- 1Laboratory of Medical Parasitology, Biotechnology, and Biomolecules, Pasteur Institute of Tunis, University Tunis El Manar, Tunis, Tunisia
- 2Faculty of Sciences Bizerte, University of Carthage, Tunis, Tunisia
- 3Laboratory Microorganisms and Active Biomolecules, Faculty of Sciences of Tunis, University Tunis El Manar, Tunis, Tunisia
- 4External Consultants Service Pasteur Institute of Tunis, Tunis, Tunisia
Background: Colorectal cancer (CRC) is a major public health problem worldwide and in Tunisia. It ranks among the main cancers in terms of incidence and cancer-related cause of death. Its pathogenesis is currently considered to be multifactorial involving genetic and environmental factors. Recent studies have suggested that the gene encoding the β1 subunit of the IL-12 receptor, an important pro-inflammatory cytokine of the anti-tumor response, could be involved in the susceptibility to inherited CRC. Hence, it would be interesting to study the role of single nucleotide polymorphisms (SNPs) within the IL-12RB1 gene (rs401502 and rs11575934) in CRC susceptibility.
Aim: Our purpose was to assess whether genetic variants IL-12RB1 +1196G/C (rs401502) and IL-12RB1 +705A/G (rs11575934) within the IL-12RB1 gene are associated with the sporadic CRC risk.
Methods: A total of 110 Tunisian patients with sporadic CRC and 141 healthy control subjects were included in this study. Genotyping was performed by high-resolution melting (HRM) analysis. All results were confirmed by direct DNA sequencing or PCR-RFLP methods. Later, the allele frequencies and genotype distribution were established and compared between the control group and CRC patients.
Results: The obtained results showed that the two target SNPs were in Hardy–Weinberg equilibrium (HWE) in both patients and controls. Minor allele frequencies of rs401502 SNP were 16.4% in CRC cases and 23.8% in controls. Mutant allele of rs11575934 SNP was present with 21.4% in CRC patients and 29.8% in control group. An association study showed a significant association of two target polymorphisms with CRC, according to the dominant genetic model with OR = 0.577, 95% CI = [0.343 to 0.972], p = 0.038 and OR = 0.547, 95% CI = [0.328 to 0.911], p = 0.02, respectively.
Conclusion: In this study, we found, for the first time, a potential protective effect of two SNPs in the IL-12RB1 gene, namely rs401502 and rs11575934, in sporadic colorectal cancer in Tunisians.
Introduction
Colorectal cancer (CRC) is one of the most prevalent gastrointestinal malignancies (Araghi et al., 2019). It is the third most common cancer diagnosed in both men and women and the second leading cause of cancer deaths worldwide in 2020 (Sung et al., 2021). Tunisia is also highly affected by this disease with an increase in the incidence these last years (Khiari et al., 2017). CRC is a complex disease that involves many factors in its tumorigenesis, namely, hereditary, genetic, and environmental factors (Lucas et al., 2017; Behrens et al., 2018; Balhareth et al., 2019; Wong et al., 2019; McNabb et al., 2020). Several studies have been carried out in order to investigate such risk factors and therefore to better understand the mechanisms of its genesis, improve early detection, and thus increase the effectiveness of treatment (Seo and Cairns 2018).
Cytokines play a major role in modulating the immune response and in organizing the control of pathological cells, including cancer cells. These cytokines act on immune cells by modulating their activation, differentiation, and multiplication (Dinarello, 2007). The presence of variations in genes encoding cytokines could impact their expression, structure, regulation, and function, thus promoting the development of pathologies. Therefore, studying the role of genetic variations within genes encoding cytokines is essential to better understand the carcinogenesis process (Stanilova, 2012). Several genetic polymorphisms of genes encoding cytokines and their receptors have been studied for their association with colon carcinogenesis, in particular IL-6, IL1β, IL-1RN, IL-10, IL17, and TNF α (Sun et al., 2015; Bedoui et al., 2018; Ibrahimi et al., 2018; Mirjalili et al., 2018; Shi et al., 2018; Huang et al., 2019; Liu et al., 2020).
Interleukin-12 (IL-12) is a pro-inflammatory cytokine secreted by activated phagocytic and dendritic cells. It plays an important role in promoting Th1-type immune responses and cell-mediated immunity (Trinchieri 1998; Miteva et al., 2009). IL-12 exerts its action by binding to its receptor (IL-12R) with high affinity. This receptor is mainly expressed by T cells and natural killer (NK) cells. It is composed of two distinct subunits IL-12Rβ1 and IL-12Rβ2 and contributes to both IL-12/IL-23 signaling pathways (Presky et al., 1996). The gene encoding IL-12Rβ1 is located on chromosome 19 at position 31.1 (19p31.1), and it has 17 exons (Yamamoto et al., 1997). A total of 70 SNPs have been reported in the IL-12RB1 gene, of which the missense mutation is the most frequent type (van de Vosse et al., 2005). Certain mutations in the IL-12RB1 gene generate an inactive IL-12Rβ1 protein. This deficiency abolishes both IL-12 and IL-23 signaling, which play a crucial role in the cytokine signaling pathways (Núñez-Marrero 2020). In fact, several studies have been carried out to explore the association between IL-12RB1 polymorphisms and certain diseases such as cervical cancer (Hussain et al., 2013), breast cancer (Quan et al., 2014), and tuberculosis (Altare et al., 2001; Remus 2004; de Beaucoudrey et al., 2010; Boisson-Dupuis et al., 2011; Alinejad Dizaj et al., 2018; Zhou et al., 2019). Hence, the IL-12RB1 gene appears interesting as a candidate gene for testing susceptibility to cancers.
