
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 16 June 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.887217
This article is part of the Research TopicGenomic Selection and Characterization in CerealsView all 20 articles
 Ying-Hua Pan1*†
Ying-Hua Pan1*† Lei Chen1†
Lei Chen1† Hai-Feng Guo2†
Hai-Feng Guo2† Rui Feng1†
Rui Feng1† Qi-Jin Lou2†
Qi-Jin Lou2† Muhammad Abdul Rehman Rashid3
Muhammad Abdul Rehman Rashid3 Xiao-Yang Zhu2
Xiao-Yang Zhu2 Dong-Jin Qing1
Dong-Jin Qing1 Hai-Fu Liang1
Hai-Fu Liang1 Li-Jun Gao4
Li-Jun Gao4 Cheng-Cui Huang1
Cheng-Cui Huang1 Yan Zhao5*
Yan Zhao5* Guo-Fu Deng1*
Guo-Fu Deng1*The NB-ARC (nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4) gene family plays a critical role in plant development. However, our understanding of the mechanisms of how NB-ARC genes regulate plant development in the plant panicle is still limited. Here, we subjected 258 NB-ARC genes in rice to genome-wide analysis to characterize their structure, function, and expression patterns. The NB-ARC genes were classified into three major groups, and group II included nine subgroups. Evolutionary analysis of NB-ARC genes in a dicotyledon plant (Arabidopsis thaliana) and two monocotyledonous plants (Oryza sativa L. and Triticum aestivum) indicated that homologous genome segments were conserved in monocotyledons and subjected to weak positive selective pressure during evolution. Dispersed and proximal replication events were detected. Expression analysis showed expression of most NB-ARC genes in roots, panicles, and leaves, and regulation at the panicle development stage in rice Ce253. The GNP12 gene encodes RGH1A protein, which regulates rice yield according to panicle length, grain number of panicle, and grain length, with eight major haplotypes. Most members of NB-ARC protein family are predicted to contain P-loop conserved domains and localize on the membrane. The results of this study will provide insight into the characteristics and evolution of NB-ARC family and suggest that GNP12 positively regulates panicle development.
NB-ARC (nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4) proteins are critical regulators of signaling pathways and play important roles in effector recognition and signal transduction in healthy plant growth and development (Jacquemin et al., 2014; Wen et al., 2017; Chen et al., 2018). NB-ARC proteins contain an NB-ARC domain that is proposed to act as a molecular switch, with a nucleotide-binding (NB) subdomain including a P-loop, a C-terminal extension that forms a four-helix bundle (ARC1), and a winged-helix fold (van der Biezen and Jones, 1998; Albrecht and Takken, 2006). The NB subdomain can bind and hydrolyze ATP in vitro (Tameling et al., 2002). NB-ARC belongs to the STAND (signal transduction ATPases with numerous domains) family of NTPases, and is proposed to work as an NTP-hydrolyzing switch by binding and hydrolyzing NTP and regulating signal transduction by conformational changes (Leipe et al., 2004). Recent studies revealed NB-ARC proteins with similar central nucleotide-binding-ARC domain architectures act in metazoan innate immunity and programmed cell death pathways (Slootweg et al., 2009). For example, NB-ARC genes share homology with human APAF-1 and C. elegans CED-4, proteins that regulate cell death (Yildirim-Ersoy et al., 2011). RPP1A belongs to the NB-ARC gene family and reduces plant growth with broad-spectrum resistance to virulent strains of H. parasitica in Arabidopsis (Michael Weaver et al., 2006). Constitutive expression of TIR-NB-ARC-LRR gene VpTNL1 in Arabidopsis resulted in either a wild-type or dwarf phenotype (Wen et al., 2017). NB-ARC genes might contribute to R. glutinosa consecutive monoculture problems (Chen et al., 2018). Domains can work together as a platform to mediate downstream signal transduction events (Lukasik and Takken, 2009). In wheat, many NB-ARC gene clusters include highly similar genes likely formed by tandem duplications (Andersen et al., 2020). Some NB genes have been reported to be regulated in response to morphological development (Igari et al., 2008). AhRAF4 was observed to correlate with morphological changes in development in maize (Dolezal et al., 2014). NB-ARC-containing sequences in wheat exhibit maximum homology with proteins from indica and Brachypodium distachyon, and distributions of NB-ARC sequences are balanced among the three wheat sub-genomes (Chandra et al., 2017). In indica, Oryza glaberrima (Slootweg et al., 2009), and Oryza brachyantha (FF), conserved NB-ARC genes were subjected to strong purifying selection but selection was more relaxed for expanded homologous genes (Jacquemin et al., 2014). NB-ARC protein function has been studied in wheat, rice, and Arabidopsis, but the mechanism of NB-ARC protein function in rice panicle development remains poorly understood.
Rice is one of the most important cereal crops in the world and feeds half population of the world (Zhao et al., 2015). In recent years, many important genes controlling rice grain yield have been isolated and functionally characterized. LP1 encodes a Remorin_C-containing protein of unknown function and the LP1 allele of Xiushui79 leads to reduced panicle length (Liu et al., 2016). Grain Number per Panicle1 (GNP1), Rice GA20ox1 encoding a cytokinin (Tameling et al., 2002), and the biosynthesis gene for gibberellins upregulate cytokinin activity to increase grain number and grain yield in rice (Wu et al., 2016). GW8 encodes an SBP-domain transcription factor that regulates grain width by binding directly to the GW7 promoter to repress its expression (Wang et al., 2015). OsMKK3 encodes a MAP kinase that controls rice grain size and chalkiness (Pan et al., 2021). Although few studies have focused on NB-ARC proteins in rice, these important plant proteins likely have multiple regulatory roles. For example, the gain-of-function mutation of NB-ARC protein RLS1 (Rapid Leaf Senescence1) causes high-light-dependent HR-like cell death in rice (Wang et al., 2020). Overall, a greater understanding of mechanisms underlying grain yield will strengthen our understanding of regulatory mechanisms for these traits and facilitate the breeding of crop varieties with high-yield potential.
In this study, members of the NB-ARC gene family were identified from analysis of the rice genome. The motif composition and gene structures of these genes were systematically analyzed, and tandem duplication and gene replication events were identified. Collinear relationships between rice, wheat, and Arabidopsis were compared. The expression levels of 11 subgroups of NB-ARC genes in different tissues and panicle development of rice Ce253 were analyzed by RNA-Seq and the expression levels of 18 genes were measured via qPCR. Functional analysis was performed of a selected NB-ARC gene, GNP12. This gene serves as a positive regulator in panicle development and panicle length. The results of this research provide expand our understanding of the function of the NB-ARC gene family and provide guidance for future efforts to improve rice breeding.
The rice variety Ce253, widely planted in Guangxi Province, China, was selected for this study and was obtained from the Rice Research Institute, Guangxi Academy of Agricultural Sciences. Rice was planted under natural field conditions at the Rice Research Institute of Guangxi Academy of Agricultural Sciences, Nanning, China in 2020. The distance between plants within rows was 16 cm, and the distance between plants in separate rows was 20 cm. Field management, including irrigation, fertilizer application, and pest control, were performed according to normal agricultural practices. Fully filled grains were subjected to grain width, length, and weight measurement with a Wanshen SC-G automatic seed test system. All trait measurements were repeated at least three times.
Genome-wide identification of NB-ARC genes from three species of monocotyledonous and dicotyledonous plants was performed. Hidden Markov Model (HMM) (Punta et al., 2012) (version 3.0) analysis was used for the search. HMM profiles of NB-ARC genes (PF00931.2) were obtained from the Pfam database (http://pfam.xfam.org/) with an e-value≤1e-3. The results of the HMMER sequence alignment were screened to remove protein sequences that were at least 45% longer than the length of the HMM model domain, while retaining the longest protein sequence in the variable shear. Simple Modular Architecture Research Tool (SMART) (version 8.0) (http://smart.embl-heidelberg.de) was used for further analysis with all non-redundant protein sequences (Schultz et al., 1998). Finally, 258 NB-ARC genes models were identified in the rice genome for further analysis. The basic information of the identified NB-ARC proteins was obtained using the tools at the ExPasy website (http://web.expasy.org/protparam/). CDS coordinate information is listed in Supplementary Table S1. Gene family data were analyzed by Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China). A BLASTP search of the NCBI nonredundant protein database was used to assign the NB-ARC domains. Multiple alignments of NB-ARC domains with 11 different plant NB-ARC domains of 11 subgroups were performed using MEME. The submotifs were analyzed through http://weblogo.berkeley.edu/logo.cgi. The 3D model of the NB-ARC genes was predicted by Jpred and SWISS-MODEL. We used the intersection results of Pfam (pfoo31, - e 1e-20) + smart (- e 0.1 -- dome 0.01) and blast (- e 1e-14 and identity >28%) as the identified homologous genes.
