- N.K. Koltsov Institute of Developmental Biology of Russian Academy of Sciences, Moscow, Russia
Tight packaging of DNA in chromatin severely constrains DNA accessibility and dynamics. In contrast, nucleosomes in active chromatin state are highly flexible, can exchange their histones, and are virtually “transparent” to RNA polymerases, which transcribe through gene bodies at rates comparable to that of naked DNA. Defining mechanisms that revert nucleosome repression, in addition to their value for basic science, is of key importance for the diagnosis and treatment of genetic diseases. Chromatin activity is largely regulated by histone posttranslational modifications, ranging from small chemical groups up to the yet understudied “bulky” ubiquitylation and sumoylation. However, it is to be revealed how histone marks are “translated” to permissive or repressive changes in nucleosomes: it is a general opinion that histone modifications act primarily as “signals” for recruiting the regulatory proteins or as a “neutralizer” of electrostatic shielding of histone tails. Here, we would like to discuss recent evidence suggesting that histone ubiquitylation, in a DNA stress–dependent manner, can directly regulate the dynamics of the nucleosome and their primary structure and can promote nucleosome decomposition to hexasome particles or additionally stabilize nucleosomes against unwrapping. In addition, nucleosome repression/ derepression studies are usually performed with single mononucleosomes as a model. We would like to review and discuss recent findings showing that internucleosomal interactions could strongly modulate the dynamics and rearrangements of nucleosomes. Our hypothesis is that bulky histone modifications, nucleosome inherent dynamics, internucleosome interactions, and DNA torsions could act in cooperation to orchestrate the formation of different dynamic states of arrayed nucleosomes and thus promote chromatin functionality and diversify epigenetic programming methods.
Introduction
Many diseases and behavioral pathologies such as cancer (Espinosa, 2008; Cao and Yan, 2012; Johnsen, 2012; Cole et al., 2015), metabolic disorders (Gluckman et al., 2009; Gao et al., 2014), cardiovascular and autoimmune diseases, and diabetes (Dieker and Muller, 2010; Zou et al., 2014) are the results of gene deregulation (Gray, 2006; Perini and Tupler, 2006; Bhaumik et al., 2007; Weake, 2014; Mirabella et al., 2016). However, despite the critical importance of gene regulatory principles for the diagnosis, prevention, and therapy of genetic diseases (and, in general, for directed manipulation of gene activity), many aspects of gene regulation have not been well-elucidated thus far and remain unclear on how DNA processing machineries overcome the tight multilevel packaging of DNA in cell nuclei.
In eukaryotes, the genetic information required to control all life processes exists in the form of chromatin, a complex hierarchical structure of DNA super-helices, stabilized by a multitude of protein–DNA and protein–protein interactions. On the first level of compaction, 147 bp of every ∼200 bp of DNA are wrapped in 1.75 turns around an octamer of histone proteins, comprising one H3-H4 tetramer flanked on each side by H2A-H2B dimers (Figure 1A), thus forming nucleosomes, the basic repeated chromatin units (Luger et al., 1997; Vasudevan et al., 2010). Nucleosome arrays fold into ‘solenoids (Kruithof et al., 2009; Kepper et al., 2011; Victor et al., 2012) or “zig-zag”-like (Dorigo et al., 2004; Grigoryev, 2004; Schalch et al., 2005; Grigoryev et al., 2009) arrangements to form the 25–34 nm chromatin fiber, stabilized by linker histones H1/H5 (Robinson and Rhodes, 2006). The 30-nm fiber further self-associates and condenses into higher-order tertiary structures.
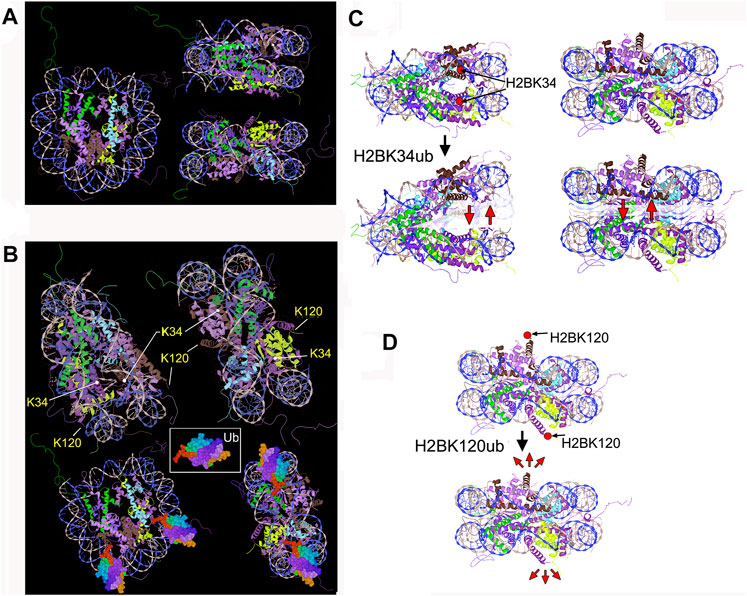
FIGURE 1. (A) Nucleosome (1kx5) front, top, and side view. •• H3.2 (chains A,E), •• H2B 1.1 (chains D,H), •• H2A type 1 (chains C,G), and •• H4 (chains B,F) (B) Positions of H2B K34 and K120 indicated by arrows. Drafts at the bottom illustrate the H2BK34ub nucleosome (Ubiquitin PDB: 1ubq). (C,D) Sketches, depicting the potential mechanisms of nucleosome-destabilizing effects by H2BK34-/ K120-ubiquitylation. (C) H2BK34ub installed in the occluded nucleosome region could act as a “wedge”, facilitating DNA gyre–gyre separation (“gaping”). (D) Mechanistic forces applied to the H2A-H2B dimer by ubiquitin deposited to H2B termini could weaken the nucleosome through stochastic motions of bulky ubiquitin and/or its steric clashes with the nucleosome surface. This could also promote nucleosome breathing and DNA-dimer opening motions.
Nucleosomes in their “canonic” state (as seen by X-ray studies (Luger et al., 1997; Richmond and Davey, 2003; Vasudevan et al., 2010) are rather robust static units, refractory to DNA-binding proteins, and thus, literary should present an “immovable barrier” even for the “irresistible force” of progressing RNA polymerases (Kornberg and Lorch, 1991). So, despite the nucleosomes being the key elements in gene regulation (Gibney and Nolan, 2010), it is still understudied how they relieve their intrinsically repressive effects on DNA expression.
Due to the structural tensions associated with the bending of the stiff (Manning, 2006) 147 bp core nucleosomal DNA around the histone globule, nucleosomes retain some degree of dynamicity and undergo spontaneous fluctuations of nucleosome wrapping, which range from 10–250 ms “breathing” and more slow “opening” motions (Koopmans et al., 2007; Armeev et al., 2018), up to the lo 1–10 min nucleosome hinge-like “gaping” openings (Zlatanova et al., 2009; Ngo and Ha, 2015). Fluctuations in nucleosome wrapping and transiently increasing DNA exposure (Polach and Widom, 1996) play important role in regulating the accessibility of transcription factors to the nucleosome DNA (Li et al., 2005) and alleviating RNA polymerases entering the nucleosome (Hodges et al., 2009; Selth et al., 2010). It is possible that any stimuli increasing the basic level of inherent nucleosome dynamics will contribute to the derepression of the nucleosome.
Nucleosomal histones are subjected to a multitude of reversible posttranslational modifications (PTMs) which, supposedly, control virtually all aspects of chromatin functioning. According to the “histone code” concept, PTMs “acting in a combinatorial or sequential fashion on one or multiple histone tails specify unique downstream function” (Strahl and Allis, 2000). Many PTMs are dynamically deposited during the cell cycle to control particular cellular processes, whereas certain histone PTMs are thought to program the transcription memory transmitted to the progeny cells. A different (though debatable (Alabert et al., 2015; Reveron-Gomez et al., 2018)) view that histone PTMs are not transmitted to progeny chromatin but instead persistently bound histone-modifiers reestablish the PTM pattern on the daughter chromatin (Petruk et al., 2012; Petruk et al., 2013) came from histone H3K4me3/ H3K27me3 “inheritance” studies, which used (probably insufficiently sensitive) proximity-ligation assay to monitor the modified histones on replicated DNA. Finally, several recent studies (Reinberg and Vales, 2018; Escobar et al., 2019; Escobar et al., 2021) provided evidence that the repressive histone modifications but not active ones are inherited upon DNA replication.
In a classic view, both “small” (methylation and acetylation) (Rothbart and Strahl, 2014) and “bulky” (ubiquitin and SUMO-1,2/3 polypeptides (Cubenas-Potts and Matunis, 2013; Weake, 2014)) histone PTMs are considered binding targets for effector proteins (Rothbart and Strahl, 2014; Andrews et al., 2016) or as regulators of chromatin higher-order folding (acting by modulation of internucleosome interactions (Pepenella et al., 2014b; Prakash and Fournier, 2018)) but not as direct triggers of primary nucleosome structure reversing DNA repression. By tuning histone charges and deposition of a modest steric bulk, “small” histone PTMs only moderately affect the spontaneous fluctuations of nucleosomes without affecting their stability (PTMs at the nucleosome entry-exit) or modestly decrease nucleosome stability without affecting nucleosome dynamics (PTMs near the nucleosome dyad axis) (Bowman and Poirier, 2015; Armeev et al., 2018).
