- 1Laboratório de Recursos Genéticos, Programa de Pós Graduação Em Biotecnologia, Universidade Federal de São João Del Rei, São João Del Rei, Brazil
- 2Departamento de Ciência da Computação, Universidade Federal de São João Del Rei, São João Del Rei, Brazil
- 3Laboratório de Recursos Genéticos, Departamento de Zootecnia, Universidade Federal de São João Del Rei, São João Del Rei, Brazil
The Neotropical region bears the most diverse freshwater fish fauna on the planet and is the stage for dramatic conservation struggles. Initiatives aiming for conservation of a single emblematic fish, a flagship species, to which different onlookers relate on a cultural/personal level, holds promise towards engagement and conservation actions benefiting whole biological communities and ecosystems. Here, we present the first comprehensive genomic resources for Salminus brasiliensis, a potential flagship Neotropical species. This fish faces pressing conservation issues, as well as taxonomic uncertainty, being a main species relevant to angling and commercial fisheries. We make available 178 million Illumina paired-end reads, 90 bases long, comprising 16 Gb (≈15X coverage) of filtered data, obtained from a primary genomic library of 500-bp fragments. We present the first de novo genomic assembly for S. brasiliensis, with ∼1 Gb (N50 = 10,889), as well as the coding genome annotation of 12,962 putative genes from assembled genomic fragments over 10 kb, most of which could be identified from the Ostariophysi GenBank database. We also provide a genome-wide panel for more than 80,000 predicted microsatellite loci for low-cost, fast and abundant DNA marker development for this species. A total of 47, among 52 candidates, empirically assayed microsatellites were confirmed as polymorphic in this fish. All genomic data produced for S. brasiliensis is hereby made publicly accessible. With the disclosure of these results, we intend to foster general biology studies and to provide tools to be applied immediately in conservation and aquaculture in this candidate flagship Neotropical species.
Introduction
The Neotropical region bears the most diverse freshwater fish fauna on the planet, apparently as the result of rapid speciation process (Melo et al., 2021). Freshwater systems and fish biodiversity face ever-increasing impacts from human activity, a global issue that often does not muster as much societal awareness (Su et al., 2021) as other pressing conservation challenges such as accelerated rates of deforestation and normative deregulation (Gonçalves et al., 2020; Gonçalves-Souza et al., 2021). Associated with its particularly rich fish fauna, South America harbors some of the largest river basins on Earth and it is the stage for dramatic conservation struggles involving conflicting stakeholder concerns. The degree of cumulative anthropogenic degradation of its aquatic systems is worrisome, as epitomized by two catastrophic mine-waste spills caused by obsolete tailing dam failures in Brazil in 2015 (Fernandes et al., 2016) and 2019 (Vergilio et al., 2020), leading to severe effects upon freshwater biodiversity (e.g., Gomes et al., 2019), in addition to social disruption and loss of human life (Polignano and Lemos, 2020).
Conservation initiatives aiming a single emblematic species, to which large numbers of diverse onlookers can relate on a personal or social level, holds promise towards engagement and successful actions for the benefit of whole biological communities and ecosystems (e.g., Leader-Williams et al., 1990; Dietz et al., 1994). For different taxonomic groups and geographic locations, some more well-known, culturally or economically important species can act as flagship species (e.g., Ochieng et al. 2021; Preston et al., 2021). Still, among those characterized as the 20 most charismatic species, none is a fin fish (Albert et al., 2021), and a recent assessment in Brazil noted only one Amazonian teleost fish, Araipamas gigas, as a flagship species, among 62 elected taxa (Wosnick et al., 2021). In this regard, the singular Neotropical migratory fish fauna, also known as piracema fish (Carolsfeld et al., 2004), are under-represented, especially given their prominent social-environmental importance, fragile conservation status and general public awareness.
