
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Genet., 22 June 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.886494
This article is part of the Research TopicPopulation Genetics in Conservation Biology: Recent AdvancesView all 5 articles
 Nicholas W. Jeffery1*
Nicholas W. Jeffery1* Sarah J. Lehnert2
Sarah J. Lehnert2 Tony Kess2
Tony Kess2 Kara K. S. Layton3
Kara K. S. Layton3 Brendan F. Wringe1
Brendan F. Wringe1 Ryan R.E. Stanley1
Ryan R.E. Stanley1A key component of the global blue economy strategy is the sustainable extraction of marine resources and conservation of marine environments through networks of marine protected areas (MPAs). Connectivity and representativity are essential factors that underlie successful implementation of MPA networks, which can safeguard biological diversity and ecosystem function, and ultimately support the blue economy strategy by balancing ocean use with conservation. New “big data” omics approaches, including genomics and transcriptomics, are becoming essential tools for the development and maintenance of MPA networks. Current molecular omics techniques, including population-scale genome sequencing, have direct applications for assessing population connectivity and for evaluating how genetic variation is represented within and among MPAs. Effective baseline characterization and long-term, scalable, and comprehensive monitoring are essential for successful MPA management, and omics approaches hold great promise to characterize the full range of marine life, spanning the microbiome to megafauna across a range of environmental conditions (shallow sea to the deep ocean). Omics tools, such as eDNA metabarcoding can provide a cost-effective basis for biodiversity monitoring in large and remote conservation areas. Here we provide an overview of current omics applications for conservation planning and monitoring, with a focus on metabarcoding, metagenomics, and population genomics. Emerging approaches, including whole-genome sequencing, characterization of genomic architecture, epigenomics, and genomic vulnerability to climate change are also reviewed. We demonstrate that the operationalization of omics tools can enhance the design, monitoring, and management of MPAs and thus will play an important role in a modern and comprehensive blue economy strategy.
The global “blue economy” strategy (BES) centers on the long-term sustainable use of ocean resources to promote economic benefits while also preserving ocean ecosystems (Smith-Godfrey 2016). This emerging approach integrates the conservation of ecosystem services, including economic activities, tourism, transportation, fishing, and resource extraction, with marine spatial planning and conservation to sustain the health of wild populations and marine ecosystems (Bennett et al., 2019). The application of the BES has been gaining momentum as has the supporting research (Figure 1). Marine protected areas (MPAs) are centered in the global effort to safeguard biological diversity and thus are integral to the BES (Gaines et al., 2010; Agardy et al., 2011). The pace of MPA establishment is increasing globally, catalyzed by international agreements and conservation targets (e.g., Aichi Target 11, the “30by30” target initiative; Global Ocean Alliance 2022). As of 2022, nearly 8% of the global ocean falls under some form of spatial-based marine biodiversity conservation measure (UNEP-WCMC and IUCN 2022) with the expansion expected to continue.
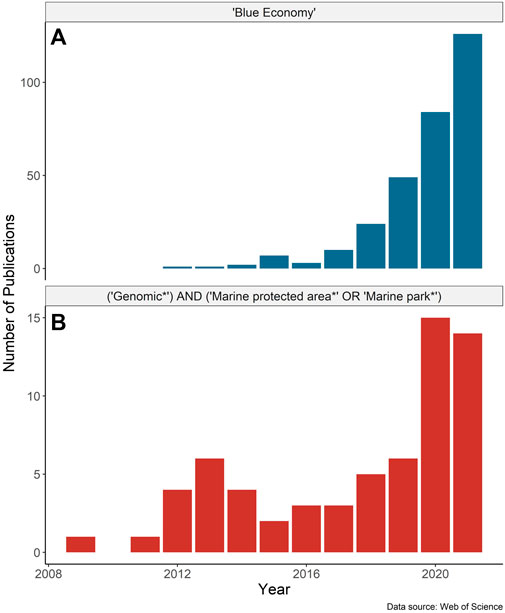
FIGURE 1. Number of publications by year based on the Web of Science search for papers with any field containing (A) “Blue Economy” and (B) “Genomic*” AND (“Marine protected area*” OR “Marine park*”). Search results were accessed on February 18, 2022, and include only publications up to the end of 2021.
Marine protected areas represent a spatial tool for the conservation of biodiversity, and fit into a broader approach for ecosystem-based management and spatial planning (Halpern et al., 2010). MPAs can contribute to the blue economy by protecting unique or vulnerable populations (e.g., Morris and Green, 2014) from anthropogenic impacts such as overharvesting (Morris et al., 2014; Sinclair-Waters et al., 2018b), by supplementing fisheries management (e.g., Gaines et al., 2010), and/or by enhancing “blue” carbon sequestration (Macreadie et al., 2021). When established as a functioning, connected network, MPAs can help to conserve important ecosystem functions and enhance population productivity beyond their legislated boundaries (Harmelin-Vivien et al., 2008; Grorud-Colvert et al., 2014; Di Lorenzo et al., 2020). Healthy marine ecosystems can also provide the indirect benefit of helping to mitigate impacts of climate change by protecting against other stressors and providing habitat refugia (e.g., Tittensor et al., 2019; Bryndum-Buchholz et al., 2022). The rapid global expansion in the global coverage of MPAs, which increasingly are being applied at a large spatial scale and in remote locations (e.g., Wilhelm et al., 2014), necessitates new, scalable technologies. Emerging omics approaches are uniquely suited to address this problem and help inform the design, monitoring, and successful implementation of MPAs; however, the incorporation of these approaches into MPA research is still in its infancy (Figure 1). Omics approaches are generally described as high-throughput technologies to holistically sequence or quantify DNA, RNA, proteins, metabolites, and other molecules, and include (meta)genomics, (meta)transcriptomics, epigenomics, lipidomics, metabolomics, and proteomics, among others (sensu Samuel et al., 2021).
