
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 13 May 2022
Sec. Statistical Genetics and Methodology
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.865449
This article is part of the Research Topic Gene by Environment Interaction and Effects on Human Health View all 5 articles
Background: In utero arsenic and cadmium exposures are linked with reduced birth weight as well as alterations in placental molecular features. However, studies thus far have focused on summarizing transcriptional activity at the gene level and do not capture transcript specification, an important resource during fetal development to enable adaptive responses to the rapidly changing in utero physiological conditions. In this study, we conducted a genome-wide analysis of the placental transcriptome to evaluate the role of differential transcript usage (DTU) as a potential marker of in utero arsenic and cadmium exposure and fetal growth restriction.
Methods: Transcriptome-wide RNA sequencing was performed in placenta samples from the Rhode Island Child Health Study (RICHS, n = 199). Arsenic and cadmium levels were measured in maternal toenails using ICP-MS. Differential transcript usage (DTU) contrasting small (SGA) and appropriate (AGA) for gestational age infants as well as above vs. below median exposure to arsenic and cadmium were assessed using the DRIMSeq R package. Genetic variants that influence transcript usage were determined using the sQTLseeker R package.
Results: We identified 82 genes demonstrating DTU in association with SGA status at an FDR <0.05. Among these, one gene, ORMDL1, also demonstrated DTU in association with arsenic exposure, and fifteen genes (CSNK1E, GBA, LAMTOR4, MORF4L1, PIGO, PSG1, PSG3, PTMA, RBMS1, SLC38A2, SMAD4, SPCS2, TUBA1B, UBE2A, YIPF5) demonstrated DTU in association with cadmium exposure. In addition to cadmium exposure and SGA status, proportions of the LAMTOR4 transcript ENST00000474141.5 also differed by genetic variants (rs10231604, rs12878, and rs3736591), suggesting a pathway by which an in utero exposure and genetic variants converge to impact fetal growth through perturbations of placental processes.
Discussion: We report the first genome-wide characterization of placental transcript usage and associations with intrauterine metal exposure and fetal growth restriction. These results highlight the utility of interrogating the transcriptome at finer-scale transcript-level resolution to identify novel placental biomarkers of exposure-induced outcomes.
Fetal growth is susceptible to varying in utero conditions and environmental exposures. In particular, trace toxic metals and metalloids are linked to reduced birth weight as well as health effects in childhood (Chung et al., 2015; Valeri et al., 2017; Wang G. et al., 2021; McRae et al., 2022). Specifically, several reports link reductions in birth weight to in utero levels of arsenic (Xu et al., 2011; Deyssenroth et al., 2018) and cadmium (Llanos and Ronco, 2009; Shirai et al., 2010; Luo et al., 2017; Sabra et al., 2017; Deyssenroth et al., 2018; Freire et al., 2019; Punshon et al., 2019). A growing body of evidence links placental programming disruptions in the pathway between exposure to these metals and fetal growth restriction (Punshon et al., 2019). For example, we and others have shown that gestational exposure to arsenic and cadmium is associated with being born small for gestational age (SGA), and levels of these metals covary with the altered expression of placental gene networks implicated with SGA status. (Deyssenroth et al., 2018).
Variations in transcripts that map to a gene, including alternative start and stop codons and skipped exons, are realized through alternative splicing and other transcriptional process. While the overall function of a protein is generally conserved across transcripts, the preferential expression of specific transcripts can inform where the encoded protein is localized within a cell, whether it is exported, its enzymatic activity, and binding capacity to interact with other proteins and nucleic acid (Kelemen et al., 2013). Modulating expression at the level of transcripts can, thereby, increase the diversity in protein function to dynamically respond to developmental stage-specific needs and environmental cues. A homeostasis in transcript-proportions is generally maintained under normal conditions, and a shift in this balance has implications for health (Kim et al., 2018).
Perinatal studies evaluating transcriptomic markers of exposure-induced birth outcomes typically evaluate gene expression (i.e., total transcript abundance for each gene) differences to identify loci of interest. While summarizing transcriptome-wide data to gene-level resolution is a convenient and useful metric to conduct differential expression analysis, it may also obscure the true underlying variability in transcriptomic regulation. Indeed, altered proportions of transcripts across conditions can result in no detectable overall differences in gene expression. Current advances in sequencing technology allow for more fine-scaled interrogations to capture transcript-level differences in expression. These include describing changes in proportions among transcripts within a gene, also referred to as differential transcript usage (DTU).
Most studies to date have linked changes in gene transcript-level expression to cancer (Zhang and Manley, 2013) and neurodegenerative outcomes (Dredge et al., 2001). In addition, in vitro and in vivo studies have also linked environmental stressors, including alcohol (Sariyer et al., 2017), methyl-mercury (Li et al., 2018) and social interaction, as well as genetic variants as sources of transcript expression profile perturbations. However, the relevance of alterations in transcript proportions as markers of environmental exposures and birth outcomes is understudied in population studies. This is despite the relevance of transcript-specification for the adaptive progression through fetal development and its potential to reflect disruptions in this process. This study seeks to evaluate the association between placental transcript proportions, arsenic and cadmium exposure and birth weight in a birth cohort study.
Mother-infant pairs were recruited at Women and Infants Hospital in Providence, Rhode Island as part of the Rhode Island Child Health Study (n = 841). Participants who met eligibility criteria included women with singleton pregnancies who were at least 18 years of age, delivered at term (≥37 weeks gestation) and did not experience major pregnancy or offspring complications (e.g., chromosomal abnormalities, congenital defects). The study population was oversampled for small for gestational age (SGA, < 10% Fenton growth curve) and large for gestational age (LGA, > 90% Fenton growth curve) infants. Appropriate for gestational age (AGA) infants were concurrently enrolled and matched for gestational and maternal age. Informed consent was obtained from all participants, and the study was reviewed and approved by the Internal Review Boards at Woman and Infants Hospital and Emory University. The current study is restricted to participants with available placental RNA seq data and no reported maternal smoke exposure during pregnancy (n = 196). The birth weight analysis focused on comparing SGA (n = 30) and AGA (n = 112) infants. Arsenic and cadmium measurements were available for 171 participants.
