
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 31 March 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.831611
Modern research has proved that the main medicinal component of Rhodiola crenulata, which has a wide range of medicinal value, is its secondary metabolite salidroside. The MYB transcription factor family is widely involved in biosynthesis of second metabolism and other roles in the stress response in plants, so a genome-wide identification and analysis for this family in R. crenulata is worth conducting. In this research, genome-wide analysis identified 139 MYB genes based on conserved domains in the R. crenulata genome, and 137 genes were used to construct a phylogenetic tree and modified with expression files to reveal evolutionary characteristics. Physical and chemical characteristics, gene structure, and conserved motif analysis were also used to further analyze RcMYBs. Additionally, cis-acting elements related to transcription, hormone, and MYB binding were found in the promoter region of the selected RcMYBs. Four RcMYBs were cloned, sequenced, and their gene expression pattern was analyzed for further analysis of their functions. The research results lay the foundation for further research on the function of RcMYB and R. crenulata.
Rhodiola is a perennial plant in the genus Rhodiola of the Crassulaceae family, and is composed of more than 90 species. Most Rhodiola species grow in high-altitude and cold regions from Eurasia to North America. Among them, 73 varieties of Rhodiola are distributed in China. Rhodiola extract has also shown anti-inflammatory and anti-radiation effects and prevent cardiovascular diseases and cancer (Qu et al., 2012; Fan et al., 2020). Salidroside, which is mainly found in roots and stems, is the most medicinal functional ingredient in Rhodiola, and its content is an important factor in evaluating the quality of Rhodiola. Previous studies have shown that the biosynthesis of salidroside is derived from tyrosine, and some enzymes, tyrosine decarboxylase (TyDC), monoamine oxidase (MAO), 4-hydroxyphenylacetaldehyde reductase (4HPAR), UGTs, and 4-HPAA synthase (4HPAAS), are involved in the pathway (Lan et al., 2013; Torrens-Spence et al., 2017).
MYB transcription factors are one of the largest transcription factor family in plants, with the characteristics of multiple species and diversified functions (Prouse and Campbell, 2012), especially playing an important role in the regulation of plant secondary metabolism, such as the synthesis of flavonoids (Lai et al., 2013; Liu et al., 2015). It is found that MYB-conserved domains form a helix-turn-helix (HTH) structure in space and the structure interacts with regulatory elements in the promoter. The number and position of repeated MYB domains and MYB transcription factors are divided into four categories, namely 1R-MYB/MYB-related, R2R3-MYB, 3R-MYB, and 4R-MYB (Jiang et al., 2004a; Dubos et al., 2010). Among them, the R2R3-MYB protein is the most studied. Many members of this family have important regulatory effects in many biological processes, such as plant growth and development, organ morphogenesis, secondary metabolism, and stress response (Butt et al., 2017).
The research on MYB genes in plants mostly covers Arabidopsis thaliana and Oryza sativa. About 126 R2R3-MYB subfamily members have been identified in Arabidopsis thaliana, and their functions have been demonstrated and verified (Yanhui et al., 2006). These characterized AtMYBs can be used to predict the functions of MYBs in other species by homology analysis. So far, no research on the MYB transcription factor family in R. crenulata has been completed.
In this research, a total of 139 RcMYB genes of R. crenulata were identified using a known MYB genome sequence from A. thaliana. Moreover, the exon-intron structure, chromosome location, phylogenetic relationships, conserved motifs, and promotor elements of RcMYB genes were clarified. Besides, a co-expression network was constructed based on the expression patterns of key genes in the biosynthetic pathway of salidroside and the identified MYB genes, aiming to speculate on the RcMYB members involved in regulating the synthesis of salidroside. The selected RcMYB genes were verified by cloning and resequencing. Finally, the expression pattern of these selected RcMYBs in different tissues was determined. Based on the current data, this research is the first report on genome-wide gene family identification in R. crenulata. Our research may be helpful for further study about the functional characterization of RcMYBs.
The tissue culture materials of R. crenulata used in the experiment were collected from unknown mountains in Wenchuan county, located in the northwest of Sichuan province, China (Supplementary Figure S1).
To identify the RcMYB genes, the whole-genome sequencing result data of R. crenulata (Fu et al., 2017) from GigaDB (http://gigadb.org/dataset/10030) and annotation files from NCBI (https://www.ncbi. nlm.nih.gov/) were downloaded. Then, the candidate RcMYB sequences were screened with the online tool Pfam (http://pfam.xfam.org/) to identify the MYB domain, and sequences containing no conserved MYB domains were deleted. Further, BLAST comparison by submitting the identified RcMYBs into the NCBI database was performed (Ron et al., 2002). All AtMYBs were downloaded from PlantTFDB v5.0 (http://planttfdb.gao-lab.org/), along with the function annotation from the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/) and NCBI databases. The secondary metabolism-related AtMYBs were selected as the comparison group for subsequent screening of target RcMYBs.
Gene ID information of the RcMYB loci on the chromosome (scaffolds) was obtained from the genome annotation files (Fu et al., 2017). The distribution of RcMYBs on the chromosome (scaffolds) was generated with TBtools (Chen et al., 2020). The gene density was calculated and outputted by TBtools, while the GC density was calculated by Biostrings (https://bioconductor.org/packages/Biostrings). Analyzed results and annotations were visualized by an online Circos drawing (Gu et al., 2014).
The phylogenetic analysis of RcMYBs was conducted using a method from a previous work (Long et al., 2021). Briefly, amino sequences of RcMYBs were aligned by Clustal W with default parameters and then a phylogenetic tree was constructed using the neighbor joining (NJ) method in MEGA X after a selection of a substitution model (Kumar et al., 2018). The parameters of NJ analysis were as follows: bootstrap method done 1,000 times for statistical testing, model with JTT + G, and 50% sites coverage cutoff. The phylogenetic tree was modified by online software Interactive Tree Of Life (iTOLv6).
The physical and chemical characteristics of RcMYB proteins like molecular weight (MW) and isoelectric point (pI) were calculated by the ProtParam tool in the ExPASy Server (https://web.expasy.org/protparam/). All the amino acid sequences of RcMYB proteins were submitted to the online tool MEME suite (http://meme-suite.org/tools/meme) where we performed motifs analysis. Conserved motifs were detected with setting the repeat number to any (anr), the maximum number of motifs to 12, and the rest of the run parameters to system default. The structure diagram and LOGO of each motif from the result were downloaded. TBtools was used to combine the motif structure diagram with the phylogenetic tree, then a gene structure diagram of RcMYBs was outputted (Chen et al., 2020).
To explore the cis-elements in the promoter region of the RcMYB genes, 2 kb upstream RcMYBs sequences were uploaded to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
RNA-seq data in three tissues (root, stem, leaf) of R. crenulata were downloaded from NCBI respectively through SRA searching (SRX2577721 for root, SRX2577722 for stem, and SRX2577723 for leaf). We followed the method described by Wu and Zhang (2013). Briefly, transcriptome data were mapped to the R. crenulata genome by HISAT2 (version 2.2.1), and results were output in the SAM format. SAMtools (version 1.6) was used to convert result into the BAM format and we inputted the BAM file as a parameter into Stringtie (version 2.1.5) to generate the input file for Ballgown (version 2.22.0). Differential expression analysis was completed through software package Ballgown in R and visualized by iTOLv6 (https://itol.embl.de/).