However, recent data suggest that currently understudied large polypeptide PTMs could play an active role in directly altering the nucleosome primary structure and dynamics. In addition, since intrinsic chromatin organization is based on a hierarchy of DNA helices (DNA double helix, nucleosome DNA wrapping, and chromatin fiber), the chromatin structure is subjected to superhelical stresses in DNA, which could significantly affect the nucleosome properties and functionality. Furthermore, due to multiple internucleosome interactions, the model describing a nucleosome array just as a polymer of individual “canonic” nucleosomes does not adequately recapitulate nucleosome functionalities. The interaction between nucleosomes via flexible histone termini could significantly affect nucleosome structural transitions (Krajewski, 2016). We would like to discuss these phenomena in view of the recent and older literature data.
Nucleosomes as Key Elements in Epigenetic Regulation of Chromatin Activity
What differentiates nucleosomes in transcriptionally active chromatin from the canonic ones? In both cases, these particles possess the same composition and share the same organizational principles. However, in transcribed chromatin regions, nucleosomes are dynamic and (Zlatanova et al., 2009; Armeev et al., 2018) exhibit high conformational flexibility (Saavedra and Huberman, 1986; Morse et al., 1987; Krajewski and Luchnik, 1991), easily exchange their histone subunits (Zlatanova et al., 2009; Venkatesh and Workman, 2015), and support fast progression of RNA polymerases (Singh and Padgett, 2009) that is accompanied by nucleosome unfolding and unshielding of histone H3 sulfhydryls which are otherwise buried at the nucleosome dyad and inaccessible in the canonic nucleosome state (Prior et al., 1983; Chen et al., 1991).
A notable hallmark of the transcribed chromatin is the dynamic monoubiquitylation of histone H2B at lysines K120 (K123 in yeast) (Batta et al., 2011; Fleming et al., 2008; Trujillo and Osley, 2012; Wright and Kao, 2015) and K34 (Li et al., 2017; Wu et al., 2011; Wu et al., 2013; Wu et al., 2014) (Figure 1B). This feature would be consistent with a series of recent findings showing that K34-ubiquitylation of histone H2B (and H2BK120ub to a lesser degree) can significantly enhance nucleosome dynamics, decrease nucleosome stability, and promote eviction of one histone H2A-H2B dimer (Krajewski et al., 2018; Krajewski W. A., 2020), especially in the presence of histone chaperons. This effect is likely due to the steric hindrances by “bulky” ubiquitin moieties, which destabilize the nucleosome (Krajewski, 2019; Krajewski WA., 2020). The resulting hexasome particle was stable, suggesting that dissociation of one ubiquitylated histone dimer is sufficient to relieve the steric stresses incurred by massive ubiquitin moieties (Krajewski et al., 2018; Krajewski W. A., 2020).
The 8.6 kD ubiquitin (Renatus et al., 2006) and 10–12 kD SUMO (Bayer et al., 1998; Huang et al., 2004) are close in size to histones that principally distinguishes these PTMs from “small” chemical modifications. A steric bulk deposited by ubiquitylation and sumoylation could act to “mechanically” alter the canonic nucleosomes. Nucleosome-destabilizing forces would be stronger when bulky PTMs are deposited within the nucleosome lateral surface and so, directly conflict with the compact nucleosome structure. For example, ubiquitylation of histone H2B at lysine K34, which is “buried” between two DNA gyres (Li et al., 2017; Wu et al., 2011) (Figure 1B), could act as a “wedge”, facilitating DNA gyre–gyre opening (Figure 1C). Bulky PTMs at histone termini (e.g., H2BK120ub, Figure 1B) disturb the nucleosome core less but could affect intra-nucleosomal interactions, for example, by electrostatic repulsion (Figure 1D). The association of H2A-H2B dimers on the nucleosome interface could be weakened by stochastic (Brownian) motions of the attached PTMs, which will tend to “tear-off” one histone dimer out from the nucleosome interface (Figure 1D). Of note, it has been shown that dynamic nucleosome conformations could be shifted to more unwrapped structures by binding bulky objects to the nucleosome periphery (Polach and Widom, 1996; Buning et al., 2015), such as the transcription factors (Polach and Widom, 1996), an adjacent nucleosome, or long linker DNA (Buning et al., 2015). Due to the interactions between histone tails and nucleosome-associated core DNA (Cutter and Hayes, 2015; Shaytan et al., 2016; Chakraborty and Loverde, 2017; Morrison et al., 2018) or linker DNA (Davey et al., 2002; Cutter and Hayes, 2015; Schunter et al., 2017), bulky PTMs of histone termini could destabilize the intra-nucleosome interactions either directly or by colliding with the nucleosome surface.
These results suggest a hypothesis (Krajewski, 2019; Krajewski WA., 2020) that in contrast to “small” histone PTMs, attachment to nucleosomes at certain positions of ubiquitin (and, supposedly, other bulky PTMs) could potentially represent an in vivo mechanism to functionalize canonic nucleosomes by strikingly increasing their dynamics and triggering the conversion of a nucleosome to a more functionally active hexasome particle.
Interestingly, recent single-molecule magnetic tweezer experiments (Xiao et al., 2020) have shown that H2AK119ub, on the contrary, dramatically prevents the peeling of the DNA from the histone octamer that stabilizes the nucleosome. The stabilizing effect of ubH2A was not a result of the enhanced stability of the octamer (ibid) but likely relies on the Ub-mediated steric clashes that prevent nucleosome unfolding. Although these results would benefit from refinement with more relevant biochemical approaches, it could be supposed that at some nucleosome positions, “hindrances” caused by bulky modifications could strongly stabilize and “lock” the nucleosome unwrapped state. With this example, one can propose that “bulky” modifications could create a stable “code” of both active and repressed chromatin states.
Histone ubiquitylation is one of the key epigenetic marks with a wide spectrum of action (Weake, 2014), so the functions of H2BK120ub and H2BK34ub (and H2A119ub) are not only limited to the proposed “direct” nucleosome-regulatory role but also involve other ubiquitylation-mediated binding events for the chromatin regulators (Vaughan et al., 2021). There are still less data available on H2BK34ub; therefore, we will just mention here two recent studies on the interactions of Dot1L and H2BK120ub nucleosomes, which are critical to direct H3K79 methylation (Anderson et al., 2019; Valencia-Sanchez et al., 2019).
Previous work showed that H2B-ubiquitylation is sufficient to directly enhance the nucleosome dynamics and nucleosome-hexasome transition in vitro (the effects were comparable to those produced by ATP-driven chromatin remodelers) and, supposedly, in vivo. But, however, the “direct” and “indirect” (via other regulatory factors) nucleosomal effects of the bulky PTMs are not self-exclusive. There also might be an interplay between histone ubiquitylation and another histone PTMs and their corresponding co-factors regulating chromatin dynamics in vivo. One example is that PRC2 co-factors JARID2 and AEBP2 play a crucial role in both the recruitment and activation of PRC2 through their recognition of H2AK119ub1 (Kasinath et al., 2021), which further orchestrates the local chromatin environment.
The tight link of H2BK120/ K34-ubiquitylation with transcription and replication shows a plausible mechanism assisting RNA and DNA polymerases to overcome the nucleosome barrier. The MOF–MSL complex, which deposits H2BK34ub (Krajewski and Vassiliev, 2019; Wu et al., 2011), plays a critical role in transcription, initiation, and elongation and is enriched at transcription start sites (Wu et al., 2014). Regardless of the exact mechanism, it could be hypothesized that H2BK34ub-facilitated destabilization and a dimer eviction in +1 nucleosome (which presents a greater transcription barrier in vivo than downstream nucleosomes (Adelman and Lis, 2012; Gilmour, 2009; Teves and Henikoff, 2014; Weber et al., 2014)) assists transient uncoiling of the promoter-proximal boundary of the +1 nucleosome and facilitates the release of Pol II from pausing and its transition to elongation step (Figures 2A,B). PAF1 associated with MOF–MSL and RNF20/40 (which deposit H2BK120ub (Hwang et al., 2003)) progresses together during transcription and elongation (Wu et al., 2014) that supposes that H2B-ubiquitylation, in cooperation with histone chaperones (Hsieh et al., 2013; Hsieh et al., 2015; Gurova et al., 2018), orchestrates unwrapping/rewrapping of transcribed nucleosomes by facilitating coordinated sequential dissociation and rebinding of the nucleosome-proximal and nucleosome-distal H2A-H2B dimer—steps required for RNA Pol II to traverse the nucleosome (Kulaeva et al., 2013). In contrast, H2AK119ub (associated with silenced genes (Meas and Mao, 2015)) could prevent Pol II progression and block remodeling activities (i.e., Swi-Snf and related) that act through peeling on DNA from the nucleosome.
