
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
DATA REPORT article
Front. Genet., 06 July 2022
Sec. Statistical Genetics and Methodology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.883199
This article is part of the Research TopicExploring Omics Tools & Data Integration Approaches to Deepen Insights into Complex DiseasesView all 5 articles
Cystic fibrosis (CF) is a common heritable genetic disorder caused by a defect in the cystic fibrosis conductance regulator gene, resulting in several complications in the human body (Kreda et al., 2012). So far, the pathological changes in the lungs are best studied due to the high mortality rates linked to poor lung function and the recurrent development of severe biofilm-related infections (Flume et al., 2009; Ciofu et al., 2015). Staphylococcus aureus and Pseudomonas aeruginosa are the most prevalent pathogens that colonize structurally abnormal airways such as those diagnosed with CF and other chronic obstructive lung diseases (Lyczak et al., 2002; Hubert et al., 2013).
Although these bacteria seem to succeed with one another, CF patients acquire coinciding P. aeruginosa and S. aureus pulmonary infections, being co-infection usually associated with decreased lung function and increased frequency of pulmonary exacerbations (Limoli et al., 2016). Furthermore, P. aeruginosa and S. aureus pathogens adopt a biofilm mode of growth, which contributes to high tolerance to antibiotic treatment (Schobert and Jahn, 2010) and the recalcitrant nature of these chronic co-infections (Burmølle et al., 2006; Lopes et al., 2012), leading to significant patient morbidity and mortality (Cox et al., 2010). Interactions between P. aeruginosa and S. aureus have been widely studied, and it is commonly admitted that P. aeruginosa outcompetes S. aureus, perhaps outcompeting S. aureus for limited nutrients (Mashburn et al., 2005) or producing anti-staphylococcal compounds (DeLeon et al., 2014; Fugère et al., 2014), having S. aureus a minimal contribution to the overall course of the CF-associated biofilm infections (Bragonzi et al., 2012; Filkins et al., 2015). However, P. aeruginosa and S. aureus have been identified in the same lobe of CF lungs (Hogan et al., 2016; Wakeman et al., 2016) and are frequently diagnosed (Limoli et al., 2016; Zolin et al., 2019) as co-infecting species in CF patients. Moreover, P. aeruginosa strains isolated from early infection outcompete S. aureus, while strains isolated from chronic infection are less aggressive and can be co-cultivated with S. aureus (Frydenlund Michelsen et al., 2016; Limoli et al., 2017), suggesting that these pathogens can interact in vivo.
In a previous study, we showed that S. aureus can grow and coexist with P. aeruginosa under dual-species biofilm conditions (Magalhães et al., 2021). Following up on these findings, and acknowledging that the molecular mechanisms behind these interactions are largely unknown, the purpose of the present study was, therefore, to identify the major transcriptomic features of P. aeruginosa–S. aureus dual-species biofilms, using high-throughput RNA-sequencing (RNA-seq). Herein, we described the full transcriptome of P. aeruginosa and S. aureus single- and dual-species biofilms and used a data analysis approach based on direct and functional gene interactions, namely gene set enrichment. These results will be invaluable for future functional studies involving P. aeruginosa–S. aureus interactions.
P. aeruginosa UCBPP-PA14 and S. aureus ATCC 25923 were used throughout this work. Both bacteria were stored at −80 ± 2°C in tryptic soy broth (TSB, Liofilchem, Italy) supplemented with 20% glycerol. Before each assay, bacteria were sub-cultured from frozen stock preparations onto plates of TSB supplemented with 2% (w/v) agar and incubated aerobically at 37°C for 24 h.