Here, we provide an overview of molecular omics approaches, focusing on genomics, transcriptomics, and environmental DNA (eDNA) metabarcoding, and emphasize how development and application of such methods can address key needs for the successful design and management of MPAs to support the goals of a blue economy. We demonstrate how omics tools can be used to characterize genetic diversity, evaluate functional connectivity, identify adaptive variation and environmental association, and to predict population-level responses to climate change. These tools can reveal fine-scale evolutionary processes required for conservation planning that were previously either not considered or not detectable (Xuereb et al., 2019). We also explore how metagenomics and eDNA metabarcoding are powerful approaches for baseline characterization of eukaryotic and microbial diversity, helping to inform ecological status and the long-term management of protected areas. We focus our perspective through the application of conservation areas within a BES. While other omics approaches (e.g., metabolomics) have applications for BES, they were outside the scope of this review.
Representativity and connectivity are core design principles when establishing MPA networks to ensure that the protection is equitably distributed through space. Genomic approaches, such as reduced representation genome sequencing (e.g., restriction site–associated DNA sequencing, RAD-seq; Davey and Blaxter, 2010), pooled sequencing (PoolSeq; Schlötterer et al., 2014), and low-coverage whole-genome resequencing (lcWGS; Lou et al., 2021), have considerably advanced our understanding of genetic diversity and population connectivity in non-model species over the last decade. Characterizing genome-wide variation through genomic sequencing has revealed cryptic intraspecific population structure where panmixia has otherwise been assumed, informing both representativity and connectivity for conservation network design (e.g., Barney et al., 2017; Van Wyngaarden et al., 2017).
MPA networks that consider connectivity and gene flow in their design will be better positioned to conserve diversity and adaptive variation within species (von der Heyden 2009), while also protecting genetically distinct populations (Andrello et al., 2022). Explicit consideration of genetically unique or diverse populations can help maintain diversity, and may allow for potential genetic rescue or the re-establishment of populations following extirpation (Xuereb et al., 2019), thus increasing resilience via the conservation of biocomplexity (Mendez et al., 2014). Movement of individuals between protected areas can occur through active migration or passive dispersal (e.g., Roberts et al., 2021), in some cases resulting in gene flow, as well as contributing to productivity and recruitment to adjacent fished stocks (e.g., Huserbråten et al., 2013). Genomic tools can provide empirical evidence of connectivity, which is difficult to quantify with conventional monitoring approaches for many taxa (Balbar et al., 2020). For example, genetic studies assessing kinship in Australasian snapper (Chrysophrys auratus) revealed that reproductive productivity within an MPA disproportionately contributed to recruitment in surrounding areas (Le Port et al., 2017). In other cases, genetic methods have revealed evidence of selection for limited dispersal, which correspondingly drove fine-scale genetic structure associated with the MPA protective measures (Sanford et al., 2006; Baskett and Barnett, 2015).
Often, fine-scale population differentiation identified in marine species is driven by adaptation to gradients in physical and chemical ocean conditions (e.g., Stanley et al., 2018). Recent genomic studies have revealed molecular underpinnings of local adaptation to the environment (e.g., temperature, salinity, and oceanography). Adaptive traits may be underlain by genetic architectures ranging from many small-effect loci (Bay R. A. et al., 2017) to single large-effect genes (Barson et al., 2015; Prince et al., 2017). The genetic architecture underlying local adaptation is predicted to affect how species respond to both current conditions and future change, making quantification of adaptive variation essential information for building resiliency into conservation planning (Bay R. A. et al., 2017; Lowen et al., 2019; Oomen et al., 2020; Layton and Bradbury 2022). Genomic data can also reveal other types of variation in the genome that play key roles in adaptation and population structure and persistence; this variation could inform conservation design and be used for monitoring. For example, Catanach et al. (2019) demonstrated that structural variants in the Australasian snapper genome outnumbered the SNP associated variation based on total bases affected. Structural variation such as large chromosomal inversions (Kess et al., 2019) and copy number variants (CNVs) (Kess et al., 2021; Layton et al., 2021) have also been identified in aquatic systems underlying divergent ecotypes and genomic signals of climate adaptation. By identifying and incorporating information on adaptive variation into conservation planning, specific areas can be prioritized to help protect biodiversity and promote long-term persistence of populations under climate change (Xuereb et al., 2021).
Recent genomic studies have also revealed the magnitudes of population decline using measures of genetic diversity such as effective population size (Ne), and have uncovered genome-wide differences between declining and stable populations (Hollenbeck et al., 2016; Lehnert et al., 2019). These types of analyses hold promise to identify species and populations of conservation concern for MPA design. Similarly, metrics of genetic diversity and connectivity, including Ne and heterozygosity, can be incorporated into conservation planning through network objectives (e.g., Gajdzik et al., 2021) or planning tools such as Marxan. In coastal Africa, Phair et al. (2021) demonstrated that conservation planning based on habitat models alone risked missing important genetic variation in the coastal seagrass (Zostera capensis) leading to design configurations that did not include evolutionarily unique populations and were thus less resilient to environmental change. These results emphasize the value of collecting genetic information a priori. Incorporating population-level stratification into the MPA network design can promote the conservation of genetic diversity and resiliency following the stewardship aspects of the BES.
While the theoretical applicability of omics tools in the MPA design and monitoring are robust, few real-world examples exist. One example of genomic application to MPA research focuses on the Gilbert Bay MPA in Labrador, Canada. With the collapse and subsequent moratorium of the Atlantic cod (Gadus morhua) came an increased pressure on nearshore cod stocks. A phenotypically and genetically distinct population of inshore cod reside within the Gilbert Bay MPA, which was established to protect this population (DFO 2010). Though the MPA protected the core habitat, genomic stock identification revealed that these genetically divergent cod were being exploited in adjacent fisheries (Sinclair-Waters et al., 2018a,b). In this case, genomic data corroborated prior tagging data (Morris et al., 2014) but also revealed the adaptive mechanism (i.e., chromosomal inversions) of divergence between the Gilbert Bay and northern cod (Sinclair-Waters et al., 2018a). In the long-term, the ability to genetically assign catch in fisheries outside the MPA will provide an important and robust basis for MPA management. Another study conducted on the California sea cucumber (Parastichopus californicus) used genomic data to identify priority areas for MPAs (Xuereb et al., 2021). The study found that focal areas for conservation differed among genomic metrics (i.e., diversity vs adaptive variation), highlighting the need to match the genetic metrics with the conservation objectives when evaluating design decisions. In each example, the application of genomic approaches revealed novel information about the conservation priorities that were directly applicable to design and monitoring.