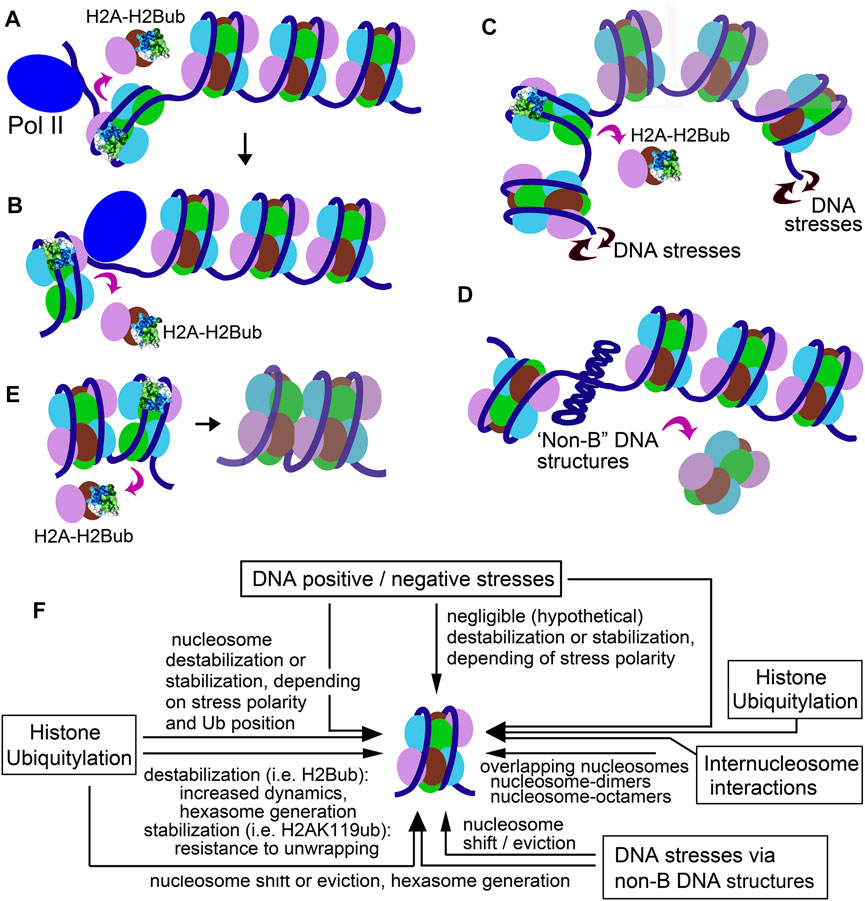
FIGURE 2. Sketches illustrating the nucleosomal effects of H2Bub. (A,B) RNA Pol II traversing the H2B-ubiquitylated nucleosome. (A) Eviction of the promoter-proximal H2A-H2B dimer promotes the polymerase complex to enter the nucleosome; (B) Eviction of the promoter-distal H2A-H2B dimer promotes RNA Pol II to successfully elongate through the nucleosome. (C) Hexasome generation by DNA stresses. (D) Nucleosome rearrangement by transition of a DNA segment to a cruciform structure. (E) Formation of “overlapping” nucleosomes. (F) Schematic figure, depicting how different mechanisms (histone ubiquitylation, DNA stress, and internucleosome interaction) could cooperate to regulate chromatin dynamics and function.
In general, the consequences of histone ubiquitylation and sumoylation on the nucleosome primary structure are still understudied, although the experimental data support the direct, destabilizing, or stabilizing effects of bulky PTMs. In vitro H4K34-monoubiquitylation moderately destabilizes nucleosomal association of the H3–H4 tetramer, supposedly, due to the clash between DNA phosphate backbone and deposited ubiquitin (Machida et al., 2016). In vivo H3K4-polyubiquitylation by RNF8 promotes nucleosome disassembly and eviction from the DNA (Xia et al., 2017), although it is not clear whether this could be a direct effect of histone ubiquitylation. UV-irradiation activates the ubiquitylation of histones H3 and H4 by CUL4-DDB, promoting the eviction of histones and stimulating the recruitment of XPC repair protein (Wang et al., 2006). In biochemical studies, H2AK119-monoubiquitylation had marginal nucleosome stabilizing/ destabilizing effects (Fierz et al., 2012) but could directly alter the nucleosome interface in vivo and protect the H3K36 residue from modification (Bi et al., 2016). Using single-molecule magnetic tweezers, it has been shown that H2AK119ub stabilizes the nucleosome from unwrapping (Xiao et al., 2020) (see above).
There is less data on histone sumoylation. In yeast cells, genetically engineered multiple sumoylation of histone H2B had only a minor structural effect on nucleosomes (Chandrasekharan et al., 2009). The H4K12su is a gene silencing marker (Shiio and Eisenman, 2003; Nathan et al., 2006), despite the H4K12 position being near the H4 basic patch where the steric bulk and hindrances by installed SUMO polypeptides could affect the critical (for chromatin compaction) interaction between H4 tails and the H2A-H2B acidic patch on the adjoining nucleosome (Allahverdi et al., 2011; Pepenella et al., 2014b). Indeed, spFRET studies have shown that H4K12su destabilizes long-range internucleosome interactions and moderately represses the formation of compact chromatin (Dhall et al., 2014).
DNA Stresses and DNA Non-Canonic Structures in Epigenetic Control
Virtually any process that manipulates DNA strands can generate positive or negative DNA torsional stress (Esposito and Sinden, 1988; Baranello et al., 2012; Gilbert and Allan, 2014; Corless and Gilbert, 2016). For example, waves of positive and negative supercoiling are generated ahead and behind the RNA polymerase, respectively (Liu and Wang, 1987), which may be directly observed in vivo (e.g., (Lee and Garrard, 1991; Ljungman and Hanawalt, 1995; Naughton et al., 2013; Gerasimova et al., 2016)) and in vitro (Pfaffle and Jackson, 1990; Jackson, 1993; Bancaud et al., 2007). It is assumed that negative DNA stresses favor DNA wrapping on the histone octamer, while positive supercoiling destabilizes nucleosomes (Esposito and Sinden, 1988; Pfaffle and Jackson, 1990; Clark et al., 1993; Jackson, 1993; Bancaud et al., 2007). It would be appealing to attribute to DNA stresses an active role in the regulation of a primary nucleosome structure. Indeed, the generation of artificially high levels of positive DNA torsions in a single chromatin fiber by magnetic tweezers can break histone dimer–tetramer docking and induce transient, reversible nucleosome reorganization (Bancaud et al., 2007); the authors assume that a wave of such nucleosome chiral transitions can propagate ahead of a transcribing polymerase in vivo. However, after decades of studies, there is still no consensus whether the “physiological” levels of DNA torsions under physiologically relevant conditions could have any substantial effect on the nucleosome structure. Both supporting (Garner et al., 1987; Jackson, 1993; Sheinin et al., 2013; Teves and Henikoff, 2014) and opposing (Clark et al., 1993; Sheinin et al., 2013) observations were published. The “physiological” levels of DNA supercoiling only marginally affected the stability of unmodified nucleosomes in vitro—such that histone octamers assembled on negatively supercoiled DNA with only a slight preference compared to that of positively supercoiled DNA (Clark and Felsenfeld, 1991; Clark et al., 1993). This question is of particular importance since years of studies accumulated mounting evidence of how cells could regulate DNA stresses. In addition, numerous “non-canonical” DNA structures have been discovered, which are capable of adopting non-B DNA conformation to absorb or enhance DNA torsions (Smith, 2008; Baranello et al., 2012; Kaushik et al., 2016).
A different situation could be if a nucleosome structure is already intrinsically destabilized by deposited bulky histone modification. We propose that DNA topology, favoring or disfavoring nucleosome wrapping, may contribute to the structural effects of histone ubiquitylation (Krajewski, 2019; Krajewski WA., 2020) (Figure 2C). In our experiments, “physiological” negative and positive supercoiling in long DNA templates had opposing (stimulating or inhibitory, respectively) effects on the hexasome generation upon assembly of H2BK34ub nucleosomes (Krajewski et al., 2018) but had no effect on unmodified nucleosomes. We suppose that nucleosome “unfolding” using moderate positive DNA stress restrains the steric hindrances in ubiquitylated nucleosomes, while nucleosome compaction by negative stresses enhances the hindrances (Krajewski et al., 2018). More strong DNA topology effects in short (298 bp) minicircle DNAs have diverse effects on unmodified, H2BK34ub and H2BK120ub nucleosomes (Krajewski, 2018), suggesting that DNA topology states can strongly and selectively (and, likely, bi-directionally) affect nucleosome stability and dynamics depending on the type of H2B-ubiquitylation. It is notable that certain DNA topologies increased the stability of H2BK120ub nucleosomes over unmodified ones (see (Xiao et al., 2020) and discussion above). The H2BK34- and H2BK120-ubiquitylated nucleosomes exhibited quite selective sensitivity and sustainability to positive and negative DNA stresses (Krajewski, 2018; Krajewski et al., 2018), implying that bulky PTMs could play an active role in amplifying or mitigating the nucleosomal effects of DNA torque (including those by translocating RNA Pol II) and, thus, highlighting the nucleosome-regulatory role of DNA stresses. It could be interesting to see how positive and negative DNA stresses could affect “DNA-peeling refractory” H2AK119ub nucleosomes.
In addition to their direct nucleosome stability effects, DNA stresses could also affect the nucleosomes “indirectly” by generating non-standard DNA structures. Even relatively short stretches of alternating (CG) pairs and inverted repeat DNA sequences can form different structural isomers (left-handed helices and cruciforms) in response to superhelical stress at low “physiological” densities (Esposito and Sinden, 1988; McLean and Wells, 1988; Smith, 2008; Wells, 1988). These structures can regulate (absorb) superhelical stresses in DNA and also can affect nucleosome distribution by “translationally shifting” histone octamers along with DNA or displacing nucleosomes from the DNA (Figure 2D). Many studies suggest that Z-DNA and cruciforms cannot be organized in the nucleosome. Deposition of nucleosomes on supercoiled DNA containing a region of Z-DNA or a cruciform leads to the exclusion of regions of Z-DNA from the interiors of nucleosome cores in vitro and in vivo (Krajewski, 1996).
