- Department Of Ophthalmology, The People’s Hospital Of Guangxi Zhuang Autonomous Region and Institute Of Ophthalmic Diseases, Guangxi Academy Of Medical Sciences, Nanning, Guangxi, China
Th17 and regulatory T cells (Tregs) play crucial roles in the pathogenesis of autoimmune diseases. Th17/Treg homeostasis is critically involved in maintaining the immune balance. Disturbed Th17/Treg homeostasis contributes to the progression of autoimmune diseases. MicroRNAs (miRNAs) have emerged as a new vital factor in the regulation of disturbed Th17/Treg homeostasis. To better understand the epigenetic mechanisms of miRNAs in regulating Treg/Th17 homeostasis, we included and evaluated 97 articles about autoimmune diseases and found that miRNAs were involved in the regulation of Treg/Th17 homeostasis from several aspects positively or negatively, including Treg differentiation and development, Treg induction, Treg stability, Th17 differentiation, and Treg function. Uveitis is one of the ocular autoimmune diseases, which is also characterized with Th17/Treg imbalance. However, our understanding of the miRNAs in the pathogenesis of uveitis is elusive and not well-studied. In this review, we further summarized miRNAs found to be involved in autoimmune uveitis and their potential role in the regulation of Th17/Treg homeostasis.
Introduction
Disturbed Th17/Treg homeostasis in autoimmune diseases and uveitis
Multiple studies have demonstrated that disturbed Th17/Treg homeostasis plays a pivotal role in the progression of many autoimmune diseases, such as rheumatoid arthritis (RA) (Wang et al., 2012), systemic lupus erythematosus (SLE) (Alunno et al., 2012), and autoimmune uveitis. Th17 cells are defined as effector T lymphocytes (Teffs) that contribute to inflammation development. They could produce pro-inflammatory cytokines, such as interleukin (IL)-17A, IL-17F, IL-21, IL-22, and TNF-a, which are pathogenic in various autoimmune diseases (Sutton et al., 2009). Overall, the high frequency and elevated function of Th17 can induce inflammation. Treg cells (Tregs) are CD4+CD25 + cells that express the transcription factor fork-head box P3 (FoxP3), which controls critical transcriptional programs for Treg cells’ function. They could regulate immune responses and maintain self-tolerance (Wan and Flavell, 2008; Fujio et al., 2010). The loss of Tregs or their function results in fatal autoimmune diseases in mice and humans (Sakaguchi et al., 2020). To summarize, Th17 cells contribute to active autoimmunity and autoimmune disease progression by secreting IL-17A and other pro-inflammatory cytokines, while Tregs control active autoimmunity and establish self-tolerance by inhibiting effector T-lymphocyte proliferation and suppressing their function (Naqvi et al., 2021; Zhou et al., 2007).
There is a consensus of opinion that the disruption of the balance between pathogenic Th17 and Tregs would result in the breakdown of self-tolerance and contribute to autoimmune disease development (Paust and Cantor, 2005; Tang and Bluestone, 2006). Similarly, maintaining Th17/Treg balance is considered a key factor for the treatment strategy of autoimmune diseases, including uveitis. Autoimmune uveitis is also a CD4+ T cell-mediated autoimmune disease (Zhou et al., 2007). It has been demonstrated that decreased frequency and diminished function of Treg cells would contribute to the inflammation progress in an animal model of experimental autoimmune uveitis (EAU). Moreover, the inflammation was significantly alleviated by shifting the imbalance with induced Tregs. Furthermore, it also had been verified that the disturbed Th17/Treg balance was closely associated with active uveitis such as Vogt-Koyanagi-Harada disease (VKH) and Behcet’s disease (BD) (Chen et al., 2008; Nanke et al., 2008). However, the regulation mechanism of the disturbed Treg/Th17 balance is still unclear.
MicroRNAs are critically involved in the regulation of Th17/Treg homeostasis
Recently, microRNAs (miRNAs) have emerged as critical regulators in Th17/Treg homeostasis. miRNAs are a novel group of small ncRNA molecules of 19–24 nucleotides (nt) in length that participate in the post-transcriptional regulation of gene expression mostly by pairing with 3′UTR of their mRNA targets and inhibition of its translation (Sato et a., 2011). Both innate and adaptive immunities are highly regulated at the post-transcriptional level with miRNA interference (Tsitsiou and Lindsay, 2009; Wei et al., 2020). Multiple studies showed that miRNAs are pivotal regulators involved in autoimmune diseases and can be used as an epigenetic regulation in immune response.
Studies showed that the miRNA network had a profound role within T-cell biology, including T-cell differentiation, T-cell proliferation, cytokine secretion, and Treg cell function. To gain functional evidence of the role of miRNAs in Treg/Teff biology, researchers have used the CD4-Cre Dicer deletion mouse models to analyze the development and function of T cells. Cobb et al. (2006) indicated that depleting some miRNAs by eliminating Dicer reduced the number of peripheral Treg cells. Chong et al. (2008) found that the lack of miRNAs resulted in a two- to three-fold decrease in the frequency of Tregs (Zhou et al., 2008). In addition, some studies with animal models reported that some miRNAs were involved in the progression of autoimmune diseases by regulating Foxp3 in Treg cells (Liston et al., 2008; Zheng et al., 2007). Overall, these studies have clearly demonstrated that the immunosuppressive mechanism in Treg cells was controlled by miRNAs. However, although this critical role has been firmly demonstrated, key mechanisms of the miRNA function in regulating Treg cell development and function remain elusive.
The potential role of miRNAs in the pathogenesis of uveitis
Autoimmune uveitis is an intraocular autoimmune eye disorder which is characterized with immune-mediated damage in the uveal, vascular, and retina tissues. The progressive damage of photoreceptors caused by autoreactive T cells eventually leads to irreversible visual impairment and even blindness (Caspi, 2010). It was estimated that it accounts for 10%–25% of blindness globally (Gritz and Wong, 2004). Although its pathogenesis is comprehensive and still not clear, immunological abnormality, especially Th17/Treg homeostasis, is widely considered as the pivotal factor for its etiology. Similarly, miRNAs have also been investigated for their roles in uveitis pathobiology as biomarkers or therapeutic targets. However, the detailed mechanisms of miRNAs for the abnormal immune system in uveitis still need to be further explored.
In the review, the miRNAs involved in Th17/Treg homeostasis and their regulatory network have been summarized first. We aimed to integrate the studies on individual miRNAs into a more global understanding of the function of the miRNAs in the regulatory network of Treg/Th17 balance. Moreover, considering the critical role of disturbed Th17/Treg homeostasis in uveitis pathology, we then reviewed the possible miRNAs in uveitis physiology by regulating Treg/Th17 hemostasis, aiming to improve the mechanistic understanding of miRNA biology in uveitis.
Methods
Search strategy and literature search
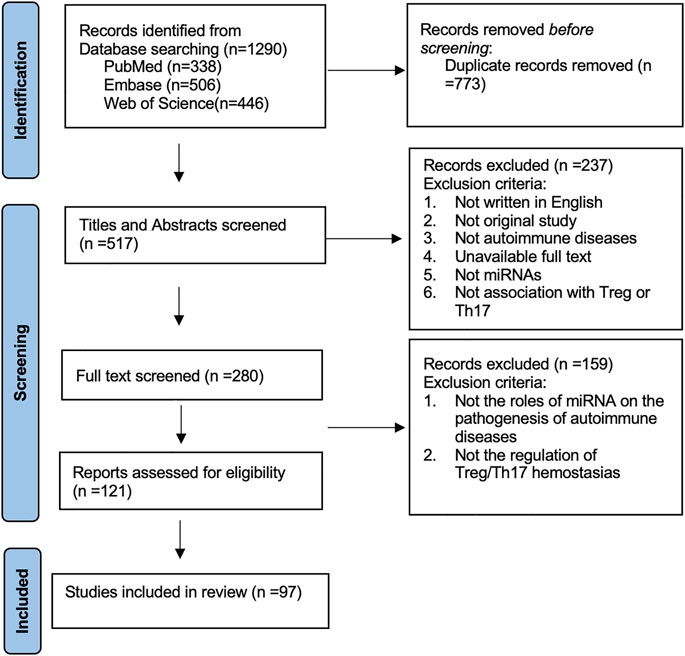
This study was conducted according to the Preferred Reporting Items for Systematic Reviews statement. We conducted a search of journal articles from several databases published from 2010 until the present day, including PubMed, Web of Science, and Embase. The terms chosen were autoimmune inflammation OR autoimmune disease and microRNA* OR miRNA* OR miR* and Th17 OR Treg. Initially, 1,290 articles were recovered. Then, duplications of articles were removed, and after removing 773 duplicates, the articles’ abstract and title were assessed subsequently. In total, 280 articles were subjected to full-text screening. Based on full-text screening and eligibility assessment, 97 articles were included into the systematic review eventually. The roles of miRNAs in the pathogenesis of autoimmune diseases by regulating Th17/Treg homeostasis were reviewed and summarized. The exclusion criteria are shown in the PRISMA flow diagram. The systematic review process is shown in Figure 1.
