
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 17 March 2022
Sec. RNA
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.866772
This article is part of the Research Topic RNA-Chromatin Interactions: Biology, Mechanism, Disease and Therapeutics View all 5 articles
 Lee O. Vaasjo1,2*
Lee O. Vaasjo1,2*New roles for RNA in mediating gene expression are being discovered at an alarming rate. A broad array of pathways control patterning of N6-methyladenosine (m6A) methylation on RNA transcripts. This review comprehensively discusses long non-coding RNAs (lncRNAs) as an additional dynamic regulator of m6A methylation, with a focus on the untranslated regions (UTRs) of mRNAs. Although there is extensive literature describing m6A modification of lncRNA, the function of lncRNA in guiding m6A writers has not been thoroughly explored. The independent control of lncRNA expression, its heterogeneous roles in RNA metabolism, and its interactions with epigenetic machinery, alludes to their potential in dynamic patterning of m6A methylation. While epigenetic regulation by histone modification of H3K36me3 has been demonstrated to pattern RNA m6A methylation, these modifications were specific to the coding and 3′UTR regions. However, there are observations that 5′UTR m6A is distinct from that of the coding and 3′UTR regions, and substantial evidence supports the active regulation of 5′UTR m6A methylation. Consequently, two potential mechanisms in patterning the UTRs m6A methylation are discussed; (1) Anti-sense lncRNA (AS-lncRNA) can either bind directly to the UTR, or (2) act indirectly via recruitment of chromatin-modifying complexes to pattern m6A. Both pathways can guide the m6A writer complex, facilitate m6A methylation and modulate protein translation. Findings in the lncRNA-histone-m6A axis could potentially contribute to the discovery of new functions of lncRNAs and clarify lncRNA-m6A findings in translational medicine.
RNA modifications and RNA-RNA interactions are some of the oldest biological building blocks of the cell (Schwartz, 1998; Higgs and Lehman, 2015). Long non-coding RNAs (lncRNAs) are an abundant type of non-protein-coding RNA that have diverse functions in the nucleus, including DNA organization, recruitment of histone proteins, RNA metabolism, and translational control via direct epigenetic interactions (Schmitz et al., 2016). LncRNAs have been described to guide DNA methylation, histone modifications, and, recently, RNA methylation (Kim et al., 2015; Marchese et al., 2017; Chen et al., 2020). While patterned by multiple mechanisms, n6-methyladenosine (m6A) methylation of RNA is the most abundant internal post-transcriptional modification and is most prevalent on the coding sequence (CDS) and 3′ untranslated region (UTR) (Meyer et al., 2012). The reversible modification of m6A methylation is catalyzed by “writer” proteins (Mettl3/Mettl14/WTAP) (Figure 1A), and demethylated by “erasers” (FTO/ALKBH5). M6A methylation has been described to be involved in alternative splicing, transport, stability of RNAs and to regulate RNA translation (B. Wu et al., 2017a; Shi H. et al., 2019). Cap-independent translation is a potent ribosome recruitment mechanism that bypasses translational control checkpoints during a rapid cellular response to environmental or physiological insults (Leppek et al., 2018). While present in low abundance, m6A methylation at the 5′UTR has been shown to selectively initiate cap-independent protein translation (Meyer et al., 2015; Zhou et al., 2015; Coots et al., 2017). Yet, the mechanisms that govern m6A patterning on the 5′UTR are poorly understood.
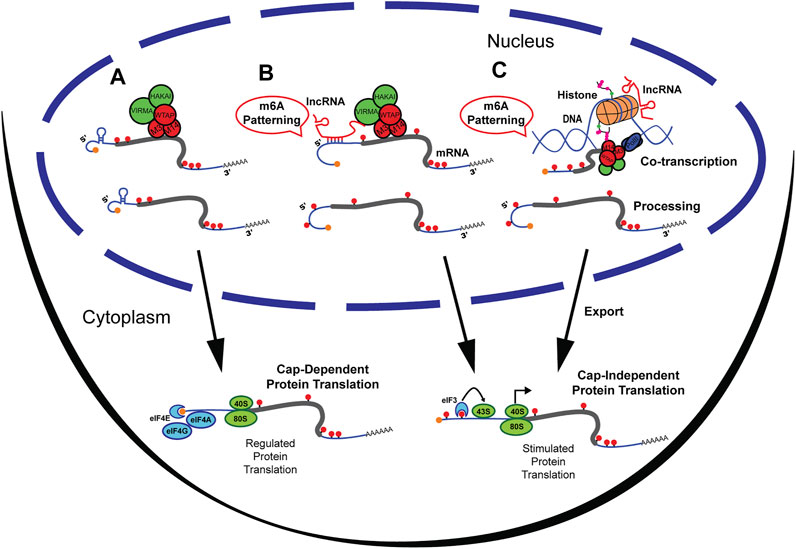
FIGURE 1. M6A methylation at the UTRs can be patterned by lncRNAs. (A) M6A methylation by writer complex occurring primarily at the CDS and 3′UTR of mRNA. Methylated mRNA is then exported from the nucleus and undergoes cap-dependent protein translation. (B) LncRNAs directly guide the m6A Writer complex by association with Virma to pattern the 5′UTR with m6A. Upon export, 5′UTR methylated mRNA undergoes Cap-independent protein translation by recruitment of eiF3 and bypassing regulatory networks. (C) LncRNAs can recruit histone modifying enzymes that result in m6A patterning. Transcripts are then exported from the nucleus and mRNAs methylated at the 5′UTR undergo Cap-independent protein translation.
The 5′UTR is a critical regulator of the final product of gene expression given it can either enhance or repress the translational state of messenger RNAs (mRNAs) (Sendoel et al., 2017; Leppek et al., 2018). Since translational control is highly regulated (Silvera et al., 2010; Buffington et al., 2014), and single mRNA transcripts can persistently generate protein products (English et al., 2016), a mechanism that can tag RNAs to bypass canonical translational control is of tremendous significance. As observed in the study of the heat shock response (Meyer et al., 2015; Zhou et al., 2015), changes in m6A methylated 5′UTR (m6A 5′UTR) can alter a cell’s biological state in response to environmental cues or perturbation (Figure 1). This prompts a significant need to understand 5′UTR m6A patterning mechanisms. However, most studies observe a scarcity of m6A methylation at the 5′UTR (Fu et al., 2014). Because 5‘UTR methylation is both WTAP-independent (Schwartz et al., 2014) and Zc3h13-independent (Wen et al., 2018), this suggests that it is regulated by other sources (Meyer et al., 2012; Dominissini et al., 2013; Schwartz et al., 2014; Koranda et al., 2018). Recently, knock-out of the Mettl14/Mettl3 associated complex component Vir-like m6A methyltransferase associated or VIRMA (a.k.a. KIAA1429), was shown to increase the amount of 5’UTR m6A. This suggests that the process may be regulated by protein participants of the Mettl14/Mettl3 complex (Yue et al., 2018). Furthermore, VIRMA upregulation has been associated with tumorigenesis and seminoma cancer, consistent with aberrant gene expression profiles (Lobo et al., 2019). Studies have demonstrated that m6A at the 5′UTR can be altered due to biological signals such as normal development (Xiao et al., 2019), neurogenesis (Yoon et al., 2017), HIV infection (Lichinchi et al., 2016), memory formation (Widagdo et al., 2016) and stress response (J. Yu F. et al., 2018), supporting dynamic regulation of m6A 5′UTR. However, the mechanism by which transcript- and methylation-site specificity at the 5′UTR is controlled remains elusive (Zhao et al., 2018).
Multiple forms of regulating m6A methylation have been described and are frequently being discovered (Huang et al., 2020). For example, the histone modification H3K36me3 (Huang et al., 2019) was found to guide m6A methylation co-transcriptionally, and microRNAs (miRNAs) (Chen et al., 2015) were found to mediate binding of Mettl3 to target sites on mRNAs. Yet, both mechanisms are preferential towards m6A patterning of the CDS and 3′UTR. Interestingly, there are in-depth descriptions of lncRNAs that recruit chromatin modifiers, and that guide DNA methylation (Savell et al., 2016; F. Yu J. et al., 2018; Mishra and Kanduri, 2019). Non-Coding RNAs are broadly known to act as guides for RNA modifications and m6A is no exception; lncRNAs are now accepted as regulators of post-transcriptional modifications (Leighton and Bredy, 2018; Chen et al., 2020). Here, lncRNAs are reviewed as guides for m6A UTR patterning and two potential non-mutually exclusive mechanisms by which lncRNAs can dynamically control m6A at the UTR are discussed. In one scenario (Figure 1B), lncRNAs bind directly to the UTR of the mRNA transcripts to regulate VIRMA binding and control UTR m6A levels, such as lncRNA GATA3-AS (Lan et al., 2019). In the second scenario (Figure 1C), lncRNA regulate epigenetic modifications of histone subunits that ultimately pattern m6A on mRNA (Huang et al., 2019). This review provides an in-depth analysis of these two non-opposing mechanisms that may guide m6A to the 3′UTR and potentially the 5′UTR, while highlighting the cross-talk between the epigenome and the epitranscriptome.
Histone modifiers, m6A writers, as well as hundreds of lncRNAs are thought to localize to the same subcellular nuclear compartment. However, whether these biological processes localize and can function simultaneously at a single active gene during transcription, e.g., co-transcriptionally, is a fundamental question in understanding the precise control of m6A methylation patterning (Perales and Bentley, 2009; Huang et al., 2020).