Internucleosome Interaction as an Additional Source of Chromatin Functionality
A “nucleosome-octamer” and “nucleosome-dimer” structure in which a nucleosome particle is associated with an additional histone octamer (Voordouw and Eisenberg, 1978; Stein, 1979; Daban and Cantor, 1982; Ausio et al., 1984; Aragay et al., 1988; Aragay et al., 1991) or another nucleosome (Tatchell and Van Holde, 1979; Ausio et al., 1984; Yager et al., 1989), respectively, was described years ago, although since then was forgotten for decades. Both the nucleosome-octamers and nucleosome-dimers are likely to be formed via trans-interactions between histone octamers. The site-directed histone-DNA and histone–histone cross-linking (Zheng and Hayes, 2003a; Zheng and Hayes, 2003b; Kan et al., 2007; Kan and Hayes, 2007; Kan et al., 2009; Pepenella et al., 2014a) revealed multitude interactions between histone tails and DNA of neighboring nucleosomes (reviewed in: (Luger et al., 2012; Pepenella et al., 2014b; Krajewski, 2016)). The nuclease digestion pattern and digestion kinetics of nucleosome-octamers and nucleosome-dimers are similar to those in single nucleosomes; therefore, it could be supposed that these particles largely retain the basic features of nucleosomal organization (Stein, 1979; Eisenberg and Felsenfeld, 1981; Krajewski and Vassiliev, 2012).
The ability of a nucleosome to bind extra histone octamers/dimers could play an important gene regulatory role during transient chromatin disassembly–reassembly through DNA replication or transcription. For example, a nucleosome behind the RNA Pol II could transiently bind a histone octamer or the evicted histone H2A/H2B dimer from the nucleosome being transcribed—this could be a possible mechanism of how the nucleosome reinstates its initial position on the DNA after the passage of the RNA Pol II complex.
The interaction between nucleosomes could, supposedly, affect chromatin remodeling and deposition of histone modifications. In polynucleosomes, human and yeast Swi/Snf complexes can generate structurally altered ‘asymmetric’ pairs of adjacent nucleosomes (Ulyanova and Schnitzler, 2005; Krajewski and Vassiliev, 2010). These “autosome” structures contain intact histone core octamers, but their nuclease cleavage pattern indicates the association of one internucleosomal and one subnucleosomal (220 and 70 bp, respectively) DNA fragment. In dinucleosomes, Isw1a/b and Isw2 generate extra structural alterations compared to mononucleosomes (Krajewski, 2013; Krajewski, 2014). Remodeling of the nucleosome-dimer particles by yeast Isw2 facilitated in vitro the association of nucleosome-dimers with the MLL SET-domain polypeptide (Krajewski and Vassiliev, 2012). SET7 and ALL-1 SET polypeptides showed binding preferences for dinucleosomes (but not mononucleosomes) remodeled with yIsw1/Isw2. The assembly of nucleosomes in oligonucleosomes promoted histone H3 methylation by the EZH2/EED, which only inefficiently modifies single mononucleosomes (Martin et al., 2006). Furthermore, reorganization of di- and oligonucleosomes (but not mononucleosomes) by binding of histone H1 further increased H3 methylation by EZH2 (Martin et al., 2006). However, there is no direct evaluation of the significance of internucleosomal interactions in promoting increased PRC2 HMTase activity as of yet. It could be that dinucleosome-enhanced PRC2 HMTase activity is largely due to the mechanism of allosteric activation (Jiao and Liu, 2015; Yu et al., 2019), and incorporation of H1 further facilitates positioning and activity of the PRC2 complex (that is indirectly supported by strong inhibition of methylation with over-stoichiometric amounts of H1 (Martin et al., 2006)). In general, the reports showed that adjusting the internucleosome spacing could affect the activity of the writers of histone PTMs including PRC2, but many of these studies were performed in an artifactual manner by changing the nucleosome spacing length.
It could be supposed that spontaneous movements of nucleosomes along the DNA, nucleosome dynamic fluctuations, and nucleosome instability incurred by histone ubiquitylation, even in absence of chromatin remodeling activities, could result in transient relocation of a histone H2A-H2B dimer from one nucleosome to the surface of the neighboring nucleosome, thus facilitating the formation of hexasomes and other subnucleosomal structures. Similarly, the hexasome particle generated by histone ubiquitylation could transiently associate with the adjacent nucleosome to form the structurally altered “autosome-like” arrangement. Owen-Hughes’ lab has shown that interactions between two nucleosomes could generate partial unwrapping of one nucleosome with the eviction of one H2A/H2B dimer and “merging” the resulting hexasome and a nucleosome into a single particle in which overlapping octamers and hexasomes invade each other’s space (Engeholm et al., 2009). The authors supposed (ibid) that nucleosome overlapping could be promoted by the eviction of H2A-H2B dimer and by exposure of the nucleosome DNA-binding surfaces. Engeholm et al. supposed that this could occur by the action of Swi-/Snf-related remodeling activities, which can reduce the stability of nucleosomal association of the histone dimer (Bruno et al., 2003; Vicent et al., 2004) and unravel up to 50 bp from the edge of the nucleosomes (Fan et al., 2003; Flaus and Owen-Hughes, 2003; Kassabov et al., 2003; Krajewski and Vassiliev, 2010), such that the nucleosomes may associate through the exposed DNA-binding surfaces to form dinucleosome-like particle (Schnitzler et al., 2001; Ulyanova and Schnitzler, 2005; Ulyanova and Schnitzler, 2007). It is possible that other pathways resulting in destabilized binding of histone dimers with the nucleosome and promoting hexasome generation, such as histone ubiquitylation and nucleosome-destabilizing DNA stresses, could facilitate nucleosome colliding and overlapping (Figure 2E).
Conclusion
Here, we tried to briefly overview the evidence showing that cooperation between bulky histone modifications, DNA stresses, DNA non-canonic structure, and internucleosomal interactions could create an additional “layer” of chromatin activity determinants Figure 4E. We hypothesize that in such manner, these factors could create a “code” of chromatin activity states, in addition to the histone code of chromatin activity signals, which could promote the formation and stabilization of a highly dynamic, accessible structure of a nucleosome array. The proposed models stress the diversity of mechanisms by which histone PTMs, DNA conformations, and internucleosomal interactions regulate chromatin functionality.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was conducted in the frame of IDB RAS government programs of basic research in 2022 No. 0088-2021-0007.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or any claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
references
Adelman, K., and Lis, J. T. (2012). Promoter-proximal Pausing of RNA Polymerase II: Emerging Roles in Metazoans. Nat. Rev. Genet. 13, 720–731. doi:10.1038/nrg3293
Alabert, C., Barth, T. K., Reverón-Gómez, N., Sidoli, S., Schmidt, A., Jensen, O. N., et al. (2015). Two Distinct Modes for Propagation of Histone PTMs across the Cell Cycle. Genes Dev. 29, 585–590. doi:10.1101/gad.256354.114
