
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 07 April 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.831656
This article is part of the Research Topic Legume Breeding in Transition: Innovation and Outlook View all 28 articles
 Uday Chand Jha1*
Uday Chand Jha1* Harsh Nayyar2
Harsh Nayyar2 Swarup K Parida3
Swarup K Parida3 Melike Bakır4
Melike Bakır4 Eric J. B. von Wettberg5,6
Eric J. B. von Wettberg5,6 Kadambot H. M. Siddique7*
Kadambot H. M. Siddique7*Legume crops, belonging to the Fabaceae family, are of immense importance for sustaining global food security. Many legumes are profitable crops for smallholder farmers due to their unique ability to fix atmospheric nitrogen and their intrinsic ability to thrive on marginal land with minimum inputs and low cultivation costs. Recent progress in genomics shows promise for future genetic gains in major grain legumes. Still it remains limited in minor legumes/underutilized legumes, including adzuki bean, cluster bean, horse gram, lathyrus, red clover, urd bean, and winged bean. In the last decade, unprecedented progress in completing genome assemblies of various legume crops and resequencing efforts of large germplasm collections has helped to identify the underlying gene(s) for various traits of breeding importance for enhancing genetic gain and contributing to developing climate-resilient cultivars. This review discusses the progress of genomic resource development, including genome-wide molecular markers, key breakthroughs in genome sequencing, genetic linkage maps, and trait mapping for facilitating yield improvement in underutilized legumes. We focus on 1) the progress in genomic-assisted breeding, 2) the role of whole-genome resequencing, pangenomes for underpinning the novel genomic variants underlying trait gene(s), 3) how adaptive traits of wild underutilized legumes could be harnessed to develop climate-resilient cultivars, 4) the progress and status of functional genomics resources, deciphering the underlying trait candidate genes with putative function in underutilized legumes 5) and prospects of novel breeding technologies, such as speed breeding, genomic selection, and genome editing. We conclude the review by discussing the scope for genomic resources developed in underutilized legumes to enhance their production and play a critical role in achieving the “zero hunger” sustainable development goal by 2030 set by the United Nations.
Burgeoning pressure from the global human population, increasing food demands, and adverse effects of global climate change are serious concerns for global food and nutrition security (Godfray et al., 2010; Foley et al., 2011; Ebi and Loladze 2019). In addition, increasing outbreaks of plant diseases and pests, loss of arable land, and increasing environmental degradation due to excessive use of chemical fertilizers and pesticides have constrained crop yields (Godfray et al., 2010; Lesk et al., 2016). Of the various approaches for sustaining global food production without deteriorating soil and environmental health, crop diversification is needed to maintain sustainable agro-ecological systems and prevent biodiversity losses (Hufnagel J. et al., 2020; Tamburini et al., 2020). Legume crops remain the third most widely grown class of crops globally (Gepts et al., 2005), providing “one third of all dietary protein nitrogen” to the human population, enriching soil fertility by fixing atmospheric nitrogen in association with symbiotically active rhizobacteria in roots (Graham and Vance, 2003), and adding rotational value to subsequent crops (Yigezu et al., 2019; Marques et al., 2020). Likewise, legume fodder and forage mitigate the rising global demand for dietary protein by livestock and provide industrial raw materials (Das and Arora 1978; Elfaki and Abdelatti 2018). Most studies have focused on major grain legumes, such as soybean, common bean, and chickpea. However, some legume crops (Supplementary Table S1) with high nutrient contents are grown in limited areas on small scales in developing countries under low input conditions and marginal land (Cullis and Kunert 2017; Kamenya et al., 2021). Despite the enormous potential of these legumes, they are neglected and known as “underutilized” legumes (Cullis and Kunert 2017; Kamenya et al., 2021). Underutilized species are rarely grown outside of a narrow geographic area, are cultivated with low chemical inputs or mechanization, are not broadly used outside of traditional cuisines, and have not been the focus of major public and private breeding companies. In the last decade, major grain legume crops have witnessed unprecedented advances in genomic resource development, including the development of reference genome sequences due to rapid advances in genome sequencing technologies, especially, next-generation sequencing (NGS). However, underutilized legume crops are lagging behind in terms of developing genomic resources. Thus, in this review we analyze the present global status of these underutilized legumes in terms of area, production, major production and nutritional quality limitation and origin (Supplementary Table S1) and discuss the available genomic resources, including their molecular marker repertoire and genome assemblies. We review the progress in genetic linkage maps and identification of trait QTLs through bi-parental mapping and genome-wide association studies of various underutilized legumes, including the downstream application of genomic assisted breeding (GAB). The discovery of various trait candidate gene(s) with putative function through transcriptome sequencing are discussed with examples. We also brief how crop wild relatives (CWRs), whole-genome resequencing (WGRS), and pangenome sequences could underpin novel structural variants across the whole genome in these crops. Finally, we propose the prospects and scope of novel breeding schemes—genomic selection, genome editing, and speed breeding—for enhancing genetic gain to achieve “zero hunger” in 2030.
Underutilized legumes generally require few inputs, are rich in protein, vitamins, and minerals, and can often withstand harsh environments, including drought, extreme temperature, and waterlogging. Furthermore, these legumes replenish soil nitrogen by fixing atmospheric nitrogen through root nodules, ameliorate soil properties, and sustain agro-ecosystem services (Bhartiya et al., 2015; Ditzler et al., 2021). In addition to their role in combating nutritional and economic security, underutilized legumes play critical roles in various human diseases as they are rich in bioactive compounds and nutraceutical and medicinal properties (Prasad and Singh 2015; Bazzano et al., 2001). However, despite these benefits, there are several constraints and challenges related to the production and productivity of these legumes due to biotic and abiotic stresses (Supplementary Table S1). Furthermore, the edible seeds of some underutilized legumes contain antinutritional elements, constraining their use (Campbell et al., 1994; Tate and Ennenking 2006; Kroc et al., 2017). Combining modern genomic and traditional breeding approaches could help develop new plant types, reduce yield losses from biotic and abiotic stresses, add value for consumer preferences, and eliminate antinutritional properties.
One of the aspects of the advances in DNA sequencing technology over the past two decades has been the potential to democratize research. Before the advent of next generation sequencing, performing molecular genetic research outside of a handful of species, such as fruit flies and Arabidopsis, was cost-prohibitive. Exponential declines in the cost of sequencing have made research in nearly any species not only feasible, but practical. Consequently, crops like chickpeas, pigeonpea and cowpea, once considered minor crops, now have a rich array of genome resources (e.g., Jha 2018; Varshney et al., 2021). However, there are still a range of crop species that have received less attention, due to being grown over a limited geographic extent or market demand that is mostly restricted to a small region.
For those crop species that still trail behind others for genomic resources, there is hope that lessons learned in other species can be applied to others. In legumes, where there is substantial genome synteny across the entire family [e.g., (Ren et al., 2019)], the potential for comparative genomics to speed up research in understudied species is particularly high. With improving databases for mining genomic information from more widespread cultivated legumes [e.g., (Bauchet et al., 2019; Berendzen et al., 2021)], this task has become easier than in the past.
In a range of minor legume crops, one of the foci for improvement are “domestication syndrome” traits, such as pod shattering, seed dormancy, seed size, and palatability. There is growing evidence that at least some of the loci controlling these traits are shared, such as for pod shattering (Ogutcen et al., 2018). With shared loci and extensive genomic synteny, either finding natural variation at these loci or using genetic modification become much easier.
In the last decade, rapid advances in genome sequencing technologies have enriched the genomic resources, including genome-wide distributed high-throughput molecular markers especially, simple sequence repeats (SSRs) and single nucleotide polymorphisms (SNPs), transcriptomes, and whole-genome assemblies, of various underutilized legumes.
Hybridization-based molecular markers, such as restriction fragment length polymorphisms (RFLP), and PCR-based molecular markers, such as RAPD, SSR markers, have been used to analyze, tag, and map trait gene(s) in various underutilized legumes (Bohra et al., 2014). However, the arrival of next-generation sequencing technology (NGS) based high-throughput (HTP) markers, especially SNPs, has replaced traditional PCR-based molecular markers for genotyping. Second- and third-generation sequencing technologies have enabled the mining of massive numbers of SSRs and SNPs through whole-genome sequencing, WGRS, and transcriptome sequencing efforts in various crops, including underutilized legumes (Edwards and Batley 2010).
Likewise, the advent of NGS-based HTP genotyping platforms, such as Illumina’s GoldenGate assay, Illumina’s HiSeq 4000 platform, and Illumina’s Infinium SNP array, enabled the discovery of copious SNPs across multiple genomes that facilitate a range of investigations, including the diversity of genebank collections (Sokolkova et al., 2020). Aiming at comprehensive mining of SSR markers for Vigna species including cowpea, mungbean and adzuki bean, microsatellite database VigSatDB has been developed (Jasrotia et al., 2019). A comprehensive list of molecular markers, mapping populations available in various underutilized legumes are in Table 1. Thus, these molecular markers will provide the foundation for implementing genomic assisted breeding for improving genetic gain in underutilized grain legumes.
Adzuki bean (Vigna angularis var. angularis) (2n = 2x = 22) is an important grain legume of Asiatic origin (Kang et al., 2015). The draft genome sequence of adzuki bean was assembled on 11 pseudo-chromosomes, estimating 612 Mb or 75% of the estimated genome and high-confidence 26,857 protein-coding genes (Kang et al., 2015) (Table 2). Yang K. et al. (2015) assembled a draft genome assembly of “Jingnong 6” cultivar covering 450 Mb of the total genome.
Bambara groundnut (Vigna subterranean) (2n = 2x = 22) is an important legume crop, rich in protein (18–26%), carbohydrate (63%), and fat (6.5%) and having inherent drought tolerance capacity (Shegro et al., 2013). It originated from West Africa and is mainly grown in sub-Saharan areas, especially Nigeria (Olukolu et al., 2012). Chang et al. (2019) assembled the genome sequence of bambara groundnut, with a genome size of ∼535.05 Mb with 31,707 protein-coding genes.
Mungbean (Vigna radiata, 2n = 2X = 22) is a warm-season legume crop, originated from India and mostly grown in South and Southeast Asian countries. Kang et al. (2014) first assembled the mungbean genome sequence, estimating 421 Mb or 80% of the total genome size and 22,427 protein-coding genes, with scaffold length 431 Mb and N50 length 35.4 Mb covering 314 Mb. Recently, a mungbean genome sequence was assembled with a total scaffold size of 475 Mb and N50 scaffold value of 5.2 Mb (Ha et al., 2021).
Urdbean (Vigna mungo, 2n = 2x = 22), native to Indian subcontinent, mostly grown in South and Southeast Asian countries (Kaewwongwal et al., 2015), is a rich source of dietary protein, vitamins, folate, and iron (Kakati et al., 2010). The genome assembly of Chai Nat 80 cultivar measured 499 Mb with an N50 length of 5.2 Mb (Pootakham et al., 2021). Subsequently, Jegadeesan et al. (2021) assembled a genome assembly of urdbean, measuring 475 Mb or 82% of the genome with scaffold N50 of 1.42 Mb and 42,115 genes with coding sequence.
Cluster bean (Cyamopsis tetragonoloba, 2n = 2x = 14), native to west Africa and India, an important commercial legume crop widely grown in India and parts of Africa, contains hetero-polysaccharide called guar gum or galactomannan used extensively in the cosmetic and pharmaceutical industries (Gillett 1958). Gaikwad et al. (2020) assembled the first genome sequence of RGC-936 cultivar, measuring 550.31 Mbp with N50 length of 78.27 Mbps and 34,680 protein-coding genes.
Dolichos bean (Lablab purpureus) (2n = 2x = 22) is a versatile legume crop of African origin, rich in seed protein and highly tolerant to various abiotic stresses (Maass et al., 2010). It is mostly cultivated in tropical and sub-tropical regions globally (Maass et al., 2010). The genome assembly of Lablab purpureus was constructed recently, with an estimated 395.47 Mb genome size and 20,946 protein-coding genes (Chang et al., 2019).
Grass pea (Lathyrus sativus) is a climate-resilient legume of Central Asia and Abyssinia origin, diploid (2n = 2x = 14), cool-season legume species (Kamphuis et al., 2015; Emmrich et al., 2020) primarily grown on the Indian subcontinent and in northern and eastern Africa, including Ethiopia (Kumar et al., 2011). The assembled genome size of EIv1 was measured at 8.12 Gbp with scaffold N50 value of 59.7 kbp and 33,819 high-confidence genes (Kamphuis et al., 2015; Emmrich et al., 2020).
Horsegram [Macrotyloma uniflorum (Lam.) Verdc.], native to tropical southern Asia, is a diploid legume (2n = 20, 22) grown in India, Africa, and Australia (Arora and Chandel 1972). The genome sequence of the HPK-4 genotype was assembled on ten pseudomolecules measuring 259.2 Mb or 89% of the total length of the assembled sequence (Shirasawa et al., 2021a). Another genome assembly of accession PHG-9, measuring 279.1 Mb with 24,521 annotated genes has recently been constructed (Mahesh et al., 2021).
Red clover (Trifolium pratense L.; Fabaceae, 2n = 2x = 14) is an important forage legume of European origin, with a genome size of 418 Mbp. Ištvánek et al. (2014) completed a de novo assembly of the red clover genome, comprising ∼314.6 Mbp.
Likewise, subterranean clover presumed to be originated from Southern Australia, belonging to Trifolium genus, is an annual diploid (2n = 2x = 16) pasture legume with 540 Mbps genome size (Kaur et al., 2017). Hirakawa et al. (2016) assembled the genome sequence of T. subterraneum L., measuring 471.8 Mb or 85.4% of the whole genome and containing 42,706 protein-coding genes. Subsequently, Kaur et al. (2017) assembled an advanced genome assembly of T. subterraneum L., estimating 512 Mb with 31,272 protein-coding genes.
Tepary bean (Phaseolus acutifolius A. Gray), native to the Sonoran Desert and a sister species of common bean, is gaining attention due to its inherent capacity for biotic and abiotic stress tolerance (Moghaddam et al., 2021) and important source traits for improving biotic and abiotic stress tolerance in common bean (Moghaddam et al., 2021). A reference genome assembly of cultivated landrace Frijol Bayo, possessing inherent heat tolerance, was constructed using Illumina X10 and HiSeq platforms and PACBIO with 101.28× sequence coverage, and measured 512,626,114 bp with 27,538 high-confidence genes (Moghaddam et al., 2021).
White lupin (Lupinus albus L. 2n = 50) originated from Mediterranean region, contains high protein content (30–40% whole seed) (Bähr et al., 2014) and can use higher soil phosphorus than other legume crops due to its special “cluster root” structure (Lambers et al., 2013). However, improving yield stability and minimizing anti-nutritional alkaloids in white lupin seed through conventional breeding remains challenging. Hence, to elucidate the function of various trait gene(s) related to quality and quantitative importance, Bárbara Hufnagel et al. (2020) assembled a high-quality genome sequence of white lupin, scaling 451 Mb and 38,258 annotated protein-coding genes.
Likewise, narrow-leafed lupin (Lupinus angustifolious) is an important grain legume of Mediterranean origin, enriched with dietary protein (40–45%) and fiber (25–30%) (Lee et al., 2006). Hane et al. (2017) assembled the draft genome sequence of Tanjil cultivar, estimating 609 Mb and 33,076 protein-coding genes. Subsequently, Wang et al. (2021a) constructed an improved genome assembly of Tanjil, measuring 615.8 Mb with contig N50 = 5.65 Mb, using a long-read whole-genome sequencing approach.
Common vetch (Vicia sativa, 2n = 14) originated from Near Eastern centre of diversity, is a wild and partially domesticated legume crop with a genome size of 1.8 Gb (Shirasawa et al., 2021b). It is used as silage and hay for livestock feeding. The reference genome assembly has been assembled, spanning 1.5 Gb and 31,146 genes (Shirasawa et al., 2021b).
As most of the traits with agricultural importance including biotic, abiotic stress tolerance and quality traits are governed by multiple gene(s)/quantitative trait loci (QTL). In order to map these traits various molecular breeding approaches are available to breeders, including family based bi-parental mapping approach, marker-assisted backcrossing. Subsequently, the availability of high-throughput molecular markers has accelerated the precise mapping of various trait QTLs through employing novel molecular breeding schemes including MutMap, multi-parental cross (MAGIC), genome-wide association mapping, genomic selection and QTL seq approach (Meuwissen et al., 2001; Cavanagh et al., 2008; Takagi et al., 2013; Takagi et al., 2015). In underutilized legumes several bi-parental mapping populations based on interspecific and intraspecific crosses have been developed aiming at constructing genetic linkage map and mapping/tagging targeted trait QTLs of agronomic importance (for details Table 3). However, mapping resolution of detected QTLs through bi-parental mapping approach remains low. Therefore, to increase the resolution of trait QTLs novel breeding scheme viz., genome-wide association study (GWAS), nested association mapping and MAGIC has been developed. We believe these approaches could be implemented in underutilized legumes to increase the resolution of trait QTLs.
Initially, morphological-based markers, isozymes, RFLP, amplified fragment length polymorphisms (AFLP), randomly amplified polymorphic DNAs (RAPD), and SSR markers were used to construct preliminary genetic linkage maps in various underutilized legumes [for details, (Bohra et al., 2014)]. However, the increasing ease of developing high-throughput SNP markers derived by GBS, restriction site-associated DNA sequencing (RAD-seq), and whole genome resequencing has facilitated developing highly dense/saturated consensus linkage maps in various underutilized legumes.
Several genetic maps of mungbean based on SSR markers have been developed (Bohra et al., 2014). Later, a genetic map measuring 1,060.2 cM was developed from an intraspecific mapping population (Wang et al., 2020) and a denser genetic map with 1,291.7 cM and harboring 538 SNPs was developed from an interspecific mapping population derived from Vigna radiata × V. umbellate cross (Mathivathana et al., 2019) (Table 4).