Recently, Belhadj and collaborators showed an association between the allele frequencies of IL-12RB1 polymorphisms in patients with hereditary CRC in a family study (Belhadj et al., 2020). These results were based on previous studies that identified IL-12RB1 as a potential candidate gene for its biological function (Chubb et al., 2016a). The rs401502 and rs11575934 SNPs are located in a protein-coding region of the IL-12RB1 gene within exon 10 and exon7, respectively (de Beaucoudrey et al., 2010). These are missense mutations characterized by a substitution of a single nucleotide with another, resulting in an amino acid sequence change (IL-12RB1+1196 G/C; G378R; ID SNP NCBI: rs401502 and IL-12RB1+705 A/G; Q214R; ID SNP NCBI: rs11575934). These mutations lead to the absence of IL-12Rβ1 expression on the surface of activated T and NK cells or, more rarely, they give rise to a non-functional protein (van de Vosse et al., 2013). To the best of our knowledge, there were no studies that involved the association between these two mutations and sporadic colorectal cancer susceptibility. These polymorphisms lead to the absence of the expression of this protein on the surface of activated T and NK cells or, more rarely, they give rise to a non-functional protein (van de Vosse et al., 2013).
In this study, we investigated for the first time the association between two single nucleotide polymorphisms (SNPs) within the IL-12RB1 gene, namely, IL-12RB1 +1196G/C (G378R, NCBI SNP ID: rs401502) and IL-12RB1 +705A/G (Q214R, NCBI SNP ID: rs11575934), and the colorectal cancer susceptibility in a Tunisian cohort.
Materials and Methods
Study Subjects
We performed a case-control association study including 110 Tunisian patients with sporadic CRC and 141 healthy volunteers as a control group. Cases were confirmed by colonoscopy and histology, and the tumors were classified according to the tumor–node–metastasis (TNM) classification. Healthy subjects with previous gastrointestinal problems (inflammatory bowel diseases or others) were excluded from this study. Our study populations were recruited from Salah-Azaïz Institute and from external consultants’ service, Pasteur Institute of Tunis.
All subjects included in this study agreed to participate and signed an informed consent form. All collected data were anonymized, and no personal data were used. Ethical approval was obtained from the Ethics Committee of the Salah-Azaïz Institute of Tunis (reference: ISA/2016/03).
Biological Samples and DNA Extraction
A whole blood sample (5 ml) was collected on an EDTA tube for both study populations. DNA extraction was performed using the salting-out method described by Miller et al. (Miller et al., 1988). The DNA concentration and purity were measured by using the NanoDrop 2000 spectrophotometer (Thermo Fisher). All DNA working concentrations were adjusted to 10 ng/μL.
SNP Genotyping
Two SNPs of interest (rs401502 and rs11575934) were genotyped using the qPCR-HRM method, as described and well-developed in our laboratory (Jelassi et al., 2020).
In order to genotype the IL-12RB1 +1196 G/C (rs401502) polymorphism, a pair of primers was designed using the Primer3 tool (v.0.4.0) (http://frodo.wi.mit.edu/primer3/) (Jelassi et al., 2020). The newly designed primers, forward 5′-GCATTGAATGGCAGCCTGTG-3′ and reverse 5′-GTAATGGCCTGGAATGGCCT-3′, amplify a specific region (107bp) containing the SNP of interest. The obtained genotypes were confirmed by direct sequencing using the same primers (Jelassi et al., 2020).
For the rs11575934 polymorphism, we used the primers described by Lee et al. (2005), 5′-GGTTAAGTGACTGGTGCCAAG-3′ forward and 5′-CTCAAACCACTGGCCTCAAG-3′ reverse, which amplify a 322bp sequence (Lee et al., 2005). The genotyping results were confirmed by PCR-RFLP, using PvuII enzyme (New England BioLabs, Beverly, MA, United States) at 37°C for 16 h.
Statistical Analysis
GraphPad Prism software was used for statistical analysis (Motulsky 2016). Hardy–Weinberg equilibrium was tested among cases and controls using the chi-squared (χ2) test. Logistic regression analysis was accomplished to investigate genotype and allele frequency differences between cases and controls and calculate specific odds ratios (ORs), 95% confidential intervals (CIs), and p values.