ICP-MS was performed at the Dartmouth Trace Metal laboratory to determine metal levels across a panel of 19 metals measured in toenail clippings obtained from RICHS mothers and infants following hospital discharge as previously described (Everson et al., 2016; Punshon et al., 2016; Everson et al., 2017; Deyssenroth et al., 2018). Briefly, toenail clippings were received from participants, on average, within 2.8 months postpartum. Study samples were cleaned, microwave digested, and analyzed alongside certified reference materials via ICP-MS at the Dartmouth Trace Element Analysis Core following guidelines outlined in EPA 6020A. Metal measurements are expressed as microgram (μG) metal per gram toenail. Values falling below the sample-specific limit of detection (LOD) were replaced with the
Placenta collection, placental transcriptome and SNP genotyping data acquisition were performed as previously described (Peng et al., 2017; Clarkson-Townsend et al., 2020). Briefly, placenta tissue was biopsied 2 cm from the umbilical cord insertion site, free from maternal decidua, within 2 h of delivery. Biopsies were placed in RNALater and maintained at 4°C for 72 h prior to storage at −80°C. RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA) following manufacturer’s instructions. Single-end 50 bp RNA sequencing reads were generated using the Illumina HiSeq 2,500 platform. SNP genotyping data was generated using the Illumina MegaEX chip and processed as previously described (Peng et al., 2017).
Transcript abundance was quantitated based on the GRCh38.12 v28 human reference genome using salmon. (Patro et al., 2017). The total detected counts (# transcripts = 203,027) were restricted to protein-coding genes (# transcripts = 147,015, # genes = 19,989). Our survey was additionally restricted to genes with at least two transcripts to detect differential transcript usage. The data was further filtered to retain transcripts and genes meeting a minimum expression threshold in at least the number of samples of our smallest phenotypic group (i.e., SGA = 30; As and Cd = 85) based on the following criteria: genes with a minimum overall gene expression of 10 estimated counts, transcripts within genes with a minimum transcript expression of 10 estimated counts, and transcripts accounting for a minimum of 10% of total expression for a given gene. The final datasets consisted of 5,660 genes (23,168 transcripts) for the SGA analysis and 5,057 genes (15,998 transcripts) for the arsenic and cadmium analyses. Differential transcript usage (DTU) contrasting SGA and AGA placenta was performed using the DRIMSeq R package (Nowicka and Robinson, 2016). For the metal DTU analysis, samples were dichotomized above and below the median (Arsenic = 0.04 ug/g; Cadmium = 0.01 ug/g). We repeated testing for metal DTU in a subsetted analysis, restricting our contrast to individuals in the 3rd (n = 57) vs. 1st tertile (n = 58) of exposure. As an additional control on false discovery rate, a posthoc filtering procedure was performed whereby p-values were set to one for transcripts with small standard deviations in per-sample proportions. Significant p-values were determined using the stageR R package (Van den Berge and Clement, 2021). In this two-stage testing procedure, a screen to identify genes exhibiting differential transcript usage is performed in the first stage, and transcripts participating in the differential transcript usage of those genes are confirmed in the second stage. Gene ontology enrichment analyses were performed using the enrichR (Kuleshov et al., 2016) R package. We profiled genetic polymorphisms associated with differential transcript usage, also known an splice quantitative trait loci (sQTLs), using the sQTLseeker R package (Monlong et al., 2014). The identified sQTLs were further queried for known disease associations reported in the NHGRI-EBI GWAS catalog. SGA differential transcript usage analyses were adjusted for maternal race/ethnicity and sex. Arsenic and cadmium differential transcript usage analyses were additionally adjusted for metal analysis batch. All analyses were conducted using R 4.1.1. The code implemented for the presented differential transcript usage analysis is available here: https://github.com/Deyssenroth-Lab/RICHS_DTU/tree/master.
The demographic characteristics contrasting SGA and AGA infants in our study are shown in Table 1. The same demographic characteristics contrasting infants above and below median arsenic and cadmium levels are shown in Supplementary Tables S1, S2. SGA and AGA infants differed by maternal race/ethnicity (greater proportion of non-white mothers among SGA infants compared to AGA infants). Infants above vs. below the median fetal cadmium exposure differed by sex (greater proportion of female infants in group with above median levels of cadmium exposure) and ICP-MS batch (Supplementary Table S2).
Out of 5,660 tested genes, we identified 82 genes that displayed differential transcript usage comparing SGA vs. AGA infants (Supplementary Table S3). Examples of genes demonstrating differential transcript usage are shown in Figure 1. This figure depicts instances where equimolar levels of transcripts are present in one condition, and preferential selection of a specific transcript is apparent in the other condition. For example, equivalent placental levels of INHBA (ENST00000242208.4; ENST00000442711.1) and ITGAV (ENST00000261023.7; ENST0000433736.6) transcripts are observed among SGA infants (Figures 1A,B). However, one transcript is preferentially expressed (INHBA: ENST00000242208.4; ITGAV: ENST00000261023.7) over the other among AGA infants. An opposing trend is observed for NAA20 and RAB1B (Figures 1C,D). Here, equivalent placental levels (NAA20: ENST000004663154.5; ENST00000480550.1) and RAB1B (ENST00000311481.0; ENST0000527397.1) transcripts are observed among AGA infants, and one placental transcript is preferentially expressed (NAA20: ENST000004663154.5; RAB1B: ENST00000311481.0) over the other among SGA infants. Gene ontology enrichment analysis indicated an overrepresentation of ubiquitin-related processes among genes displaying differential transcript usage by SGA status (Supplementary Figure S1).