A comprehensive genetic map of urd bean (V. mungo) covering 1,588.7 cM with 3,675 SNPs was developed (Somta et al., 2019). Based on a F2 population, Kai Yang et al. (2015) developed an initial genetic map in adzuki bean measuring 1,031.17 cM. Wang et al. (2021b) presented a denser genetic map measuring 1,365.0 cM in adzuki bean (V angularis). In zombi pea (V. vexillata), a high-density linkage map spanning 1740.9 cM harboring 6,529 SNPs with an average distance of 0.27 cM between markers was developed from an F2 mapping population of TVNu 240’ × “TVNu 1,623” (Amkul et al., 2019).
Hane et al. (2017) presented a high-density linkage map of narrow-leafed lupin measuring 2,500.8 cM with 9,972 loci and Iqbal et al. (2019) developed a high-density linkage map of yellow lupin measuring 2,261.3 cM. Santos et al. (2018) developed a genetic map of lathryus covering 724.2 cM with 307 loci. Chahota et al. (2020) presented a genetic map for horse gram measuring 1,423.4 cM with 211 loci (Table4).
The above linkage maps can be used to identify various traits of biotic, abiotic stress tolerance, agronomic, and culinary importance in numerous underutilized legumes. The selected major trait QTLs identified in the last decade based on bi-parental mapping populations are listed in (Table 4). Biotic stress remains the most significant yield stress in underutilized grain legumes globally. The increased availability of genomic resources, especially molecular markers, has identified/tagged various disease-resistant QTLs/gene(s); for example, one major QTL qCc_PDS6.1 against Callosobruchus chinensis (bean weevil) and another QTL qCm_PDS6.1against Callosobruchus maculatus (cowpea weevil) have been identified (Amkul et al., 2019). Likewise, four major QTLs (antr04_1, antr05_1, antr04_2 and antr05_2) controlling anthracnose resistance explaining 14–25% (Rychel-Bielska et al., 2020) of the phenotypic variation in white lupin. Restriction site-associated DNA sequencing derived SNP markers were used as candidate markers for the R gene of phomopsis stem blight disease resistance in narrow-leafed lupin (Yang et al., 2013b). Recently, one major QTL with LOC109334326, LOC109334327 underlying candidate genes was deciphered for gray leaf spot disease in narrow-leafed lupin (Zhou et al., 2021).
Like biotic stresses, abiotic stresses, particularly drought, causes significant yield losses in underutilized legumes (Liu et al., 2017; Chahota et al., 2020). Several QTLs contributing to drought tolerance have been discovered in mungbean (Liu et al., 2017), and horse gram (Chahota et al., 2020).
Low seed-alkaloid content (<0.02%) is a prime objective of quality improvement in lupin. In lupin the iucundus allele is a major gene regulating seed alkaloid content. Several mapping populations have been developed for identifying low alkaloid controlling QTLs and gene(s). Li et al. (2011) identified a microsatellite-anchored fragment length polymorphism-derived PCR marker (lucLi) linked to the low-alkaloid locus iucundus (0.9 cM). Likewise, Lin et al. (2009) developed a sequence-specific PCR marker (PauperM1) closely linked (1.4 cM) to the low-alkaloid locus pauper in white lupin (Lupinus albus L.). Moreover, of five SNP markers co-segregating the pauper locus in a set of 140 lupin accessions, the LAGI01_35805_F1_R1 marker was highly linked with this gene and could be used in low seed alkaloid lupin breeding programs (Rychel and Książkiewicz, 2019). Subsequently, Kroc et al. (2019) developed a co-dominant derived cleaved amplified polymorphic sequence (dCAPS) marker (iuc_RAP2-7) from the RAP2-7 candidate gene of alkaloid locus iucundus responsible for seed alkaloid content in narrow-leafed lupin, which could be used in marker-assisted breeding for low alkaloid content in lupin. Furthermore, fine mapping of this seed alkaloid controlling genomic region unveiled four candidate gene(s)—LOC109339893, LOC109339862, LOC109339875 and LOC109339876—on LG7 in the interval of 20.70–20.89 Mb (Wang et al., 2021a).
GWAS is gaining popularity for uncovering genotype–phenotype associations in various plant species, including underutilized legumes (Huang and Han 2014; Liu and Yan 2019), by establishing the genetic basis of the genotype–phenotype association for the trait of interest in a large panel of diverse accessions based on multiple crossing-over events over the recent demographic history of a taxa (Huang and Han 2014). Due to the unprecedented advances in NGS technology, an increasing repertoire of HTP markers in several underutilized legumes have helped to identify loci associated with aspects of complex trait architecture. GWAS has been assisted by the subsequent availability of genome-wide SNP markers for various traits, including phenological traits, quality/nutritional traits, biotic and abiotic stresses, and yield and yield-related traits, in many underutilized legumes (Plewiński et al., 2020). In narrow-leafed lupin, a GWAS incorporating massive analysis of cDNA ends (MACE) markers in 126 gentoypes uncovered significant MTAs related to flower initiation, maturity, plant height, and yield traits (Plewiński et al., 2020). The underlying candidate genes were Lup019134, Lup015264, Lup021911, and Lup021909 for flower initiation, Lup015264 and Lup004734 for maturity, Medtr1g030750 for plant height, and Lup021835 and Lup022535 for yield traits (Plewiński et al., 2020).
GWAS has been used increasingly for dissecting complex QTLs controlling various abiotic stresses in crop plants, including underutilized legumes. To elucidate the underlying genomic regions attributing macro- and micro-nutrients in mungbean seeds, Wu et al. (2020) identified 43 MTAs related to calcium, iron, manganese, phosphorus, sulfur, and zinc using inductively coupled plasma (ICP) spectroscopy and GBS-derived SNPs in a set of 95 global mungbean accessions. The explained phenotypic variation ranged from 1 to 38%. Further, Reddy et al. (2021) used a GBS-based GWAS study to dissect the molecular basis of phosphorus uptake efficiency and phosphorus utilization efficiency in 120 mungbean genotypes. The authors uncovered 116 SNPs in 61 protein-coding genes related to phosphorus uptake efficiency and phosphorus utilization efficiency traits. The significantly associated SNPs explained phenotypic variation ranging from 17 to 20% for total phosphorus utilization (under low phosphorus) and it ranged from 15 to 21% for phosphorus utilization efficiency. Six candidate genes—VRADI01G04370, VRADI05G20860, VRADI06G12490, VRADI08G20910, VRADI08G00070 and VRADI09G09030—regulating phosphorus uptake efficiency and phosphorus utilization efficiency were deciphered (Reddy et al., 2021).
Recently, recruiting 5,041 SNPs in a minicore collection of 293 mungbean accessions identified four significant MTAs for maturation and hypocotyl color within the Vradi02g04380 gene on chromosome 2 encoding zinc finger A20 and AN1 domain stress-associated protein (Sokolkova et al., 2020). Despite the popularity of GWAS for elucidating marker-trait associations, it has some drawbacks regarding population structure and low-frequency causal alleles causing false negative results (Korte and Farlow 2013). To minimize and overcome the population structure related problems, artificially designed populations such as MAGIC and nested association mapping, could be used [for details (Alseekh et al., 2021)].
CWRs, including those of underutilized legumes, are a hidden reservoir of novel trait gene(s), offering scope for broadening genetic diversity in crop breeding programs (Warschefsky et al., 2014; Zhang and Batley, 2020). In the past, during domestication process, several genes associated with adaptive traits conferring abiotic stress tolerance were lost rendering modern cultivated crop plants to adapt poorly under stress condition (Warschefsky et al., 2014; Zhang and Batley, 2020). However, CWRs serve as reservoir of these biotic and abiotic stress adaptive genes. Thus, recapturing these genes from CWRs through introgression and novel breeding tools could facilitate in increasing the fitness of genepool (Burgarella et al., 2019). Several CWRs of underutilized legumes. e.g., V. nakashimae, are potential sources of bruchid resistance (Somta et al., 2006) and salinity tolerance (Yoshida et al., 2016) in adzuki bean. Likewise, harnessing bruchid resistance genes/genomic regions from Vigna radiata var. sublobata can improve bruchid resistance in mungbean (Schafleitner et al., 2016) (Table 5). In urd bean, V. mungo var. silvestris could be promising for transferring bruchid and mungbean yellow mosaic India virus resistance genes into high-yielding urd bean breeding lines (Souframanien and Gopalakrishna, 2006; Souframanien et al., 2010). Further, the genomic sequences of wild underutilized legumes have been assembled to gain insight into the novel trait genes of CWRs. Whole-genome sequencing of M. ruthenica offered novel insights into many genes, including the FHY3/FAR1 gene family conferring higher drought tolerance in cultivated M. sativa (Wang et al., 2021c). Mou Yin et al. (2021) advocated evidence for multiple family genes and TF family genes, viz., C2H2, CAMTA and NAC attributing various abiotic stress tolerances through chromosome-scale genome sequencing of M. ruthenica. Novel SNP and InDel markers were recovered from genome sequencing of V. radiata var. sublobata; the wild relative accession TC1966 of mungbean could be useful for exploring biotic and abiotic stress tolerant genomic regions through comparative mapping of cultivated mung bean (Kang et al., 2014). Thus, these CWR genomic resources could be used to develop climate-resilient grain legume cultivars.
Current advances in genomic resource development in underutilized legumes have enabled breeders to develop improved cultivars. For example, tagging various traits in narrow-leafed lupin, such as LanFTc1 PCR-based INDEL markers for vernalization responsiveness locus Ku/Julius (Nelson et al., 2017; Plewiński et al., 2019; Taylor et al., 2019), InDel2, InDel10, and PhtjM7 for PhtjR (Yang et al., 2013b; Yang H. et al., 2015), Anseq3 and Anseq4 for Lanr1 (Yang et al., 2012), and TP222136 and TP47110 markers for antr04_1/antr05_1 and TP338761 for antr04_2/antr05_2 (anthracnose resistance) (Rychel-Bielska et al., 2020), the iucLi co-dominant marker (Li et al., 2011) and RAP2-7 PCR-based dCAPS marker for major alkaloid content locus iucundus (Kroc et al., 2019) are available. Likewise, a diagnostic marker LAGI01_35805_F1_R1 linked to pauper locus controlling low alkaloid content in white lupin could be used for practicing MAS of white lupin lines with low-alkaloid content (Rychel and Książkiewicz 2019). Moreover, co-dominant markers linked to the tardus (Li et al., 2010) and lentus (Li et al., 2012) genes, attributed to low pod shattering, could be of interest for developing zero shattering narrow-leafed lupin using marker-assisted breeding.
Similarly, CEDG261 and DMB-SSR160 markers linked to bruchid resistance could be used in GAB in moth bean breeding programs (Somta et al., 2018). Downstream application of GAB in concert with other novel breeding approaches for enhancing genetic gain in various underutilized legumes is depicted in Figure 1.
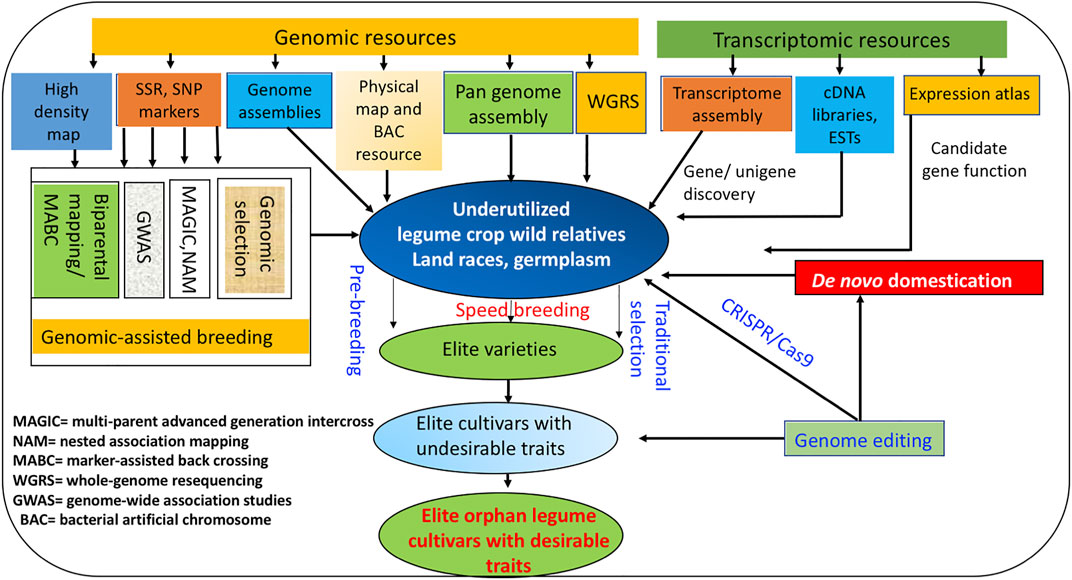
FIGURE 1. “Omics” and emerging novel breeding approaches for improving genetic gain in underutilized legumes.
The advent of NGS-based RNA-seq technology assessing global gene expression has offered a platform for the discovery of functional markers, including EST-SSRs and SNPs, capturing gene space and shedding light on a myriad of trait candidate genes and their plausible functions (O’Rourke et al., 2013; Yang et al., 2017; Glazińska et al., 2019). Previously, EST markers, microarrays, and cDNA libraries were the major functional genomic resources for investigating the function of various trait genes. For example, cDNA library sequencing identified 125,821 unique sequences (O’Rourke et al., 2013) in white lupin.
Subsequently, advances in transcriptome sequencing facilitated the discovery of many unigenes and differentially expressed genes for various traits of importance for details (see Table 6). Transcriptome studies have also shed light on the functional role of various underlying candidate gene(s) controlling seed biology, plant phenology, biotic and abiotic stress tolerance, yield traits, and nutritional quality traits, including alkaloid regulation in narrow-leafed lupin, β-N-oxalyl-L-α, β-diaminopropionic acid (β-ODAP) in grass pea and condensed tannin in winged bean (Kroc et al., 2019; Yang et al., 2017; Xu et al., 2018).
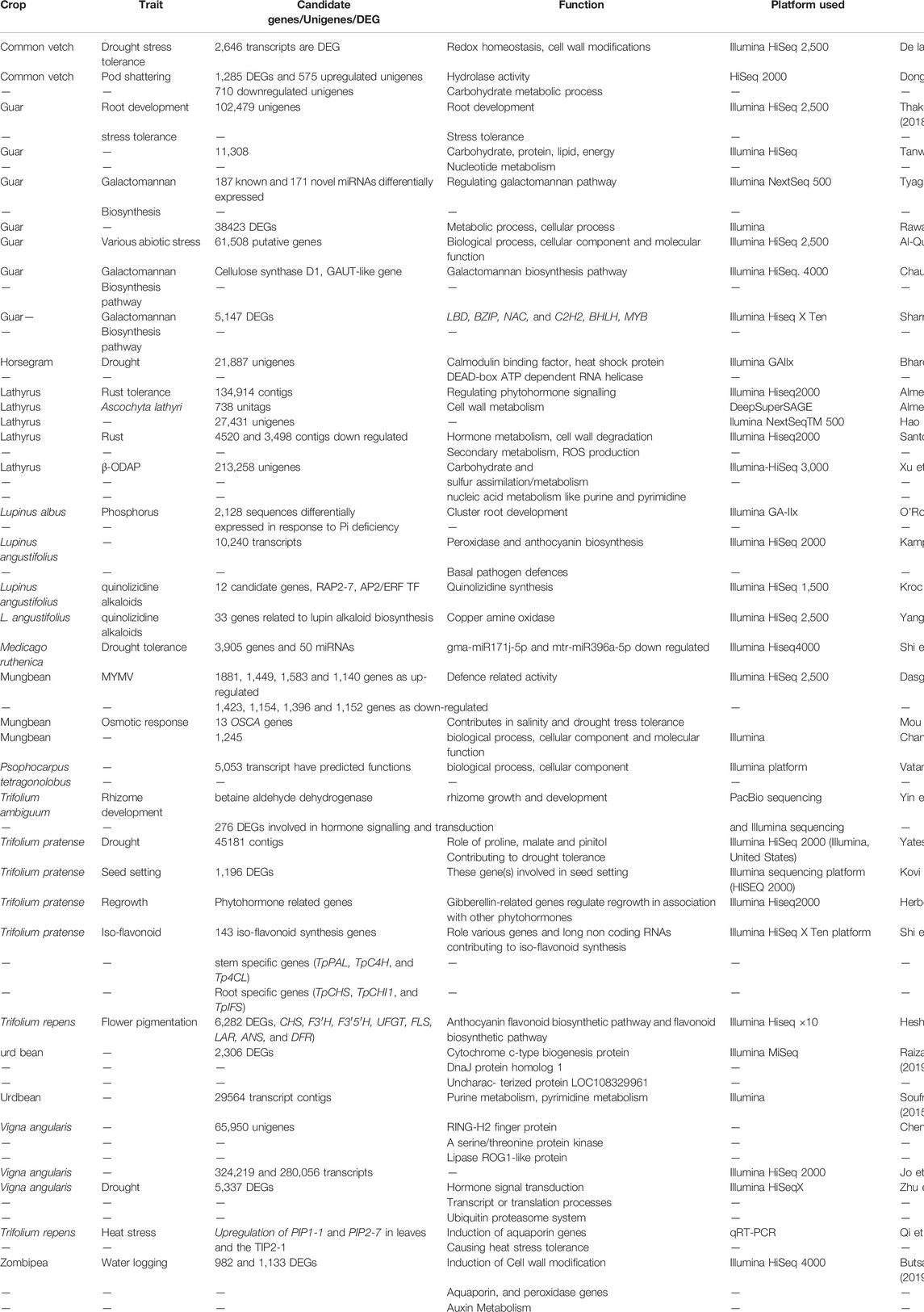
TABLE 6. List of various differentially expressed genes/candidate traits genes with putative function in underutilised legumes.
