- 1Department of Gastroenterology, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Department of Gastroenterology, Qingdao Women and Children’s Hospital, Qingdao, China
Gastric cancer (GC) has a high incidence worldwide, and when detected, the majority of patients have already progressed to advanced stages. Long non-coding RNAs (lncRNAs) have a wide range of biological functions and affect tumor occurrence and development. However, the potential role of lncRNAs in GC diagnosis remains unclear. We selected five high-quality samples from each group of chronic non-atrophic gastritis, gastric mucosal intraepithelial neoplasia, and GC tissues for analysis. RNA-seq was used to screen the differentially expressed lncRNAs, and we identified 666 differentially expressed lncRNAs between the chronic non-atrophic gastritis and GC groups, 13 differentially expressed lncRNAs between the gastric mucosal intraepithelial neoplasia and GC groups, and 507 differentially expressed lncRNAs between the chronic non-atrophic gastritis and gastric mucosal intraepithelial neoplasia groups. We also identified six lncRNAs (lncRNA H19, LINC00895, lnc-SRGAP2C-16, lnc-HLA-C-2, lnc-APOC1-1, and lnc-B3GALT2-1) which not only differentially expressed between the chronic non-atrophic gastritis and GC groups, but also differentially expressed between the gastric mucosal intraepithelial neoplasia and GC groups. Furthermore, RT-qPCR was used to verify the differentially co-expressed lncRNAs. LncSEA was used to conduct a functional analysis of differentially expressed lncRNAs. We also predicted the target mRNAs of the differentially expressed lncRNAs through bioinformatics analysis and analyzed targeting correlations between three differentially co-expressed lncRNAs and mRNAs (lncRNA H19, LINC00895, and lnc-SRGAP2C-16). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases were used to explore the functions of target mRNAs of differentially expressed lncRNAs. In conclusion, our study provides a novel perspective on the potential functions of differentially expressed lncRNAs in GC occurrence and development, indicating that the differentially expressed lncRNAs might be new biomarkers for early GC diagnosis.
Introduction
According to global malignant tumor statistics released by the WHO in 2018, there are more than one million new cases of gastric cancer (GC) worldwide annually. GC ranks fifth in cancer incidence and is the third leading cause of cancer-related deaths (Ferlay et al., 2019). The incidence of GC in China is approximately six times that of the United States, which poses a serious threat to human life and health, and places a considerable burden on public health (Verma and Sharma, 2018). Due to the lack of specific clinical manifestations in the early stage of GC, more than 60% of patients have undergone local or distant metastasis when the diagnosis is confirmed, and the best treatment opportunity has been missed (Thrift and El-Serag, 2020). At present, GC diagnosis primarily relies on endoscopy, pathology, and serum tumor markers, such as carcinoembryonic antigen. The pathological results of gastroscopic biopsy are the gold standard for GC diagnosis, however, it remains difficult for certain patients to undergo early GC screening under endoscopy because of its high cost and invasiveness. Moreover, the effect of tumor markers is limited because of the lack of high specificity and sensitivity (Yang et al., 2016; Yu and Zheng, 2018). Therefore, it is crucial to investigate potential and effective biomarkers for early GC diagnosis.
Research on GC initially focused on the biological functions of protein-encoding genes. With the advancement of genome high-throughput sequencing technology, it has been revealed that <2% of the genome has the protein encoding function (Saw et al., 2021). The transcriptional products of the remaining 98% of genes are named non-coding RNAs (ncRNAs), which are involved as regulatory factors in the occurrence and development of many diseases (Wang et al., 2014; Booy et al., 2021). Long noncoding RNAs (lncRNAs) found in the nucleus and cytoplasm are novel ncRNAs. With the development of molecular biology technology, lncRNAs have become a research hotspot in the occurrence and progression of GC and other cancers (Luis et al., 2013). Although lncRNAs do not directly participate in the protein expression process, they can combine with RNA in several biological processes, such as gene expression regulation, cell cycle modulation, transcription process interference, and protein activation function (Sigova et al., 2013; Laurent et al., 2015). In recent years, various lncRNAs have been confirmed to be involved in the occurrence, invasion, metastasis, and prognosis of multiple cancers, including GC. For example, Endo et al. found that lncRNA HOTAIR was highly expressed in GC tissues and was positively correlated with tumor malignancy and poor prognosis (Endo et al., 2013). It has been reported that MALAT-1 might inhibit E-cadherin expression by regulating the epithelial-mesenchymal transition (EMT), thereby promoting the malignant proliferation of GC cells (Chen et al., 2017). Furthermore, the expression of lncRNA DANCR in GC tissues was significantly upregulated, which was correlated with tumor volume, clinical stage, lymph node metastasis, and depth of invasion. Simultaneously, it has been shown that SALL4 in GC cells could activate the expression of DANCR through the β-catenin pathway, thereby exerting a carcinogenic effect in vivo and vitro (Pan et al., 2018). LncRNA UCA1 was found to promote GC development by regulating the PI3K-Akt-mTOR axis (Li et al., 2017). lncRNA CCAT2 silencing could inhibit the PI3K/mTOR axis, further resist GC cell malignant development, induce autophagy, and initiate cancer cell apoptosis (Zhang et al., 2017; Yu et al., 2018). Gao et al. (2015) used the receiver operating characteristic curve to evaluate the sensitivity and specificity of UCA1 as a tumor marker, and discovered that plasma UCA1 had a high diagnostic value in GC. Various lncRNAs have been found to be closely related to GC and are expected to become molecular markers for early GC prediction. However, the functions and mechanisms of most lncRNAs have not yet been elucidated. In the future, a comprehensive and systematic exploration of differentially expressed lncRNAs will be required.
RNA sequencing (RNA-seq), also called transcriptome sequencing technology, is based on second-generation high-throughput sequencing technology to obtain almost all transcript expression information, including mRNA and ncRNA. This technology has the characteristics of high efficiency, low species restriction, and high coverage of the obtained information (Wang et al., 2009). In this study, high-quality samples were subjected to lncRNA library construction, comparison and splicing, and fragment screening after strict quality inspection of all samples, and then Illumina NovaSeq 6000 was used to sequence the lncRNA libraries. The data obtained were used for differential expression analysis of lncRNAs. We screened the differentially expressed lncRNAs between every two groups in chronic non-atrophic gastritis tissues, gastric mucosal low-grade/high-grade intraepithelial neoplasia tissues, and GC tissues using edgeR, and identified lncRNAs H19, LINC00895, lnc-SRGAP2C-16, lnc-B3GALT2-1, lnc-HLA-C-2, and lnc-APOC1-1 as research focus. Finally, we used RT-qPCR to verify the above six differentially expressed lncRNAs and explore new biomarkers for early GC diagnosis.
Methods
Sample Collection and Inclusion Criteria
All patients included in this study were treated with endoscopy or surgery at the Affiliated Hospital of Qingdao University from September 2019 to September 2020. The inclusion criteria were as follows: 1) all samples were confirmed by postoperative pathology; 2) no history of other tumors; 3) no tumor treatment was applied before surgery, such as radiotherapy, chemotherapy, or immunotherapy; 4) the histopathological classification diagnosis met the WHO diagnostic criteria for digestive system tumors (2019); and 5) all selected patients signed an informed consent form after the study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University.