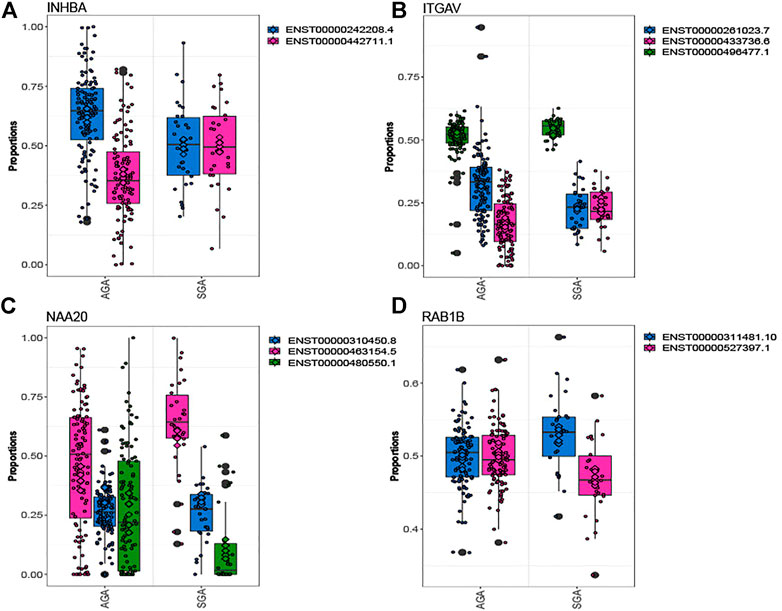
FIGURE 1. Genes depicting differential transcript usage between SGA and AGA infants. Eighty-two genes demonstrated significant differential transcript usage. Shown are examples of four genes, INHBA (A), ITGAV (B), NAA20 (C), and RAB1B (D). The x-axis indicates the proportion of total measured gene expression captured by individual transcripts.
Comparing infants above and below the median for fetal arsenic and cadmium exposure, we identified 16 and 160 genes demonstrating differential transcript usage, respectively (Supplementary Tables S4, S5). Similarly, we identified 13 and 155 genes demonstrating differential transcript usage comparing infants in the 3rd vs. 1st tertile of arsenic and cadmium exposure, respectively (Supplementary Tables S6, S7). Overlapping these results with the SGA analysis, one gene (ORMDL1) demonstrated differential transcript usage across both the SGA and the dichotomized arsenic analyses. As seen in Figure 2, the direction of the association is consistent across both contrasts (i.e., upregulation of the ORMDL1 transcript ENST00000392349.8 is observed among both SGA infants and infants above the median for fetal arsenic exposure). However, ORMDL1 was not among the genes differentially expressed by arsenic exposure when comparing individuals in the 3rd vs. 1st tertile of exposure.
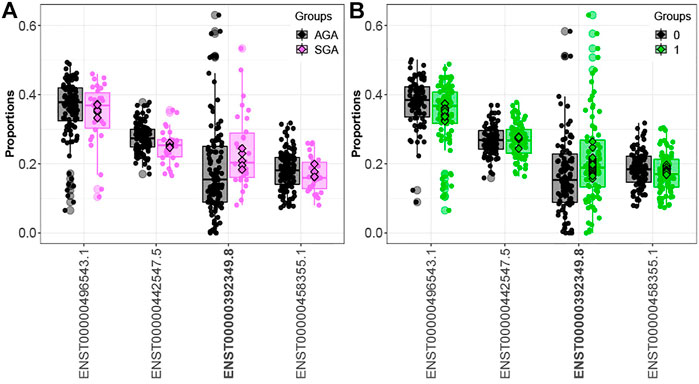
FIGURE 2. ORMDL1 differential transcript usage. Elevated expression of the ORMDL1 transcript ENST00000392349.8 is observed in SGA infants compared to AGA infants (A) and infants above vs. below the median fetal exposure to arsenic (B).
Fifteen genes (CSNK1E, GBA, LAMTOR4, MORF4L1, PIGO, PSG1, PSG3, PTMA, RBMS1, SLC38A2, SMAD4, SPCS2, TUBA1B, UBE2A, YIPF5) demonstrated differential transcript usage across both the SGA and dichotomized cadmium analyses. Out of these fifteen genes, fourteen were also identified when comparing individuals in the 3rd. vs. 1st tertile of exposure (CSNK1E, GBA, LAMTOR4, PIGO, PSG1, PSG3, PTMA, RBMS1, SLC38A2, SMAD4, SPCS2, TUBA1B, UBE2A, YIPF5). Ten genes (CSNK1E, GBA, LAMTOR4, PIGO, PSG1, PSG3, PTMA, SPCS2, TUBA1B, and UBE2A) displayed differential transcript usage of the same transcript and in a consistent direction in association with SGA and above the median fetal cadmium exposure levels. Figure 3 focuses on the overlapping trends for LAMTOR4. Here, downregulation of the LAMTOR4 transcript ENST00000474141.5 is observed among both SGA infants and infants above the median for fetal cadmium exposure.
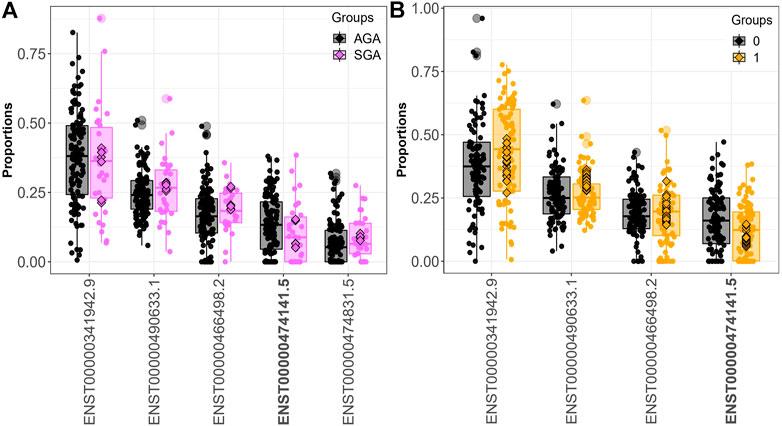
FIGURE 3. LAMTOR4 differential transcript usage. Decreased expression of the LAMTOR4 transcript ENST00000474141.5 is observed in SGA infants compared to AGA infants (A) and infants above vs. below the median fetal exposure to (B).