In association with small RNA sequencing, degradome sequencing and transcriptome sequencing helped unravel key molecular players, including various phytohormones and metabolic pathways involved in floral development and organ abscission of L. luteus (Glazinska et al., 2017; Glazińska et al., 2019). Moreover, participation of small RNA related to seed biology and the conglutin gene encoding seed storage protein was demonstrated in a transcriptome study in narrow-leafed lupin (DeBoer et al., 2019).
Transcriptome studies could improve our understanding of the regulatory mechanisms of the complex network of gene(s), pathogenesis-related genes, phytohormone signaling response, and non-coding RNAs mediating plant immune responses to attacking pathogens (Almeida et al., 2014; Dasgupta et al., 2021). To gain insight into the molecular mechanisms involved in conferring rust resistance in grasspea, an RNA-seq study in rust-responsive grasspea (resistant vs. susceptible) revealed the upregulation of salicylic acid and abscisic acid in the rust-resistant genotype and downregulation of jasmonate and ethylene pathways in the susceptible genotype (Almeida et al., 2014) (Table 6). Additionally, several pathogenesis-related genes and the mildew resistance locus O (MLO)-like resistance gene were discovered in this study.
An RNA-seq study offered insight into the participatory role of WRKY, NAC and MYB transcription factors, phytoene synthase, cytochrome P450, and JAZ and LOX genes attributing to mungbean yellow mosaic virus (MYMV) resistance (Dasgupta et al., 2021).
Likewise, transcriptome studies can decipher the complex molecular mechanisms and underlying possible candidate gene(s) networks during perceiving abiotic stress signaling and mediate various abiotic stress tolerances by activating antioxidant mechanisms and other cellular protective mechanisms, enabling plants to acclimate to abiotic stress (Bhardwaj et al., 2013; Butsayawarapat et al., 2019; De la Rosa et al., 2020).
A de novo transcriptome analysis of two contrasting horse gram genotypes for drought tolerance revealed the involvement of various TFs (NAC, MYB, and WRKY families) in conferring drought stress tolerance (Bhardwaj et al., 2013). De novo transcriptome sequencing of contrasting drought tolerant and sensitive genotypes of common vetch revealed a plethora of differentially expressed genes under water stress (De la Rosa et al., 2020). Most of the genes mediating drought tolerance are associated with cell wall modification, oxidative stress response and ABA response (De la Rosa et al., 2020). In zombi pea, a comparative transcriptome analysis revealed up-regulatory activity of glycolysis and fermentative genes in the waterlogging-sensitive genotype; in contrast, the waterlogging-tolerant genotype had enhanced activity of auxin-regulated lateral root initiation, aquaporin, and peroxidase genes (Butsayawarapat et al., 2019) (Table 6).
Deciphering the underlying genes and molecular function of quality parameters, including nutritional and industrially important parameters, using transcriptomic studies could improve these traits (Yang et al., 2017, Xu et al., 2018; Tyagi et al., 2018). Small RNA sequencing indicated the involvement of several miRNAs and their target genes coding for carbohydrate metabolism, kinase, and enzymes for regulating galactomannan biosynthesis in cluster bean (Tyagi et al., 2018) (Table 6). The authors also discovered two novel unigenes, mannosyltransferase/mannan synthase (ManS) and UDP- D-glucose 4-epimerase (UGE), targeted by Ct-miR3130, Ct-miR3135, and Ct-miR3157 miRNAs. Likewise, an RNA-seq study revealed preferential expression of 2,535 and 2,724 genes in endosperm and 3,720 and 2,530 genes in the embryo involved in guar gum biosynthesis (Hu et al., 2019).
Transcriptome assembly through RNA-seq identified several candidate genes regulating quinolizidine alkaloids (QAs) biosynthesis, an anti-nutritional factor in narrow-leafed lupin (Kamphuis et al., 2015; Yang et al., 2017; Kroc et al., 2019). Short-read sequencing using Illumina HiSeq2500 in association with long-read sequencing using PacBio technology of high QA-containing genotypes identified 33 candidate genes associated with QA biosynthesis in narrow-leafed lupin (Yang et al., 2017). Furthermore, transcriptome profiling offered insight into the genes involved in the accumulation and degradation of β-N-oxalyl-L-α, β-diaminopropionic acid (β-ODAP), a neurotoxin found in grasspea (Xu et al., 2018). Similarly, RNA-seq analysis of high- and low-tannin-containing lines of winged bean, using Illumina Nextseq 500, revealed 1,235 differentially expressed contigs in these two lines. Several genes related to condensed tannin were elucidated, including anthocyanidin 3-O-glucosyltransferase (A-3GOT), anthocyanidin synthase (ANS), chalcone synthase (CHS) phenylalanine ammonia-lyase (PAL) (Singh et al., 2017).
The decoding of various underutilized legume genome sequences and resequencing efforts have made SNP markers accessible, providing great opportunities to perform genomic selection (GS). This approach has been used for estimating the genomic breeding value of tested individuals without any prior phenotypic information by measuring the genome-wide marker effect based on various prediction models (Meuwissen et al., 2001). Thus, the benefits of GS could be harnessed for the selection of progenies with known genotypic scores with high “genetic merit” for improving genetic gain.
Assessing anthracnose resistance in white lupin using GS based on GBS-derived SNPs in the ridge regression BLUP model, Rychel-Bielska et al. (2020) reported a moderately high predictive ability (0.56). Application of GS is very limited in minor legumes; however, increasing repertoire of genome wide SNP markers will greatly assist in implementing GS for improving future genetic gain in these legumes.
Speed breeding could be used to increase breeding efficiency by shortening the breeding cycle and reducing plant space, cost, and labor resources, thereby increasing genetic gain (Watson et al., 2018; Hickey et al., 2019). Speed breeding protocols have been established by optimizing photoperiod, daylength, and temperature in various legume crops, including soybean (Fang et al., 2021), chickpea (Samineni et al., 2019), pigeonpea (Saxena et al., 2019), and pea (Mobini and Warkentin, 2016). However, this approach has not been implemented in any underutilized legumes. Thus, the establishment of a speed breeding protocol could open up new avenues for improving genetic gain in various underutilized legumes more quickly than traditional breeding methods.
With the declining costs of genome assembly construction, whole genome resequencing is gaining popularity for uncovering genomic regions controlling traits of agronomic importance in a large set of global crop germplasm (Hufnagel B. et al., 2020).
The WGRS approach can elucidate the causal candidate gene(s)/genomic regions associated with traits of interest. Like other major grain legumes, WGRS has been used in underutilized legume crops (Yang H. et al., 2015; Hufnagel B. et al., 2020). The resequencing of nine lupin cultivars discovered 180,596–795,735 SNP markers and 243 candidate diagnostic markers linked to the PhtjR (phomopsis stem blight disease) gene (Yang H. et al., 2015). Of these candidate diagnostic markers, nine were validated in commercial cultivars, offering an opportunity to practice marker-assisted breeding for phomopsis stem blight disease resistance in narrow-leafed lupin.
Resequencing 11 modern cultivars, two landraces, and one wild relative of white lupin and comparing them with the reference genome sequence revealed the recent breeding history of white lupin (Hufnagel B. et al., 2020). Similarly, 38 narrow-leafed lupin accessions, including 19 wild and 19 cultivated types, with 19× coverage of the genome were resequenced to reveal the genomic signal for domestication and genes associated with the domestication process (Wang et al., 2021a). A selective sweep analysis in the same study identified 303 genomic regions under strong selection, with 8.2% of the genome under selection associated with domestication. Further, these selective sweeps harbored nine key domestication-related traits, including early flowering, reduced pod shattering, white flower, and low alkaloid (Wang et al., 2021a). WGRS efforts of three mungbean accessions using the Ion Torrent Personal Genome MachineTM (PGMTM) platform identified 233,799 SNPs and 9,544 insertions and deletions in coding and non-coding regions, revealing great opportunity for future mung bean improvement using genomic-assisted breeding (Bangar et al., 2021).
Previously, molecular biologists and geneticists have relied mainly on the “single reference genome sequence” of a species for genetic analyses within and across species (Sherman and Salzberg, 2020; Della Coletta et al., 2021). However, the single reference genome sequence does not explain all of the genomic variation/structural variants available within and across species; “pangenomics” studies can capture all of the genomic information in a species. The pangenome refers to the entire non-redundant DNA sequences existing in a species, constituting the “core” genome common to all individuals in a species, with “dispensable” genome the variable fraction or “accessory” genome (Tettelin et al., 2005; Sherman and Salzberg, 2020; Della Coletta et al., 2021; Lei et al., 2021). In the context, Hufnagel et al. (2021) constructed the pangenome of white lupin using a “map to pan” approach (Hu et al., 2017) by sequencing 39 accessions, which identified 32,068 core genes and 14,822 dispensable genes. They also identified 333 selection sweeps related to low alkaloid content and candidate genes (LaDHDPS, LaHLT, and LaAT) controlling alkaloid content. Pangenome analyses of other underutilized legumes could provide novel insights into genomic variation for future trait discovery.
Several legume genera have multiple domesticated species. For example, Vigna has 10 domesticated taxa, Phaseolus seven, and Lupinus four. Super pan-genomes across these genera might have immense power to provide insight into similarities in domestication syndromes, the genetic basis of traits influencing geographic distribution, and disease and pest resistance.
Despite the success of transferring gene(s) of interest into high-yielding cultivars, environmental biosafety and regulatory governing bodies have not allowed the widespread adoption of transgenic technology (Zhang Y. et al., 2018).
Genome editing tools, especially the CRISPR/Cas9 based technique, has revolutionized functional genomics and plant breeding, creating novel genetic variation in plants by editing targeted genes of interest with precision and efficiency (Chen and Gao, 2014). Examples of genome editing in various crops are increasing (Chen and Gao 2014; Zhang Y. et al., 2018); however, there has been limited success in legume species. Notable instances of CRISPR/Cas9 mediated genome editing have been reported in soybean (Cai et al., 2015; Sun et al., 2015; Han et al., 2019), cowpea (Ji et al., 2019) and Medicago trancatula (Michno et al., 2015). In case of cowpea, Ji et al. (2019) employed CRISPR/Cas9 based genome editing tool in the symbiosis receptor -like kinase target gene VuSYMRK that controls nodule symbiosis in cowpea. The edited plant exhibited complete inhibition in nodule formation and consequently, the mutant plants were unable to synthesise nodules in association with Sinorhizobium sp. strain NGR234. Furthermore, complete male and female sterile plants were generated by editing SPO11-1 gene through CRISPR/Cas9 technology in cowpea (Juranić et al., 2020). In the context of underutilized legume, the CRISPR/Cas9 genome engineering technique was used to edit the isoflavone synthase gene contributing to rhizobial defense signaling in red clover (Dinkins et al., 2021). Furthermore, gene-editing technology in association with base editors and prime-editing could be harnessed for de novo domestication of CWRs of underutilized legumes and “reengineering of metabolism” to increase resilience and enhance nutritive value (Gasparini et al., 2021; Nasti and Voytas 2021).
Crop wild relatives are the richest reservoir of genetic diversity for improving various biotic and abiotic stress resistance in crop plants and could therefore be used as new crops through “de novo domestication” or “redomestication” process (Fernie and Yan 2019; Von Wettberg et al., 2021). Domestication of new legume underutilized crops from their wild relatives could strengthen crop diversity, and thus be vital for sustainable agriculture (Zhang et al., 2018b). Among the various underutilized grain legume species, Vigna stipulaceae could be targeted for de novo domestication due to its inherent capacity for drought and salinity stress tolerance and reduced pod shattering (Takahashi et al., 2019). Likewise, being an “incompletely domesticated species” and having inherent stress tolerance ability against biotic and abiotic stress, hairy vetch (Vicia villosa) is an ideal legume crop for de novo domestication (Renzi et al., 2020).
Of the various approaches, mutagenesis and forward screening and CRISPR/Cas9 based gene editing are important techniques for introducing domestication-related traits in wild relatives for de novo domestication (Shapter et al., 2013; Li et al., 2018). Ethyl methanesulfonate mutagenesis and forward screening enabled the domestication of Vigna stipulacea Kuntze by selecting mutants with reduced pod shattering and reduced seed dormancy (Takahashi et al., 2019). Likewise, CRISPR/Cas9 genome editing technology could be used to eliminate g-glutamyl-b-cyano-alanine (GBCA) toxin from seeds of common vetch (Vicia sativa), providing a zero-toxin vetch variety for combating the rising global protein demand (Nguyen et al., 2020).
Given the rising demand for food, feed, and forage, there is an urgent need to develop sustainable food resources. Underutilized legumes are versatile crops with great potential for mitigating global food security challenges, but they are lagging behind major legumes in terms of genomic resource development. More genomic sequencing of CWRs, landraces, and improved breeding lines will provide novel insights into genomic variations for investigating evolution, domestication events, and the diversification of underutilized legumes. Increasing genomic resources will allow increased genome-assisted breeding of these legumes. Likewise, WGRS in association with GWAS and pangenome integration with GWAS could underpin the causal genes/haplotypes of complex traits of interest. Emerging genome editing techniques could play a critical role in minimizing toxins or negative parameters associated with various nutritional quality traits, such as editing GBCA encoding gene(s) in common vetch, BOAA encoding gene(s) in grasspea, and genes involved in producing QAs in white lupin. These technologies also have great potential for introducing de novo domestication in CWRs by removing phenotypically undesired traits in various CWRs of underutilized legumes.
Moreover, genomic selection and speed breeding approaches could enhance genetic gain in underutilized legumes. The rich diversity in these underutilized legumes needs proper collection, conservation, and characterization (Kamenya et al., 2021). Furthermore, the establishment of sound varietal releases and seed distribution systems could play a central role in popularizing these climate-smart underutilized legumes among farmers (Bohra et al., 2020). Disseminating knowledge on the global demand and profitability of these legumes needs strengthening via extension services, especially in developing countries (Kamenya et al., 2021). Hence, collective genomics, novel breeding knowledge, and sound seed system approaches could improve underutilized legume productivity for securing global food security.
UJ and HN developed the conceptual structure. UJ and KS prepared the original draft. EvW, SP, and MB contributed specific sections. KS edited manuscript. All authors read and approved the final manuscript.
EvW is partially supported in work on underutilized legumes by USDA Hatch funding and by the Ministry of Science and Higher Education of the Russian Federation as part of World-class Research Center program: Advanced Digital Technologies (Contract No. 075-15-2020-934 dated 17 November 2020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
UJ acknowledges support from Indian Council of Agricultural Research (ICAR), New Delhi.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.831656/full#supplementary-material
Abdallah, F., Kumar, S., Amri, A., Mentag, R., Kehel, Z., Mejri, R. K., et al. (2021). Wild Lathyrus Species as a Great Source of Resistance for Introgression into Cultivated Grass Pea (Lathyrus sativus L.) against Broomrape Weeds (Orobanche crenata Forsk. and Orobanche foetida Poir.). Crop Sci. 61, 263–276. doi:10.1002/csc2.20399
Adegboyega, T. T., Abberton, M. T., AbdelGadir, A. H., Dianda, M., Maziya-Dixon, B., Oyatomi, O. A., et al. (2019). Nutrient and Antinutrient Composition of Winged Bean (Psophocarpus tetragonolobus (L.) DC.) Seeds and Tubers. J. Food Qual. 2019, 1–8. doi:10.1155/2019/3075208
Al-Qurainy, F., Alshameri, A., Gaafar, A. R., Khan, S., Nadeem, M., Alameri, A. A., et al. (2019). Comprehensive Stress-Based De Novo Transcriptome Assembly and Annotation of Guar (Cyamopsis tetragonoloba (L.) Taub.): An Important Industrial and Forage Crop. Int. J. Genomics 2019, 7295859. doi:10.1155/2019/7295859
Almeida, N. F., Krezdorn, N., Rotter, B. r., Winter, P., Rubiales, D., and Vaz Patto, M. C. (2015). Lathyrus Sativus Transcriptome Resistance Response to Ascochyta lathyri Investigated by deepSuperSAGE Analysis. Front. Plant Sci. 6, 178. doi:10.3389/fpls.2015.00178
Almeida, N. F., Leitão, S. T., Krezdorn, N., Rotter, B., Winter, P., Rubiales, D., et al. (2014). Allelic Diversity in the Transcriptomes of Contrasting Rust-Infected Genotypes of Lathyrus sativus, a Lasting Resource for Smart Breeding. BMC Plant Biol. 14 (1), 1–15. doi:10.1186/s12870-014-0376-2
Alseekh, S., Kostova, D., Bulut, M., and Fernie, A. R. (2021). Genome-wide Association Studies: Assessing Trait Characteristics in Model and Crop Plants. Cell. Mol. Life Sci. 78, 5743–5754. doi:10.1007/s00018-021-03868-w
Amarowicz, R., Estrella, I., Hernández, T., and Troszyńska, A. (2008). Antioxidant Activity of Extract of Adzuki Bean and its Fractions. J. Food Lipids 15 (1), 119–136. doi:10.1111/j.1745-4522.2007.00106.x
Amkul, K., Wang, L., Somta, P., Wang, S., and Cheng, X. (2019). Construction of a High Density Linkage Map and Genome Dissection of Bruchid Resistance in Zombi Pea (Vigna vexillata (L.) A. Rich). Sci. Rep. 9 (1), 1–10. doi:10.1038/s41598-019-48239-5
Arora, R. K., and Chandel, K. P. S. (1972). Botanical Source Area of Wild Herbage Legumes in India. Trop. Grassland 6, 213–221.