Moreover, we searched human or animal studies about the miRNAs in the pathogenesis of uveitis by regulating Th17 or Treg cells. Terms chosen were uveitis and microRNA* OR miRNA* OR miR* and Th17 OR Treg. The exclusion criteria were as follows: 1) Full text unavailable; 2) not written in English. Finally, eight animal studies about experimental autoimmune uveitis (EAU) and nine human studies about Behçet’s disease and Vogt-Koyanagi-Harada (VKH) syndrome were included for further review. The following data were extracted: first name of the author, year of publication, study species, miRNA, and the potential role of miRNA in the Th17/Treg homeostasis.
Results
miRNAs involved in Th17/Treg homeostasis by regulating Treg differentiation and development
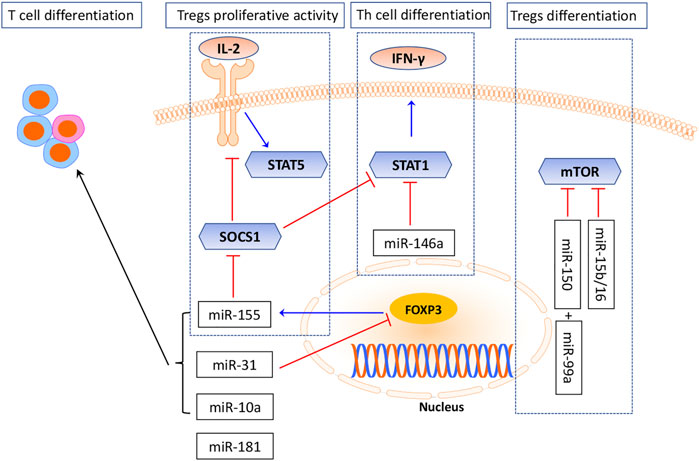
Several miRNAs had been demonstrated to regulate Treg differentiation and development, particularly miR-155 and miR-146a (Rodriguez et al., 2007). miR-155 is processed from an exon of non-coding RNA. Its expression was greatly upregulated in active T cells (Eis et al., 2005; van den Berg et al., 2003). The critical role of miR-155 in the differentiation of T cells and its subtype Tregs was obtained from studies in miR-155-knockout mice, which failed to develop a protective response after immunization (Thai et al., 2007). The miR-155-knockout mice were found to display a bias toward T helper 2 (Th2) differentiation, indicating that miR-155 promotes differentiation into Th1 cells (Naqvi et al., 2021). Furthermore, miR-155 was also involved in regulating Treg cell differentiation, maintenance, and function by targeting FoxP3 (Lu et al., 2009; Mahesh and Biswas, 2019), which binds to an intron within the DNA sequence encoding the miR-155 precursor mRNA and thereby maintaining the high levels of miR-155 expression in Tregs (Heyn et al., 2016; Hippen et al., 2018). The number of Tregs in the thymus and periphery is decreased significantly if miR-155 is downregulated (Kohlhaas et al., 2009). Moreover, Lu et al. (2009) demonstrated that Foxp3-dependent miR-155 sustained IL-2R signaling by targeting the suppressor of cytokine signaling 1 (SOCS1) protein. In the absence of miR-155, increased amounts of SOCS1 attenuate the IL-2R pathway, leading to reduced activating transcription factor signal transducer and the activator of transcription 5 (STAT5) phosphorylation and reduced competitive fitness. Conclusively, as is shown in Figure 2, these studies suggested a positive role of miR-155 in sustaining Treg proliferative activity and numbers via its inhibition on SOCS1, a negative regulator of the IL-2 pathway (Yao et al., 2012; Yin et al., 2018). However, several studies have demonstrated that SOCS1 is necessary for the functions of Tregs and SOCS1 knockout in Tregs would lose Foxp3 expression, which seems controversial with the role of miR-155. Nevertheless, like a double-edged sword, SOCS1 may have negative or positive effects under different circumstances.
Another key miRNA is miR-146a. Its deficiency would result in an increased number of Treg cells and could cause a defect in their immunosuppressive response (Holmstrom et al., 2017; Tang et al., 2009).miR-146a was mainly expressed in Treg cells and affected Treg cell’s ability to suppress the Th1 response by targeting the signal transducer and activator of transcription 1(STAT1), which is an important factor for Th1 differentiation (Lu et al., 2010). STAT1 is a key transcription factor in the IFN-γ response (Lu et al., 2010). In addition, SOCS1 is also a negative regulator of STAT1; STAT1 activation with ablation of SOCS1 resulted in a similar Th1-mediated pathology and differentiation (Lu et al., 2009; Lu et al., 2010).
Except that, there are some other miRNAs which are critically involved in Treg differentiation and development. As is shown in Figure 2, miR-31 was shown to inhibit T-cell differentiation into Tregs by binding to a potential target site within 3’ UTR of FoxP3 mRNA and downregulating its synthesis (Allantaz et al., 2012). miR-10a negatively regulates the plasticity of peripheral Tregs by differentiating them into T follicular helper (Tfh) cells (Sethi et al., 2013; Wu et al., 2019). mTOR exerts a critical role in inhibiting Treg differentiation via miRNA mediation (Delgoffe et al., 2009; Warth et al., 2015). Studies showed that miR-99 and miR-150 work in concert to repress mTOR levels. miR-99 represses the expression of mTOR by directly binding to 3′UTR of its mRNA. miR-150-mediated silencing of mTOR was observed only when co-expressed with miR-99a, suggesting a functional synergy between miRNAs (Warth et al., 2015).
To understand the miRNA function in Treg development, Yogesh Singh and his colleagues searched for important miRNAs and their relevant target genes. miR-15b/16, miR-24, and miR-29a were found to impact induced Tregs (iTregs) in vitro via the mTOR signaling pathway (Singh et al., 2015; Singh et al., 2015). miR-181 also played an important role in T-cell development, while the mechanisms remain only partially explored (Li et al., 2007; Neilson et al., 2007). miR-181 is a family composed of six miRNAs, miR-181a/b-1, miR-181a/b-2, and miR-181c/d. Investigations suggested that miR-181a overexpression augmented T-cell receptor (TCR) signaling strength by targeting several protein tyrosine phosphatases, which displayed negative regulatory functions in TCR signaling (Li et al., 2007). In the absence of miR-181a/b-1, TCR signals are insufficient to produce Foxp3+precursors. Nevertheless, it has been suggested that miR-181 acts as an important regulator of T-cell development (Kim et al., 2021).
miRNAs involved in Th17/Treg homeostasis by regulating Treg induction and stability
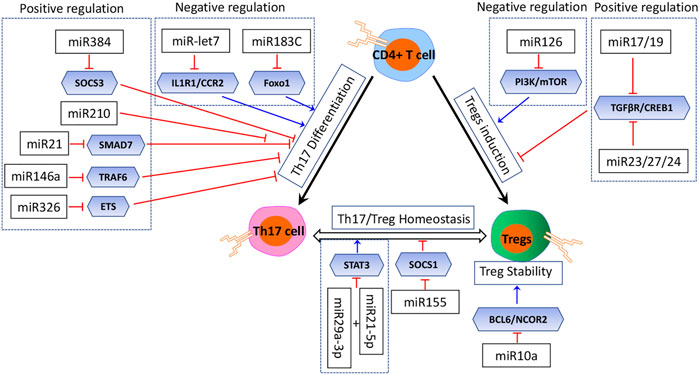
Treg induction in vivo is most likely controlled by multiple miRNAs, which might act in concert or in an isolated manner (Figure 3) (Curotto de Lafaille and Lafaille, 2009). The in vitro Treg induction in the presence of TGFβ is also dependent on proper regulation by miRNAs (Cobb et al., 2006; Chong et al., 2008). Multiple studies had identified miRNAs with both positive and negative regulatory effects on Treg induction (Warth et al., 2015). The PI3K/Akt/mTOR pathway with several miRNAs forms a network which positively regulates Treg induction cooperatively. Qin reported that miR-126 could upregulate Treg induction by inhibiting the activity of PI3K/Akt/mTOR, while in the absence of miR126, the activity of the PI3K/Akt/mTOR pathway was enhanced, and Foxp3 expression and Treg induction were decreased (Qin et al., 2013). On the other hand, several miRNAs have a negative effect on Treg induction (Warth et al., 2015). For example, miR17 and miR19, members of the miR17∼92 cluster, function as negative regulators of Treg induction while being dispensable for thymic Treg development (Jiang et al., 2011). miR17 directly targets TGFβ-receptor II and the cAMP-responsive element-binding protein 1 (CREB1), both involved in proper Treg induction. The TGFβ signaling pathway is also a target of the miR23-miR27-miR24 cluster, and an overexpression of this cluster impairs Treg induction (Cho et al., 2016). miR-31 was also reported to negatively regulate the Treg induction by inhibiting retinoic acid-inducible protein 3, which was validated in an experimental autoimmune encephalomyelitis (EAE) model in mice (Zhang et al., 2015).