We identified 16,528 sQTLs mapping to 1,041 individual genes. Overlaying the sQTL and differential transcript usage findings, we observed one gene, LAMTOR4, with a transcript differentially expressed with respect to a SNP variant, SGA status and fetal cadmium exposure. Three SNPs are associated with the expression of LAMTOR4 transcripts ENST00000474141.5 and ENST00000341942.9. The impact of SNPs rs10231604 [7:100148578], rs12878 [7:100149507], and rs3736591 [7:100153394] are shown in Figure 4, Supplementary Figures S2, S3, respectively. Consistent trends in relative transcript expression patterns are observed for all three SNPs. Increasing dosage of the alternate allele (e.g., rs10231604-A) increases the relative expression of LAMTOR4 transcript ENST00000341942.9 and decreases the relative expression of LAMTOR4 transcript ENST00000474141.5. Reductions in the relative expression level of LAMTOR4 transcript ENST00000474141.5 in the presence of the sQTL variant alleles is consistent with the reductions in relative expression levels of the same transcript observed in association with SGA status and among infants above the median level for fetal cadmium exposure (Figures 3A,B).
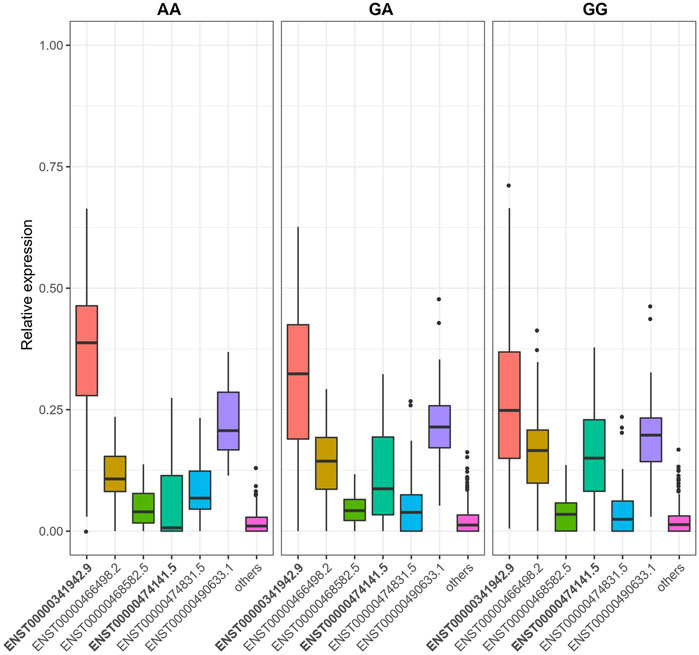
FIGURE 4. SNP rs10231604 regulates the expression of LAMTOR4 transcripts. Relative expression of transcript ENST00000341942.9 decreases with increasing dosage of the wildtype allele (G). Relative expression of transcript ENST00000474141.5 increases with increasing dosage of the wildtype allele (G).
Figure 5 shows the coding exons for the two transcripts under sQTL regulation as well as the location of the sQTL SNPs with respect to these transcripts. SNP rs10231604 is located upstream of the start codon, SNP rs12878 is located near exon 2, and rs3736591 is located near exon 3. A GWAS-associated SNP (rs77686669) linked with venous thromboembolism in African Americans (Heit et al., 2017) that maps to this region is also shown.
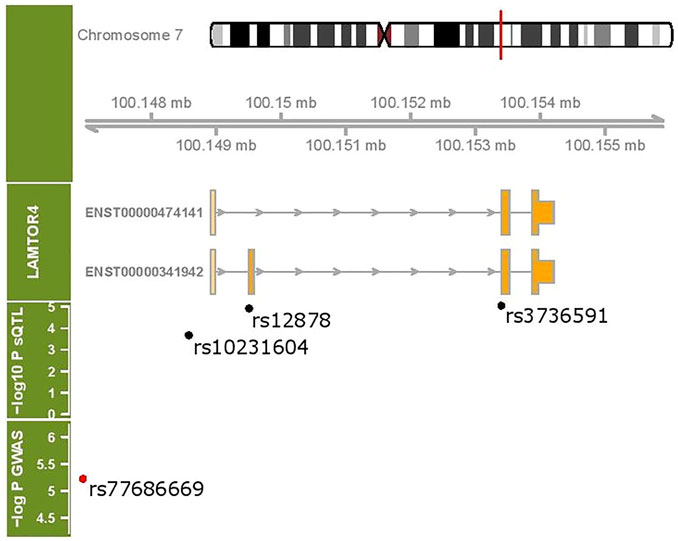
FIGURE 5. LAMTOR4 genomic region (chr 7:100148907- 100155944). Two transcripts under sQTL regulation are shown. The location of three sQTL variants influencing the expression of the LAMTOR4 transcripts are labeled. Shown in red is a GWAS-identified SNP associated with venous thromboembolism.
We report here the first study characterizing placental DTU in association with birth weight and in utero metal exposure. Several genes we identified in relation to SGA status were previously identified as relevant to placental function based on analyzing overall gene expression differences, including GBA (Jebbink et al., 2015), GCLC (Hannan et al., 2018), INHA (Brew et al., 2016), INHBA (Deyssenroth et al., 2018), ITGAV (Wang et al., 2019), NFKBIA (Mahany et al., 2018), PSG1 (Whitehead et al., 2013), RBFOX2 (Goldman- Wohl et al., 2020; Freitag et al., 2021), SLC38A2 (Vaughan et al., 2021), and SPIRE2 (Azar et al., 2021).