Arumuganathan, K., and Earle, E. D. (1991). Nuclear DNA Content of Some Important Plant Species. Plant Mol. Biol. Rep. 9, 208–218. doi:10.1007/bf02672069
Awurum, A. N., and Emechebe, A. M. (2001). The Effects of Leaf Spot Diseases and Staking on Yield and Yield Attributes of Winded Bean Psophocarpus Tetragonolobus (L. Dc.). J. Appl. Chem. Agric. Res. 7, 42–47. doi:10.4314/jacar.v7i1.41123
Bähr, M., Fechner, A., Hasenkopf, K., Mittermaier, S., and Jahreis, G. (2014). Chemical Composition of Dehulled Seeds of Selected Lupin Cultivars in Comparison to Pea and Soya Bean. Lwt-food Sci. Technol. 59, 587–590. doi:10.1016/j.lwt.2014.05.026
Bangar, P., Tyagi, N., Tiwari, B., Kumar, S., Barman, P., Kumari, R., et al. (2021). Identification and Characterization of SNPs in Released, Landrace and Wild Accessions of Mungbean (Vigna radiata (L.) Wilczek) Using Whole Genome Re-sequencing. J. Crop Sci. Biotechnol. 24, 153–165. doi:10.1007/s12892-020-00067-0
Barbetti, M. J. (2007). Resistance in Annual Medicago Spp. To Phoma Medicaginis and Leptosphaerulina Trifolii and its Relationship to Induced Production of a Phytoestrogen. Plant Dis. 91, 239–244. doi:10.1094/pdis-91-3-0239
Bassal, H., Merah, O., Ali, A. M., Hijazi, A., and El Omar, F. (2020). Psophocarpus Tetragonolobus: An Underused Species with Multiple Potential Uses. Plants 9, 1730. doi:10.3390/plants9121730
Bauchet, G. J., Bett, K. E., Cameron, C. T., Campbell, J. D., Cannon, E. K., Cannon, S. B., et al. (2019). The Future of Legume Genetic Data Resources: Challenges, Opportunities, and Priorities. Legume Sci. 1 (1), e16. doi:10.1002/leg3.16
Bazzano, L. A., He, J., Ogden, L. G., Loria, C., Vupputuri, S., Myers, L., et al. (2001). Legume Consumption and Risk of Coronary Heart Disease in US Men and Women. Arch. Intern. Med. 161, 2573–2578. doi:10.1001/archinte.161.21.2573
Bennett, M. D., and Leitch, I. J. (2012). Angiosperm DNA C-Values Database. Available online at: http://data.kew.org/cvalues/.
Berendzen, J., Brown, A. V., Cameron, C. T., Campbell, J. D., Cleary, A. M., Dash, S., et al. (2021). The Legume Information System and Associated Online Genomic Resources. Legume Sci. 2021, e74. doi:10.1002/leg3.74
Bhardwaj, J., Chauhan, R., Swarnkar, M. K., Chahota, R. K., Singh, A. K., Shankar, R., et al. (2013). Comprehensive Transcriptomic Study on Horse Gram (Macrotyloma uniflorum): De Novo Assembly, Functional Characterization and Comparative Analysis in Relation to Drought Stress. BMC Genomics 14, 647–648. doi:10.1186/1471-2164-14-647
Bhartiya, A., Aditya, J. P., and Kant, L. (2015). Nutritional and Remedial Potential of an Underutilized Food Legume Horsegram [Macrotyloma Uni Orum (Lam.) Verdc.] : a Review. J. Anim. Plant Sci 25, 908–920.
Blink, M., and Jansen, P. C. M. (2006). “Vigna Aconitifolia (Jacq.) Maréchal,” in Plant Genetic Resources of Tropical Africa. 1. Cereals and Pulses. Editors M. Blink, and G. Belay (Wageningen, Netherlands, Leiden, Netherlands: Backhuys Publishers), 200–203. Prota Foundation.
Bohra, A., Chand Jha, U., Godwin, I. D., and Kumar Varshney, R. (2020). Genomic Interventions for Sustainable Agriculture. Plant Biotechnol. J. 18, 2388–2405. doi:10.1111/pbi.13472
Bohra, A., Jha, U. C., Kishor, P. B. K., Pandey, S., and Singh, N. P. (2014). Genomics and Molecular Breeding in Lesser Explored Pulse Crops: Current Trends and Future Opportunities. Biotechnol. Adv. 32, 1410–1428. doi:10.1016/j.biotechadv.2014.09.001
Boschin, G., and Arnoldi, A. (2011). Legumes Are Valuable Sources of Tocopherols. Food Chem. 127, 1199–1203. doi:10.1016/j.foodchem.2011.01.124
Boschin, G., D’Agostina, A., Annicchiarico, P., and Arnoldi, A. (2008). Effect of Genotype and Environment on Fatty Acid Composition of Lupinus Albus L. Seed. Food Chem. 108, 600–606. doi:10.1016/j.foodchem.2007.11.016
Bounejmate, M., Robson, A., and Beale, P. (1992). Annual Medicago Species in Morocco. II. Distribution in Relation to Soil and Climate. Aust. J. Agric. Res. 43, 751–763. doi:10.1071/ar9920751
Burgarella, C., Barnaud, A., Kane, N. A., Jankowski, F., Scarcelli, N., Billot, C., et al. (2019). Adaptive Introgression: an Untapped Evolutionary Mechanism for Crop Adaptation. Front. Plant Sci. 10, 4. doi:10.3389/fpls.2019.00004
Butsayawarapat, P., Juntawong, P., Khamsuk, O., and Somta, P. (2019). Comparative Transcriptome Analysis of Waterlogging-Sensitive and Tolerant Zombi Pea (Vigna vexillata) Reveals Energy Conservation and Root Plasticity Controlling Waterlogging Tolerance. Plants 8, 264. doi:10.3390/plants8080264
Cai, Y., Chen, L., Liu, X., Sun, S., Wu, C., Jiang, B., et al. (2015). CRISPR/Cas9-Mediated Genome Editing in Soybean Hairy Roots. PLoS One 10, e0136064. doi:10.1371/journal.pone.0136064
Campbell, C. G. (1997). Grass Pea: Lathyrus Sativus L. Promoting the Conservation and Use of Underutilized and Neglected Crops. International Plant Genetic Resources Institute, Vol. 18.
Campbell, C. G., Mehra, R. B., Agrawal, S. K., Chen, Y. Z., Abdel Moneim, A. M., Khawaja, H. I. T., et al. (1994). “Current Status and Future Strategy in Breeding Grasspea (Lathyrus sativus),” in Expanding the Production and Use of Cool Season Food Legumes (Dordrecht: Springer), 617–630. doi:10.1007/978-94-011-0798-3_37
Cavanagh, C., Morell, M., Mackay, I., and Powell, W. (2008). From Mutations to MAGIC: Resources for Gene Discovery, Validation and Delivery in Crop Plants. Curr. Opin. Plant Biol. 11, 215–221. doi:10.1016/j.pbi.2008.01.002
Cerda‐Hurtado, I. M., Mayek‐Pérez, N., Hernández‐Delgado, S., Muruaga‐Martínez, J. S., Reyes‐Lara, M. A., Reyes‐Valdés, M. H., et al. (2018). Climatic Adaptation and Ecological Descriptors of Wild Beans from Mexico. Ecol. Evol. 8, 6492–6504.
Chahota, R. K., Sharma, V., Rana, M., Sharma, R., Choudhary, S., Sharma, T. R., et al. (2020). Construction of a Framework Linkage Map and Genetic Dissection of Drought-and Yield-Related QTLs in Horsegram (Macrotyloma uniflorum). Euphytica 216, 1. doi:10.1007/s10681-020-02583-0
Chandel, K. P. S., Lester, R. N., and Starling, R. J. (1984). The Wild Ancestors of Urid and Mung Beans (Vigna mungo (L.) Hepper and V. radiata (L.) Wilczek). Bot. J Linn. Soc. 89, 85–96. doi:10.1111/j.1095-8339.1984.tb01002.x
Chang, Y., Liu, H., Liu, M., Liao, X., Sahu, S. K., Fu, Y., et al. (2019). The Draft Genomes of Five Agriculturally Important African Orphan Crops. GigaScience 8, giy152. doi:10.1093/gigascience/giy152
Chapman, M. A. (2015). Transcriptome Sequencing and Marker Development for Four Underutilized Legumes. Appl. Plant Sci. 3, 1400111. doi:10.3732/apps.1400111
Chaudhury, A., Kaila, T., and Gaikwad, K. (2019). Elucidation of Galactomannan Biosynthesis Pathway Genes through Transcriptome Sequencing of Seeds Collected at Different Developmental Stages of Commercially Important Indian Varieties of Cluster Bean (Cyamopsis tetragonoloba L.). Sci. Rep. 9 (1), 1–17. doi:10.1038/s41598-019-48072-w
Chen, K., and Gao, C. (2014). Targeted Genome Modification Technologies and Their Applications in Crop Improvements. Plant Cel. Rep. 33, 575–583. doi:10.1007/s00299-013-1539-6
Chen, H., Chen, X., Tian, J., Yang, Y., Liu, Z., Hao, X., et al. (2016). Development of Gene-Based SSR Markers in rice Bean (Vigna umbellata L.) Based on Transcriptome Data. PloS One 11, e0151040. doi:10.1371/journal.pone.0151040
Chen, H., Liu, L., Wang, L., Wang, S., Somta, P., and Cheng, X. (2015a). Development and Validation of EST-SSR Markers from the Transcriptome of Adzuki Bean (Vigna angularis). PloS One 10, e0131939. doi:10.1371/journal.pone.0131939
Chen, H., Wang, L., Wang, S., Liu, C., Blair, M. W., and Cheng, X. (2015b). Transcriptome Sequencing of Mung Bean (Vigna radiate L.) Genes and the Identification of EST-SSR Markers. PLoS ONE 10, e0120273. doi:10.1371/journal.pone.0120273
Cheng, A., Raai, M. N., Zain, N. A. M., Massawe, F., Singh, A., and Wan-Mohtar, W. A. A. Q. I. (2019). In Search of Alternative Proteins: Unlocking the Potential of Underutilized Tropical Legumes. Food Sec. 11 (6), 1205–1215. doi:10.1007/s12571-019-00977-0
Cowling, W. A., Hamblin, J., Wood, P. M., and Gladstones, J. S. (1987). Resistance to Phomopsis Stem Blight in Lupinus Angustifolius L. 1. Crop Sci. 27, 648–652. doi:10.2135/cropsci1987.0011183x002700040007x
Cui, J., Lu, Z., Wang, T., Chen, G., Mostafa, S., Ren, H., et al. (2021). The Genome of Medicago Polymorpha Provides Insights into its Edibility and Nutritional Value as a Vegetable and Forage Legume. Hort. Res. 8, 1–2. doi:10.1038/s41438-021-00483-5
Cullis, C., and Kunert, K. J. (2017). Unlocking the Potential of Underutilised Legumes. J. Expt. Bot. 68, 1895–1903. doi:10.1093/jxb/erw437
Dachapak, S., Tomooka, N., Somta, P., Naito, K., Kaga, A., and Srinives, P. (2018). QTL Analysis of Domestication Syndrome in Zombi Pea (Vigna vexillata), an Underutilized Legume Crop. PLoS ONE 13, e0200116. doi:10.1371/journal.pone.0200116
Das, B., and Arora, S. K. (1978). ‘Guar Seed–Its Chemistry and Industrial Utilization of gum.’ Guar– its Improvement and Management. Forage Res. 4, 79–101.
Dasgupta, U., Mishra, G. P., Dikshit, H. K., Mishra, D. C., Bosamia, T., Roy, A., et al. (2021). Comparative RNA-Seq Analysis Unfolds a Complex Regulatory Network Imparting Yellow Mosaic Disease Resistance in Mungbean [Vigna radiata (L.) R. Wilczek]. PLoS ONE 16, e0244593. doi:10.1371/journal.pone.0244593
De la Rosa, L., López-Román, M. I., González, J. M., Zambrana, E., Marcos-Prado, T., and Ramírez-Parra, E. (2021). Common Vetch Valuable Germplasm for Resilient Agriculture: Genetic Characterization and Spanish Core Collection Development. Front. Plant Sci. 12, 617873. doi:10.3389/fpls.2021.617873
De la Rosa, L., Zambrana, E., and Ramirez-Parra, E. (2020). Molecular Bases for Drought Tolerance in Common Vetch: Designing New Molecular Breeding Tools. BMC Plant Biol. 20, 1–8. doi:10.1186/s12870-020-2267-z
De Vega, J. J., Ayling, S., Hegarty, M., Kudrna, D., Goicoechea, J. L., Ergon, Å., et al. (2015). Red clover (Trifolium pratense L.) Draft Genome Provides a Platform for Trait Improvement. Sci. Reps. 5 (1), 1–0. doi:10.1038/srep17394
DeBoer, K., Melser, S., Sperschneider, J., Kamphuis, L. G., Garg, G., Gao, L. L., et al. (2019). Identification and Profiling of Narrow-Leafed Lupin (Lupinus angustifolius) microRNAs during Seed Development. BMC Genome 20, 135. doi:10.1186/s12864-019-5521-8
Della Coletta, R., Qiu, Y., Ou, S., Hufford, M. B., and Hirsch, C. N. (2021). How the Pan-Genome is Changing Crop Genomics and Improvement. Genome Biol. 22, 1–9. doi:10.1186/s13059-020-02224-8
Dinkins, R. D., Hancock, J., Coe, B. L., May, J. B., Goodman, J. P., Bass, W. T., et al. (2021). Isoflavone Levels, Nodulation and Gene Expression Profiles of a CRISPR/Cas9 Deletion Mutant in the Isoflavone Synthase Gene of Red clover. Plant Cell Reps 40, 517–528. doi:10.1007/s00299-020-02647-4
Dong, R., Dong, D., Luo, D., Zhou, Q., Chai, X., Zhang, J., et al. (2017). Transcriptome Analyses Reveal Candidate Pod Shattering-Associated Genes Involved in the Pod Ventral Sutures of Common Vetch (Vicia sativa L.). Front. Plant Sci. 8, 649. doi:10.3389/fpls.2017.00649
Drummond, C. S., Eastwood, R. J., Miotto, S. T., and Hughes, C. E. (2012). Multiple continental Radiations and Correlates of Diversification in Lupinus (Leguminosae): Testing for Key Innovation with Incomplete Taxon Sampling. Syst. Biol. 61, 443–460. doi:10.1093/sysbio/syr126
Ditzler, L., van Apeldoorn, D. F., Pellegrini, F., Antichi, D., Bàrberi, P., and Rossing, W. A. H. (2021). Current Research on the Ecosystem Service Potential of Legume Inclusive Cropping Systems in Europe. A Review. Agron. Sustain. Dev. 41, 26. doi:10.1007/s13593-021-00678-z
Ebi, K. L., and Loladze, I. (2019). Elevated Atmospheric CO2 Concentrations and Climate Change Will Affect Our Food's Quality and Quantity. Lancet Planet. Health 3, e283–e284. doi:10.1016/s2542-5196(19)30108-1
Edwards, D., and Batley, J. (2010). Plant Genome Sequencing: Applications for Crop Improvement. Plant Biotechnol. J. 8, 2–9. doi:10.1111/j.1467-7652.2009.00459.x
Elfaki, M. O., and Abdelatti, K. A. (2018). Rumen Content as Animal Feed: a Review. J. Vet. Med. Anim. Prod. 20 (2), 7.
Emmrich, P. M., Sarkar, A., Njaci, I., Kaithakottil, G. G., Ellis, N., Moore, C., et al. (2020). A Draft Genome of Grass Pea (Lathyrus sativus), a Resilient Diploid Legume. bioRxiv. doi:10.1101/2020.04.24.058164
Etheridge, J. V., and Bateman, G. L. (1999). Fungicidal Control of Foliar Diseases of White Lupin (Lupinus Albus). Crop Prot. 18, 349–354. doi:10.1016/s0261-2194(99)00031-9
Fang, Y., Wang, L., Sapey, E., Fu, S., Wu, T., Zeng, H., et al. (2021). Speed-Breeding System in Soybean: Integrating Off-Site Generation Advancement, Fresh Seeding, and Marker-Assisted Selection. Front. Plant Sci. 12, 717077. doi:10.3389/fpls.2021.717077
FAO (2013). FAOStat 2013 Food and Agriculture Organization of the United Nations Statistics Division.
FAO (2018). Definitions and Standards Used in FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (Accessed on August 18, 2018).
FAO (2017). FAOStat 2017 Food and Agriculture Organization of the United Nations Statistics Division.