A gene co-expression network was constructed by using the WGCNA R package to further find RcMYB genes which might be involved in the metabolism of salidroside (Langfelder and Horvath, 2008). The FPKM values of 407 genes in three tissues were used to construct the network (Supplementary Table S1). The soft thresholding power β of nine was selected in order for the networks to exhibit an approximate scale-free topology. The adjacency matrix was transformed into a topological overlap matrix (TOM) to calculate the corresponding dissimilarity (1-TOM similarity). Afterward, a gene dendrogram was produced based on genes which were clustered using the TOM dissimilarity measure (Zhang and Horvath, 2005). Genes with co-expression patterns were grouped in the same module, and modules with salidroside metabolism pathway genes were visualized by Cytoscape (version 3.8.2; Shannon et al., 2003).
DNA and RNA were extracted from the leaf and root tissues of R. crenulata individually. DNA extraction of materials was undertaken with CTAB methods (Minas et al., 2011), whereas RNA extraction followed the protocol of Trizol (TIANGEN, Beijing, China) reagent. RNA was reverse-transcribed using the PrimeScript™ RT Reagent Kit with a genomic DNA Eraser (Takara, Dalian, China), and quantitative RT-PCRs (qRT-PCR) were performed using a ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China). All the experiments were performed following the manufacturer’s instructions. Data were normalized by the expression level of 18S rRNA (Lan et al., 2013), and the primers used for qRT-PCR are listed in Supplementary Table S2.
Four RcMYB candidate genes were isolated from different cDNAs using specific primers. The full-length cDNA sequence of the four RcMYB genes excluding the terminator codon were amplified by PCR. The PCR products were sub-cloned into the pEASY-Blunt cloning vector (TransGen Biotech, Beijing, China) and sequenced (Sangon Biotech, Shanghai, China). The primers used for vector contractor and sequencing are listed in Supplementary Table S2.
To identify MYB genes in the R. crenulata genome, the candidate RcMYBs were screened based on annotation and were uploaded to the Pfam34.0 database. Then two hidden Markov models (HMM) of MYB-DNA-binding domain (PF00249) and MYB-DNA-Bind 6 domain (PF13921) were used in the identified RcMYBs whereas the sequences without MYB domains were deleted. Identified RcMYBs were compared with MYBs in other species through NCBI BLAST for further verification. Finally, 139 MYB genes were confirmed from the R. crenulata reference genome, and then designated as RcMYB1 to RcMYB139 (Supplementary Table S6). Multiple sequence alignment of the RcMYBs was carried out by Clustal W, and showed that RcMYB138 and RcMYB139 had few common sites for alignment and were deleted from the phylogenetic tree. The length of most RcMYB protein sequences ranged from 150 to 550 amino acids (AA), and the theoretical isoelectric point (pI) ranged from 3.31 to 10.38 (Supplementary Table S6). The length of the two domains in RcMYBs were shown in Pfam: DNA-binding domain with a length of 46AA and DNA-bind 6 domain with a length of 60AA (Supplementary Figure S2A). Detailed information of RcMYB genes is shown in Supplementary Table S6.
To figure out the phylogenetic relationship of MYB family genes, 137 RcMYBs were used to construct a neighbor joining phylogenetic tree by MEGA X (Figure 1). Gene expression level analysis needed to be added. To predict the function of RcMYBs, we selected the RcMYB protein with a relatively conserved sequence which was used as a BLASTP query in the TAIR database. Then genes clustered in five groups were predicted and marked by different colors. The RcMYBs in each group shared a high degree of similarity and were predicted to participate in certain biological processes. The RcMYB genes clustered in group 5 were conserved, though the BLAST results of RcMYB11 and RcMYB76 were extremely conservative in the process of nucleosome assembly, whereas the rest of the RcMYBs were predicted to mainly participate in circadian rhythm. Particularly, RcMYB35 and RcMYB84 were also speculated to be involved in the biosynthesis of auxin, while RcMYB82 was thought to be involved in the biosynthesis of chlorophyll. Then we found that RcMYB101 and AtCCA1 have an extremely high sequence alignment score, which indicated that RcMYB101 was very likely to be involved in the regulation of protein-containing complex assembly and response to organonitrogen compound, if RcMYB101 retains the most primitive evolutionary information in this gene group (Eric et al., 2007; Wu and Zhang, 2013; Ye et al., 2016). The same situation also occurred in group 1 and the corresponding gene was RcMYB28, which showed a highly similarity to the left cluster of genes predicted to participate in embryo development and right cluster of genes involved in cell population proliferation, therefore it was predicted to participate in both processes in the database. The RcMYB genes clustered in group 2 were relatively conserved, and similar to AtMYB96, AtMYB94, and AtMYB30, were assumed to be involved in the response to auxin (Cui et al., 2016; Liao et al., 2017; Lee and Seo, 2019). The RcMYBs in group 3 may be involved in various biological processes through BLAST, such as wounding response, cell expansion, cutide deposition. Among them, RcMYB71 was most likely to participate in suberin biosynthesis progress. The phylogenetic relationship revealed that RcMYBs in group 4 had extremely high comparison scores with the AtMYB proteins involved in secondary metabolism, such as cinnamic acid biosynthesis and flavonoid biosynthesis (Figure 1; Mehrtens et al., 2005; Liu et al., 2015).
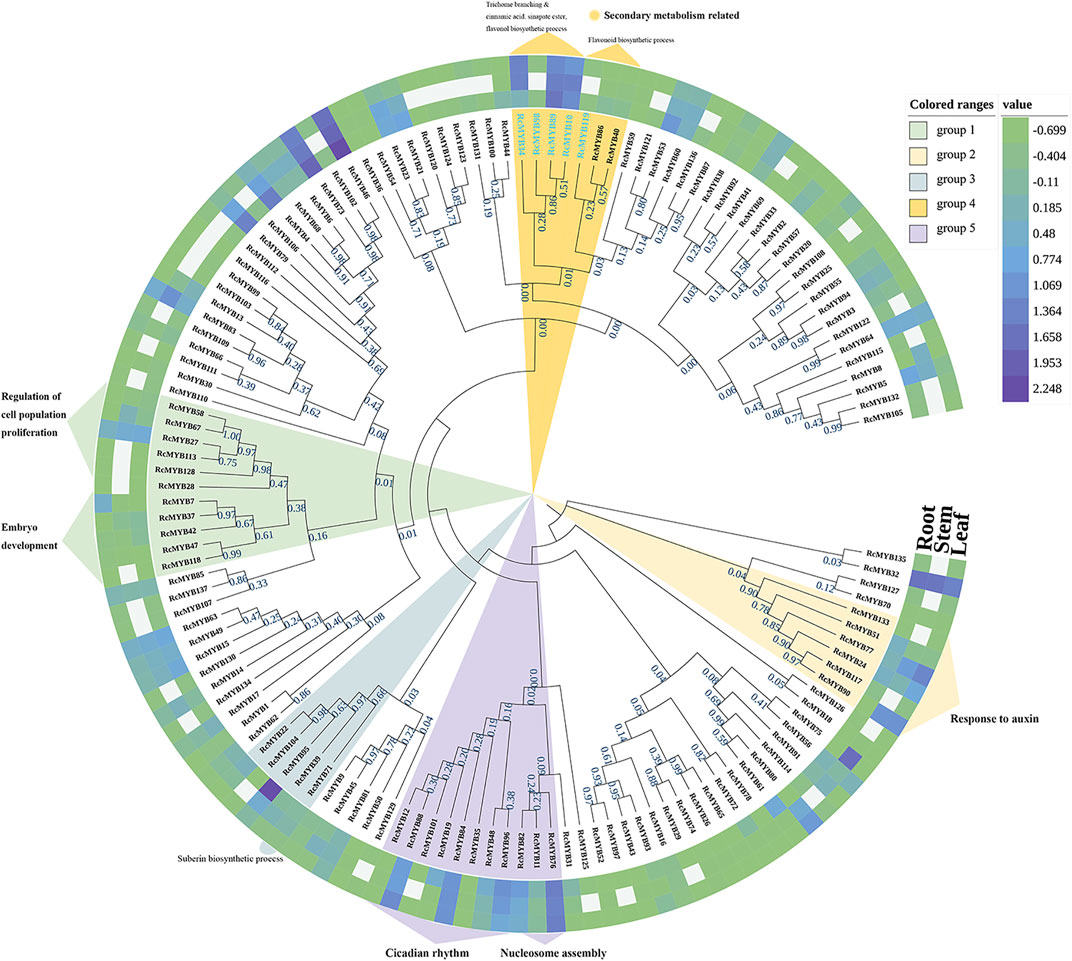
FIGURE 1. Phylogenetic tree of RcMYB genes with their relative expression levels in three tissues modified by iTOLv6.0. Functional annotations were predicted by BLAST in the TAIR database. Blank cells indicated data gaps and the labels of the selected RcMYBs were dyed blue. Value is equal to log10(FPKM+0.2).