One particular migratory species arguably fits the flagship bill better than most: Salminus brasiliensis Cuvier 1816 (Characiformes, Bryconidae), also known as dourado, dorado, pirayú, saipé, pirajuba, dama and picudo among other common names throughout South America (Froese and Pauley, 2000). It is an emblematic, large (females may reach as much as 26 kg), long-distance swimmer, top-predator with formidable aesthetic appeal (Figure 1). It also enjoys wide recognition from the general public and has established importance in angling, artisanal and commercial fisheries (Feitosa et al., 2004; Gagne et al., 2017). It commonly occurs in the La Plata River Basin, a vast hydrographic system which spans the Paraná, Paraguay and Uruguay Rivers and also in the Patos Lagoon in southern Brazil, being native to a total of five countries, including Argentina, Bolivia, Paraguay and Uruguay. It also has been targeted for aquaculture due to its favorable growth rate, size and gastronomic quality (Crescêncio et al., 2005; Zaniboni-Filho and Schulz, 2003; Zaniboni-Filho et al. 2017), despite being a voracious piscivorous species, which could pose challenges to commercial cultivation. Salminus brasiliensis is spawned mainly for stock supplementation purposes, along efforts to mitigate the environmental impact of hydroelectric dams upon migratory fish. It has suffered a sharp reduction of natural stocks also because of removal of the riparian zone along rivers, land and water pollution, introduction of invasive species and overfishing. The species has not yet been included, however, in the Red Book of Threatened Species (Rosa and Lima, 2008), mainly given its relatively high abundance in the Pantanal region of the Paraguay Basin. Yet, it has been considered vulnerable in the Paraná Basin (Marques et al., 2002; Abilhoa, 2004) and highly vulnerable in the La Plata River (Zayas and Cordiviola, 2007). It also has been classified as virtually extinct, with population size below a viable threshold, in some major Paraná Basin locations, such as the Grande, Tietê and Paranapanema Rivers (Rosa and Lima, 2008).

FIGURE 1. Salminus brasiliensis in its natural habitat, in Bonito, MS, Brazil (Author: André Seale).
Rosso et al. (2012) observed the differentiation among S. brasiliensis stocks from Argentina’s Pampa Plain and other parts of South America. According to Machado et al. (2017), the species displays distinct haplogroups, with the Upper Paraná containing a distinct Evolutionary Significant Unit from the remaining La Plata River Basin. Thus, there are taxonomic uncertainties associated with this species and further work is needed to inform its conservation and management plans.
Genetic and genomic resources can be applied to resolve conservation and taxonomic issues. So far, eight polymorphic microsatellites were described for this species by Rueda et al. (2011) and another 47 by our own group (in Cao et al., 2016) from next-generation sequencing (NGS) data first fully disclosed herein. Brandão-Dias et al. (2014) provided the first cytoplasmic genomic resources for S. brasiliensis with publication of its assembled and annotated mitogenome (17,721 bp long). Here, we present the first comprehensive genomic resources for Salminus brasiliensis, contributing to basic molecular genetics knowledge, and to resolution of conservation and taxonomic challenges.
Materials and Methods
DNA Extraction and Sequencing
The genomic DNA sample from S. brasiliensis was obtained from a single female specimen from the Itutinga Hatchery at Grande River, in the Upper Paraná Basin, located in Minas Gerais–Brazil (21°17′040″S, 44°37′023″W). Following euthanasia, the fish was held on in ice during transport to the laboratory, where a muscle tissue sample was removed and placed in liquid nitrogen until DNA extraction. The individual was kept as a voucher (LARGE1305471). All procedures were authorized by UFSJ’s Ethics Committee on Animal Research (CEUA-UFSJ); specimen collection was conducted under SISBIO license number 37222 and genetic patrimony access was granted through license CGEN - A9D0E51. No experimentation on live animals was conducted in this research. DNA extraction, library preparation and NGS steps were conducted as presented in Yazbeck et al. (2018). In summary, a single primary library of random genomic fragments (∼500 bp) was sequenced using an Illumina HiSeq 2000, generating 90-bp paired-end short reads. Raw data was filtered for quality and removal of duplicates and stored as two corresponding FASTQ files.