Feng, X., Orellana, G. E., Green, J. C., Melzer, M. J., Hu, J. S., and Karasev, A. V. (2019). A New Strain of Bean Common Mosaic Virus from lima Bean (Phaseolus Lunatus): Biological and Molecular Characterization. Plant Dis. 103, 1220–1227. doi:10.1094/pdis-08-18-1307-re
Fernie, A. R., and Yan, J. (2019). De Novo domestication: an Alternative Route toward New Crops for the Future. Mol. Plant 12, 615–631. doi:10.1016/j.molp.2019.03.016
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a Cultivated Planet. Nature 478, 337–342. doi:10.1038/nature10452
Fontanari, G. G., Batistuti, J. P., da Cruz, R. J., Saldiva, P. H., and Arêas, J. A. (2012). Cholesterol-lowering Effect of Whole Lupin (Lupinus Albus) Seed and its Protein Isolate. Food Chem. 132, 1521–1526. doi:10.1016/j.foodchem.2011.11.145
Francis, C. M., Enneking, D., and Abd El Moneim, A. M. (2000). “When and Where Will Vetches Have an Impact as Grain Legumes?,” in Linking Research and Marketing Opportunities for Pulses in the 21st Century 2000 (Dordrecht: Springer), 375–384. doi:10.1007/978-94-011-4385-1_34
Frick, K. M., Kamphuis, L. G., Siddique, K. H., Singh, K. B., and Foley, R. C. (2017). Quinolizidine Alkaloid Biosynthesis in Lupins and Prospects for Grain Quality Improvement. Front. Plant Sci. 8, 87. doi:10.3389/fpls.2017.00087
Fuller, D. Q. (2007). Contrasting Patterns in Crop Domestication and Domestication Rates: Recent Archaeobotanical Insights from the Old World. Ann. Bot. (Lond.) 2007 (100), 903–924. doi:10.1093/aob/mcm048
Gaikwad, K., Ramakrishna, G., Srivastava, H., Saxena, S., Kaila, T., Tyagi, A., et al. (2020). Chromosome Scale Reference Genome of Cluster Bean (Cyamopsis tetragonoloba (L.) Taub.). bioRxiv. 2020 Jan 1. doi:10.1101/2020.05.16.098434
Garcia, T., Duitama, J., Zullo, S. S., Gil, J., Ariani, A., Dohle, S., et al. (2021). Comprehensive Genomic Resources Related to Domestication and Crop Improvement Traits in Lima Bean. Nat. Commun. 12, 1–7. doi:10.1038/s41467-021-20921-1
Gasparini, K., dos Reis Moreira, J., Peres, L. E., and Zsögön, A. (2021). De Novo domestication of Wild Species to Create Crops with Increased Resilience and Nutritional Value. Curr. Opin. Plant Biol. 60, 102006. doi:10.1016/j.pbi.2021.102006
Gautam, N. K., Kumar, K., and Prasad, M. (2016). Leaf Crinkle Disease in Urdbean (Vigna mungo L. Hepper): An Overview on Causal Agent, Vector and Host. Protoplasma 253, 729–746. doi:10.1007/s00709-015-0933-z
Gepts, P., Beavis, W. D., Brummer, E. C., Shoemaker, R. C., Stalker, H. T., Weeden, N. F., et al. (2005). Legumes as a Model Plant Family. Genomics for Food and Feed Report of the Cross-Legume Advances through Genomics Conference. Plant Physiol. 137 (4), 1228–1235. doi:10.1104/pp.105.060871
Gillett, J. B. (1958). Indigofera (Microcharis) in Tropical Africa with the Related Genera Cyamopsis and Rhynchotropis, 1–166. H.M.S.O Kew Bull.
Gladstones, J. S. (1998). in Lupins as Crop Plants: Biology, Production and Utilization. Editors J. S. Gladstones, C. Atkins, and J. Hamblin (Oxon, New York: CAB International), 1–39.
Glazińska, P., Kulasek, M., Glinkowski, W., Wojciechowski, W., and Kosiński, J. (2019). Integrated Analysis of Small RNA, Transcriptome and Degradome Sequencing Provides New Insights into floral Development and Abscission in Yellow Lupine (Lupinus luteus L.). Intl. J. Mol. Sci. 20, 5122. doi:10.3390/ijms20205122
Glazinska, P., Wojciechowski, W., Kulasek, M., Glinkowski, W., Marciniak, K., Klajn, N., et al. (2017). De Novo transcriptome Profiling of Flowers, Flower Pedicels and Pods of Lupinus Luteus (Yellow Lupine) Reveals Complex Expression Changes during Organ Abscission. Front. Plant Sci. 8, 641. doi:10.3389/fpls.2017.00641
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food Security: The Challenge of Feeding 9 Billion People. Science 327, 812–818. doi:10.1126/science.1185383
Graham, P. H., and Vance, C. P. (2003). Legumes: Importance and Constraints to Greater Use. Plant Physiol. 131, 872–877. doi:10.1104/pp.017004
Gresta, F. A., Wink, M. I., Prins, U., Abberton, M. I., Capraro, J. E., Scarafoni, A. L., et al. (2017). “Lupins in European Cropping Systems,” in Legumes in Cropping Systems, 88–108. doi:10.1079/9781780644981.0088
Grigoreva, E., Ulianich, P., Ben, C., Gentzbittel, L., and Potokina, E. (2019). First Insights into the Guar (Cyamopsis tetragonoloba (L.) Taub.) Genome of the ‘Vavilovskij 130’Accession, Using Second and Third-Generation Sequencing Technologies. Russ. J. Genet. 55, 1406–1416. doi:10.1134/s102279541911005x
Ha, J., Satyawan, D., Jeong, H., Lee, E., Cho, K. H., Kim, M. Y., et al. (2021). A Near‐complete Genome Sequence of Mungbean (Vigna radiata L.) Provides Key Insights into the Modern Breeding Program. Plant Genome 18, e20121. doi:10.1002/tpg2.20121
Han, J., Guo, B., Guo, Y., Zhang, B., Wang, X., and Qiu, L. J. (2019). Creation of Early Flowering Germplasm of Soybean by CRISPR/Cas9 Technology. Front. Plant Sci. 10, 1446. doi:10.3389/fpls.2019.01446
Hane, J. K., Ming, Y., Kamphuis, L. G., Nelson, M. N., Garg, G., Atkins, C. A., et al. (2017). A Comprehensive Draft Genome Sequence for Lupin (Lupinus angustifolius), an Emerging Health Food: Insights into Plant–Microbe Interactions and Legume Evolution. Plant Biotechnol. J. 15, 318–330. doi:10.1111/pbi.12615
Hao, X., Yang, T., Liu, R., Hu, J., Yao, Y., Burlyaeva, M., et al. (2017). An RNA Sequencing Transcriptome Analysis of Grasspea (Lathyrus sativus L.) and Development of SSR and KASP Markers. Front. Plant Sci. 8, 1873. doi:10.3389/fpls.2017.01873
Hepper, F. N. (1963). The Bambara Groundnut (Voandzeia subterranea) and Kersting’s Groundnut (Kerstingiella geocarpa) Wild in West Africa. Kew Bull. 16, 395–407. doi:10.2307/4114681
Herbert, D. B., Gross, T., Rupp, O., and Becker, A. (2021). Transcriptome Analysis Reveals Major Transcriptional Changes during Regrowth after Mowing of Red clover (Trifolium pratense). BMC Plant Biol. 21, 1–20. doi:10.1186/s12870-021-02867-0
Hickey, L. T., Hafeez, N. A., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 37, 744–754. doi:10.1038/s41587-019-0152-9
Hirakawa, H., Kaur, P., Shirasawa, K., Nichols, P., Nagano, S., Appels, R., et al. (2016). Draft Genome Sequence of Subterranean Clover, a Reference for Genus Trifolium. Sci. Reps. 22, 1–9. doi:10.1038/srep30358
Hillocks, R. J., Bennett, C., and Mponda, O. M. (2012). BambaraNut:A Review of Utilisation, Market Potential and Crop Improvement. Afr. Crop Sci. J. 20, 1–16.
Ho, W. K., Chai, H. H., Kendabie, P., Ahmad, N. S., Jani, J., Massawe, F., et al. (2017). Integrating Genetic Maps in Bambara Groundnut [Vigna subterranea (L) Verdc.] and Their Syntenic Relationships Among Closely Related Legumes. BMC Genome 18, 1–9. doi:10.1186/s12864-016-3393-8
Hu, H., Wang, H., Zhang, Y., Kan, B., Ding, Y., and Huang, J. (2019). Characterization of Genes in Guar Gum Biosynthesis Based on Quantitative RNA-Sequencing in Guar Bean (Cyamopsis tetragonoloba). Sci. Reps. 9, 1–0. doi:10.1038/s41598-019-47518-5
Hu, Z., Sun, C., Lu, K. C., Chu, X., Zhao, Y., Lu, J., et al. (2017). EUPAN Enables Pan-Genome Studies of a Large Number of Eukaryotic Genomes. Bioinformatics 33, 2408–2409. doi:10.1093/bioinformatics/btx170
Huang, X., and Han, B. (2014). Natural Variations and Genome-wide Association Studies in Crop Plants. Annu. Rev. Plant Biol. 65, 531–551. doi:10.1146/annurev-arplant-050213-035715
Hufnagel, B., Marques, A., Soriano, A., Marquès, L., Divol, F., Doumas, P., et al. (2020). High-quality Genome Sequence of white Lupin Provides Insight into Soil Exploration and Seed Quality. Nat. Commun. 11, 1–2. doi:10.1038/s41467-019-14197-9
Hufnagel, B., Soriano, A., Taylor, J., Divol, F., Kroc, M., Sanders, H., et al. (2021). Pangenome of white Lupin Provides Insights into the Diversity of the Species. bioRxiv. 2021 Jan 1. doi:10.1111/pbi.13678
Hufnagel, J., Reckling, M., and Ewert, F. (2020). Diverse Approaches to Crop Diversification in Agricultural Research. A Review. Agron. Sust Dev. 40, 1–17. doi:10.1007/s13593-020-00617-4
Huyghe, C. (1997). White Lupin (Lupinus albus L.). Field Crops Res. 53, 147–160. doi:10.1016/s0378-4290(97)00028-2
Hymowitz, T., and Boyd, J. (1977). Origin, Ethnobotany and Agricultural Potential of the Winged Bean: Psophocarpus Tetragonolobus. Econ. Bot. 31, 180–188. doi:10.1007/bf02866589
Iizuka, N. (1990). Studies on Virus Diseases of Adzuki Bean (Vigna Angularis Wight) in Japan. Bull. Tohoku Natl. Agric. Exp. Station 82, 77–113.
Iqbal, M. M., Erskine, W., Berger, J. D., and Nelson, M. N. (2020). Phenotypic Characterisation and Linkage Mapping of Domestication Syndrome Traits in Yellow Lupin (Lupinus luteus L.). Theor. Appl. Genet. 133, 2975–2987. doi:10.1007/s00122-020-03650-9
Iqbal, M. M., Huynh, M., Udall, J. A., Kilian, A., Adhikari, K. N., Berger, J. D., et al. (2019). The First Genetic Map for Yellow Lupin Enables Genetic Dissection of Adaptation Traits in an Underutilized Grain Legume Crop. BMC Genet. 20, 68. doi:10.1186/s12863-019-0767-3
Isemura, T., Kaga, A., Tomooka, N., Shimizu, T., and Vaughan, D. A. (2010). Construction of a Genetic Linkage Map and Genetic Analysis of the Domestication Related Traits in rice Bean (Vigna umbellata). Ann. Bot. 106, 927–944. doi:10.1093/aob/mcq188
Isemura, T., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., et al. (2012). Construction of a Genetic Linkage Map and Genetic Analysis of Domestication Related Traits in Mungbean (Vigna radiata). PLoS One 7, e41304. doi:10.1371/journal.pone.0041304
Isobe, S., Kölliker, R., Hisano, H., Sasamoto, S., Wada, T., Klimenko, I., et al. (2009). Construction of a Consensus Linkage Map for Red Clover (Trifolium pratense L.). BMC Plant Biol. 9 (1), 1–11.
Ištvánek, J., Dluhošová, J., Dluhoš, P., Pátková, L., Nedělník, J., and Řepková, J. (2017). Gene Classification and Mining of Molecular Markers Useful in Red clover (Trifolium pratense) Breeding. Front. Plant Sci. 8, 367. doi:10.3389/fpls.2017.00367
Ištvánek, J., Jaroš, M., Křenek, A,., and Řepková, J. (2014). Genome Assembly and Annotation for Red clover (Trifolium Pratense; Fabaceae). Am. J. Bot. 101, 327–337.
Jasrotia, R. S., Yadav, P. K., Iquebal, M. A., Bhatt, S. B., Arora, V., Angadi, U. B., et al. (2019). VigSatDB: Genome-wide Microsatellite DNA Marker Database of Three Species of Vigna for Germplasm Characterization and Improvement. Database. 2019 Jan 1. doi:10.1093/database/baz055
Jegadeesan, S., Raizada, A., Dhanasekar, P., and Suprasanna, P. (2021). Draft Genome Sequence of the Pulse Crop Blackgram [Vigna mungo (L.) Hepper] Reveals Potential R-Genes. Sci. Reps. 11, 1–0. doi:10.1038/s41598-021-90683-9
Jha, U. C. (2018). Current Advances in Chickpea Genomics: Applications and Future Perspectives. Plant Cell Reps 37, 947–965. doi:10.1007/s00299-018-2305-6
Ji, J., Zhang, C., Sun, Z., Wang, L., Duanmu, D., and Fan, Q. (2019). Genome Editing in Cowpea Vigna Unguiculata Using CRISPR-Cas9. Int. J. Mol. Sci. 20 (10), 2471. doi:10.3390/ijms20102471
Jo, Y., Lian, S., Cho, J. K., Choi, H., Kim, S. M., Kim, S. L., et al. (2016). De Novo transcriptome Assembly of Two Vigna Angularis Varieties Collected from Korea. Genomics Data 8, 119–120. doi:10.1016/j.gdata.2016.05.004
Jones, C., De Vega, J., Lloyd, D., Hegarty, M., Ayling, S., Powell, W., et al. (2020). Population Structure and Genetic Diversity in Red clover (Trifolium pratense L.) Germplasm. Sci. Reps. 10, 1–2. doi:10.1038/s41598-020-64989-z
Jones, D. B., Gersdorff, C. E., Johns, C. O., and Finks, A. J. (1922). The Proteins of the Lima Bean, Phaseolus Lunatus. J. Biol. Chem. 53, 231–240. doi:10.1016/s0021-9258(18)85778-7
Juranić, M., Nagahatenna, D. S., Salinas-Gamboa, R., Hand, M. L., Sánchez-León, N., Leong, W. H., et al. (2020). A Detached Leaf Assay for Testing Transient Gene Expression and Gene Editing in Cowpea (Vigna unguiculata [L.] Walp.). Plant Methods 16 (1), 1–7. doi:10.1186/s13007-020-00630-4
Kadam, S. S., Salunkhe, D. K., and Luh, B. S. (1984). Winged Bean in Human Nutrition. Crit. Rev. Food Sci. Nutr. 21, 1–40. doi:10.1080/10408398409527395
Kaewwongwal, A., Kongjaimun, A., Somta, P., Chankaew, S., Yimram, T., and Srinives, P. (2015). Genetic Diversity of the Black Gram [Vigna mungo (L.) Hepper] Gene Pool as Revealed by SSR Markers. Breed. Sci. 65, 127–137. doi:10.1270/jsbbs.65.127
Kaga, A., Isemura, T., Tomooka, N., and Vaughan, D. A. (2008). The Genetics of Domestication of the Azuki Bean (Vigna angularis). Genetics 178, 1013–1036. doi:10.1534/genetics.107.078451
Kakati, P., Deka, S., Kotoki, D., and Saikia, S. (2010). Effect of Traditional Methods of Processing on the Nutrient Contents and Some Antinu- Tritional Factors in Newly Developed Cultivars of green Gram [Vigna radiata (L.) Wilezek] and Black Gram [Vigna Mungo (L.) Hepper] of Assam. India. Int. Food Res. J. 17, 377–384.
Kamenya, S. N., Mikwa, E. O., Song, B., and Odeny, D. A. (2021). Genetics and Breeding for Climate Change in Underutilised Crops. Theor. Appl. Genet. 23, 1–29.
Kamphuis, L. G., Hane, J. K., Nelson, M. N., Gao, L., Atkins, C. A., and Singh, K. B. (2015). Transcriptome Sequencing of Different Narrow‐leafed Lupin Tissue Types Provides a Comprehensive Uni‐gene Assembly and Extensive Gene‐based Molecular Markers. Plant Biotechnol. J. 13, 14–25. doi:10.1111/pbi.12229
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome Sequence of Mungbean and Insights into Evolution within Vigna Species. Nat. Commun. 5, 1–9. doi:10.1038/ncomms6443
Kang, Y. J., Satyawan, D., Shim, S., Lee, T., Lee, J., Hwang, W. J., et al. (2015). Draft Genome Sequence of Adzuki Bean, Vigna Angularis. Sci. Reps. 5, 1–8. doi:10.1038/srep08069
Kasprzak, A., Šafář, J., Janda, J., Doležel, J., Wolko, B., and Naganowska, B. (2006). The Bacterial Artificial Chromosome (BAC) Library of the Narrow-Leafed Lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Lett. 11, 396–407. doi:10.2478/s11658-006-0033-3
Katoch, R. (2020). “Common Diseases and Insect–Pests of Rice Bean,” in Ricebean (Singapore: Springer), 347–353. doi:10.1007/978-981-15-5293-9_18
Kaul, T., Eswaran, M., Thangaraj, A., Meyyazhagan, A., Nehra, M., Raman, N. M., et al. (2019). Rice Bean (Vigna umbellata) Draft Genome Sequence: Unravelling the Late Flowering and Unpalatability Related Genomic Resources for Efficient Domestication of This Underutilized Crop. bioRxiv. 2019 Jan 1. doi:10.1101/816595
Kaur, P., Bayer, P. E., Milec, Z., Vrána, J., Yuan, Y., Appels, R., et al. (2017). An Advanced Reference Genome of Trifolium Subterraneum L. Reveals Genes Related to Agronomic Performance. Plant Biotechnol. J. 15, 1034–1046. doi:10.1111/pbi.12697
Keatinge, J. D., Easdown, W. J., Yang, R. Y., Chadha, M. L., and Shanmugasundaram, S. (2011). Overcoming Chronic Malnutrition in a Future Warming World: the Key Importance of Mungbean and Vegetable Soybean. Euphytica 180, 129–141. doi:10.1007/s10681-011-0401-6
Kim, T. S., Raveendar, S., Suresh, S., Lee, G. A., Lee, J. R., Cho, J. H., et al. (2015). Transcriptome Analysis of Two Vicia Sativa Subspecies: Mining Molecular Markers to Enhance Genomic Resources for Vetch Improvement. Genes 6 (4), 1164–1182. doi:10.3390/genes6041164
Kislev, M. E. (1989). Origins of the Cultivation of Lathyrus Sativus and L. Cicera (Fabaceae). Econ. Bot. 43, 262–270. doi:10.1007/bf02859868
Kitano‐Okada, T., Ito, A., Koide, A., Nakamura, Y., Han, K. H., Shimada, K., et al. (2012). Anti‐obesity Role of Adzuki Bean Extract Containing Polyphenols: In Vivo and In Vitro Effects. J. Sci. Food Agric. 92 (13), 2644–2651. doi:10.1002/jsfa.5680
Konduri, V., Godwin, I. D., and Liu, C. J. (2000). Genetic Mapping of the Lablab Purpureus Genome Suggests the Presence of “Cuckoo” Gene(s) in This Species. Theor. Appl. Genet. 100, 866–871. doi:10.1007/s001220051363
Korte, A., and Farlow, A. (2013). The Advantages and Limitations of Trait Analysis with GWAS: a Review. Plant Methods 9, 29. doi:10.1186/1746-4811-9-29
Kovi, M. R., Amdahl, H., Alsheikh, M., and Rognli, O. A. (2017). De novo and Reference Transcriptome Assembly of Transcripts Expressed During Flowering Provide Insight into Seed Setting in Tetraploid Red Clover. Sci. Rep. 7 (1), 1–11.