To explore the structural diversity of the RcMYB members, the exon-intron organization of each RcMYB gene was analyzed by comparing their cDNA sequences with the corresponding genomic DNA sequences. As shown in Figures 2A, 3A, most RcMYB genes contained one or two introns, whereas some RcMYB genes consisted of no introns. Particularly, RcMYB107 contained 12 introns, while RcMYB18 and RcMYB120 contained a long intron in their gene structures. Notably, the gene structure of four genes (RcMYB10, RcMYB34, RcMYB89, and RcMYB98) clustered in group 4 showed a high similarity.
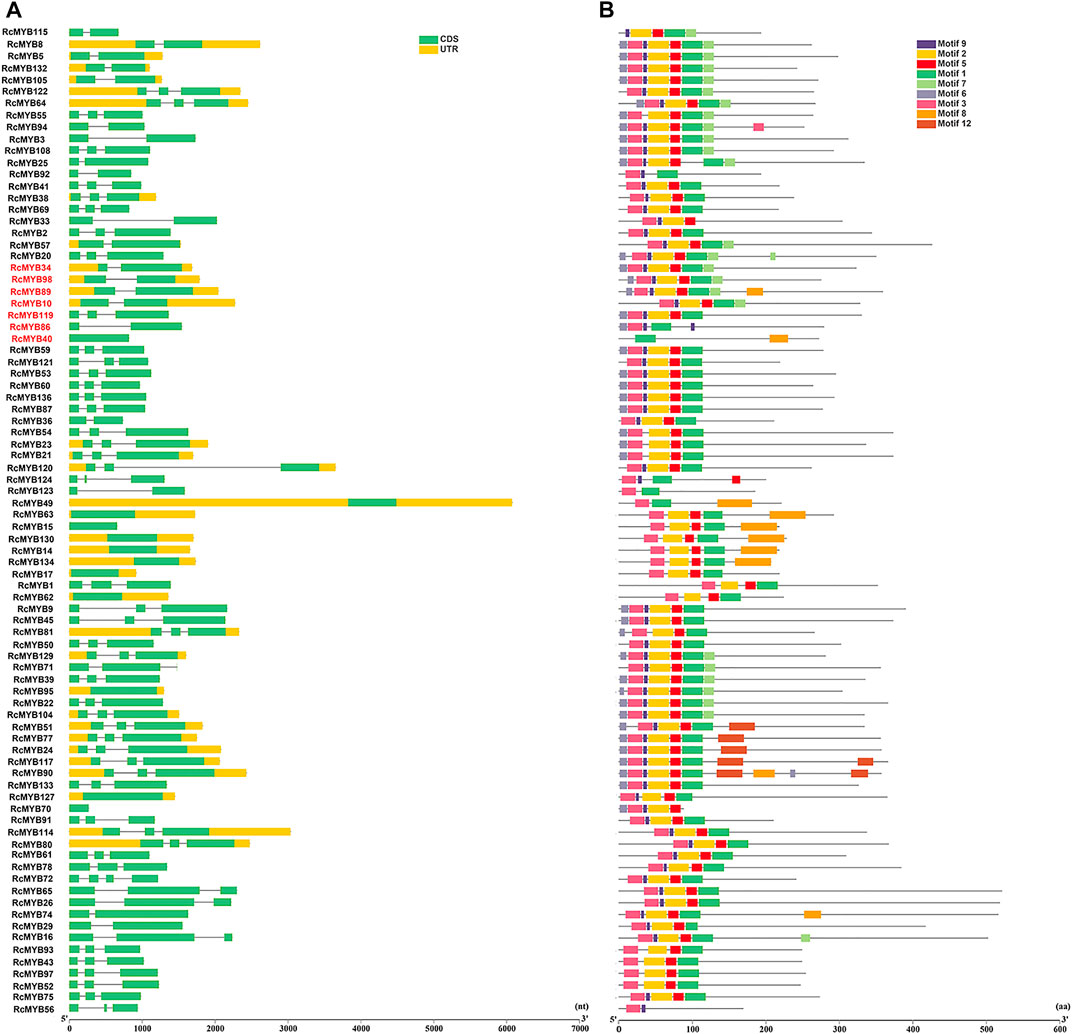
FIGURE 2. The gene structures and conserved motifs of R2R3 MYB proteins in R. crenulata. (A) The coding sequence (CDS) and untranslated region (UTR) were displayed in different colors, and the lines between boxes represented introns. (B) The phylogenetic tree constructed with RcMYB proteins and conserved domains of the RcMYB family was analyzed by MEME suite. Different colors indicated different motifs.
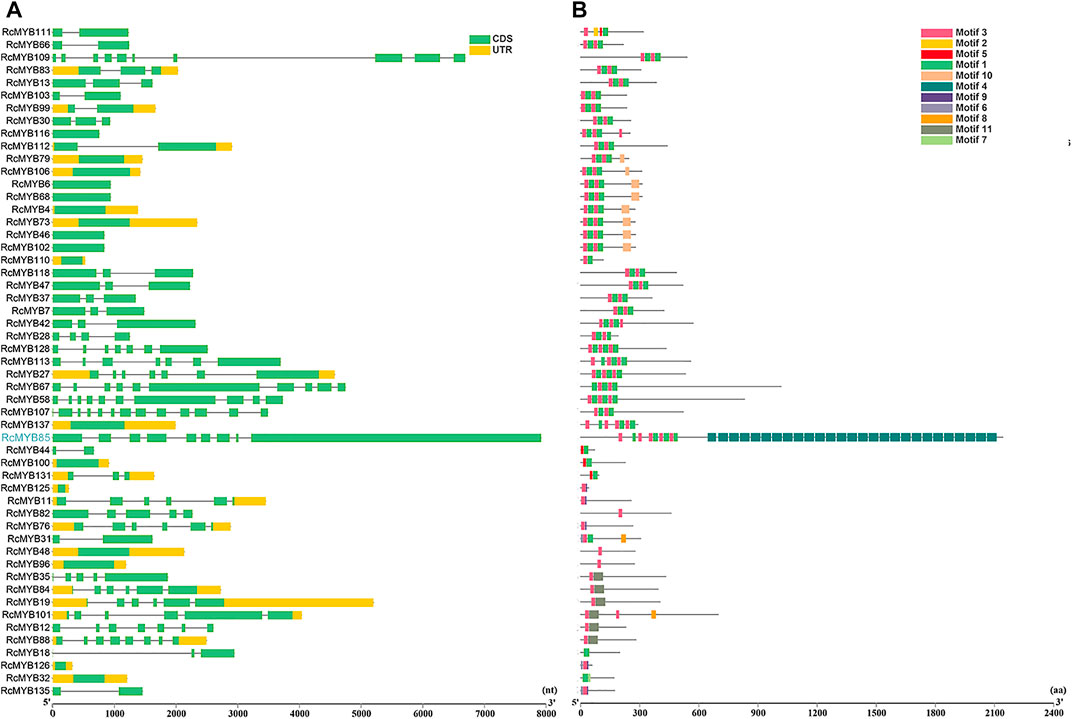
FIGURE 3. The gene structures and conserved motifs of RcMYB genes (1R-, 3R-, and 4R-) (A) The coding sequence (CDS) and untranslated region (UTR) were displayed in different colors, and the lines between boxes represented introns. (B) The phylogenetic tree constructed with RcMYB proteins and conserved domains of the RcMYB family was analyzed by MEME suite. Different colors indicated different motifs.