Allahverdi, A., Yang, R., Korolev, N., Fan, Y., Davey, C. A., Liu, C.-F., et al. (2011). The Effects of Histone H4 Tail Acetylations on Cation-Induced Chromatin Folding and Self-Association. Nucleic Acids Res. 39, 1680–1691. doi:10.1093/nar/gkq900
Anderson, C. J., Baird, M. R., Hsu, A., Barbour, E. H., Koyama, Y., Borgnia, M. J., et al. (2019). Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase. Cel Rep. 26, 1681–1690. doi:10.1016/j.celrep.2019.01.058
Andrews, F. H., Strahl, B. D., and Kutateladze, T. G. (2016). Insights into Newly Discovered marks and Readers of Epigenetic Information. Nat. Chem. Biol. 12, 662–668. doi:10.1038/nchembio.2149
Aragay, A. M., Diaz, P., and Daban, J.-R. (1988). Association of Nucleosome Core Particle DNA with Different Histone Oligomers. J. Mol. Biol. 204, 141–154. doi:10.1016/0022-2836(88)90605-5
Aragay, A. M., Fernandez-Busquets, X., and Daban, J. R. (1991). Different Mechanisms for In Vitro Formation of Nucleosome Core Particles. Biochemistry 30, 5022–5032. doi:10.1021/bi00234a026
Armeev, G. A., Gribkova, A. K., Pospelova, I., Komarova, G. A., and Shaytan, A. K. (2018). Linking Chromatin Composition and Structural Dynamics at the Nucleosome Level. Curr. Opin. Struct. Biol. 56, 46–55. doi:10.1016/j.sbi.2018.11.006
Ausio, J., Seger, D., and Eisenberg, H. (1984). Nucleosome Core Particle Stability and Conformational Change. J. Mol. Biol. 176, 77–104. doi:10.1016/0022-2836(84)90383-8
Bancaud, A., Wagner, G., Conde e Silva, N., Lavelle, C., Wong, H., Mozziconacci, J., et al. (2007). Nucleosome Chiral Transition under Positive Torsional Stress in Single Chromatin Fibers. Mol. Cel 27, 135–147. doi:10.1016/j.molcel.2007.05.037
Baranello, L., Levens, D., Gupta, A., and Kouzine, F. (2012). The Importance of Being Supercoiled: How DNA Mechanics Regulate Dynamic Processes. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1819, 632–638. doi:10.1016/j.bbagrm.2011.12.007
Batta, K., Zhang, Z., Yen, K., Goffman, D. B., and Pugh, B. F. (2011). Genome-wide Function of H2B Ubiquitylation in Promoter and Genic Regions. Genes Dev. 25, 2254–2265. doi:10.1101/gad.177238.111
Bayer, P., Arndt, A., Metzger, S., Mahajan, R., Melchior, F., Jaenicke, R., et al. (1998). Structure Determination of the Small Ubiquitin-Related Modifier SUMO-1. J. Mol. Biol. 280, 275–286. doi:10.1006/jmbi.1998.1839
Bhaumik, S. R., Smith, E., and Shilatifard, A. (2007). Covalent Modifications of Histones during Development and Disease Pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016. doi:10.1038/nsmb1337
Bi, X., Yang, R., Feng, X., Rhodes, D., and Liu, C.-F. (2016). Semisynthetic UbH2A Reveals Different Activities of Deubiquitinases and Inhibitory Effects of H2A K119 Ubiquitination on H3K36 Methylation in Mononucleosomes. Org. Biomol. Chem. 14, 835–839. doi:10.1039/c5ob02323h
Bowman, G. D., and Poirier, M. G. (2015). Post-translational Modifications of Histones that Influence Nucleosome Dynamics. Chem. Rev. 115, 2274–2295. doi:10.1021/cr500350x
Bruno, M., Flaus, A., Stockdale, C., Rencurel, C., Ferreira, H., and Owen-Hughes, T. (2003). Histone H2A/H2B Dimer Exchange by ATP-dependent Chromatin Remodeling Activities. Mol. Cel 12, 1599–1606. doi:10.1016/s1097-2765(03)00499-4
Buning, R., Kropff, W., Martens, K., and van Noort, J. (2015). spFRET Reveals Changes in Nucleosome Breathing by Neighboring Nucleosomes. J. Phys. Condens. Matter 27, 064103. doi:10.1088/0953-8984/27/6/064103
Cao, J., and Yan, Q. (2012). Histone Ubiquitination and Deubiquitination in Transcription, DNA Damage Response, and Cancer. Front. Oncol. 2, 26. doi:10.3389/fonc.2012.00026
Chakraborty, K., and Loverde, S. M. (2017). Asymmetric Breathing Motions of Nucleosomal DNA and the Role of Histone Tails. J. Chem. Phys. 147, 065101. doi:10.1063/1.4997573
Chandrasekharan, M. B., Huang, F., and Sun, Z.-W. (2009). Ubiquitination of Histone H2B Regulates Chromatin Dynamics by Enhancing Nucleosome Stability. Proc. Natl. Acad. Sci. U.S.A. 106, 16686–16691. doi:10.1073/pnas.0907862106
Chen, T. A., Smith, M. M., Le, S. Y., Sternglanz, R., and Allfrey, V. G. (1991). Nucleosome Fractionation by Mercury Affinity Chromatography. Contrasting Distribution of Transcriptionally Active DNA Sequences and Acetylated Histones in Nucleosome Fractions of Wild-type Yeast Cells and Cells Expressing a Histone H3 Gene Altered to Encode a Cysteine 110 Residue. J. Biol. Chem. 266, 6489–6498. doi:10.1016/s0021-9258(18)38145-6
Clark, D. J., and Felsenfeld, G. (1991). Formation of Nucleosomes on Positively Supercoiled DNA. EMBO J. 10, 387–395. doi:10.1002/j.1460-2075.1991.tb07960.x
Clark, D. J., Ghirlando, R., Felsenfeld, G., and Eisenberg, H. (1993). Effect of Positive Supercoiling on DNA Compaction by Nucleosome Cores. J. Mol. Biol. 234, 297–301. doi:10.1006/jmbi.1993.1585
Cole, A. J., Clifton-Bligh, R., and Marsh, D. J. (2015). Histone H2B Monoubiquitination: Roles to Play in Human Malignancy. Endocr. Relat. Cancer 22, T19–T33. doi:10.1530/erc-14-0185
Corless, S., and Gilbert, N. (2016). Effects of DNA Supercoiling on Chromatin Architecture. Biophys. Rev. 8, 245–258. doi:10.1007/s12551-016-0210-1
Cubeñas-Potts, C., and Matunis, M. J. (2013). SUMO: a Multifaceted Modifier of Chromatin Structure and Function. Develop. Cel 24, 1–12. doi:10.1016/j.devcel.2012.11.020
Cutter, A. R., and Hayes, J. J. (2015). A Brief Review of Nucleosome Structure. FEBS Lett. 589, 2914–2922. doi:10.1016/j.febslet.2015.05.016
Daban, J.-R., and Cantor, C. R. (1982). Structural and Kinetic Study of the Self-Assembly of Nucleosome Core Particles. J. Mol. Biol. 156, 749–769. doi:10.1016/0022-2836(82)90140-1
Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W., and Richmond, T. J. (2002). Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9Å Resolution. J. Mol. Biol. 319, 1097–1113. doi:10.1016/s0022-2836(02)00386-8
Dhall, A., Wei, S., Fierz, B., Woodcock, C. L., Lee, T.-H., and Chatterjee, C. (2014). Sumoylated Human Histone H4 Prevents Chromatin Compaction by Inhibiting Long-Range Internucleosomal Interactions. J. Biol. Chem. 289, 33827–33837. doi:10.1074/jbc.m114.591644
Dieker, J., and Muller, S. (2010). Epigenetic Histone Code and Autoimmunity. Clinic Rev. Allerg Immunol. 39, 78–84. doi:10.1007/s12016-009-8173-7
Dorigo, B., Schalch, T., Kulangara, A., Duda, S., Schroeder, R. R., and Richmond, T. J. (2004). Nucleosome Arrays Reveal the Two-Start Organization of the Chromatin Fiber. Science 306, 1571–1573. doi:10.1126/science.1103124
Eisenberg, H., and Felsenfeld, G. (1981). Hydrodynamic Studies of the Interaction between Nucleosome Core Particles and Core Histones. J. Mol. Biol. 150, 537–552. doi:10.1016/0022-2836(81)90379-x
Engeholm, M., de Jager, M., Flaus, A., Brenk, R., van Noort, J., and Owen-Hughes, T. (2009). Nucleosomes Can Invade DNA Territories Occupied by Their Neighbors. Nat. Struct. Mol. Biol. 16, 151–158. doi:10.1038/nsmb.1551
Escobar, T. M., Loyola, A., and Reinberg, D. (2021). Parental Nucleosome Segregation and the Inheritance of Cellular Identity. Nat. Rev. Genet. 22, 379–392. doi:10.1038/s41576-020-00312-w
Escobar, T. M., Oksuz, O., Saldaña-Meyer, R., Descostes, N., Bonasio, R., and Reinberg, D. (2019). Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication. Cell 179, 953–963. doi:10.1016/j.cell.2019.10.009
Espinosa, J. M. (2008). Histone H2B Ubiquitination: the Cancer Connection. Genes Dev. 22, 2743–2749. doi:10.1101/gad.1732108
Esposito, F., and Sinden, R. R. (1988). DNA Supercoiling and Eukaryotic Gene Expression. Oxf Surv. Eukaryot. Genes 5, 1–50.