We collected a total of 22 cases of chronic non-atrophic gastritis (gastritis group, gas1), 14 cases of gastric mucosal low-grade or high-grade intraepithelial neoplasia (gastric precancerous lesion group, gas2), of which 9 cases were of gastric mucosal low-grade intraepithelial neoplasia, 5 cases of gastric mucosal high-grade intraepithelial neoplasia, and 9 cases of GC (GC group, gas3). From the above three groups, five high-quality samples were screened to analyze the differentially expressed lncRNAs between every two groups and to explore lncRNAs with the same expression trend among the groups.
RNA Extraction and lncRNA Library Construction
An RNA extraction kit (Aidlab, Beijing, China) was used to extract RNA from 45 patient tissue samples, which were diluted to a working concentration of 100 ng/ml and stored at −80°C. RNA (1 μl) was extracted and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, United States) was used to conduct initial RNA sample inspection. Finally, 5 cases were selected from each group, and 15 samples that met the conditions were used for lncRNA library construction and sequencing analysis. The strand-specific library construction kit NEBNext® (NEB, MA, United States) was used for library construction, and the library was quantified using Qubit2.0 (Life Technologies, CA, United States). Agilent 2100 (Agilent Technologies, CA, United States) was used to detect the length of the inserted library. qPCR was used to quantify the effective concentration of the library. Different libraries that met the quality control standards were sequenced using the NovaSeq 6000 high-throughput sequencing platform (Illumina, CA, United States). LncRNA sequencing was conducted by Berry Hekang Biotechnology (Beijing, China).
Screening of the Differentially Expressed lncRNAs
The original sequencing data were normalized and then compared with the human reference gene sequence using HISAT2 software (Pertea et al., 2016). StringTie software was used for transcript assembly (Pertea et al., 2016). Cuffmerge software was used to splice and concatenate the transcripts of each sample and obtain complete transcriptome information of the sequence (Trapnell et al., 2012). Subsequently, we used Cuffcompare software to remove transcripts that overlapped with the database annotated exon region and used Cuffquant software to select transcripts with a value of FPKM≥0.5. Moreover, CPC, CNCI, CPAT, PLEK, and PFAM software were used to analyze and identify non-coding transcripts (Kong et al., 2007; Mistry et al., 2007; Sun et al., 2013; Wang et al., 2013; Li et al., 2014; Finn et al., 2016). Then, the overlapping data of the software screening results were selected as the lncRNA datasets for analysis and prediction. Cuffdiff software was used to calculate the FPKM values of the transcripts (Trapnell et al., 2012). edgeR was used to perform a differential analysis of the transcripts (Robinson et al., 2010). The screening criteria for differentially expressed lncRNAs were |logFC|>1 and FDR<0.05.
Real-Time Quantitative PCR
Total RNA was isolated from patient tissues in three groups using TRIzol reagent (Shanghai Biological Engineering Co., Ltd., Shanghai, China). Extracted total RNA (1 μl) was used for quality identification. When the OD260/280 value of the RNA sample was between 1.8 and 2.0, the sample was considered to be of high purity. The genome removal DNA reaction system included gDNA Eraser, 5x gDNA Eraser Buffer, RNA, and RNase Free dH2O. The reverse transcription system included PrimeScript RT Enzyme Mix I, RT Prime Mix, 5xPrimeScript Buffer 2, and RNase Free dH2O. Primer sequences used for RT-qPCR are listed in Table 1. TB Green™ Premix Ex Taq™ II kit (Takara, Shiga, Japan) and Pikoreal96 fluorescence quantitative PCR instrument (Thermo Fisher Scientific) for RT-qPCR. GAPDH was used as an internal reference gene, and the relative expression of each gene was displayed by the result of 2−ΔΔCT. The experiment for each group was repeated three times.
Bioinformatic Analysis
We used two methods to predict targeted mRNAs: co-location and co-expression target mRNA prediction. The first method was to use the complementary pairing relationship between lncRNAs and known mRNAs and the location of lncRNAs on the chromosome relative to the mRNAs to predict target mRNAs. The second method was through correlation analysis of lncRNA and mRNA expression using the Pearson correlation coefficient method. The target mRNAs with Cor>0.99 were selected. We drew the interaction networks of lncRNA-mRNA pairs using Cytoscape software (v. 3.7.2).
GeneCards is an integrative database that provides comprehensive information on all annotated and predicted human genes (Barshir et al., 2021). LncSEA is an extensive human lncRNA sets resource and enrichment analysis platform (Chen et al., 2021). We attempted to find the gene symbols (e.g., H19, lnc-SRGAP2C-16) of the differentially expressed lncRNAs obtained by RNA-seq (e.g., NONHSAG007409.2, NONHSAG002625.2) through the GeneCard database, then imported them into the LncSEA website to conduct functional enrichment analysis.
Gene Ontology (GO) analysis and pathway analysis were used to determine the roles of target mRNAs of differentially expressed lncRNAs in the development of GC. We uploaded all target mRNAs into the Database for Annotation, Visualization, and Integrated Discovery (DAVID) for annotation and functional analysis. GO terms with a p value < 0.05 were selected. The top 10 enriched GO terms associated with the targeted mRNAs were presented. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was also performed to identify the significant pathways of the target genes.
Statistics
In this study, GraphPad Prism 6.01 software was used for data processing, statistical analysis, and graphing. The data in this study conform to a normal distribution. We used Student’s t-test for comparison between the two groups. Statistical significance was set at p < 0.05.
Results
Screening of Differentially Expressed lncRNAs
We collected 45 clinical samples from the chronic non-atrophic gastritis group, gastric mucosal intraepithelial neoplasia group, and GC group and selected five high-quality samples from each group according to the quality control standards. The quality inspection results of the 15 samples are listed in Supplementary Table S1.
After the samples were sequenced, we used edgeR to analyze the difference in FPKM of lncRNA transcripts between samples according to the screening criteria of differentially expressed genes (|logFC|>1, FDR<0.05). The volcano figures display the upregulated and downregulated lncRNAs (Figure 1). Using the gastritis group (gas1) as the control group and GC group (gas3) as the experimental group for comparison, we found that a total of 666 lncRNAs with differential expression were identified, of which 188 were upregulated and 478 were downregulated (Supplementary Table S2). Using the gastric precancerous lesion group (gas2) as the control group and GC group (gas3) as the experimental group, we discovered that a total of 13 lncRNAs with differential expression were screened, including six with elevated expression levels and seven with decreased expression levels (Supplementary Table S3). Considering the gastritis group (gas1) as the control group and the gastric precancerous lesion group (gas2) as the experimental group, a total of 507 differential lncRNAs were screened, of which 236 increased and 271 decreased (Supplementary Table S4).