Single- and dual-species biofilms formed by P. aeruginosa and S. aureus were prepared as described previously (Magalhães et al., 2017), with minor modifications. Briefly, overnight cultures of each species, grown in TSB at 37°C and 120 rpm in air conditions, were washed in sterile water and diluted in TSB to obtain 1 × 107 CFU/ml. Bacterial numbers were estimated using optical density at 620 nm. Calibrations were previously performed for each bacterial strain to correlate the absorbance at 620 nm with the number of colony-forming units (CFUs) (Magalhães et al., 2021). For dual-species cultures, the suspended inoculum of each species was combined in a 1:1 ratio. Bacterial suspensions were dispensed in 24-well polystyrene plates (Orange Scientific, Braine-l`Alleud, Belgium) and incubated at 37°C on a horizontal shaker (120 rpm) for 24 h. Twenty-four-hour biofilms were then washed once with 0.9% NaCl, scraped from the bottom and the wall of the plates in 1 ml of RNA protect bacteria reagent (QIAGEN), which was diluted 2:1 in RNase-free water, as indicated by the manufacturer. After 5 min of incubation at room temperature, biofilm cells were harvested by centrifugation (20 min, 3,132×g) and RNA isolation was then performed. This assay was repeated six independent times.
Total RNA was extracted using the RNeasy mini kit (QIAGEN) as optimized before (França et al., 2012). In brief, cells were suspended in 600 µl of the lysis buffer provided by the kit, plus 500 µl of phenol and 12 µl of β-mercaptoethanol. This suspension was transferred to a safe lock tube (2 ml) with 0.4 g of acid-washed 150–212 mm glass beads (Sigma) and using a BeadBug™ 6 (Benchmark Scientific) cell disruptor, the cells were lysed (4 × 4,500 rpm for 35 s, with 5 min intervals on ice between cycles). Finally, the tubes were centrifuged, and the suspension was transferred into a new tube. An equal volume of 70% of ethanol was added, the suspension was transferred into the RNeasy mini kit columns, and the manufacturer’s instructions were strictly followed. RNA quality was determined using the Agilent TapeStation 4200 (Agilent) and RNA quality indicators were above eight for all samples. Thereafter, total RNA obtained from three independent experiments was mixed and treated with TURBO DNase (Ambion) to degrade genomic DNA. Additionally, before the library construction, bacterial ribosomal RNA was removed using the NEBNext rRNA Depletion Kit (Bacteria). RNA libraries were prepared by strictly following the instructions of the kit KAPA HyperPrep (Roche). Libraries’ quality was determined using Agilent TapeStation 4200, and data were generated using Illumina NovaSeq 6000 from paired-end reads (2 × 150 bp).
After sequencing, Bcl2fastq version 2 (Illumina) was used for base calling and to convert the data to FASTQs files. CLC Genomics Workbench version 21 (QIAGEN) was then used for quality, ambiguity, and length trimming, using default settings. Thereafter, CLC was used for alignment using S. aureus (GenBank accession number: CP009361) and P. aeruginosa (GenBank accession number: NZ_CP034244) genomes, normalization of the reads (to transcripts per million—TPM), and for the analysis of differential gene expression (using single-species biofilms as control). Baggerley’s test (Pawitan et al., 2005) was applied to identify statistically significant alterations in single- vs. dual-species biofilms. Alterations with fold changes below two and p-values above 0.05 were discarded. Raw and analysed datasets have been deposited in NCBI’s Gene Expression Omnibus database and are accessible through GEO series accession number GSE195909.
Gene function was annotated based on the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, version 11.5) (Szklarczyk et al., 2021), BLAST, Clusters of Orthologous Groups of proteins (COGs) (Tatusov et al., 2000; Galperin et al., 2015), Gene Ontology (GO) (Ashburner et al., 2000; Carbon et al., 2021), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000; Kanehisa, 2019; Kanehisa et al., 2021) databases. The functional annotations were all determined based on the highest sequence similarity in these databases. GO enrichment analysis of differentially expressed genes was performed using standard GO terms from the Gene Ontology Resource and a Fisher’s exact test with FDR p-value <0.05 to estimate the statistical significance of the enrichment. Similarly, KEGG pathway analysis of differentially expressed genes was performed using KOBAS v2.1.1 (p < 0.05, hypergeometric test/Fisher’s exact test with FDR) (Bu et al., 2021).