Results
Patients’ and Healthy Subjects’ Profile
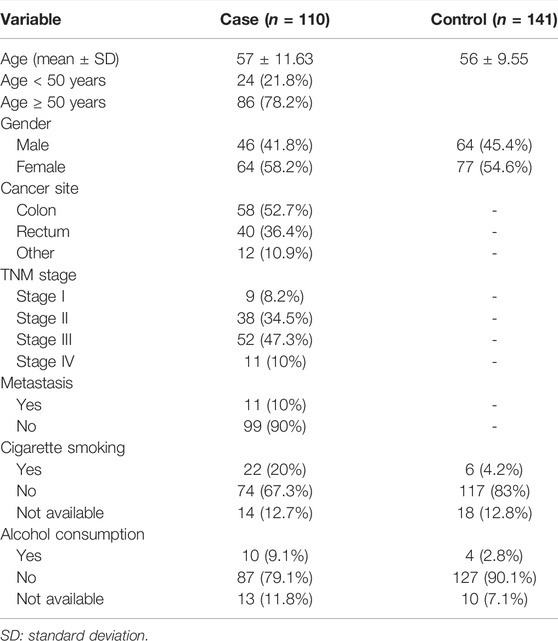
The clinical characteristics of study participants are summarized in Table 1. The sex and age distribution of subjects were very close in the patients and control groups. In fact, men represented 41.8% (46/110) and 45.4% (64/141), respectively, in the two groups, whereas mean ages were 57 ± 11.6 and 56 ± 9.6 years, respectively.
Different stages were observed in our group of patients, of which 8.2% of cases (9/110) were in stage I, 34.5% of cases (38/110) were in stage II, 47.3% of cases (52/110) were in stage III, and 10% of cases (11/110) were in stage IV. Metastasis was reported in 10% of cases (11/110).
The site of the tumor was the colon in 58 patients (52.7%) and the rectum in 40 patients (36.4%), and the 12 (10.9%) other patients’ locations were reported in the sigmoid.
Tobacco and alcohol consumption was low (less than 20%) in both patients and controls.
IL-12RB1+1196G/C Polymorphism
The genotypic and allelic distributions of rs401502 SNP in case and control groups are reported in Table 2. The distribution of rs401502 SNP was in Hardy–Weinberg equilibrium (HWE) in both CRC patients and controls (p = 0.17 and p = 0.06, respectively) (Table 2). The minor allele frequencies (MAF) were 16.4% in CRC cases and 23.8% in control group. The frequencies of IL-12RB1 +1196G/C genotypes in cases and controls were as follows: GG 68.2% and 55.3%, GC 30.9% and 41.9%, and CC 0.9% and 2.8%, respectively (Table 2).
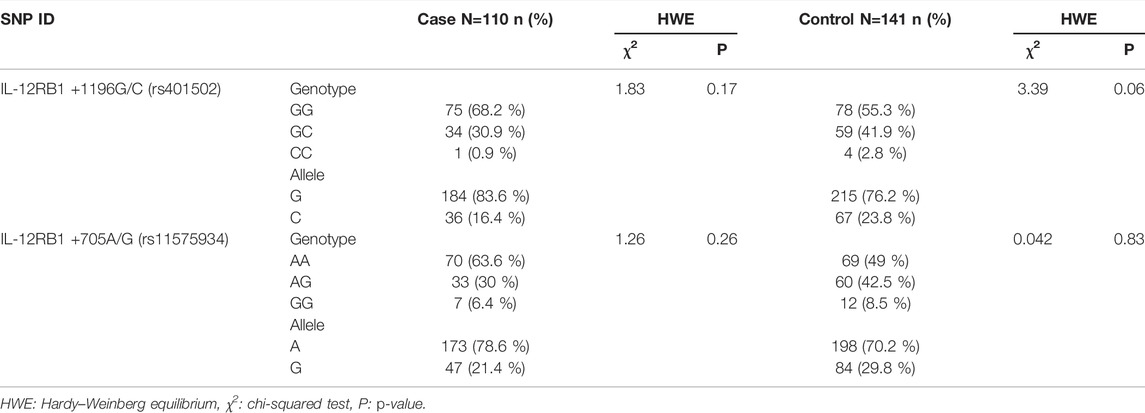
TABLE 2. Hardy–Weinberg test for allelic and genotypic frequencies in patients with CRC and the control group.
An association study of the rs401502 polymorphism with CRC risk showed a significant association in the dominant model (Table 3). In fact, we found that the frequency of the G/G genotype was 55.3% (78/141), and the frequency of G/C + C/C genotypes was 44.7% (63/141) in healthy controls, whereas these rates were 68.2% (75/110) and 31.8% (35/110), respectively, in patients.