Kozak, B., Galek, R., Zalewski, D., and Sawicka-Sienkiewicz, E. (2019). Preliminary Genetic Map of a New Recombinant Inbred Line Population for Narrow-Leafed Lupin (Lupinus angustifolius L.). Agronomy 9, 653. doi:10.3390/agronomy9100653
Kroc, M., Rybiński, W., Wilczura, P., Kamel, K., Kaczmarek, Z., Barzyk, P., et al. (2017). Quantitative and Qualitative Analysis of Alkaloids Composition in the Seeds of a white Lupin (Lupinus albus L.) Collection. Genet. Resour. Crop Evol. 64, 1853–1860. doi:10.1007/s10722-016-0473-1
Kroc, M., Czepiel, K., Wilczura, P., Mokrzycka, M., and Święcicki, W. (2019). Development and Validation of a Gene-Targeted dCAPS Marker for Marker-Assisted Selection of Low-Alkaloid Content in Seeds of Narrow-Leafed Lupin (Lupinus angustifolius L.). Genes 10, 428. doi:10.3390/genes10060428
Książkiewicz, M., Nazzicari, N., Nelson, M. N., Renshaw, D., Rychel, S., Ferrari, B., et al. (2017). A High-Density Consensus Linkage Map of white Lupin Highlights Synteny with Narrow-Leafed Lupin and Provides Markers Tagging Key Agronomic Traits. Sci. Reps. 7, 1–5.
Kumar, S., Bejiga, G., Ahmed, S., Nakkoul, H., and Sarker, A. (2011). Genetic Improvement of Grass Pea for Low Neurotoxin (β-ODAP) Content. Food Chem. Toxicol. 49, 589–600. doi:10.1016/j.fct.2010.06.051
Kumar, S., Palve, A. S., Patel, S. K., Selvanayagam, S., Sharma, R., and Rathore, A. (2020). Development of Genomic Microsatellite Markers in Cluster Bean Using Next-Generation DNA Sequencing and Their Utility in Diversity Analysis. Curr. Plant Biol. 21, 100134. doi:10.1016/j.cpb.2019.100134
Lambers, H., Clements, J. C., and Nelson, M. N. (2013). How a Phosphorus‐acquisition Strategy Based on Carboxylate Exudation powers the success and Agronomic Potential of Lupines (Lupinus, Fabaceae). Am. J. Bot. 100, 263–288. doi:10.3732/ajb.1200474
Lee, G. A. (2013). Archaeological Perspectives on the Origins of Azuki (Vigna Angularis). Holocene 23, 453–459. doi:10.1177/0959683612460788
Lee, Y. P., Mori, T. A., Sipsas, S., Barden, A., Puddey, I. B., Burke, V., et al. (2006). Lupin-enriched Bread Increases Satiety and Reduces Energy Intake Acutely. Amer J. Clinic. Nutr. 84, 975–980. doi:10.1093/ajcn/84.5.975
Lei, L., Goltsman, E., Goodstein, D., Wu, G. A., Rokhsar, D. S., and Vogel, J. P. (2021). Plant Pan-Genomics Comes of Age. Annu. Rev. Plant Biol. 72, 411–435. doi:10.1146/annurev-arplant-080720-105454
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of Extreme Weather Disasters on Global Crop Production. Nature 529, 84–87. doi:10.1038/nature16467
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018). Domestication of Wild Tomato Is Accelerated by Genome Editing. Nat. Biotechnol. 36, 1160–1163. doi:10.1038/nbt.4273
Li, W., Riday, H., Riehle, C., Edwards, A., and Dinkins, R. (2019). Identification of Single Nucleotide Polymorphism in Red clover (Trifolium pratense L.) Using Targeted Genomic Amplicon Sequencing and RNA-Seq. Front. Plant Sci. 10, 1257. doi:10.3389/fpls.2019.01257
Li, X., Yan, G., Renshaw, D., and Yang, H. (2010). Development of a Co-dominant DNA Marker Tightly Linked to Gene Tardus Conferring Reduced Pod Shattering in Narrow-Leafed Lupin (Lupinus angustifolius L.). Euphytica 176, 49–58. doi:10.1007/s10681-010-0212-1
Li, X., Yang, H., and Yan, G. (2012). Development of a Co-dominant DNA Marker Linked to the Gene Lentus Conferring Reduced Pod Shattering for Marker-Assisted Selection in Narrow-Leafed Lupin (Lupinus angustifolius) Breeding. Plant Breed 131, 540–544. doi:10.1111/j.1439-0523.2012.01978.x
Li, X., Yang, H., Buirchell, B., and Yan, G. (2011). Development of a DNA Marker Tightly Linked to Low-Alkaloid Gene Iucundus in Narrow-Leafed Lupin (Lupinus angustifolius L.). Crop Pasture Sci. 62, 218–224. doi:10.1071/cp10352
Lichtin, N., Salvo-Garrido, H., Till, B., Caligari, P. D., Rupayan, A., Westermeyer, F., et al. (2020). Genetic and Comparative Mapping of Lupinus Luteus L. Highlight Syntenic Regions with Major Orthologous Genes Controlling Anthracnose Resistance and Flowering Time. Sci. Reps 10, 1–5. doi:10.1038/s41598-020-76197-w
Lin, R., Renshaw, D., Luckett, D., Clements, J., Yan, G., Adhikari, K., et al. (2009). Development of a Sequence-specific PCR Marker Linked to the Gene “Pauper” Conferring Low-Alkaloids in white Lupin (Lupinus albus L.) for Marker Assisted Selection. Mol. Breed. 23, 153–161. doi:10.1007/s11032-008-9222-2
Liu, C., Fan, B., Cao, Z., Su, Q., Wang, Y., Zhang, Z., et al. (2016). Development of a High-Density Genetic Linkage Map and Identification of Flowering Time QTLs in Adzuki Bean (Vigna angularis). Sci. Reps. 6, 1–0. doi:10.1038/srep39523
Liu, C., Fan, B., Cao, Z., Su, Q., Wang, Y. A., Zhang, Z., et al. (2016). A Deep Sequencing Analysis of Transcriptomes and the Development of EST-SSR Markers in Mungbean (Vigna radiata). J. Genet. 95, 527–535. doi:10.1007/s12041-016-0663-9
Liu, C., Wu, J., Wang, L., Fan, B., Cao, Z., Su, Q., et al. (2017). Quantitative Trait Locus Mapping under Irrigated and Drought Treatments Based on a Novel Genetic Linkage Map in Mungbean (Vigna radiata L.). Theor. Appl. Genet. 130, 2375–2393. doi:10.1007/s00122-017-2965-6
Liu, H. J., and Yan, J. (2019). Crop Genome‐wide Association Study: a Harvest of Biological Relevance. Plant J. 97, 8–18. doi:10.1111/tpj.14139
Liu, L., Bestel, S., Shi, J., Song, Y., and Chen, X. (2013). Paleolithic Human Exploitation of Plant Foods during the Last Glacial Maximum in North China. Proc. Natl. Acad. Sci. USA 110, 5380–5385. doi:10.1073/pnas.1217864110
Liu, M. S., Kuo, T. C., Ko, C. Y., Wu, D. C., Li, K. Y., Lin, W. J., et al. (2016). Genomic and Transcriptomic Comparison of Nucleotide Variations for Insights into Bruchid Resistance of Mungbean (Vigna radiata [L.] R. Wilczek). BMC Plant Biol. 16, 1–6. doi:10.1186/s12870-016-0736-1
Lonardi, S., Muñoz‐Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S. I., Lo, S., et al. (2019). The Genome of Cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98, 767–782. doi:10.1111/tpj.14349
Maass, B. L., Knox, M. R., Venkatesha, S. C., Angessa, T. T., Ramme, S., and Pengelly, B. C. (2010). Lablab Purpureus–a Crop Lost for Africa? Trop. Plant Biol. 3 (3), 123–135. doi:10.1007/s12042-010-9046-1
Mahesh, H. B., Prasannakumar, M. K., Manasa, K. G., Perumal, S., Khedikar, Y., Kagale, S., et al. (2021). Genome, Transcriptome, and Germplasm Sequencing Uncovers Functional Variation in the Warm-Season Grain Legume Horsegram Macrotyloma Uni Orum (Lam.) Verdc. Front. Plant Sci. 12, 758119. doi:10.3389/fpls.2021.758119
Marques, E., Kur, A., Bueno, E., and von Wettberg, E. (2020). Defining and Improving the Rotational and Intercropping Value of a Crop Using a Plant–Soil Feedbacks Approach. Crop Sci. 60 (5), 2195–2203. doi:10.1002/csc2.20200
Mathivathana, M. K., Murukarthick, J., Karthikeyan, A., Jang, W., Dhasarathan, M., Jagadeeshselvam, N., et al. (2019). Detection of QTLs Associated with Mungbean Yellow Mosaic Virus (MYMV) Resistance Using the Interspecific Cross of Vigna radiata × Vigna umbellata. J. Appl. Genet. 60, 255–268. doi:10.1007/s13353-019-00506-x
Mayes, S., Ho, W. K., Chai, H. H., Gao, X., Kundy, A. C., Mateva, K. I., et al. (2019). Bambara Groundnut: An Exemplar Underutilised Legume for Resilience under Climate Change. Planta 250, 803–820. doi:10.1007/s00425-019-03191-6
Mercado-Ruaro, P., and Delgado-Salinas, A. (1998). Karyotypic Studies on Species of Phaseolus (Fabaceae: Phaseolinae). Am. J. Bot. 85, 1–9. doi:10.2307/2446547
Meuwissen, T. H., Hayes, B. J., and Goddard, M. E. (2001). Prediction of Total Genetic Value Using Genome-wide Dense Marker Maps. Genetics 157, 1819–1829. doi:10.1093/genetics/157.4.1819
Michno, J. M., Wang, X., Liu, J., Curtin, S. J., Kono, T. J., and Stupar, R. M. (2015). CRISPR/Cas Mutagenesis of Soybean and Medicago Truncatula Using a New Web-Tool and a Modified Cas9 Enzyme. GM crops & Food 6, 243–252. doi:10.1080/21645698.2015.1106063
Mishra, S. K., Macedo, M. L. R., Panda, S. K., and Panigrahi, J. (2018). Bruchid Pest Management in Pulses: Past Practices, Present Status and Use of Modern Breeding Tools for Development of Resistant Varieties. Ann. Appl. Biol. 172, 4–19. doi:10.1111/aab.12401
Mobini, S. H., and Warkentin, T. D. (2016). A Simple and Eficient Method of In Vivo Rapid Generation Technology in Pea (Pisum sativum L.) In Vitro. Cell Dev-Plant 52, 530–536. doi:10.1007/s11627-016-9772-7
Moghaddam, S. M., Oladzad, A., Koh, C., Ramsay, L., Hart, J. P., Mamidi, S., et al. (2021). The Tepary Bean Genome Provides Insight into Evolution and Domestication under Heat Stress. Nat. Commun. 12 (1), 1–4. doi:10.1038/s41467-021-22858-x
Mousavi-Derazmahalleh, M., Bayer, P. E., Nevado, B., Hurgobin, B., Filatov, D., Kilian, A., et al. (2018). Exploring the Genetic and Adaptive Diversity of a Pan-Mediterranean Crop Wild Relative: Narrow-Leafed Lupin. Theor. Appl. Genet. 131, 887–901. doi:10.1007/s00122-017-3045-7
Nair, R. M., Schafleitner, R., Kenyon, L., Srinivasan, R., Easdown, W., Ebert, A. W., et al. (2012). Genetic Improvement of Mungbean. SABRAO J. Breed. Genet. 44, 177–190.
Nair, R. M., Pandey, A. K., War, A. R., Hanumantharao, B., Shwe, T., Alam, A. K. M. M., et al. (2019). Biotic and Abiotic Constraints in Mungbean Production—Progress in Genetic Improvement. Front. Plant Sci. 10, 1340. doi:10.3389/fpls.2019.01340
Nasti, R. A., and Voytas, D. F. (2021). Attaining the Promise of Plant Gene Editing at Scale. Proc. Nat. Acad. Sci. 118 (22), e2004846117. doi:10.1073/pnas.2004846117
Nelson, M. N., Książkiewicz, M., Rychel, S., Besharat, N., Taylor, C. M., Wyrwa, K., et al. (2017). The Loss of Vernalization Requirement in Narrow‐leafed Lupin Is Associated with a Deletion in the Promoter and De‐repressed Expression of a Flowering Locus T (FT) Homologue. New Phytol. 213, 220–232. doi:10.1111/nph.14094
Nguyen, V., Riley, S., Nagel, S., Fisk, I., and Searle, I. R. (2020). Common Vetch: A Drought Tolerant, High Protein Neglected Leguminous Crop with Potential as a Sustainable Food Source. Front. Plant Sci. 11, 818. doi:10.3389/fpls.2020.00818
Nichols, P. G., Revell, C. K., Humphries, A. W., Howie, J. H., Hall, E. J., Sandral, G. A., et al. (2012). Temperate Pasture Legumes in Australia—their History, Current Use, and Future Prospects. Crop Pasture Sci. 63, 691–725. doi:10.1071/cp12194
O’Rourke, J. A., Fu, F., Bucciarelli, B., Yang, S. S., Samac, D. A., Lamb, J. F., et al. (2015). The Medicago Sativa Gene index 1.2: a Web-Accessible Gene Expression Atlas for Investigating Expression Differences between Medicago Sativa Subspecies. BMC Genom 16, 1–7. doi:10.1186/s12864-015-1718-7
O’Rourke, J. A., Yang, S. S., Miller, S. S., Bucciarelli, B., Liu, J., Rydeen, A., et al. (2013). An RNA-Seq Transcriptome Analysis of Orthophosphate-Deficient white Lupin Reveals Novel Insights into Phosphorus Acclimation in Plants. Plant Physiol. 161, 705–724. doi:10.1104/pp.112.209254
Ogutcen, E., Pandey, A., Khan, M. K., Marques, E., Penmetsa, R. V., Kahraman, A., et al. (2018). Pod Shattering: a Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement. Agronomy 8, 137. doi:10.3390/agronomy8080137
Olukolu, B. A., Mayes, S., Stadler, F., Ng, N. Q., Fawole, I., Dominique, D., et al. (2012). Genetic Diversity in Bambara Groundnut (Vigna subterranea (L.) Verdc.) as Revealed by Phenotypic Descriptors and DArT Marker Analysis. Genet. Resour. Crop Evol. 59, 347–358. doi:10.1007/s10722-011-9686-5
Parida, A., Raina, S. N., and Narayan, R. K. J. (1990). Quantitative DNA Variation between and within Chromosome Complements of Vigna Species (Fabaceae). Genetica 82, 125–133. doi:10.1007/bf00124642
Parimala, K., Meenakumari, K. V. S., Sudhakar, R., and Durga, K. K. (2011). Screening of Horsegram Genotypes against Yellow Mosaic Virus and Powdery Mildew Diseases. Indian. J. Plant Prot. 39, 160–161.
Patel, D. P., Dabas, B. S., Sapra, R. S., and Mandal, S. (1995). Evaluation of Horsegram (Macrotyloma uniflorum) (Lam.) Germplasm. New Delhi, India: National Bureau of Plant Genetic Resources Publication.