Bioinformatics
We verified FASTQ quality values with FastQC (ver. 0.11.09–Andrews, 2010) and we used KmerGenie (Chikhi and Medvedev, 2014) to assess k values from 41 to 61, in order to search which k-mer would result in a more diverse dataset. Then, different preliminary de novo genomic assemblies were attempted from the generated short reads, assaying best k-mer values with Minia (Chikhi and Rizk, 2012). The chosen k-mer was used for a de novo assembly using SOAPdenovo 2 (Luo et al., 2012). Assembly qualities were inferred by search for single-copy conserved core eukaryotic ortholog genes with BUSCO (Benchmarking universal Single-Copy Orthologs) (Simão et al., 2015), using the Zebrafish, Danio rerio, database (BuscoDB). The procedure to map perfect-repeat microsatellite loci (potential candidates for new molecular markers) in the chosen assembly was also conducted according to the methodology detailed in Yazbeck et al. (2018). In summary, an unknown genomic assembly performed with these short reads was used for potential microsatellite loci mining by a service provider. We used the data produced by this step, namely predicted PCR products, as queries to be searched in the new chosen assembly, using BLAST (Boratyn et al., 2013). The filtered short reads were mapped onto the chosen assembly using the SOAP aligner, and the resulting file was converted into BAM format with the aid of SAMtools (Li et al., 2009).
Based on an estimate of the average eukaryotic gene sequence length of 12,000 bp (Cooper and Hausman, 2016), due to the fragmented nature of the assemblies and computational power restrictions, we proceeded to perform functional annotation of contigs and scaffolds larger than 10,000 bases only, using the MAKER pipeline (Cantarel et al., 2008) using the Characiformes database2 (peptide sequences and expressed sequence tags) from NCBI as a training set. Annotated sequences were then characterized by BLAST, from the superorder Ostariophysi data available from NCBI3 and functionally classified with the aid of PANTHER (Mi et al., 2010). The mitogenome was obtained according to the steps presented in Yazbeck et al. (2014).
Results
The library sequencing resulted in a pair of twin FASTQ files consisting of 90-bp paired-end reads amounting to around 16 Gb of filtered data, with 97.4% exhibiting PHRED Q quality values of 20 or more (1% or less of wrong base calls). The average quality value per read was Q = 38 and CG content was 41.05%. This data is now available at NCBI’s Sequence Read Archive (SRA) (Supplementary Material S1-under accession number PRJNA792751).
Even though k-mer analysis indicated k = 41 as a best candidate, three different assembly attempts were assayed, with k = 41, 47 and 55, using MINIA and SOAPdenovo 2. Table 1 shows the number of complete core eukaryotic genes retrieved from each assembly, with the k = 41 assembly performing less well than k = 47 (98 and 194 complete genes, respectively). Both attempts performed with SOAPdenovo 2 (k = 47 and 55) resulted in 0.92 and 0.96 Gb assemblies (Table 2), with the former presenting a superior N50 value (10,889). Thus, the k = 47 assembly was selected for the annotation process and used as the basis for microsatellite mapping. It consisted of 560,090 contigs or scaffolds varying from 100 to 134135 bases (average 1,643) (Supplementary Material S2). Only 28 scaffolds were larger than 100 kb, 24,106 were larger than 10 kb, and 115,292 larger than 1 kb. This assembly showed a CG content of 41.25%.

TABLE 1. Results from the search of a set of 303 core eukaryotic orthologous genes in alternative assemblies performed with different k-mer values, according to BUSCO.
An undisclosed assembly with these short reads by a service provider, followed by microsatellite search, revealed 86,832 potential microsatellite loci. These loci were searched with BLAST using the predicted PCR products and mapped in our new chosen k = 47 assembly, and the results are available in Supplementary Table S1. Around 76% of predicted loci could be found with a 100% match (66,206 microsatellites), another ∼18% (14,527) were found with similarity of 90% or above and ∼0.9% 742) with similarity within the 77–89% range. Around 6% of microsatellite loci determined from the unknown assembly (5,357) could not be mapped back on the assembly we utilized, including 13 loci empirically validated (out of 47) as polymorphic markers (Cao et al., 2016).
The MAKER prediction pipeline resulted in the annotation of 12,962 genes from around 485 Mb of analysed sequences from S. brasiliensis, with gene, exon and intron average sizes of 6,125, 194 and 760 bp, respectively. These data were consolidated as a general feature format file (GFF3), with detailed description of coding sequences (CDS), hereby made available through FigShare (Supplementary Material S3). The genes found then were searched through BLAST against the Ostariophysi databank from NCBI, allowing the characterization of 10,211 genes (Supplementary Table S2), which were functionally classified according to Gene Ontology regarding their potential biological role, resulting in the elucidation of function of 3,245 S. brasiliensis genes (Figure 2).