The co-transcriptional nature of m6A deposition on RNA molecules was described early in the re-invigoration of the m6A modification field (Shi X. et al., 2019). M6A writers interact with transcription factors, like FoxO6 (Zong et al., 2020), with transcriptional machinery, like Poll2, along with nascent transcribed RNA (Zhou et al., 2019). Furthermore, the writer Mettl3 can bind directly with both promoter regions (Barbieri et al., 2017) and transcription start sites (TSS) (Xiao et al., 2019), and even with epigenetic machinery like histone methyltransferases (Xu et al., 2021). For example, during TGF-β pathway activation, the transcription factors SMAD2/3 promotes writer complex Mettl3, Mettl14 and WTAP activity to selectively methylate transcripts associated with cell fate specification (Bertero et al., 2018). Additionally, RNA binding proteins that bind to m6A sites, e.g. m6A “readers,” such as YTHDC1, can also interact with epigenetic machinery (Li et al., 2020). Pivotal findings have been made so far uncovering the co-transcriptional landscape of m6A methylation, however, these are likely only the first of many interactions with transcriptional machinery to be discovered. Overall, it is still unclear what patterning mechanisms prime the gene/transcript at the epigenetic level.
LncRNAs have long been observed to interact with genomic machinery within the nucleus. These lncRNAs have been described to have direct interactions with DNA enhancer regions [e.g. Pvt1 lncRNA to MYC enhancer (Olivero et al., 2020)], transcription factors (Z. Wang et al., 2018a) (e.g., EPIC1), histones, pre-mRNA, and RNA-binding proteins within the nucleus (Yao et al., 2019). Over 120,000 species of lncRNA have been described to date (Volders et al., 2015), with thousands of lncRNAs identified within the nucleus (Frankish et al., 2019) using sequencing and fluorescent in situ hybridization (Cabili et al., 2015) (FISH). Specific lncRNAs demonstrate subcellular localization at nuclear speckles (Quinodoz et al., 2021), paraspeckles (Bond and Fox, 2009), and other nuclear regions such as nuclear bodies (Chujo and Hirose, 2017). Nuclear localization studies highlight how speckle-associated genomic domains tend to be rich in open-reading frames (ORFs) and highly transcriptionally active (van Steensel and Furlong, 2019). Importantly, nuclear speckles is where m6A methylation has been described to occur (Jia et al., 2011; Schöller et al., 2018), and where Mettl14 is known to localize via direct interaction with laminin-A (Zhang M. et al., 2020). While this evidence suggests nuclear speckle localizing lncRNAs could play a regulatory role in m6A methylation patterning, more studies are necessary to elucidate the function of lncRNAs within specific compartments of the nucleus.
In the complex 3D environment of the nucleus, epigenetic machinery regulates gene transcription and repression. The histone proteins H2A, H2B, H3, and H4 are fundamental constituents of the nucleosome, which are modified on their N-terminal tails with reversible chromatin modifications. The best studied modifications occur on H3 and H4, which include histone acetylation (H3K27ac) and various forms of lysine methylation (H3K4me1, H3K27me3 and H3K36me3) (Zhao et al., 2021). Proteins that read these histone modifications can activate or repress DNA accessibility and bind with RNA transcription machinery (Zhao et al., 2021). Conversely, histone proteins respond to signals generated during transcription and pre-mRNA processing. The pre-mRNA processing mechanisms known to interact with histone modifications and transcription machinery include: splicing, RNA editing, 5′ end capping, and, most recently, m6A methylation (Bentley, 2002; Huang et al., 2020; Kan et al., 2022). Given the novelty, only a few studies have identified epigenetic-epitranscriptomic network interactions. As described in the following sections, H3K36me3 and H3K27me3 were found to bind with m6A writers, suggesting this new branch in the field of RNA modifications is likely to continue to expand (Huang et al., 2019; Wu et al., 2020).
Many biological processes dynamically modulate lncRNA expression, m6A patterning, and the chromatin landscape (see Table 1). This review presents many of the typical physiological and pathological cell states in which all three of these epigenetic-epitranscriptomic mechanisms exhibit dynamic expression patterns. While this section lists correlational observations, many of the examples delineated here have already been described to exhibit bidirectional regulatory relationships that involve lncRNAs, histone modifications and/or m6A methylation.
The dynamic mechanisms that govern the precise control of m6A methylation is of particular interest in the growing field of RNA modifications (Shi H. et al., 2019). Given that patterns in m6A can change rapidly, it has been proposed that 5′UTR m6A methylation may be a means of coordinated rapid response to environmental perturbation (Zhou et al., 2015). Differential and often rapid m6A methylation of specific transcripts has been described in multiple biological systems such as cancer, development, stress, learning and memory, infection, and cellular reprogramming (See Table 1).
The complexity of the nervous system has generated great interest in the epitranscriptome. A pioneering study of m6A in the brain observed dynamic changes in m6A levels during cortical neurogenesis and was found to be critical in mediating RNA decay during neuronal maturation (Yoon et al., 2017). In another study, the m6A levels at the 5′UTR of the synaptic protein DSCR1.4 increased with BDNF stimulation resulting in axon growth, confirming m6A involvement in central nervous system plasticity (Seo et al., 2019) and axon regeneration (Weng et al., 2018a). Interestingly, a slight increase in 5′UTR m6A-modified transcripts was observed within synaptosome fractions when compared to whole cell lysate (Merkurjev et al., 2018). Among the noteworthy synaptic RNAs identified by Merkurjev et al. were CaMKIIa and Shank1, that have been previously suggested to undergo non-canonical Cap-independent protein translation (Pinkstaff et al., 2001; Studtmann et al., 2014). The mammalian stress response represents another potent example of a physiological process that exhibits dynamic changes in the epitranscriptome. During stress response, changes in readers (YTHDC1), writers (Mettl3), erasers (FTO) as well as global changes in m6A patterns are observed. Specifically, 5′UTR m6A increased with response to fasting (Zhou et al., 2018), and exhibited brain region-specific dynamics in stress regulation in rodents (Engel et al., 2018). These studies fortify the notion that 5′ UTR m6A methylation acts as a rapid-response mechanism to physiological and environmental change.
Understanding m6A methylation patterns during epithelial mesenchymal transition (EMT) of oncogenes is a rapidly expanding field (Yue et al., 2019; Bera and Lewis, 2020). Increases in 5′UTR m6A were observed during EMT of cancer cells and during metastasis (Lin et al., 2019). The cross-talk of histone methylation and m6A methylation was described in great mechanistic detail and is suggested to be important during pathogen infection and the host immune response (Wu et al., 2020), as well as in playing a significant role in maintaining the pluripotency of stem cells (Huang et al., 2019). However, generally low levels of m6A methylation are observed during early phases of development and throughout pluripotency (Aguilo et al., 2015), but this phenomenon is poorly understood. Nevertheless, these lines of evidence support that 5′UTR m6A methylation exhibits context dependent patterning and coordinated rapid response.
LncRNAs are well described to exhibit differential and cell-type specific expression patterns across multiple biological systems and during cell state changes including cancer (Terashima et al., 2017), stress (Carrieri et al., 2012), development (Pillay et al., 2021) and memory formation (Wang et al., 2017a) (see Table 1).
Production of anti-sense (AS) RNAs is abundant in the human brain (Mills et al., 2016). For instance, AS RNAs are integral to the epigenetic regulation of the activity dependent neuronal cFos gene during memory formation. The anti-sense FOS (AS-Fos) RNA was found to be temporally co-expressed in an activity-dependent manner with cFos mRNA. Upon cFos open reading frame activation, a transcript produced from the 3′UTR, AS-fos RNA, binds to the CpG promoter region of the Fos gene, inhibiting DNA methylation and promoting gene transcription (Savell et al., 2016). Savell et al. found AS-Fos to be essential for long-term memory formation but not short-term memory in the hippocampus during fear learning. This study alludes to the importance of temporarily precise transcriptional control by lncRNAs in the context of memory formation (Savell et al., 2016).
LncRNAs have commonly been studied in the context of stroke. One report found about 80 lncRNAs were differentially expressed during ischemic stroke, including the upregulation of the antisense lncRNA-N1LR(Z. Wu et al., 2017b). LncRNA upregulation is associated with stroke risk and recurrence (Bao et al., 2018), including antisense noncoding RNA in the INK4 locus (ANRILs) (Zhang et al., 2012). Interestingly, the expression of ANRILs is also associated with inflammation and oxidative stress (Cai and Jiang, 2020), as well as melanoma and neural tumors (Pasmant et al., 2007). This suggests lncRNA ANRILs respond to multiple cellular stressors.
Deep-sequencing studies of tumor biopsies and cancer cell lines have identified hundreds and occasionally thousands of differentially expressed lncRNAs. Among these studies, lncRNA EPIC1 (epigenetically-induced lncRNA1) was identified. EPIC1 directly interacts with the oncogene MYC and enhances MYC binding to target gene promoters resulting cell-cycle progression (Wang Y. et al., 2018). The lncRNA MEG3 is differentially expressed in during EMT transition and in multiple forms of cancer (Du et al., 2013; Terashima et al., 2017). MEG3 was found to associate with JARED2, to recruit PRC2, and induce histone H3K27 methylation on the regulatory regions of CDH1 gene. In summary, lncRNAs exhibit dynamic roles in cancer progression, many of which entail direct interactions with genes and histone modifying enzymes.
Epigenetic machinery is an essential core regulator and stabilizer of gene expression programs during both normal physiological and pathological states. The biological processes that regulate changes in histone modifications are heavily reviewed (Zhao et al., 2021). The epigenetic landscape is generally thought to include DNA methylation, nucleosome remodeling, 3D DNA organization, and reversible histone modifications. This review focuses on the nature of histone modifications and their potential m6A pattering capabilities during changes in cellular physiology.