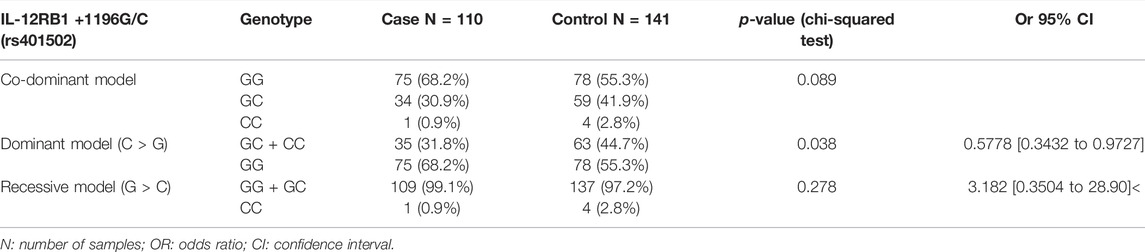
TABLE 3. Allelic and genotypic distributions of the IL12RB1 +1196G/C (rs401502) polymorphism in patients with CRC and controls.
This corresponds to a significant decrease in the risk of CRC with G/C + C/C, with OR = 0.577, 95% CI = [0.343 to 0.972], and p = 0.038. The frequencies of heterozygous and recessive homozygous genotypes (G/C + CC), according to the dominant model, were higher in the control group (44.7%) than those in CRC patients (31.8%), showing a potential protective effect of the rs401502 polymorphism among the Tunisian population.
On the other hand, the genetic co-dominant and recessive models showed no association with CRC risk (p = 0.08 and 0.27, respectively) (Table 3).
IL-12RB1+705A/G Polymorphism
The frequency distribution of the rs11575934 SNP was in Hardy–Weinberg equilibrium (HWE) in both cases and controls (p = 0.26 and p = 0.83, respectively) (Table 2). The minor allele frequencies were 21.4% and 29.8% in CRC patients and the control group, respectively. The IL-12RB1 +705A/G polymorphism frequencies in cases and control groups were as follows: AA 63.6% and 49%, AG 30% and 42.5%, and GG 6.4% and 8.5%.
A significant association of the IL-12RB1 +705A/G polymorphism was observed with CRC cases in the genetic dominant model, as shown in Table 4. The frequency of the A/A genotype was 49.9% (69/141), and the rate of A/G + G/G genotypes was 51.1% (72/141) in healthy controls. In CRC patients, these frequencies were 64.6% (70/110) and 36.4% (40/110). This corresponds to a decreased risk of CRC in association with G/C + C/C with an OR = 0.547, 95% CI = [0.328 to 0.911], and p = 0.02.
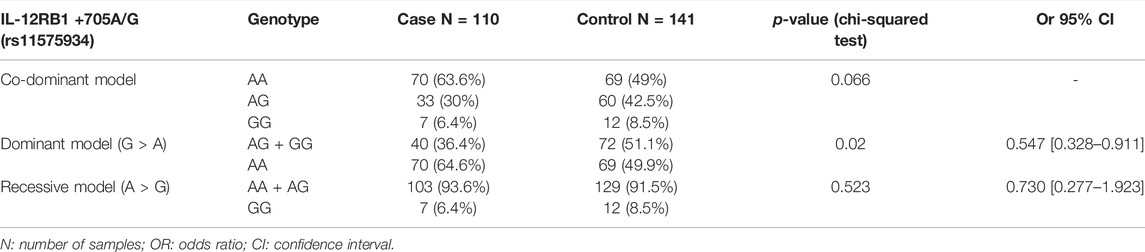
TABLE 4. Allelic and genotypic distributions of the IL-12RB1+705 A/G polymorphism in patients with CRC and controls.
According to the obtained results, the frequencies of the heterozygous and recessive homozygous genotypes, under the genetic dominant model, were higher in control groups (51.1%) than those in cases (36.4%). This could suggest a possible protective effect of the rs11575934 polymorphism. However, the co-dominant and recessive models did not show any significant difference (p = 0.66 and 0.52, respectively).
Discussion
Colorectal cancer (CRC) is a major public health problem worldwide and in Tunisia. Many SNPs, among various genes, either involved in the immune response or the signaling pathway, have been identified as high- or low-risk variations associated with CRC. Cytokines expressed in CRC cells or in the tumor microenvironment seem to play an important role in local immunoregulation (Terzic et al., 2010; Klampfer 2011).
To the best of our knowledge, there are limited studies in a Tunisian population on whether IL-12RB1 gene polymorphisms can affect CRC risk and long-term survival in Tunisian patients with CRC.