Fan, H.-Y., He, X., Kingston, R. E., and Narlikar, G. J. (2003). Distinct Strategies to Make Nucleosomal DNA Accessible. Mol. Cel 11, 1311–1322. doi:10.1016/s1097-2765(03)00192-8
Fierz, B., Kilic, S., Hieb, A. R., Luger, K., and Muir, T. W. (2012). Stability of Nucleosomes Containing Homogenously Ubiquitylated H2A and H2B Prepared Using Semisynthesis. J. Am. Chem. Soc. 134, 19548–19551. doi:10.1021/ja308908p
Flaus, A., and Owen-Hughes, T. (2003). Dynamic Properties of Nucleosomes during thermal and ATP-Driven Mobilization. Mol. Cel Biol. 23, 7767–7779. doi:10.1128/mcb.23.21.7767-7779.2003
Fleming, A. B., Kao, C.-F., Hillyer, C., Pikaart, M., and Osley, M. A. (2008). H2B Ubiquitylation Plays a Role in Nucleosome Dynamics during Transcription Elongation. Mol. Cel 31, 57–66. doi:10.1016/j.molcel.2008.04.025
Gao, C., Huang, W., Kanasaki, K., and Xu, Y. (2014). The Role of Ubiquitination and Sumoylation in Diabetic Nephropathy. Biomed. Res. Int. 2014, 160692. doi:10.1155/2014/160692
Garner, M. M., Felsenfeld, G., O'Dea, M. H., and Gellert, M. (1987). Effects of DNA Supercoiling on the Topological Properties of Nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 84, 2620–2623. doi:10.1073/pnas.84.9.2620
Gerasimova, N. S., Pestov, N. A., Kulaeva, O. I., Clark, D. J., and Studitsky, V. M. (2016). Transcription-induced DNA Supercoiling: New Roles of Intranucleosomal DNA Loops in DNA Repair and Transcription. Transcription 7, 91–95. doi:10.1080/21541264.2016.1182240
Gibney, E. R., and Nolan, C. M. (2010). Epigenetics and Gene Expression. Heredity 105, 4–13. doi:10.1038/hdy.2010.54
Gilbert, N., and Allan, J. (2014). Supercoiling in DNA and Chromatin. Curr. Opin. Genet. Develop. 25, 15–21. doi:10.1016/j.gde.2013.10.013
Gilmour, D. S. (2009). Promoter Proximal Pausing on Genes in Metazoans. Chromosoma 118, 1–10. doi:10.1007/s00412-008-0182-4
Gluckman, P. D., Hanson, M. A., Buklijas, T., Low, F. M., and Beedle, A. S. (2009). Epigenetic Mechanisms that Underpin Metabolic and Cardiovascular Diseases. Nat. Rev. Endocrinol. 5, 401–408. doi:10.1038/nrendo.2009.102
Gray, S. G. (2006). DMMC Workshop: Unravelling Chromatin and the Role of Epigenetics in Disease. Epigenetics 1, 187–189. doi:10.4161/epi.1.4.3399
Grigoryev, S. A., Arya, G., Correll, S., Woodcock, C. L., and Schlick, T. (2009). Evidence for Heteromorphic Chromatin Fibers from Analysis of Nucleosome Interactions. Proc. Natl. Acad. Sci. U.S.A. 106, 13317–13322. doi:10.1073/pnas.0903280106
Grigoryev, S. A. (2004). Keeping Fingers Crossed: Heterochromatin Spreading through Interdigitation of Nucleosome Arrays. FEBS Lett. 564, 4–8. doi:10.1016/s0014-5793(04)00258-3
Gurova, K., Chang, H. W., Valieva, M. E., Sandlesh, P., and Studitsky, V. M. (2018). Structure and Function of the Histone Chaperone FACT - Resolving FACTual Issues. Biochim. Biophys. Acta Gene Regul. Mech. 1861 (9), 892–904. doi:10.1016/j.bbagrm.2018.07.008
Hodges, C., Bintu, L., Lubkowska, L., Kashlev, M., and Bustamante, C. (2009). Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II. Science 325, 626–628. doi:10.1126/science.1172926
Hsieh, F.-K., Kulaeva, O. I., Patel, S. S., Dyer, P. N., Luger, K., Reinberg, D., et al. (2013). Histone Chaperone FACT Action during Transcription through Chromatin by RNA Polymerase II. Proc. Natl. Acad. Sci. U.S.A. 110, 7654–7659. doi:10.1073/pnas.1222198110
Hsieh, F.-K., Kulaeva, O. I., and Studitsky, V. M. (2015). Experimental Analysis of hFACT Action during Pol II Transcription In Vitro. Methods Mol. Biol. 1276, 315–326. doi:10.1007/978-1-4939-2392-2_19
Huang, W.-C., Ko, T.-P., Li, S. S.-L., and Wang, A. H.-J. (2004). Crystal Structures of the Human SUMO-2 Protein at 1.6 Å and 1.2 Å Resolution. Eur. J. Biochem. 271, 4114–4122. doi:10.1111/j.1432-1033.2004.04349.x
Hwang, W. W., Venkatasubrahmanyam, S., Ianculescu, A. G., Tong, A., Boone, C., and Madhani, H. D. (2003). A Conserved RING finger Protein Required for Histone H2B Monoubiquitination and Cell Size Control. Mol. Cel 11, 261–266. doi:10.1016/s1097-2765(02)00826-2
Jackson, V. (1993). Influence of Positive Stress on Nucleosome Assembly. Biochemistry 32, 5901–5912. doi:10.1021/bi00073a024
Jiao, L., and Liu, X. (2015). Structural Basis of Histone H3K27 Trimethylation by an Active Polycomb Repressive Complex 2. Science 350, aac4383. doi:10.1126/science.aac4383
Johnsen, S. A. (2012). The Enigmatic Role of H2Bub1 in Cancer. FEBS Lett. 586, 1592–1601. doi:10.1016/j.febslet.2012.04.002
Kan, P.-Y., Caterino, T. L., and Hayes, J. J. (2009). The H4 Tail Domain Participates in Intra- and Internucleosome Interactions with Protein and DNA during Folding and Oligomerization of Nucleosome Arrays. Mol. Cel Biol. 29, 538–546. doi:10.1128/mcb.01343-08
Kan, P.-Y., Lu, X., Hansen, J. C., and Hayes, J. J. (2007). The H3 Tail Domain Participates in Multiple Interactions during Folding and Self-Association of Nucleosome Arrays. Mol. Cel Biol. 27, 2084–2091. doi:10.1128/mcb.02181-06
Kan, P., and Hayes, J. (2007). Detection of Interactions between Nucleosome Arrays Mediated by Specific Core Histone Tail Domains. Methods 41, 278–285. doi:10.1016/j.ymeth.2006.08.012
Kasinath, V., Beck, C., Sauer, P., Poepsel, S., Kosmatka, J., Faini, M., et al. (2021). JARID2 and AEBP2 Regulate PRC2 in the Presence of H2AK119ub1 and Other Histone Modifications. Science 371, abc3393. doi:10.1126/science.abc3393
Kassabov, S. R., Zhang, B., Persinger, J., and Bartholomew, B. (2003). SWI/SNF Unwraps, Slides, and Rewraps the Nucleosome. Mol. Cel 11, 391–403. doi:10.1016/s1097-2765(03)00039-x
Kaushik, M., Kaushik, S., Roy, K., Singh, A., Mahendru, S., Kumar, M., et al. (2016). A Bouquet of DNA Structures: Emerging Diversity. Biochem. Biophys. Rep. 5, 388–395. doi:10.1016/j.bbrep.2016.01.013
Kepper, N., Ettig, R., Stehr, R., Marnach, S., Wedemann, G., and Rippe, K. (2011). Force Spectroscopy of Chromatin Fibers: Extracting Energetics and Structural Information from Monte Carlo Simulations. Biopolymers 95, 435–447. doi:10.1002/bip.21598
Koopmans, W. J. A., Brehm, A., Logie, C., Schmidt, T., and van Noort, J. (2007). Single-pair FRET Microscopy Reveals Mononucleosome Dynamics. J. Fluoresc. 17, 785–795. doi:10.1007/s10895-007-0218-9
Kornberg, R. D., and Lorch, Y. (1991). Irresistible Force Meets Immovable Object: Transcription and the Nucleosome. Cell 67, 833–836. doi:10.1016/0092-8674(91)90354-2
Krajewski, W. A. (2020a). "Direct" and "Indirect" Effects of Histone Modifications: Modulation of Sterical Bulk as a Novel Source of Functionality. Bioessays 42, e1900136. doi:10.1002/bies.201900136
Krajewski, W. A. (2013). Comparison of the Isw1a, Isw1b, and Isw2 Nucleosome Disrupting Activities. Biochemistry 52, 6940–6949. doi:10.1021/bi400634r
Krajewski, W. A. (2018). Effects of DNA Superhelical Stress on the Stability of H2B-Ubiquitylated Nucleosomes. J. Mol. Biol. 430, 5002–5014. doi:10.1016/j.jmb.2018.09.014
Krajewski, W. A. (1996). Enhancement of Transcription by Short Alternating CG Tracts Incorporated within a Rous Sarcoma Virus-Based Chimeric Promoter: In Vivo Studies. Mol. Gen. Genet. 252, 249–254. doi:10.1007/s004380050226
Krajewski, W. A., Li, J., and Dou, Y. (2018). Effects of Histone H2B Ubiquitylation on the Nucleosome Structure and Dynamics. Nucleic Acids Res. 46, 7631–7642. doi:10.1093/nar/gky526
Krajewski, W. A., and Luchnik, A. N. (1991). High Rotational Mobility of DNA in Animal Cells and its Modulation by Histone Acetylation. Mol. Gen. Genet. 231, 17–21. doi:10.1007/bf00293816
Krajewski, W. A. (2016). On the Role of Inter-nucleosomal Interactions and Intrinsic Nucleosome Dynamics in Chromatin Function. Biochem. Biophys. Rep. 5, 492–501. doi:10.1016/j.bbrep.2016.02.009
Krajewski, W. A. (2020b). The Intrinsic Stability of H2B-Ubiquitylated Nucleosomes and Their In Vitro Assembly/disassembly by Histone Chaperone NAP1. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1864, 129497. doi:10.1016/j.bbagen.2019.129497
Krajewski, W. A. (2019). Ubiquitylation: How Nucleosomes Use Histones to Evict Histones. Trends Cel Biol. 29, 689–694. doi:10.1016/j.tcb.2019.06.002