There are hundreds of examples that describe the dynamic regulation and necessity of precise epigenetic control of chromatin remodeling during brain plasticity, stress response and development (see Table 1) (Mossink et al., 2021). Histone modifications such as H3K27ac have been extensively studied in the context of learning and memory formation (Campbell and Wood, 2019). Additionally, histone deacetylase 2 (HDAC2) is activated by glucocorticoid stress hormone and essential in regulating physiological stress response (Wang S. E. et al., 2017). Histone methyltransferases, like KMT2A and KMT2B, that regulate H3K4me are required for working memory and long-term memory formation to occur (Kerimoglu et al., 2013; Jakovcevski et al., 2015). Furthermore, increases in H3K9me2 were observed to exacerbate the anxiolytic response to withdrawal from cocaine addiction (Anderson et al., 2018). These examples highlight the capability of histone modifying enzymes to respond relatively quickly to changes in physiological state, a necessary characteristic for timely regulation of m6A patterning.
This review only briefly examines many types of changes in cell state that depend on the epitranscriptome and epigenome for down-stream physiological processes to occur. Importantly, for many of these, lncRNAs play essential roles. Next, many relevant mechanisms by which lncRNA act co-transcriptionally and during RNA pre-processing are discussed, as to further highlight the potential of lncRNA to pattern m6A methylation via multiple mechanisms.
Non-Coding RNAs are some of the oldest biological building blocks of the cell. This section reviews ncRNAs and lncRNAs interacting directly with RNA transcripts and as guides in RNA modification. Furthermore, given the regulatory implications of m6A at the 5′UTR, instances of lncRNAs binding to the untranslated regions of mRNAs are discussed. Additionally, functional categorizations of lncRNAs in terms of biogenesis and mode of action are reviewed. This section serves to contrast lncRNAs that bind with histone modifying enzymes and focuses of lncRNAs binding directly with RNA transcripts.
Non-coding RNAs (ncRNAs) have been studied in great depth for their ability to act as guides in RNA methylation, acetylation and pseudouridylation. These ncRNAs serve as case studies in the analysis of lncRNA-guided m6A methylation in the complex nuclear environment. Small nucleolar RNAs (snoRNAs) are abundant ancient ncRNAs that range between 80 and 1,000 nucleotides in length. There are at least 200 guide snoRNAs in humans, necessary for multiple post-transcriptional modifications in eukaryotic rRNAs and tRNAs(Dieci et al., 2009). SnoRNAs guide the methylation (Kiss-Laszlo, 1998; van Nues et al., 2011), acetylation (Sharma et al., 2017), and pseudouridylation (Kiss et al., 2004) of ncRNAs in order to generate functional and mature RNA species. Another example are small Cajal-body-associated RNAs (scaRNAs) that guide the post-transcriptional modification of spliceosomal small nuclear RNA (snRNAs). ScaRNA have been found to bind directly via RNA:RNA interactions with snRNA to guide 2’-O’methylation and pseudouridylation of the transcript (Darzacq et al., 2002). This line of evidence supports nc-RNAs and lncRNAs interacting with target RNAs in complex nuclear environments (Engreitz et al., 2016), acting on multiple RNA metabolism pathways to facilitate post-transcriptional events. However, ncRNAs binding specifically to the 5′ UTR of mRNA transcripts is significant, given the effect on translational control.
LncRNAs are well known to bind directly with target RNA transcripts causing alternative splicing, scaffolding to RNA binding proteins and change in protein translation dynamics (Yao et al., 2019). While less than 10% of developmentally active As-lncRNAs exhibit complimentary sequence overlap with 3′ UTR or 5′ UTRs of protein coding mRNA transcripts (Pillay et al., 2021), there are multiple examples of AS-ncRNAs binding to 5′UTRs. This section highlights examples of lncRNAs binding specifically to 5′UTRs.
The discovery of the antisense lncRNA for ubiquitin carboxyterminal hydrolase L1 (AS-Uchl1) was significant, given it was the first description of a lncRNA regulating protein translation at the ribosomal level (Carrieri et al., 2012). AS-Uchl1 is nuclear enriched, and upon binding with the 5′UTR of UCHL1 mRNA, both are exported to the cytoplasm. AS-Uchl1 then recruit ribosomes to initiate the translation of UCHL1 protein. Given AS-Uchl1 expression was found to be regulated by stress signaling in neurons, this alludes to fast-acting lncRNAs that can alter gene regulatory networks in response to physiological change in state (Carrieri et al., 2012).
Few studies have deciphered the mechanisms of lncRNA and 5′UTR binding. For instance, the ZEB2-AS1 was reported to bind to the 5′UTR of Zeb2 pre-mRNA after EMT. Upon binding, ZEB2-AS1 acts on the spliceosome, facilitating the retention of an internal ribosome entry site (IRES) containing intron in Zeb2 mRNA. The IRES promotes cap-independent protein translation of Zeb2 and down regulates E-cadherin (Beltran et al., 2008). Others have implicated expression of ZEB2-AS1 with shorter overall survival in patients with acute myeloid leukemia (Shi X. et al., 2019). Overall, the description of ZEB2-AS1 is a clear example of lncRNA binding to 5‘UTRs during mRNA co-transcriptional events.
These examples specifically highlight and support how antisense lncRNAs can function in different locations of the cell. AS-Uchl1 is trafficked to the cytoplasm and is an example of lncRNAs functioning outside the nucleus. In contrast, ZEB2-AS1 was an example of a lncRNA that acts within the area it was transcribed. Next, the nomenclature and functional implications of lncRNAs acting near or distant from the site of its transcription is reviewed.
The specificity of lncRNAs targeting individual mRNAs (or DNA/Chromatin) depends in part on its transcriptional origin within the genome. This review utilizes a broad classification of lncRNAs dependent on their origin and site of action; Cis-acting lncRNAs that act near the site of transcription (Figure 2A), and Trans-acting lncRNAs that act at distant sites from their locus of transcription (Figure 2B), for example, in the cytoplasm (Marchese et al., 2017; Kopp and Mendell, 2018). This classification of lncRNA facilitates interpreting the mechanism by which lncRNAs might guide m6A patterning, given the co-transcriptional nature of m6A methylation and known nuclear functions in RNA binding of distinct lncRNAs.
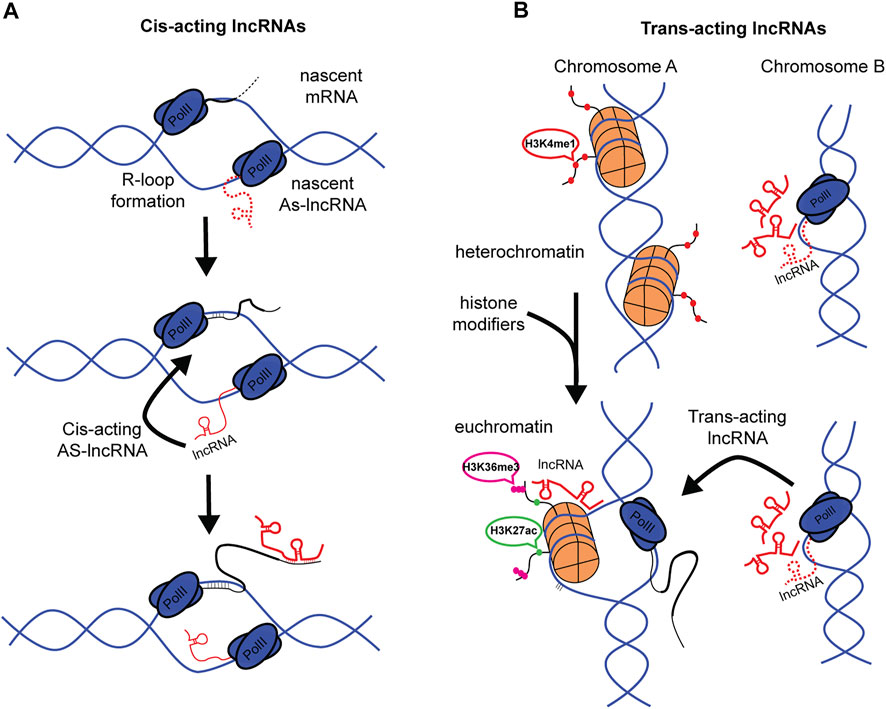
FIGURE 2. Cis- and Trans-acting lncRNAs in m6A patterning. (A) Cis-acting lncRNA can be generated by bidirectional transcription via R-loop formation. AS-lncRNA can then bind directly with nascent mRNA. (B) Representation of Trans-acting lncRNAs. Histones are shown to be repressed in Chromosome A. Change in physiological state opens chromatin to facilitate gene expression, simultaneously, lncRNAs at Chromosome B are being transcribed. LncRNAs are then trafficked to Chromosome A to guide histone modifications. (Red dots, H3K4me1. Green dots, H3K27ac. Magenta dots, H3K36me3).
Cis-acting lncRNAs, or cis-antisense lncRNAs, are well known to function in gene regulation. These can be generated in a variety of ways, including bi-directional transcription during R-Loop formation (Tan-Wong et al., 2019) or presence of bi-directional promoters (Uesaka et al., 2014) (Figure 2A). These local lncRNAs are quite stable and exhibit long half-lives, with an average of 4.8 h, many exceeding 12 h, though of less duration than the mRNAs they regulate (Tani et al., 2015). Most studies agree that AS-lncRNAs mostly localize, and likely function, near their transcriptional loci. Some estimates suggest around 93% of nuclear lncRNAs are Cis-acting lncRNAs (Quinodoz et al., 2021). Given the anti-sense nature of cis-acting AS-lncRNAs, the long half-life, and the immediate proximity to target mRNAs, these AS-lncRNAs make suitable candidates as direct binding partners with the UTR and guides of m6A writer machinery. This hypothesis is supported by the observation that GATA3-AS lncRNA binds with GATA3 mRNA to regulate m6A patterning (Lan et al., 2019).