The features of MYB transcription factors were typically determined by their conserved motifs and domains. To have a more comprehensive understanding of the conserved domains of the RcMYB genes, motif elicitation (MEME) analysis was performed. That result showed that 12 conserved motifs of RcMYB proteins were predicted through the MEME suite (Figures 2B, 3B). In addition to what is shown in Figure 2B, specific amino acid sequences of each motif are also provided in Supplementary Figure S2B. Based on the number of the MYB domains and duplications of conserved sequences (Ambawat et al., 2013; Feng et al., 2017), MYB proteins in R. crenulata were classified into three types and as expected, R2R3-MYBs were in the majority (Supplementary Table S3). As shown in Figures 2B, 3B, motif 3 was found in most RcMYB proteins except RcMYB18, RcMYB32, RcMYB40, RcMYB44, RcMYB100, and RcMYB131. Interestingly, motif 4 in RcMYB85 repeated 27 times, which might suggest a unique function (Figure 3B). Some motifs like motif 8, motif 10, motif 11, and motif 12 were only found in genes clustered in the same group, indicating a similar function of these genes.
The evolutionary relationships within a gene family are typically analyzed according to their chromosomal distributions. Though the R. crenulata chromosomes were not assembled, we determined the corresponding scaffold information of the RcMYB genes based on the genome database. It was found that the RcMYB gene family was randomly distributed on 126 scaffolds, and only a few scaffolds contained multiple genes (Supplementary Figure S3). The exceptions were RcMYB14 and RcMYB15, RcMYB59 and RcMYB60, RcMYB62 and RcMYB63, RcMYB110 and RcMYB111, and RcMYB123 and RcMYB124, which were located within 6 kb with each other on chromosomes, suggesting duplication events. To remove the influence of the high content of repetitive sequences in the draft genome, the 50 longest scaffolds with MYB-annotated genes were screened and identified. While the length of these scaffolds only accounted for 7.56% of the total length of scaffolds, the loaded structural genes size exceeded 26.18% of the total DNA size, which suggested that the genome was well-assembled (Supplementary Figure S4).
Considering the important of glycosides and flavonoids in R. crenulata for its medical use, RcMYBs involved in the secondary metabolism process, including RcMYB10, RcMYB34, RcMYB89, RcMYB98, and RcMYB119, were paid more attention (Figure 1). A phylogenetic tree was constructed by using the five RcMYBs with genes identified in tomato (Solanum lycopersicum), wheat (Triticum aestivum), Arabidopsis (Arabidopsis thaliana), and rice (Oryza sativa) that were involved in the secondary metabolism process (Matus et al., 2008; Ballester et al., 2010; Shen et al., 2019). The analysis showed that RcMYB10, RcMYB34, RcMYB89, and RcMYB98 were clustered into the same group with AtMYB5, which functions in regulating flavonoid biosynthesis (Matus et al., 2008). RcMYB119, though, was clustered into the same group with OsCYP93G1 and Ta4CL2; the similarity was not significant (Supplementary Figure S5).
To analyze the potential cis-elements, the 2 kb upstream sequences of the selected RcMYB genes were submitted to online tool PlantCARE. The type and position of cis-elements were marked in different colors (Figure 4), and their potential functions are annotated in Supplementary Table S4. Numerous cis-acting elements were detected and mainly divided into promoter element type and plant growth and environmental responsive type. Among these cis-elements, TATA-box, CAAT-box, and ABRE were conspicuous, which were involved in transcription initiation and abscisic acid response, indicating that these RcMYB genes are mainly involved in regulating the initiation of transcription and abiotic responses. The promoter region of RcMYB118 contained 10 CGTCA-motifs and 10 TGACG-motifs, which are involved in MeJA-responsiveness, suggesting a hormone-regulating role of RcMYB118. The auxin-responsive element (Aux-RR core) were found in promotors of RcMYB6, RcMYB9, RcMYB11, RcMYB12, RcMYB14, and RcMYB15.
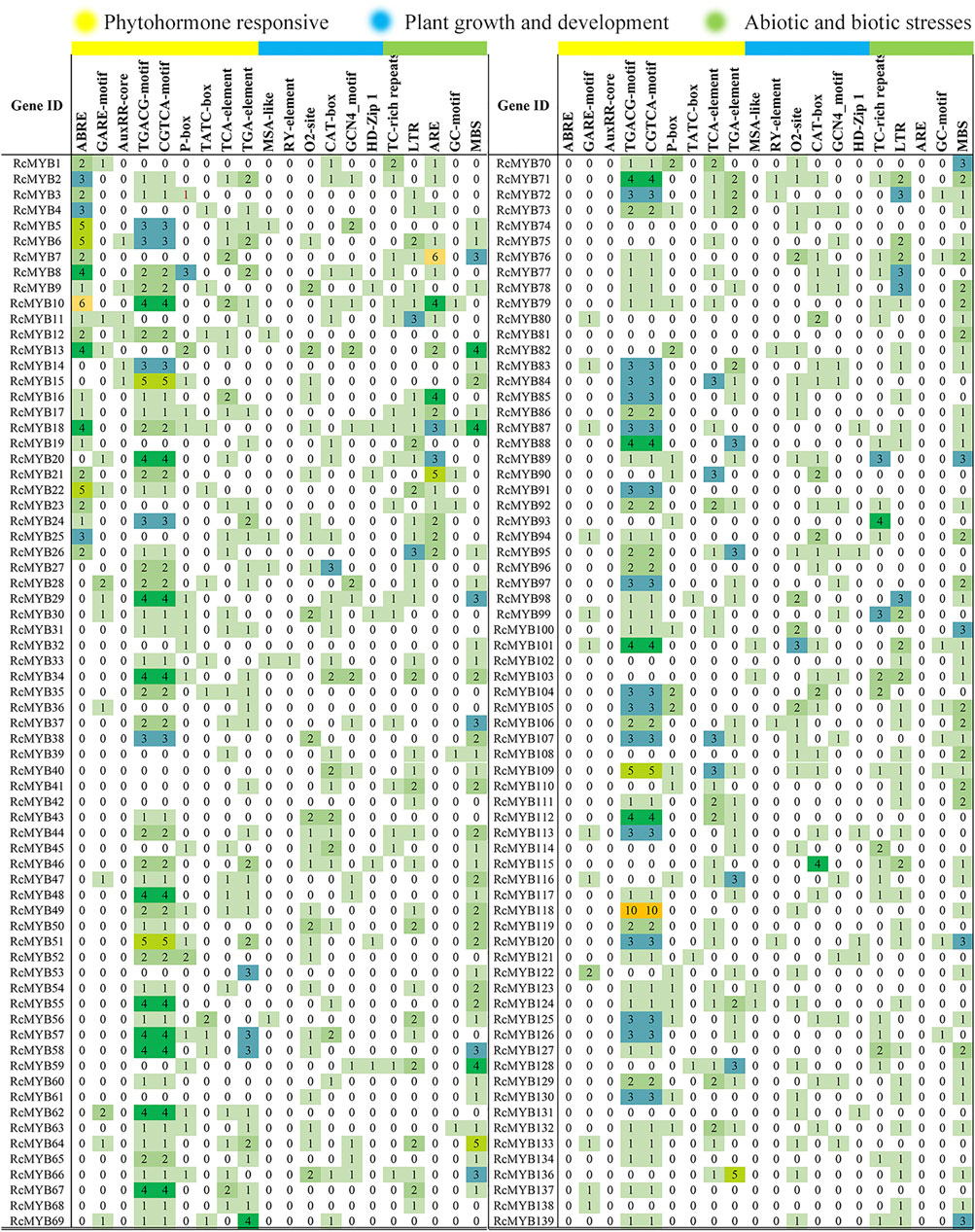
FIGURE 4. Number of each cis-acting element in the RcMYB genes in the promoter region. The detailed cis-element is listed in Supplementary Table S4.