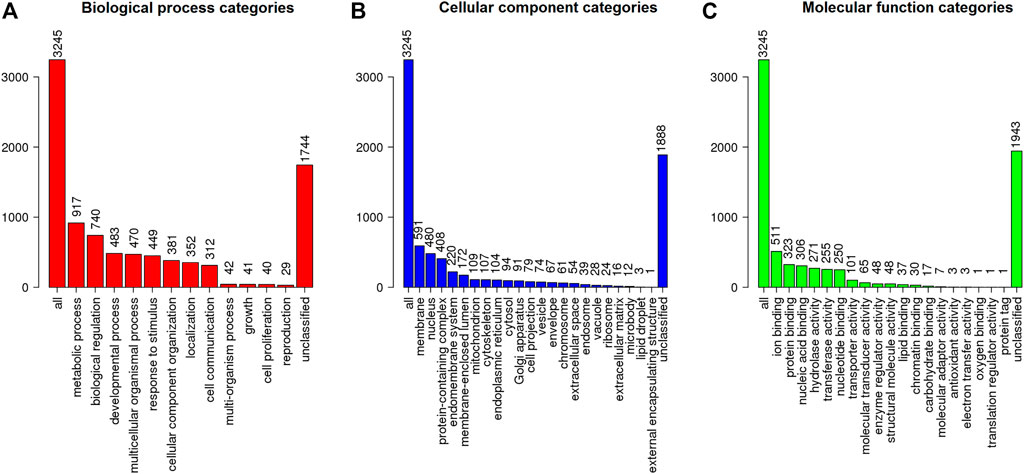
FIGURE 2. Functional classification (according to Gene Ontology) regarding the (A) biological process, (B) cellular component, and (C) molecular function for predicted genes from S. brasiliensis.
The sequenced individual’s mitogenome is deposited in GenBank under accession KY825190 and amounts to 17,998 bp.
Discussion
Genomics can produce powerful tools for fish conservation (Bernos et al., 2020), and we here provide the first comprehensive genomic data on S. brasiliensis. This study generated a massive set of short DNA reads with associated base-call quality scores, which is now publicly available at NCBI’s Sequence Read Archive, allowing its application by other research groups. This data set, even though of low genomic coverage, is considered of high quality (average of one error per each 6,250 bp). It allowed the exploration of different de novo genomic assemblies, which showed that the adoption of different k-mer parameters resolve different genomic regions, as judged by complete core eukaryotic genes analysis. We make available one assembly (k = 47), while very fragmented, as shown by its N50 and short contigs/scaffolds, it still allowed the annotation of over 12,000 protein-encoding sequences, whereas only 13 mitochondrial genes were previously available (Brandão-Dias et al., 2014). Among arbitrary examples of fully annotated genes from this assembly is the Leucine rich repeat containing 10 gene (LRRC10) involved in heart-tissue regeneration in Astyanax mexicanus (Stockdale et al., 2018) and a gonadotrophin-releasing hormone (GnRH) gene, which could support biotechnological development for induced hatchery spawning of this species, as it is involved in gonadal maturation. This functional annotation is made available as a GFF3 file. The number of annotated genes found (almost 13,000) leads us to believe that the presented assembly resolves approximately 40–50% of this species’ genome, judging from comparison with completely characterized genomes from other fishes (e.g. 30,741 coding sequences from Danio rerio - Howe et al., 2013).
We also provided a panel of tens of thousands of candidate microsatellite loci, which can serve as a starting point for fast and inexpensive molecular marker development in this species, allowing development of marker loci for stock delimitation, linkage mapping, QTL characterization and association studies. Previously, Cao et al. (2016) validated 47 polymorphic microsatellite loci from this panel, attesting to its utility for efficient molecular marker development. Some microsatellite loci determined from a genomic assembly produced with the short reads presented here, which however could not be traced back to the k = 47 assembly, were among these empirically validated markers. We thus vouch for the retention of the non-mapped loci, since it has been shown to produce practical results. There is inherent difficulty in resolving repetitive regions with short reads, which explains the varying results from assembly to assembly. Furthermore, among the more than 5,000 loci not found, it can be shown with other alignment tools (e.g. Swipe–Rognes, 2011), that many microsatellite loci are indeed present in different repetition resolutions, searching and matching primer pairs (results not shown), since BLAST has limited power working with low-complexity sequences, such as those present in microsatellite motifs.