Frequencies of rs401502 and rs11575934 SNPs were established for the first time in the general Tunisian population, using the qPCR-HRM technique (Jelassi et al., 2020). In our study, these two SNPs were genotyped by qPCR-HRM in 110 Tunisian patients with sporadic CRC. The case-control study showed a statistically significant difference between these two groups (patients and controls) for the two polymorphisms rs401502 and rs11575934 with OR = 0.577, 95% CI = [0.343 to 0.972], p = 0.038 and OR = 0.547, 95% CI = [0.328 to 0.911], p = 0.02, respectively. Frequencies of the mutant allele the heterozygous and recessive homozygous genotypes for two target SNPs were higher in the control group than those in patients with CRC. This suggests that these mutations did not present a risk factor for sporadic CRC and that they would rather be associated with a potential protective effect. We observed that genetic studies increasingly show that mutations have a potential protective role in certain multifactorial diseases (Yamamoto-Furusho et al., 2011), in particular cancerous pathologies (Tong et al., 2012; Núñez-Marrero 2020). Globally, association studies of these SNPs with sporadic CRC are rare. Our results agree with those already reported by Chubb et al. that have shown the importance of IL-12RB1 as a candidate gene for inherited CRC (Chubb et al., 2016a). Recently, a GWAS study identified three SNPs, c.94C > T (p.Gln32Ter), c.1624C > T (p.Gln542Ter), and c.1237T > C (p.Cys413Arg), linked to inherited CRC in the frame of a family study (Belhadj et al., 2020). These results were based on previous studies that identified IL-12RB1 as a potential candidate gene for hereditary CRC due to its biological importance (Chubb et al., 2016a).
The IL-12RB1 polymorphisms have been explored in other cancers including cervical cancer (Hussain et al., 2013); breast cancer (Quan et al., 2014); esophageal cancer (Tao et al., 2012); and gastric cancer (Vogelaar et al., 2015). The role of mutations in the IL-12RB1 gene in the carcinogenesis process and its consequences is controversial. Núñez-Marrero et al. suggested that certain polymorphisms (such as rs3761041, rs401502, and rs404733 SNPs) increase the risk of breast cancer through a loss-of-function effect, thereby leading to a reduced transduction signal of the IL-12 cytokine. Indeed, certain mutations in the IL-12RB1 gene generate an inactive IL-12Rβ1 protein. This deficit abolishes both IL-12 and IL-23 signaling pathways (Núñez-Marrero 2020). On the other hand, still concerning breast cancer, certain variations (in particular the rs438421 SNP) would be protective by improving the activity of the IL-12R receptor, thus promoting the production of IFN-γ especially in cytotoxic T cells (van de Vosse et al., 2013; Núñez-Marrero 2020). Furthermore, beyond the mutations favorable to the development of the disease or those which would be protective of it, rare variations and other common ones with an unknown functional effect have been reported for IL-12RB1 (van de Vosse et al., 2013). Many studies have investigated the involvement of certain SNPs within genes encoding cytokines and their receptors such as IL-6, IL1β, IL-1RN, IL-10, IL17, and TNFα. Indeed, interleukin-6 (IL-6) and tumor necrosis factor (TNF) are the most widely studied cytokines in CRC and other malignancies. These two cytokines actively participate in the signal transduction of the STAT3 pathway and NF-κB, respectively. These cytokines also promote tumor progression by enhancing proliferation, invasion, and resistance to apoptosis (Uchiyama et al., 2012; West et al., 2015).
Concerning the IL-12RB1 gene, the target SNPs lead to the absence of the IL-12Rβ1 expression on the surface of activated T and NK cells or, more rarely, they give rise to a non-functional IL12RB1 surface protein. This alteration of the protein expression could alter the inflammatory processes, leading to a protective effect through a decrease in chronic inflammation. In all cases, additional proteomics and functional studies are necessary in order to clarify their precise impact.
Conclusion
This study shows for the first time an association between two SNPs in IL-12RB1 gene (rs401502 and rs11575934) and the sporadic CRC, with a potential protective effect. Furthermore, it would be interesting to study the correlation between these polymorphisms and protein expression.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Biomedical Ethics Committee of the Salah-Azaïz Institute. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
RJ, HC, and IZ conceived and designed the study. SD, IZ, and RA collected the samples and related data. RJ, HB, and NM performed the experiments. RJ, HB, NS, and HC analyzed the data. RJ wrote the draft. HC, KA, and AB revised the manuscript. All authors approved the final version of the manuscript submitted.
Funding
This study was supported by the Ministry of Higher Education and Scientific Research, Tunisia, and by the Internal collaborative project PCI-06 of the Pasteur Institute of Tunis.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer KH declared a shared affiliation with the authors SD, HBS, NS to the handling editor at the time of review.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the individuals involved in this study for providing their general information and biological samples.