To study the responses of P. aeruginosa and S. aureus biofilm cells during interspecies interaction, we compared the gene expression profiles of both bacteria after 24 h of dual-species versus single-species growth. Earlier studies by Magalhães et al. (2021) have shown that S. aureus is present at high cell numbers in the P. aeruginosa-dominated 24 h dual-species biofilm consortia, indicating that the effects of interspecies interactions had not translated into significant changes in the population dynamics. Despite the evident coexistence interaction displayed after 24 h of co-culture, the transcriptome in each bacterium was affected by the presence of the other one when compared to the single-species transcriptome.
The total number of sequencing reads obtained ranged between 79,325,532 and 139,925,400 (Supplementary Table S1). A principal component analysis (PCA) of all samples showed a clear separation between the conditions under study (Supplementary Figure S1). Furthermore, heat maps revealed marked differences in the expression profile of either P. aeruginosa or S. aureus when grown as single- or dual-species biofilms (Supplementary Figure S2).
In the differential gene expression analysis of our RNA-seq data, single-species biofilms were used as the control, so the genes upregulated in P. aeruginosa could be interpreted as those positively regulated by S. aureus, whereas genes downregulated in P. aeruginosa would represent those negatively regulated by S. aureus, and vice-versa. We identified a total of 262 (6 upregulated and 246 downregulated) and 1,905 (101 upregulated and 1,804 downregulated) genes differentially expressed (fold-change ≥ 2 and p < 0.05) by P. aeruginosa and S. aureus, respectively (Supplementary Figure S3). The list of the 10 most highly up and downregulated genes in P. aeruginosa and S. aureus dual-species biofilms, as well as their annotated functions and COG families, are shown in Table 1.
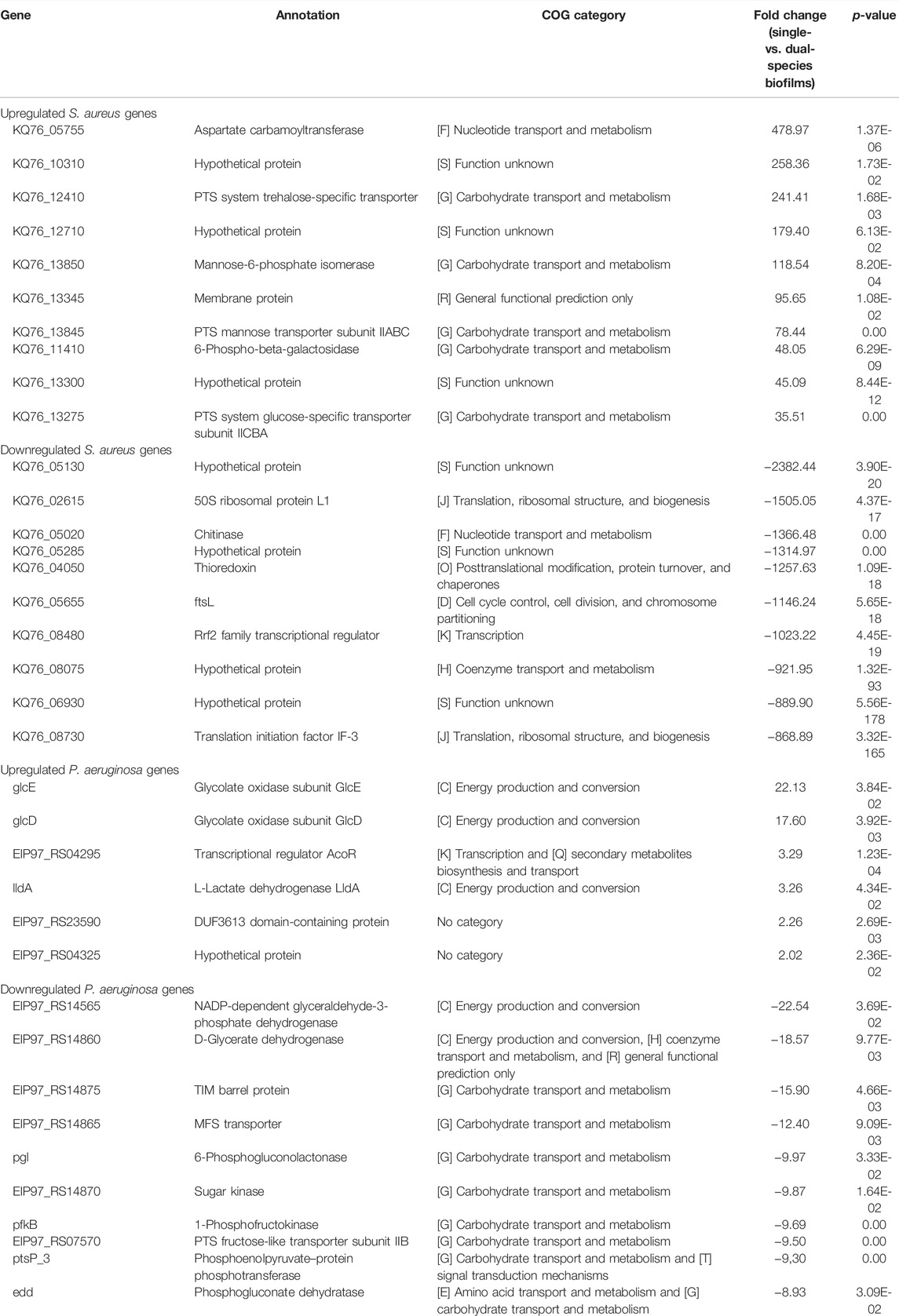
TABLE 1. List of the 10 genes with higher fold-change among the differentially expressed genes (fold-change ≥ 2, and p < 0.05) in P. aeruginosa and S. aureus biofilms cultured under single- vs. dual-species conditions. COGs, Clusters of Orthologous Groups of proteins.