The mitogenome of the sequenced specimen showed a larger size (277 bp) than the one previously published (Brandão-Dias et al., 2014), with extra DNA at the D-loop region, allowing rapid identification of putative variable sites to be explored in population genetics and other studies.
Together, the results presented here have the potential to be applied in conservation initiatives and taxonomic investigation of this important large fish, a candidate Neotropical migratory flagship species, aiding resolution of ongoing scientific and environmental issues faced by this organism. It will certainly aid future telomere-to-telomere genome characterization of S. brasiliensis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/, PRJNA792751, SRR17407720; https://www.ncbi.nlm.nih.gov/sra/, SRR17486808; https://figshare.com/, 10.6084/m9.figshare.18131495; https://figshare.com/, 10.6084/m9.figshare.11796468; https://figshare.com/, 10.6084/m9.figshare.17754188; https://figshare.com/, 10.6084/m9.figshare.19169756.
Ethics Statement
The animal study was reviewed and approved by the Comissão de Ética no Uso de Animais, Universidade Federal de São João Del Rei.
Author Contributions
RG processed and analyzed NGS data and results, planned and conducted bioinformatic analysis, conducted quality control of data and results and wrote the manuscript. RO mobilized computational infrastructure, developed pipeline scripts, planned and conducted bioinformatic analysis. IS supported and conducted bioinformatic analysis. GY supervised the project, planned the NGS approach, secured funding, directed DNA extraction, conducted bioinformatic analysis, analyzed results and co-wrote the manuscript. All authors have approved the final version of the manuscript.
Funding
This work was developed with resources from The Brazilian National Council for Scientific and Technological Development (CNPq-308124/2020-0), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES - Code 001), Companhia Energética de Minas Gerais and The Brazilian Electricity Regulatory Agency (CEMIG-ANEEL - GT345) and Minas Gerais State Agency for Research and Development (FAPEMIG - APQ-04569-10).
Conflict of Interest
GY filled for a patent request involving certain combinations of specific microsatellites, from the set previously described in Cao et al. (2016), at INPI (Instituto Nacional da Propriedade Industrial), Brazil (BR1020180128558), to be used as PCR multiplexes in commercial genotyping operations. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank UFSJ (Pró Reitoria de Pesquisa e Pós Graduação, Programa de Pós Graduação em Biotecnologia and Programa de Pós Graduação em Ciência da Computação), CNPq, FAPEMIG and CAPES. We also thank José Mauro Ribeiro for conducting mtDNA assembly and Fausto Moreira da Silva Carmo for helping in collecting and conducting DNA extraction from the sequenced specimen. We also acknowledge André and Lucia Seale for their courtesy in providing a specimen photograph.
Footnotes
1https://www.ufsj.edu.br/large.
2https://www.ncbi.nlm.nih.gov/protein/?term=txid7991%5bOrganism:exp%5d.
3https://www.ncbi.nlm.nih.gov/nuccore/?term=txid32519[Organism:exp].
References
Abilhoa, V. E. L. F. (2004). “Duboc. Peixes,” in Livro vermelho da fauna ameaçada no Estado do Paraná. Editors S. B. Mikich, and R. S. Bérnils (Curitiba: Instituto Ambiental do Paraná), 581–677.
Albert, J. S., Destouni, G., Duke-Sylvester, S. M., Magurran, A. E., Oberdorff, T., Reis, R. E., et al. (2021). Scientists' Warning to Humanity on the Freshwater Biodiversity Crisis. Ambio 50, 85–94. doi:10.1007/s13280-020-01318-8
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed January 29, 2022).