Spatial conservation tools like MPAs help to regulate anthropogenic impacts on populations, and these impacts can be monitored using molecular omics approaches. Short-term human impacts, including overfishing, aquaculture escape events, and pollution, have produced measurable differences in adaptive variation among populations (Wringe et al., 2018; Therkildsen et al., 2019; Phair et al., 2020), demonstrating that omics have utility in quantifying anthropogenic impacts within and outside of MPAs. Transcriptomic methods (i.e., RNA-sequencing) can identify differences in gene expression among individuals, including variation in the transcript level and splicing, and can be used to identify mechanisms of rapid adaptation (Jacobs and Elmer, 2021). For example, Baratti et al. (2022) demonstrated higher concentrations of contaminants in marbled crab (Pachygrapsus marmoratus) from a polluted port compared to those in an adjacent MPA, and RNA-sequencing supported corresponding differences in the expression of stress-related genes supporting the conservation efficacy of the MPA. Epigenetic changes, such as DNA methylation, can provide another heritable means by which organisms may alter their phenotype and rapidly respond to environmental or anthropogenic stressors without changes to their underlying DNA sequence (Anastasiadi et al., 2021). As technologies evolve to accurately characterize genomic variation associated with phenotype, greater capacity to quantify and mitigate impacts of stressors, predict population responses, and ultimately conserve genetic biodiversity will improve.
A growing area of omics application and research is eDNA metabarcoding, which has revolutionized biomonitoring in aquatic ecosystems through detection of invasive and/or rare species (Weltz et al., 2017; Matejusova et al., 2021) and the characterization of community-level biodiversity (Bani et al., 2020; Djurhuus et al., 2020). This method involves the collection of extracellular DNA from aquatic samples and is often more taxonomically and monetarily efficient than traditional monitoring approaches (Fediajevaite et al., 2021; He et al., 2022). eDNA collection is minimally invasive and can be implemented in various environments and conditions, and thus represents a useful tool for MPA monitoring (Sawaya et al., 2019; Gold et al., 2021). Globally, MPAs cover millions of square kilometers, and scalable, cost-effective tools such as metabarcoding represent a tractable efficient solution for monitoring at large spatial extents and in remote areas. Traditional survey-based methods (e.g., trawling) are time-intensive and often limited by depth or topography, whereas eDNA sampling, while not without its own biases, is relatively simple, cost-effective, can be scaled across large MPAs (Gold et al., 2021), and provides a standardized biomonitoring platform deployable across ecosystem types (e.g., coastal to remote deep ocean).
New eDNA studies are developing PCR-free methods that employ direct sequencing of genomic DNA through shotgun metagenomic sequencing or “genome skimming” (Cowart et al., 2018), which can provide a more accurate measure of species abundance and prevent amplification biases (Peel et al., 2019). In addition, it is now common practice to sequence DNA directly in the field, using long-read Nanopore sequencing which can generate full-length gene or genome sequences to improve taxonomic resolution (Baloğlu et al., 2021). For reliable identifications, comprehensive and well-curated reference databases are critical; while such databases are publicly available and growing, many remain incomplete and can be difficult to curate (Weigand et al., 2019). In light of this, new bioinformatic approaches make use of reference-free identification algorithms, employing density-based clustering to detect both known and unknown species (Baloğlu et al., 2021). Exploration of the use of automated eDNA collection and processing systems, either mounted statically in key areas, or onboard automated vehicles is underway, providing a platform for real-time, continuous monitoring (e.g., Hansen et al., 2020). The use of marine invertebrates as natural eDNA samplers has also gained recent attention (e.g., sponges, Mariani et al., 2019). Though more work remains to test the applicability of eDNA as a surrogate for long-term monitoring approaches (Antich et al., 2021; Gold et al., 2021; He et al., 2022), it shows promise to be an adaptable technology for biomonitoring, which can inform management, detect species distribution shifts under climate change, and be a core part of the larger effort to sustainably manage marine activities under a BES (Curry and Ausubel 2021).
Interest in the blue economy and research involving genomics in MPAs has expanded markedly since 2018 (Figure 1). The proliferation of marine omics studies and the move to open-access data repositories have the potential to contribute to the planning and monitoring of MPAs and be a key tool in fisheries management for the blue economy. Advances in genome assembly approaches (e.g., chromosome level assemblies using chromatin capture, optical mapping, and long-read sequencing) add to a growing number of high quality reference genomes, through larger initiatives such as the Vertebrate Genomes Project. As the number of resources for species grows, so does the ability to integrate datasets into larger-scale single-species studies (Jeffery et al., 2018), or multi-species research (Stanley et al., 2018; Gajdzik et al., 2021). We recommend the regular incorporation of genomic and other omics data for species of interest (i.e., at the very least, those that are conservation priorities for the network) in the design and ongoing monitoring of marine conservation areas, to understand population structure, connectivity, and species diversity (see Table 1 for a list of applications and recommendations).

TABLE 1. Examples of molecular omics tools and their applications in various stages of MPA design and management, including baseline data acquisition, network design to incorporate genetic diversity and connectivity, and monitoring MPAs. Recommendations for which methods or tools to use are provided, with example references. We note that this does not represent an exhaustive list of methods and applications, but can be used to guide the process of MPA design and implementation.
Multi-species genomic approaches can provide information for understanding interactions between various levels of the ecosystem within an MPA network (Andrello et al., 2022). For example, comparative genomic approaches can help prioritize species (or populations) needing protection by identifying those that are most vulnerable to threats (Zoonomia Consortium 2020) or those with the greatest likelihood of recovery (Beichman et al., 2019). In contrast to the current genomic approaches relying on single reference genomes, a pangenomics approach which focuses not only on sequence diversity but also on the structural diversity of genomes would improve the quantification of adaptive diversity and predictions of future changes (Brockhurst et al., 2019).