GO biologic process (including the three main categories: cellular component, molecular function, and biological process) and KEGG pathway analyses were performed on this cohort of genes (fold-change ≥ 2 and p < 0.05), and the GO terms and pathways enriched are reported in Figure 1. For S. aureus genes, in the biological process category, GO terms associated with metabolism were found significantly enriched among the downregulated genes. In the cellular component, “cytoplasm” and “intracellular anatomical structure” were the two enriched categories. In the molecular function, “catalytic activity” was the dominant category. Among the upregulated genes, “carbohydrate transmembrane transporter activity” in molecular function, “intrinsic component of plasma membrane” and “integral component of plasma membrane” in the cellular component, and “carbohydrate transport” in the biological process were the most enriched GO terms. As could be expected, many of the most significantly upregulated S. aureus genes listed in Table 1 have functions related to these pathways. Regarding P. aeruginosa GO analysis, only six different subcategories were found to be significantly enriched (Figure 1A). Among these subcategories, “cytosol”, “cytoplasm,” and “intracellular anatomical structure” in the cellular component and “generation of precursor metabolites and energy” for biological processes were those receiving the most abundant annotations for P. aeruginosa downregulated genes. “Lactate dehydrogenase activity” was the only GO term significantly enriched for P. aeruginosa upregulated genes in the molecular function category. In addition, the two annotated genes (glcE and lldA) of this pathway comprised the P. aeruginosa top 10 upregulated genes shown in Table 1. In the KEGG annotations, significantly expressed genes were divided into 24 subcategories, with major alterations occurring in the downregulated genes of both species (Figure 1B). Among the downregulated genes in the S. aureus group, the enriched KEGG pathways were related to “vancomycin resistance,” followed by “peptidoglycan biosynthesis” and “pyrimidine metabolism.” Within the group of DOWNREGULATED genes in P. aeruginosa “protein export” and “aminobenzoate degradation” pathways were the most enriched pathways. The COG analysis showed that the majority of the differentially expressed genes have no assigned category based on the categories of Clusters of Orthologous Groups (COGs) (Supplementary Figure S3). However, there is also a high number of downregulated genes differentially expressed in P. aeruginosa that belong to energy production and conversion (category C; 32 genes) and, in the case of S. aureus, genes that belong to the amino acid transport and metabolism (category E; 163 genes).