Krajewski, W. A., and Vassiliev, O. L. (2019). Analysis of Histone Ubiquitylation by MSL1/MSL2 Proteins In Vitro. Arch. Biochem. Biophys. 666, 22–30. doi:10.1016/j.abb.2019.03.015
Krajewski, W. A., and Vassiliev, O. L. (2012). Remodeling of Nucleosome-Dimer Particles with yIsw2 Promotes Their Association with ALL-1 SET Domain In Vitro. Biochemistry 51, 4354–4363. doi:10.1021/bi201645c
Krajewski, W. A., and Vassiliev, O. L. (2010). The Saccharomyces cerevisiae Swi/Snf Complex Can Catalyze Formation of Dimeric Nucleosome Structures In Vitro. Biochemistry 49, 6531–6540. doi:10.1021/bi1006157
Krajewski, W. A. (2014). Yeast Isw1a and Isw1b Exhibit Similar Nucleosome Mobilization Capacities for Mononucleosomes, but Differently Mobilize Dinucleosome Templates. Arch. Biochem. Biophys. 546, 72–80. doi:10.1016/j.abb.2014.02.003
Kruithof, M., Chien, F.-T., Routh, A., Logie, C., Rhodes, D., and van Noort, J. (2009). Single-molecule Force Spectroscopy Reveals a Highly Compliant Helical Folding for the 30-nm Chromatin Fiber. Nat. Struct. Mol. Biol. 16, 534–540. doi:10.1038/nsmb.1590
Kulaeva, O. I., Hsieh, F.-K., Chang, H.-W., Luse, D. S., and Studitsky, V. M. (2013). Mechanism of Transcription through a Nucleosome by RNA Polymerase II. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1829, 76–83. doi:10.1016/j.bbagrm.2012.08.015
Lee, M. S., and Garrard, W. T. (1991). Positive DNA Supercoiling Generates a Chromatin Conformation Characteristic of Highly Active Genes. Proc. Natl. Acad. Sci. U.S.A. 88, 9675–9679. doi:10.1073/pnas.88.21.9675
Li, G., Levitus, M., Bustamante, C., and Widom, J. (2005). Rapid Spontaneous Accessibility of Nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53. doi:10.1038/nsmb869
Li, J., He, Q., Liu, Y., Liu, S., Tang, S., Li, C., et al. (2017). Chemical Synthesis of K34-Ubiquitylated H2B for Nucleosome Reconstitution and Single-Particle Cryo-Electron Microscopy Structural Analysis. Chembiochem 18, 176–180. doi:10.1002/cbic.201600551
Liu, L. F., and Wang, J. C. (1987). Supercoiling of the DNA Template during Transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027. doi:10.1073/pnas.84.20.7024
Ljungman, M., and Hanawalt, P. C. (1995). Presence of Negative Torsional Tension in the Promoter Region of the Transcriptionally Poised Dihydrofolate Reductase Gene In Vivo. Nucleic Acids Res. 23, 1782–1789. doi:10.1093/nar/23.10.1782
Luger, K., Dechassa, M. L., and Tremethick, D. J. (2012). New Insights into Nucleosome and Chromatin Structure: an Ordered State or a Disordered Affair? Nat. Rev. Mol. Cel Biol. 13, 436–447. doi:10.1038/nrm3382
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 389, 251–260. doi:10.1038/38444
Machida, S., Sekine, S., Nishiyama, Y., Horikoshi, N., and Kurumizaka, H. (2016). Structural and Biochemical Analyses of Monoubiquitinated Human Histones H2B and H4. Open. Biol. 6, 1–9. doi:10.1098/rsob.160090
Manning, G. S. (2006). The Persistence Length of DNA Is Reached from the Persistence Length of its Null Isomer through an Internal Electrostatic Stretching Force. Biophysical J. 91, 3607–3616. doi:10.1529/biophysj.106.089029
Martin, C., Cao, R., and Zhang, Y. (2006). Substrate Preferences of the EZH2 Histone Methyltransferase Complex. J. Biol. Chem. 281, 8365–8370. doi:10.1074/jbc.m513425200
McLean, M. J., and Wells, R. D. (1988). The Role of DNA Sequence in the Formation of Z-DNA versus Cruciforms in Plasmids. J. Biol. Chem. 263, 7370–7377. doi:10.1016/s0021-9258(18)68652-1
Meas, R., and Mao, P. (2015). Histone Ubiquitylation and its Roles in Transcription and DNA Damage Response. DNA Repair 36, 36–42. doi:10.1016/j.dnarep.2015.09.016
Mirabella, A. C., Foster, B. M., and Bartke, T. (2016). Chromatin Deregulation in Disease. Chromosoma 125, 75–93. doi:10.1007/s00412-015-0530-0
Morrison, E. A., Bowerman, S., Sylvers, K. L., Wereszczynski, J., and Musselman, C. A. (2018). The Conformation of the Histone H3 Tail Inhibits Association of the BPTF PHD finger with the Nucleosome. Elife 7. doi:10.7554/eLife.31481
Morse, R. H., Pederson, D. S., Dean, A., and Simpson, R. T. (1987). Yeast Nudeosomes Allow thermal Untwisting of DNA. Nucl. Acids Res. 15, 10311–10330. doi:10.1093/nar/15.24.10311
Nathan, D., Ingvarsdottir, K., Sterner, D. E., Bylebyl, G. R., Dokmanovic, M., Dorsey, J. A., et al. (2006). Histone Sumoylation Is a Negative Regulator in Saccharomyces cerevisiae and Shows Dynamic Interplay with Positive-Acting Histone Modifications. Genes Dev. 20, 966–976. doi:10.1101/gad.1404206
Naughton, C., Avlonitis, N., Corless, S., Prendergast, J. G., Mati, I. K., Eijk, P. P., et al. (2013). Transcription Forms and Remodels Supercoiling Domains Unfolding Large-Scale Chromatin Structures. Nat. Struct. Mol. Biol. 20, 387–395. doi:10.1038/nsmb.2509
Ngo, T. T. M., and Ha, T. (2015). Nucleosomes Undergo Slow Spontaneous Gaping. Nucleic Acids Res. 43, 3964–3971. doi:10.1093/nar/gkv276
Pepenella, S., Murphy, K. J., and Hayes, J. J. (2014a). A Distinct Switch in Interactions of the Histone H4 Tail Domain upon Salt-dependent Folding of Nucleosome Arrays. J. Biol. Chem. 289, 27342–27351. doi:10.1074/jbc.m114.595140
Pepenella, S., Murphy, K. J., and Hayes, J. J. (2014b). Intra- and Inter-nucleosome Interactions of the Core Histone Tail Domains in Higher-Order Chromatin Structure. Chromosoma 123, 3–13. doi:10.1007/s00412-013-0435-8
Perini, G., and Tupler, R. (2006). Altered Gene Silencing and Human Diseases. Clin. Genet. 69, 1–7. doi:10.1111/j.1399-0004.2005.00540.x
Petruk, S., Black, K. L., Kovermann, S. K., Brock, H. W., and Mazo, A. (2013). Stepwise Histone Modifications Are Mediated by Multiple Enzymes that Rapidly Associate with Nascent DNA during Replication. Nat. Commun. 4, 2841. doi:10.1038/ncomms3841
Petruk, S., Sedkov, Y., Johnston, D. M., Hodgson, J. W., Black, K. L., Kovermann, S. K., et al. (2012). TrxG and PcG Proteins but Not Methylated Histones Remain Associated with DNA through Replication. Cell 150, 922–933. doi:10.1016/j.cell.2012.06.046
Pfaffle, P., and Jackson, V. (1990). Studies on Rates of Nucleosome Formation with DNA under Stress. J. Biol. Chem. 265, 16821–16829. doi:10.1016/s0021-9258(17)44835-6
Polach, K. J., and Widom, J. (1996). A Model for the Cooperative Binding of Eukaryotic Regulatory Proteins to Nucleosomal Target Sites. J. Mol. Biol. 258, 800–812. doi:10.1006/jmbi.1996.0288
Prakash, K., and Fournier, D. (2018). Evidence for the Implication of the Histone Code in Building the Genome Structure. Biosystems 164, 49–59. doi:10.1016/j.biosystems.2017.11.005
Prior, C. P., Cantor, C. R., Johnson, E. M., Littau, V. C., and Allfrey, V. G. (1983). Reversible Changes in Nucleosome Structure and Histone H3 Accessibility in Transcriptionally Active and Inactive States of rDNA Chromatin. Cell 34, 1033–1042. doi:10.1016/0092-8674(83)90561-5
Reinberg, D., and Vales, L. D. (2018). Chromatin Domains Rich in Inheritance. Science 361, 33–34. doi:10.1126/science.aat7871
Renatus, M., Parrado, S. G., D'Arcy, A., Eidhoff, U., Gerhartz, B., Hassiepen, U., et al. (2006). Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2. Structure 14, 1293–1302. doi:10.1016/j.str.2006.06.012
Reverón-Gómez, N., González-Aguilera, C., Stewart-Morgan, K. R., Petryk, N., Flury, V., Graziano, S., et al. (2018). Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication. Mol. Cel 72, 239–249. doi:10.1016/j.molcel.2018.08.010
Robinson, P. J., and Rhodes, D. (2006). Structure of the '30nm' Chromatin Fibre: A Key Role for the Linker Histone. Curr. Opin. Struct. Biol. 16, 336–343. doi:10.1016/j.sbi.2006.05.007
Rothbart, S. B., and Strahl, B. D. (2014). Interpreting the Language of Histone and DNA Modifications. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1839, 627–643. doi:10.1016/j.bbagrm.2014.03.001
Saavedra, R. A., and Huberman, J. A. (1986). Both DNA Topoisomerases I and II Relax 2 μm Plasmid DNA in Living Yeast Cells. Cell 45, 65–70. doi:10.1016/0092-8674(86)90538-6
Schalch, T., Duda, S., Sargent, D. F., and Richmond, T. J. (2005). X-ray Structure of a Tetranucleosome and its Implications for the Chromatin Fibre. Nature 436, 138–141. doi:10.1038/nature03686