The non-invasive nature of eDNA metabarcoding sampling makes it an ideal alternative to direct, in some cases destructive, monitoring techniques, and a strong candidate for real-time autonomous sampling (e.g., Hansen et al., 2020). Similarly, metagenomic sequencing of microorganisms provides a tool for monitoring ecological quality (Fruehe et al., 2021) and ecosystem services within MPAs (Curry and Ausubel 2021). For example, eDNA of benthic microbial community composition coupled with machine learning approaches has shown promise for improving indices of environmental impacts (Fruehe et al., 2021). While metabarcoding is now routinely applied by researchers to answer ecological questions, its uptake by managers and commercial industries is lagging behind (Curry and Ausubel 2021). Yet eDNA has great potential to complement existing biodiversity sampling approaches such as trawling and electrofishing (e.g., Afzali et al., 2021; Gold et al., 2021; He et al., 2022), and eventually, potentially replace these traditional methods in sensitive habitats often associated with protection by MPAs.
To contribute to real-time monitoring for the blue economy, it has even been proposed that ships could be designed to filter water along their transport routes, potentially covering large geographic areas and making the data readily available to resource managers and the public (Curry and Ausubel 2021). The “real-time” inference drawn from eDNA research can be further enhanced using environmental RNA (eRNA), which offers additional power to these eDNA approaches due to the high turnover rate of RNA compared to DNA (Yates et al., 2021). eRNA can enhance spatio-temporal resolution compared to traditional eDNA methods, as it primarily reflects physiologically active organisms that are in close proximity to sampling locations, and it can provide novel monitoring approaches, such as the ability to assess the health status of organisms and communities within a more focal area (Cristescu 2019; Yates et al., 2021).
Molecular omics technologies offer significant potential to improve and transform marine conservation planning, and recognition of its importance for monitoring and providing real-time information for the blue economy is quickly taking hold. The ability to use millions of genome-wide markers to delineate populations, assess connectivity, and determine environmental drivers is invaluable when designing networks of MPAs and subsequently managing and monitoring them. While the fields of genomics and eDNA are perpetually evolving, this should not be seen as a reason to refrain from their operationalization now. The data and sequences generated can be incorporated into future or expanded studies, be used as the basis for monitoring programs, or provide historical baselines as we enter an era of unprecedented biodiversity change. The adoption of these techniques toward marine conservation will allow targeted planning of protected areas and effective monitoring of changes in populations from anthropogenic and climate impacts, together enabling targeted and adaptive marine conservation.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
NJ and RS conceived the study. SL compiled the data and created Figure 1. All authors wrote, edited, and approved the manuscript.
This work was supported by funding provided to the Marine Conservation Targets Program with Fisheries and Oceans Canada.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Geneviève Faille (DFO Québec) and Kristen Westfall (DFO Pacific) for their helpful review of this article. We also thank the two reviewers for their helpful comments in improving the drafts of this manuscript.
Afzali, S. F., Bourdages, H., Laporte, M., Mérot, C., Normandeau, E., Audet, C., et al. (2021). Comparing Environmental Metabarcoding and Trawling Survey of Demersal Fish Communities in the Gulf of St. Lawrence, Canada. Environ. DNA 3, 22–42. doi:10.1002/edn3.111
Agardy, T., Di Sciara, G. N., and Christie, P. (2011). Mind the Gap: Addressing the Shortcomings of Marine Protected Areas through Large Scale Marine Spatial Planning. Mar. Policy 35, 226–232. doi:10.1016/j.marpol.2010.10.006
Anastasiadi, D., Venney, C. J., Bernatchez, L., and Wellenreuther, M. (2021). Epigenetic Inheritance and Reproductive Mode in Plants and Animals. Trends Ecol. Evol. 36, 1124–1140. doi:10.1016/j.tree.2021.08.006
Andrello, M., D’Aloia, C., Dalongeville, A., Escalante, M. A., Guerrero, J., Perrier, C., et al. (2022). Evolving Spatial Conservation Prioritization with Intraspecific Genetic Data. Trends Ecol. Evol. 37, 553–564. doi:10.1016/j.tree.2022.03.003
Antich, A., Palacín, C., Cebrian, E., Golo, R., Wangensteen, O. S., and Turon, X. (2021). Marine Biomonitoring with eDNA: Can Metabarcoding of Water Samples Cut it as a Tool for Surveying Benthic Communities? Mol. Ecol. 30, 3175–3188. doi:10.1111/mec.15641
Balbar, A. C., Daigle, R. M., Heaslip, S. G., Jeffery, N. W., Proudfoot, B., Robb, C. K., et al. (2020). Approaches for Assessing and Monitoring Representation, Replication, and Connectivity in Marine Conservation Networks. DFO Can. Sci. Advis. Sec. Res. Doc. 2020. 050. vii + 57 p.
Baloğlu, B., Chen, Z., Elbrecht, V., Braukmann, T., MacDonald, S., and Steinke, D. (2021). A Workflow for Accurate Metabarcoding Using Nanopore MinION Sequencing. Methods Ecol. Evol. 12, 794–804. doi:10.1111/2041-210x.13561