Trans-acting lncRNAs, in contrast to cis-acting lncRNAs, function at distant nuclear or cytoplasmic sites from their transcriptional loci of origin (Figure 2B). Common examples of trans-acting lncRNAs might be transcribed from pseudogenes (Muro and Andrade-Navarro, 2010; Johnsson et al., 2013) and large intergenic non-coding RNAs (lincRNAs) (Guttman et al., 2011). Trans-acting lncRNAs are known to interact with epigenetic machinery (Zhao et al., 2010), and it is this involvement in chromatin remodeling that is likely to contribute to a trans-acting pathway that alters UTR methylation patterns. This proposal is enticing, given that trans-acting lncRNAs can affect multiple gene/mRNA species through “multi-way contract” with histone remodeling complexes. This classification of lncRNAs provides insight into how different, sometimes parallel pathways might converge on RNA expression mechanisms.
Since the first observation that lncRNAs undergo m6A methylation (Meyer et al., 2012), a multitude of studies have expanded the repertoire and importance of m6A modified lncRNAs(Fazi and Fatica, 2019; Lv et al., 2020; Xue et al., 2020). Conversely, a few yet pivotal studies have identified role of lncRNAs in guiding the m6A writer complex, readers, and erasers to mRNA targets (Figure 3A). A particular example is that of the cis-acting lncRNA GATA3-AS and its ability to recruit VIRMA and facilitate the m6A modification of the 3′UTR of GATA3 pre-mRNA. The downstream effect of GATA3 m6A methylation was disrupted binding of HuR protein, down regulation of GATA3, and increased metastasis of liver cancer (Lan et al., 2019). More studies are necessary to elucidate the mechanism by which lncRNA recruits VIRMA and the structural changes induced by lncRNA-mRNA binding that would alter writer complex activity to pattern m6A.
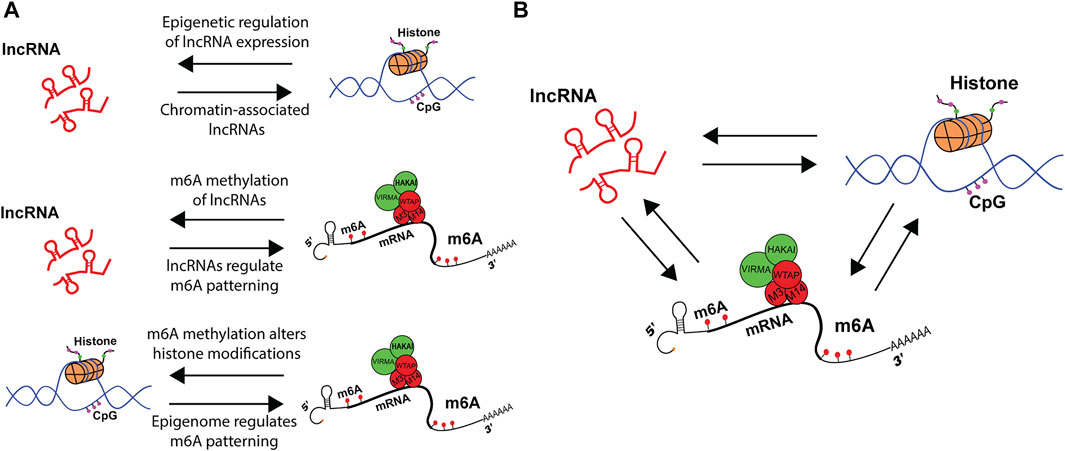
FIGURE 3. Epigenetic crosstalk among lncRNAs, histones and m6A regulate gene expression. (A) schematic representation of bi-directional regulation in co-transcriptional machinery. LncRNAs can change histone dynamics, while histones control lncRNA expression. M6A on lncRNAs modulate RNA metabolism, while lncRNAs guide m6A patterning. Finally, m6A alters histone modifications, while histone modifications pattern m6A modification. (B) Crosstalk between lncRNAs, histone modifications and m6A integrate distinct signals that alter upstream epigenetic landscape and downstream RNA metabolism.
M6A readers and erasers have been described to utilize both cis- and trans-acting lncRNAs as guides. LINC00857 was observed to cooperate with reader YTHDC1 to increase the stability of SLC7A5 mRNA in colorectal cancer cells (Tang et al., 2021). The lncRNA KB 1980E6.3 was found to form an RNA: protein complex with the m6A reader IGF2BP1 to facilitate the recognition and mRNA stability of m6A modified c-Myc in breast cancer stem cells (Zhu et al., 2021). LncRNAs have been found to interact with both m6A FTO and ALKBH5 Eraser proteins. FOXM1-AS increases the interaction of FOXM1 and ALKBH5, promoting demethylation of FOXM1 decreasing both FOXM1 expression and tumor growth (Zhang et al., 2017). In a similar study, the lncRNA GAS-AS1 was found to promote the ALKBH5-dependent demethylation of GAS mRNA and inhibit cervical cancer proliferation (Wang et al., 2019; Chen et al., 2020). Additionally, the lincRNA CASC15 is thought to recruit the demethylase FTO to SIM2, decreasing SIM2 mRNA stability and promoting esophageal cancer progression (Qin et al., 2020). Furthermore, specific lncRNAs such as CACNA1G-AS1 and ACAP2-IT1 have been predicted to regulate m6A readers and writers expression (Zheng et al., 2021). These initial studies provide substantial evidence that lncRNAs have dynamic interactions with m6A proteins, and additional research is likely to provide further examples.
There is a growing body of literature that describes bi-directional interactions between the epigenome and the epitranscriptome (Figure 3A). This was first observed in the context of m6A methylation upon knock-down of m6A writer Mettl14, which altered the expression of histone modifying proteins (Y. Wang Z. et al., 2018). Since then, manipulations of readers, writers, and erasers, as well as the m6A modification itself, have been found to impact histone modifications. See Kan et al. for recent review (Kan et al., 2022). A clear example was the observation that m6A could co-transcriptionally direct the demethylation of histone H3K9me2 (Li et al., 2020). This occurs by m6A reader YTHDC1 physically interacting with the H3K9me2 demethylase KDM3B at m6A-associated chromatin regions, promoting H3K9me2 demethylation and increasing overall gene expression. In another example, H3K27me3 was described as a barrier for m6A modification during transcription. Furthermore, the histone demethylase KDM6B that targets H3K27me3 directly recruits writers Mettl3 and Mettl14 to facilitate m6A methylation of co-transcribing mRNAs while simultaneously promoting transcription (Wu et al., 2020).
Recently, chromatin remodeling by H3K36me3 was observed to pattern m6A at the CDS and 3′UTR regions of RNA (Huang et al., 2019). Specifically, H3K36me3 scantly effected m6A levels in the 5′UTR in contrast to the CDS and 3′UTR. Furthermore, the repressive histone mark H3K9me3 was negatively correlated with m6A peaks, and metagene profiles of m6A at H3K36me3-negative sites correlated with increased 5′UTR methylation (Huang et al., 2019). Additionally, all the members of the core m6A writer complex, Mettl14, Mettl3 and WTAP, were found to bind with H3K36me3 and not with H3K9me3. However, members of the associated writer complex, VIRMA, Zc3h13, and Hakai were not tested. Interestingly, individual shRNA silencing of Mettl14, Mettl3 or WTAP did not dissociate the remaining m6A writer complex proteins from H3K36me3, which warrants future investigation.
As described, H3K36me3 peaks were anti-correlated with m6A at the 5′UTR (Huang et al., 2019). This discrepancy H3K36me3 relative to m6A patterning can be rationalized by considering the “histone code.” It is generally accepted that a gene is occupied by multiple nucleosomes, given that a nucleosome repeat consists of 140–200 bp of DNA. While the length of the mammalian 5′UTR can range between few nucleotides to several thousand, the median length of the 5′UTR in humans and mice is of 218 and 175, respectively (Leppek et al., 2018). Additionally, the first nucleosome immediately after the transcriptional start site (TSS), e.g., the one that may occupy the 5′UTR, exhibits distinct regulatory dynamics when compared to those of the CDS (Zhang and Pugh, 2011). These correlations warrant further exploration of how the epigenetic landscape patterns m6A on the 5′UTRs co-transcriptionally. Consequently, other histone post-translational modifications and the role of 3D DNA organization need to be explored in the context of m6A methylation.
There is an extensive body of literature that describes lncRNAs interacting with the histone modifiers (Yao et al., 2019) (Figure 3A). Interestingly, lncRNA databases predict that at least 20% of lncRNAs guide DNA/protein and chromatin interactions within the nucleus (Volders et al., 2015). This is impressive, given over 10,000 have been predicted to exist (Volders et al., 2015). This account supports the abundant discovery of lncRNAs that interact with chromatin modifiers. This section reviews major findings of lncRNAs interacting with histone methylation proteins, as to highlight the potential of lncRNAs to interact with histone modifiers, enabling m6A patterning of mRNA transcripts.
As previously mentioned, H3K36me3 can guide m6A methylation co-transcriptionally (Huang et al., 2019). Multiple lncRNAs such as MEG3 (Terashima et al., 2017), Kcnq1ot1 (Pandey et al., 2008) and Air (Nagano et al., 2008) interact directly with histone methyltransferases for H3K36, and specifically regulate H3K36me3. LncRNAs have been found to interact with a variety of histone methyltransferases. An interesting example is that of HOTTIP, a divergently expressed lncRNA that promotes entire gene-expression programs by H3K4me3 patterning (Wang et al., 2011). In addition, the lncRNA Hotair that binds to G-A base pair rich DNA, correlates with H3K27me3 peaks (Chu et al., 2011). Deep-sequencing has also revealed both cis- and trans-acting lncRNAs, with 218 confirmed lincRNAs that bind directly with the Polycomb repressive complex 2 (PRC2), a protein complex that exhibits histone methyltransferase activity primarily on H3K27me3 (Zhao et al., 2010).