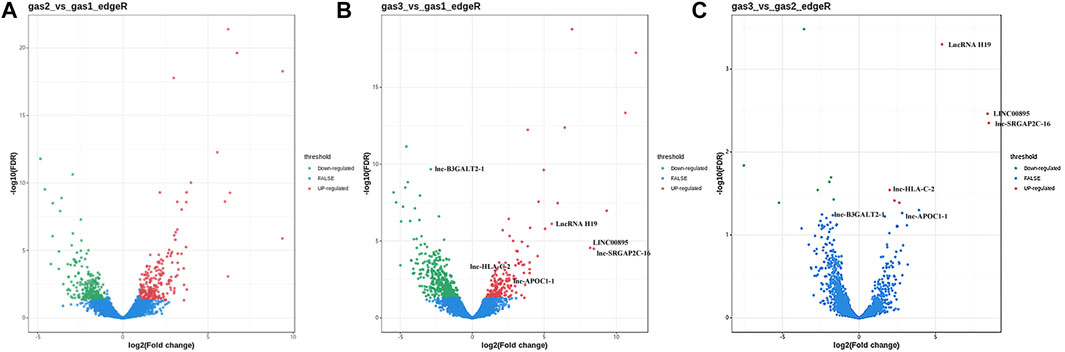
FIGURE 1. The volcano figures of upregulated and downregulated lncRNAs. (A) The differential lncRNAs between gastritis group (gas1) and gastric precancerous lesion group (gas2). (B) The differential lncRNAs between gastritis group (gas1) and gastric cancer group (gas3). (C) The differential lncRNAs between gastric precancerous lesion group (gas2) and gastric cancer group (gas3).
It was also found that six lncRNAs not only differentially expressed between gas1 and gas3, but also differentially expressed between gas2 and gas3 (Table 2). Among them, there were five upregulated lncRNAs, namely, lncRNA H19, LINC00895, lnc-SRGAP2C-16, lnc-HLA-C-2, and lnc-APOC1-1, and one downregulated lncRNA, lnc-B3GALT2-1.
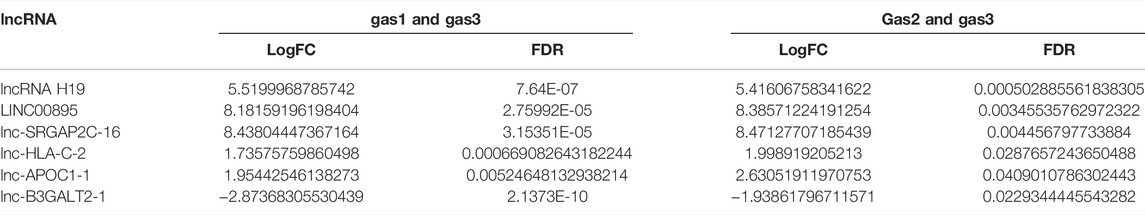
TABLE 2. The LogFC and FDR of six differentially expressed lncRNAs between the gastritis group (gas1) and the gastric cancer group (gas3) as well as between the gastric precancerous lesions group (gas2) and the gastric cancer group (gas3).
Expression Verification of Differentially Expressed lncRNAs
To verify the expression of the six differentially expressed lncRNAs identified by sequencing analysis, we used RT-qPCR to evaluate the expression levels of lncRNA H19, LINC00895, lnc-SRGAP2C-16, lnc-HLA-C-2, lnc-APOC1-1, and lnc-B3GALT2-1 in 15 samples of chronic non-atrophic gastritis (gastritis), gastric mucosal low-grade/high-grade intraepithelial neoplasia (GIN), and GC (Gastric Cancer) tissues. The results are shown in Figure 2. Compared with the chronic non-atrophic gastritis group, the expression of lncRNA H19 (n = 10, p < 0.01, Figure 2A), LINC00895 (n = 10, p < 0.001, Figure 2B), lnc-SRGAP2C-16 (n = 10, p < 0.01, Figure 2C), lnc-HLA-C-2 (n = 10, p < 0.001, Figure 2D), and lnc-APOC1-1 (n = 10, p < 0.0001, Figure 2E) was significantly increased in the GC group, whereas lnc-B3GALT2-1 expression (n = 10, p < 0.001, Figure 2F) was significantly decreased. Compared with the gastric mucosal low-grade/high-grade intraepithelial neoplasia group, the expression level of lncRNA H19 (n = 10, p < 0.05, Figure 2A), LINC00895 (n = 10, p < 0.001, Figure 2B), lnc-SRGAP2C-16 (N = 10, p < 0.001, Figure 2C), lnc-HLA-C-2 (n = 10, p < 0.01, Figure 2D), lnc-APOC1-1 (n = 10, p < 0.0001, Figure 2E) in the GC group was significantly upregulated, while the expression level of lnc-B3GALT2-1 (n = 10, p < 0.01, Figure 2F) was significantly downregulated. The above results were consistent with the expression of the six differentially expressed lncRNAs obtained by sequencing.
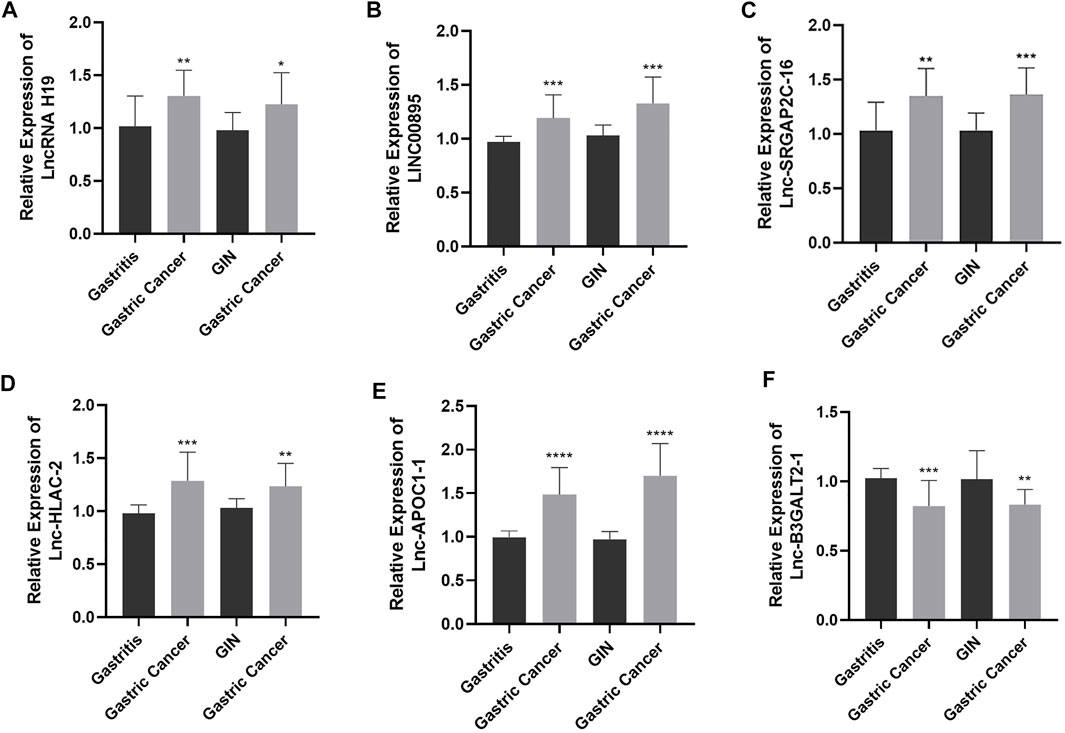
FIGURE 2. The expression levels of lncRNA H19 (A), LINC00895 (B), lnc-SRGAP2C-16 (C), lnc-HLA-C-2 (D), lnc-APOC1-1 (E), and lnc-B3GALT2-1 (F) in chronic non-atrophic gastritis (Gastritis) vs. gastric cancer (Gastric Cancer) tissues and in gastric mucosal low-grade/high-grade intraepithelial neoplasia (GIN) vs. gastric cancer (Gastric Cancer) tissues.