Bani, A., De Brauwer, M., Creer, S., Dumbrell, A. J., Limmon, G., Jompa, J., et al. (2020). Informing Marine Spatial Planning Decisions with Environmental DNA. Adv. Ecol. Res. 62, 375–407. doi:10.1016/bs.aecr.2020.01.011
Baratti, M., Pinosio, S., Gori, M., Biricolti, S., Chini, G., Fratini, S., et al. (2022). Differential Gene Expression and Chemical Patterns of an Intertidal Crab Inhabiting a Polluted Port and an Adjacent Marine Protected Area. Sci. Total Environ. 822, 153463. doi:10.1016/j.scitotenv.2022.153463
Barney, B. T., Munkholm, C., Walt, D. R., and Palumbi, S. R. (2017). Highly Localized Divergence within Supergenes in Atlantic Cod (Gadus morhua) within the Gulf of Maine. BMC Genomics 18, 271. doi:10.1186/s12864-017-3660-3
Barson, N. J., Aykanat, T., Hindar, K., Baranski, M., Bolstad, G. H., Fiske, P., et al. (2015). Sex-dependent Dominance at a Single Locus Maintains Variation in Age at Maturity in Salmon. Nature 528, 405–408. doi:10.1038/nature16062
Baskett, M. L., and Barnett, L. A. K. (2015). The Ecological and Evolutionary Consequences of Marine Reserves. Annu. Rev. Ecol. Evol. Syst. 46, 49–73. doi:10.1146/annurev-ecolsys-112414-054424
Bay, R. A., Rose, N. H., Logan, C. A., and Palumbi, S. R. (2017b). Genomic Models Predict Successful Coral Adaptation if Future Ocean Warming Rates Are Reduced. Sci. Adv. 3, e1701413. doi:10.1126/sciadv.1701413
Bay, R. A., and Palumbi, S. R. (2014). Multilocus Adaptation Associated with Heat Resistance in Reef-Building Corals. Curr. Biol. 24, 2952–2956. doi:10.1016/j.cub.2014.10.044
Bay, R. A., Rose, N., Barrett, R., Bernatchez, L., Ghalambor, C. K., Lasky, J. R., et al. (2017a). Predicting Responses to Contemporary Environmental Change Using Evolutionary Response Architectures. Am. Nat. 189, 463–473. doi:10.1086/691233
Beichman, A. C., Koepfli, K.-P., Li, G., Murphy, W., Dobrynin, P., Kliver, S., et al. (2019). Aquatic Adaptation and Depleted Diversity: a Deep Dive into the Genomes of the Sea Otter and Giant Otter. Mol. Biol. Evol. 36, 2631–2655. doi:10.1093/molbev/msz101
Bennett, N. J., Cisneros-Montemayor, A. M., Blythe, J., Silver, J. J., Singh, G., Andrews, N., et al. (2019). Towards a Sustainable and Equitable Blue Economy. Nat. Sustain. 2, 991–993. doi:10.1038/s41893-019-0404-1
Bravington, M. V., Grewe, P. M., and Davies, C. R. (2016). Absolute Abundance of Southern Bluefin Tuna Estimated by Close-Kin Mark-Recapture. Nat. Commun. 7, 13162–13274. doi:10.1038/ncomms13162
Brockhurst, M. A., Harrison, E., Hall, J. P. J., Richards, T., McNally, A., and MacLean, C. (2019). The Ecology and Evolution of Pangenomes. Curr. Biol. 29, R1094–R1103. doi:10.1016/j.cub.2019.08.012
Bruce, T., Meirelles, P. M., Garcia, G., Paranhos, R., Rezende, C. E., de Moura, R. L., et al. (2012). Abrolhos Bank Reef Health Evaluated by Means of Water Quality, Microbial Diversity, Benthic Cover, and Fish Biomass Data. PloS One 7 (6), e36687. doi:10.1371/journal.pone.0036687
Bryndum-Buchholz, A., Boerder, K., Stanley, R. R. E., Hurley, I., Boyce, D. G., Dunmall, K. M., et al. (2022). A Climate-Resilient Marine Conservation Network for Canada. FACETS 7, 571–590. doi:10.1139/facets-2021-0122
Catanach, A., Crowhurst, R., Deng, C., David, C., Bernatchez, L., and Wellenreuther, M. (2019). The Genomic Pool of Standing Structural Variation Outnumbers Single Nucleotide Polymorphism by Threefold in the Marine Teleost Chrysophrys Auratus. Mol. Ecol. 28 (6), 1210–1223. doi:10.1111/mec.15051
Cowart, D. A., Murphy, K. R., and Cheng, C.-H. C. (2018). Metagenomic Sequencing of Environmental DNA Reveals Marine Faunal Assemblages from the West Antarctic Peninsula. Mar. Genomics 37, 148–160. doi:10.1016/j.margen.2017.11.003
Cristescu, M. E. (2019). Can Environmental RNA Revolutionize Biodiversity Science? Trends Ecol. Evol. 34, 694–697. doi:10.1016/j.tree.2019.05.003
Cros, A., Toonen, R. J., Donahue, M. J., and Karl, S. A. (2017). Connecting Palau's Marine Protected Areas: a Population Genetic Approach to Conservation. Coral Reefs 36, 735–748. doi:10.1007/s00338-017-1565-x
Curry, A., and Ausubel, J. H. (2021). “Biological Information for the New Blue Economy and the Emerging Role of eDNA,” in Preparing a Workforce for the New Blue Economy (Elsevier), 249–258. doi:10.1016/b978-0-12-821431-2.00030-5
D'Aloia, C. C., Bogdanowicz, S. M., Andrés, J. A., and Buston, P. M. (2022). Population Assignment Tests Uncover Rare Long-Distance Marine Larval Dispersal Events. Ecology 103, e03559. doi:10.1002/ecy.3559
D’Aloia, C. C., Bogdanowicz, S. M., Harrison, R. G., and Buston, P. M. (2017). Cryptic Genetic Diversity and Spatial Patterns of Admixture within Belizean Marine Reserves. Conserv. Genet. 18, 211–223. doi:10.1007/s10592-016-0895-5
Davey, J. W., and Blaxter, M. L. (2010). RADSeq: Next-Generation Population Genetics. Briefings Funct. Genomics 9, 416–423. doi:10.1093/bfgp/elq031
Devloo-Delva, F., Maes, G. E., Hernández, S. I., Mcallister, J. D., Gunasekera, R. M., Grewe, P. M., et al. (2019). Accounting for Kin Sampling Reveals Genetic Connectivity in Tasmanian and New Zealand School Sharks, Galeorhinus galeus. Ecol. Evol. 9, 4465–4472. doi:10.1002/ece3.5012
DFO (2010). Review of the Gilbert Bay Marine Protected Area Monitoring Indicators, Protocols, and Strategies, and as Assessment of the Gilbert Bay Cod Population. Can. Sci. Advis. Sec. Sci. Advis. Rep. 2010, 12. 027.