The exon-intron structural information for the collected rice NB-ARC genes was acquired from reference genome annotation files (Os-Nipponbare-Reference-IRGSP-1.0 pseudomolecules) to compare genomic and coding sequences (http://rice.uga.edu/). The cDNAs were aligned with their corresponding genomic DNA sequences. To map all NB-ARC genes, the chromosome distribution and conserved regions were confirmed by analysis of reference genome annotation files. A chromosome distribution diagram was drawn using the SVG package in Perl. Gene duplication in rice was identified using the replicate gene classifier program of MCScanX software (Wang et al., 2012). All protein-coding sequences were aligned using blastp, and the alignment results were used as input files for MCScanX software to predict gene replication. A gene was identified as a replication gene according to e-value<1e-5 or e-value<1e-10. Five replication events were identified: segmental, tandem, proximal, and decentralized (dispersed).
Conserved motifs in the gene family sequences were identified using the MEME program (http://alternate.meme-suite.org/tools/meme) with statistical significance (Bailey et al., 2015). The MEME program was run with default settings, maximum motif search value of 15, and an optimum motif width of 10–100 amino acid residues. A phylogenetic tree was constructed with the neighbor-joining algorithm in MEGA (version 7.0) with bootstrap test of 1,000 times and drawn with iTOL (https://itol.embl.de/) (Kumar et al., 2016). ML (maximum likelihood method) evolutionary tree was showed in Supplementary Figure S1. Genome annotations and corresponding protein sequences were downloaded from EnsemblPlants (Brassica_rapa. IVFCAAS v1.36, Brassica_oleracea.v2.1.36, Arabidopsis_thaliana.TAIR10) and GenoScope (Brassica_napus.annotation_v5). The gene structure and motifs were analyzed by systematic evolutionary relationships. Synteny detection was performed by McScanX and drawn with Circos software.
We compared the Ka/Ks ratios as a proxy of the selective pressures acting on gene pairs for reciprocal best match gene pairs from Arabidopsis, rice, and wheat. Ka/Ks was calculated as the ratio of the number of nonsynonymous substitutions per non-synonymous site (Ka) in a period to the number of synonymous substitutions per synonymous site (Ks) in the same period. Ka, Ks, and Ka/Ks values are based on coding sequence alignment and calculated using the KaKs_calculator software package based on the Nei and Gojobori model (Nei and Gojobori, 1986). Positively or negatively selected sites were identified based on Ka/Ks ratios with a confidence interval for each ratio given by a p value and an adjusted p value (Adj.Pval) from multiple comparisons (Massingham and Goldman, 2005). Homologous genes with a Ka/Ks ratio above 1 were under strong positive selection, between 0.5 and 1 were considered to be under weak positive selection, and below 0.1 were considered to be under negative selection (purifying selection).
To assess subcellular localization, sgRNA-Cas9 plant expression vectors were constructed as described previously (Mao et al., 2013). The targeting sequence (5′- TAGTCGACGACAATGCTGCCAGG-3′), corresponded to +400 to +422 within the third exon encoding the C-terminal end of GNP12 (starting at amino acid residue 134). To construct the GFP plasmids, the cDNAs of LOC_Os101g458510 were amplified from Nipponbare and the cDNA for LOC_Os12g36720 was amplified from Ce253. The cDNAs were cloned into the pBWA(V)HS-ccdb-GLos-GFP vector to generate the insertion. Plasmids CaMV35S:GFP, CaMV35S:LOC_Os101g458510-GFP and CaMV35S:GNP12-GFP were transformed into rice leaf protoplasts for subcellular localization. GFP was excited with a 488 nm laser and imaging was performed.
Panicles of Ce253 of different lengths (3, 5, 10, 15, 20, and 25 cm) at the booting stage were flash-frozen in liquid nitrogen. Total RNA was extracted from these tissues using a EasyPure® Plant RNA Kit (Trans, Catalog no. ER301-01). RNA purity was determined by assaying 1 µl of total RNA extract on a NanoDrop 1,000. We measured the optical density (OD) ratio between 260 and 280 nm from samples, where pure RNA eluted in H20 (pH 7.0–8.5) or TE (pH 8.0) is expected to exhibit a ratio of 2.0–2.1. Total RNA (2.0 μg) was used for cDNA synthesis with a PrimeScript™ RT Reagent Kit with gDNA Eraser (TransScript® II One-Step RT-PCR SuperMix, Tran, Catalog no. AH411-02). The resulting cDNA samples were diluted five-fold and used as templates for qRT-PCR using a TransStart® Green qPCR SuperMix and a CFX96 RealTime system (Bio-RAD, Hercules, CA, United States) following the manufacturer’s instructions. The qRT-PCR reactions were performed as 10 µl mixtures containing 5 µl of 2× Green qPCR MasterMix, 1 µl of cDNA, 0.25 µl of each primer (10 µM), and 3.5 µl of ddH2O. Amplification steps were 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s, followed by 65°C for 5 s, 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. Each experiment was repeated at least three times. The qRT-PCR analysis was performed using the ΔΔCt method. Details on gene-specific primers used for real-time PCR are provided in Supplementary Table S1. The ubiquitin gene (LOC_Os03g13170) (Chen et al., 2019) was used as a control (*p < 0.05; **p < 0.01; Student’s t-test). RNA samples used for RNA-seq analysis were prepared from different panicles of Ce253 grown under normal field conditions with three biological replicates. RNA library sequencing was performed on an Illumina Hiseq™ 2,500/4,000 platform by Majorbio. Sequence analysis was performed using the method provided by Majorbio (http://www.majorbio.com/). RNA-Seq data for NB-ARC genes in different tissues were obtained from http://expression.ic4r.org/.
The single-nucleotide polymorphisms (SNPs) of 827 accessions used for haplotype analysis were acquired from the 3K rice genomes (3K-RG) dataset. The grain length data for 827 accessions were downloaded from the Rice SNP-Seek Database. One-way ANOVA followed by Duncan’s new multiple-range test were performed using SPSS 21.0 software. The genomic sequences of 1,400 cultivated and 58 wild accessions were obtained from the 3K rice genomes (3K-RG) dataset and OryzaGenome (http://viewer.shigen.info/oryzagenome/), respectively, and were used to construct a minimum spanning tree for RGH1A. Arlequin version 3.5 software was used to calculate a haplotype network and the distance matrix output was used in Hapstar-0.6 to draw a minimum spanning tree. The average nucleotide diversity (π) and Tajima’s D for each subpopulation in RGH1A and 40-kb flanking regions were calculated using DnaSP 5.10 software (Librado and Rozas, 2009). The nucleotide diversity curves were generated using 60 bp window and 15 bp step length.
A total of 258 rice NB-ARC genes were identified in the Nipponbare genome (Supplementary Table S2). The lengths of the CDS for these genes range from 624 (LOC_Os02g27680) to 7,773 (LOC_Os06g41690) bp. Analysis revealed that 49.22% of these NB-ARC genes contain 2–6 introns and 86.04% NB-ARC genes contain 1–4 exons (Supplementary Table S2). Most of the NB-ARC genes have a long CDS and show a highly conserved structure (Figure 1). To explore the expansion of NB-ARC family members in rice, these sequences were used to generate an unrooted phylogenetic tree. The NB-ARC family members were divided into three major groups. Group I contained 29 genes, Group II contained 226 genes, and group III contained only three genes. The NB-ARC genes in group II were further classified into nine subgroups (groups IIa, IIb, IIc, IId, IIe, IIf, IIg, IIh, and IIi). Of these, the two groups with the most members were IIf with 35 genes and IIi with 37 genes. Group IIc contained the fewest number of genes, 13. A total of 15 motifs were identified by MEME, and two of these, M2 and M6, were present in 98.83 and 97.28%, respectively, of the NB-ARC genes. Genes on a single branch contained similar numbers of introns and similar distribution and number of motifs. In a given subtribe of genes, the positions of motifs were highly conserved. In group IIf, the genes contained 1-4 introns and 2–12 exons. Most genes in this group share 10 similar motifs (motifs 1, 2, 3, 4, 6, 8, 9, 10, 11, and 12) (Figure 1). Group IIf was representative in motifs and numbers of NB-ARC genes. Overall, NB-ARC genes were unevenly distributed across all 12 chromosomes, with 29 (11.24%), 61 (23.64%), and 34 (13.18%) genes located on Chr 8, 11, and 12, respectively. (Figure 2A). Gene analysis revealed duplication events including segmental, tandem, proximal, and dispersed, but not singleton duplications. Dispersed (110) and proximal replication 82) most frequently occurred in NB-ARC genes across all chromosomes (Figure 2B). Segmental replication was detected only on Chr2, Chr4, Chr7, and Chr11 (Figure 2B). Pairwise replication of genes was also detected in NB-ARC genes (Figure 2C). The genes in Group IIf, mapped to eight chromosomes, with dispersed, tandem, and proximal replication. In general, genes in subgroup IIf of group II showed typical structure with main motif and contain more genes, and analysis suggested that replication events were the main driving force of NB-ARC evolution.