The essential role of miRNAs in Treg stability had also been highlighted by the Treg-specific ablation in mouse models; these miRNAs deficiency resulted in disturbed Treg stability (Cobb et al., 2006; Liston et al., 2008; Zhou et al., 2008). As described before, miR-155 is highly expressed in Tregs (Naqvi et al., 2021). miR155 deficiency not only resulted in a reduced frequency of Tregs and impaired Treg development but also resulted in disturbed Treg stability by targeting SOCS1, a negative regulator of STAT5 signaling which determines the responsiveness to IL2 (Kohlhaas et al., 2009; Lu et al., 2009; Yao et al., 2012). miR-10a is a Treg-specific miRNA and had been identified to stabilize the Treg-specific gene expression by targeting effector T-cell genes Bcl6 and Ncor2 (Jeker et al., 2012; Takahashi et al., 2012). Also, miR-10a controls the stability of FoxP3 expression in peripherally derived and in vitro-induced Tregs (Takahashi et al., 2012). Takahashi showed that miR-10a was induced synergistically with TGF-β and retinoic acid in conventional CD4+ T cells and was required for the stability of FoxP3 expression rather than the function of Tregs (Takahashi et al., 2012). However, Jeker et al. (2012) suggested that miR-10a may be indispensable for Treg stabilization and function. To summarize, miR-155 had been identified as one of the most predominant miRNAs in Treg induction, while it is still controversial that whether miR-10a is involved in Treg induction.
miRNAs involved in Th17/Treg homeostasis by regulating Th17 differentiation
Th17 differentiation is also regulated tightly by miRNAs (Figure 3) (Ghoreschi et al., 2010; Lee et al., 2012). Naïve CD4+ T cells are differentiated to the Th17 phenotype with the treatment of TGF-β, IL-6, and IL-1 (Ivanov et al., 2006; Yang et al., 2008; Korn et al., 2009). One potential mechanism for the fine tuning of this balance is miRNAs; however, the roles of miRNAs that intrinsically control Th17 differentiation remain elusive. Based on the current investigation, several miRNAs were reported to play an inhibitory role. Ichiyama et al. (2016) reported five upregulated miRNAs (miR-183, miR-96, miR-182, miR-10b, and miR-351) in Th17 cells compared with the other Th subsets by RNA sequencing, and the upregulated miR-183C cluster (miR-183, miR-96, and miR-182) with its direct target (Foxo1) could suppress Th17 differentiation by RORγt. Additionally, a study by Angelou demonstrated that let-7 miRNAs negatively regulated Th17 proliferation and differentiation via directly targeting the cytokine receptors Il1r1, Il23r, Ccr2, and Ccr5 (Angelou et al., 2019). Another study by Wu et al., (2018) reported that the overexpression of miR-210 in psoriasis had a negative effect on Th17 differentiation.
However, they also had some miRNAs which played an inducing role in Th17 differentiation (Figure 3). Qu and his colleagues revealed that miR-384 was critically involved in EAE. They had also identified that miR-384-favored naïve T cells toward Th17 differentiation in vitro by targeting the suppressor of cytokine signaling 3 (SOCS3) (Liu et al., 2015; Qu et al., 2012) Hosseini et al. (2016) showed that miR-223 could modulate chemokine signaling to promote Th17 differentiation and suppress Treg differentiation, highlighting its potential role in maintaining the Treg/Th17 balance. A study by Howard L.Weiner had characterized that miR-21 was involved in a T-cell–intrinsic miRNA pathway that enhanced TGF-β signaling and then promoted Th17 differentiation by targeting SMAD-7, a negative regulator of TGF-β signaling (Murugaiyan et al., 2015). Furthermore, the essential role of miR-21 was highlighted in a mouse model of EAE. miR-21 expression was elevated in Th17 cells, and the mice lacking miR-21 had a defect in Th17 differentiation and were resistant to EAE; except that, miR-146a was demonstrated to be involved in Th17 differentiation via targeting TRAF6 (Li et al., 2017; Li et al., 2019). miR-326 also impacts Th17 differentiation by targeting the ETS transcription factor family (Du et al., 2009). Additionally, J. Zhou identified that miR-29a-3p and miR-21-5p had a synergistic effect on STAT3 inhibition and regulated Treg/Th17 cells, therefore inducing an imbalance (Zhou et al., 2018).
Furthermore, Kamran Ghaedi developed a network of autoimmune-deregulated miRNAs in Th17 differentiation. Several miRNAs and their downstream regulators involved in Th17 differentiation have been discovered by using the integrative miRWalk database. They nominated several miRNAs which probably may have a strong possibility for inducing an inhibitory role in Th17 differentiation, respectively (positive miRNAs include miR-27b, miR-27a, miR-30c, miR-1, miR-141, and miR-20b; negative miRNAs include miR-93, miR-20a, miR-152, miR-21, and miR-106a) (Honardoost et al., 2015).
miRNAs involved in Th17/Treg homeostasis by regulating Treg function and cytokine secretion
Evidence suggests that miRNAs played a crucial role in cytokine secretion and Treg function in autoimmune diseases.(Valencia et al., 2006; Bayry et al., 2007; Nadkarni et al., 2007; Buckner, 2010). Here, we summarize these critical miRNAs involved in Treg function. It had been elucidated that miR-155 and miR-146a are two essential regulators in regulating Treg cell function in many autoimmune diseases. miR-155 was required for the release of cytokine production such as IL-2 and IFN-γ (Vigorito et al., 2007; Lu et al., 2009). Moreover, its mimics induced cytokine production, while the miR-155 knockout attenuated cytokine release in response to antigen stimulation (Rodriguez et al., 2007; Thai et al., 2007; Sharma et al., 2016). miR-146a is highly expressed in Tregs and exerts the orchestration of immunosuppressive signaling events in T effector cells and Tregs (Lu et al., 2010). miR-146a restrains IFN-γ mediated pathogenic Th1 responses. In addition, it maintains the Treg function by regulating pro-inflammatory cytokines, such as TNF-a, IFN-γ, and IL-17, and its target is STAT1, which is a key transcription factor required for T-cell response. Its critical role had been verified that miR-146a deficiency resulted in an impaired Treg function in the mouse model (Lu et al., 2010). In addition, the miR17-92 cluster, which includes miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92, is reported to play an important role in Treg function under the inflammatory status by preserving antigen-specific Tregs and regulating immunosuppressive IL-10 secretion in Tregs (Jiang et al., 2011). Yang et al. (2016) reported that miR-17 could regulate the suppressive function of Tregs and effector T cells via inhibiting Eos and other transcriptional co-regulators in an IL-6-enriched environment. However, it is not essential for Treg regulation under non-inflammatory status.