References
Alinejad Dizaj, M., Mortaz, E., Mahdaviani, S. A., Mansouri, D., Mehrian, P., Verhard, E. M., et al. (2018). Susceptibility to Mycobacterial Disease Due to Mutations in IL-12Rβ1 in Three Iranian Patients. Immunogenetics 70, 373–379. doi:10.1007/s00251-017-1041-3
Altare, F., Ensser, A., Breiman, A., Reichenbach, J., Baghdadi, J. E., Fischer, A., et al. (2001). Interleukin‐12 Receptor β1 Deficiency in a Patient with Abdominal Tuberculosis. J. Infect. Dis. 184, 231–236. doi:10.1086/321999
Araghi, M., Soerjomataram, I., Jenkins, M., Brierley, J., Morris, E., Bray, F., et al. (2019). Global Trends in Colorectal Cancer Mortality: Projections to the Year 2035. Int. J. Cancer 144, 2992–3000. doi:10.1002/ijc.32055
Balhareth, A., Aldossary, M. Y., and McNamara, D. (2019). Impact of Physical Activity and Diet on Colorectal Cancer Survivors' Quality of Life: a Systematic Review. World J. Surg. Onc 17, 153. doi:10.1186/s12957-019-1697-2
Bedoui, S. A., Barbirou, M., Stayoussef, M., Dallel, M., Mokrani, A., Makni, L., et al. (2018). Association of interleukin-17A Polymorphisms with the Risk of Colorectal Cancer: A Case-Control Study. Cytokine 110, 18–23. doi:10.1016/j.cyto.2018.04.017
Behrens, G., Gredner, T., Stock, C., Leitzmann, M. F., Brenner, H., and Mons, U. (2018). Cancers Due to Excess Weight, Low Physical Activity, and Unhealthy Diet. Dtsch Arztebl Int. 115, 578–585. doi:10.3238/arz10.3238/arztebl.2018.0578
Belhadj, S., Terradas, M., Munoz-Torres, P. M., Aiza, G., Navarro, M., Capella, G., et al. (2020). Candidate Genes for Hereditary Colorectal Cancer: Mutational Screening and Systematic Review. Hum. Mutat. 41, 1563–1576. doi:10.1002/humu.24057
Boisson-Dupuis, S., El Baghdadi, J., Parvaneh, N., Bousfiha, A., Bustamante, J., Feinberg, J., et al. (2011). IL-12Rbeta1 Deficiency in Two of Fifty Children With Severe Tuberculosis From Iran, Morocco, and Turkey. PLoS One 6, e18524. doi:10.1371/journal.pone.0018524
Chubb, D., Broderick, P., Dobbins, S. E., Frampton, M., Kinnersley, B., Penegar, S., et al. (2016a). Rare Disruptive Mutations and Their Contribution to the Heritable Risk of Colorectal Cancer. Nat. Commun. 7, 11883. doi:10.1038/ncomms11883
de Beaucoudrey, L., Samarina, A., Bustamante, J., Cobat, A., Boisson-Dupuis, S., Feinberg, J., et al. (2010). Revisiting Human IL-12Rβ1 Deficiency. Medicine (Baltimore) 89, 381–402. doi:10.1097/MD.0b013e3181fdd832
Dinarello, C. A. (2007). Historical Insights Into Cytokines. Eur. J. Immunol. 37 (Suppl 1), S34–S45. doi:10.1002/eji.200737772
Huang, X., Qin, S., Liu, Y., Tao, L., and Jiang, H. (2019). Associations of Tumor Necrosis Factor-α Polymorphisms with the Risk of Colorectal Cancer: a Meta-Analysis. Biosci. Rep. 39, BSR20181750. doi:10.1042/BSR20181750
Hussain, S. K., Madeleine, M. M., Johnson, L. G., Du, Q., Galloway, D. A., Daling, J. R., et al. (2013). Nucleotide Variation in IL-10 and IL-12 and Their Receptors and Cervical and Vulvar Cancer Risk: A Hybrid Case-Parent Triad and Case-Control Study. Int. J. Cancer 133, 201–213. doi:10.1002/ijc.28000
Ibrahimi, M., Moossavi, M., Mojarad, E. N., Musavi, M., Mohammadoo-khorasani, M., and Shahsavari, Z. (2018). Positive Correlation between Interleukin-1 Receptor Antagonist Gene 86bp VNTR Polymorphism and Colorectal Cancer Susceptibility: a Case-Control Study. Immunol. Res. 67, 151–156. doi:10.1007/s12026-018-9034-3
Jelassi, R., Ben Salah, H., Jouini, A., Hedhli, A., Mahmoud, W., Ammi, R., et al. (2020). High-resolution Melting to Identify Single Nucleotide Polymorphisms of IL-12 Receptor B Gene (IL-12RB1) in the Tunisian Population. Ijhg 20, 32–40. doi:10.31901/24566330.2020/20.01.747