Consistent literature support also exists for several of the genes we identified in relation to metal exposure. For example, among our arsenic-responsive regions, cord blood methylation of CD151 was previously associated with drinking water arsenic exposure in a prospective cohort study in Bangladesh (Kile et al., 2014). In a mouse study, altered lung gene expression of CLIC5 was observed comparing unexposed mice and mice exposed to 10 ppb arsenic in chow or drinking water over a 5–6 week period (Kozul et al., 2009). In an in vitro study, VAV3 was identified as a trivalent arsenical-responsive gene based on gene expression differences comparing treated and untreated HUC-1 cells (Su et al., 2006).
We also identified placental DTU among known cadmium-responsive genes. For example, differential gene expression based on cadmium exposure was observed in vitro studies across human breast cancer cell lines (PECAM1, DAB2, PGK1)23 (Lubovac-Pilav et al., 2013), human renal epithelial cells (SETD2, TBC1D15, TNPO1) (Garrett et al., 2013), rat liver cells [ANGPTL4 (Permenter et al., 2011), ADM. (Hsiao and Stapleton, 2009)], and mouse embryonic fibroblast cell lines (EFEMP1) (Oldani et al., 2020). Similarly, in vivo studies also indicate an impact on gene expression patterns in rat hypothalamus (PSMA1, RPLP0, SHC1) (Saedi et al., 2021), mouse testes (LYE6) (Hu et al., 2014), mouse pup heart (ALDOA). (Hudson et al., 2019), mouse liver (PRKCE) (Jackson et al., 2020), and mouse femur (EEF1A1) (Ohba et al., 2007). Our study adds to this body of literature by demonstrating that the expression pattern of these genes is also disrupted due to exposure to these metals in human placenta.
Importantly, we also identified many loci previously unreported in relation to SGA status, arsenic and cadmium exposure in this study. This suggests that surveying the placental transcriptome at transcript-level resolution may reveal an additional layer of insight beyond assessing overall gene expression differences. Indeed, as seen in Figure 1, while the distribution in the proportion of individual transcripts within genes can shift across conditions, this change may not translate to a change in overall gene expression. Our findings, therefore, suggest the relevance of transcript-level expression as a more nuanced placental marker of in utero exposures and postnatal outcomes.
We identified a lysosomal signaling gene, LAMTOR4 (Mu et al., 2017), with altered transcript-level expression comparing SGA and AGA infants, infants above vs. below the median for fetal cadmium exposure, and across SNP alleles (rs10231604, rs12878, rs3736591). Consistent with our finding, these three SNPs are also known to inform LAMTOR4 splicing patterns across adult tissues (i.e., whole blood, brain, skin adipose tissue, breast, colon, muscle, etc) based on reports by the GTEx biobank. This suggests that genetic regulation of LAMTOR4 transcript-level expression is a systemic phenomenon that extends beyond the in utero period in the placenta.
Specifically, the placental proportion of LAMTOR4 transcript ENST00000474141.5 is reduced in SGA infants, among infants above the median for fetal cadmium exposure, and infants with greater dosage of the alternate alleles for rs10231604, rs12878 and rs3736591. Interestingly, ENST00000474141.5 does not include exon 2, a feature that distinguishes it from all the other known LAMTOR4 transcripts. The functional consequence of skipping this exon at the protein level and, more broadly, placental tissue level remains unclear. Additional experimental studies are needed to better understand the impact of transcript selection on placental phenotype.
While the focus of our analysis is on birth weight, a prenatally determined outcome, the perturbations on placental function we identified may also increase susceptibility to health effects experienced later in life. Indeed, SNP variants neighboring the sQTLs we identified are implicated with venous thromboembolism. SNPs in this region also confer genetic risk in developing Alzheimer’s disease (Wang Z. et al., 2021), a disorder that can similarly arise due to vascular pathology. Therefore, our findings suggest that genetic variants and in utero environmental exposures (i.e., cadmium) can converge on common transcriptomic targets to disrupt placental programming. These impact growth of the developing fetus prenatally, with potential implications on vascular health extending into the postnatal period.
Several limitations inherent in our study warrant considerations. While a major strength of our study is the availability of in utero metal measurements, genotyping and placental RNAseq data, the small sample size of the study precludes more in-depth evaluations. For example, our findings suggest that genetic variants and environmental exposures converge on common targets to disrupt placental programming, suggesting that genetic factors may underly variation in the susceptibility to exposure-induced diseases of prenatal origin. The current study was not powered to formally examine the presence of such effect modification in cadmium-related effects on birth weight due to genotype. Similarly, while we identified genes with concordant transcript usage differences in our birth weight and exposure analyses, this study was not sufficiently powered to formally test whether changes in the expression of specific transcripts mediate the exposure-related effects on birth weight. Finally, RNAseq data was generated on bulk placenta tissue, which entails a mixed composition of cells. The biopsy protocol at the time of tissue collection ensured representative sampling across all study samples. However, we cannot exclude the possibility that differences in cell type abundances across conditions underly the differential expression patterns by birth weight and exposure detected in this study. This impacts the interpretation of the findings from one pointing to direct disruption of protein function at the cellular level to more indirect effects on overall placental function through a shift of cell types. While we cannot specify which effects predominate in the current study, both are biologically relevant.
In summary, we report the first genome-wide characterization of placental transcript usage and associations with intrauterine metal exposure and fetal growth restriction. These results highlight the utility of interrogating the transcriptome at finer-scale transcript-level resolution to identify novel placental biomarkers of exposure-induced outcomes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/gap/cgi-bin/study.cgi?study_id=phs001586.v1.p1.
The studies involving human participants were reviewed and approved by the Internal Review Boards at Woman and Infants Hospital and Emory University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
MD contributed to the conception and statistical analysis of study as well as manuscript preparation. SP and KH generated the sequencing data that formed the premise of the analysis. JC and CM provided the resources, infrastructure, original premise of the parent study and contributed to manuscript preparation. All authors reviewed and approved the submitted manuscript.