Di Lorenzo, M., Guidetti, P., Di Franco, A., Calò, A., and Claudet, J. (2020). Assessing Spillover from Marine Protected Areas and its Drivers: A Meta‐analytical Approach. Fish. Fish. 21, 906–915. doi:10.1111/faf.12469
Djurhuus, A., Closek, C. J., Kelly, R. P., Pitz, K. J., Michisaki, R. P., Starks, H. A., et al. (2020). Environmental DNA Reveals Seasonal Shifts and Potential Interactions in a Marine Community. Nat. Commun. 11, 254. doi:10.1038/s41467-019-14105-1
Dorant, Y., Benestan, L., Rougemont, Q., Normandeau, E., Boyle, B., Rochette, R., et al. (2019). Comparing Pool‐seq, Rapture, and GBS Genotyping for Inferring Weak Population Structure: The American Lobster ( Homarus americanus ) as a Case Study. Ecol. Evol. 9, 6606–6623. doi:10.1002/ece3.5240
Fediajevaite, J., Priestley, V., Arnold, R., and Savolainen, V. (2021). Meta-analysis Shows that Environmental DNA Outperforms Traditional Surveys, but Warrants Better Reporting Standards. Ecol. Evol. 11, 4803–4815. doi:10.1002/ece3.7382
Frühe, L., Cordier, T., Dully, V., Breiner, H. W., Lentendu, G., Pawlowski, J., et al. (2021). Supervised Machine Learning Is Superior to Indicator Value Inference in Monitoring the Environmental Impacts of Salmon Aquaculture Using eDNA Metabarcodes. Mol. Ecol. 30, 2988–3006. doi:10.1111/mec.15434
Gaines, S. D., White, C., Carr, M. H., and Palumbi, S. R. (2010). Designing Marine Reserve Networks for Both Conservation and Fisheries Management. Proc. Natl. Acad. Sci. U.S.A. 107, 18286–18293. doi:10.1073/pnas.0906473107
Gajdzik, L., DeCarlo, T. M., Aylagas, E., Coker, D. J., Green, A. L., Majoris, J. E., et al. (2021). A Portfolio of Climate‐tailored Approaches to Advance the Design of Marine Protected Areas in the Red Sea. Glob. Change Biol. 27, 3956–3968. doi:10.1111/gcb.15719
Global Ocean Alliance (2022). Global Ocean Alliance: 30by30 Initiative. https://www.gov.uk/government/topical-events/global-ocean-alliance-30by30-initiative (Accessed January, 2022).
Gold, Z., Sprague, J., Kushner, D. J., Zerecero Marin, E., and Barber, P. H. (2021). eDNA Metabarcoding as a Biomonitoring Tool for Marine Protected Areas. PLoS One 16, e0238557. doi:10.1371/journal.pone.0238557
Grorud-Colvert, K., Claudet, J., Tissot, B. N., Caselle, J. E., Carr, M. H., Day, J. C., et al. (2014). Marine Protected Area Networks: Assessing whether the Whole Is Greater Than the Sum of its Parts. PloS One 9, e102298. doi:10.1371/journal.pone.0102298
Halpern, B. S., Lester, S. E., and McLeod, K. L. (2010). Placing Marine Protected Areas onto the Ecosystem-Based Management Seascape. Proc. Natl. Acad. Sci. U.S.A. 107, 18312–18317. doi:10.1073/pnas.0908503107
Hansen, B. K., Jacobsen, M. W., Middelboe, A. L., Preston, C. M., Marin, R., Bekkevold, D., et al. (2020). Remote, Autonomous Real-Time Monitoring of Environmental DNA from Commercial Fish. Sci. Rep. 10, 13272–13278. doi:10.1038/s41598-020-70206-8
Harmelin-Vivien, M., Le Diréach, L., Bayle-Sempere, J., Charbonnel, E., García-Charton, J. A., Ody, D., et al. (2008). Gradients of Abundance and Biomass across Reserve Boundaries in Six Mediterranean Marine Protected Areas: Evidence of Fish Spillover? Biol. Conserv. 141, 1829–1839. doi:10.1016/j.biocon.2008.04.029
He, X., Stanley, R. R. E., Jeffery, N. W., Rubidge, E. M., Hamilton, L. C., Westfall, K. M., et al. (2022). Fish Community Surveys in Eelgrass Beds Using Both eDNA Metabarcoding and Seining: Implications for Biodiversity Monitoring in the Coastal Zone. Can. J. Fish. Aquat. Sci., 1–12. doi:10.1139/cjfas-2021-0215
Hollenbeck, C. M., Portnoy, D. S., and Gold, J. R. (2016). A Method for Detecting Recent Changes in Contemporary Effective Population Size from Linkage Disequilibrium at Linked and Unlinked Loci. Heredity 117, 207–216. doi:10.1038/hdy.2016.30
Huserbråten, M. B., Moland, E., Knutsen, H., Olsen, E. M., André, C., and Stenseth, N. C. (2013). Conservation, Spillover and Gene Flow within a Network of Northern European Marine Protected Areas. PLOS one 8 (9), e73388. doi:10.1371/journal.pone.0073388
Jacobs, A., and Elmer, K. R. (2021). Alternative Splicing and Gene Expression Play Contrasting Roles in the Parallel Phenotypic Evolution of a Salmonid Fish. Mol. Ecol. 30 (20), 4955–4969. doi:10.1111/mec.15817
Jeffery, N. W., Wringe, B. F., McBride, M. C., Hamilton, L. C., Stanley, R. R. E., Bernatchez, L., et al. (2018). Range-wide Regional Assignment of Atlantic Salmon (Salmo salar) Using Genome Wide Single-Nucleotide Polymorphisms. Fish. Res. 206, 163–175. doi:10.1016/j.fishres.2018.05.017
Kess, T., Bentzen, P., Lehnert, S. J., Sylvester, E. V. A., Lien, S., Kent, M. P., et al. (2019). A Migration-Associated Supergene Reveals Loss of Biocomplexity in Atlantic Cod. Sci. Adv. 5, eaav2461. doi:10.1126/sciadv.aav2461
Kess, T., Dempson, J. B., Lehnert, S. J., Layton, K. K. S., Einfeldt, A., Bentzen, P., et al. (2021). Genomic Basis of Deep‐water Adaptation in Arctic Charr ( Salvelinus alpinus ) Morphs. Mol. Ecol. 30, 4415–4432. doi:10.1111/mec.16033
Layton, K. K. S., and Bradbury, I. R. (2022). Harnessing the Power of Multi‐omics Data for Predicting Climate Change Response. J. Anim. Ecol. doi:10.1111/1365-2656.13619
Layton, K. K. S., Snelgrove, P. V. R., Dempson, J. B., Kess, T., Lehnert, S. J., Bentzen, P., et al. (2021). Genomic Evidence of Past and Future Climate-Linked Loss in a Migratory Arctic Fish. Nat. Clim. Change 11 (2), 158–165. doi:10.1038/s41558-020-00959-7
Lehnert, S. J., Kess, T., Bentzen, P., Kent, M. P., Lien, S., Gilbey, J., et al. (2019). Genomic Signatures and Correlates of Widespread Population Declines in Salmon. Nat. Commun. 10, 2996. doi:10.1038/s41467-019-10972-w
Lou, R. N., Jacobs, A., Wilder, A. P., and Therkildsen, N. O. (2021). A Beginner's Guide to Low‐coverage Whole Genome Sequencing for Population Genomics. Mol. Ecol. 30, 5966–5993. doi:10.1111/mec.16077