It is unlikely any specific pathway will be found to exclusively regulate m6A methylation patterns. This is perhaps due to the diversity of proteins within the writer complex contributing to a combinatorial mechanism to dictate m6A deposition. While lncRNAs may not be the exclusive mechanism that guides UTR m6A methylation, it is a contributor of m6A patterning in RNA, as it is for DNA and histones. A continuum of interesting phenomena hasbeen described to pattern the RNA modifications, and future research will likely describe these multiple mechanisms as cofactors in the crosstalk of the epigenome and the epitranscriptome (Figure 3B). Such findings will elucidate previously undescribed RNA interactions to which disease or single nucleotide polymorphisms (SNPs) may be attributed. Future research will provide more examples of extensive cross talk between the epigenome and epitranscriptome. Most likely positive and negative feedback systems, as well as sources of illness and targets of intervention.
LOV: selected topic of review, wrote the manuscript, generated the table, generated figures, compiled bibliography, edited and proof-read manuscript, compiled, and submitted manuscript.
This work was supported by the Brain Institute at Tulane University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Gratitude for early comments by J. Morgan, D. M. Diaz-Morales, I. Hoefakar. Extended gratitude to E. Munoz Buitrago, K. A. Fulton, X. Zhuang and M. J. Galazo for overall support. L. Earls for comments. Special thanks to R. A. Aponte Rivera and A. S. Tiemroth for later revisions of this manuscript. Several topics discussed here originate from the Tulane graduate course Genome Biology lead by L. Earls.
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of M 6 A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell 17 (6), 689–704. doi:10.1016/j.stem.2015.09.005
Anderson, E. M., Larson, E. B., Guzman, D., Wissman, A. M., Neve, R. L., Nestler, E. J., et al. (2018). Overexpression of the Histone Dimethyltransferase G9a in Nucleus Accumbens Shell Increases Cocaine Self-Administration, Stress-Induced Reinstatement, and Anxiety. J. Neurosci. 38 (4), 803–813. doi:10.1523/JNEUROSCI.1657-17.2017
Aprea, J., and Calegari, F. (2015). Long Non‐coding RNA S in Corticogenesis: Deciphering the Non‐coding Code of the Brain. Embo J. 34 (23), 2865–2884. doi:10.15252/embj.201592655
Aprea, J., Lesche, M., Massalini, S., Prenninger, S., Alexopoulou, D., Dahl, A., et al. (2015). Identification and Expression Patterns of Novel Long Non-coding RNAs in Neural Progenitors of the Developing Mammalian Cortex. Neurogenesis 2 (1), e995524. doi:10.1080/23262133.2014.995524
Audas, T. E., and Lee, S. (2016). Stressing Out over Long Noncoding RNA. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1859 (1), 184–191. doi:10.1016/j.bbagrm.2015.06.010
Bao, M.-H., Szeto, V., Yang, B. B., Zhu, S.-z., Sun, H.-S., and Feng, Z.-P. (2018). Long Non-coding RNAs in Ischemic Stroke. Cell Death Dis 9 (3), 281. doi:10.1038/s41419-018-0282-x
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millán-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 Maintains Myeloid Leukaemia by m6A-dependent Translation Control. Nature 552 (7683), 126–131. doi:10.1038/nature24678
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell 15 (6), 707–719. doi:10.1016/j.stem.2014.09.019
Beltran, M., Puig, I., Peña, C., García, J. M., Álvarez, A. B., Peña, R., et al. (2008). A Natural Antisense Transcript Regulates Zeb2/Sip1 Gene Expression during Snail1-Induced Epithelial-Mesenchymal Transition. Genes Dev. 22 (6), 756–769. doi:10.1101/gad.455708
Bentley, D. (2002). The mRNA Assembly Line: Transcription and Processing Machines in the Same Factory. Curr. Opin. Cel Biol. 14 (3), 336–342. doi:10.1016/S0955-0674(02)00333-2
Bera, A., and Lewis, S. M. (2020). Regulation of Epithelial-To-Mesenchymal Transition by Alternative Translation Initiation Mechanisms and its Implications for Cancer Metastasis. Ijms 21 (11), 4075. doi:10.3390/ijms21114075
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 Interactome Reveals that TGFβ Controls m6A mRNA Methylation in Pluripotency. Nature 555 (7695), 256–259. doi:10.1038/nature25784
Bond, C. S., and Fox, A. H. (2009). Paraspeckles: Nuclear Bodies Built on Long Noncoding RNA. J. Cel Biol. 186 (5), 637–644. doi:10.1083/jcb.200906113
Buffington, S. A., Huang, W., and Costa-Mattioli, M. (2014). Translational Control in Synaptic Plasticity and Cognitive Dysfunction. Annu. Rev. Neurosci. 37 (1), 17–38. doi:10.1146/annurev-neuro-071013-014100
Cabili, M. N., Dunagin, M. C., McClanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and Abundance Analysis of Human lncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol. 16 (1), 20. doi:10.1186/s13059-015-0586-4
Cai, R., and Jiang, J. (2020). LncRNA ANRIL Silencing Alleviates High Glucose-Induced Inflammation, Oxidative Stress, and Apoptosis via Upregulation of MME in Podocytes. Inflammation 43 (6), 2147–2155. doi:10.1007/s10753-020-01282-1
Campbell, R. R., and Wood, M. A. (2019). How the Epigenome Integrates Information and Reshapes the Synapse. Nat. Rev. Neurosci. 20 (3), 133–147. doi:10.1038/s41583-019-0121-9
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long Non-coding Antisense RNA Controls Uchl1 Translation through an Embedded SINEB2 Repeat. Nature 491 (7424), 454–457. doi:10.1038/nature11508
Chen, T., Hao, Y.-J., Zhang, Y., Li, M.-M., Wang, M., Han, W., et al. (2015). m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 16 (3), 289–301. doi:10.1016/j.stem.2015.01.016
Chen, Y., Lin, Y., Shu, Y., He, J., and Gao, W. (2020). Interaction between N6-Methyladenosine (m6A) Modification and Noncoding RNAs in Cancer. Mol. Cancer 19 (1), 94. doi:10.1186/s12943-020-01207-4
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E., and Chang, H. Y. (2011). Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cel 44 (4), 667–678. doi:10.1016/j.molcel.2011.08.027
Chujo, T., and Hirose, T. (2017). Nuclear Bodies Built on Architectural Long Noncoding RNAs: Unifying Principles of Their Construction and Function. Mol. Cell 40 (12), 889–896. doi:10.14348/molcells.2017.0263
Coots, R. A., Liu, X.-M., Mao, Y., Dong, L., Zhou, J., Wan, J., et al. (2017). m6A Facilitates eIF4F-independent mRNA Translation. Mol. Cel 68 (3), 504–514. e7. doi:10.1016/j.molcel.2017.10.002
Darzacq, X., Jády, B. E., Verheggen, C., Kiss, A. M., Bertrand, E., Kiss, T., et al. (2002). Cajal Body-specific Small Nuclear RNAs: a Novel Class of 2'-O-Methylation and Pseudouridylation Guide RNAs. EMBO J. 21 (11), 2746–2756. doi:10.1093/emboj/21.11.2746
Devaux, Y., Zangrando, J., Schroen, B., Creemers, E. E., Pedrazzini, T., Chang, C. P., et al. (2015). Long Noncoding RNAs in Cardiac Development and Ageing. Nat. Rev. Cardiol. 12 (7), 415–425. doi:10.1038/nrcardio.2015.55
Dieci, G., Preti, M., and Montanini, B. (2009). Eukaryotic snoRNAs: A Paradigm for Gene Expression Flexibility. Genomics 94 (2), 83–88. doi:10.1016/j.ygeno.2009.05.002
Dominissini, D., Moshitch-Moshkovitz, S., Salmon-Divon, M., Amariglio, N., and Rechavi, G. (2013). Transcriptome-wide Mapping of N6-Methyladenosine by m6A-Seq Based on Immunocapturing and Massively Parallel Sequencing. Nat. Protoc. 8 (1), 176–189. doi:10.1038/nprot.2012.148
Du, Z., Fei, T., Verhaak, R. G. W., Su, Z., Zhang, Y., Brown, M., et al. (2013). Integrative Genomic Analyses Reveal Clinically Relevant Long Noncoding RNAs in Human Cancer. Nat. Struct. Mol. Biol. 20 (7), 908–913. doi:10.1038/nsmb.2591
Engel, M., Eggert, C., Kaplick, P. M., Eder, M., Röh, S., Tietze, L., et al. (2018). The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 99 (2), 389–403. e9. doi:10.1016/j.neuron.2018.07.009