Khiari, H., Ben Ayoub, H. W., Ben Khadhra, H., and Hsairi, M. (2017). Colorectal Cancer Incidence Trend and Projections in Tunisia (1994–2024). Asian Pac. J. Cancer Prev. 18, 2733–2739. doi:10.22034/APJCP.2017.18.10.2733
Klampfer, L. (2011). Cytokines, Inflammation and Colon Cancer. Ccdt 11, 451–464. doi:10.2174/156800911795538066
Lee, H. W., Lee, H. S., Kim, D. K., Ko, D. S., Han, S. K., Shim, Y.-S., et al. (2005). Lack of an Association between Interleukin-12 Receptor β1 Polymorphisms and Tuberculosis in Koreans. Respiration 72, 365–368. doi:10.1159/000086249
Liu, Y., Li, B., Wang, L., and Kong, D. (2020). A Functional Polymorphism in the Promoter Region of Interleukin-12B Increases the Risk of Colorectal Cancer. Biomed. Res. Int. 2020, 1–6. doi:10.1155/2020/2091781
Lucas, C., Barnich, N., and Nguyen, H. (2017). Microbiota, Inflammation and Colorectal Cancer. Ijms 18, 1310. doi:10.3390/ijms18061310
McNabb, S., Harrison, T. A., Albanes, D., Berndt, S. I., Brenner, H., Caan, B. J., et al. (2020). Meta‐analysis of 16 Studies of the Association of Alcohol with Colorectal Cancer. Int. J. Cancer 146, 861–873. doi:10.1002/ijc.32377
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A Simple Salting Out Procedure for Extracting DNA from Human Nucleated Cells. Nucl. Acids Res. 16, 1215. doi:10.1093/nar/16.3.1215
Mirjalili, S. A., Moghimi, M., Aghili, K., Jafari, M., Abolbaghaei, S. M., Neamatzadeh, H., et al. (2018). Association of Promoter Region Polymorphisms of Interleukin-10 Gene with Susceptibility to Colorectal Cancer: a Systematic Review and Meta-Analysis. Arq. Gastroenterol. 55, 306–313. doi:10.1590/S0004-2803.201800000-66
Miteva, L., Stanilov, N., Deliysky, T., Mintchev, N., and Stanilova, S. (2009). Association of Polymorphisms in Regulatory Regions of Interleukin-12p40 Gene and Cytokine Serum Level with Colorectal Cancer. Cancer Invest. 27, 924–931. doi:10.3109/07357900902918486
Núñez-Marrero, A., Arroyo, N., Godoy, L., Rahman, M. Z., Matta, J. L., and Dutil, J. (2020). SNPs in the Interleukin-12 Signaling Pathway Are Associated with Breast Cancer Risk in Puerto Rican Women. Oncotarget 11, 3420–3431. doi:10.18632/oncotarget.27707
Presky, D. H., Yang, H., Minetti, L. J., Chua, A. O., Nabavi, N., Wu, C. Y., et al. (1996). A Functional Interleukin 12 Receptor Complex is Composed of Two B-Type Cytokine Receptor Subunits. Proc. Natl. Acad. Sci. USA 93, 14002–14007. doi:10.1073/pnas.93.24.14002
Quan, L., Gong, Z., Yao, S., Bandera, E. V., Zirpoli, G., Hwang, H., et al. (2014). Cytokine and Cytokine Receptor Genes of the Adaptive Immune Response are Differentially Associated With Breast Cancer Risk in American Women of African and European Ancestry. Int. J. Cancer 134, 1408–1421. doi:10.1002/ijc.28458
Remus, N., Chentoufi, M., El Baghdadi, J., Schurr, E., Fieschi, C., Feinberg, J., et al. (2004). Association of IL12RB1 Polymorphisms With Pulmonary Tuberculosis in Adults in Morocco. J. Infect. Dis. 190, 580–587. doi:10.1086/422534
Seo, M. K., and Cairns, J. (2018). Do cancer Biomarkers Make Targeted Therapies Cost-Effective? A Systematic Review in Metastatic Colorectal Cancer. PLoS One 13, e0204496. doi:10.1371/journal.pone.0204496
Shi, X., Jia, Y., Xie, X., and Li, S. (2017). Single-nucleotide Polymorphisms of the IL-12 Gene lead to a Higher Cancer Risk: a Meta-Analysis Based on 22,670 Subjects. Genes Genet. Syst. 92, 173–187. doi:10.1266/ggs.16-00024
Stanilova, S. (2012). “Cytokine Gene Polymorphisms in Colorectal Cancer,” in Colorectal Cancer Biology–From Genes to Tumor. Editors R. Ettarh , 59–78.