Plewiński, P., Ćwiek‐Kupczyńska, H., Rudy, E., Bielski, W., Rychel‐Bielska, S., Stawiński, S., et al. (2020). Innovative Transcriptome‐based Genotyping Highlights Environmentally Responsive Genes for Phenology, Growth and Yield in a Non‐model Grain Legume. Plant Cel. Environ. 43, 2680–2698. doi:10.1111/pce.13880
Plewiński, P., Książkiewicz, M., Rychel-Bielska, S., Rudy, E., and Wolko, B. (2019). Candidate Domestication-Related Genes Revealed by Expression Quantitative Trait Loci Mapping of Narrow-Leafed Lupin (Lupinus angustifolius L.). Int. J. Mol. Sci. 20 (22), 5670. doi:10.3390/ijms20225670
Pootakham, W., Nawae, W., Naktang, C., Sonthirod, C., Yoocha, T., Kongkachana, W., et al. (2021). A Chromosome‐scale Assembly of the Black Gram (Vigna mungo) Genome. Mol. Ecol. Resour. 21, 238–250. doi:10.1111/1755-0998.13243
Porch, T. G., Beaver, J. S., Debouck, D. G., Jackson, S. A., Kelly, J. D., and Dempewolf, H. (2013). Use of Wild Relatives and Closely Related Species to Adapt Common Bean to Climate Change. Agronomy 3, 433–461. doi:10.3390/agronomy3020433
Prasad, S. K., and Singh, M. K. (2015). Horse Gram-An Underutilized Nutraceutical Pulse Crop: a Review. J. Food Sci. Technol. 52, 2489–2499. doi:10.1007/s13197-014-1312-z
Qi, H., Kang, D., Zeng, W., Jawad Hassan, M., Peng, Y., Zhang, X., et al. (2021). Alterations of Endogenous Hormones, Antioxidant Metabolism, and Aquaporin Gene Expression in Relation to γ-Aminobutyric Acid-Regulated Thermotolerance in White Clover. Antioxidants 10, 1099. doi:10.3390/antiox10071099
Raizada, A., and Souframanien, J. (2019). Transcriptome Sequencing, De Novo Assembly, Characterisation of Wild Accession of Blackgram (Vigna mungo Var. silvestris) as a Rich Resource for Development of Molecular Markers and Validation of SNPs by High Resolution Melting (HRM) Analysis. BMC Plant Biol. 191-6. doi:10.1186/s12870-019-1954-0
Ramtekey, V., Bhuriya, A., Ayer, D., Parekh, V., Modha, K., Kale, B., et al. (2019). Molecular Tagging of Photoperiod Responsive Flowering in Indian Bean [Lablab purpureus (L.) Sweet]. Indian J. Genet. 79, 269. doi:10.31742/ijgpb.79s.1.17
Rawal, H. C., Kumar, S., Amitha Mithra, S. V., Solanke, A. U., Nigam, D., Saxena, S., et al. (2017). High Quality Unigenes and Microsatellites Markers from Tissue Specific Transcriptome and Development of a Database in Clusterbean (Cymopsis tetragonoloba (L.) Taub.). Genes 8, 313. doi:10.3390/genes8110313
Reddy, V. R., Das, S., Dikshit, H. K., Mishra, G. P., Aski, M. S., Singh, A., et al. (2021). Genetic Dissection of Phosphorous Uptake and Utilization Efficiency Traits Using GWAS in Mungbean. Agronomy 11, 1401. doi:10.3390/agronomy11071401
Ren, L., Huang, W., and Cannon, S. B. (2019). Reconstruction of Ancestral Genome Reveals Chromosome Evolution History for Selected Legume Species. New Phytol. 223, 2090–2103. doi:10.1111/nph.15770
Ren, Y. Z., Yue, Y. L., Jin, G. X., and Du, Q. (2014). First Report of Bacterial Blight of Guar Caused by Xanthomonas Axonopodis Pv. Cyamopsidis in China. Plant Dis. 98, 840. doi:10.1094/pdis-09-13-0983-pdn
Renzi, J. P., Chantre, G. R., Smýkal, P., Presotto, A. D., Zubiaga, L., Garayalde, A. F., et al. (2020). Diversity of Naturalized Hairy Vetch (Vicia Villosa Roth) Populations in Central Argentina as a Source of Potential Adaptive Traits for Breeding. Front. Plant Sci. 11, 189. doi:10.3389/fpls.2020.00189
Riday, H., and Krohn, A. L. (2010). Genetic Map-Based Location of the Red clover (Trifolium pratense L.) Gametophytic Self-Incompatibility Locus. Theor. Appl. Genet. 121, 761–767. doi:10.1007/s00122-010-1347-0
Robotham, O., and Chapman, M. (2017). Population Genetic Analysis of Hyacinth Bean (Lablab purpureus (L.) Sweet, Leguminosae) Indicates an East African Origin and Variation in Drought Tolerance. Genet. Resour. Crop Evol. 64, 139–148. doi:10.1007/s10722-015-0339-y
Rubatzky, V. E., and Yamaguchi, M. (1997). World Vegetables Principles, Production, and Nutritive Values. Fruits 5 (51), 381.
Rychel-Bielska, S., Nazzicari, N., Plewiński, P., Bielski, W., Annicchiarico, P., and Książkiewicz, M. (2020). Development of PCR-Based Markers and Whole-Genome Selection Model for Anthracnose Resistance in white Lupin (Lupinus albus L.). J. Appl. Genet. 61, 531–545. doi:10.1007/s13353-020-00585-1
Rychel, S., and Książkiewicz, M. (2019). Development of Gene-Based Molecular Markers Tagging Low Alkaloid Pauper Locus in white Lupin (Lupinus albus L.). J. Appl. Genet. 60, 269–281. doi:10.1007/s13353-019-00508-9
Samineni, S., Sen, M., Sajja, S. B., and Gaur, P. M. (2019). Rapid Generation advance (RGA) in Chickpea to Produce up to Seven Generations Per Year and Enable Speed Breeding. Crop J. 8, 164–169. doi:10.1016/j.cj.2019.08.003
Sammour, R. H. (2014). Genetic Diversity in Lathyrus sativus L. Germplasm. Res. Rev. Biosci. 8, 325–336.
Sankar, A. U., Anitha, K., Sivaraj, N., Kumari, K. M., Sunil, N., and Chakrabarty, S. K. (2015). Screening of Horsegram Germplasm Collected from Andhra Pradesh against Anthracnose. Legume Res. 38, 753–757. doi:10.18805/lr.v38i6.6719
Santos, C., Almeida, N. F., Alves, M. L., Horres, R., Krezdorn, N., Leitão, S. T., et al. (2018). First Genetic Linkage Map of Lathyrus Cicera Based on RNA Sequencing-Derived Markers: Key Tool for Genetic Mapping of Disease Resistance. Hort. Res. 5, 1–4. doi:10.1038/s41438-018-0047-9
Sato, S., Isobe, S., Asamizu, E., Ohmido, N., Kataoka, R., Nakamura, Y., et al. (2005). Comprehensive Structural Analysis of the Genome of Red clover (Trifolium pratense L.). DNA Res. 12, 301–364. doi:10.1093/dnares/dsi018
Saxena, K. B., Saxena, R. K., Hickey, L. T., and Varshney, R. K. (2019). Can a Speed Breeding Approach Accelerate Genetic Gain in Pigeonpea? Euphytica 215, 202. doi:10.1007/s10681-019-2520-4
Schafleitner, R., Huang, S. M., Chu, S. H., Yen, J. Y., Lin, C. Y., Yan, M. R., et al. (2016). Identification of Single Nucleotide Polymorphism Markers Associated with Resistance to Bruchids (Callosobruchus spp.) in Wild Mungbean (Vignaradiata Var. sublobata) and Cultivated V. radiata through Genotyping by Sequencing and Quantitative Trait Locus Analysis. BMC Plant Biol. 16, 1–5. doi:10.1186/s12870-016-0847-8
Sengooba, T. N., Spence, N. J., Walkey, D. G. A., Allen, D. J., and Femi Lana, A. (1997). The Occurrence of Bean Common Mosaic Necrosis Virus in Wild and Forage Legumes in Uganda. Plant Pathol. 46, 95–103. doi:10.1046/j.1365-3059.1997.d01-12.x
Shapter, F. M., Cross, M., Ablett, G., Malory, S., Chivers, I. H., King, G. J., et al. (2013). High-throughput Sequencing and Mutagenesis to Accelerate the Domestication of Microlaena Stipoides as a New Food Crop. PLoS One 8, e82641. doi:10.1371/journal.pone.0082641
Sharma, S., Tyagi, A., Srivastava, H., Ramakrishna, G., Sharma, P., Sevanthi, A. M., et al. (2021). Exploring the Edible Gum (Galactomannan) Biosynthesis and its Regulation during Pod Developmental Stages in Clusterbean Using Comparative Transcriptomic Approach. Sci. Reps. 11, 1–2. doi:10.1038/s41598-021-83507-3
Shegro, A., Van Rensburg, W. J., and Adebola, P. O. (2013). Assessment of Genetic Variability in Bambara Groundnut (Vigna subterrenea L. Verdc.) Using Morphological Quantitative Traits. Academia J. Agric. Res. 1 (3), 045–051.
Sherman, R. M., and Salzberg, S. L. (2020). Pan-genomics in the Human Genome Era. Nat. Rev. Genet. 21, 243–254. doi:10.1038/s41576-020-0210-7
Shi, R., Jiao, W., Yun, L., Zhang, Z., Zhang, X., Wang, Q., et al. (2021). Utilization of Transcriptome, Small RNA, and Degradome Sequencing to Provide Insights into Drought Stress and Rewatering Treatment in Medicago Ruthenica. Front. Plant Sci. 12, 675903. doi:10.3389/fpls.2021.675903
Shirasawa, K., Kosugi, S., Sasaki, K., Ghelfi, A., Okazaki, K., Toyoda, A., et al. (2021b). Genome Features of Common Vetch (Vicia sativa) in Natural Habitats. bioRxiv. doi:10.1101/2021.03.09.434686
Shirasawa, K., Chahota, R., Hirakawa, H., Nagano, S., Nagasaki, H., Sharma, T. R., et al. (2021a). A Chromosome-Scale Draft Genome Sequence of Horsegram (Macrotyloma uniflorum). bioRxiv. 2021a Jan 1. doi:10.1101/2021.01.18.427074
Singh, T., Ramakrishnan, S., Mahanta, S. K., Tyagi, V. C., and Roy, A. K. (2018). Tropical Forage Legumes in India: Status and Scope for Sustaining Livestock Production. London, UK: Intech Open, 123–143.
Singh, V., Goel, R., Pande, V., Asif, M. H., and Mohanty, C. S. (2017). De Novo sequencing and Comparative Analysis of Leaf Transcriptomes of Diverse Condensed Tannin-Containing Lines of Underutilized Psophocarpus tetragonolobus (L.) DC. Sci. Reps. 7, 1–3. doi:10.1038/srep44733
Sokolkova, A., Burlyaeva, M., Valiannikova, T., Vishnyakova, M., Schafleitner, R., Lee, C. R., et al. (2020). Genome-wide Association Study in Accessions of the Mini-Core Collection of Mungbean (Vigna radiata) from the World Vegetable Gene Bank (Taiwan). BMC Plant Biol. 20, 1–9. doi:10.1186/s12870-020-02579-x
Somta, P., Chen, J., Yundaeng, C., Yuan, X., Yimram, T., Tomooka, N., et al. (2019). Development of an SNP-Based High-Density Linkage Map and QTL Analysis for Bruchid (Callosobruchus maculatus F.) Resistance in Black Gram (Vigna mungo (L.) Hepper). Sci. Reps 9 (1), 1–9. doi:10.1038/s41598-019-40669-5
Somta, P., Jomsangawong, A., Yundaeng, C., Yuan, X., Chen, J., Tomooka, N., et al. (2018). Genetic Dissection of Azuki Bean Weevil (Callosobruchus chinensis L.) Resistance in Moth Bean (Vigna aconitifolia [Jaqc.] Maréchal). Genes 9, 555. doi:10.3390/genes9110555
Somta, P., Kaga, A., Tomooka, N., Kashiwaba, K., Isemura, T., Chaitieng, B., et al. (2006). Development of an Interspecific Vigna Linkage Map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its Use in Analysis of Bruchid Resistance and Comparative Genomics. Plant Breed 125, 77–84. doi:10.1111/j.1439-0523.2006.01123.x
Souframanien, J., and Gopalakrishna, T. (2006). ISSR and SCAR Markers Linked to the Mungbean Yellow Mosaic Virus (MYMV) Resistance Gene in Blackgram [Vigna mungo (L.) Hepper]. Plant Breed 125, 619–622. doi:10.1111/j.1439-0523.2006.01260.x
Souframanien, J., Gupta, S. K., and Gopalakrishna, T. (2010). Identification of Quantitative Trait Loci for Bruchid (Callosobruchus maculatus) Resistance in Black Gram [Vigna mungo (L.) Hepper]. Euphytica 176, 349–356. doi:10.1007/s10681-010-0210-3
Souframanien, J., and Reddy, K. S. (2015). De Novo Assembly, Characterization of Immature Seed Transcriptome and Development of Genic-SSR Markers in Black Gram [Vigna mungo (L.) Hepper]. PLoS One 10, e0128748. doi:10.1371/journal.pone.0128748
Soylu, S., Soylu, E. M., Kara, M., Kurt, Ş., and Choi, Y. J. (2020). First Report of Downy Mildew Disease Caused by Peronospora Viciae on Common Vetch (Vicia Sativa) in Turkey. Plant Dis. 104, 1876. doi:10.1094/pdis-12-19-2568-pdn
Sserumaga, J. P., Kayondo, S. I., Kigozi, A., Kiggundu, M., Namazzi, C., Walusimbi, K., et al. (2021). Genome-wide Diversity and Structure Variation Among Lablab [Lablab purpureus (L.) Sweet] Accessions and Their Implication in a Forage Breeding Program. Genet. Resour. Crop Evol. 19, 1–4.
Sudha, M., Karthikeyan, A., Shobhana, V. G., Nagarajan, P., Raveendran, M., Senthil, N., et al. (2015). Search for Vigna Species Conferring Resistance to Mungbean Yellow Mosaic Virus in Mungbean. Plant Genet. Resour. 13, 162. doi:10.1017/s1479262114000859
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted Mutagenesis in Soybean Using the CRISPR-Cas9 System. Sci. Reps. 5, 10342. doi:10.1038/srep10342
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL‐seq: Rapid Mapping of Quantitative Trait Loci in rice by Whole Genome Resequencing of DNA from Two Bulked Populations. Plant J. 74, 174–183. doi:10.1111/tpj.12105
Takagi, H., Tamiru, M., Abe, A., Yoshida, K., Uemura, A., Yaegashi, H., et al. (2015). MutMap Accelerates Breeding of a Salt-Tolerant Rice Cultivar. Nat. Biotechnol. 33, 445–449. doi:10.1038/nbt.3188
Takahashi, Y., Sakai, H., Yoshitsu, Y., Muto, C., Anai, T., Pandiyan, M., et al. (2019). Domesticating Vigna Stipulacea: A Potential Legume Crop with Broad Resistance to Biotic Stresses. Front. Plant Sci. 10, 1607. doi:10.3389/fpls.2019.01607
Talhinhas, P., Baroncelli, R., and Le Floch, G. (2016). Anthracnose of Lupins Caused by Colletotrichum Lupini: A Recent Disease and a Successful Worldwide Pathogen. J. Plant Pathol. 5, 10342. doi:10.1038/srep10342
Tamburini, G., Bommarco, R., Wanger, T. C., Kremen, C., van der Heijden, M. G., Liebman, M., et al. (2020). Agricultural Diversification Promotes Multiple Ecosystem Services Without Compromising Yield. Sci. Adv. 6, eaba1715. doi:10.1126/sciadv.aba1715
Tan, L. X., Azam-Ali, S., Goh, E. V., Mustafa, M. A., Chai, H. H., Kuan, Ho. W., et al. (2020). Bambara Groundnut: An Underutilized Leguminous Crop for Global Food Security and Nutrition. Front. Nutr. 7, 276. doi:10.3389/fnut.2020.601496
Tanwar, U. K., Pruthi, V., and Randhawa, G. S. (2017). RNA-seq of Guar (Cyamopsis tetragonoloba, L. Taub.) Leaves: De Novo Transcriptome Assembly, Functional Annotation and Development of Genomic Resources. Front. Plant Sci. 8, 91. doi:10.3389/fpls.2017.00091
Tate, M., and Enneking, D. (2006). Common Vetch (Vicia Sativa Ssp. Sativa): Feed or Future Food? Grain Legumes 47, 16–17.
Taylor, C. M., Kamphuis, L. G., Zhang, W., Garg, G., Berger, J. D., Mousavi‐Derazmahalleh, M., et al. (2019). INDEL Variation in the Regulatory Region of the Major Flowering Time Gene LanFTc1 Is Associated with Vernalization Response and Flowering Time in Narrow‐leafed Lupin (Lupinus angustifolius L.). Plant Cel Environ 42, 174–187. doi:10.1111/pce.13320
Taylor, C. M., Garg, G., Berger, J. D., Ribalta, F. M., Croser, J. S., Singh, K. B., et al. (2021). A Trimethylguanosine Synthase1-like (TGS1) Homologue Is Implicated in Vernalisation and Flowering Time Control. Theor. Appl. Genet. 134, 3411–3426. doi:10.1007/s00122-021-03910-2
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome Analysis of Multiple Pathogenic Isolates of Streptococcus Agalactiae: Implications for the Microbial “Pan-genome”. Proc. Nat. Acad. Sci. 102, 13950–13955. doi:10.1073/pnas.0506758102
Thakur, O., and Randhawa, G. S. (2018). Identification and Characterization of SSR, SNP and InDel Molecular Markers from RNA-Seq Data of Guar (Cyamopsis tetragonoloba, L. Taub.) Roots. BMC Genome 19, 951. doi:10.1186/s12864-018-5205-9
Thirumaran, A. S., and Kanchana, S. (2000). “Role of Pulses in Human Diets,” in Pulses Production Strategies in Tamil Nadu (Coimbatore, India: Centre for Plant Breeding and Genetics, Tamil Nadu Agricultural University), 129. Available at: http://www.tnau.ac.in/tech/pulsesbook. pdf (Accessed July 13, 2012).
Tian, J., Isemura, T., Kaga, A., Vaughan, D. A., and Tomooka, N. (2013). Genetic Diversity of the rice Bean (Vigna umbellata) Genepool as Assessed by SSR Markers. Genome 56, 717–727. doi:10.1139/gen-2013-0118
Tomooka, N., Vaughan, D. A., Moss, H., and Maxted, N. (2002). The Asian Vigna: Genus Vigna Subgenus Ceratotropis Genetic Resources. New York: Kluwer Academic Publishers.