Lowen, J. B., Hart, D. R., Stanley, R. R. E., Lehnert, S. J., Bradbury, I. R., and DiBacco, C. (2019). Assessing Effects of Genetic, Environmental, and Biotic Gradients in Species Distribution Modelling. ICES J. Mar. Sci. 76, 1762–1775. doi:10.1093/icesjms/fsz049
Macreadie, P. I., Costa, M. D. P., Atwood, T. B., Friess, D. A., Kelleway, J. J., Kennedy, H., et al. (2021). Blue Carbon as a Natural Climate Solution. Nat. Rev. Earth Environ. 2, 826–839. doi:10.1038/s43017-021-00224-1
Mariani, S., Baillie, C., Colosimo, G., and Riesgo, A. (2019). Sponges as Natural Environmental DNA Samplers. Curr. Biol. 29, R401–R402. doi:10.1016/j.cub.2019.04.031
Matejusova, I., Graham, J., Bland, F., Lacaze, J.-P., Herman, G., Brown, L., et al. (2021). Environmental DNA Based Surveillance for the Highly Invasive Carpet Sea Squirt Didemnum Vexillum: A Targeted Single-Species Approach. Front. Mar. Sci. 8, 728456. doi:10.3389/fmars.2021.728456
Mendez, M., Kershaw, F., Palumbi, S., Pinsky, M., Ray, C., Rosenbaum, H., et al. (2014). Marine Spatial Planning 2.0: Genes and Satellites to Conserve Seascape Dynamics. Aquat. Conserv. Mar. Freshw. Ecosyst. 24 (6), 742–744. doi:10.1002/aqc.2533
Morris, C. J., and Green, J. M. (2014). MPA Regulations Should Incorporate Adaptive Management-The Case of Gilbert Bay Labrador Atlantic Cod (Gadus morhua). Mar. Policy 49, 20–28. doi:10.1016/j.marpol.2014.03.025
Morris, C. J., Green, J. M., Snelgrove, P. V. R., Pennell, C. J., and Ollerhead, L. M. N. (2014). Temporal and Spatial Migration of Atlantic Cod (Gadus morhua) inside and outside a Marine Protected Area and Evidence for the Role of Prior Experience in Homing. Can. J. Fish. Aquat. Sci. 71 (11), 1704–1712. doi:10.1139/cjfas-2014-0036
Oomen, R. A., Kuparinen, A., and Hutchings, J. A. (2020). Consequences of Single-Locus and Tightly Linked Genomic Architectures for Evolutionary Responses to Environmental Change. J. Hered. 111, 319–332. doi:10.1093/jhered/esaa020
Peel, N., Dicks, L. V., Clark, M. D., Heavens, D., Percival‐Alwyn, L., Cooper, C., et al. (2019). Semi‐quantitative Characterisation of Mixed Pollen Samples Using MinION Sequencing and Reverse Metagenomics (RevMet). Methods Ecol. Evol. 10, 1690–1701. doi:10.1111/2041-210x.13265
Phair, N. L., Nielsen, E. S., and von der Heyden, S. (2021). Applying Genomic Data to Seagrass Conservation. Biodivers. Conserv. 30, 2079–2096. doi:10.1007/s10531-021-02184-w
Phair, N. L., Toonen, R. J., Knapp, I. S. S., and von der Heyden, S. (2020). Anthropogenic Pressures Negatively Impact Genomic Diversity of the Vulnerable Seagrass Zostera Capensis. J. Environ. Manag. 255, 109831. doi:10.1016/j.jenvman.2019.109831
Prince, D. J., O'Rourke, S. M., Thompson, T. Q., Ali, O. A., Lyman, H. S., Saglam, I. K., et al. (2017). The Evolutionary Basis of Premature Migration in Pacific Salmon Highlights the Utility of Genomics for Informing Conservation. Sci. Adv. 3 (8), e1603198. doi:10.1126/sciadv.1603198
Roberts, K. E., Cook, C. N., Beher, J., and Treml, E. A. (2021). Assessing the Current State of Ecological Connectivity in a Large Marine Protected Area System. Conserv. Biol. 35, 699–710. doi:10.1111/cobi.13580
Samuel, R. M., Meyer, R., Buttigieg, P. L., Davies, N., Jeffery, N. W., Meyer, C., et al. (2021). Toward a Global Public Repository of Community Protocols to Encourage Best Practices in Biomolecular Ocean Observing and Research. Front. Mar. Sci. 8, 758694. doi:10.3389/fmars.2021.758694
Sanford, E., Holzman, S. B., Haney, R. A., Rand, D. M., and Bertness, M. D. (2006). Larval Tolerance, Gene Flow, and the Northern Geographic Range Limit of Fiddler Crabs. Ecology 87, 2882–2894. doi:10.1890/0012-9658(2006)87[2882:ltgfat]2.0.co;2
Sawaya, N. A., Djurhuus, A., Closek, C. J., Hepner, M., Olesin, E., Visser, L., et al. (2019). Assessing Eukaryotic Biodiversity in the Florida Keys National Marine Sanctuary through Environmental DNA Metabarcoding. Ecol. Evol. 9, 1029–1040. doi:10.1002/ece3.4742
Schlötterer, C., Tobler, R., Kofler, R., and Nolte, V. (2014). Sequencing Pools of Individuals - Mining Genome-wide Polymorphism Data without Big Funding. Nat. Rev. Genet. 15, 749–763. doi:10.1038/nrg3803
Sinclair-Waters, M., Bradbury, I. R., Morris, C. J., Lien, S., Kent, M. P., and Bentzen, P. (2018a). Ancient Chromosomal Rearrangement Associated with Local Adaptation of a Postglacially Colonized Population of Atlantic Cod in the Northwest Atlantic. Mol. Ecol. 27, 339–351. doi:10.1111/mec.14442
Sinclair-Waters, M., Bentzen, P., Morris, C. J., Ruzzante, D. E., Kent, M. P., Lien, S., et al. (2018b). Genomic Tools for Management and Conservation of Atlantic Cod in a Coastal Marine Protected Area. Can. J. Fish. Aquat. Sci. 75, 1915–1925. doi:10.1139/cjfas-2017-0254
Smith-Godfrey, S. (2016). Defining the Blue Economy. Marit. Aff. J. Natl. Marit. Found. India 12, 58–64. doi:10.1080/09733159.2016.1175131
Stanley, R. R. E., DiBacco, C., Lowen, B., Beiko, R. G., Jeffery, N. W., Van Wyngaarden, M., et al. (2018). A Climate-Associated Multispecies Cryptic Cline in the Northwest Atlantic. Sci. Adv. 4 (3), eaaq0929. doi:10.1126/sciadv.aaq0929
Therkildsen, N. O., Wilder, A. P., Conover, D. O., Munch, S. B., Baumann, H., and Palumbi, S. R. (2019). Contrasting Genomic Shifts Underlie Parallel Phenotypic Evolution in Response to Fishing. Science 365, 487–490. doi:10.1126/science.aaw7271
Tittensor, D. P., Beger, M., Boerder, K., Boyce, D. G., Cavanagh, R. D., Cosandey-Godin, A., et al. (2019). Integrating Climate Adaptation and Biodiversity Conservation in the Global Ocean. Sci. Adv. 5 (11), eaay9969. doi:10.1126/sciadv.aay9969
Unep-Wcmc, IUCN (2022). Protected Planet: The World Database on Protected Areas (WDPA) and World Database on Other Effective Area-Based Conservation Measures (WD-OECM) [Online], February 2022. UNEP-WCMC and IUCN. Cambridge, UK. Available at: www.protectedplanet.net.