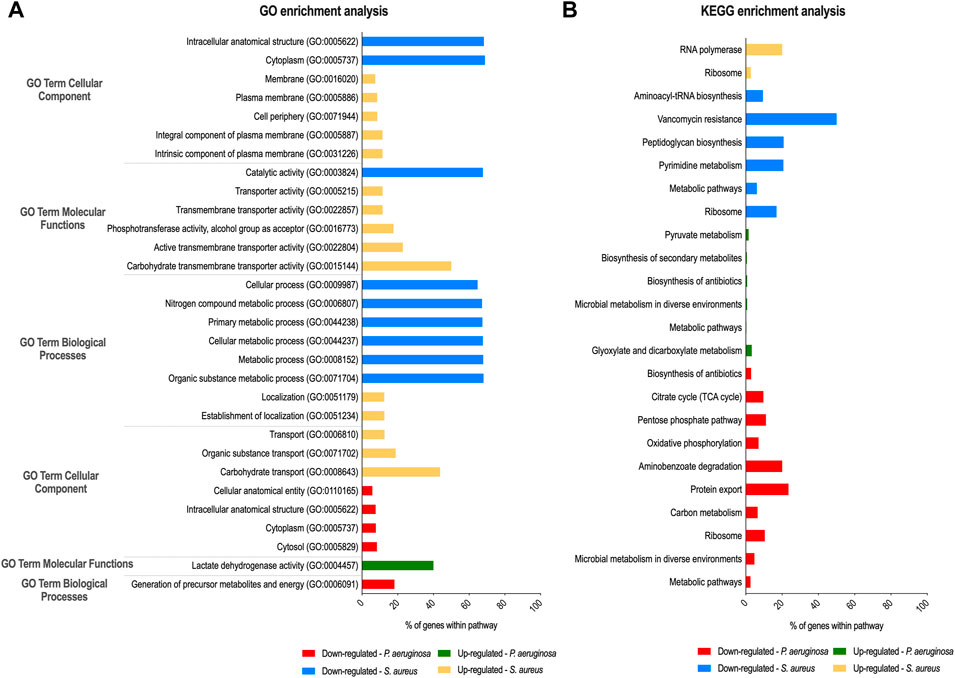
FIGURE 1. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of P. aeruginosa and S. aureus after dual-species biofilm growth. GO (A) and KEGG pathways (B) analyses were performed to identify, respectively, biological processes and pathways significantly enriched within differentially expressed genes in S. aureus and P. aeruginosa.
Our dual-transcriptome analysis of P. aeruginosa and S. aureus reveals that the adaptations are dominated by metabolic changes since the largest number of differentially expressed genes belongs to the functional classes “metabolism” and “transport.” In particular, our data also confirmed previous observations that P. aeruginosa drives S. aureus into fermentation (Filkins et al., 2015; Tognon et al., 2019). The lactate dehydrogenase (ldh1 and KQ76_13385), L-lactate permease (KQ76_12340), and acetolactate synthase (KQ76_11515) genes were upregulated 2- to 11-fold in the dual-species biofilms, indicating that S. aureus preferentially converted pyruvate into lactate. In line with these findings, one of the most upregulated genes of P. aeruginosa in response to S. aureus was the membrane-bound L-lactate dehydrogenase lldA (3-fold increase), suggesting that P. aeruginosa takes up lactate secreted by S. aureus to use it as a carbon and energy source. A study performed by Camus et al. (2020) suggests that acetoin may also play a role in metabolic interactions between P. aeruginosa and S. aureus. P. aeruginosa demonstrated an enhanced ability to catabolize acetoin produced by S. aureus as an alternative carbon source, resulting in increased survival during co-culture and avoiding the toxic accumulation of acetoin on S. aureus (Camus et al., 2020). Interestingly, we observed that the gene encoding the transcriptional regulator AcoR (EIP97_RS04295), described to be responsible for acetoin catabolism (Liu et al., 2018), was 3-fold upregulated in P. aeruginosa dual-species biofilms (Table 1). Moreover, we also found no evidence for the induction of anti-staphylococcal molecules in P. aeruginosa, suggesting that no direct competition prevails during dual-species biofilm growth. Additionally, a relatively low number of genes were significantly differentially expressed in the P. aeruginosa dual-species biofilm when compared to S. aureus, which had a more marked transcriptomic response, further suggesting that this species is less affected by S. aureus as we have phenotypically shown earlier (Magalhães et al., 2021). A similar trend was observed in other studies comparing mono- and co-cultures of P. aeruginosa and S. aureus (Filkins et al., 2015; Miller et al., 2017), indicating that P. aeruginosa appears to easily maintain itself as a dominant organism in various in vitro systems.