Bernos, T. A., Jeffries, K. M., and Mandrak, N. E. (2020). Linking Genomics and Fish Conservation Decision Making: a Review. Rev. Fish. Biol. Fish. 30, 587–604. doi:10.1007/s11160-020-09618-8
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a More Efficient Report with Usability Improvements. Nucleic Acids Res. 41, W29–W33. doi:10.1093/nar/gkt282
Brandão-Dias, P. F. P., Carmo, A. O. d., Martins, A. P. V., Pimenta, R. J. G., Alves, C. B. M., and Kalapothakis, E. (2014). Complete Mitochondrial Genome of Salminus brasiliensis (Characiformes, Characidae). Mitochondrial DNA, 1–2. doi:10.3109/19401736.2014.958676
Cantarel, B. L., Korf, I., Robb, S. M. C., Parra, G., Ross, E., Moore, B., et al. (2008). MAKER: An Easy-To-Use Annotation Pipeline Designed for Emerging Model Organism Genomes. Genome Res. 18, 188–196. doi:10.1101/gr.6743907
Cao, Y.-L., Caputo, L. I., Cheng, H., da Silva Carmo, F. M., de Carvalho, L. C., Yazbeck, G. M., et al. (2016). Microsatellite Records for Volume 8, Issue 3. Conservation Genet. Resour. 8, 359–370. doi:10.1007/s12686-016-0581-4
Carolsfeld, J., Harvey, B., Ross, C., and Anton, B. (2004). Migratory Fishes of South America: Biology, Fisheries and Conservation Status. Ottawa: IDRC and the World Bank.
Chikhi, R., and Medvedev, P. (2014). Informed and Automated K-Mer Size Selection for Genome Assembly. Bioinformatics 30, 31–37. doi:10.1093/bioinformatics/btt310
Chikhi, R., and Rizk, G. (2012). “Space-efficient and exact de Bruijn graph representation based on a Bloom filter,” in Algorithms in Bioinformatics Lecture Notes in Computer Science. Editors B. Raphael, and J. Tang (Berlin, Heidelberg: Springer), 236–248. doi:10.1007/978-3-642-33122-0_19
Cooper, G. M., and Hausman, R. E. (2016). A Célula - 3ed: Uma Abordagem Molecular. Porto Alegre: Artmed.
Crescêncio, R., Ituassú, D. R., Roubach, R., Pereira Filho, M., Cavero, B. A. S., and Gandra, A. L. (2005). Influência Do período de alimentação no consumo e ganho de peso Do pirarucu. Pesq. Agropec. Bras. 40, 1217–1222. doi:10.1590/S0100-204X2005001200009
Dietz, J. M., Dietz, L. A., and Nagagata, E. Y. (1994). “The Effective Use of Flagship Species for Conservation of Biodiversity: the Example of Lion Tamarins in Brazil,” in Creative Conservation. Editors P. J. S. Olney, G. M. Mace, and A. T. C. Feistner (Dordrecht: Springer Netherlands), 32–49. doi:10.1007/978-94-011-0721-1_2
Fernandes, G. W., Goulart, F. F., Ranieri, B. D., Coelho, M. S., Dales, K., Boesche, N., et al. (2016). Deep into the Mud: Ecological and Socio-Economic Impacts of the Dam Breach in Mariana, Brazil. Natureza & Conservação 14, 35–45. doi:10.1016/j.ncon.2016.10.003
Froese, R., and Pauley, D. (2000). FishBase 2000: Concepts, Design and Data Sources. Los Baños, Laguna, Philippines: ICLARM.