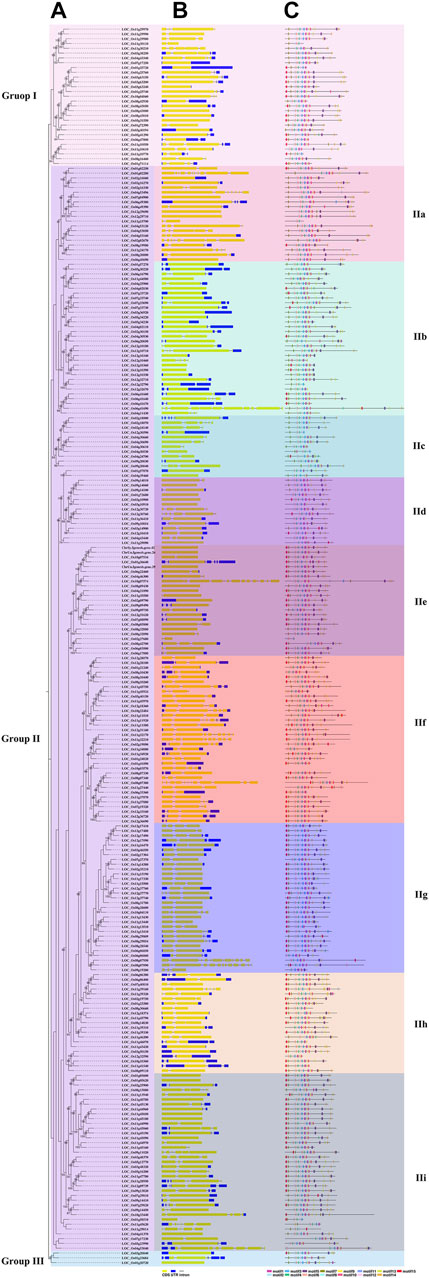
FIGURE 1. Phylogenetic tree representing the relationships among 258 genes of rice. Phylogenetic tree (A), exon/intron structure. Number is bootstrap values. (B), and motif composition (C) of NB-ARC genes in rice. Neighbor-Joining (NJ)-Phylogenetic trees shown in a and b were prepared using the same methods. The widths of Gy bars at the bottom indicate relative lengths of genes and proteins. Yellow boxes and blue lines in b represent exons and introns, respectively. Different boxes in c represent different motifs. Different background colors represent different groups and subgroups.
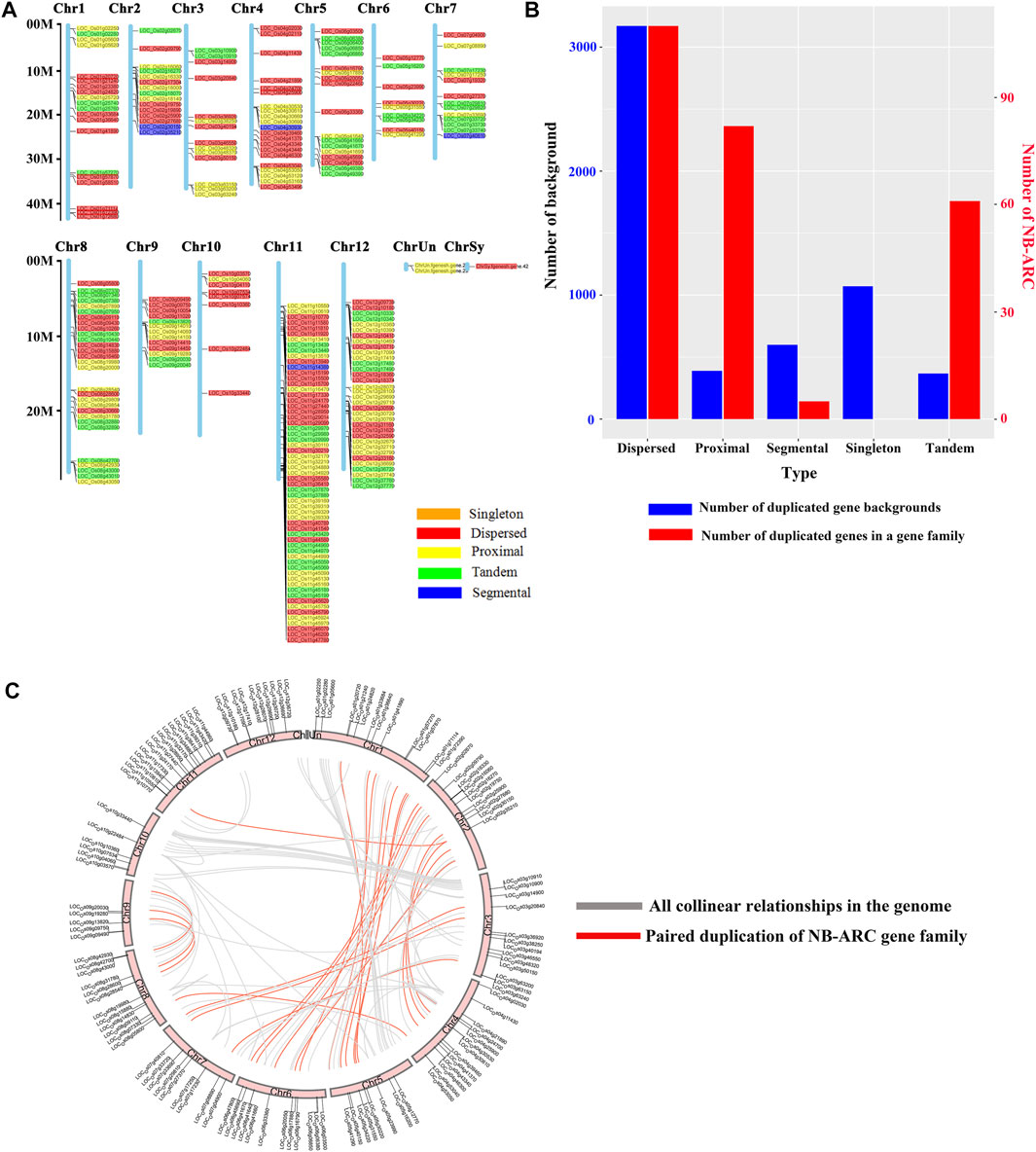
FIGURE 2. Chromosomal distribution of NB-ARC gene duplication events. (A) Distribution of NB-ARC genes with indicated duplication types. (B) Numbers of duplicated genes for different duplication types. (C) Collinearity of replicative genes in the protein family.
To study the relationships between the NB-ARC genes of rice and other model plant species, including gramineous plants, we performed cluster analysis (Figure 3A). The NB-ARC genes of a dicotyledon plant, Arabidopsis, and two monocotyledonous plants, rice and wheat, were compared. Pfam was used to compare 1,052 and 50 NB-ARC genes in wheat and Arabidopsis, respectively. Next, based on the highly conserved NB-ARC domains of rice, wheat, and Arabidopsis, a phylogenetic tree was built. The 50 genes of Arabidopsis belonged to a single subgroup, and every subgroup contained rice and wheat genes (Figure 3A). Within each subgroup, genes from the homologous chromosome group generally clustered into a clade (Figure 3A); for example, LocOs12g36690, LocOs12g36720, and LocOs12g33160 were clustered in a subgroup. To further study the relationships between the NB-ARC genes of rice and other plants, we next performed whole genome synteny analysis (Figure 3B). A total of 65 homologous genome segments were distributed on 12 chromosomes in wheat (Supplementary Table S3). The highest number of homologous genome segments, 19, mapped to the syntenic locus in chromosome 4 and wheat chromosomes 2A, 2B, 2D. Two homologous genome segments mapped to chromosome 5 in Arabidopsis (Figure 3B) For further evolutionary analysis, the Ka, Ks, and Ka/Ks values of homologous gene pairs were calculated based on the comparative synteny map (Figure 3C). Ka/Ks ratios of 2,571 gene pairs were evaluated in rice and wheat in protein coding genes. Fifty-two gene pairs had Ka/Ks > 1, 2,412 gene pairs had 0.5 < Ka/Ks < 1, and no gene pairs had <0.1. Ka/Ks ratios were similarly evaluated for 70 gene pairs in rice and Arabidopsis thaliana to examine in protein coding genes. Five gene pairs had Ka/Ks > 1, 59 gene pairs had 0.5 < Ka/Ks < 1, no gene pairs had <0.1, and the remaining had no data so could not be calculated. In rice, 15 gene pairs had Ka/Ks > 1, 431 gene pairs had 0.5 < Ka/Ks < 1, no gene pairs had <0.1, and five gene pairs had 0.1 < Ka/Ks < 0.5 (Supplementary Table S4). Because the majority of homologous NB-ARC gene pairs had 0.5 < Ka/Ks < 1, this suggests that the NB-ARC gene family in these three plant species experienced weak positive selective pressure during evolution. Evolution of the few NB-ARC genes in rice was governed by strong constraints that may have contributed to their structural and functional stability (Figure 2C and Supplementary Table S5). Interestingly, some homologous genome segments mapped between rice and wheat were not observed between rice and Arabidopsis, which may indicate that these homologous pairs formed after the divergence of dicotyledonous and monocotyledonous plants. Overall, the analysis shows that the NB-ARC gene family is highly conserved in monocotyledons and homologous genome segments suggest that segmental duplications may be the main cause of the extension of this gene family. Multiple copies of genes have arisen during the evolution of dicotyledons and monocotyledons.