Functional Enrichment Analysis of Differentially Expressed lncRNAs
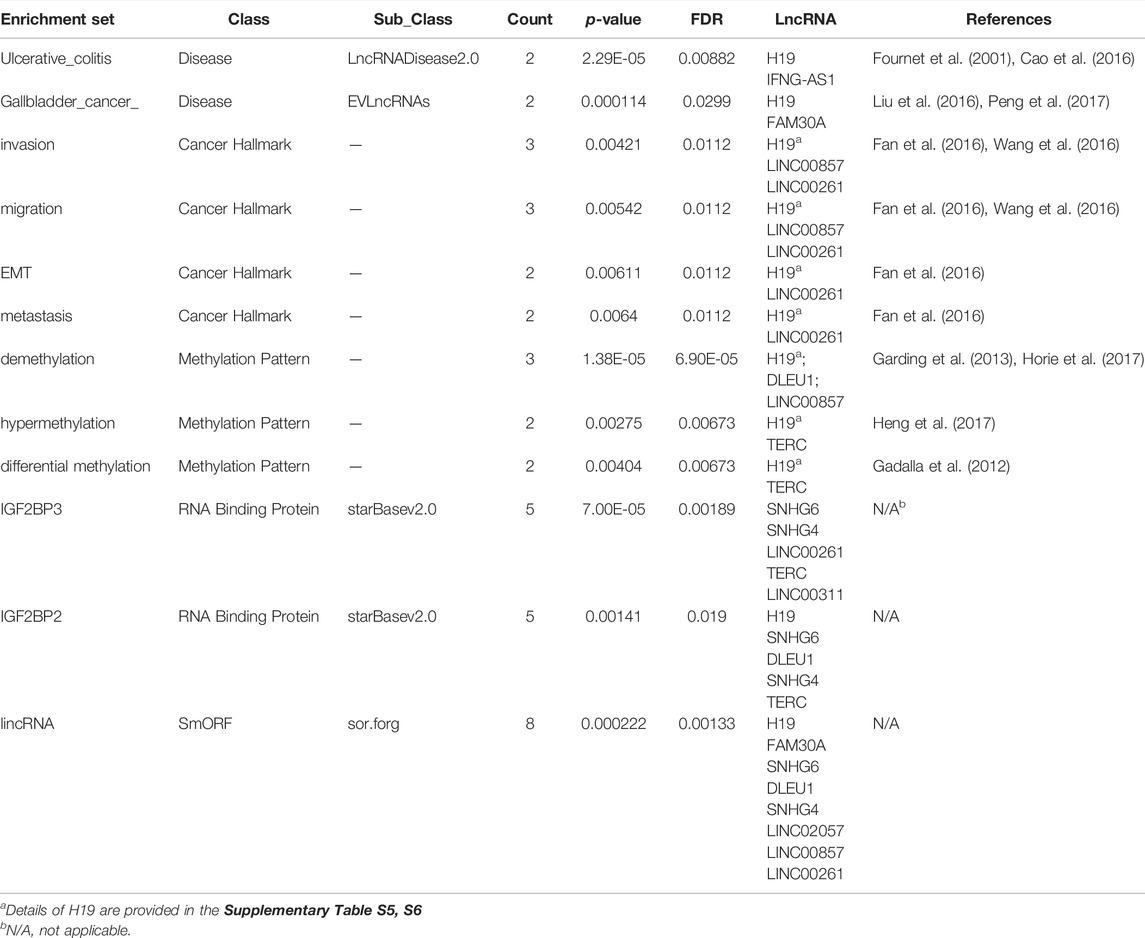
We conducted enrichment analysis for all the differentially expressed lncRNAs with corresponding gene symbols in GeneCard using LncSEA. Although the number of lncRNAs involved was limited, significant functional enrichment sets were still found (Table 3). H19 and IFNG-AS1 might be associated with ulcerative colitis (Cao et al., 2016; Fournet et al., 2001). Gallbladder cancer development might be related to H19 and FAM30A (Liu et al., 2016; Peng et al., 2017). H19 is involved in the invasion, migration, epithelial-mesenchymal transition (EMT), and metastasis of various cancers, such as hepatic cancer, pancreatic cancer, and gastric cancer (Supplementary Table S5). LINC00857 and LINC00261 are also related to cancer invasion and migration (Wang et al., 2016; Fan et al., 2016). lncRNAs are crucial in some essential epigenetic regulation processes, such as DNA methylation. Many studies have experimentally supported the associations between lncRNA H19 and DNA methylation, including demethylation, hypermethylation, and differential methylation (Supplementary Table S6). DLEU1 is linked to demethylation in leukemia, and LINC00857 is correlated to demethylation in NSCLC (Garding et al., 2013; Horie et al., 2017). Previous research found that TERC is associated with hypermethylation in breast cancer and differential methylation in dyskeratosis congenita (Gadalla et al., 2012; Heng et al., 2017). IGF2BP3 and IGF2BP2 are two classes of RNA binding proteins of the differentially expressed lncRNAs. Many short or small ORFs (sORFs/smORFs) can encode small peptides in the body. It has been found that eight differentially expressed lncRNAs have the smORF named lincRNA.
Relationship Between Differentially Expressed lncRNAs and mRNAs
We utilized the complementary pairing relationship and the position of the differentially expressed lncRNAs relative to the mRNAs on the chromosome to predict target mRNAs. We also predicted lncRNA-targeted genes by co-expression analysis. The Pearson correlation coefficient method was used to analyze the correlation between the differentially expressed lncRNAs and mRNAs. We found 278 target genes of differentially expressed lncRNAs between gas1 and gas3 after deleting duplicate genes (Supplementary Table S7). The target genes of differentially expressed lncRNAs between gas2 and gas3 were only identified by co-expression analysis, with a total of 54 genes (Supplementary Table S8). Furthermore, we identified 196 target genes of differentially expressed lncRNAs between gas1 and gas2 (Supplementary Table S9).
Furthermore, we found that three of the six co-expressed differentially expressed lncRNAs had targeting correlation with mRNAs, namely lncRNA H19, LINC00895, lnc-SRGAP2C-16 (Figure 3). From the network, LINC00895 and lnc-SRGAP2C-16 had 3 same target mRNAs (BTN1A1, C1orf61, and IFNL2). lnc-SRGAP2C-16 had 46 target mRNAs, indicating that lnc-SRGAP2C-16 might play an important role in the occurrence and development of GC. This provides us with new insights for further exploring the biological mechanisms of lncRNA H19, LINC00895, and lnc-SRGAP2C-16 in the process of chronic non-atrophic gastritis and gastric mucosal low-grade or high-grade intraepithelial neoplasia to GC.