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long Non-coding RNAs: Spatial Amplifiers that Control Nuclear Structure and Gene Expression. Nat. Rev. Mol. Cel Biol 17 (12), 756–770. doi:10.1038/nrm.2016.126
Fazi, F., and Fatica, A. (2019). Interplay between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cel Dev. Biol. 7 (June), 1–11. doi:10.3389/fcell.2019.00116
Frankish, A., Diekhans, M., Ferreira, A.-M., Johnson, R., Jungreis, I., Loveland, J., et al. (2019). GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 47 (D1), D766–D773. doi:10.1093/nar/gky955
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene Expression Regulation Mediated through Reversible m6A RNA Methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Goff, L. A., Groff, A. F., Sauvageau, M., Trayes-Gibson, Z., Sanchez-Gomez, D. B., Morse, M., et al. (2015). Spatiotemporal Expression and Transcriptional Perturbations by Long Noncoding RNAs in the Mouse Brain. Proc. Natl. Acad. Sci. USA 112 (22), 6855–6862. doi:10.1073/pnas.1411263112
Golden, S. A., Christoffel, D. J., Heshmati, M., Hodes, G. E., Magida, J., Davis, K., et al. (2013). Epigenetic Regulation of RAC1 Induces Synaptic Remodeling in Stress Disorders and Depression. Nat. Med. 19 (3), 337–344. doi:10.1038/nm.3090
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., et al. (2011). lincRNAs Act in the Circuitry Controlling Pluripotency and Differentiation. Nature 477 (7364), 295–300. doi:10.1038/nature10398
Higgs, P. G., and Lehman, N. (2015). The RNA World: Molecular Cooperation at the Origins of Life. Nat. Rev. Genet. 16 (1), 7–17. doi:10.1038/nrg3841
Huang, H., Weng, H., and Chen, J. (2020). The Biogenesis and Precise Control of RNA m6A Methylation. Trends Genet. 36 (1), 44–52. doi:10.1016/j.tig.2019.10.011
Huang, H., Weng, H., Zhou, K., Wu, T., Zhao, B. S., Sun, M., et al. (2019). Histone H3 Trimethylation at Lysine 36 Guides m6A RNA Modification Co-transcriptionally. Nature 567 (7748), 414–419. doi:10.1038/s41586-019-1016-7
Jakovcevski, M., Ruan, H., Shen, E. Y., Dincer, A., Javidfar, B., Ma, Q., et al. (2015). Neuronal Kmt2a/Mll1 Histone Methyltransferase Is Essential for Prefrontal Synaptic Plasticity and Working Memory. J. Neurosci. 35 (13), 5097–5108. doi:10.1523/JNEUROSCI.3004-14.2015
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Johnsson, P., Ackley, A., Vidarsdottir, L., Lui, W.-O., Corcoran, M., Grandér, D., et al. (2013). A Pseudogene Long-Noncoding-RNA Network Regulates PTEN Transcription and Translation in Human Cells. Nat. Struct. Mol. Biol. 20 (4), 440–446. doi:10.1038/nsmb.2516
Kan, R. L., Chen, J., and Sallam, T. (2022). Crosstalk between Epitranscriptomic and Epigenetic Mechanisms in Gene Regulation. Trends Genet. 38 (2), 182–193. doi:10.1016/j.tig.2021.06.014
Kerimoglu, C., Agis-Balboa, R. C., Kranz, A., Stilling, R., Bahari-Javan, S., Benito-Garagorri, E., et al. (2013). Histone-Methyltransferase MLL2 (KMT2B) Is Required for Memory Formation in Mice. J. Neurosci. 33 (8), 3452–3464. doi:10.1523/JNEUROSCI.3356-12.2013
Kim, D. H., Marinov, G. K., Pepke, S., Singer, Z. S., He, P., Williams, B., et al. (2015). Single-Cell Transcriptome Analysis Reveals Dynamic Changes in lncRNA Expression during Reprogramming. Cell Stem Cell 16 (1), 88–101. doi:10.1016/j.stem.2014.11.005
Kiss, A. M., Jády, B. E., Bertrand, E., and Kiss, T. (2004). Human Box H/ACA Pseudouridylation Guide RNA Machinery. Mol. Cel Biol 24 (13), 5797–5807. doi:10.1128/MCB.24.13.5797-5807.2004
Kiss-Laszlo, Z. (1998). Sequence and Structural Elements of Methylation Guide snoRNAs Essential for Site-specific Ribose Methylation of Pre-rRNA. EMBO J. 17 (3), 797–807. doi:10.1093/emboj/17.3.797
Kopp, F., and Mendell, J. T. (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172 (3), 393–407. doi:10.1016/j.cell.2018.01.011
Koranda, J. L., Dore, L., Shi, H., Patel, M. J., Vaasjo, L. O., Rao, M. N., et al. (2018). Mettl14 Is Essential for Epitranscriptomic Regulation of Striatal Function and Learning. Neuron 99 (2), 283–292. doi:10.1016/j.neuron.2018.06.007
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 Contributes to Liver Cancer Progression through N6-methyladenosine-dependent post-transcriptional Modification of GATA3. Mol. Cancer 18 (1), 186. doi:10.1186/s12943-019-1106-z
Leighton, L., and Bredy, T. (2018). Functional Interplay between Small Non-coding RNAs and RNA Modification in the Brain. ncRNA 4 (2), 15. doi:10.3390/ncrna4020015
Leppek, K., Das, R., and Barna, M. (2018). Functional 5′ UTR mRNA Structures in Eukaryotic Translation Regulation and How to Find Them. Nat. Rev. Mol. Cel Biol 19 (3), 158–174. doi:10.1038/nrm.2017.103
Li, Y., Xia, L., Tan, K., Ye, X., Zuo, Z., Li, M., et al. (2020). N6-Methyladenosine Co-transcriptionally Directs the Demethylation of Histone H3K9me2. Nat. Genet. 52 (9), 870–877. doi:10.1038/s41588-020-0677-3
Liang, G., He, J., and Zhang, Y. (2012). Kdm2b Promotes Induced Pluripotent Stem Cell Generation by Facilitating Gene Activation Early in Reprogramming. Nat. Cel Biol 14 (5), 457–466. doi:10.1038/ncb2483
Lichinchi, G., Gao, S., Saletore, Y., Gonzalez, G. M., Bansal, V., Wang, Y., et al. (2016). Dynamics of the Human and Viral m6A RNA Methylomes during HIV-1 Infection of T Cells. Nat. Microbiol. 1 (4), 16011. doi:10.1038/nmicrobiol.2016.11
Lin, X., Chai, G., Wu, Y., Li, J., Chen, F., Liu, J., et al. (2019). RNA m6A Methylation Regulates the Epithelial Mesenchymal Transition of Cancer Cells and Translation of Snail. Nat. Commun. 10 (1), 2065. doi:10.1038/s41467-019-09865-9
Lobo, J., Costa, A. L., Cantante, M., Guimarães, R., Lopes, P., Antunes, L., et al. (2019). M6A RNA Modification and its Writer/reader VIRMA/YTHDF3 in Testicular Germ Cell Tumors: A Role in Seminoma Phenotype Maintenance. J. Transl Med. 17 (1), 1–13. doi:10.1186/s12967-019-1837-z
Lv, Z., Sun, L., Xu, Q., Xing, C., and Yuan, Y. (2020). Joint Analysis of lncRNA m6A Methylome and lncRNA/mRNA Expression Profiles in Gastric Cancer. Cancer Cel Int 20 (1), 464. doi:10.1186/s12935-020-01554-8
Marazzi, I., Greenbaum, B. D., Low, D. H. P., and Guccione, E. (2018). Chromatin Dependencies in Cancer and Inflammation. Nat. Rev. Mol. Cel Biol 19 (4), 245–261. doi:10.1038/nrm.2017.113
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The Multidimensional Mechanisms of Long Noncoding RNA Function. Genome Biol. 18 (1), 1–13. doi:10.1186/s13059-017-1348-2
Merkurjev, D., Hong, W.-T., Iida, K., Oomoto, I., Goldie, B. J., Yamaguti, H., et al. (2018). Synaptic N6-Methyladenosine (m6A) Epitranscriptome Reveals Functional Partitioning of Localized Transcripts. Nat. Neurosci. 21 (July), 1004–1014. doi:10.1038/s41593-018-0173-6
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mills, J. D., Chen, B. J., Ueberham, U., Arendt, T., and Janitz, M. (2016). The Antisense Transcriptome and the Human Brain. J. Mol. Neurosci. 58 (1), 1–15. doi:10.1007/s12031-015-0694-3
Mishra, K., and Kanduri, C. (2019). Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. ncRNA 5 (4), 54. doi:10.3390/ncrna5040054
Mossink, B., Negwer, M., Schubert, D., and Nadif Kasri, N. (2021). The Emerging Role of Chromatin Remodelers in Neurodevelopmental Disorders: a Developmental Perspective. Cell. Mol. Life Sci. 78 (6), 2517–2563. doi:10.1007/s00018-020-03714-5
Muro, E. M., and Andrade-Navarro, M. A. (2010). Pseudogenes as an Alternative Source of Natural Antisense Transcripts. BMC Evol. Biol. 10 (1), 338. doi:10.1186/1471-2148-10-338
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R., et al. (2008). The Air Noncoding RNA Epigenetically Silences Transcription by Targeting G9a to Chromatin. Science 322 (5908), 1717–1720. doi:10.1126/science.1163802
Olivero, C. E., Martínez-Terroba, E., Zimmer, J., Liao, C., Tesfaye, E., Hooshdaran, N., et al. (2020). p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cel 77 (4), 761–774. e8. doi:10.1016/j.molcel.2019.12.014