WGCNA analysis enabled the grouping of genes into seven co-expression networks (modules) with 407 genes (Figure 5A). The identification of the hub genes in each module was calculated by cytoHubba (version 0.1) through different algorithms, and the top 10 genes based on gene connection degrees were selected (Supplementary Table S5). These RcMYBs genes were selected as hub genes in each module and their connected genes were analyzed and visualized by Cytoscape to construct the network diagram (Figure 5C and Supplementary Figure S6). It was found that the expression pattern of RcMYB10 showed a positive correlation with RcTyDC9 (CCG026415.1) and UDPGT4 (CCG024532.1), two genes encoding the key enzymes involved in salidroside biosynthesis (Supplementary Figure S6C). RcMYB34, however, showed a negative correlation with UDPGTs and RcMAOA (CCG009333.1, Figure 5C). This analysis suggested that RcMYB10 and RcMYB34 might play roles in regulating salidroside biosynthesis.
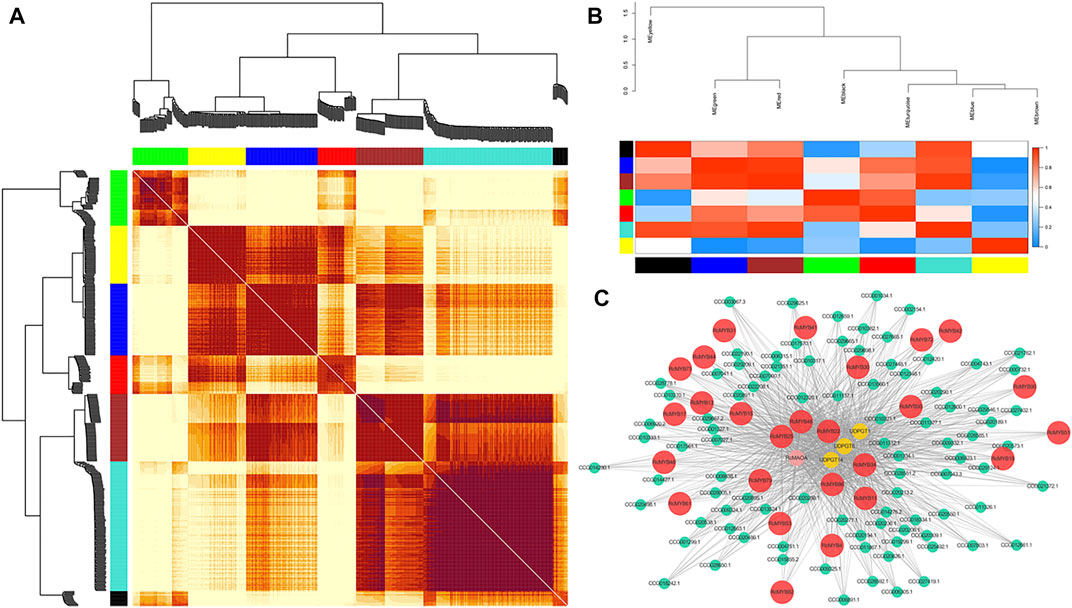
FIGURE 5. Co-expression network of RcMYB genes in R. crenulata. (A) The heatmap of correlation degree of modules. (B) Clustering of modules based on eigengenes. The colors ranging from blue to red represent Pearson correlation coefficients ranging from 0 to 1, indicating low to high correlations, respectively. (C) Visualization of connections of genes in the turquoise module. The RcMYB genes are colored in red. Connections of genes in other modules are shown in Supplementary Figure S6.
To identify the accuracy of RcMYB genes and further experimental analysis, the genomic sequence and coding regions of RcMYB10, RcMYB34, RcMYB89, and RcMYB98 were cloned and sequenced. It was found that the genomic sequence and coding regions of RcMYB34 and RcMYB89 showed a consistency with reference sequences (CCG031502.1 and CCG008637.1), whereas some single nucleotide polymorphisms (SNPs) were detected in the coding region of RcMYB98 (Figure 6A). The SNPs in the coding region of RcMYB98 only resulted in an amino acid change out of the R2R3 MYB domain. Besides, a 6 bp deletion and some SNPs were detected in the coding region of sequenced RcMYB10, resulted in three amino acids changed out of the R2R3 MYB domain (Figure 6B and Supplementary Figure S7).
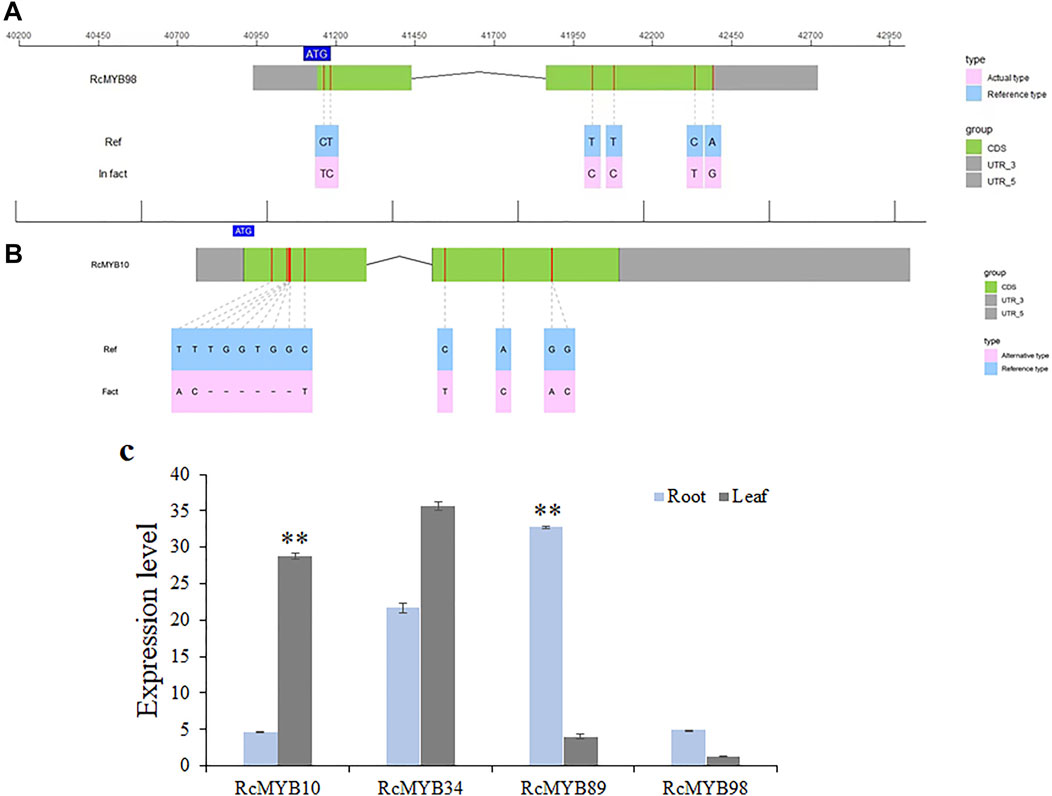
FIGURE 6. Gene cloning and expression level analysis of clustered RcMYBs. (A) Cloning and sequencing of RcMYB98. (B) Cloning and sequencing of RcMYB10. (C) Expression level analysis of RcMYB10, RcMYB34, RcMYB89, and RcMYB98 in root and leaf. Values are the mean ± standard deviation of three biological replicates per treatment. “**” above each column indicates a significant difference (p < 0.01; n = 3).