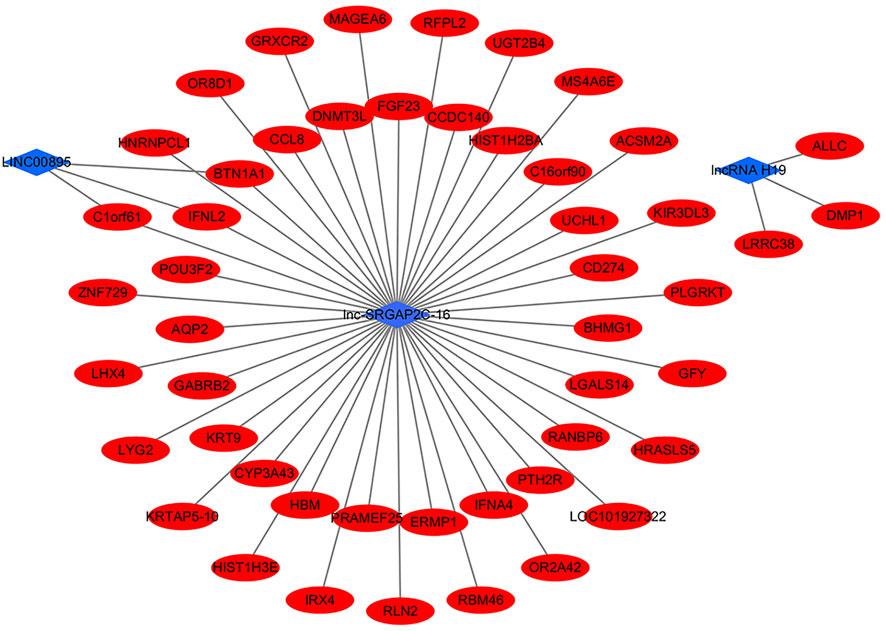
FIGURE 3. The interaction networks of differentially expressed lncRNAs (lncRNA H19, LINC00895, lnc-SRGAP2C-16) and target mRNAs.
Functional Enrichment Analysis of Target mRNAs
We also performed functional enrichment analysis on all the target mRNAs of differentially expressed lncRNAs. The enriched items and pathways of the differentially expressed lncRNA-targeted mRNAs are shown in Figure 4. The top 10 enriched items of targeted mRNAs in biological processes (BP) were oxygen transport, neutrophil chemotaxis, chemotaxis, innate immune response in mucosa, peptide cross-linking, sensory perception of smell, inflammatory response, innate immune response, bicarbonate transport, and G-protein coupled receptor signaling pathway. Furthermore, certain important GO functions might be related to oxygen transporter activity, structural molecule activity, haptoglobin binding, oxygen binding, sequence-specific DNA binding, heme binding, iron ion binding, RAGE receptor binding, neuropeptide receptor activity, and lipid binding in molecular function (MF). In addition, the extracellular region, hemoglobin complex, blood microparticle, haptoglobin-hemoglobin complex, endocytic vesicle lumen, nuclear nucleosome, integral component of plasma membrane, keratin filament, extracellular space, and cornified envelope were significant components of cellular components (CC). However, we found no significant KEGG pathways related to the target genes through the DAVID database.
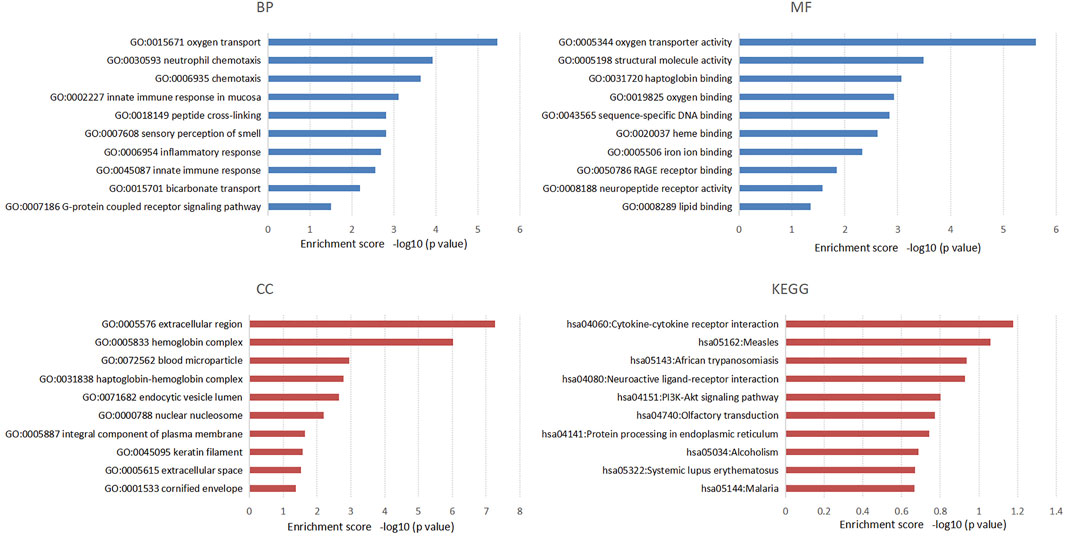
FIGURE 4. The top 10 enriched items of differential lncRNAs targeted mRNAs in the process of chronic non-atrophic gastritis and gastric mucosal intraepithelial neoplasia to gastric cancer in GO and KEGG enrichment analysis.
Discussion
Numerous studies have compared the gene expression profiles between tumor and normal tissues and found that various lncRNAs were differentially expressed in tumors, which may become potential tumor markers and targets for tumor intervention, providing new ideas for tumor diagnosis and treatment (Gu et al., 2015; Renganathan and Felley-Bosco, 2017). One study screened differential genes from GC and its adjacent normal tissues by microarray analysis and discovered that the expression levels of 33 lncRNAs were significantly different. The researchers further verified that the expression levels of HIF1A-AS1 and PVT1 were significantly downregulated in GC tissues, while the expression levels of CBR3-AS1 and UCA1 were significantly upregulated (Gao et al., 2015). Zhang et al. quantitatively evaluated the lncRNAs in GC cell lines and plasma and found that the expression of five lncRNAs in cancer cells was significantly increased, including TINCR (Zhang et al., 2017). More and more lncRNAs were found to act as carcinogens to promote the occurrence and development of GC.
In this study, we used RNA-seq technology to perform quantitative differential lncRNA expression analysis in the development of GC. We selected five high-quality samples from the chronic non-atrophic gastritis, gastric mucosal intraepithelial neoplasia, and GC groups for the construction of lncRNA libraries. We found that, compared with the chronic non-atrophic gastritis group, the GC group showed a total of 666 differential lncRNAs. Compared with the gastric mucosa low-grade/high-grade intraepithelial neoplasia group, the GC group showed a total of 13 differential lncRNAs. Taken together, there were 6 differential lncRNAs (lncRNA H19, LINC00895, lnc-SRGAP2C-16, lnc-HLA-C-2, lnc-APOC1-1, and lnc-B3GALT2-1) that were co-expressed in the same direction in the above two comparison results. Subsequently, we verified the difference in lncRNA expression levels between groups through RT-qPCR, which was consistent with the sequencing results, suggesting that the progression of chronic non-atrophic gastritis and gastric mucosal low-grade or high-grade intraepithelial neoplasia to GC was associated with the expression of the above six differentially expressed lncRNAs.