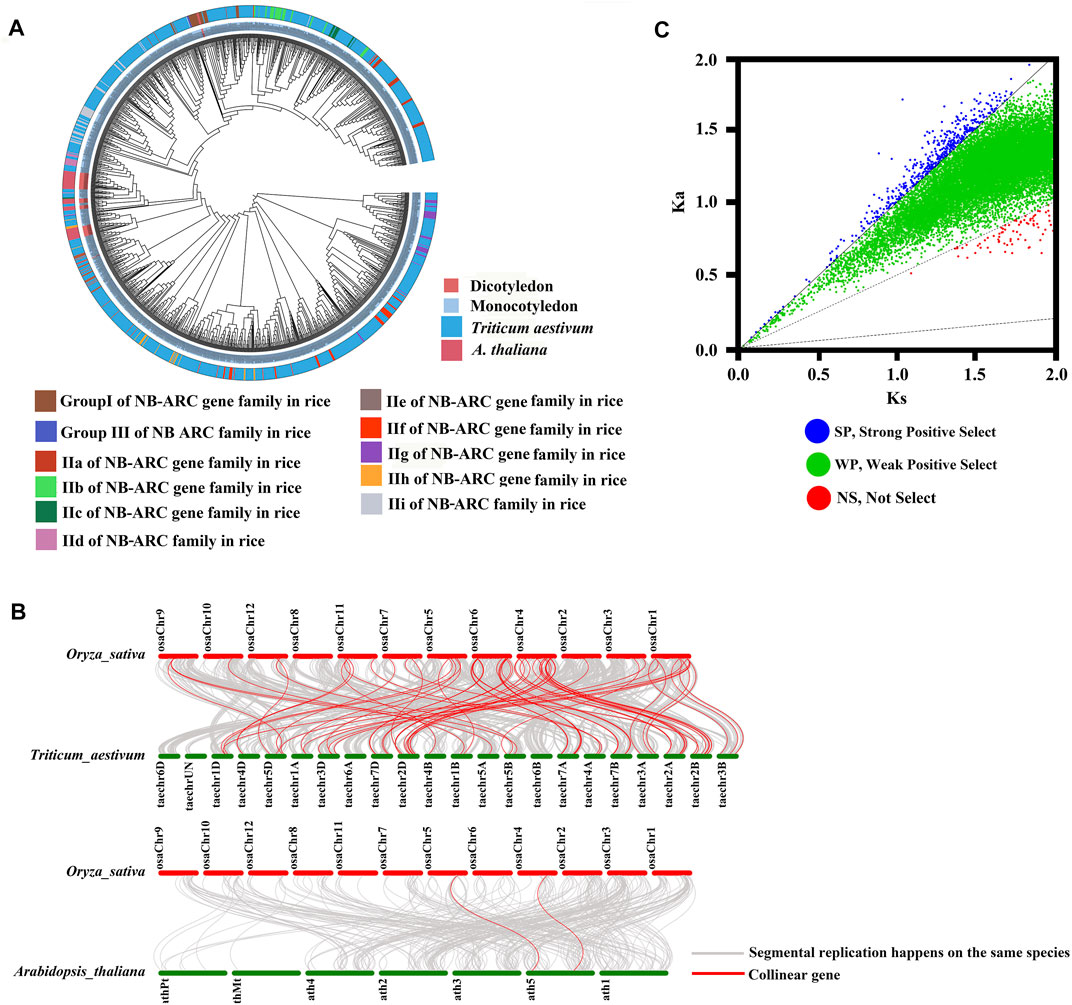
FIGURE 3. Phylogenetic, Ka/Ks, and synteny analysis of NB-ARC genes from rice, Arabidopsis, and Triticum aestivum. (A) Group I, Group II, Group III, IIa, IIb, IIc, IId, IIe, IIf, IIg, IIh, and IIi were identified in the rice genome; 1,052 and 50 NB-ARC genes were identified in Triticum aestivum and Arabidopsis thaliana. Genes from rice, Arabidopsis, Triticum aestivum are indicated with the prefixes Os, At, and Tran, respectively. (B) Synteny analysis of Oryza sativa, Arabidopsis, and Triticum aestivum. Gray lines in the background indicate collinear blocks of plant genomes. Different color bars represent the chromosomes of different species. The chromosome number is labeled at the top or bottom of each chromosome. (C) Ka/Ks analysis of NB-ARC genes from rice, Arabidopsis and Triticum aestivum.
To study the spatial and temporal expression patterns of NB-ARC genes, transcriptomic profiling was profiled across eight different tissues (anther, callus, leaf, panicle, pistil, root, seed, and shoot) to analyze the role of these genes in rice organ development according to the Rice Expression Database (Supplementary Table S6 and Figure 4A). In our findings, 23 genes exhibited no expression in any tissues, five genes showed expression in eight tissues, and the remaining 88 genes showed FPKM values higher than 1 in more than three tissues. LOC_Os02g25900 and LOC_Os12g36720 exhibited FPKM values higher than 1 in anther, callus, leaf, panicle, pistil, root, seed, and shoot. Expression of these genes varied for different tissues, with expression of 141 genes in root, 94 genes in panicle, and only nine genes in anther.
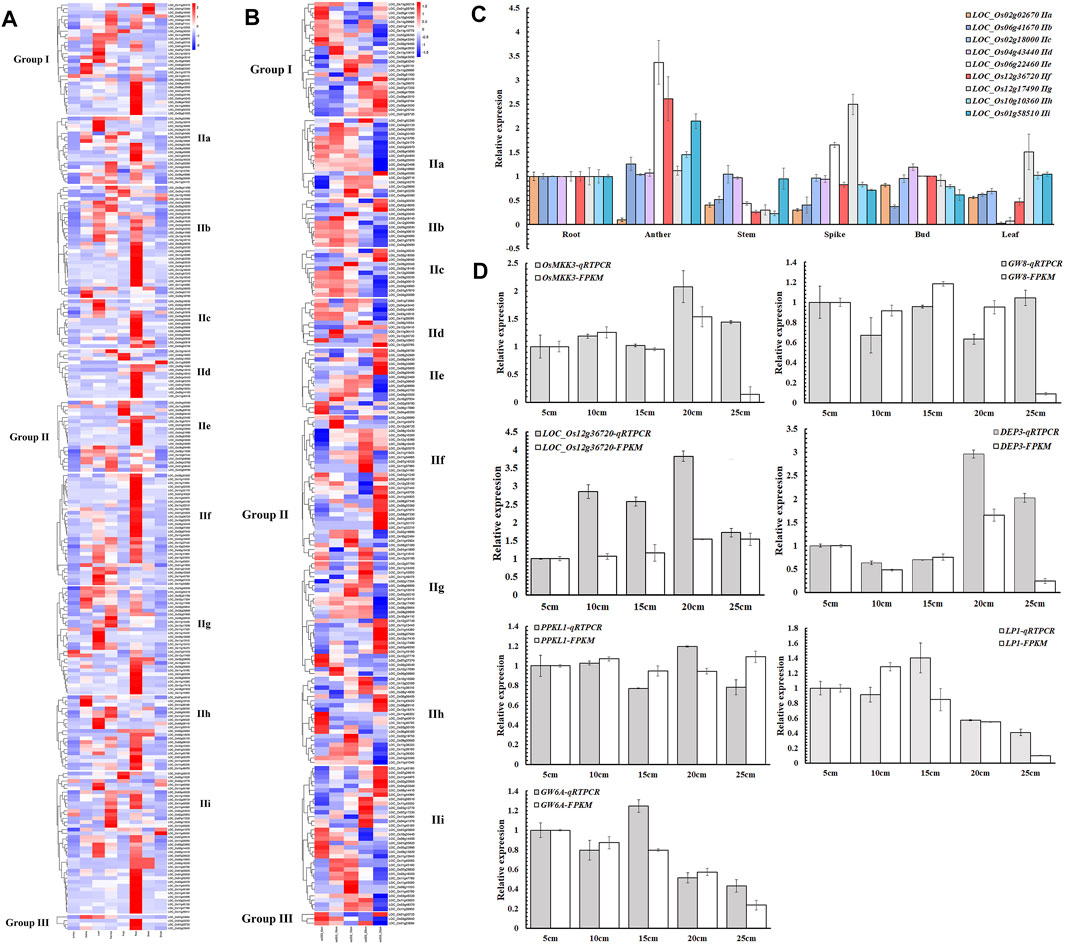
FIGURE 4. Expression profiles of NB-ARC genes. (A) Expression patterns of rice NB-ARC genes. The expression data are RNA-Seq data from the Rice Expression Database. The white box indicates no expression (zero fragments per kb of exon per million mapped reads (FPKM)) in this tissue. The transcript abundances in different tissues in the heat map were estimated by Log2 (FPKM) values. (B) Expression patterns of rice NB-ARC genes in Ce253 at 5, 10, 15, 20, and 25 cm panicle lengths. (C) Expression of rice NB-ARC genes in root, anther, shoot, bud, leaf, and panicle of Ce253. (D) Expression of key genes that control panicle development in 5, 10, 15, 20, and 25 cm panicle lengths in Ce253. Data are given as mean ± s. e.m. (n = 3). UBQ (ubiquitin) was used as a control. Error bar indicates 95% confidence interval.
For the IIf subgroup, one gene was most expressed in callus and 14 genes were most expressed in panicle. Six genes of IIf were most expressed in pistil and 23 genes of IIf were most expressed in root. IIf, a typical subgroup in the NB-ARC gene family, includes genes such as LOC_Os12g36720 that was expressed in eight different tissues (Figure 4A). To confirm the expression patterns, expression levels of 11 genes of different subgroups were detected by qRT-PCR. The results showed that six of these genes exhibited similar expression patterns to the transcriptomic data (Figure 4B). Most of these genes showed high expression in root. LOC_Os12g36720 in IIf was most highly expressed in anther and panicle.