Overall, these data enabled us to identify key pathways and genes involved in the interaction of both bacteria during dual-species biofilm growth that warrant further investigations. Hence, these results may help unveil key molecular mechanisms driving the coexistence of these pathogens that might impact infection progression and the selection of potential targets for future studies aiming to develop preventive and/or therapeutic strategies for P. aeruginosa–S. aureus biofilm infections in CF, as well as in other diseases involving co-infection with these pathogens.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
MOP and NC conceived the study and participated in its design and coordination. APM performed the bacterial cultures, RNA extraction, and data analysis. AF performed the RNA-sequencing data trimming, alignment, analysis, and deposited the data. APM prepared the draft, and AF, MOP, and NC proofread the final draft. All authors read and approved the manuscript.
This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UID/BIO/04469/2020 unit and the BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2020–Programa Operacional Regional do Norte. The authors also acknowledge the support, through the Programa Operacional Competitividade e Internacionalização (COMPETE 2020) and by national funds, through FCT, of the PhD Grant of APM (SFRH/BD/132165/2017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.883199/full#supplementary-material
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25, 25–29. doi:10.1038/75556
Bragonzi, A., Farulla, I., Paroni, M., Twomey, K. B., Pirone, L., Lorè, N. I., et al. (2012). Modelling Co-infection of the Cystic Fibrosis Lung by Pseudomonas aeruginosa and Burkholderia Cenocepacia Reveals Influences on Biofilm Formation and Host Response. PLoS One 7, e52330. doi:10.1371/journal.pone.0052330
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 49, W317–W325. doi:10.1093/nar/gkab447
Burmølle, M., Webb, J. S., Rao, D., Hansen, L. H., Sørensen, S. J., and Kjelleberg, S. (2006). Enhanced Biofilm Formation and Increased Resistance to Antimicrobial Agents and Bacterial Invasion Are Caused by Synergistic Interactions in Multispecies Biofilms. Appl. Environ. Microbiol. 72, 3916–3923. doi:10.1128/AEM.03022-05
Camus, L., Briaud, P., Bastien, S., Elsen, S., Doléans-Jordheim, A., Vandenesch, F., et al. (2020). Trophic Cooperation Promotes Bacterial Survival of Staphylococcus aureus and Pseudomonas aeruginosa. ISME J. 14, 3093–3105. doi:10.1038/s41396-020-00741-9
Carbon, S., Carbon, S., Douglass, E., Good, B. M., Unni, D. R., Harris, N. L., et al. (2021). The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 49, D325–D334. doi:10.1093/nar/gkaa1113
Ciofu, O., Tolker-Nielsen, T., Jensen, P. Ø., Wang, H., and Høiby, N. (2015). Antimicrobial Resistance, Respiratory Tract Infections and Role of Biofilms in Lung Infections in Cystic Fibrosis Patients. Adv. Drug Deliv. Rev. 85, 7–23. doi:10.1016/j.addr.2014.11.017
Cox, M. J., Allgaier, M., Taylor, B., Baek, M. S., Huang, Y. J., Daly, R. A., et al. (2010). Airway Microbiota and Pathogen Abundance in Age-Stratified Cystic Fibrosis Patients. PLoS One 5, e11044. doi:10.1371/journal.pone.0011044
DeLeon, S., Clinton, A., Fowler, H., Everett, J., Horswill, A. R., and Rumbaugh, K. P. (2014). Synergistic Interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an In Vitro Wound Model. Infect. Immun. 82, 4718–4728. doi:10.1128/IAI.02198-14