LncRNA H19, a star molecule in GC research, has already been reported to have carcinogenic mechanisms. Researchers analyzed the expression of lncRNAs in GC and normal tissues, indicating that H19 expression increased most significantly among 71 upregulated lncRNAs, approximately 9 times higher than that in normal tissues (Song et al., 2013). Yang et al. also found that the expression level of H19 in various GC cell lines and tissues was significantly higher than that in normal controls. Abnormal expression of H19 promoted the proliferation of AGS GC cell lines, while silencing H19 induced cell apoptosis. Mechanically, upregulated H19 participates in tumorigenesis by inhibiting p53 activation in GC (Yang et al., 2012). Zhuang et al. found that H19 predominantly targeted RUNX1 through miR-675 to regulate GC cell proliferation, providing a potential target for GC treatment (Zhuang et al., 2014). It has been reported that miR-141 binds to H19 to inhibit H19-induced proliferation and invasion. Furthermore, miR-141 and H19 could directly affect each other, and also competitively bind to each other’s target genes to affect the development of GC (Zhou et al., 2015). According to our verified experiments of differential lncRNAs, the expression level of lncRNA H19 in the GC group was significantly higher than that in the chronic non-atrophic gastritis group, which was consistent with the results of previous studies. In addition, our sequencing results also showed that LINC00895, lnc-SRGAP2C-16, lnc-HLA-C-2, and lnc-APOC1-1 were significantly increased in the GC compared to that of the chronic non-atrophic gastritis and gastric mucosal intraepithelial neoplasia groups, while lnc-B3GALT2-1 expression decreased significantly. However, currently, only LINC00895 has been found to be a high-frequency integration site of the HPV gene, which is related to HPV carcinogenesis (Yang-Chun et al., 2020). Thus, the mechanism of LINC00895 in GC requires further exploration. Notably, six lncRNAs co-expressed in the process of chronic non-atrophic gastritis and gastric mucosal low-grade or high-grade intraepithelial neoplasia to GC, and the expression levels were significantly different, indicating that the six differentially expressed lncRNAs might achieve the purpose of early GC diagnosis. In the future, we need to further investigate the six lncRNAs in body fluids to provide a new method for early GC diagnosis.
Through the LncSEA database, we directedly performed functional enrichment analysis for differentially expressed lncRNAs. In addition to H19, other differentially expressed lncRNAs are involved in disease occurrence, cancer invasion, metastasis, EMT, and methylation. However, the number of gene symbols of the differentially expressed lncRNAs found by GeneCard and the number of these lncRNAs involved in the LncSEA functional analysis were limited. Therefore, the results obtained by direct enrichment analysis of lncRNAs need further exploration. We are still a long way from unraveling the mystery of lncRNAs.
In addition, lncRNAs can bind to mRNAs and exhibit a wide range of biological functions. For example, lnc-LBCS directly interacts with hnRNPK and forms a complex with hnRNPK and AR mRNA to inhibit AR translation efficiency and cancer progression in castrated prostate cancer research (Gu et al., 2019). In short, analyzing the co-expression relationship of lncRNA-mRNA and identifying potential target genes of lncRNAs will help predict the function of lncRNAs. In this study, we found targeting correlations between three differentially expressed lncRNAs and mRNAs using the Pearson correlation coefficient method. The function of lncRNAs could be predicted through their target genes. It has been reported that BTN1A1 expression was essential for the regulation and secretion of milk fat droplets. Mechanistically, BTN1A1 might act as a structural protein or signal receptor by binding to xanthine dehydrogenase/oxidase (Ogg et al., 2004). C1orf61 acted as a tumor activator in hepatocellular carcinoma tumorigenesis and regulated diverse genes related to cell growth, migration, invasion and epithelial-mesenchymal transition (EMT) (Yu et al., 2021). At present, there is a lack of research on the mechanism of the three lncRNAs and target mRNAs in the tumorigenesis of GC and a lack of in vivo and in vitro experiments for verification. Therefore, we will focus on the underlying mechanism in our future research.
In previous studies, researchers focused on the expression of lncRNAs between GC and adjacent chronic non-atrophic gastritis but lacked attention to the differential lncRNAs between the gastric mucosa low-grade/high-grade intraepithelial neoplasia and GC. Therefore, we added the gastric mucosal low-grade/high-grade intraepithelial neoplasia group to discover the same differentially expressed lncRNAs in the three groups in this study. However, we only found differentially expressed lncRNAs during the development of GC. We should continue to expand the sample size to further analyze the specificity and sensitivity of six differential lncRNAs as markers for monitoring tumor progression and their correlation with clinicopathological parameters. Moreover, we should conduct long-term follow-up of patients and analyze the relationship between differentially expressed lncRNAs and survival. Subsequent research is expected to further explore the specific mechanisms of six lncRNAs in the development of GC and investigate their effects on the biological behaviors of GC cells, such as proliferation, invasion, induction of cell apoptosis, and EMT through in vivo and in vitro experiments, which will enrich the evidence of lncRNAs as biomarkers for early GC diagnosis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository and accession number can be found below: CNGB Sequence Archive (CNSA) (Guo et al., 2020) of China National GeneBank DataBase (CNGBdb) (Chen et al., 2020) with accession number CNP0002510.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Affiliated Hospital of Qingdao University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
X-YLiu and T-QZ was responsible for the collection and assembly of data, data analysis and interpretation, and manuscript writing. QZ, JG and PZ was responsible for the provision of study material, collection of data, and technical expertise. TM, Z-BT and PZ was responsible for the conception and design, revising the manuscript, and final approval of the manuscript. X-YLi was responsible for revising the manuscript, financial support, and final approval of the manuscript. All authors have read and approved the final version of the manuscript.
Funding
The study was supported by the National Natural Science Foundation (No. 81802777), the National Natural Science Foundation (No. 81800496), the “Clinical medicine + X” scientific research project of Affiliated Hospital of Qingdao University, and Qingdao Chinese Medicine Technology Project (2021-zyym26).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing. And we thank all the authors for helping with the writing and publication of this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.833857/full#supplementary-material
Supplementary Table 1 | The quality inspection results and reads quality of the 15 samples.
Supplementary Table 2 | The differentially expressed lncRNAs between the chronic non-atrophic gastritis and GC groups.
Supplementary Table 3 | The differentially expressed lncRNAs between the gastric mucosal intraepithelial neoplasia and GC groups.
Supplementary Table 4 | The differentially expressed lncRNAs between the chronic non-atrophic gastritis and gastric mucosal intraepithelial neoplasia groups.
Supplementary Table 5 | The association between H19 and various cancers in invasion, migration, EMT, and metastasis through LncSEA.
Supplementary Table 6 | The association between H19 and Methylation Pattern through LncSEA.
Supplementary Table 7 | The target genes of differentially expressed lncRNAs between the chronic non-atrophic gastritis and GC groups.
Supplementary Table 8 | The target genes of differentially expressed lncRNAs between the gastric mucosal intraepithelial neoplasia and GC groups.
Supplementary Table 9 | The target genes of differentially expressed lncRNAs between the chronic non-atrophic gastritis and gastric mucosal intraepithelial neoplasia groups.
Supplementary Table 10 | The download links of softwares used in screening the differentially expressed lncRNAs.