Sun, R., Jia, F., Liang, Y., Li, L., Bai, P., Yuan, F., et al. (2015). Interaction Analysis of IL-12A and IL-12B Polymorphisms with the Risk of Colorectal Cancer. Tumor Biol. 36, 9295–9301. doi:10.1007/s13277-015-3685-7
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tao, Y.-P., Wang, W.-L., Li, S.-Y., Zhang, J., Shi, Q.-Z., Zhao, F., et al. (2012). Associations Between Polymorphisms in IL-12A, IL-12B, IL-12Rβ1, IL-27 gene and Serum Levels of IL-12p40, IL-27p28 With Esophageal Cancer. J. Cancer Res. Clin. Oncol. 138, 1891–1900. doi:10.1007/s00432-012-1269-0
Terzić, J., Grivennikov, S., Karin, E., and Karin, M. (2010). Inflammation and Colon Cancer. Gastroenterology 138, 2101–2114. doi:10.1053/j.gastro.2010.01.058
Tong, Z., Yang, X. O., Yan, H., Liu, W., Niu, X., Shi, Y., et al. (2012). A Protective Role by Interleukin-17F in Colon Tumorigenesis. PLoS One 7, e34959. doi:10.1371/journal.pone.0034959.g001
Trinchieri, G. (1998). Proinflammatory and Immunoregulatory Functions of Interleukin-12. Int. Rev. Immunol. 16, 365–396. doi:10.3109/08830189809043002
Uchiyama, T., Takahashi, H., Endo, H., Sakai, E., Hosono, K., Nagashima, Y., et al. (2012). IL-6 Plays Crucial Roles in Sporadic Colorectal Cancer through the Cytokine Networks Including CXCL7. Jct 03, 874–879. doi:10.4236/jct.2012.326112
van de Vosse, E., de Paus, R. A., van Dissel, J. T., and Ottenhoff, T. H. M. (2005). Molecular Complementation of IL-12Rβ1 Deficiency Reveals Functional Differences between IL-12Rβ1 Alleles Including Partial IL-12Rβ1 Deficiency. Hum. Mol. Genet. 14, 3847–3855. doi:10.1093/hmg/ddi409
van de Vosse, E., Haverkamp, M. H., Ramirez-Alejo, N., Martinez-Gallo, M., Blancas-Galicia, L., Metin, A., et al. (2013). IL-12Rβ1 Deficiency: Mutation Update and Description of the IL12RB1 Variation Database. Hum. Mutat. 34, 1329–1339. doi:10.1002/humu.22380
Vogelaar, I. P., van der Post, R. S., van de Vosse, J. H., Hoogerbrugge, N., Ligtenberg, M. J., Gomez Garcia, E., et al. (2015). Gastric Cancer in Three Relatives of a Patient With a Biallelic IL12RB1 Mutation. Fam. Cancer 14, 89–94. doi:10.1007/s10689-014-9764-x
West, N. R., McCuaig, S., Franchini, F., and Powrie, F. (2015). Emerging Cytokine Networks in Colorectal Cancer. Nat. Rev. Immunol. 15, 615–629. doi:10.1038/nri3896
Wong, M. C., Ding, H., Wang, J., Chan, P. S., and Huang, J. (2019). Prevalence and Risk Factors of Colorectal Cancer in Asia. Intest Res. 17, 317–329. doi:10.5217/ir.2019.00021
Yamamoto, K., Kobayashi, H., Miura, O., Hirosawa, S., and Miyasaka, N. (1997). Assignment of IL12RB1 and IL12RB2, Interleukin-12 Receptor β1 and β2 Chains, to Human Chromosome 19 Band p13.1 and Chromosome 1 Band p31.2, Respectively, by In Situ Hybridization. Cytogenet. Genome Res. 77, 257–258. doi:10.1159/000134589
Yamamoto-Furusho, J. K., Álvarez-León, E., Fragoso, J. M., Gozalishvilli, A., Vallejo, M., and Vargas-Alarcón, G. (2011). Protective Role of Interleukin-19 Gene Polymorphisms in Patients with Ulcerative Colitis. Hum. Immunol. 72, 1029–1032. doi:10.1016/j.humimm.2011.08.013
Keywords: colorectal cancer, IL-12RB1, rs401502, rs11575934, single nucleotide polymorphism (SNP)
Citation: Jelassi R, Dhouioui S, Ben Salah H, Saidi N, Mzoughi N, Ammi R, Bouratbine A, Aoun K, Zidi I and Chelbi H (2022) rs401502 and rs11575934 Polymorphisms of the IL-12 Receptor Beta 1 Gene are Protective Against Colorectal Carcinogenesis. Front. Genet. 13:864419. doi: 10.3389/fgene.2022.864419
Received: 28 January 2022; Accepted: 11 April 2022;
Published: 13 May 2022.
Edited by:
Mahmood Rasool, King Abdulaziz University, Saudi ArabiaReviewed by:
Kamel Hamzaoui, Tunis El Manar University, TunisiaRukhsana Nawaz, United Arab Emirates University, United Arab Emirates
Jakaria Jakaria, Bogor Agricultural University, Indonesia
Copyright © 2022 Jelassi, Dhouioui, Ben Salah, Saidi, Mzoughi, Ammi, Bouratbine, Aoun, Zidi and Chelbi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanen Chelbi, Y2hlbGJpX2hhQHlhaG9vLmZy
 Refka Jelassi
Refka Jelassi Sabrine Dhouioui3
Sabrine Dhouioui3 Hamza Ben Salah
Hamza Ben Salah Nasreddine Saidi
Nasreddine Saidi Nabiha Mzoughi
Nabiha Mzoughi Karim Aoun
Karim Aoun Ines Zidi
Ines Zidi Hanen Chelbi
Hanen Chelbi