Schnitzler, G. R., Cheung, C. L., Hafner, J. H., Saurin, A. J., Kingston, R. E., and Lieber, C. M. (2001). Direct Imaging of Human SWI/SNF-remodeled Mono- and Polynucleosomes by Atomic Force Microscopy Employing Carbon Nanotube Tips. Mol. Cel Biol. 21, 8504–8511. doi:10.1128/mcb.21.24.8504-8511.2001
Schunter, S., Villa, R., Flynn, V., Heidelberger, J. B., Classen, A.-K., Beli, P., et al. (2017). Ubiquitylation of the Acetyltransferase MOF in Drosophila melanogaster. Plos. One. 12, e0177408. doi:10.1371/journal.pone.0177408
Selth, L. A., Sigurdsson, S., and Svejstrup, J. Q. (2010). Transcript Elongation by RNA Polymerase II. Annu. Rev. Biochem. 79, 271–293. doi:10.1146/annurev.biochem.78.062807.091425
Shaytan, A. K., Armeev, G. A., Goncearenco, A., Zhurkin, V. B., Landsman, D., and Panchenko, A. R. (2016). Coupling between Histone Conformations and DNA Geometry in Nucleosomes on a Microsecond Timescale: Atomistic Insights into Nucleosome Functions. J. Mol. Biol. 428, 221–237. doi:10.1016/j.jmb.2015.12.004
Sheinin, M. Y., Li, M., Soltani, M., Luger, K., and Wang, M. D. (2013). Torque Modulates Nucleosome Stability and Facilitates H2A/H2B Dimer Loss. Nat. Commun. 4, 2579. doi:10.1038/ncomms3579
Shiio, Y., and Eisenman, R. N. (2003). Histone Sumoylation Is Associated with Transcriptional Repression. Proc. Natl. Acad. Sci. U.S.A. 100, 13225–13230. doi:10.1073/pnas.1735528100
Singh, J., and Padgett, R. A. (2009). Rates of In Situ Transcription and Splicing in Large Human Genes. Nat. Struct. Mol. Biol. 16, 1128–1133. doi:10.1038/nsmb.1666
Smith, G. R. (2008). Meeting DNA Palindromes Head-To-Head. Genes Dev. 22, 2612–2620. doi:10.1101/gad.1724708
Stein, A. (1979). DNA Folding by Histones: the Kinetics of Chromatin Core Particle Reassembly and the Interaction of Nucleosomes with Histones. J. Mol. Biol. 130, 103–134. doi:10.1016/0022-2836(79)90421-2
Strahl, B. D., and Allis, C. D. (2000). The Language of Covalent Histone Modifications. Nature 403, 41–45. doi:10.1038/47412
Tatchell, K., and Van Holde, K. E. (1979). Nucleosome Reconstitution: Effect of DNA Length on Nucleosome Structure. Biochemistry 18, 2871–2880. doi:10.1021/bi00580a031
Teves, S. S., and Henikoff, S. (2014). Transcription-generated Torsional Stress Destabilizes Nucleosomes. Nat. Struct. Mol. Biol. 21, 88–94. doi:10.1038/nsmb.2723
Trujillo, K. M., and Osley, M. A. (2012). A Role for H2B Ubiquitylation in DNA Replication. Mol. Cel 48, 734–746. doi:10.1016/j.molcel.2012.09.019
Ulyanova, N. P., and Schnitzler, G. R. (2005). Human SWI/SNF Generates Abundant, Structurally Altered Dinucleosomes on Polynucleosomal Templates. Mol. Cel Biol. 25, 11156–11170. doi:10.1128/mcb.25.24.11156-11170.2005
Ulyanova, N. P., and Schnitzler, G. R. (2007). Inverted Factor Access and Slow Reversion Characterize SWI/SNF-altered Nucleosome Dimers. J. Biol. Chem. 282, 1018–1028. doi:10.1074/jbc.m609473200
Valencia-Sanchez, M. I., De, I. P., Wang, M., Vasilyev, N., Chen, R., Nudler, E., et al. (2019). Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. Mol. Cel 74 (5), 1010–1019. doi:10.1016/j.molcel.2019.03.029
Vasudevan, D., Chua, E. Y. D., and Davey, C. A. (2010). Crystal Structures of Nucleosome Core Particles Containing the '601' strong Positioning Sequence. J. Mol. Biol. 403, 1–10. doi:10.1016/j.jmb.2010.08.039
Vaughan, R. M., Kupai, A., and Rothbart, S. B. (2021). Chromatin Regulation through Ubiquitin and Ubiquitin-like Histone Modifications. Trends Biochem. Sci. 46, 258–269. doi:10.1016/j.tibs.2020.11.005
Venkatesh, S., and Workman, J. L. (2015). Histone Exchange, Chromatin Structure and the Regulation of Transcription. Nat. Rev. Mol. Cel Biol. 16, 178–189. doi:10.1038/nrm3941
Vicent, G. P., Nacht, A. S., Smith, C. L., Peterson, C. L., Dimitrov, S., and Beato, M. (2004). DNA Instructed Displacement of Histones H2A and H2B at an Inducible Promoter. Mol. Cel 16, 439–452. doi:10.1016/j.molcel.2004.10.025
Victor, J. M., Zlatanova, J., Barbi, M., and Mozziconacci, J. (2012). Pulling Chromatin Apart: Unstacking or Unwrapping? BMC. Biophys. 5, 21. doi:10.1186/2046-1682-5-21
Voordouw, G., and Eisenberg, H. (1978). Binding of Additional Histones to Chromatin Core Particles. Nature 273, 446–448. doi:10.1038/273446a0
Wang, H., Zhai, L., Xu, J., Joo, H.-Y., Jackson, S., Erdjument-Bromage, H., et al. (2006). Histone H3 and H4 Ubiquitylation by the CUL4-DDB-ROC1 Ubiquitin Ligase Facilitates Cellular Response to DNA Damage. Mol. Cel 22, 383–394. doi:10.1016/j.molcel.2006.03.035
Weake, V. M. (2014). “Histone Ubiquitylation Control of Gene Expression,” in Fundamentals of Chromatin. Editors J. Workman, and S. Abmayr (New York, NY: Springer), 257–307. doi:10.1007/978-1-4614-8624-4_6
Weber, C. M., Ramachandran, S., and Henikoff, S. (2014). Nucleosomes Are Context-specific, H2A.Z-Modulated Barriers to RNA Polymerase. Mol. Cel 53, 819–830. doi:10.1016/j.molcel.2014.02.014
Wells, R. D. (1988). Unusual DNA Structures. J. Biol. Chem. 263, 1095–1098. doi:10.1016/s0021-9258(19)57268-4
Wright, D. E., and Kao, C.-F. (2015). (Ubi)quitin' the H2bit: Recent Insights into the Roles of H2B Ubiquitylation in DNA Replication and Transcription. Epigenetics 10, 122–126. doi:10.1080/15592294.2014.1003750
Wu, L., Lee, S. Y., Zhou, B., Nguyen, U. T. T., Muir, T. W., Tan, S., et al. (2013). ASH2L Regulates Ubiquitylation Signaling to MLL: Trans-regulation of H3 K4 Methylation in Higher Eukaryotes. Mol. Cel 49, 1108–1120. doi:10.1016/j.molcel.2013.01.033
Wu, L., Li, L., Zhou, B., Qin, Z., and Dou, Y. (2014). H2B Ubiquitylation Promotes RNA Pol II Processivity via PAF1 and pTEFb. Mol. Cel 54, 920–931. doi:10.1016/j.molcel.2014.04.013
Wu, L., Zee, B. M., Wang, Y., Garcia, B. A., and Dou, Y. (2011). The RING finger Protein MSL2 in the MOF Complex Is an E3 Ubiquitin Ligase for H2B K34 and Is Involved in Crosstalk with H3 K4 and K79 Methylation. Mol. Cel 43, 132–144. doi:10.1016/j.molcel.2011.05.015
Xia, Y., Yang, W., Fa, M., Li, X., Wang, Y., Jiang, Y., et al. (2017). RNF8 Mediates Histone H3 Ubiquitylation and Promotes Glycolysis and Tumorigenesis. J. Exp. Med. 214, 1843–1855. doi:10.1084/jem.20170015
Xiao, X., Liu, C., Pei, Y., Wang, Y.-Z., Kong, J., Lu, K., et al. (2020). Histone H2A Ubiquitination Reinforces Mechanical Stability and Asymmetry at the Single-Nucleosome Level. J. Am. Chem. Soc. 142, 3340–3345. doi:10.1021/jacs.9b12448
Yager, T. D., McMurray, C. T., and Van Holde, K. E. (1989). Salt-induced Release of DNA from Nucleosome Core Particles. Biochemistry 28, 2271–2281. doi:10.1021/bi00431a045
Yu, J.-R., Lee, C.-H., Oksuz, O., Stafford, J. M., and Reinberg, D. (2019). PRC2 Is High Maintenance. Genes Dev. 33, 903–935. doi:10.1101/gad.325050.119
Zheng, C., and Hayes, J. J. (2003a). Intra- and Inter-nucleosomal Protein-DNA Interactions of the Core Histone Tail Domains in a Model System. J. Biol. Chem. 278, 24217–24224. doi:10.1074/jbc.m302817200
Zheng, C., and Hayes, J. J. (2003b). Structures and Interactions of the Core Histone Tail Domains. Biopolymers 68, 539–546. doi:10.1002/bip.10303
Zlatanova, J., Bishop, T. C., Victor, J.-M., Jackson, V., and Van Holde, K. (2009). The Nucleosome Family: Dynamic and Growing. Structure 17, 160–171. doi:10.1016/j.str.2008.12.016
Keywords: nucleosomes, histones, hexasomes, ubiquitylation, histone modifications, histone code, DNA stresses
Citation: Krajewski WA (2022) Histone Modifications, Internucleosome Dynamics, and DNA Stresses: How They Cooperate to “Functionalize” Nucleosomes. Front. Genet. 13:873398. doi: 10.3389/fgene.2022.873398
Received: 10 February 2022; Accepted: 28 March 2022;
Published: 28 April 2022.
Edited by:
Dileep Vasudevan, Institute of Life Sciences (ILS), IndiaReviewed by:
Li Wei, Institute of Physics (CAS), ChinaSanxiong Liu, NYU Grossman School of Medicine, United States
Copyright © 2022 Krajewski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wladyslaw A. Krajewski, d2tyYWpld3NraUBob3RtYWlsLmNvbQ==
 Wladyslaw A. Krajewski
Wladyslaw A. Krajewski