References
Barshir, R., Fishilevich, S., Iny-Stein, T., Zelig, O., Mazor, Y., Guan-Golan, Y., et al. (2021). GeneCaRNA: A Comprehensive Gene-Centric Database of Human Non-Coding RNAs in the GeneCards Suite. J. Mol. Biol. 433, 166913. doi:10.1016/j.jmb.2021.166913
Booy, E. P., Gussakovsky, D., Choi, T., and McKenna, S. A. (2021). The Noncoding RNA BC200 Associates with Polysomes to Positively Regulate mRNA Translation in Tumor Cells. J. Biol. Chem. 296, 100036. doi:10.1074/jbc.RA120.015775
Cao, B., Song, N., Zhang, M., Di, C., Yang, Y., Lu, Y., et al. (2016). Systematic Study of Novel lncRNAs in Different Gastrointestinal Cancer Cells. Discov. Med. 21, 159–171.
Chen, D., Liu, L., Wang, K., Yu, H., Wang, Y., Liu, J., et al. (2017). The Role of MALAT-1 in the Invasion and Metastasis of Gastric Cancer. Scand. J. Gastroenterol. 52, 790–796. doi:10.1080/00365521.2017.1280531
Chen, F. Z., You, L. J., Yang, F., Wang, L. N., Guo, X. Q., Gao, F., et al. (2020). CNGBdb: China National GeneBank DataBase. Yi Chuan 42, 799–809. doi:10.16288/j.yczz.20-080
Chen, J., Zhang, J., Gao, Y., Li, Y., Feng, C., Song, C., et al. (2021). LncSEA: a Platform for Long Non-Coding RNA Related Sets and Enrichment Analysis. Nucleic Acids Res. 49, D969–D980. doi:10.1093/nar/gkaa806
Endo, H., Shiroki, T., Nakagawa, T., Yokoyama, M., Tamai, K., Yamanami, H., et al. (2013). Enhanced Expression of Long Non-Coding RNA HOTAIR Is Associated with the Development of Gastric Cancer. PLoS One 8, e77070. doi:10.1371/journal.pone.0077070
Fan, Y., Wang, Y.-F., Su, H.-F., Fang, N., Zou, C., Li, W.-F., et al. (2016). Decreased Expression of the Long Noncoding RNA LINC00261 Indicate Poor Prognosis in Gastric Cancer and Suppress Gastric Cancer Metastasis by Affecting the Epithelial-Mesenchymal Transition. J. Hematol. Oncol. 9, 57. doi:10.1186/s13045-016-0288-8
Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D. M., Piñeros, M., et al. (2019). Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 144, 1941–1953. doi:10.1002/ijc.31937
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 44, D279–D285. doi:10.1093/nar/gkv1344
Fournet, J.-C., Mayaud, C., de Lonlay, P., Gross-Morand, M.-S., Verkarre, V., Castanet, M., et al. (2001). Unbalanced Expression of 11p15 Imprinted Genes in Focal Forms of Congenital Hyperinsulinism. Am. J. Pathol. 158, 2177–2184. doi:10.1016/s0002-9440(10)64689-5
Gadalla, S. M., Katki, H. A., Shebl, F. M., Giri, N., Alter, B. P., and Savage, S. A. (2012). The Relationship between DNA Methylation and Telomere Length in Dyskeratosis Congenita. Aging Cell 11, 24–28. doi:10.1111/j.1474-9726.2011.00755.x
Gao, J., Cao, R., and Mu, H. (2015). Long Non-Coding RNA UCA1 May Be a Novel Diagnostic and Predictive Biomarker in Plasma for Early Gastric Cancer. Int. J. Clin. Exp. Pathol. 8, 12936–12942.
Garding, A., Bhattacharya, N., Claus, R., Ruppel, M., Tschuch, C., Filarsky, K., et al. (2013). Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the in Cis Downregulation of a Gene Cluster that Targets NF-kB. Plos Genet. 9, e1003373. doi:10.1371/journal.pgen.1003373
Gu, P., Chen, X., Xie, R., Xie, W., Huang, L., Dong, W., et al. (2019). A Novel AR Translational Regulator lncRNA LBCS Inhibits Castration Resistance of Prostate Cancer. Mol. Cancer 18, 109. doi:10.1186/s12943-019-1037-8
Gu, Y., Chen, T., Li, G., Yu, X., Lu, Y., Wang, H., et al. (2015). LncRNAs: Emerging Biomarkers in Gastric Cancer. Future Oncol. 11, 2427–2441. doi:10.2217/fon.15.175
Guo, X., Chen, F., Gao, F., Li, L., Liu, K., You, L., et al. (2020). CNSA: A Data Repository for Archiving Omics Data. Database (Oxford) 2020, baaa055. doi:10.1093/database/baaa055
Heng, J., Zhang, F., Guo, X., Tang, L., Peng, L., Luo, X., et al. (2017). Integrated Analysis of Promoter Methylation and Expression of Telomere Related Genes in Breast Cancer. Oncotarget 8, 25442–25454. doi:10.18632/oncotarget.16036
Horie, M., Kaczkowski, B., Ohshima, M., Matsuzaki, H., Noguchi, S., Mikami, Y., et al. (2017). Integrative CAGE and DNA Methylation Profiling Identify Epigenetically Regulated Genes in NSCLC. Mol. Cancer Res. 15, 1354–1365. doi:10.1158/1541-7786.Mcr-17-0191
Kong, L., Zhang, Y., Ye, Z.-Q., Liu, X.-Q., Zhao, S.-Q., Wei, L., et al. (2007). CPC: Assess the Protein-Coding Potential of Transcripts Using Sequence Features and Support Vector Machine. Nucleic Acids Res. 35, W345–W349. doi:10.1093/nar/gkm391
Laurent, G. S., Wahlestedt, C., and Kapranov, P. (2015). The Landscape of Long Noncoding RNA Classification. Trends Genet. 31, 239–251. doi:10.1016/j.tig.2015.03.007
Li, A., Zhang, J., and Zhou, Z. (2014). PLEK: A Tool for Predicting Long Non-Coding RNAs and Messenger RNAs Based on an Improved K-Mer Scheme. BMC Bioinform. 15, 311. doi:10.1186/1471-2105-15-311
Li, C., Liang, G., Yang, S., Sui, J., Yao, W., Shen, X., et al. (2017). Dysregulated lncRNA-UCA1 Contributes to the Progression of Gastric Cancer through Regulation of the PI3K-Akt-mTOR Signaling Pathway. Oncotarget 8, 93476–93491. doi:10.18632/oncotarget.19281
Liu, H., Chen, P., Jiang, C., Han, J., Zhao, B., Ma, Y., et al. (2016). Screening for the Key lncRNA Targets Associated with Metastasis of Renal Clear Cell Carcinoma. Medicine (Baltimore) 95, e2507. doi:10.1097/md.0000000000002507
Luis, M., Tavares, A., Carvalho, L. S., Lara-Santos, L., Araújo, A., and de Mello, R. A. (2013). Personalizing Therapies for Gastric Cancer: Molecular Mechanisms and Novel Targeted Therapies. World J. Gastroenterol. 19, 6383–6397. doi:10.3748/wjg.v19.i38.6383
Mistry, J., Bateman, A., and Finn, R. D. (2007). Predicting Active Site Residue Annotations in the Pfam Database. BMC Bioinformatics 8, 298. doi:10.1186/1471-2105-8-298