This research was funded by NIH-NIEHS P30ES009089, NIH-NIEHS R00ES029571, NIH-NIEHS R01ES022223, NIH-NIEHS R01ES029212 and NIH-NIEHS R24ES028507.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.865449/full#supplementary-material
Azar, C., Valentine, M. C., Trausch-Azar, J., Rois, L., Mahjoub, M., Nelson, D. M., et al. (2021). RNA-seq Identifies Genes Whose Proteins Are Upregulated during Syncytia Development in Murine C2C12 Myoblasts and Human BeWo Trophoblasts. Physiol. Rep. 9 (1), e14671. doi:10.14814/phy2.14671
Brew, O., Sullivan, M. H. F., and Woodman, A. (2016). Comparison of Normal and Pre-eclamptic Placental Gene Expression: A Systematic Review with Meta-Analysis. PLoS One 11 (8), e0161504. doi:10.1371/journal.pone.0161504
Chung, S. E., Cheong, H.-K., Ha, E.-H., Kim, B.-N., Ha, M., Kim, Y., et al. (2015). Maternal Blood Manganese and Early Neurodevelopment: The Mothers and Children's Environmental Health (MOCEH) Study. Environ. Health Perspect. 123 (7), 717–722. doi:10.1289/ehp.1307865
Clarkson-Townsend, D. A., Kennedy, E., Everson, T. M., Deyssenroth, M. A., Burt, A. A., Hao, K., et al. (2020). Seasonally Variant Gene Expression in Full-Term Human Placenta. FASEB J. 34 (8), 10431–10442. doi:10.1096/fj.202000291R
Deyssenroth, M. A., Gennings, C., Liu, S. H., Peng, S., Hao, K., Lambertini, L., et al. (2018). Intrauterine Multi-Metal Exposure Is Associated with Reduced Fetal Growth through Modulation of the Placental Gene Network. Environ. Int. 120, 373–381. doi:10.1016/j.envint.2018.08.010
Dredge, B. K., Polydorides, A. D., and Darnell, R. B. (2001). The Splice of Life: Alternative Splicing and Neurological Disease. Nat. Rev. Neurosci. 2 (1), 43–50. doi:10.1038/35049061
Everson, T. M., Armstrong, D. A., Jackson, B. P., Green, B. B., Karagas, M. R., and Marsit, C. J. (2016). Maternal Cadmium, Placental PCDHAC1 , and Fetal Development. Reprod. Toxicol. 65, 263–271. doi:10.1016/j.reprotox.2016.08.011
Everson, T. M., Kappil, M., Hao, K., Jackson, B. P., Punshon, T., Karagas, M. R., et al. (2017). Maternal Exposure to Selenium and Cadmium, Fetal Growth, and Placental Expression of Steroidogenic and Apoptotic Genes. Environ. Res. 158, 233–244. doi:10.1016/j.envres.2017.06.016
Freire, C., Amaya, E., Gil, F., Murcia, M., LLop, S., Casas, M., et al. (2019). Placental Metal Concentrations and Birth Outcomes: The Environment and Childhood (INMA) Project. Int. J. Hyg. Environ. Health 222 (3), 468–478. doi:10.1016/j.ijheh.2018.12.014
Freitag, N., Xie, Y., Adam, L. M., Borowski, S., Blois, S. M., and Barrientos, G. (2021). Expression of the Alternative Splicing Regulator Rbfox2 during Placental Development Is Differentially Regulated in Preeclampsia Mouse Models. Am. J. Reprod. Immunol. 86 (6), e13491. doi:10.1111/aji.13491
Garrett, S. H., Clarke, K., Sens, D. A., Deng, Y., Somji, S., and Zhang, K. K. (2013). Short and Long Term Gene Expression Variation and Networking in Human Proximal Tubule Cells when Exposed to Cadmium. BMC Med. Genomics 6 (1), S2. doi:10.1186/1755-8794-6-S1-S2
Goldman-Wohl, D., Greenfield, C., Eisenberg-Loebl, I., Denichenko, P., Jbara, A., Karni, R., et al. (2020). Trophoblast Lineage Specific Expression of the Alternative Splicing Factor RBFOX2 Suggests a Role in Placental Development. Placenta 100, 142–149. doi:10.1016/j.placenta.2020.07.004
Hannan, N. J., Binder, N. K., Beard, S., Nguyen, T.-V., Kaitu’u-Lino, T. u. J., and Tong, S. (2018). Melatonin Enhances Antioxidant Molecules in the Placenta, Reduces Secretion of Soluble Fms-like Tyrosine Kinase 1 (sFLT) from Primary Trophoblast but Does Not rescue Endothelial Dysfunction: An Evaluation of its Potential to Treat Preeclampsia. PLoS One 13 (4), e0187082. doi:10.1371/journal.pone.0187082
Heit, J. A., Armasu, S. M., McCauley, B. M., Kullo, I. J., Sicotte, H., Pathak, J., et al. (2017). Identification of Unique Venous Thromboembolism-Susceptibility Variants in African-Americans. Thromb. Haemost. 117 (4), 758–768. doi:10.1160/TH16-08-0652
Hsiao, C.-j. J., and Stapleton, S. R. (2009). Early Sensing and Gene Expression Profiling under a Low Dose of Cadmium Exposure. Biochimie 91 (3), 329–343. doi:10.1016/j.biochi.2008.10.006
Hu, H., Lu, X., Cen, X., Chen, X., Li, F., and Zhong, S. (2014). RNA-seq Identifies Key Reproductive Gene Expression Alterations in Response to Cadmium Exposure. Biomed. Res. Int. 2014, 529271. doi:10.1155/2014/529271
Hudson, K. M., Belcher, S. M., and Cowley, M. (2019). Maternal Cadmium Exposure in the Mouse Leads to Increased Heart Weight at Birth and Programs Susceptibility to Hypertension in Adulthood. Sci. Rep. 9 (1), 13553. doi:10.1038/s41598-019-49807-5