Gagne, T. O., Ovitz, K. L., Griffin, L. P., Brownscombe, J. W., Cooke, S. J., and Danylchuk, A. J. (2017). Evaluating the Consequences of Catch-And-Release Recreational Angling on golden Dorado (Salminus Brasiliensis) in Salta, Argentina. Fish. Res. 186, 625–633. doi:10.1016/j.fishres.2016.07.012
Gomes, L. C., Chippari-Gomes, A. R., Miranda, T. O., Pereira, T. M., Merçon, J., Davel, V. C., et al. (2019). Genotoxicity Effects on Geophagus Brasiliensis Fish Exposed to Doce River Water after the Environmental Disaster in the City of Mariana, MG, Brazil. Braz. J. Biol. 79, 659–664. doi:10.1590/1519-6984.188086
Gomes, L. C., Feitosa, L. A., Fernandes, R., Da Costa, R. S., and Agostinho, A. A. (2004). Parâmetros populacionais e simulação Do rendimento por recruta de Salminus brasiliensis (Cuvier, 1816) Do alto rio Paraná. Acta Sci. Biol. Sci. 26, 317–323. doi:10.4025/actascibiolsci.v26i3.1593
Gonçalves-Souza, D., Vilela, B., Phalan, B., and Dobrovolski, R. (2021). The Role of Protected Areas in Maintaining Natural Vegetation in Brazil. Sci. Adv. 7, eabh2932. doi:10.1126/sciadv.abh2932
Gonçalves, P. R., Di Dario, F., Petry, A. C., Martins, R. L., da Fonseca, R. N., Henry, M. D., et al. (2020). Brazil Undermines parks by Relocating Staff. Science 368, 1199. doi:10.1126/science.abc8297
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The Zebrafish Reference Genome Sequence and its Relationship to the Human Genome. Nature 496, 498–503. doi:10.1038/nature12111
Leader-Williams, N., Harrison, J., and Green, M. J. B. (1990). Designing Protected Areas to Conserve Natural Resources. Sci. Prog. 74, 189–204.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an Empirically Improved Memory-Efficient Short-Read De Novo Assembler. GigaSci 1, 18. doi:10.1186/2047-217X-1-1810.1186/2047-217X-1-18
Machado, C. D. B., Ishizuka, T. K., Freitas, P. D. D., Valiati, V. H., and Galetti, P. M. (2017). DNA Barcoding Reveals Taxonomic Uncertainty in Salminus (Characiformes). Syst. Biodiversity 15, 372–382. doi:10.1080/14772000.2016.1254390
Marques, A. A. B., Fontana, C. S., Vélez, E., Bencke, G. A., Schneider, M., and dos Reis, R. E. (2002). Lista das Espécies da Fauna Ameaçadas de Extinção no Rio Grande Do Sul. Porto Alegre: FZB/MCT–PUCRS/PANGEA, 52p. (Publicações Avulsas FZB, nº11). Available at: http://www.tecniflora.com.br/fauna_ameacada_RS.pdf (Accessed January 29, 2022).
Melo, B. F., Sidlauskas, B. L., Near, T. J., Roxo, F. F., Ghezelayagh, A., Ochoa, L. E., et al. (2021). Accelerated Diversification Explains the Exceptional Species Richness of Tropical Characoid Fishes. Syst. Biol. 71, 78–92. doi:10.1093/sysbio/syab040
Mi, H., Dong, Q., Muruganujan, A., Gaudet, P., Lewis, S., and Thomas, P. D. (2010). PANTHER Version 7: Improved Phylogenetic Trees, Orthologs and Collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210. doi:10.1093/nar/gkp1019
Polignano, M. V., and Lemos, R. S. (2020). Rompimento da barragem da Vale em Brumadinho: impactos socioambientais na Bacia Do Rio Paraopeba. Cienc. Cult. 72, 37–43. doi:10.21800/2317-66602020000200011
Preston, S. D., Liao, J. D., Toombs, T. P., Romero-Canyas, R., Speiser, J., and Seifert, C. M. (2021). A Case Study of a Conservation Flagship Species: the Monarch Butterfly. Biodivers Conserv 30, 2057–2077. doi:10.1007/s10531-021-02183-x
Rognes, T. (2011). Faster Smith-Waterman Database Searches with Inter-sequence SIMD Parallelisation. BMC Bioinformatics 12, 221. doi:10.1186/1471-2105-12-221
Rosa, R. S., and Lima, F. C. (2008). Os peixes brasileiros ameaçados de extinção. Livro vermelho da fauna brasileira ameaçada de extinção 2, 9–275.