Few studies have explored the roles of NB-ARC genes in normal plant growth and development. To investigate a possible function of these genes to regulate plant panicle development, the expression of selected NB-ARC genes was analyzed at the panicle development stage of cultivated rice (Figure 4B, c). In rice Ce253, 40, 34,32, 46, and 64 genes exhibited FPKM values higher than 1 at 5, 10, 15, 20, and 25 cm panicle length stages, respectively, and 215, 221, 223, 209, and 191 genes had FPKM values lower than 1 in 5, 10, 15, 20, and 25 cm panicle length stages, respectively. Most genes of the first subgroup of IIf had FPKM values lower than 1 at the early stage of young panicle differentiation (5, 10 cm), 12 and 11 genes exhibited FPKM values higher than 1 at 20 and 25 cm panicle length stages, respectively, and two, four, and four genes at 5, 10, and 15 cm panicle length stages, respectively (Figure 4B). The observed expressional activation in panicle development for LOC_Os12g36690, LOC_Os11g45970, and LOC_Os12g36720 suggests these genes play a crucial role in panicle development of rice.
To test the ability of NB-ARC genes to regulate plant panicle development, LOC_Os12g36720 was selected as a representative IIf gene. Expression of this gene was detected in anther, callus, leaf, panicle, pistil, root, seed, and shoot, and exhibited a remarkably high level of expression in Ce253 during panicle development. Interestingly, LOC_Os12g36720 encodes GNP12, a Resistance Gene Homologs (RGH1A) gene (Supplementary Table S7). Therefore, LOC_Os12g36720, GNP12, was selected for further characterization (Supplementary Table S6). To further characterize the function of this gene, we used a CRISPR-Cas9 system for targeted gene mutation of GNP12 in Ce253 (Cr line) (Figure 5A). The resulting deletions lead to frameshifting mutations that result in incomplete peptides of GNP12 that exhibit a loss of NB-ARC domain function. Compared to Ce253, the three Cr lines Cr-1, Cr-2, and Cr-3, were reduced 13.68, 3.71, and 9.39%, respectively, in panicle length (Figure 5B); 23.80, 8.50, and 24%, respectively, in panicle grain number; 2.76, 0.7, and 2.51%, respectively, in grain length; and 26.58, 8.79, and 10.98% in setting percentage, respectively. The three Cr lines showed significant reductions in panicle length, grain number of panicle, grain length, and setting percentage (Figure 5A, b, e, f, h, j). To investigate the tissue structures affected by GNP12, microstructures of grain chaff were observed by scanning electron microscopy (Figure 5D). The chaff of Cr was shorter than that of Ce253 (Figure 5D), but the Cr lines did not differ in length-width grain ratio or grain width (Figure 5C, i, k).
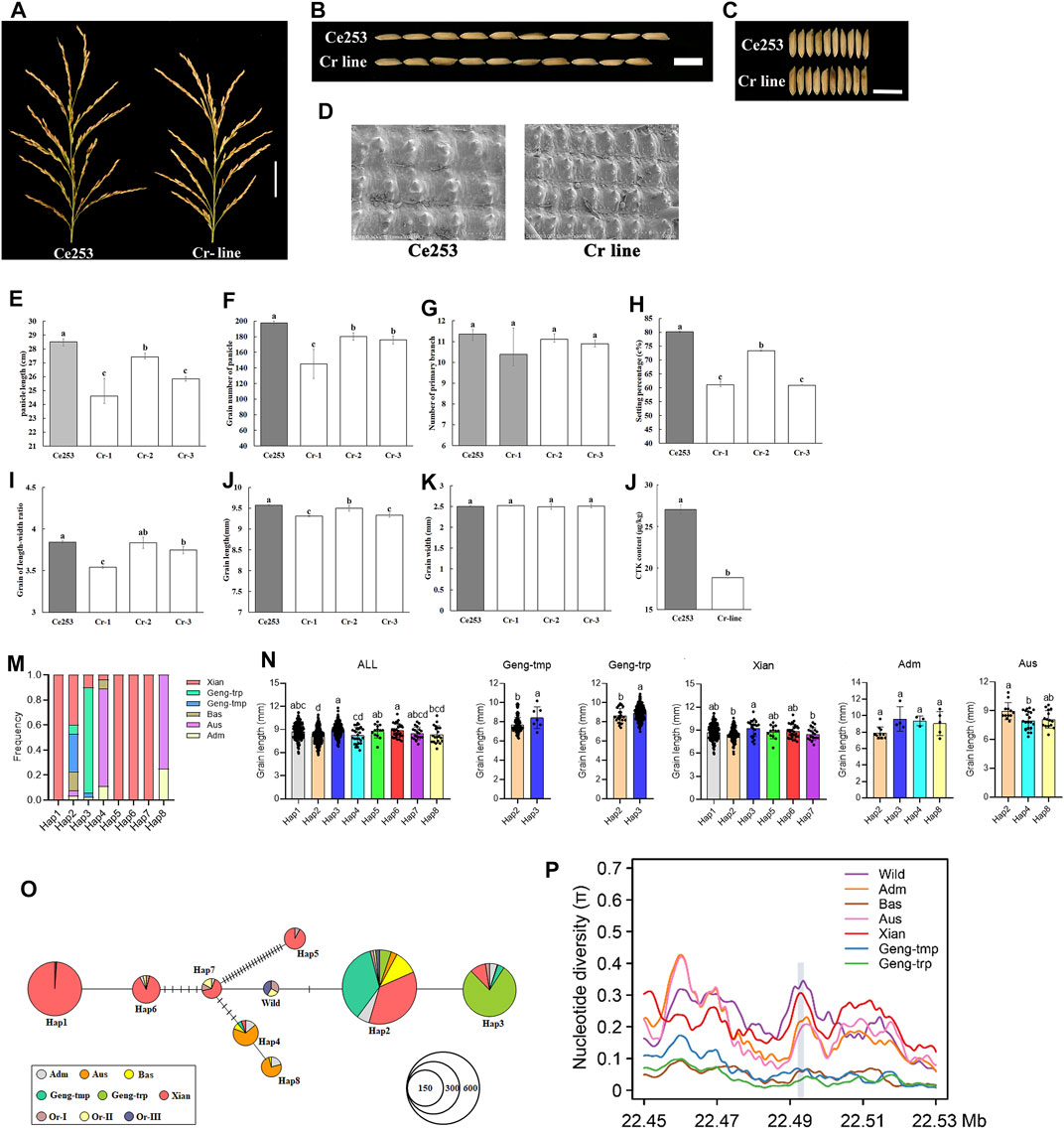
FIGURE 5. The effect of GNP12 on cell proliferation length of panicle, grain number of panicle and grain size. (A) Length of panicle of Ce253 and Cr-lines. Scale bars, 5 cm. (B) Length of grains from Ce253 and Cr-line. (C) Width of grains from Ce253 and Cr-line. Scale bars, 1 cm. (D) Scanning electron microscopic images of chalkiness in transgenic plants and Ce253. Bar, 50 μm. e-l Panicle length (cm) (E), grain number of panicle (F), primary branch number (G), setting percentage (%) (H), grain of length-width ratio (I), grain length (mm) (J), grain width (mm) (K), CTK content in Ce253 and Cr-line (l). Data are given as mean ± s. e.m. (n = 12). Error bar indicates 95% confidence interval. Student’s t tests were used to generate p values. Haplotype and evolutionary analysis of GNP12 in rice (M) Major haplotypes of GNP12 using 32 single-nucleotide polymorphisms (SNPs) in the coding sequence region in 827 accessions (N) Phenotypic evaluation of haplotype grain length in different subgroups (O) Haplotype network analysis of GNP12 (P) Nucleotide diversity of GNP12.
Cytokinin (CTK) is a primary determinant of plant architecture (Tameling et al., 2002). To investigate whether GNP12 influences CTK levels, we compared the CTK content in wild-type and Cr lines. The CTK content in Cr lines was reduced to 30.28% of that in the wild-type (Figure 5l). The results suggest that GNP12 may affect panicle development and grain production in rice through changes in CTK content.
To investigate natural variation in GNP12, we analyzed the polymorphism of GNP12 in the 3K rice genomes (3K-RG) dataset (Wang et al., 2018). We identified eight major haplotypes of GNP12 with 32 single-nucleotide polymorphisms (SNPs) in the coding sequence region in 827 accessions of the 3K-RG dataset (Figure 5m and Table 1). Except for Hap2, the frequencies of the major haplotypes differed significantly among different subgroups. Hap1, Hap5, Hap6, and Hap7 were specifically found in indica rice cultivars, with Hap1 being the most prevalent. Hap2 was mainly identified in indica, temperate japonica, tropical japonica, and Bas rice cultivars. Hap3 was mainly present in tropical japonica and indica rice cultivars. Hap4 and Hap8 were mainly found in Aus rice cultivars (Figure 5n). Phenotypic evaluation showed that Hap1, Hap3, Hap5, and Hap6 in the indica subgroup and Hap3 in the japonica subgroup showed longer grain length than other haplotypes. Haplotype network analysis showed that favorable haplotypes of GNP12 may have different origins. Favorable indica haplotypes Hap3 and Hap6 may have come directly from O. rufipogon I and favorable indica haplotypes Hap1 and Hap5 may have come from novel favorable mutations during indica domestication. Favorable haplotype Hap3 in japonica may have arisen during the differentiation of temperate and tropical japonica (Figure 5o).