High cAMP concentration maintenance is essential for the suppressive function of Tregs. Graham M and his colleagues demonstrated that miR-142-5p acts as an immune regulator of intracellular cAMP, thereby controlling Treg suppressive function. Deletion of miR-142-5p in Treg would result in multisystem autoimmunity (Anandagoda et al., 2019). On the other hand, miR-142-3p was also demonstrated to regulate Tregs’ suppressive function by inhibiting the expression of adenylyl cyclase 9, which is responsible for generating cAMP (Huang et al., 2009). The miR-106b-25 cluster, which includes mir-25, mir-106b, and mir-93, is involved in TGF-beta production which is essential for Treg maturation (Ha, 2011; Liu et al., 2020). miR-15a/16 was shown to regulate Tregs’ suppressive function by inhibiting CTLA-4 expression (Liu et al., 2014). miRNA let-7d was shown to enhance Tregs’ suppressive function by inhibiting Th1 proliferation and cytokine secretion (Okoye et al., 2014; Geng et al., 2020).
miRNAs involved in uveitis pathogenesis by regulating Treg/Th17 homeostasis in animals
As summarized previously, accumulating evidence had revealed the critical significance of miRNAs in the Treg/Th17 balance for autoimmune diseases, which share similar pathological characteristics with autoimmune-mediated eye diseases, including autoimmune uveitis. However, it also had a personalized pathology because of the fact that the eyeball is an immune-privileged organ and has adapted several negative regulators to suppress inflammation by mediating Treg cells (Benhar et al., 2012; Taylor and Ng, 2018). Epigenetic mechanisms in autoimmune uveitis had been investigated in several studies; however, it has not been fully explored at present. Here, we reviewed miRNAs involved in uveitis pathogenesis by regulating Th17 or Treg cells.
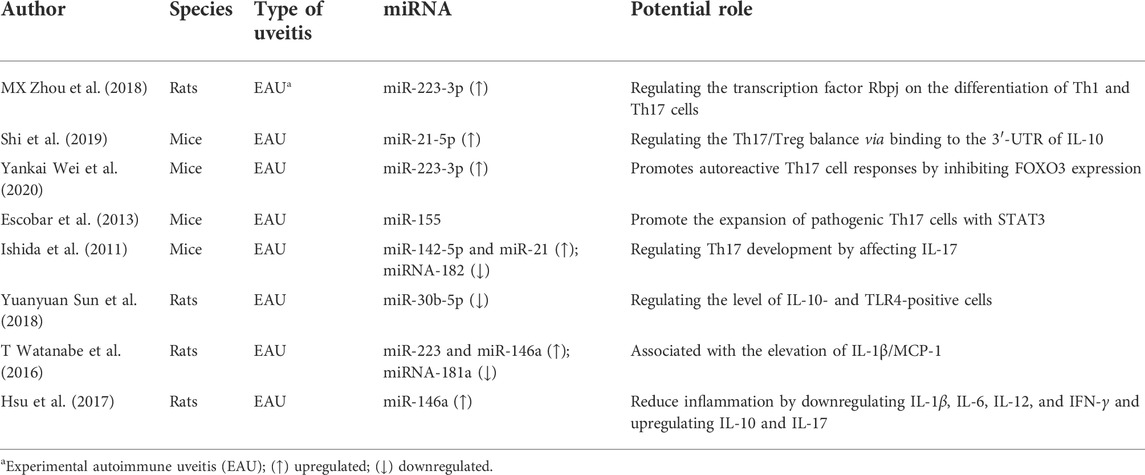
We included eight animal studies about experimental autoimmune uveitis (EAU) which is the classic animal model of uveitis (Caspi et al., 2008) (Bansal et al., 2015). As is summarized in Table 1, several miRNAs had been demonstrated to be regulated and play a vital role in EAU. Among them, miR-223-3p has been reported as an important miRNA in the Tregs/Th17 homeostasis via several different mechanisms. A study by MX Zhou found that upregulated miR-223-3p regulated Th1 and Th17 differentiations by the transcription factor Rbpj. Another study by Yankai Wei found that miR-223-3p promoted autoreactive Th17 cell responses by inhibiting the FOXO3 expression. In addition, T Watanabe and his colleague found that the overexpression of miR-223-3p was closely associated with the elevation of L-1β/MCP-1. In the EAU animal model, miR-155 and miR-146a had also been verified as two essential regulators in Tregs/Th17 homeostasis. Escobar et al. (2014) demonstrated that miR-155 and STAT3/miR-155 axes contributed to EAU development by modulating the Th17 cell differentiation. miR-146a was found to be upregulated in EAU retina; moreover, its overexpression was closely associated with the inflammation score of EAU by regulating IL-1β/MCP-1 and IL-10 and IL-17 (Watanabe et al., 2016). In EAU, miR-30b-5p was reported to be involved in disease development by targeting IL-10 and TLR4 in T cells (Sun et al., 2018). It was well demonstrated that miR-142-5p could control Tregs’ suppressive function by regulating intracellular cAMP (Talebi et al., 2017; Anandagoda et al., 2019), and miR-21 could promote Th17 differentiation by regulating TGF-β signaling. In EAU, both miR-142-5p and miR-21 were found to be overexpressed in ocular tissues and corresponding with the dynamic expression of IL-17, indicating the involvement of miR-142-5p and miR-21 in the development of EAU by regulating Treg/Th17 hemostasis (Murugaiyan et al., 2015; Watanabe et al., 2016; Anandagoda et al., 2019). miR-181a, which could regulate T-cell and Treg differentiation by targeting TCR signaling, was observed to be downregulated in the rat model of EAU and was corresponding with the score of EAU (Li et al., 2007; Watanabe et al., 2016). miR-30b-5p had been reported as T-cell-associated miRNAs; however, its role in Treg/Th17 hemostasis had not been clearly elucidated. A study by YY Sun reported that downregulated miR-30b-5p could regulate the levels of IL-10- and TLR4-positive cells (Sun et al., 2018; Torri et al., 2017). Collectively, these observations indicate that miRNAs are vital regulators of EAU development by regulating the Treg/Th17 balance.
Furthermore, differentially expressed miRNAs had been identified between diseased animal models and healthy control. Guo et al. had reported 36 upregulated miRNAs and 31 downregulated miRNAs in peripheral blood lymphocytes from EAU; moreover, these candidate miRNAs were closely associated with immune signaling and contributed to EAU development (Ishida et al., 2011; Watanabe et al., 2016; Wei et al., 2020). Hsu et al. (2015) had reported three upregulated miRNAs (miR- 9-3p, miR-182-5p, and miR-183-5p) and four downregulated miRNAs (miR-146a-5p, miR-155-5p, miR-147b, and miR-223-3p) in the retina from EAAU (Hsu et al., 2015).
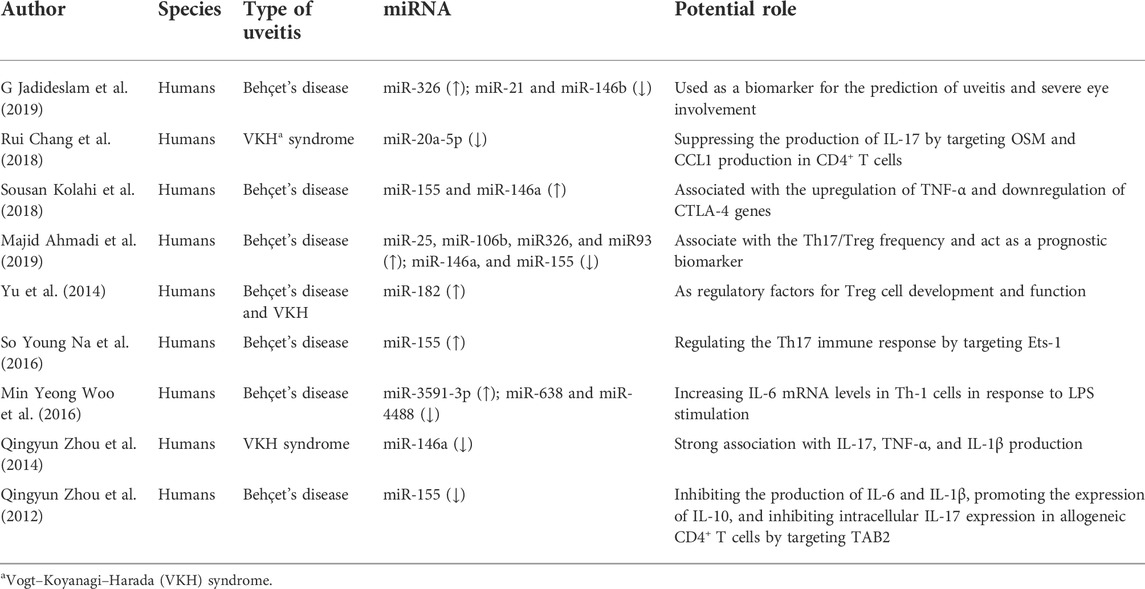
miRNAs involved in uveitis pathogenesis by regulating Tregs/Th17 in humans
Nine studies on uveitis patients were included, and the critical implications of miRNAs are summarized in Table 2. The types of uveitis are mainly Behçet’s disease (BD) and Vogt-Koyanagi-Harada (VKH) syndrome, which are systematic autoimmune diseases with eye involvement (Woo et al., 2016; Wei et al., 2020; Vega-Tapia et al., 2021). Extra ophthalmic involvement usually concerns the skin, central nervous system, gastrointestinal, and mucocutaneous disorders (Paovic et al., 2013; Baltmr et al., 2016).