Rosso, J. J., Mabragaña, E., González Castro, M., and Díaz de Astarloa, J. M. (2012). DNA Barcoding N Eotropical Fishes: Recent Advances from the P Ampa P Lain, A Rgentina. Mol. Ecol. Resour. 12, 999–1011. doi:10.1111/1755-0998.12010
Rueda, E. C., Amavet, P., Brancolini, F., Sommer, J., and Ortí, G. (2011). Isolation and Characterization of Eight Polymorphic Microsatellite Markers for the Migratory Characiform Fish, Salminus Brasiliensis. J. Fish Biol. 79, 1370–1375. doi:10.1111/j.1095-8649.2011.03109.x
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 31, 3210–3212. doi:10.1093/bioinformatics/btv351
Stockdale, W. T., Lemieux, M. E., Killen, A. C., Zhao, J., Hu, Z., Riepsaame, J., et al. (2018). Heart Regeneration in the Mexican Cavefish. Cell Rep. 25, 1997–2007. e7. doi:10.1016/j.celrep.2018.10.072
Su, G., Logez, M., Xu, J., Tao, S., Villéger, S., and Brosse, S. (2021). Human Impacts on Global Freshwater Fish Biodiversity. Science 371, 835–838. doi:10.1126/science.abd3369
Tobias Ochieng, N., Elizabeth, K. N., and Nigel, L.-W. (2021). Measuring the Conservation Attitudes of Local Communities towards the African Elephant Loxodonta africana, a Flagship Species in the Mara Ecosystem. PLOS ONE 16, e0253234. doi:10.1371/journal.pone.0253234
Vergilio, C. d. S., Lacerda, D., Oliveira, B. C. V. d., Sartori, E., Campos, G. M., Pereira, A. L. d. S., et al. (2020). Metal Concentrations and Biological Effects from One of the Largest Mining Disasters in the World (Brumadinho, Minas Gerais, Brazil). Sci. Rep. 10, 5936. doi:10.1038/s41598-020-62700-w
Wosnick, N., Leite, R. D., Giareta, E. P., Nunes, A. R. O. P., Nunes, J. L. S., Charvet, P., et al. (2021). Evaluating Conservation Status and Governmental Efforts towards Regional Flagship Species in Brazil. J. Environ. Manage. 292, 112732. doi:10.1016/j.jenvman.2021.112732
Yazbeck, G. M., Oliveira, R. S., Ribeiro, J. M., Carmo, F. M. S., and Carvalho, M. B. (2016). First complete mitochondrial genome for any anostomid fish: Leporinus piavussu, a recently described piracema species. Mitochondrial DNA Part A 27, 2293–2294. doi:10.3109/19401736.2014.987239
Yazbeck, G. M., Oliveira, R. S., Ribeiro, J. M., Graciano, R. D., Santos, R. P., Carmo, F. M. S., et al. (2018). A Broad Genomic Panel of Microsatellite Loci from Brycon orbignyanus (Characiformes: Bryconidae) an Endangered Migratory Neotropical Fish. Sci. Rep. 8, 8511. doi:10.1038/s41598-018-26623-x
Zaniboni-Filho, E., Ribolli, J., Hermes-Silva, S., and Nuñer, A. P. O. (2017). Wide Reproductive Period of a Long-Distance Migratory Fish in a Subtropical River, Brazil. Neotrop. Ichthyol. 15, 1. doi:10.1590/1982-0224-20160135
Zaniboni-Filho, E., and Schulz, U. H. (2003). “Migratory Fishes of the Uruguay River,” in Migratory Fishes of the South America: Biology, Social Importance and Conservation Status (Victoria: World Fisheries Trust), 157–194.
Zayas, M. A., and Cordiviola, E. (2007). The conservation state of Characidaefish (Pisces: Characiformes) in anareaofthe Plata basin, Argentina estado de conservación de pecesCharacidae (Pisces: Characiformes) enunárea de lacuenca del plata, argentina. Gayana 71 (2), 178–186. doi:10.4067/s0717-65382007000200006
Keywords: bioinformatics, sequencing-by-synthesis, population genetics resources, hatchery, environmental management, characiformes
Citation: Graciano RCD, Oliveira RS, Santos IM and Yazbeck GM (2022) Genomic Resources for Salminus brasiliensis. Front. Genet. 13:855718. doi: 10.3389/fgene.2022.855718
Received: 15 January 2022; Accepted: 18 February 2022;
Published: 28 March 2022.
Edited by:
Tony Silveira, Federal University of Rio Grande, BrazilReviewed by:
Eric M. Hallerman, Virginia Tech, United StatesCarolina Machado, Federal University of São Carlos, Brazil
Copyright © 2022 Graciano, Oliveira, Santos and Yazbeck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriel M. Yazbeck, ZG5hQHVmc2ouZWR1LmJy
 Raissa Cristina Dias Graciano
Raissa Cristina Dias Graciano Rafael Sachetto Oliveira
Rafael Sachetto Oliveira Isllas Miguel Santos3
Isllas Miguel Santos3 Gabriel M. Yazbeck
Gabriel M. Yazbeck