To determine whether selection acted on GNP12, we analyzed the nucleotide diversity of GNP12 among different rice subgroups. On average, the nucleotide diversities of GNP12 in tropical japonica and indica subgroups were respectively much lower and higher than those in other haplotypes. To assess whether the DNA sequence evolved randomly or under a non-random process, Tajima’s D values were calculated for GNP12 in tropical japonica and indica and were negative and positive, respectively. Both values deviated significantly from zero, implying potential positive selection and balancing selection acting on GNP12 in tropical japonica and indica, respectively (Figure 5p). To further test whether the observed reduction in nucleotide diversity in tropical japonica was due to positive selection or a bottleneck effect, we calculated the nucleotide diversity of 40-kb flanking regions of GNP12. We found that the average nucleotide diversity of the GNP12 flanking regions in tropical japonica was significantly lower than that in other regions, suggesting that the decrease of nucleotide diversity of GNP12 in tropical japonica may be largely caused by positive selection (Table 2).
To further investigate the subdomains and functions of NB-ARC genes, 11 predicted proteins from group I, group II, and group III were selected for further analysis. Strong conservation was identified for several subdomains: αC, αM, αS, β3, β2-β5, and P-loop (Figure 6A, b). The most conserved residues in the putative NB-ARC domains were found in the P-loop (G193 and S223) and αS (L468) subdomains (Figure 6A). Analysis of the gene structure and conserved motifs revealed high conservation of rice NB ARC genes with no obvious variation in the amino acid residues of the P-loop region among homologous genes (Figure 6C). We observed few variants of the 11 amino acids in the P-loop core, with substantial variation in genic frequency of these variants in rice. These results indicate the significant conservation of the P-loop in most rice NB-ARC proteins. The predicted 3D structures of the identified rice NB-ARC proteins were conserved, which was consistent with the phylogenetic, gene structure, and conserved domain analysis. LOC_Os02g02670, LOC_Os06g41670, LOC_Os06g22460, LOC_Os012g36720, LOC_Os12g17490.1, and LOC_Os10g10360 were predicted to share similar structures (Figure 6C). Predicted models were constructed to heuristically maximize alignment coverage, identity percentage, and confidence score for the tested sequences. For the 11 predicted NB-ARC proteins, α-helix was most highly predicted as secondary structure (37.41–63.56%), followed by random coil (20.03–35.64%), extended strands (7.3–20.84%) and β-strands (0–5.29%) (Figure 6C and Supplementary Table S8). The 3D modeling results revealed similarity of predicted tertiary structures, implying that NB-ARC proteins may have evolved from a shared ancestor and/or under purification selection force for stabilization during long-term acclimation after initial divergence.
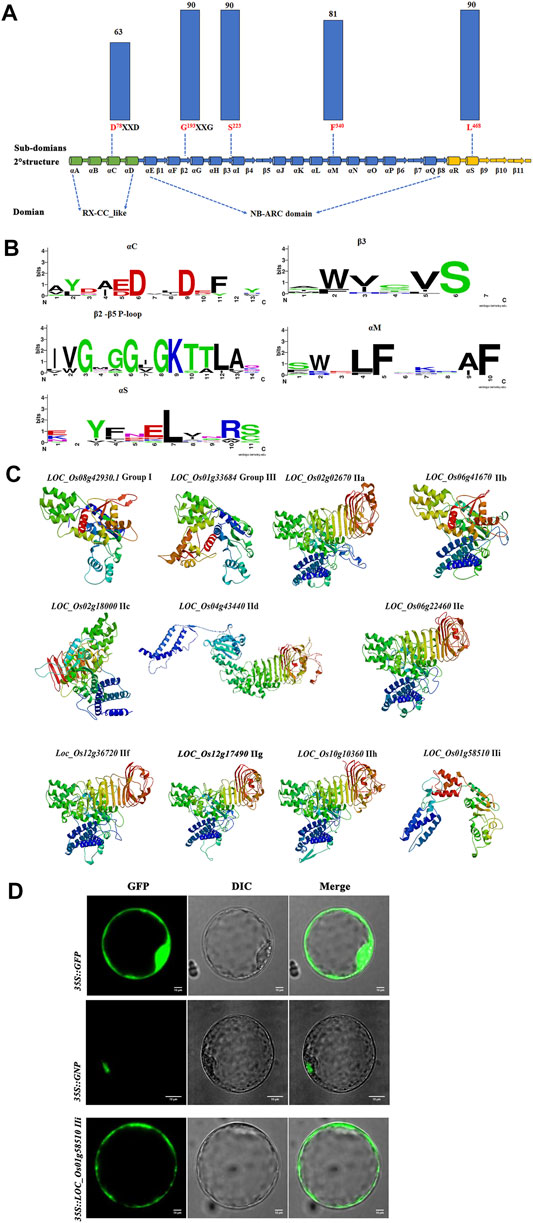
FIGURE 6. Predicted structures and subcellular localization of putative NB-ARC protein domains based on 11 proteins from each 11 subgroups. (A) Conservation of key motifs, residues, and secondary structure between putative NB-ARC domains. Dashed lines indicate positions within secondary structure elements. The histograms above the motifs represent the degree of conservation (% of identical to conserved residuals) for putative NB-ARC domains (blue columns) among 11 proteins from 11 subgroups. (B) Sequence logos representing the conservation of key motifs and neighboring sequences. The size of the letters corresponds to information content. (C) 3D model of 11 proteins from 11 subgroups in NB-ARC family. (D) Subcellular localization of three proteins from groups I, IIf, and IIi.
To further characterize the function of the NB-ARC genes, subcellular localization studies were performed. We randomly selected two NB-ARC genes, LOC_Os12g36720 (IIf), LOC_Os101g458510 (IIi), for subcellular localization in rice protoplast cells. LOC_Os12g36720-GFP and LOC_Os101g458510-GFP expression vectors were constructed and transformed into rice protoplasts for transient expression. Control GFP expression was observed throughout the membrane, cytoplasm and nucleus, and the GNP12 and LOC_Os101g458510 proteins were mainly expressed in the nucleus and membrane, respectively. This result indicated that these proteins are widely present in various organelles.
NB-ARC proteins play central roles in recognizing pathogens, initiating defense cascades, and maintaining cell development. The NB-ARC conserved domain maintains stable similar structures in different species, likely due to convergent evolution, suggesting these proteins may perform similar recognition and activation mechanisms (Slootweg et al., 2009). In the Fabaceae family, eight conserved motifs of NB-ARC domains have been identified (Pal et al., 2007). Plant NOD-like receptor (NLR) proteins contain NB-ARC domains with structural similarities to their mammalian homologues (Steele et al., 2019). However, there is high polymorphism in some NB-ARC genes in some species. The combination of conserved motifs and the evolution of rich polymorphisms may allow response to environmental signal stimulation to achieve coevolution by replication events. In Arabidopsis, analysis of the NLR family revealed that positive selection and recombination occurred frequently in the leucine-rich repeat (LRR) domain but there was negative selection in the nucleotide-binding (NB-ARC) domain (Mondragón-Palomino et al., 2017). In peanut, AhRAF4 of NB-ARC proteins evolved by recombination with duplications and point mutations from Arachis duranensis (Deng et al., 2018). Among O. sativa (Slootweg et al., 2009), O. glaberrima (Slootweg et al., 2009), and O. brachyantha (FF), a high number of paralogs suggests that the NB-ARC family experienced highly dynamic evolution, with a large number of tandem arrays and duplicated genes observed in the O. sativa subspecies (Jacquemin et al., 2014). NB-ARC genes diversified through duplication to encode receptors adapted to external signals (Andersen et al., 2020). IPm21 encodes a coiled-coil, nucleotide-binding site, leucine-rich repeat (CC-NBS-LRR) protein, and evolutionary analysis of 38 non-redundant Pm21 alleles indicated that the nucleotide diversity of the LRR domain was significantly higher than those of the CC and NB-ARC domains (He et al., 2020). NB-ARC genes can form genomic clusters adjacent to LRR-RLK-XIIs by mechanisms other than genomic clustering (Ngou et al., 2022). Overall, the evolution of NB-ARC genes allows these genes to maintain key function through conservation of the NB-ARC domain while the C-terminal regions of these proteins exhibit polymorphisms to allow these genes to co-evolve with the environment through replication events.