Then, we analyzed the expression profiles of selected RcMYB genes in the root and leaf of R. crenulata by qRT-PCR (Figure 6C). As shown in Figure 6D, the expression level of RcMYB34 in root and leaf was consistent, whereas the expression pattern of RcMYB10, RcMYB89, and RcMYB98 was contrary. The expression level of RcMYB10 in leaf was significantly higher than in root, but RcMYB89 and RcMYB98 expressed higher in root than leaf.
R. crenulata has broad application prospects in the field of modern medicine, and the second metabolites of its extract, such as flavonoids and glycosides, have been well-studied and utilized (Yousef et al., 2006; Yang et al., 2012; Zhou and Jiang, 2014). High altitude, with a lower oxygen and higher-pressure situation, contributed the most to the content of salidroside and other secondary metabolites in R. crenulate. Previous studies about the biosynthesis of secondary metabolites in R. crenulate usually pay more attention to the genes encoding the key enzyme of the synthesis pathway (Lan et al., 2013; Torrens-Spence et al., 2017), therefore ignoring the role of upstream regulation by transcription factors under abiotic stress conditions. In this research, the first systematic study of the transcription factor families in R. crenulata using bioinformatics tools and expression profiles based on the sequenced R. crenulata genome was performed. Here, a total of 139 RcMYB genes were identified in the R. crenulata genome, and their characteristics, gene structure, and conserved motifs were analyzed and predicted.
MYB transcription factor family members are widely involved in many biological processes in plants, such as plant growth, development, differentiation, response to abiotic and biotic stresses, and secondary metabolism (Ambawat et al., 2013; Liu et al., 2015). Whole-genome identification of MYB TFs has been conducted in many sequenced plants. Since its definitions in Arabidopsis (Stracke et al., 2001) and rice (Jiang et al., 2004b), MYB genes had been identified and cloned in many species, including grape (Matus et al., 2008; Fournier-Level et al., 2009), tobacco (Pattanaik et al., 2010), cauliflower (Chiu et al., 2010), soybean (Du et al., 2012), pear (Feng et al., 2015), Casuarina equisetifolia (Wang et al., 2021), and Liriodendron chinense (Wu et al., 2021). In this research, 101 R2R3-MYBs were identified, accounting for 72.6% of all RcMYBs (Supplementary Table S3). Most R2R3-MYB proteins are largely conserved and usually divided into the same subgroups as Arabidopsis MYB proteins, though, divergence between them exists. Comparative analysis of R2R3-MYBs from different plant species revealed that this TF family has experienced extensive expansion during evolution. The expansion of the MYB family in plants fits well with the results that many MYB members participate in various biological processes and in plant-specific processes. The MYB TFs were extensively duplicated during the process of plant evolution, leading to new members of the MYB TF family controlling specific functions. Previous studies in other plants confirmed that the expansion of the MYB multigene family was a result of genome-wide duplication or region-specific tandem duplication (Du et al., 2012; Lin and Rausher, 2021). In this study, a series of tandem gene duplications were observed in five scaffolds involving 10 RcMYB genes (Supplementary Figure S3), which suggested that tandem duplication events happened and played a role in the expansion of the large RcMYB family. Gene duplication is an important mechanism to generate new genes. A duplicate gene copy can lead to genetic and morphological diversification by evolving new gene functions (neo-functionalization), whereas the other copy can maintain the ancestral function (Rensing 2014). In this study, RcMYB63 was closed to RcMYB62 in the chromosome and considered as a duplicate gene copy, its C-terminal region contained motif 8 compared to RcMYB62 (Figure 2 and Supplementary Figure S2). The same was observed between RcMYB123 and RcMYB124, in which two motifs were identified in C- and N- terminal regions of RcMYB124. Besides, as a plant grown in a high-altitude and harsh environment, it seems that R. crenulata generates specific-function MYB genes during the process of adaptation to the environment and evolution. It was interesting that RcMYB85 contained a motif with 27 repeats and showed no alignment in Arabidopsis and rice. Although the phylogenetic analysis indicated that the homology of RcMYB85 and RcMYB137 was high and their motifs at N-terminal region were consistent, the repeated motifs at the C-terminal of RcMYB85 were unique (Figure 1 and Supplementary Figure S3). Further research about the function of RcMYB85 will be needed. It is interesting that this plant can survive in these extremely growing environments. The roles of MYB transcription factors in drought (Bi et al., 2016; Butt et al., 2017; Wang et al., 2017), heat (Zhao et al., 2017), cold (An et al., 2012; Yan et al., 2014; Yang et al., 2022), and other stress conditions (An et al., 2012; Guo et al., 2017; Liu et al., 2021) have been well-studied. In this research, the cis-acting elements of the upstream sequence of the RcMYB genes were analyzed, and the results showed that in addition to numerous score promoter elements, there were indeed many environmental response elements and hormone response elements, such as the light-responsive elements GT1-motif, abscisic acid responsiveness element ABRE, MeJA-responsiveness TGACG-motif, auxin-responsive element TGA-motif, and anaerobic induced response element ARE (Figure 4 and Supplementary Table S4). These motifs indicated that the expression of RcMYB genes would be induced by many abiotic factors to enhance or inhibit transcription. Besides, there were MYB binding sites involved in some elements like MRE, which suggested some RcMYBs could cooperate with other RcMYBs to perform or emphasize functions. These cis-elements might be helpful for the adaptation of R. crenulata to the tough environment.
The gene expression patterns can provide important clues to explore gene function. In this research, WGCNA analysis indicated that RcMYB10 and RcMYB34 correlated with the genes encoding key enzymes involved in the biosynthesis of salidroside. The distribution of second metabolites was different in root and leaf of R. crenulata. Although RcMYB10, RcMYB34, RcMYB89, and RcMYB98 were predicted to be involved in the secondary metabolism process, their regulation of secondary metabolites was probably varied.
In this study, the MYB transcription factor family in R. crenulata was identified, and a series of bioinformatics analyses were carried out to reveal the characteristics of RcMYBs. Further, RcMYB members that may be involved in the secondary metabolic process of R. crenulata were cloned and analyzed. After sequencing and analysis, the understanding of its function and structure increased, and information such as cis-acting elements and target genes were predicted. This research has provided the data basis for further research on MYB transcription factors of R. crenulata.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
BX and XM designed the experiment and amended the manuscript. BX, BC, and XQ performed the experiments. SL, YZ, and CT contributed to the vector construction. BX and BC wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (No. 81973569) and the Xinglin Talent Program of Chengdu University of TCM (No. 030058042).