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 30, 207–210. doi:10.1093/nar/30.1.207
Filkins, L. M., Graber, J. A., Olson, D. G., Dolben, E. L., Lynd, L. R., Bhuju, S., et al. (2015). Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards Fermentative Metabolism and Reduced Viability in a Cystic Fibrosis Model. J. Bacteriol. 197, 2252–2264. doi:10.1128/JB.00059-15
Flume, P. A., Mogayzel, P. J., Robinson, K. A., Goss, C. H., Rosenblatt, R. L., Kuhn, R. J., et al. (2009). Cystic Fibrosis Pulmonary Guidelines. Am. J. Respir. Crit. Care Med. 180, 802–808. doi:10.1164/rccm.200812-1845PP
França, A., Freitas, A. I., Henriques, A. F., and Cerca, N. (2012). Optimizing a qPCR Gene Expression Quantification Assay for S. Epidermidis Biofilms: A Comparison between Commercial Kits and a Customized Protocol. PLoS One 7, e37480. doi:10.1371/journal.pone.0037480
Frydenlund Michelsen, C., Hossein Khademi, S. M., Krogh Johansen, H., Ingmer, H., Dorrestein, P. C., and Jelsbak, L. (2016). Evolution of Metabolic Divergence in Pseudomonas aeruginosa during Long-Term Infection Facilitates a Proto-Cooperative Interspecies Interaction. ISME J. 10, 1323–1336. doi:10.1038/ismej.2015.220
Fugère, A., Lalonde Séguin, D., Mitchell, G., Déziel, E., Dekimpe, V., Cantin, A. M., et al. (2014). Interspecific Small Molecule Interactions between Clinical Isolates of Pseudomonas aeruginosa and Staphylococcus aureus from Adult Cystic Fibrosis Patients. PLoS One 9, e86705. doi:10.1371/journal.pone.0086705
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded Microbial Genome Coverage and Improved Protein Family Annotation in the COG Database. Nucleic Acids Res. 43, D261–D269. doi:10.1093/nar/gku1223
Hogan, D. A., Willger, S. D., Dolben, E. L., Hampton, T. H., Stanton, B. A., Morrison, H. G., et al. (2016). Analysis of Lung Microbiota in Bronchoalveolar Lavage, Protected Brush and Sputum Samples from Subjects with Mild-To-Moderate Cystic Fibrosis Lung Disease. PLoS One 11, e0149998. doi:10.1371/journal.pone.0149998
Hubert, D., Réglier-Poupet, H., Sermet-Gaudelus, I., Ferroni, A., Le Bourgeois, M., Burgel, P.-R., et al. (2013). Association between Staphylococcus aureus Alone or Combined with Pseudomonas aeruginosa and the Clinical Condition of Patients with Cystic Fibrosis. J. Cyst. Fibros. 12, 497–503. doi:10.1016/j.jcf.2012.12.003
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. doi:10.1093/nar/28.1.27
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., and Tanabe, M. (2021). KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 49, D545–D551. doi:10.1093/nar/gkaa970
Kanehisa, M. (2019). Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 28, 1947–1951. doi:10.1002/pro.3715
Kreda, S. M., Davis, C. W., and Rose, M. C. (2012). CFTR, Mucins, and Mucus Obstruction in Cystic Fibrosis. Cold Spring Harb. Perspect. Med. 2, a009589. doi:10.1101/cshperspect.a009589
Limoli, D. H., Yang, J., Khansaheb, M. K., Helfman, B., Peng, L., Stecenko, A. A., et al. (2016). Staphylococcus aureus and Pseudomonas aeruginosa Co-infection Is Associated with Cystic Fibrosis-Related Diabetes and Poor Clinical Outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 35, 947–953. doi:10.1007/s10096-016-2621-0
Limoli, D. H., Whitfield, G. B., Kitao, T., Ivey, M. L., Davis, M. R., Grahl, N., et al. (2017). Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. MBio 8, e00186–17. doi:10.1128/mBio.00186-17
Liu, Q., Liu, Y., Kang, Z., Xiao, D., Gao, C., Xu, P., et al. (2018). 2,3-Butanediol Catabolism inPseudomonas aeruginosaPAO1. Environ. Microbiol. 20, 3927–3940. doi:10.1111/1462-2920.14332