Onder, T. T., Kara, N., Cherry, A., Sinha, A. U., Zhu, N., Bernt, K. M., et al. (2012). Chromatin-modifying Enzymes as Modulators of Reprogramming. Nature 483 (7391), 598–602. doi:10.1038/nature10953
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-specific Transcriptional Silencing through Chromatin-Level Regulation. Mol. Cel 32 (2), 232–246. doi:10.1016/j.molcel.2008.08.022
Pasmant, E., Laurendeau, I., Héron, D., Vidaud, M., Vidaud, D., and Bièche, I. (2007). Characterization of a Germ-Line Deletion, Including the Entire INK4/ARF Locus, in a Melanoma-Neural System Tumor Family: Identification of ANRIL, an Antisense Noncoding RNA Whose Expression Coclusters with ARF. Cancer Res. 67 (8), 3963–3969. doi:10.1158/0008-5472.CAN-06-2004
Perales, R., and Bentley, D. (2009). "Cotranscriptionality": The Transcription Elongation Complex as a Nexus for Nuclear Transactions. Mol. Cel 36 (2), 178–191. doi:10.1016/j.molcel.2009.09.018
Pillay, S., Takahashi, H., Carninci, P., and Kanhere, A. (2021). Antisense RNAs during Early Vertebrate Development Are Divided in Groups with Distinct Features. Genome Res. 31 (6), 995–1010. doi:10.1101/gr.262964.120
Pinkstaff, J. K., Chappell, S. A., Mauro, V. P., Edelman, G. M., and Krushel, L. A. (2001). Internal Initiation of Translation of Five Dendritically Localized Neuronal mRNAs. Proc. Natl. Acad. Sci. 98 (5), 2770–2775. doi:10.1073/pnas.051623398
Pirogov, S. A., Gvozdev, V. A., and Klenov, M. S. (2019). Long Noncoding RNAs and Stress Response in the Nucleolus. Cells 8 (7), 668. doi:10.3390/cells8070668
Qin, B., Dong, M., Wang, Z., Wan, J., Xie, Y., Jiao, Y., et al. (2020). Long Non-coding RNA CASC15 F-acilitates E-sophageal S-quamous C-ell C-arcinoma T-umorigenesis via D-ecreasing SIM2 S-tability via FTO-mediated D-emethylation. Oncol. Rep. 45 (3), 1059–1071. doi:10.3892/or.2020.7917
Quinodoz, S. A., Jachowicz, J. W., Bhat, P., Ollikainen, N., Banerjee, A. K., Goronzy, I. N., et al. (2021). RNA Promotes the Formation of Spatial Compartments in the Nucleus. Cell 184 (23), 5775–5790. e30. doi:10.1016/j.cell.2021.10.014
Savell, K. E., Gallus, N. V. N., Simon, R. C., Brown, J. A., Revanna, J. S., Osborn, M. K., et al. (2016). Extra-coding RNAs Regulate Neuronal DNA Methylation Dynamics. Nat. Commun. 7 (May). doi:10.1038/ncomms12091
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of Long Noncoding RNA Function in Development and Disease. Cel. Mol. Life Sci. 73 (13), 2491–2509. doi:10.1007/s00018-016-2174-5
Schöller, E., Weichmann, F., Treiber, T., Ringle, S., Treiber, N., Flatley, A., et al. (2018). Interactions, Localization, and Phosphorylation of the m6A Generating METTL3-METTL14-WTAP Complex. RNA 24 (4), 499–512. doi:10.1261/rna.064063.117
Schwartz, A. W. (1998). Origins of the RNA World in the Molecular Origins of Life. Nat. Rev. Genet. 16, 237–254. doi:10.1017/CBO9780511626180.013
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cel Rep. 8 (1), 284–296. doi:10.1016/j.celrep.2014.05.048
Sendoel, A., Dunn, J. G., Rodriguez, E. H., Naik, S., Gomez, N. C., Hurwitz, B., et al. (2017). Translation from Unconventional 5′ Start Sites Drives Tumour Initiation. Nature 541 (7638), 494–499. doi:10.1038/nature21036
Seo, J.-Y., Jung, Y., Kim, D.-Y., Ryu, H. G., Lee, J., Kim, S. W., et al. (2019). DAP5 Increases Axonal Outgrowth of Hippocampal Neurons by Enhancing the Cap-independent Translation of DSCR1.4 mRNA. Cel Death Dis 10 (2), 49. doi:10.1038/s41419-018-1299-x
Sharma, S., Yang, J., van Nues, R., Watzinger, P., Kötter, P., Lafontaine, D. L. J., et al. (2017). Specialized Box C/D snoRNPs Act as Antisense Guides to Target RNA Base Acetylation. Plos Genet. 13 (5), e1006804. doi:10.1371/journal.pgen.1006804
Shi, H., Wei, J., and He, C. (2019a). Where, when, and How: Context-dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cel 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Shi, X., Li, J., Ma, L., Wen, L., Wang, Q., Yao, H., et al. (2019b). Overexpression of ZEB2-AS1 lncRNA I-s A-ssociated with P-oor C-linical O-utcomes in A-cute M-yeloid L-eukemia. Oncol. Lett. 17 (6), 4935–4947. doi:10.3892/ol.2019.10149
Shirahama, S., Miki, A., Kaburaki, T., and Akimitsu, N. (2020). Long Non-coding RNAs Involved in Pathogenic Infection. Front. Genet. 11 (May), 3389. doi:10.3389/fgene.2020.00454
Silvera, D., Formenti, S. C., and Schneider, R. J. (2010). Translational Control in Cancer. Nat. Rev. Cancer 10 (4), 254–266. doi:10.1038/nrc2824
Sridharan, R., Gonzales-Cope, M., Chronis, C., Bonora, G., McKee, R., Huang, C., et al. (2013). Proteomic and Genomic Approaches Reveal Critical Functions of H3K9 Methylation and Heterochromatin Protein-1γ in Reprogramming to Pluripotency. Nat. Cel Biol 15 (7), 872–882. doi:10.1038/ncb2768
Studtmann, K., Ölschläger-Schütt, J., Buck, F., Richter, D., Sala, C., Bockmann, J., et al. (2014). A Non-canonical Initiation Site Is Required for Efficient Translation of the Dendritically Localized Shank1 mRNA. PLoS ONE 9 (2), e88518. doi:10.1371/journal.pone.0088518
Sun, L., and Fang, J. (2016). Epigenetic Regulation of Epithelial-Mesenchymal Transition. Cel. Mol. Life Sci. 73 (23), 4493–4515. doi:10.1007/s00018-016-2303-1
Tan-Wong, S. M., Dhir, S., and Proudfoot, N. J. (2019). R-loops Promote Antisense Transcription across the Mammalian Genome. Mol. Cel 76 (4), 600–616. e6. doi:10.1016/j.molcel.2019.10.002
Tang, S., Liu, Q., and Xu, M. (2021). LINC00857 Promotes Cell Proliferation and Migration in Colorectal Cancer by Interacting with YTHDC1 and Stabilizing SLC7A5. Oncol. Lett. 22 (2), 578. doi:10.3892/ol.2021.12839
Tani, H., Imamachi, N., Mizutani, R., Imamura, K., Kwon, Y., Miyazaki, S., et al. (2015). Genome-Wide Analysis of Long Noncoding RNA Turnover. Methods Mol. Biol. 1262, 305–320. doi:10.1007/978-1-4939-2253-6_19
Terashima, M., Tange, S., Ishimura, A., and Suzuki, T. (2017). MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J. Biol. Chem. 292 (1), 82–99. doi:10.1074/jbc.M116.750950
Uesaka, M., Nishimura, O., Go, Y., Nakashima, K., Agata, K., and Imamura, T. (2014). Bidirectional Promoters Are the Major Source of Gene Activation-Associated Non-coding RNAs in Mammals. BMC Genomics 15 (1), 35. doi:10.1186/1471-2164-15-35
van Nues, R. W., Granneman, S., Kudla, G., Sloan, K. E., Chicken, M., Tollervey, D., et al. (2011). Box C/D snoRNP Catalysed Methylation Is Aided by Additional Pre-rRNA Base-Pairing. EMBO J. 30 (12), 2420–2430. doi:10.1038/emboj.2011.148
van Steensel, B., and Furlong, E. E. M. (2019). The Role of Transcription in Shaping the Spatial Organization of the Genome. Nat. Rev. Mol. Cel Biol 20 (6), 327–337. doi:10.1038/s41580-019-0114-6
Volders, P.-J., Verheggen, K., Menschaert, G., Vandepoele, K., Martens, L., Vandesompele, J., et al. (2015). An Update on LNCipedia: a Database for Annotated Human lncRNA Sequences. Nucleic Acids Res. 43 (D1), D174–D180. doi:10.1093/nar/gku1060
Walther, K., and Schulte, L. N. (2021). The Role of lncRNAs in Innate Immunity and Inflammation. RNA Biol. 18 (5), 587–603. doi:10.1080/15476286.2020.1845505
Wang, A., Wang, J., Liu, Y., and Zhou, Y. (2017a). Mechanisms of Long Non-coding RNAs in the Assembly and Plasticity of Neural Circuitry. Front. Neural Circuits 11 (October). doi:10.3389/fncir.2017.00076
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 472 (7341), 120–124. doi:10.1038/nature09819
Wang, S. E., Ko, S. Y., Jo, S., Choi, M., Lee, S. H., Jo, H.-R., et al. (2017b). TRPV1 Regulates Stress Responses through HDAC2. Cel Rep. 19 (2), 401–412. doi:10.1016/j.celrep.2017.03.050
Wang, X., Zhang, J., and Wang, Y. (2019). Long Noncoding RNA GAS5-AS1 Suppresses Growth and Metastasis of Cervical Cancer by Increasing GAS5 Stability. Am. J. Transl Res. 11 (8), 4909–4921.