Author BX was employed by the company China Resources Sanjiu (Ya’an) Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Fanbo Meng and Yili Wang, Chengdu University of TCM, for their help in data analysis and their support in gene expression analysis, respectively.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.831611/full#supplementary-material
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). MYB Transcription Factor Genes as Regulators for Plant Responses: An Overview. Physiol. Mol. Biol. Plants 19 (3), 307–321. doi:10.1007/s12298-013-0179-1
An, Y., Dai, X., and Zhang, W.-H. (2012). A R2R3-Type MYB Gene, OsMYB2, Is Involved in Salt, Cold, and Dehydration Tolerance in rice. J. Exp. Bot. 63 (7), 2541–2556. doi:10.1093/jxb/err431
Ballester, A.-R., Molthoff, J., de Vos, R., Hekkert, B. t. L., Orzaez, D., Fernaݩndez-Moreno, J.-P., et al. (2010). Biochemical and Molecular Analysis of Pink Tomatoes: Deregulated Expression of the Gene Encoding Transcription Factor SlMYB12 Leads to Pink Tomato Fruit Color. Plant Physiol. 152 (1), 71–84. doi:10.1104/pp.109.147322
Bi, H., Luang, S., Li, Y., Bazanova, N., Morran, S., Song, Z., et al. (2016). Identification and Characterization of Wheat Drought-Responsive MYB Transcription Factors Involved in the Regulation of Cuticle Biosynthesis. J. Exp. Bot. 67 (18), 5363–5380. doi:10.1093/jxb/erw29810.1093/jxb/erw298
Butt, H. I., Yang, Z., Gong, Q., Chen, E., Wang, X., Zhao, G., et al. (2017). GaMYB85, an R2R3 MYB Gene, in Transgenic Arabidopsis Plays an Important Role in Drought Tolerance. BMC Plant Biol. 17 (1), 142. doi:10.1186/s12870-017-1078-3
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). Tbtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Chiu, L.-W., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The Purple Cauliflower Arises from Activation of a MYB Transcription Factor. Plant Physiol. 154 (3), 1470–1480. doi:10.1104/pp.110.164160
Cui, F., Brosché, M., Lehtonen, M. T., Amiryousefi, A., Xu, E., Punkkinen, M., et al. (2016). Dissecting Abscisic Acid Signaling Pathways Involved in Cuticle Formation. Mol. Plant 9 (6), 926–938. doi:10.1016/j.molp.2016.04.001
Du, H., Yang, S.-S., Liang, Z., Feng, B.-R., Liu, L., Huang, Y.-B., et al. (2012). Genome-Wide Analysis of the MYB Transcription Factor Superfamily in Soybean. BMC Plant Biol. 12, 106. doi:10.1186/1471-2229-12-106
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi:10.1016/j.tplants.2010.06.005
Fan, F., Yang, L., Li, R., Zou, X., Li, N., Meng, X., et al. (2020). Salidroside as a Potential Neuroprotective Agent for Ischemic Stroke: A Review of Sources, Pharmacokinetics, Mechanism and Safety. Biomed. Pharmacother. 129, 110458. doi:10.1016/j.biopha.2020.110458
Feng, G., Burleigh, J. G., Braun, E. L., Mei, W., and Barbazuk, W. B. (2017). Evolution of the 3R-MYB Gene Family in Plants. Genome Biol. Evol. 9 (4), 1013–1029. doi:10.1093/gbe/evx056
Feng, S., Xu, Y., Yang, L., Sun, S., Wang, D., and Chen, X. (2015). Genome-Wide Identification and Characterization of R2R3-MYB Transcription Factors in Pear. Scientia Horticulturae 197, 176–182. doi:10.1016/j.scienta.2015.09.033
Fournier-Level, A., Lacombe, T., Le Cunff, L., Boursiquot, J.-M., and This, P. (2009). Evolution of the VvMybA Gene Family, the Major Determinant of Berry Colour in Cultivated Grapevine (Vitis vinifera L.). Heredity 104 (4), 351–362. doi:10.1038/hdy.2009.148
Fu, Y., Li, L., Hao, S., Guan, R., Fan, G., Shi, C., et al. (2017). Draft Genome Sequence of the Tibetan Medicinal Herb Rhodiola Crenulata. GigaScience 6 (6), 1–5. doi:10.1093/gigascience/gix033
Ganko, E. W., Meyers, B. C., and Vision, T. J. (2007). Divergence in Expression between Duplicated Genes in Arabidopsis. Mol. Biol. Evol. 24 (10), 2298–2309. doi:10.1093/molbev/msm158
Gu, Z., Gu, L., Eils, R., Schlesner, M., Brors, B., and Benedikt, B. (2014). Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 30 (19), 2811–2812. doi:10.1093/bioinformatics/btu393
Guo, H., Wang, Y., Wang, L., Hu, P., Wang, Y., Jia, Y., et al. (2017). Expression of the MYB Transcription Factor Gene BplMYB46 Affects Abiotic Stress Tolerance and Secondary Cell wall Deposition in Betula Platyphylla. Plant Biotechnol. J. 15 (1), 107–121. doi:10.1111/pbi.12595
Jiang, C., Gu, J., Chopra, S., Gu, X., and Peterson, T. (2004a). Ordered Origin of the Typical Two- and Three-Repeat Myb Genes. Gene 326, 13–22. doi:10.1016/j.gene.2003.09.049
Jiang, C., Gu, X., and Peterson, T. (2004b). Identification of Conserved Gene Structures and Carboxy-Terminal Motifs in the Myb Gene Family of Arabidopsis and Oryza Sativa L. Ssp. Indica. Genome Biol. 5 (7), R46. doi:10.1186/gb-2004-5-7-r46
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lai, Y., Li, H., and Yamagishi, M. (2013). A Review of Target Gene Specificity of Flavonoid R2R3-MYB Transcription Factors and a Discussion of Factors Contributing to the Target Gene Selectivity. Front. Biol. 8 (6), 577–598. doi:10.1007/s11515-013-1281-z
Lan, X., Chang, K., Zeng, L., Liu, X., Qiu, F., Zheng, W., et al. (2013). Engineering Salidroside Biosynthetic Pathway in Hairy Root Cultures of Rhodiola Crenulata Based on Metabolic Characterization of Tyrosine Decarboxylase. Plos One 8, e75459. doi:10.1371/journal.pone.0075459
Langfelder, P., and Horvath, S. (2008). WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinformatics 9 (1), 559. doi:10.1186/1471-2105-9-559
Lee, H. G., and Seo, P. J. (2019). MYB96 Recruits the HDA15 Protein to Suppress Negative Regulators of ABA Signaling in Arabidopsis. Nat. Commun. 10 (1), 1713. doi:10.1038/s41467-019-09417-1
Liao, C., Zheng, Y., and Guo, Y. (2017). MYB30 Transcription Factor Regulates Oxidative and Heat Stress Responses through ANNEXIN‐Mediated Cytosolic Calcium Signaling in Arabidopsis. New Phytol. 216 (1), 163–177. doi:10.1111/nph.14679
Lin, R.-C., and Rausher, M. D. (2021). Ancient Gene Duplications, Rather Than Polyploidization, Facilitate Diversification of Petal Pigmentation Patterns in Clarkia Gracilis (Onagraceae). Mol. Biol. Evol. 38 (12), 5528–5538. doi:10.1093/molbev/msab242
Liu, J., Osbourn, A., and Ma, P. (2015). MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 8 (5), 689–708. doi:10.1016/j.molp.2015.03.012