Jackson, T. W., Ryherd, G. L., Scheibly, C. M., Sasser, A. L., Guillette, T. C., and Belcher, S. M. (2020). Gestational Cd Exposure in the CD-1 Mouse Induces Sex-specific Hepatic Insulin Insensitivity, Obesity, and Metabolic Syndrome in Adult Female Offspring. Toxicol. Sci. 178 (2), 264–280. doi:10.1093/toxsci/kfaa154
Jebbink, J. M., Boot, R. G., Keijser, R., Moerland, P. D., Aten, J., Veenboer, G. J. M., et al. (2015). Increased Glucocerebrosidase Expression and Activity in Preeclamptic Placenta. Placenta 36 (2), 160–169. doi:10.1016/j.placenta.2014.12.001
Kelemen, O., Convertini, P., Zhang, Z., Wen, Y., Shen, M., Falaleeva, M., et al. (2013). Function of Alternative Splicing. Gene 514 (1), 1–30. doi:10.1016/j.gene.2012.07.083
Kile, M. L., Rodrigues, E. G., Mazumdar, M., Dobson, C. B., Diao, N., Golam, M., et al. (2014). A Prospective Cohort Study of the Association between Drinking Water Arsenic Exposure and Self-Reported Maternal Health Symptoms during Pregnancy in Bangladesh. Environ. Health 13 (1), 29. doi:10.1186/1476-069x-13-29
Kim, H. K., Pham, M. H. C., Ko, K. S., Rhee, B. D., and Han, J. (2018). Alternative Splicing Isoforms in Health and Disease. Pflugers Arch. - Eur. J. Physiol. 470 (7), 995–1016. doi:10.1007/s00424-018-2136-x
Kozul, C. D., Hampton, T. H., Davey, J. C., Gosse, J. A., Nomikos, A. P., Eisenhauer, P. L., et al. (2009). Chronic Exposure to Arsenic in the Drinking Water Alters the Expression of Immune Response Genes in Mouse Lung. Environ. Health Perspect. 117 (7), 1108–1115. doi:10.1289/ehp.0800199
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 44 (W1), W90–W97. doi:10.1093/nar/gkw377
Li, Y., He, B., Gao, J., Liu, Q. S., Liu, R., Qu, G., et al. (2018). Methylmercury Exposure Alters RNA Splicing in Human Neuroblastoma SK-N-SH Cells: Implications from Proteomic and post-transcriptional Responses. Environ. Pollut. 238, 213–221. doi:10.1016/j.envpol.2018.03.019
Llanos, M. N., and Ronco, A. M. (2009). Fetal Growth Restriction Is Related to Placental Levels of Cadmium, lead and Arsenic but Not with Antioxidant Activities. Reprod. Toxicol. 27 (1), 88–92. doi:10.1016/j.reprotox.2008.11.057
Lubovac-Pilav, Z., Borràs, D. M., Ponce, E., and Louie, M. C. (2013). Using Expression Profiling to Understand the Effects of Chronic Cadmium Exposure on MCF-7 Breast Cancer Cells. PLoS One 8 (12), e84646. doi:10.1371/journal.pone.0084646
Luo, Y., McCullough, L. E., Tzeng, J.-Y., Darrah, T., Vengosh, A., Maguire, R. L., et al. (2017). Maternal Blood Cadmium, lead and Arsenic Levels, Nutrient Combinations, and Offspring Birthweight. BMC Public Health 17 (1), 354. doi:10.1186/s12889-017-4225-8
Mahany, E. B., Han, X., Borges, B. C., da Silveira Cruz-Machado, S., Allen, S. J., Garcia-Galiano, D., et al. (2018). Obesity and High-Fat Diet Induce Distinct Changes in Placental Gene Expression and Pregnancy Outcome. Endocrinology 159 (4), 1718–1733. doi:10.1210/en.2017-03053
McRae, N., Gennings, C., Rivera Rivera, N., Tamayo-Ortiz, M., Pantic, I., Amarasiriwardena, C., et al. (2022). Association between Prenatal Metal Exposure and Adverse Respiratory Symptoms in Childhood. Environ. Res. 205, 112448. doi:10.1016/j.envres.2021.112448
Monlong, J., Calvo, M., Ferreira, P. G., and Guigó, R. (2014). Identification of Genetic Variants Associated with Alternative Splicing Using sQTLseekeR. Nat. Commun. 5 (1), 4698. doi:10.1038/ncomms5698
Mu, Z., Wang, L., Deng, W., Wang, J., and Wu, G. (2017). Structural Insight into the Ragulator Complex Which Anchors mTORC1 to the Lysosomal Membrane. Cell Discov. 3, 17049. doi:10.1038/celldisc.2017.49
Nowicka, M., and Robinson, M. D. (2016). DRIMSeq: a Dirichlet-Multinomial Framework for Multivariate Count Outcomes in Genomics. F1000Res 5, 1356. doi:10.12688/f1000research.8900.1
Ohba, K.-i., Okawa, Y., Matsumoto, Y., Nakamura, Y., and Ohta, H. (2007). A Study of Investigation of Cadmium Genotoxicity in Rat Bone Cells Using DNA Microarray. J. Toxicol. Sci. 32 (1), 107–109. doi:10.2131/jts.32.107
Oldani, M., Fabbri, M., Melchioretto, P., Callegaro, G., Fusi, P., Gribaldo, L., et al. (2020). In Vitro and Bioinformatics Mechanistic-Based Approach for Cadmium Carcinogenicity Understanding. Toxicol. Vitro 65, 104757. doi:10.1016/j.tiv.2020.104757
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 14 (4), 417–419. doi:10.1038/nmeth.4197
Peng, S., Deyssenroth, M. A., Di Narzo, A. F., Lambertini, L., Marsit, C. J., Chen, J., et al. (2017). Expression Quantitative Trait Loci (eQTLs) in Human Placentas Suggest Developmental Origins of Complex Diseases. Hum. Mol. Genet. 26 (17), 3432–3441. doi:10.1093/hmg/ddx265
Permenter, M. G., Lewis, J. A., and Jackson, D. A. (2011). Exposure to Nickel, Chromium, or Cadmium Causes Distinct Changes in the Gene Expression Patterns of a Rat Liver Derived Cell Line. PLoS One 6 (11), e27730. doi:10.1371/journal.pone.0027730