Among these studies on BD, we found that miR-155 and miR-146a were significantly differentially expressed in the PBMCs; however, the result was controversial and its epigenic mechanism is still elusive (Kolahi et al., 2018; Woo et al., 2016; Zhou et al., 2012). Zhou et al. (2012) found that downregulated miR-155 inhibited the Th17 responses by targeting TGF-beta-activated kinase 1 binding protein 2 (TAB2), while Na et al. (2016) reported that miR-155 was upregulated and promoted Th17 responses via the suppression of E26 transformation- specific-1 (Ets-1) in BD. Kolahi et al. (2018) reported that upregulated miR-155 was associated with TNF-α and CTLA-4D. Another vital miRNA is miR-146a; its role in BD patient is also unelucidated and need to be further explored. Jadideslam et al. (2019) reported that the miR-146a expression had no significant difference between BD patients and healthy control. However, a study had identified that there is a strong correlation between upregulated miR-146a and TNF-α/CTLA-4 (Kolahi et al., 2018), whereas another study had identified that downregulated miR-146a was associated with the Th17/Treg frequency and could act as a prognostic biomarker (Ahmadi et al., 2019). In addition, miR-326, miR-21, miR-146b, miR-25, and miR-106b had been revealed to be differentially expressed in PBMCs from BD patients and were suggested to be biomarkers for predicting uveitis (Woo et al., 2016; Ahmadi et al., 2019; Jadideslam et al., 2019; Wei et al., 2020).
In addition to BD, VKH is also one of the main autoimmune uveitis with severe vision loss. Studies showed that VKH patients had a lower expression of miR-20a in CD4 + T cells, and upregulated miR-20a negatively regulated IL-17 expression via regulating OSM and CCL1 (Chang et al., 2018). Moreover, an analysis based on miRNA–mRNA interactions found that miR-20a may suppress Th17 differentiation through the targeting of several regulators (Honardoost et al., 2015). These findings highlighted the involvement of miR-20a in impaired Treg/Th17 balance during VKH. Several studies had indicated that differentially expressed miR-146a was linked to VKH closely. Evidence showed that miR-146a had a strong association with IL-17, TNF-α, and IL-1β production (Zhou et al., 2014). However, the exact mechanism of the miR-146a gene involved in VKH is still unknown. Additionally, abnormal encoding gene copy numbers of miRNAs (miR-23a, miR-301a, miR-182, and let-7g-3p) have been revealed to have a strong link with disease development (Vega-Tapia et al., 2021).
Conclusion
Impaired Treg/Th17 homeostasis can impact immune tolerance and trigger inflammation that can eventually result in autoimmune diseases’ development. Preserving Treg/Th17 balance is a pivotal treatment strategy. Given the importance of miRNAs for the proper function of the immune system, a number of studies have investigated their epigenic mechanisms on Treg/Th17 hemostasis and their potential role in disease pathogenesis. Collectively, differentially expressed miRNAs were involved in Th17/Treg homeostasis by regulating Treg differentiation and development, Treg induction, Treg stability, Th17 differentiation and Treg function, and cytokine secretion. Considering the similar immunological characteristics in uveitis, we reviewed and summarized miRNAs and their molecular targets in uveitis for further investigation. Several miRNAs and their potential role in regulating Treg/Th17 balance have been identified in animal studies and human studies, respectively. Overall, establishing how miRNAs contribute to Treg/Th17 homeostasis will help define the epigenetic regulation in uveitis and other autoimmune diseases and open new avenues for treatment.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
FT and ZZ wrote the manuscript drafts, and FX and ML designed the framework and revised the manuscript. KH, WD, JL, and RC drew the figures and revised the manuscript.
Funding
The work was funded by the Guangxi Science and Technology Base and Talent Special Fund (No. Guike AD19245007), the Natural Science Foundation of Guangxi Zhuang Autonomous Region (No. 2018GXNSFBA050057), the Guangxi Clinical Ophthalmic Research center (No. Guike AD19245193), and the National Natural Science Foundation of China (No. 82000925).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmadi, M., Yousefi, M., Abbaspour-Aghdam, S., Dolati, S., Aghebati-Maleki, L., Eghbal-Fard, S., et al. (2019). Disturbed Th17/Treg balance, cytokines, and miRNAs in peripheral blood of patients with Behcet's disease. J. Cell. Physiol. 234 (4), 3985–3994. doi:10.1002/jcp.27207
Allantaz, F., Cheng, D. T., Bergauer, T., Ravindran, P., Rossier, M. F., Ebeling, M., et al. (2012). Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One 7 (1), e29979. doi:10.1371/journal.pone.0029979
Alunno, A., Bartoloni, E., Bistoni, O., Nocentini, G., Ronchetti, S., Caterbi, S., et al. (2012). Balance between regulatory T and Th17 cells in systemic lupus erythematosus: The old and the new. Clin. Dev. Immunol. 2012, 823085. doi:10.1155/2012/823085
Anandagoda, N., Willis, J. C., Hertweck, A., Roberts, L. B., Jackson, I., Gökmen, M. R., et al. (2019). microRNA-142-mediated repression of phosphodiesterase 3B critically regulates peripheral immune tolerance. J. Clin. Invest. 129 (3), 1257–1271. doi:10.1172/jci124725
Angelou, C. C., Wells, A. C., Vijayaraghavan, J., Dougan, C. E., Lawlor, R., Iverson, E., et al. (2019). Differentiation of pathogenic Th17 cells is negatively regulated by let-7 MicroRNAs in a mouse model of multiple sclerosis. Front. Immunol. 10, 3125. doi:10.3389/fimmu.2019.03125
Baltmr, A., Lightman, S., and Tomkins-Netzer, O. (2016). Vogt-Koyanagi-Harada syndrome - current perspectives. Clin. Ophthalmol. 10, 2345–2361. doi:10.2147/OPTH.S94866
Bansal, S., Barathi, V. A., Iwata, D., and Agrawal, R. (2015). Experimental autoimmune uveitis and other animal models of uveitis: An update. Indian J. Ophthalmol. 63 (3), 211–218. doi:10.4103/0301-4738.156914
Bayry, J., Siberil, S., Triebel, F., Tough, D. F., and Kaveri, S. V. (2007). Rescuing CD4+CD25+ regulatory T-cell functions in rheumatoid arthritis by cytokine-targeted monoclonal antibody therapy. Drug Discov. Today 12 (13-14), 548–552. doi:10.1016/j.drudis.2007.05.002
Benhar, I., London, A., and Schwartz, M. (2012). The privileged immunity of immune privileged organs: The case of the eye. Front. Immunol. 3, 296. doi:10.3389/fimmu.2012.00296
Buckner, J. H. (2010). Mechanisms of impaired regulation by CD4(+)CD25(+)FOXP3(+) regulatory T cells in human autoimmune diseases. Nat. Rev. Immunol. 10 (12), 849–859. doi:10.1038/nri2889
Caspi, R. R. (2010). A look at autoimmunity and inflammation in the eye. J. Clin. Invest. 120 (9), 3073–3083. doi:10.1172/JCI42440
Caspi, R. R., Silver, P. B., Luger, D., Tang, J., Cortes, L. M., Pennesi, G., et al. (2008). Mouse models of experimental autoimmune uveitis. Ophthalmic Res. 40 (3-4), 169–174. doi:10.1159/000119871