Van Wyngaarden, M., Snelgrove, P. V. R., DiBacco, C., Hamilton, L. C., Rodríguez-Ezpeleta, N., Jeffery, N. W., et al. (2017). Identifying Patterns of Dispersal, Connectivity and Selection in the Sea scallop,Placopecten magellanicus,using RADseq-Derived SNPs. Evol. Appl. 10, 102–117. doi:10.1111/eva.12432
Von der Heyden, S. (2009). Why Do We Need to Integrate Population Genetics into South African Marine Protected Area Planning? Afr. J. Mar. Sci. 31 (2), 263–269. doi:10.2989/ajms.2009.31.2.14.886
Walsh, C. A. J., Momigliano, P., Boussarie, G., Robbins, W. D., Bonnin, L., Fauvelot, C., et al. (2022). Genomic Insights into the Historical and Contemporary Demographics of the Grey Reef Shark. Heredity 128, 225–235. doi:10.1038/s41437-022-00514-4
Weigand, H., Beermann, A. J., Čiampor, F., Costa, F. O., Csabai, Z., Duarte, S., et al. (2019). DNA Barcode Reference Libraries for the Monitoring of Aquatic Biota in Europe: Gap-analysis and Recommendations for Future Work. Sci. Total Environ. 678, 499–524. doi:10.1016/j.scitotenv.2019.04.247
Weltz, K., Lyle, J. M., Ovenden, J., Morgan, J. A. T., Moreno, D. A., and Semmens, J. M. (2017). Application of Environmental DNA to Detect an Endangered Marine Skate Species in the Wild. PLoS One 12, e0178124. doi:10.1371/journal.pone.0178124
West, K. M., Stat, M., Harvey, E. S., Skepper, C. L., DiBattista, J. D., Richards, Z. T., et al. (2020). eDNA Metabarcoding Survey Reveals Fine‐scale Coral Reef Community Variation across a Remote, Tropical Island Ecosystem. Mol. Ecol. 29, 1069–1086. doi:10.1111/mec.15382
Wilhelm, T. A., Sheppard, C. R. C., Sheppard, A. L. S., Gaymer, C. F., Parks, J., Wagner, D., et al. (2014). Large Marine Protected Areas - Advantages and Challenges of Going Big. Aquat. Conserv. Mar. Freshw. Ecosyst. 24, 24–30. doi:10.1002/aqc.2499
Wringe, B. F., Jeffery, N. W., Stanley, R. R. E., Hamilton, L. C., Anderson, E. C., Fleming, I. A., et al. (2018). Extensive Hybridization Following a Large Escape of Domesticated Atlantic Salmon in the Northwest Atlantic. Commun. Biol. 1, 108–109. doi:10.1038/s42003-018-0112-9
Xuereb, A., D'Aloia, C. C., Andrello, M., Bernatchez, L., and Fortin, M. J. (2021). Incorporating Putatively Neutral and Adaptive Genomic Data into Marine Conservation Planning. Conserv. Biol. 35, 909–920. doi:10.1111/cobi.13609
Xuereb, A., D’Aloia, C. C., Daigle, R. M., Andrello, M., Dalongeville, A., Manel, S., et al. (2019). “Marine Conservation and Marine Protected Areas,” in Population Genomics: Marine Organisms (Cham: Springer), 423–446. doi:10.1007/13836_2018_63
Yates, M. C., Derry, A. M., and Cristescu, M. E. (2021). Environmental RNA: a Revolution in Ecological Resolution? Trends Ecol. Evol. 36, 601–609. doi:10.1016/j.tree.2021.03.001
Keywords: marine conservation, population genomics, environmental DNA (eDNA), metabarcoding, connectivity, conservation planning
Citation: Jeffery NW, Lehnert SJ, Kess T, Layton KKS, Wringe BF and Stanley RRE (2022) Application of Omics Tools in Designing and Monitoring Marine Protected Areas For a Sustainable Blue Economy. Front. Genet. 13:886494. doi: 10.3389/fgene.2022.886494
Received: 28 February 2022; Accepted: 16 May 2022;
Published: 22 June 2022.
Edited by:
Simo Njabulo Maduna, Norwegian Institute of Bioeconomy Research (NIBIO), NorwayReviewed by:
Helene Magalon, Université de la Réunion, FranceCopyright © 2022 Jeffery, Lehnert, Kess, Layton, Wringe and Stanley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas W. Jeffery, Tmljay5KZWZmZXJ5QGRmby1tcG8uZ2MuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.