Punshon, T., Li, Z., Jackson, B. P., Parks, W. T., Romano, M., Conway, D., et al. (2019). Placental Metal Concentrations in Relation to Placental Growth, Efficiency and Birth Weight. Environ. Int. 126, 533–542. doi:10.1016/j.envint.2019.01.063
Punshon, T., Li, Z., Marsit, C. J., Jackson, B. P., Baker, E. R., and Karagas, M. R. (2016). Placental Metal Concentrations in Relation to Maternal and Infant Toenails in a U.S. Cohort. Environ. Sci. Technol. 50 (3), 1587–1594. doi:10.1021/acs.est.5b05316
Sabra, S., Malmqvist, E., Saborit, A., Gratacós, E., and Gomez Roig, M. D. (2017). Heavy Metals Exposure Levels and Their Correlation with Different Clinical Forms of Fetal Growth Restriction. PLoS One 12 (10), e0185645. doi:10.1371/journal.pone.0185645
Saedi, S., Jafarzadeh Shirazi, M. R., Niazi, A., Tahmasebi, A., and Ebrahimie, E. (2021). Prepubertal Exposure to High Dose of Cadmium Induces Hypothalamic Injury through Transcriptome Profiling Alteration and Neuronal Degeneration in Female Rats. Chemico-Biological Interactions 337, 109379. doi:10.1016/j.cbi.2021.109379
Sariyer, R., De-Simone, F. I., Donadoni, M., Hoek, J. B., Chang, S. L., and Sariyer, I. K. (2017). Alcohol-Mediated Missplicing of Mcl-1 Pre-mRNA Is Involved in Neurotoxicity. Alcohol. Clin. Exp. Res. 41 (10), 1715–1724. doi:10.1111/acer.13474
Shirai, S., Suzuki, Y., Yoshinaga, J., and Mizumoto, Y. (2010). Maternal Exposure to Low-Level Heavy Metals during Pregnancy and Birth Size. J. Environ. Sci. Health A 45 (11), 1468–1474. doi:10.1080/10934529.2010.500942
Su, P.-F., Hu, Y.-J., Ho, I.-C., Cheng, Y.-M., and Lee, T.-C. (2006). Distinct Gene Expression Profiles in Immortalized Human Urothelial Cells Exposed to Inorganic Arsenite and its Methylated Trivalent Metabolites. Environ. Health Perspect. 114 (3), 394–403. doi:10.1289/ehp.8174
Valeri, L., Mazumdar, M. M., Bobb, J. F., Claus Henn, B., Rodrigues, E., Sharif, O. I. A., et al. (2017). The Joint Effect of Prenatal Exposure to Metal Mixtures on Neurodevelopmental Outcomes at 20-40 Months of Age: Evidence from Rural Bangladesh. Environ. Health Perspect. 125 (6), 067015. doi:10.1289/ehp614
Van den Berge, K., and Clement, L. (2021). stageR: Stage-wise Analysis of High Throughput Gene Expression Data in R.
Vaughan, O. R., Maksym, K., Silva, E., Barentsen, K., Anthony, R. V., Brown, T. L., et al. (2021). Placenta-specific Slc38a2/SNAT2 Knockdown Causes Fetal Growth Restriction in Mice. Clin. Sci. (Lond). 135 (17), 2049–2066. doi:10.1042/cs20210575
Wang, G., Tang, W.-Y., Ji, H., and Wang, X. (2021). Prenatal Exposure to Mercury and Precocious Puberty: a Prospective Birth Cohort Study. Hum. Reprod. 36 (3), 712–720. doi:10.1093/humrep/deaa315
Wang, Q., Lu, X., Li, C., Zhang, W., Lv, Y., Wang, L., et al. (2019). Down-regulated Long Non-coding RNA PVT1 Contributes to Gestational Diabetes Mellitus and Preeclampsia via Regulation of Human Trophoblast Cells. Biomed. Pharmacother. 120, 109501. doi:10.1016/j.biopha.2019.109501
Wang, Z., Zhang, Q., Lin, J.-R., Jabalameli, M. R., Mitra, J., Nguyen, N., et al. (2021). Deep post-GWAS Analysis Identifies Potential Risk Genes and Risk Variants for Alzheimer's Disease, Providing New Insights into its Disease Mechanisms. Sci. Rep. 11 (1), 20511. doi:10.1038/s41598-021-99352-3
Whitehead, C. L., Walker, S. P., Ye, L., Mendis, S., Kaitu'u-Lino, T. u. J., Lappas, M., et al. (2013). Placental Specific mRNA in the Maternal Circulation Are Globally Dysregulated in Pregnancies Complicated by Fetal Growth Restriction. J. Clin. Endocrinol. Metab. 98 (3), E429–E436. doi:10.1210/jc.2012-2468
Xu, L., Yokoyama, K., Tian, Y., Piao, F. Y., Kitamura, F., Kida, H., et al. (2011). Decrease in Birth Weight and Gestational Age by Arsenic Among the Newborn in Shanghai, China. Nihon Koshu Eisei Zasshi 58 (2), 89–95.
Keywords: placenta, arsenic, cadmium, birth weight, sQTL, DTU
Citation: Deyssenroth MA, Peng S, Hao K, Marsit CJ and Chen J (2022) Placental Gene Transcript Proportions are Altered in the Presence of In Utero Arsenic and Cadmium Exposures, Genetic Variants, and Birth Weight Differences. Front. Genet. 13:865449. doi: 10.3389/fgene.2022.865449
Received: 29 January 2022; Accepted: 07 April 2022;
Published: 13 May 2022.
Edited by:
Michelle Lacey, Tulane University, United StatesReviewed by:
Jing Chen, Cincinnati Children’s Hospital Medical Center, United StatesCopyright © 2022 Deyssenroth, Peng, Hao, Marsit and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maya A. Deyssenroth, bWsyNTgzQGN1bWMuY29sdW1iaWEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.