Chang, R., Yi, S., Tan, X., Huang, Y., Wang, Q., Su, G., et al. (2018). MicroRNA-20a-5p suppresses IL-17 production by targeting OSM and CCL1 in patients with Vogt-Koyanagi-Harada disease. Br. J. Ophthalmol. 102 (2), 282–290. doi:10.1136/bjophthalmol-2017-311079
Chen, L., Yang, P., Zhou, H., He, H., Ren, X., Chi, W., et al. (2008). Diminished frequency and function of CD4+CD25high regulatory T cells associated with active uveitis in Vogt-Koyanagi-Harada syndrome. Invest. Ophthalmol. Vis. Sci. 49 (8), 3475–3482. doi:10.1167/iovs.08-1793
Cho, S., Wu, C. J., Yasuda, T., Cruz, L. O., Khan, A. A., Lin, L. L., et al. (2016). miR-23∼27∼24 clusters control effector T cell differentiation and function. J. Exp. Med. 213 (2), 235–249. doi:10.1084/jem.20150990
Chong, M. M., Rasmussen, J. P., Rudensky, A. Y., and Littman, D. R. (2008). The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205 (9), 2005–2017. doi:10.1084/jem.20081219
Cobb, B. S., Hertweck, A., Smith, J., O'Connor, E., Graf, D., Cook, T., et al. (2006). A role for Dicer in immune regulation. J. Exp. Med. 203 (11), 2519–2527. doi:10.1084/jem.20061692
Curotto de Lafaille, M. A., and Lafaille, J. J. (2009). Natural and adaptive foxp3+ regulatory T cells: More of the same or a division of labor? Immunity 30 (5), 626–635. doi:10.1016/j.immuni.2009.05.002
Delgoffe, G. M., Kole, T. P., Zheng, Y., Zarek, P. E., Matthews, K. L., Xiao, B., et al. (2009). The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30 (6), 832–844. doi:10.1016/j.immuni.2009.04.014
Du, C., Liu, C., Kang, J., Zhao, G., Ye, Z., Huang, S., et al. (2009). MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10 (12), 1252–1259. doi:10.1038/ni.1798
Eis, P. S., Tam, W., Sun, L., Chadburn, A., Li, Z., Gomez, M. F., et al. (2005). Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. U. S. A. 102 (10), 3627–3632. doi:10.1073/pnas.0500613102
Escobar, T., Yu, C. R., Muljo, S. A., and Egwuagu, C. E. (2013). STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest Ophthalmol Vis. Sci. 54 (6), 4017–4025. doi:10.1167/iovs.13-11937
Escobar, T. M., Kanellopoulou, C., Kugler, D. G., Kilaru, G., Nguyen, C. K., Nagarajan, V., et al. (2014). miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity 40 (6), 865–879. doi:10.1016/j.immuni.2014.03.014
Fujio, K., Okamura, T., and Yamamoto, K. (2010). The Family of IL-10-secreting CD4+ T cells. Adv. Immunol. 105, 99–130. doi:10.1016/S0065-2776(10)05004-2
Geng, L., Tang, X., Wang, S., Sun, Y., Wang, D., Tsao, B. P., et al. (2020). Reduced let-7f in bone marrow-derived mesenchymal stem cells triggers Treg/Th17 imbalance in patients with systemic lupus erythematosus. Front. Immunol. 11, 233. doi:10.3389/fimmu.2020.00233
Ghoreschi, K., Laurence, A., Yang, X. P., Tato, C. M., McGeachy, M. J., Konkel, J. E., et al. (2010). Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467 (7318), 967–971. doi:10.1038/nature09447
Gritz, D. C., and Wong, I. G. (2004). Incidence and prevalence of uveitis in northern California; the northern California epidemiology of uveitis study. Ophthalmology 111 (3), 491–500. doi:10.1016/j.ophtha.2003.06.014
Ha, T. Y. (2011). The role of MicroRNAs in regulatory T cells and in the immune response. Immune Netw. 11 (1), 11–41. doi:10.4110/in.2011.11.1.11
Heyn, J., Luchting, B., Hinske, L. C., Hubner, M., Azad, S. C., and Kreth, S. (2016). miR-124a and miR-155 enhance differentiation of regulatory T cells in patients with neuropathic pain. J. Neuroinflammation 13 (1), 248. doi:10.1186/s12974-016-0712-6
Hippen, K. L., Loschi, M., Nicholls, J., MacDonald, K. P. A., and Blazar, B. R. (2018). Effects of MicroRNA on regulatory T cells and implications for adoptive cellular therapy to ameliorate graft-versus-host disease. Front. Immunol. 9, 57. doi:10.3389/fimmu.2018.00057
Holmstrom, K., Pedersen, A. E., and Gad, M. (2017). Analysis of miR-146a and miR-142-3p as potential markers of freshly isolated or in vitro-expanded human Treg cells. Scand. J. Immunol. 85 (2), 113–121. doi:10.1111/sji.12517
Honardoost, M. A., Naghavian, R., Ahmadinejad, F., Hosseini, A., and Ghaedi, K. (2015). Integrative computational mRNA-miRNA interaction analyses of the autoimmune-deregulated miRNAs and well-known Th17 differentiation regulators: An attempt to discover new potential miRNAs involved in Th17 differentiation. Gene 572 (2), 153–162. doi:10.1016/j.gene.2015.08.043
Hosseini, A., Ghaedi, K., Tanhaei, S., Ganjalikhani-Hakemi, M., Teimuri, S., Etemadifar, M., et al. (2016). Upregulation of cd4+t-cell derived MiR-223 in the relapsing phase of multiple sclerosis patients. Cell J. 18 (3), 371–380. doi:10.22074/cellj.2016.4565
Hsu, Y. R., Chang, S. W., Lin, Y. C., and Yang, C. H. (2015). Expression of MicroRNAs in the eyes of lewis rats with experimental autoimmune anterior uveitis. Mediat. Inflamm. 2015, 457835. doi:10.1155/2015/457835
Hsu, Y. R., Chang, S. W., Lin, Y. C., and Yang, C. H. (2017). MicroRNA-146a Alleviates Experimental Autoimmune Anterior Uveitis in the Eyes of Lewis Rats. Mediat. Inflamm. 2017, 9601349. doi:10.1155/2017/9601349
Huang, B., Zhao, J., Lei, Z., Shen, S., Li, D., Shen, G. X., et al. (2009). miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 10 (2), 180–185. doi:10.1038/embor.2008.224
Ichiyama, K., Gonzalez-Martin, A., Kim, B. S., Jin, H. Y., Jin, W., Xu, W., et al. (2016). The MicroRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity 44 (6), 1284–1298. doi:10.1016/j.immuni.2016.05.015
Ishida, W., Fukuda, K., Higuchi, T., Kajisako, M., Sakamoto, S., and Fukushima, A. (2011). Dynamic changes of microRNAs in the eye during the development of experimental autoimmune uveoretinitis. Invest. Ophthalmol. Vis. Sci. 52 (1), 611–617. doi:10.1167/iovs.10-6115
Ivanov, I. I., McKenzie, B. S., Zhou, L., Tadokoro, C. E., Lepelley, A., Lafaille, J. J., et al. (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126 (6), 1121–1133. doi:10.1016/j.cell.2006.07.035
Jadideslam, G., Ansarin, K., Sakhinia, E., Babaloo, Z., Abhari, A., Alipour, S., et al. (2019). Expression levels of miR-21, miR-146b and miR-326 as potential biomarkers in Behcet's disease. Biomark. Med. 13 (16), 1339–1348. doi:10.2217/bmm-2019-0098
Jeker, L. T., Zhou, X., Gershberg, K., de Kouchkovsky, D., Morar, M. M., Stadthagen, G., et al. (2012). MicroRNA 10a marks regulatory T cells. PLoS One 7 (5), e36684. doi:10.1371/journal.pone.0036684
Jiang, S., Li, C., Olive, V., Lykken, E., Feng, F., Sevilla, J., et al. (2011). Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood 118 (20), 5487–5497. doi:10.1182/blood-2011-05-355644
Kim, C., Ye, Z., Weyand, C. M., and Goronzy, J. J. (2021). miR-181a-regulated pathways in T-cell differentiation and aging. Immun. Ageing. 18 (1), 28. doi:10.1186/s12979-021-00240-1
Kohlhaas, S., Garden, O. A., Scudamore, C., Turner, M., Okkenhaug, K., and Vigorito, E. (2009). Cutting edge: The Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 182 (5), 2578–2582. doi:10.4049/jimmunol.0803162
