang, S. L. (2005). Study of the Susceptibility Genes of Type 2 Diabetic Nephropathy in Northern Chinese. Shandong: Shandong University.
Wang, X. L., Mahaney, M. C., Sim, A. S., Wang, J., and Wang, J. (1997). Genetic Contribution of the Endothelial Constitutive Nitric Oxide Synthase Gene to Plasma Nitric Oxide Levels. Arterioscler. Thromb. V. asc Biol. 17, 3147–3153. doi:10.1161/01.atv.17.11.3147
Wang, Y., Kikuchi, S., Suzuki, H., Nagase, S., and Koyama, A. (1999). Endothelial Nitric Oxide Synthase Gene Polymorphism in Intron 4 Affects the Progression of Renal Failure in Non-Diabetic Renal Diseases. Nephrol. Dial. Transplant. 14 (12), 2898–2902. doi:10.1093/ndt/14.12.2898
Wells, B. S. G. A., O’Connell, D., Peterson, J., Welch, V., and Losos, M. P. (2014). The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottawa: Professor GA Wells.
Wetterslev, J., Thorlund, K., Brok, J., and Gluud, C. (2009). Estimating Required Information Size by Quantifying Diversity in Random-Effects Model Meta-Analyses. Bmc Med. Res. Methodol. 9, 86. doi:10.1186/1471-2288-9-86
Wu, Y., Ge, J., and Sheng, J. (2007). The Ralationship between Polymorphism of Intron 4 (4b/a) of the Endothelial Con-Stitutive Nitric Oxide Synthase Gene and Type 2 Diabe-Tes Millitus Complicated with Coronary Heart Disease. He Bei Yi Yao 29, 539–541.
Yigit, S., Nursal, A. F., Uzun, S., Rustemoglu, H., Dashatan, P. A., Soylu, H., et al. (2020). Impact of Endothelial NOS VNTR Variant on Susceptibility to Diabetic Neuropathy and Type 2 Diabetes Mellitus. Curr. Neurovasc Res. 17 (5), 700–705. doi:10.2174/1567202617999201214230147
Yu, L. H., Tian, G., Cai, J., and Wang, X. (2009). Study on Rela-Tionship between the Endothelial Constitutive Nitric Oxide Synthase Gene Polymorphism and Hyperuricemia in Type 2 Diabetes Mellitus. Tian Jin Yi Yao 37, 347–349.
Zhang, M., Liu, H., Yang, H. Y., Wang, Y. M., Song, D. P., and Li, H. (2005). Relationship of Endothelial Nitric Oxide Synthase (eNOS) 4a/b Gene Polymorphism with Diabetic Nephropathy. Chin. J. Diabetes 11 (4), 284–285.
Zhang, M., Song, D. P., Liu, H., Wang, Y. M., Duan, Y., and Li, H. (2003). The Polymorphism of eNOS Gene Is Associated with Higher Triglyceride Levels. Chin. J. Diab. (1), 37–39.
Zhang, X., Yang, Z., and Chen, X. (2017). Endothelial Nitric Oxide Synthase 27VNTR (4b/4a) Gene Polymorphism and the Risk of Diabetic Microvascular Complications in Chinese Populations. Cell. Mol. Biol. (Noisy-le-grand) 63 (10), 54–58. doi:10.14715/cmb/2017.63.10.8
Keywords: eNOS, polymorphism, T2DM, meta-analysis, BFDP, FPRP
Citation: Wang D, Liu L, Zhang C, Lu W, Wu F and He X (2022) Evaluation of Association Studies and Meta-Analyses of eNOS Polymorphisms in Type 2 Diabetes Mellitus Risk. Front. Genet. 13:887415. doi: 10.3389/fgene.2022.887415
Received: 01 March 2022; Accepted: 27 May 2022;
Published: 27 June 2022.
Edited by:
Manal S. Fawzy, Suez Canal University, EgyptReviewed by:
Rami M. Elshazli, Horus University, EgyptUmamaheswaran Gurusamy, University of California San Francisco, United States
Copyright © 2022 Wang, Liu, Zhang, Lu, Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wensheng Lu, THdzMjYxMzY3NkBzb2h1LmNvbQ==; Feifei Wu, MTM5OTQ2MTAwNjZAMTYzLmNvbQ==; Xiaofeng He, MzkzMTIwODIzQHFxLmNvbQ==
 Di Wang
Di Wang Liangshu Liu1
Liangshu Liu1 Xiaofeng He
Xiaofeng He