Lopes, S. P., Ceri, H., Azevedo, N. F., and Pereira, M. O. (2012). Antibiotic Resistance of Mixed Biofilms in Cystic Fibrosis: Impact of Emerging Microorganisms on Treatment of Infection. Int. J. Antimicrob. Agents 40, 260–263. doi:10.1016/j.ijantimicag.2012.04.020
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2002). Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 15, 194–222. doi:10.1128/cmr.15.2.194-222.2002
Magalhães, A. P., Lopes, S. P., and Pereira, M. O. (2017). Insights into Cystic Fibrosis Polymicrobial Consortia: The Role of Species Interactions in Biofilm Development, Phenotype, and Response to In-Use Antibiotics. Front. Microbiol. 7, 1–11. doi:10.3389/fmicb.2016.02146
Magalhães, A. P., Grainha, T., Sousa, A. M., França, Â., Cerca, N., and Pereira, M. O. (2021). Viable but Non‐cultivable State: a Strategy for Staphylococcus aureus Survivable in Dual‐species Biofilms with Pseudomonas aeruginosa ? Environ. Microbiol. 23, 5639–5649. doi:10.1111/1462-2920.15734
Mashburn, L. M., Jett, A. M., Akins, D. R., and Whiteley, M. (2005). Staphylococcus aureus Serves as an Iron Source for Pseudomonas aeruginosa during In Vivo Coculture. J. Bacteriol. 187, 554–566. doi:10.1128/JB.187.2.554-566.2005
Miller, C. L., Van Laar, T. A., Chen, T., Karna, S. L. R., Chen, P., You, T., et al. (2017). Global Transcriptome Responses Including Small RNAs during Mixed‐species Interactions with Methicillin‐resistant Staphylococcus aureus and Pseudomonas aeruginosa. Microbiologyopen 6, 1–22. doi:10.1002/mbo3.427
Pawitan, Y., Michiels, S., Koscielny, S., Gusnanto, A., and Ploner, A. (2005). False Discovery Rate, Sensitivity and Sample Size for Microarray Studies. Bioinformatics 21, 3017–3024. doi:10.1093/bioinformatics/bti448
Schobert, M., and Jahn, D. (2010). Anaerobic Physiology of Pseudomonas aeruginosa in the Cystic Fibrosis Lung. Int. J. Med. Microbiol. 300, 549–556. doi:10.1016/j.ijmm.2010.08.007
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/measurement Sets. Nucleic Acids Res. 49, D605–D612. doi:10.1093/nar/gkaa1074
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 28, 33–36. doi:10.1093/nar/28.1.33
Tognon, M., Köhler, T., Luscher, A., and Van Delden, C. (2019). Transcriptional Profiling of Pseudomonas aeruginosa and Staphylococcus aureus during In Vitro Co-culture. BMC Genomics 20, 1–15. doi:10.1186/s12864-018-5398-y
Wakeman, C. A., Moore, J. L., Noto, M. J., Zhang, Y., Singleton, M. D., Prentice, B. M., et al. (2016). The Innate Immune Protein Calprotectin Promotes Pseudomonas aeruginosa and Staphylococcus aureus Interaction. Nat. Commun. 7, 11951. doi:10.1038/ncomms11951
Keywords: Pseudomonas aeruginosa, Staphylococcus aureus, RNA sequencing, dual-species biofilms, cystic fibrosis
Citation: Magalhães AP, França A, Pereira MO and Cerca N (2022) Unveiling Co-Infection in Cystic Fibrosis Airways: Transcriptomic Analysis of Pseudomonas aeruginosa and Staphylococcus aureus Dual-Species Biofilms. Front. Genet. 13:883199. doi: 10.3389/fgene.2022.883199
Received: 24 February 2022; Accepted: 26 May 2022;
Published: 06 July 2022.
Edited by:
Gong Zhang, Jinan University, ChinaReviewed by:
Luis Caetano Martha Antunes, Oswaldo Cruz Institute (Fiocruz), BrazilCopyright © 2022 Magalhães, França, Pereira and Cerca. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela França, YWZyYW5jYUBjZWIudW1pbmhvLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.