Ogg, S. L., Weldon, A. K., Dobbie, L., Smith, A. J. H., and Mather, I. H. (2004). Expression of Butyrophilin (Btn1a1) in Lactating Mammary Gland Is Essential for the Regulated Secretion of Milk-Lipid Droplets. Proc. Natl. Acad. Sci. U.S.A. 101, 10084–10089. doi:10.1073/pnas.0402930101
Pan, L., Liang, W., Gu, J., Zang, X., Huang, Z., Shi, H., et al. (2018). Long Noncoding RNA DANCR Is Activated by SALL4 and Promotes the Proliferation and Invasion of Gastric Cancer Cells. Oncotarget 9, 1915–1930. doi:10.18632/oncotarget.23019
Peng, W., Wang, Z., and Fan, H. (2017). LncRNA NEAT1 Impacts Cell Proliferation and Apoptosis of Colorectal Cancer via Regulation of Akt Signaling. Pathol. Oncol. Res. 23, 651–656. doi:10.1007/s12253-016-0172-4
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi:10.1038/nprot.2016.095
Renganathan, A., and Felley-Bosco, E. (2017). Long Noncoding RNAs in Cancer and Therapeutic Potential. Adv. Exp. Med. Biol. 1008, 199–222. doi:10.1007/978-981-10-5203-3_7
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Saw, P. E., Xu, X., Chen, J., and Song, E.-W. (2021). Non-Coding RNAs: The New Central Dogma of Cancer Biology. Sci. China Life Sci. 64, 22–50. doi:10.1007/s11427-020-1700-9
Sigova, A. A., Mullen, A. C., Molinie, B., Gupta, S., Orlando, D. A., Guenther, M. G., et al. (2013). Divergent Transcription of Long Noncoding RNA/mRNA Gene Pairs in Embryonic Stem Cells. Proc. Natl. Acad. Sci. U.S.A. 110, 2876–2881. doi:10.1073/pnas.1221904110
Song, H., Sun, W., Ye, G., Ding, X., Liu, Z., Zhang, S., et al. (2013). Long Non-Coding RNA Expression Profile in Human Gastric Cancer and its Clinical Significances. J. Transl Med. 11, 225. doi:10.1186/1479-5876-11-225
Sun, L., Luo, H., Bu, D., Zhao, G., Yu, K., Zhang, C., et al. (2013). Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-Coding Transcripts. Nucleic Acids Res. 41, e166. doi:10.1093/nar/gkt646
Thrift, A. P., and El-Serag, H. B. (2020). Burden of Gastric Cancer. Clin. Gastroenterol. Hepatol. 18, 534–542. doi:10.1016/j.cgh.2019.07.045
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi:10.1038/nprot.2012.016
Verma, R., and Sharma, P. C. (2018). Next Generation Sequencing-Based Emerging Trends in Molecular Biology of Gastric Cancer. Am. J. Cancer Res. 8, 207–225.
Wang, J., Su, L., Chen, X., Li, P., Cai, Q., Yu, B., et al. (2014). MALAT1 Promotes Cell Proliferation in Gastric Cancer by Recruiting SF2/ASF. Biomed. Pharmacother. 68, 557–564. doi:10.1016/j.biopha.2014.04.007
Wang, L., He, Y., Liu, W., Bai, S., Xiao, L., Zhang, J., et al. (2016). Non-Coding RNA LINC00857 Is Predictive of Poor Patient Survival and Promotes Tumor Progression via Cell Cycle Regulation in Lung Cancer. Oncotarget 7, 11487–11499. doi:10.18632/oncotarget.7203
Wang, L., Park, H. J., Dasari, S., Wang, S., Kocher, J.-P., and Li, W. (2013). CPAT: Coding-Potential Assessment Tool Using an Alignment-Free Logistic Regression Model. Nucleic Acids Res. 41, e74. doi:10.1093/nar/gkt006
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 10, 57–63. doi:10.1038/nrg2484
Yang, F., Bi, J., Xue, X., Zheng, L., Zhi, K., Hua, J., et al. (2012). Up-Regulated Long Non-coding RNA H19 Contributes to Proliferation of Gastric Cancer Cells. Febs j 279, 3159–3165. doi:10.1111/j.1742-4658.2012.08694.x
Yang, L., Wang, J., Li, J., Zhang, H., Guo, S., Yan, M., et al. (2016). Identification of Serum Biomarkers for Gastric Cancer Diagnosis Using a Human Proteome Microarray. Mol. Cel Proteomics 15, 614–623. doi:10.1074/mcp.M115.051250
Yang-Chun, F., Sen-Yu, W., Yuan, Z., and Yan-Chun, H. (2020). Genome-Wide Profiling of Human Papillomavirus DNA Integration into Human Genome and its Influence on PD-L1 Expression in Chinese Uygur Cervical Cancer Women. J. Immunol. Res. 2020, 6284960. doi:10.1155/2020/6284960
Yu, J., and Zheng, W. (2018). An Alternative Method for Screening Gastric Cancer Based on Serum Levels of CEA, CA19-9, and CA72-4. J. Gastrointest. Canc 49, 57–62. doi:10.1007/s12029-016-9912-7
Yu, Y., Wang, Z., Huang, Z., Tang, X., Huang, X., and Li, W. (2021). C1orf61 Promotes Hepatocellular Carcinoma Metastasis and Increases the Therapeutic Response to Sorafenib. Biochim. Biophys. Acta Mol. Cel Res 1868, 119048. doi:10.1016/j.bbamcr.2021.119048
Yu, Z. Y., Wang, Z., Lee, K. Y., Yuan, P., and Ding, J. (2018). Effect of Silencing Colon Cancer-Associated Transcript 2 on the Proliferation, Apoptosis and Autophagy of Gastric Cancer BGC-823 Cells. Oncol. Lett. 15, 3127–3132. doi:10.3892/ol.2017.7677
Zhang, K., Shi, H., Xi, H., Wu, X., Cui, J., Gao, Y., et al. (2017). Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics 7, 213–227. doi:10.7150/thno.16044
Zhou, X., Ye, F., Yin, C., Zhuang, Y., Yue, G., and Zhang, G. (2015). The Interaction between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell Physiol Biochem 36, 1440–1452. doi:10.1159/000430309
Keywords: long non-coding RNAs, chronic non-atrophic gastritis, gastric mucosal intraepithelial neoplasia, gastric cancer, gene expression
Citation: Liu X-Y, Zhang T-Q, Zhang Q, Guo J, Zhang P, Mao T, Tian Z-B, Zhang C-P and Li X-Y (2022) Differential Long Non-Coding RNA Expression Analysis in Chronic Non-Atrophic Gastritis, Gastric Mucosal Intraepithelial Neoplasia, and Gastric Cancer Tissues. Front. Genet. 13:833857. doi: 10.3389/fgene.2022.833857
Received: 12 December 2021; Accepted: 25 March 2022;
Published: 27 April 2022.
Edited by:
Y.-H. Taguchi, Chuo University, JapanReviewed by:
Ehsan Nazemalhosseini-Mojarad, Shahid Beheshti University of Medical Sciences, IranManoj Kandpal, The Rockefeller University, United States
Copyright © 2022 Liu, Zhang, Zhang, Guo, Zhang, Mao, Tian, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Yu Li, bGl4aWFveXUwNUAxNjMuY29t
 Xin-Yuan Liu
Xin-Yuan Liu Tian-Qi Zhang2
Tian-Qi Zhang2 Xiao-Yu Li
Xiao-Yu Li