Kolahi, S., Farajzadeh, M. J., Alipour, S., Abhari, A., Farhadi, J., Bahavarnia, N., et al. (2018). Determination of mir-155 and mir-146a expression rates and its association with expression level of TNF-alpha and CTLA4 genes in patients with Behcet's disease. Immunol. Lett. 204, 55–59. doi:10.1016/j.imlet.2018.10.012
Korn, T., Bettelli, E., Oukka, M., and Kuchroo, V. K. (2009). IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517. doi:10.1146/annurev.immunol.021908.132710
Lee, Y., Awasthi, A., Yosef, N., Quintana, F. J., Xiao, S., Peters, A., et al. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13 (10), 991–999. doi:10.1038/ni.2416
Li, B., Wang, X., Choi, I. Y., Wang, Y. C., Liu, S., Pham, A. T., et al. (2017). miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J. Clin. Invest. 127 (10), 3702–3716. doi:10.1172/JCI94012
Li, Q. J., Chau, J., Ebert, P. J., Sylvester, G., Min, H., Liu, G., et al. (2007). miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129 (1), 147–161. doi:10.1016/j.cell.2007.03.008
Li, T., Li, M., Xu, C., Xu, X., Ding, J., Cheng, L., et al. (2019). miR‑146a regulates the function of Th17 cell differentiation to modulate cervical cancer cell growth and apoptosis through NF‑κB signaling by targeting TRAF6. Oncol. Rep. 41 (5), 2897–2908. doi:10.3892/or.2019.7046
Liston, A., Lu, L. F., O'Carroll, D., Tarakhovsky, A., and Rudensky, A. Y. (2008). Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205 (9), 1993–2004. doi:10.1084/jem.20081062
Liu, C., Li, N., and Liu, G. (2020). The role of MicroRNAs in regulatory T cells. J. Immunol. Res. 2020, 3232061. doi:10.1155/2020/3232061
Liu, X., Ren, S., Qu, X., Ge, C., Cheng, K., and Zhao, R. C. (2015). Mesenchymal stem cells inhibit Th17 cells differentiation via IFN-gamma-mediated SOCS3 activation. Immunol. Res. 61 (3), 219–229. doi:10.1007/s12026-014-8612-2
Liu, X., Robinson, S. N., Setoyama, T., Tung, S. S., D'Abundo, L., Shah, M. Y., et al. (2014). FOXP3 is a direct target of miR15a/16 in umbilical cord blood regulatory T cells. Bone Marrow Transpl. 49 (6), 793–799. doi:10.1038/bmt.2014.57
Lu, L. F., Boldin, M. P., Chaudhry, A., Lin, L. L., Taganov, K. D., Hanada, T., et al. (2010). Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142 (6), 914–929. doi:10.1016/j.cell.2010.08.012
Lu, L. F., Thai, T. H., Calado, D. P., Chaudhry, A., Kubo, M., Tanaka, K., et al. (2009). Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30 (1), 80–91. doi:10.1016/j.immuni.2008.11.010
Mahesh, G., and Biswas, R. (2019). MicroRNA-155: A master regulator of inflammation. J. Interferon Cytokine Res. 39 (6), 321–330. doi:10.1089/jir.2018.0155
Murugaiyan, G., da Cunha, A. P., Ajay, A. K., Joller, N., Garo, L. P., Kumaradevan, S., et al. (2015). MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J. Clin. Invest. 125 (3), 1069–1080. doi:10.1172/JCI74347
Na, S. Y., Park, M. J., Park, S., and Lee, E. S. (2016). MicroRNA-155 regulates the Th17 immune response by targeting Ets-1 in Behcet's disease. Clin. Exp. Rheumatol. 34, S56–S63.
Nadkarni, S., Mauri, C., and Ehrenstein, M. R. (2007). Anti-TNF-alpha therapy induces a distinct regulatory T cell population in patients with rheumatoid arthritis via TGF-beta. J. Exp. Med. 204 (1), 33–39. doi:10.1084/jem.20061531
Nanke, Y., Kotake, S., Goto, M., Ujihara, H., Matsubara, M., and Kamatani, N. (2008). Decreased percentages of regulatory T cells in peripheral blood of patients with behcet's disease before ocular attack: A possible predictive marker of ocular attack. Mod. Rheumatol. 18 (4), 354–358. doi:10.1007/s10165-008-0064-x
Naqvi, R. A., Datta, M., Khan, S. H., and Naqvi, A. R. (2021). Regulatory roles of MicroRNA in shaping T cell function, differentiation and polarization. Semin. Cell Dev. Biol. 124, 34–47. doi:10.1016/j.semcdb.2021.08.003
Neilson, J. R., Zheng, G. X., Burge, C. B., and Sharp, P. A. (2007). Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21 (5), 578–589. doi:10.1101/gad.1522907
Okoye, I. S., Coomes, S. M., Pelly, V. S., Czieso, S., Papayannopoulos, V., Tolmachova, T., et al. (2014). MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41 (1), 89–103. doi:10.1016/j.immuni.2014.05.019
Paovic, J., Paovic, P., and Sredovic, V. (2013). Behcet's disease: Systemic and ocular manifestations. Biomed. Res. Int. 42, 247345. doi:10.1155/2013/247345
Paust, S., and Cantor, H. (2005). Regulatory T cells and autoimmune disease. Immunol. Rev. 204, 195–207. doi:10.1111/j.0105-2896.2005.00247.x
Qin, A., Wen, Z., Zhou, Y., Li, Y., Li, Y., Luo, J., et al. (2013). MicroRNA-126 regulates the induction and function of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J. Cell. Mol. Med. 17 (2), 252–264. doi:10.1111/jcmm.12003
Qu, X., Liu, X., Cheng, K., Yang, R., and Zhao, R. C. (2012). Mesenchymal stem cells inhibit Th17 cell differentiation by IL-10 secretion. Exp. Hematol. 40 (9), 761–770. doi:10.1016/j.exphem.2012.05.006
Rodriguez, A., Vigorito, E., Clare, S., Warren, M. V., Couttet, P., Soond, D. R., et al. (2007). Requirement of bic/microRNA-155 for normal immune function. Science 316 (5824), 608–611. doi:10.1126/science.1139253
Sakaguchi, S., Mikami, N., Wing, J. B., Tanaka, A., Ichiyama, K., and Ohkura, N. (2020). Regulatory T cells and human disease. Annu. Rev. Immunol. 38, 541–566. doi:10.1146/annurev-immunol-042718-041717
Sato, F., Tsuchiya, S., Meltzer, S. J., and Shimizu, K. (2011). MicroRNAs and epigenetics. FEBS J. 278 (10), 1598–1609. doi:10.1111/j.1742-4658.2011.08089.x
Sethi, A., Kulkarni, N., Sonar, S., and Lal, G. (2013). Role of miRNAs in CD4 T cell plasticity during inflammation and tolerance. Front. Genet. 4, 8. doi:10.3389/fgene.2013.00008
Sharma, V. K., Kaveri, S. V., and Bayry, J. (2016). Impaired regulatory T cell function in autoimmune diseases: Are microRNAs the culprits? Cell. Mol. Immunol. 13 (2), 135–137. doi:10.1038/cmi.2015.98
Shi, L., Guo, H., Li, Z., Wang, Y., Wang, Y., and Cui, Y. (2019). Adenovirus-mediated down-regulation of miR-21-5p alleviates experimental autoimmune uveoretinitis in mice. Int. Immunopharmacol. 74, 105698. doi:10.1016/j.intimp.2019.105698
Singh, Y., Garden, O. A., Lang, F., and Cobb, B. S. (2015). MicroRNA-15b/16 enhances the induction of regulatory T cells by regulating the expression of rictor and mTOR. J. Immunol. 195 (12), 5667–5677. doi:10.4049/jimmunol.1401875
Sun, Y., Guo, D., Liu, B., Yin, X., Wei, H., Tang, K., et al. (2018). Regulatory role of rno-miR-30b-5p in IL-10 and toll-like receptor 4 expressions of T lymphocytes in experimental autoimmune uveitis in vitro. Mediat. Inflamm. 38, 2574067. doi:10.1155/2018/2574067
Sutton, C. E., Lalor, S. J., Sweeney, C. M., Brereton, C. F., Lavelle, E. C., and Mills, K. H. (2009). Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31 (2), 331–341. doi:10.1016/j.immuni.2009.08.001
Takahashi, H., Kanno, T., Nakayamada, S., Hirahara, K., Sciume, G., Muljo, S. A., et al. (2012). TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 13 (6), 587–595. doi:10.1038/ni.2286