Liu, X., Zhao, C., Gao, Y., Xu, Y., Wang, S., Li, C., et al. (2021). A Multifaceted Module of BRI1 ETHYLMETHANE SULFONATE SUPRESSOR1 (BES1)-MYB88 in Growth and Stress Tolerance of Apple. Plant Physiol. 185 (4), 1903–1923. doi:10.1093/plphys/kiaa116
Long, T., Xu, B., Hu, Y., Wang, Y., Mao, C., Wang, Y., et al. (2021). Genome-wide Identification of ZmSnRK2 Genes and Functional Analysis of ZmSnRK2.10 in ABA Signaling Pathway in maize (Zea mays L). BMC Plant Biol. 21 (1), 309. doi:10.1186/s12870-021-03064-9
Matus, J. T., Aquea, F., and Arce-Johnson, P. (2008). Analysis of the Grape MYB R2R3 Subfamily Reveals Expanded Wine Quality-Related Clades and Conserved Gene Structure Organization across Vitis and Arabidopsis Genomes. BMC Plant Biol. 8, 83. doi:10.1186/1471-2229-8-83
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis Transcription Factor MYB12 Is a Flavonol-Specific Regulator of Phenylpropanoid Biosynthesis. Plant Physiol. 138 (2), 1083–1096. doi:10.1104/pp.104.058032
Minas, K., McEwan, N. R., Newbold, C. J., and Scott, K. P. (2011). Optimization of a High-Throughput CTAB-Based Protocol for the Extraction of qPCR-Grade DNA from Rumen Fluid, Plant and Bacterial Pure Cultures. FEMS MICROBIOL. LETT. 325 (2), 162–169. doi:10.1111/j.1574-6968.2011.02424.x
Pattanaik, S., Kong, Q., Zaitlin, d., Werkman, J. R., Xie, C. H., Patra, B., et al. (2010). Isolation and Functional Characterization of a floral Tissue-Specific R2R3 MYB Regulator from Tobacco. Planta 231 (5), 1061–1076. doi:10.1007/s00425-010-1108-y
Prouse, M. B., and Campbell, M. M. (2012). The Interaction between MYB Proteins and Their Target DNA Binding Sites. Biochim. Biophys. Acta 1819 (1), 67–77. doi:10.1016/j.bbagrm.2011.10.010
Qu, Z.-Q., Zhou, Y., Zeng, Y.-S., Lin, Y.-K., Li, Y., Zhong, Z.-Q., et al. (2012). Protective Effects of a Rhodiola Crenulata Extract and Salidroside on Hippocampal Neurogenesis against Streptozotocin-Induced Neural Injury in the Rat. Plos One 7 (1), e29641. doi:10.1371/journal.pone.0029641
Rensing, S. A. (2014). Gene Duplication as a Driver of Plant Morphogenetic Evolution. Curr. Opin. Plant Biol. 17, 43–48. doi:10.1016/j.pbi.2013.11.002
Ron, E., Michael, D., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 30 (1), 207–210. doi:10.1093/nar/30.1.207
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shen, Y., Sun, T., Pan, Q., Anupol, N., Chen, H., Shi, J., et al. (2019). Rr MYB 5‐ and Rr MYB 10‐Regulated Flavonoid Biosynthesis Plays a Pivotal Role in Feedback Loop Responding to Wounding and Oxidation in Rosa Rugosa. Plant Biotechnol. J. 17 (11), 2078–2095. doi:10.1111/pbi.13123
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB Gene Family in Arabidopsis T. Curr. Opin. Plant Biol. 4 (5), 447–456. doi:10.1016/s1369-5266(00)00199-0
Torrens-Spence, M. P., Pluskal, T., Li, F.-S., Carballo, V., and Weng, J.-K. (2017). Complete Pathway Elucidation and Heterologous Reconstitution of Rhodiola Salidroside Biosynthesis. Mol. Plant 11 (1), 205–217. doi:10.1016/j.molp.2017.12.007
Wang, Y., Wang, Q., Liu, M., Bo, C., Wang, X., Ma, Q., et al. (2017). Overexpression of a maize MYB48 Gene Confers Drought Tolerance in Transgenic Arabidopsis Plants. J. Plant Biol. 60 (6), 612–621. doi:10.1007/s12374-017-0273-y
Wang, Y., Zhang, Y., Fan, C., Wei, Y. C., and Zhong, C. L. (2021). Genome-Wide Analysis of MYB Transcription Factors and Their Responses to Salt Stress in Casuarina E. BMC Plant Biol. 21 (1), 328. doi:10.1186/s12870-021-03083-6
Wu, D.-D., and Zhang, Y.-P. (2013). Evolution and Function of De Novo Originated Genes. Mol. Phylogenet. Evol. 67 (2), 541–545. doi:10.1016/j.ympev.2013.02.013
Wu, W., Zhu, S., Zhu, L., Wang, D., Liu, Y., Liu, S., et al. (2021). Characterization of the Liriodendron Chinense MYB Gene Family and Its Role in Abiotic Stress Response. Front. Plant Sci. 12, 641280. doi:10.3389/fpls.2021.641280
Yan, Y., Shen, L., Chen, Y., Bao, S., Thong, Z., and Yu, H. (2014). A MYB-Domain Protein EFM Mediates Flowering Responses to Environmental Cues in Arabidopsis. Develop. Cel 30 (4), 437–448. doi:10.1016/j.devcel.2014.07.004
Yang, X., Luo, Y., Bai, H., Li, X., Tang, S., Liao, X., et al. (2022). DgMYB2 Improves Cold Resistance in chrysanthemum by Directly Targeting DgGPX1. Hortic. Res. 9, uhab028. doi:10.1093/hr/uhab028
Yang, Y.-N., Liu, Z.-Z., Feng, Z.-M., Jiang, J.-S., and Zhang, P.-C. (2012). Lignans from the Root of Rhodiola Crenulata. J. Agric. Food Chem. 60 (4), 964–972. doi:10.1021/jf204660c
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 60 (1), 107–124. doi:10.1007/s11103-005-2910-y
Ye, L., Wang, B., Zhang, W., Shan, H., and Kong, H. (2016). Gains and Losses of Cis-Regulatory Elements Led to Divergence of the Arabidopsis APETALA1 and CAULIFLOWER Duplicate Genes in the Time, Space, and Level of Expression and Regulation of One Paralog by the Other. Plant Physiol. 171 (2), 1055. doi:10.1104/pp.16.00320
Yousef, G. G., Grace, M. H., Cheng, D. M., Belolipov, I. V., Raskin, I., and Lila, M. A. (2006). Comparative Phytochemical Characterization of Three Rhodiola Species. Phytochemistry 67 (21), 2380–2391. doi:10.1016/j.phytochem.2006.07.026
Zhang, B., and Horvath, S. (2005). A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 4 (1), 17. doi:10.2202/1544-6115.1128
Zhao, Y., Tian, X., Wang, F., Zhang, L., Xin, M., Hu, Z., et al. (2017). Characterization of Wheat MYB Genes Responsive to High Temperatures. BMC Plant Biol. 17 (1), 208. doi:10.1186/s12870-017-1158-4
Keywords: Rhodiola crenulata, MYB transcription factors family, genome-wide identification, gene expression analysis, WGCNA analysis
Citation: Xu B, Chen B, Qi X, Liu S, Zhao Y, Tang C and Meng X (2022) Genome-wide Identification and Expression Analysis of RcMYB Genes in Rhodiola crenulata. Front. Genet. 13:831611. doi: 10.3389/fgene.2022.831611
Received: 08 December 2021; Accepted: 21 February 2022;
Published: 31 March 2022.
Edited by:
Zefeng Yang, Yangzhou University, ChinaReviewed by:
Jianqiang Zhang, Shaanxi Normal University, ChinaCopyright © 2022 Xu, Chen, Qi, Liu, Zhao, Tang and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binjie Xu, YmluamlleHVAb3V0bG9vay5jb20=; Xianli Meng, eGxtOTk5QGNkdXRjbS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.