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018b). N6-methyladenosine RNA Modification Regulates Embryonic Neural Stem Cell Self-Renewal through Histone Modifications. Nat. Neurosci. 21 (2), 195–206. doi:10.1038/s41593-017-0057-1
Wang, Z., Yang, B., Zhang, M., Guo, W., Wu, Z., Wang, Y., et al. (2018a). lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 33 (4), 706–e9. e9. doi:10.1016/j.ccell.2018.03.006
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cel 69 (6), 1028–1038. e6. doi:10.1016/j.molcel.2018.02.015
Weng, Y.-L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018a). Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 97 (2), 313–325. e6. doi:10.1016/j.neuron.2017.12.036
Widagdo, J., Zhao, Q.-Y., Kempen, M.-J., Tan, M. C., Ratnu, V. S., Wei, W., et al. (2016). Experience-Dependent Accumulation of N 6 -Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J. Neurosci. 36 (25), 6771–6777. doi:10.1523/JNEUROSCI.4053-15.2016
Wu, B., Li, L., Huang, Y., Ma, J., and Min, J. (2017a). Readers, Writers and Erasers of N6-Methylated Adenosine Modification. Curr. Opin. Struct. Biol. 47, 67–76. doi:10.1016/j.sbi.2017.05.011
Wu, C., Chen, W., He, J., Jin, S., Liu, Y., Yi, Y., et al. (2020). Interplay of M 6 A and H3K27 Trimethylation Restrains Inflammation during Bacterial Infection. Sci. Adv. 6 (34), eaba0647. doi:10.1126/sciadv.aba0647
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of Long Noncoding RNAs in Brain Development, Functional Diversification and Neurodegenerative Diseases. Brain Res. Bull. 97, 69–80. doi:10.1016/j.brainresbull.2013.06.001
Wu, Z., Wu, P., Zuo, X., Yu, N., Qin, Y., Xu, Q., et al. (2017b). LncRNA-N1LR Enhances Neuroprotection against Ischemic Stroke Probably by Inhibiting P53 Phosphorylation. Mol. Neurobiol. 54 (10), 7670–7685. doi:10.1007/s12035-016-0246-z
Xiao, S., Cao, S., Huang, Q., Xia, L., Deng, M., Yang, M., et al. (2019). The RNA N6-Methyladenosine Modification Landscape of Human Fetal Tissues. Nat. Cel Biol 21 (5), 651–661. doi:10.1038/s41556-019-0315-4
Xu, W., Li, J., He, C., Wen, J., Ma, H., Rong, B., et al. (2021). METTL3 Regulates Heterochromatin in Mouse Embryonic Stem Cells. Nature 591 (7849), 317–321. doi:10.1038/s41586-021-03210-1
Xue, L., Li, J., Lin, Y., Liu, D., Yang, Q., Jian, J., et al. (2020). m 6 A Transferase METTL3‐induced lncRNA ABHD11‐AS1 Promotes the Warburg Effect of Non‐small‐cell Lung Cancer. J. Cel Physiol 236, 2649–2658. doi:10.1002/jcp.30023
Yao, R.-W., Wang, Y., and Chen, L.-L. (2019). Cellular Functions of Long Noncoding RNAs. Nat. Cel Biol 21 (5), 542–551. doi:10.1038/s41556-019-0311-8
Yoon, K.-J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 171 (4), 877–889. e17. doi:10.1016/j.cell.2017.09.003
Yoon, Y. J., Wu, B., Buxbaum, A. R., Das, S., Tsai, A., English, B. P., et al. (2016). Glutamate-induced RNA Localization and Translation in Neurons. Proc. Natl. Acad. Sci. USA 113 (44), E6877–E6886. doi:10.1073/pnas.1614267113
Yu, F., Zhang, G., Shi, A., Hu, J., Li, F., Zhang, X., et al. (2018a). LnChrom: A Resource of Experimentally Validated lncRNA-Chromatin Interactions in Human and Mouse. Database 2018, 1–7. doi:10.1093/database/bay039
Yu, J., Li, Y., Wang, T., and Zhong, X. (2018b). Modification of N6-Methyladenosine RNA Methylation on Heat Shock Protein Expression. PLOS ONE 13 (6), e0198604. doi:10.1371/journal.pone.0198604
Yue, B., Song, C., Yang, L., Cui, R., Cheng, X., Zhang, Z., et al. (2019). METTL3-mediated N6-Methyladenosine Modification Is Critical for Epithelial-Mesenchymal Transition and Metastasis of Gastric Cancer. Mol. Cancer 18 (1), 1–15. doi:10.1186/s12943-019-1065-4
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov 4 (1), 10. doi:10.1038/s41421-018-0019-0
Zhang, B., Zheng, H., Huang, B., Li, W., Xiang, Y., Peng, X., et al. (2016). Allelic Reprogramming of the Histone Modification H3K4me3 in Early Mammalian Development. Nature 537 (7621), 553–557. doi:10.1038/nature19361
Zhang, J., Ao, Y., Zhang, Z., Mo, Y., Peng, L., Jiang, Y., et al. (2020a). Lamin A Safeguards the M 6 A Methylase METTL14 Nuclear Speckle Reservoir to Prevent Cellular Senescence. Aging Cell 19 (10), 1–9. doi:10.1111/acel.13215
Zhang, M., Zhai, Y., Zhang, S., Dai, X., and Li, Z. (2020b). Roles of N6-Methyladenosine (m6A) in Stem Cell Fate Decisions and Early Embryonic Development in Mammals. Front. Cel Dev. Biol. 8 (August), 1–15. doi:10.3389/fcell.2020.00782
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31 (4), 591–606. e6. doi:10.1016/j.ccell.2017.02.013
Zhang, W., Chen, Y., Liu, P., Chen, J., Song, L., Tang, Y., et al. (2012). Variants on Chromosome 9p21.3 Correlated with ANRIL Expression Contribute to Stroke Risk and Recurrence in a Large Prospective Stroke Population. Stroke 43 (1), 14–21. doi:10.1161/STROKEAHA.111.625442
Zhang, Z., and Pugh, B. F. (2011). High-Resolution Genome-wide Mapping of the Primary Structure of Chromatin. Cell 144 (2), 175–186. doi:10.1016/j.cell.2011.01.003
Zhao, B. S., Nachtergaele, S., Roundtree, I. A., and He, C. (2018). Our Views of Dynamic N6-Methyladenosine RNA Methylation. Rna 24 (3), 268–272. doi:10.1261/rna.064295.117
Zhao, J., Ohsumi, T. K., Kung, J. T., Ogawa, Y., Grau, D. J., Sarma, K., et al. (2010). Genome-wide Identification of Polycomb-Associated RNAs by RIP-Seq. Mol. Cel 40 (6), 939–953. doi:10.1016/j.molcel.2010.12.011
Zhao, S., Allis, C. D., and Wang, G. G. (2021). The Language of Chromatin Modification in Human Cancers. Nat. Rev. Cancer 21 (7), 413–430. doi:10.1038/s41568-021-00357-x
Zheng, H., Huang, B., Zhang, B., Xiang, Y., Du, Z., Xu, Q., et al. (2016). Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol. Cel 63 (6), 1066–1079. doi:10.1016/j.molcel.2016.08.032
Zheng, J., Guo, J., Cao, B., Zhou, Y., and Tong, J. (2021). Identification and Validation of lncRNAs Involved in m6A Regulation for Patients with Ovarian Cancer. Cancer Cel Int 21 (1), 363. doi:10.1186/s12935-021-02076-7
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S.-B. (2015). Dynamic m6A mRNA Methylation Directs Translational Control of Heat Shock Response. Nature 526 (7574), 591–594. doi:10.1038/nature15377
Zhou, J., Wan, J., Shu, X. E., Mao, Y., Liu, X.-M., Yuan, X., et al. (2018). N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cel 69 (4), 636–647. e7. doi:10.1016/j.molcel.2018.01.019
Zhou, K. I., Shi, H., Lyu, R., Wylder, A. C., Matuszek, Ż., Pan, J. N., et al. (2019). Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol. Cel 76 (1), 70–81. e9. doi:10.1016/j.molcel.2019.07.005
Zhu, P., He, F., Hou, Y., Tu, G., Li, Q., Jin, T., et al. (2021). A Novel Hypoxic Long Noncoding RNA KB-1980E6.3 Maintains Breast Cancer Stem Cell Stemness via Interacting with IGF2BP1 to Facilitate C-Myc mRNA Stability. Oncogene 40 (9), 1609–1627. doi:10.1038/s41388-020-01638-9
Zong, X., Wang, H., Xiao, X., Zhang, Y., Hu, Y., Wang, F., et al. (2020). Enterotoxigenic Escherichia coli Infection Promotes Enteric Defensin Expression via FOXO6-METTL3-m6A-Gpr161 Signalling axis. RNA Biol. 18 (00), 576–586. doi:10.1080/15476286.2020.1820193
ANRIL antisense noncoding RNA in the INK4 locus
AS anti-sense
AS-lncRNAs anti sense long non-coding RNAs
Bp base pairs
CDS coding sequence
EMT epithelial mesenchymal transition
FISH fluorescent in situ hybridization
H2A histone H2A
H2B histone H2B
H3 histone H3
H3K4me1 histone H3 lysine 4 methylation
H3K9me2 histone H3 lysine 9 di-methylation
H3K27ac histone H3 lysine 27 acetylation
H3K27me3 histone H3 lysine 27 tri-methylation
H3K36me3 histone H3 lysine 36 tri-methylation
H4 histone H4
HDAC2 histone deacetylase 2
IRES- internal ribosome entry site
lincRNAs large intergenic non-coding RNAs
lncRNA long non-coding RNAs
m6A N6-methyladenosine
miRNA micro RNAs
mRNA messenger RNA
ncRNA non-coding RNA
ORFs open-reading frames
PRC2 polycomb repressive complex 2
rRNAs ribosomal RNAs
scaRNAs cajal-body-associated RNAs
snoRNAs small nucleolar RNAs
Keywords: lncRNA, M6A, histone methlyation, RNA modification, antisense lncRNA, RNA guide, UTRs
Citation: Vaasjo LO (2022) LncRNAs and Chromatin Modifications Pattern m6A Methylation at the Untranslated Regions of mRNAs. Front. Genet. 13:866772. doi: 10.3389/fgene.2022.866772
Received: 31 January 2022; Accepted: 28 February 2022;
Published: 17 March 2022.
Edited by:
Sam El-Osta, Monash University, AustraliaReviewed by:
Terisha Ghazi, University of KwaZulu-Natal, South AfricaCopyright © 2022 Vaasjo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lee O. Vaasjo, bHZhYXNqb211bm96QHR1bGFuZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.