Talebi, F., Ghorbani, S., Chan, W. F., Boghozian, R., Masoumi, F., Ghasemi, S., et al. (2017). MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J. Neuroinflammation 14 (1), 55. doi:10.1186/s12974-017-0832-7
Tang, Q., and Bluestone, J. A. (2006). Regulatory T-cell physiology and application to treat autoimmunity. Immunol. Rev. 212, 217–237. doi:10.1111/j.0105-2896.2006.00421.x
Tang, Y., Luo, X., Cui, H., Ni, X., Yuan, M., Guo, Y., et al. (2009). MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 60 (4), 1065–1075. doi:10.1002/art.24436
Taylor, A. W., and Ng, T. F. (2018). Negative regulators that mediate ocular immune privilege. J. Leukoc. Biol. 103, 1179–1187. doi:10.1002/JLB.3MIR0817-337R
Thai, T. H., Calado, D. P., Casola, S., Ansel, K. M., Xiao, C., Xue, Y., et al. (2007). Regulation of the germinal center response by microRNA-155. Science 316 (5824), 604–608. doi:10.1126/science.1141229
Torri, A., Carpi, D., Bulgheroni, E., Crosti, M. C., Moro, M., Gruarin, P., et al. (2017). Extracellular MicroRNA signature of human helper T cell subsets in Health and autoimmunity. J. Biol. Chem. 292 (7), 2903–2915. doi:10.1074/jbc.M116.769893
Tsitsiou, E., and Lindsay, M. A. (2009). microRNAs and the immune response. Curr. Opin. Pharmacol. 9 (4), 514–520. doi:10.1016/j.coph.2009.05.003
Valencia, X., Stephens, G., Goldbach-Mansky, R., Wilson, M., Shevach, E. M., and Lipsky, P. E. (2006). TNF downmodulates the function of human CD4+CD25hi T-regulatory cells. Blood 108 (1), 253–261. doi:10.1182/blood-2005-11-4567
van den Berg, A., Kroesen, B. J., Kooistra, K., de Jong, D., Briggs, J., Blokzijl, T., et al. (2003). High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosom. Cancer 37 (1), 20–28. doi:10.1002/gcc.10186
Vega-Tapia, F., Bustamante, M., Valenzuela, R. A., Urzua, C. A., and Cuitino, L. (2021). miRNA landscape in pathogenesis and treatment of vogt-koyanagi-harada disease. Front. Cell Dev. Biol. 9, 658514. doi:10.3389/fcell.2021.658514
Vigorito, E., Perks, K. L., Abreu-Goodger, C., Bunting, S., Xiang, Z., Kohlhaas, S., et al. (2007). microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27 (6), 847–859. doi:10.1016/j.immuni.2007.10.009
Wan, Y. Y., and Flavell, R. A. (2008). TGF-beta and regulatory T cell in immunity and autoimmunity. J. Clin. Immunol. 28 (6), 647–659. doi:10.1007/s10875-008-9251-y
Wang, W., Shao, S., Jiao, Z., Guo, M., Xu, H., and Wang, S. (2012). The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol. Int. 32 (4), 887–893. doi:10.1007/s00296-010-1710-0
Warth, S. C., Hoefig, K. P., Hiekel, A., Schallenberg, S., Jovanovic, K., Klein, L., et al. (2015). Induced miR-99a expression represses Mtor cooperatively with miR-150 to promote regulatory T-cell differentiation. EMBO J. 34 (9), 1195–1213. doi:10.15252/embj.201489589
Watanabe, T., Keino, H., Kudo, A., Sato, Y., and Okada, A. A. (2016). MicroRNAs in retina during development of experimental autoimmune uveoretinitis in rats. Br. J. Ophthalmol. 100 (3), 425–431. doi:10.1136/bjophthalmol-2015-306924
Wei, Y., Li, N., Zhao, L., Yang, C., Ma, B., Li, X., et al. (2020). MicroRNAs and autoimmune-mediated eye diseases. Front. Cell Dev. Biol. 8, 818. doi:10.3389/fcell.2020.00818
Woo, M. Y., Yun, S. J., Cho, O., Kim, K., Lee, E. S., and Park, S. (2016). MicroRNAs differentially expressed in Behcet disease are involved in interleukin-6 production. J. Inflamm. 13, 22. doi:10.1186/s12950-016-0130-7
Wu, C. J., Cho, S., Huang, H. Y., Lu, C. H., Russ, J., Cruz, L. O., et al. (2019). MiR-23∼27∼24-mediated control of humoral immunity reveals a TOX-driven regulatory circuit in follicular helper T cell differentiation. Sci. Adv. 5 (12), eaaw1715. doi:10.1126/sciadv.aaw1715
Wu, R., Zeng, J., Yuan, J., Deng, X., Huang, Y., Chen, L., et al. (2018). MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J. Clin. Invest. 128 (6), 2551–2568. doi:10.1172/JCI97426
Yang, H. Y., Barbi, J., Wu, C. Y., Zheng, Y., Vignali, P. D., Wu, X., et al. (2016). MicroRNA-17 modulates regulatory T cell function by targeting Co-regulators of the Foxp3 transcription factor. Immunity 45 (1), 83–93. doi:10.1016/j.immuni.2016.06.022
Yang, X. O., Pappu, B. P., Nurieva, R., Akimzhanov, A., Kang, H. S., Chung, Y., et al. (2008). T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28 (1), 29–39. doi:10.1016/j.immuni.2007.11.016
Yao, R., Ma, Y. L., Liang, W., Li, H. H., Ma, Z. J., Yu, X., et al. (2012). MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One 7 (10), e46082. doi:10.1371/journal.pone.0046082
Yin, Z., He, W., Chen, X., Luo, X., Zhang, Y., and Ye, Z. (2018). AB0037 Mir-155 promotes the differentiation of th17 cell by targeting ets-1 in rheumatoid arthritis. Ann. Rheumatic Dis. 77, 1219. doi:10.1136/annrheumdis-2018-eular.3877
Yu, H., Liu, Y., Bai, L., Kijlstra, A., and Yang, P. (2014). Predisposition to Behcet’s disease and VKH syndrome by genetic variants of miR-182. J. Mol. Med. (Berl) 92 (9), 961–967. doi:10.1007/s00109-014-1159-9
Zhang, L., Ke, F., Liu, Z., Bai, J., Liu, J., Yan, S., et al. (2015). MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 6, 7639. doi:10.1038/ncomms8639
Zheng, Y., Josefowicz, S. Z., Kas, A., Chu, T. T., Gavin, M. A., and Rudensky, A. Y. (2007). Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445 (7130), 936–940. doi:10.1038/nature05563
Zhou, J., Li, X., Wu, X., Zhang, T., Zhu, Q., Wang, X., et al. (2018). Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol. Res. 6 (12), 1578–1592. doi:10.1158/2326-6066.CIR-17-0479
Zhou, L., Spolski, R., Min, R., Shenderov, K., Egawa, T., Littman, D. R., et al. (2007). Ivanov, IIIL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8 (9), 967–974. doi:10.1038/ni1488
Zhou, Q., Hou, S., Liang, L., Li, X., Tan, X., Wei, L., et al. (2014). MicroRNA-146a and Ets-1 gene polymorphisms in ocular Behcet's disease and Vogt-Koyanagi-Harada syndrome. Ann. Rheum. Dis. 73 (1), 170–176. doi:10.1136/annrheumdis-2012-201627
Zhou, Q., Xiao, X., Wang, C., Zhang, X., Li, F., Zhou, Y., et al. (2012). Decreased microRNA-155 expression in ocular Behcet's disease but not in Vogt Koyanagi Harada syndrome. Invest. Ophthalmol. Vis. Sci. 53 (9), 5665–5674. doi:10.1167/iovs.12-9832
Keywords: microRNAs, autoimmunity, Treg cells, Th17 cells, intermediate, posterior, uveitis (MeSH)
Citation: Tang F, Zhou Z, Huang K, Deng W, Lin J, Chen R, Li M and Xu F (2022) MicroRNAs in the regulation of Th17/Treg homeostasis and their potential role in uveitis. Front. Genet. 13:848985. doi: 10.3389/fgene.2022.848985
Received: 05 January 2022; Accepted: 24 August 2022;
Published: 14 September 2022.
Edited by:
Shigeo Yoshida, Kurume University, JapanReviewed by:
Francesco Puppo, University of Genoa, ItalyKoh-Hei Sonoda, Kyushu University, Japan
Yingyos Jittayasothorn, National Institutes of Health (NIH), United States
Copyright © 2022 Tang, Zhou, Huang, Deng, Lin, Chen, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan Xu, b3BoX2ZhbkAxNjMuY29t; Min Li, ZXllbWlubGlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Fen Tang
Fen Tang Zhou Zhou†
Zhou Zhou† Wen Deng
Wen Deng Fan Xu
Fan Xu