Panicle grain number and length of panicle are important agronomic characteristics, but the genetic determinants of these traits remain unclear. Many genes regulate grain number per panicle, length of panicle, and grain length by influencing hormone activity. ERECTA1 (OsER1) is a cytokinin oxidase/dehydrogenase that negatively regulates grain number per panicle (Ashikari et al., 2005). ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade, and the OsER1-OsMKKK10-OsMKK4-OsMPK6 pathway is required to maintain cytokinin homeostasis (Guo et al., 2020). OsASP1 encodes a TOPLESS-related transcriptional co-repressor, is closely associated with auxin action, and regulates spikelet development (Yoshida et al., 2012). TGW6 encodes a protein with indole-3-acetic acid (IAA)-glucose hydrolase activity that can control IAA supply at the transition from the syncytial to the cellular phase to limit cell number and grain length (Ishimaru et al., 2013). GW6 encodes a GA-regulated GAST family protein and positively regulates grain width and weight (Shi et al., 2020).
NB-ARC genes regulate plant development by catalyzing ADP through ARC structure. NB-ARC proteins carry a central nucleotide-binding-ARC domain that binds ADP/ATP rather than GDP/GTP (Chattopadhyaya and Pal, 2008). The NB-ARC domain may be a molecular switch between ADP (repressed) and ATP (active) binding forms (Steele et al., 2019). A tomato (Lycopersicon esculentum) R protein I-2 with NB-ARC domain is impaired in ATP hydrolysis, but not in ATP binding, suggesting a molecular switch whose state (on/off) depends on nucleotide binding (ATP/ADP) (Tameling et al., 2006). NRTP1 encodes a CC-NB-LRR type protein, and semi-dominant mutant nrtp1-D contains an amino acid substitution in the NB-ARC domain that causes constitutive auto-activation of the NRTP1 protein for a short-root phenotype in rice (Yu et al., 2018). ATP/ADP opentenyltransferases are likely responsible for most isopentenyladenine- and tZ-type cytokinin synthesis (Miyawaki et al., 2006) and catalyze prenylation of adenosine diphosphate (ADP) or triphosphate (ATP) biosynthesis (Kieber and Schaller, 2014). The hydrolysis of adenosine triphosphate (ATP) is directly coupled to the primary active transport of CTK (Nedvěd et al., 2021). In peach, ATP/ADP PpIPT genes are key genes for cytokinin biosynthesis in nodal stems (Li et al., 2018).
The NB-ARC domain of GNP12 protein may act in panicle development by influencing hormone activity to control grain number of panicle, length of panicle, and grain length. Future work is required to test this hypothesis and investigate the regulation of these important genes to better develop molecular tools for improved genetic breeding.
In summary, we successfully analyzed the NB-ARC family of genes with conserved P-loop domains and identified conservation of NB-ARC homologous genome segments in monocotyledons. Most NB-ARC genes regulate panicle development in Ce253. We identified GNP12 as an important regulator of panicle length, panicle grain number, and grain length.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/ PRJNA815477.
Y-HP, YZ, and G-FD designed and supervised the research. LC, RF, C-CH, and D-JQ performed the experiments. Y-HP, H-FG, Q-JL, MR, X-YZ, H-FL, and L-JG analyzed the data. Y-HP and LC wrote and revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (32060452, 32160447, 32160645, 31760428, U20A2032), the key project of the Guangxi Natural Science Foundation (2018GXNSFDA281053, 2019GXNSFBA245006), Natural Science Foundation of Shandong Province (ZR2020MC096), the Base Business Project of Guangxi Academy of Agricultural Sciences (GuiNongKe 2022YM01), the project of the central government guides the development of local science and technology (Guike ZY20198015, Guike AD17129064, Guike AD20297093, Guike ZY19183020, and Guike ZY18057004), and the project of the base and talent (GuiKe 2018JZ24 and 2018-15-Z06-KF10).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.887217/full#supplementary-material
Chr: chromosome; CTK: Cytokinin; GNP: Grain Number per Panicle; IAA: Indole-3-acetic acid; MEME: Multiple EM for motif elicitation; NB-ARC: nucleotide-binding adaptor shared by APAF-1, R proteins and CED-4; ORF: Open reading frame; qRT-PCR: quantitative Reverse-Transcription Polymerase Chain Reaction; SNPs: Single-nucleotide polymorphisms.
Albrecht, M., and Takken, F. L. W. (2006). Update on the Domain Architectures of NLRs and R Proteins. Biochem. Biophysical Res. Commun. 339 (2), 459–462. doi:10.1016/j.bbrc.2005.10.074
Andersen, E. J., Lindsey, L. E., and Nepal, M. P. (2020). Genome-wide Identification of Disease Resistance Genes (R Genes) in Wheat. New York: bioRxiv.
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin Oxidase Regulates Rice Grain Production. Science 309 (5735), 741–745. doi:10.1126/science.1113373
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43 (W1), W39–W49. doi:10.1093/nar/gkv416
Chandra, S., Kazmi, A. Z., Ahmed, Z., Roychowdhury, G., Kumari, V., Kumar, M., et al. (2017). Genome-wide Identification and Characterization of NB-ARC Resistant Genes in Wheat (Triticum aestivum L.) and Their Expression during Leaf Rust Infection. Plant Cell Rep. 36 (7), 1097–1112. doi:10.1007/s00299-017-2141-0
Chattopadhyaya, R., and Pal, A. (2008). Three-dimensional Models of NB-ARC Domains of Disease Resistance Proteins in Tomato, Arabidopsis, and Flax. J. Biomol. Struct. Dyn. 25 (4), 357–371. doi:10.1080/07391102.2008.10507184
Chen, A., Gu, L., Li, M., Xu, N., Feng, F., Liu, G., et al. (2018). Identification of Rehmannia Glutinosa L. NB-ARC Family Proteins and Their Typical Changes under Consecutive Monoculture Stress. Acta Physiol. Plant 40, 95. doi:10.1007/s11738-018-2672-1
Chen, W., Cheng, Z., Liu, L., Wang, M., You, X., Wang, J., et al. (2019). Small Grain and Dwarf 2, Encoding an HD-Zip II Family Transcription Factor, Regulates Plant Development by Modulating Gibberellin Biosynthesis in Rice. Plant Sci. 288, 110208. doi:10.1016/j.plantsci.2019.110208
Deng, Y., Chen, H., Zhang, C., Cai, T., Zhang, B., Zhou, S., et al. (2018). Evolution and Characterisation of the AhRAF4 NB-ARC Gene Family Induced by Aspergillus flavus Inoculation and Abiotic Stresses in Peanut. Plant Biol. J. 20 (4), 737–750. doi:10.1111/plb.12726
Dolezal, A. L., Shu, X., OBrian, G. R., Nielsen, D. M., Woloshuk, C. P., Boston, R. S., et al. (2014). Aspergillus flavus Infection Induces Transcriptional and Physical Changes in Developing Maize Kernels. Front. Microbiol. 5, 384. doi:10.3389/fmicb.2014.00384
Guo, T., Lu, Z.-Q., Shan, J.-X., Ye, W.-W., Dong, N.-Q., and Lin, H.-X. (2020). ERECTA1 Acts Upstream of the OsMKKK10-OsMKK4-OsMPK6 Cascade to Control Spikelet Number by Regulating Cytokinin Metabolism in Rice. Plant Cell 32 (9), 2763–2779. doi:10.1105/tpc.20.00351
He, H., Ji, J., Li, H., Tong, J., Feng, Y., Wang, X., et al. (2020). Genetic Diversity and Evolutionary Analyses Reveal the Powdery Mildew Resistance Gene Pm21 Undergoing Diversifying Selection. Front. Genet. 11, 489. doi:10.3389/fgene.2020.00489
Igari, K., Endo, S., Hibara, K.-i., Aida, M., Sakakibara, H., Kawasaki, T., et al. (2008). Constitutive Activation of a CC-NB-LRR Protein Alters Morphogenesis through the Cytokinin Pathway in Arabidopsis. Plant J. 55 (1), 14–27. doi:10.1111/j.1365-313x.2008.03466.x
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of Function of the IAA-Glucose Hydrolase Gene TGW6 Enhances Rice Grain Weight and Increases Yield. Nat. Genet. 45 (6), 707–711. doi:10.1038/ng.2612
Jacquemin, J., Ammiraju, J. S. S., Haberer, G., Billheimer, D. D., Yu, Y., Liu, L. C., et al. (2014). Fifteen Million Years of Evolution in the Oryza Genus Shows Extensive Gene Family Expansion. Mol. Plant 7 (4), 642–656. doi:10.1093/mp/sst149
Kieber, J. J., and Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12, e0168. doi:10.1199/tab.0168
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). STAND, a Class of P-Loop NTPases Including Animal and Plant Regulators of Programmed Cell Death: Multiple, Complex Domain Architectures, Unusual Phyletic Patterns, and Evolution by Horizontal Gene Transfer. J. Mol. Biol. 343 (1), 1–28. doi:10.1016/j.jmb.2004.08.023