Tribhuvan, K. U., Amitha Mithra, S. V., Sharma, P., Das, A., Kumar, K., Tyagi, A., et al. (2019). Identification of Genomic SSRs in Cluster Bean (Cyamopsis tetragonoloba) and Demonstration of Their Utility in Genetic Diversity Analysis. Ind. Crops Prod. 133, 221–231. doi:10.1016/j.indcrop.2019.03.028
Tripathy, S., and Das, M. K. (2013). Guar Gum: Present Status and Applications. J. Pharm. Sci. Innov. 4, 24–28. doi:10.7897/2277-4572.02447
Tyagi, A., Nigam, D., Sv, A. M., Solanke, A. U., Singh, N. K., Sharma, T. R., et al. (2018). Genome‐wide Discovery of Tissue‐Specific miRNAs in Clusterbean (Cyamopsis tetragonoloba) Indicates Their Association with Galactomannan Biosynthesis. Plant Biotechnol. J. 16, 1241–1257. doi:10.1111/pbi.12866
Tyagi, A., Sharma, P., Saxena, S., Sharma, R., Mithra, S. A., Solanke, A. U., et al. (2019). The Genome Size of Clusterbean (Cyamopsis tetragonoloba) Is Significantly Smaller Compared to its Wild Relatives as Estimated by Flow Cytometry. Gene 707, 205–211. doi:10.1016/j.gene.2019.02.090
Uba, C. U., Oselebe, H. O., Tesfaye, A. A., and Abtew, W. G. (2021). Genetic Diversity and Population Structure Analysis of Bambara Groundnut (Vigna subterrenea L) Landraces Using DArT SNP Markers. PLoS One 16, e0253600. doi:10.1371/journal.pone.0253600
Vaijayanthi, P. V., Ramesh, S., Gowda, M. B., and Mohan, R. A. (2015). Genetic Variability for Morpho-Metric Traits in Dolichos Bean (Lablab purpureus L. Sweet) Germplasm. J. Food Legumes 28 (1), 5–10.
van Oers, C. C. C. M. (1989). “Vigna Aconitifolia (Jacq.) Maréchal,” in Plant Resources of South-East Asia No 1. Pulses. Editors L. J. G. van der Maesen, and S. Somaatmadja (Wageningen, Netherlands: Pudoc), 66–67.
Varshney, R. K., Roorkiwal, M., Sun, S., Bajaj, P., Chitikineni, A., Thudi, M., et al. (2021). A Chickpea Genetic Variation Map Based on the Sequencing of 3,366 Genomes. Nature 599 (7886), 622–627. doi:10.1038/s41586-021-04066-1
Vatanparast, M., Shetty, P., Chopra, R., Doyle, J. J., Sathyanarayana, N., and Egan, A. N. (2016). Transcriptome Sequencing and Marker Development in Winged Bean (Psophocarpus Tetragonolobus; Leguminosae). Sci. Reps. 6, 1–4. doi:10.1038/srep29070
Vaughan, D. A., Tomooka, N., and Kaga, A. (2005). “Azuki Bean [Vigna Angularis (L.) Ohwi and Ohashi],” in Genetic Resources, Chromosome Engineering and Crop Improvement. Editors R. J. Singh, and P. P. Jauhar (Boca Roca, FL: CRC Press), 1, 347–359. Grain Legumes. doi:10.1201/9780203489284-15
Vaz Patto, M. C., Skiba, B., Pang, E. C. K., Ochatt, S. J., Lambein, F., and Rubiales, D. (2006). Lathyrus Improvement for Resistance against Biotic and Abiotic Stresses: From Classical Breeding to Marker Assisted Selection. Euphytica 147, 133–147. doi:10.1007/s10681-006-3607-2
Vleugels, T., Baert, J., and Van Bockstaele, E. (2013). Evaluation of a Diverse Red clover Collection for Clover Rot Resistance (Sclerotinia trifoliorum). Commun. Agricult. Appl. Biol. Sci. 78, 519–522.
Von Wettberg, E., Davis, T. M., and Smýkal, P. (2020). Wild Plants as Source of New Crops. Front. Plant Sci. 11, 1426. doi:10.3389/fpls.2020.591554
Wang, J., Li, J., Liu, Z., Yuan, X., Wang, S., Chen, H., et al. (2020). Construction of a High-Density Genetic Map and its Application for QTL Mapping of Leaflet Shapes in Mung Bean (Vigna radiata L.). Front. Genet. 11. doi:10.3389/fgene.2020.01032
Wang, L., Do Kim, K., Gao, D., Chen, H., Wang, S., Lee, S., et al. (2016). Analysis of Simple Sequence Repeats in rice Bean (Vigna umbellata) Using an SSR-Enriched Library. Crop J. 4, 40–47. doi:10.1016/j.cj.2015.09.004
Wang, L. X., Jie, W. A., Luo, G. L., Yuan, X. X., Dan, G. O., Hu, L. L., et al. (2021b). Construction of a High-Density Adzuki Bean Genetic Map and Evaluation of its Utility Based on a QTL Analysis of Seed Size. J. Integr. Agric. 20, 1753–1761. doi:10.1016/s2095-3119(20)63343-3
Wang, M. L., Morris, J. B., Barkley, N. A., Dean, R. E., Jenkins, T. M., and Pederson, G. A. (2007). Evaluation of Genetic Diversity of the USDALablab Purpureus Germplasm Collection Using Simple Sequence Repeat Markers. J. Hortic. Sci. Biotech. 82, 571–578. doi:10.1080/14620316.2007.11512275
Wang, P., Zhou, G., Jian, J., Yang, H., Renshaw, D., Aubert, M. K., et al. (2021a). Whole‐Genome Assembly and Resequencing Reveal Genomic Imprint and Key Genes of Rapid Domestication in Narrow‐Leafed Lupin. Plant J. 105, 1192–1210. doi:10.1111/tpj.15100
Wang, T., Ren, L., Li, C., Zhang, D., Zhang, X., Zhou, G., et al. (2021c). The Genome of a Wild Medicago Species Provides Insights into the Tolerant Mechanisms of Legume Forage to Environmental Stress. BMC Plant Biol. 19, 1–7. doi:10.1186/s12915-021-01033-0
Warschefsky, E., Penmetsa, R. V., Cook, D. R., and Von Wettberg, E. J. (2014). Back to the Wilds: Tapping Evolutionary Adaptations for Resilient Crops through Systematic Hybridization with Crop Wild Relatives. Amer. J. Bot. 101, 1791–1800. doi:10.3732/ajb.1400116
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M.-D., et al. (2018). Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 4, 23–29. doi:10.1038/s41477-017-0083-8
Wei, Z. W., Hailong, R., Zinian, W., Guozhi, L., Xiang, C., and Zhihong, Q. (2015). A New Medicago Polymorpha Cultivar ‘Huaiyang Jinhuacai. Acta Hortic. Sin 42, 2335–2336.
Wu, X., Islam, A. S., Limpot, N., Mackasmiel, L., Mierzwa, J., Cortés, A. J., et al. (2020). Genome-wide SNP Identification and Association Mapping for Seed Mineral Concentration in Mung Bean (Vigna radiata L.). Front. Genet. 11, 656. http://www.fao.org/faostat. doi:10.3389/fgene.2020.00656
Xu, Q., Liu, F., Qu, R., Gillman, J. D., Bi, C., Hu, X., et al. (2018). Transcriptomic Profiling of Lathyrus Sativus L. Metabolism of β-ODAP, a Neuroexcitatory Amino Acid Associated with Neurodegenerative Lower Limb Paralysis. Plant Mol. Biol. Rep. 36, 832–843. doi:10.1007/s11105-018-1123-x
Yadav, S. S., Bejiga, G., Brink, M., and Belay, G. (2006). ‘Lathyrus Sativus L.’ PROTA4U. Wageningen, Netherlands: PROTA (Plant Resources of Tropical Africa/Ressources végétales de l’Afrique tropicale). http://www.prota4u.org/search.asp.
Yang, K., Tian, Z., Chen, C., Luo, L., Zhao, B., Wang, Z., et al. (2015). Genome Sequencing of Adzuki Bean (Vigna angularis) Provides Insight into High Starch and Low Fat Accumulation and Domestication. Proc. Nat. Acad. Sci. 112 (43), 13213–13218. doi:10.1073/pnas.1420949112
Yang, H., Jian, J., Li, X., Renshaw, D., Clements, J., Sweetingham, M. W., et al. (2015). Application of Whole Genome Re-sequencing Data in the Development of Diagnostic DNA Markers Tightly Linked to a Disease-Resistance Locus for Marker-Assisted Selection in Lupin (Lupinus angustifolius). BMC Genome 16, 1–7. doi:10.1186/s12864-015-1878-5
Yang, H., Tao, Y., Zheng, Z., Li, C., Sweetingham, M. W., and Howieson, J. G. (2012). Application of Next-Generation Sequencing for Rapid Marker Development in Molecular Plant Breeding: a Case Study on Anthracnose Disease Resistance in Lupinus angustifolius L. BMC Genom. 13, 1–2. doi:10.1186/1471-2164-13-318
Yang, H., Tao, Y., Zheng, Z., Shao, D., Li, Z., Sweetingham, M. W., et al. (2013b). Rapid Development of Molecular Markers by Next-Generation Sequencing Linked to a Gene Conferring Phomopsis Stem Blight Disease Resistance for Marker-Assisted Selection in Lupin (Lupinus angustifolius L.) Breeding. Theor. Appl. Genet. 126, 511–522. doi:10.1007/s00122-012-1997-1
Yang, H., Tao, Y., Zheng, Z., Zhang, Q., Zhou, G., Sweetingham, M. W., et al. (2013a). Draft Genome Sequence, and a Sequence-Defined Genetic Linkage Map of the Legume Crop Species Lupinus angustifolius L. PLoS One 8, e64799. doi:10.1371/journal.pone.0064799
Yang, T., Jiang, J., Burlyaeva, M., Hu, J., Coyne, C. J., Kumar, S., et al. (2014). Large-Scale Microsatellite Development in Grasspea (Lathyrus sativus L.), an Underutilized Legume of the Arid Areas. BMC Plant Biol. 14, 1–2. doi:10.1186/1471-2229-14-65
Yang, T., Nagy, I., Mancinotti, D., Otterbach, S. L., Andersen, T. B., Motawia, M. S., et al. (2017). Transcript Profiling of a Bitter Variety of Narrow-Leafed Lupin to Discover Alkaloid Biosynthetic Genes. J. Expt Bot. 68, 5527–5537. doi:10.1093/jxb/erx362
Yao, L. M., Wang, B., Cheng, L. J., and Wu, T. L . (2013). Identification of Key Drought Stress-Related Genes in the Hyacinth Bean. PLoS ONE 8, e58108. doi:10.1371/journal.pone.0058108
Yaqoob, M., Khan, N. U., Mansoor, M., Gul, S., Khalil, I. A., and Afridi, K. (2015). Moth Bean Germplasm Screening against Yellow Mosaic Virus, II. Developmentof Moth Bean High-Yielding Seed and Fodder Cultivars. Turk. J. Agric. For. 39, 212–226. doi:10.3906/tar-1406-3
Yates, S. A., Swain, M. T., Hegarty, M. J., Chernukin, I., Lowe, M., Allison, G. G., et al. (2014). De novo Assembly of Red Clover Transcriptome Based on RNA-Seq Data Provides Insight into Drought Response, Gene Discovery and Marker Identification. BMC Genom. 15 (1), 1–15.
Yigezu, Y. A., El-Shater, T., Boughlala, M., Bishaw, Z., Niane, A. A., Maalouf, F., et al. (2019). Legume-based Rotations Have clear Economic Advantages over Cereal Monocropping in Dry Areas. Agron. Sust. Dev. 39, 1–14. doi:10.1007/s13593-019-0602-2
Yin, L., Zhang, M., Wu, R., Chen, X., Liu, F., and Xing, B. (2021). Genome-wide Analysis of OSCA Gene Family Members in Vigna Radiata and Their Involvement in the Osmotic Response. BMC Plant Biol. 21, 1–2. doi:10.1186/s12870-021-03184-2
Yin, M., Zhang, S., Du, X., Mateo, R. G., Guo, W., Li, A., et al. (2021). Genomic Analysis of Medicago Ruthenica Provides Insights into its Tolerance to Abiotic Stress and Demographic History. Mol. Ecol. Resour. 21 (5), 1641–1657. doi:10.1111/1755-0998.13363
Yin, X., Yi, K., Zhao, Y., Hu, Y., Li, X., He, T., et al. (2020). Revealing the Full-Length Transcriptome of Caucasian clover Rhizome Development. BMC Plant Biol. 20, 1–5. doi:10.1186/s12870-020-02637-4
Yoshida, Y., Marubodee, R., Ogiso-Tanaka, E., Iseki, K., Isemura, T., Takahashi, Y., et al. (2016). Salt tolerance in wild relatives of adzuki bean, Vigna angularis (Willd.) Ohwi et Ohashi. Genet. Resour. Crop Evol. 63, 627–637. doi:10.1007/s10722-015-0272-0
Yoshida, J., Tomooka, N., Yee Khaing, T., Shantha, P. S., Naito, H., Matsuda, Y., et al. (2020). Unique Responses of Three Highly Salt-Tolerant Wild Vigna Species against Salt Stress. Plant Prod. Sci. 23, 114–128. doi:10.1080/1343943x.2019.1698968
You, M. P., Nichols, P. G., Katusiime, R., and Barbetti, M. J. (2021). Novel Disease Host Resistances in the World Core Collection of Trifolium Subterraneum. Plant Dis.. PDIS-09. doi:10.1094/pdis-09-20-1985-re
Yuan, J., Wang, B., and Wu, T. L. (2011). Quantitative Trait Loci (QTL) Mapping for Inflorescence Length Traits in Lablab purpureus (L.) Sweet. Afr. J. Biotechnol. 10, 3558–3566.
Yundaeng, C., Somta, P., Amkul, K., Kongjaimun, A., Kaga, A., and Tomooka, N. (2019). Construction of Genetic Linkage Map and Genome Dissection of Domestication-Related Traits of Moth Bean (Vigna aconitifolia), a Legume Crop of Arid Areas. Mol. Genet. Genome 294, 621–635. doi:10.1007/s00438-019-01536-0
Zhang, F., and Batley, J. (2020). Exploring the Application of Wild Species for Crop Improvement in a Changing Climate. Curr. Opin. Plant Biol. 56, 218–222. doi:10.1016/j.pbi.2019.12.013
Zhang, H., Li, Y., and Zhu, J. K. (2018). Developing Naturally Stress-Resistant Crops for as Sustainable Agriculture. Nat. Plants 4, 989–996. doi:10.1038/s41477-018-0309-4
Zhang, H., Tian, H., Chen, M., Xiong, J., Cai, H., and Liu, Y. (2018). Transcriptome Analysis Reveals Potential Genes Involved in Flower Pigmentation in a Red-Flowered Mutant of white clover (Trifolium repens L.). Genomics 110, 191–200. doi:10.1016/j.ygeno.2017.09.011
Zhang, Q., Yan, Q., Yuan, X., Lin, Y., Chen, J., Wu, R., et al. (2021). Two Polygalacturonase-Inhibiting Proteins (VrPGIP) of Vigna Radiata Confer Resistance to Bruchids (Callosobruchus Spp). J. Plant Physiol. 258, 153376. doi:10.1016/j.jplph.2021.153376
Zhang, Y., Massel, K., Godwin, I. D., and Gao, C. (2018). Applications and Potential of Genome Editing in Crop Improvement. Genome Biol. 19, 1. doi:10.1186/s13059-018-1586-y
Zhou, G., Jian, J., Wang, P., Li, C., Tao, Y., Li, X., et al. (2018). Construction of an Ultra-high-density Consensus Genetic Map, and Enhancement of the Physical Map from Genome Sequencing in Lupinus Angustifolius. Theor. Appl. Genet. 131, 209–223. doi:10.1007/s00122-017-2997-y
Zhou, G., Yang, H., Renshaw, D., Zou, M., Thomas, G., and Li, C. (2021). Identification of Gray Leaf Spot Disease Candidate Gene in Narrow-Leafed Lupin (Lupinus angustifolius L.). Front. Genet. 2021, 1375. doi:10.3389/fgene.2021.695791
Zhu, Z., Chen, H., Xie, K., Liu, C., Li, L., Liu, L., et al. (2020). Characterization of Drought-Responsive Transcriptome during Seed Germination in Adzuki Bean (Vigna angularis L.) by PacBio SMRT and Illumina Sequencing. Front. Genet. 11, 996. doi:10.3389/fgene.2020.00996
Keywords: underutilized legumes, genomics, molecular marker, food security, transcriptomics
Citation: Jha UC, Nayyar H, Parida SK, Bakır M, von Wettberg EJB and Siddique KHM (2022) Progress of Genomics-Driven Approaches for Sustaining Underutilized Legume Crops in the Post-Genomic Era. Front. Genet. 13:831656. doi: 10.3389/fgene.2022.831656
Received: 08 December 2021; Accepted: 24 February 2022;
Published: 07 April 2022.
Edited by:
Aamir Raina, Aligarh Muslim University, IndiaReviewed by:
Leif Skot, Aberystwyth University, United KingdomCopyright © 2022 Jha, Nayyar, Parida, Bakır, von Wettberg and Siddique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uday Chand Jha, dWRheV9nZW5lQHlhaG9vLmNvLmlu; Kadambot H. M. Siddique, a2FkYW1ib3Quc2lkZGlxdWVAdXdhLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.