Li, M., Wei, Q., Xiao, Y., and Peng, F. (2018). The Effect of Auxin and Strigolactone on ATP/ADP Isopentenyltransferase Expression and the Regulation of Apical Dominance in Peach. Plant Cell Rep. 37 (12), 1693–1705. doi:10.1007/s00299-018-2343-0
Librado, P., and Rozas, J. (2009). DnaSP V5: a Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 25 (11), 1451–1452. doi:10.1093/bioinformatics/btp187
Liu, E., Liu, Y., Wu, G., Zeng, S., Tran Thi, T. G., Liang, L., et al. (2016). Identification of a Candidate Gene for Panicle Length in Rice (Oryza Sativa L.) via Association and Linkage Analysis. Front. Plant Sci. 7, 596. doi:10.3389/fpls.2016.00596
Lukasik, E., and Takken, F. L. (2009). STANDing Strong, Resistance Proteins Instigators of Plant Defence. Curr. Opin. Plant Biol. 12 (4), 427–436. doi:10.1016/j.pbi.2009.03.001
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., and Zhu, J.-K. (2013). Application of the CRISPR-Cas System for Efficient Genome Engineering in Plants. Mol. Plant 6 (6), 2008–2011. doi:10.1093/mp/sst121
Massingham, T., and Goldman, N. (2005). Detecting Amino Acid Sites under Positive Selection and Purifying Selection. Genetics 169 (3), 1753–1762. doi:10.1534/genetics.104.032144
Michael, Weaver, L., Swiderski, M. R., Li, Y., and Jones, J. D. G. (2006). TheArabidopsis thalianaTIR-NB-LRR R-Protein, RPP1A; Protein Localization and Constitutive Activation of Defence by Truncated Alleles in Tobacco and Arabidopsis. Plant J. 47 (6), 829–840. doi:10.1111/j.1365-313x.2006.02834.x
Miyawaki, K., Tarkowski, P., Matsumoto-Kitano, M., Kato, T., Sato, S., Tarkowska, D., et al. (2006). Roles of Arabidopsis ATP/ADP Isopentenyltransferases and tRNA Isopentenyltransferases in Cytokinin Biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103 (44), 16598–16603. doi:10.1073/pnas.0603522103
Mondragón-Palomino, M., Stam, R., John-Arputharaj, A., and Dresselhaus, T. (2017). Diversification of Defensins and NLRs in Arabidopsis Species by Different Evolutionary Mechanisms. BMC Evol. Biol. 17 (1), 255. doi:10.1186/s12862-017-1099-4
Nedvěd, D., Hošek, P., Klíma, P., and Hoyerová, K. (2021). Differential Subcellular Distribution of Cytokinins: How Does Membrane Transport Fit into the Big Picture? Int. J. Mol. Sci. 22 (7), 3428. doi:10.3390/ijms22073428
Nei, M., and Gojobori, T. (1986). Simple Methods for Estimating the Numbers of Synonymous and Nonsynonymous Nucleotide Substitutions. Mol. Biol. Evol. 3 (5), 418–426. doi:10.1093/oxfordjournals.molbev.a040410
Ngou, B. P. M., Heal, R., Wyler, M., Schmid, M. W., and Jones, J. D. G. (2022). Concerted Expansion and Contraction of Immune Receptor Gene Repertoires in Plant Genomes. New York: bioRxiv. doi:10.1101/2022.1101.1101.474684
Pal, A., Chakrabarti, A., and Basak, J. (2007). New Motifs within the NB-ARC Domain of R Proteins: Probable Mechanisms of Integration of Geminiviral Signatures within the Host Species of Fabaceae Family and Implications in Conferring Disease Resistance. J. Theor. Biol. 246 (3), 564–573. doi:10.1016/j.jtbi.2007.01.013
Pan, Y., Chen, L., Zhao, Y., Guo, H., Li, J., Rashid, M. A. R., et al. (2021). Natural Variation in OsMKK3 Contributes to Grain Size and Chalkiness in Rice. Front. Plant Sci. 12, 784037. doi:10.3389/fpls.2021.784037
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam Protein Families Database. Nucleic Acids Res. 40, D290–D301. doi:10.1093/nar/gkr1065
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a Simple Modular Architecture Research Tool: Identification of Signaling Domains. Proc. Natl. Acad. Sci. U.S.A. 95 (11), 5857–5864. doi:10.1073/pnas.95.11.5857
Shi, C. L., Dong, N. Q., Guo, T., Ye, W. W., Shan, J. X., and Lin, H. X. (2020). A Quantitative Trait Locus GW6 Controls Rice Grain Size and Yield through the Gibberellin Pathway. Plant J. 103 (3), 1174–1188. doi:10.1111/tpj.14793
Slootweg, E., Tameling, W., Roosien, J., Lukasik, E., Joosten, M., Takken, F., et al. (2009). An Outlook on the Localisation and Structure-Function Relationships of R Proteins in Solanum. Potato Res. 52 (3), 229–235. doi:10.1007/s11540-009-9130-9
Steele, J. F. C., Hughes, R. K., and Banfield, M. J. (2019). Structural and Biochemical Studies of an NB-ARC Domain from a Plant NLR Immune Receptor. PLoS ONE 14 (8), e0221226. doi:10.1371/journal.pone.0221226
Tameling, W. I. L., Elzinga, S. D. J., Darmin, P. S., Vossen, J. H., Takken, F. L. W., Haring, M. A., et al. (2002). The Tomato R Gene Products I-2 and MI-1 Are Functional ATP Binding Proteins with ATPase Activity. Plant Cell 14 (11), 2929–2939. doi:10.1105/tpc.005793
Tameling, W. I. L., Vossen, J. H., Albrecht, M., Lengauer, T., Berden, J. A., Haring, M. A., et al. (2006). Mutations in the NB-ARC Domain of I-2 that Impair ATP Hydrolysis Cause Autoactivation. Plant Physiol. 140 (4), 1233–1245. doi:10.1104/pp.105.073510
van der Biezen, E. A., and Jones, J. D. G. (1998). The NB-ARC Domain: a Novel Signalling Motif Shared by Plant Resistance Gene Products and Regulators of Cell Death in Animals. Curr. Biol. 8 (7), R226–R228. doi:10.1016/s0960-9822(98)70145-9
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 Regulatory Module Determines Grain Shape and Simultaneously Improves Rice Yield and Grain Quality. Nat. Genet. 47 (8), 949–954. doi:10.1038/ng.3352
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature 557 (7703), 43–49. doi:10.1038/s41586-018-0063-9
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wang, Y., Tang, J., Li, H., Teng, Z., Wang, W., Xu, F., et al. (2020). A NB-ARC-CRRSP Signaling Module Triggers HR-like Cell Death and Associated Disease Resistance in Rice by Suppressing Antioxidant Defense Systems. SSRN J., 50. doi:10.2139/ssrn.3565037
Wen, Z., Yao, L., Singer, S. D., Muhammad, H., Li, Z., and Wang, X. (2017). Constitutive Heterologous Overexpression of a TIR-NB-ARC-LRR Gene Encoding a Putative Disease Resistance Protein from Wild Chinese Vitis Pseudoreticulata in Arabidopsis and Tobacco Enhances Resistance to Phytopathogenic Fungi and Bacteria. Plant Physiology Biochem. 112, 346–361. doi:10.1016/j.plaphy.2017.01.017
Wu, Y., Wang, Y., Mi, X.-F., Shan, J.-X., Li, X.-M., Xu, J.-L., et al. (2016). The QTL GNP1 Encodes GA20ox1, Which Increases Grain Number and Yield by Increasing Cytokinin Activity in Rice Panicle Meristems. PLoS Genet. 12 (10), e1006386. doi:10.1371/journal.pgen.1006386
Yildirim-Ersoy, F., Ridout, C. J., and Akkaya, M. S. (2011). Detection of Physically Interacting Proteins with the CC and NB-ARC Domains of a Putative Yellow Rust Resistance Protein, Yr10, in Wheat. J. Plant Dis. Prot. 118 (3/4), 119–126. doi:10.1007/bf03356391
Yoshida, A., Ohmori, Y., Kitano, H., Taguchi-Shiobara, F., and Hirano, H.-Y. (2012). ABERRANT SPIKELET and PANICLE1, Encoding a TOPLESS-Related Transcriptional Co-repressor, Is Involved in the Regulation of Meristem Fate in Rice. Plant J. 70 (2), 327–339. doi:10.1111/j.1365-313x.2011.04872.x
Yu, Z., Dong, L., Jiang, Z., Yi, K., Zhang, J., Zhang, Z., et al. (2018). A Semi-dominant Mutation in a CC-NB-LRR-type Protein Leads to a Short-Root Phenotype in Rice. Rice 11 (1), 54. doi:10.1186/s12284-018-0250-1
Keywords: NB-ARC family, evolution, expression, panicle development, rice
Citation: Pan Y-H, Chen L, Guo H-F, Feng R, Lou Q-J, Rashid MAR, Zhu X-Y, Qing D-J, Liang H-F, Gao L-J, Huang C-C, Zhao Y and Deng G-F (2022) Systematic Analysis of NB-ARC Gene Family in Rice and Functional Characterization of GNP12. Front. Genet. 13:887217. doi: 10.3389/fgene.2022.887217
Received: 01 March 2022; Accepted: 10 May 2022;
Published: 16 June 2022.
Edited by:
Genlou Sun, Saint Mary’s University, CanadaReviewed by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Pan, Chen, Guo, Feng, Lou, Rashid, Zhu, Qing, Liang, Gao, Huang, Zhao and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Hua Pan, cGFueWluZ2h1YTIwMDhAMTYzLmNvbQ==; Yan Zhao, emhhb3lhbjEyMTZAMTYzLmNvbQ==; Guo-Fu Deng, ZGVuZ2d1b2Z1MTYzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.