
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Genet., 02 February 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.827808
This article is part of the Research TopicCurrent Advances in Genomics and Gene Editing Tools for Crop Improvement in a Changing Climate ScenarioView all 20 articles
 Jagesh Kumar Tiwari1*
Jagesh Kumar Tiwari1* Tanuja Buckseth1
Tanuja Buckseth1 Clarissa Challam2
Clarissa Challam2 Rasna Zinta1,3
Rasna Zinta1,3 Nisha Bhatia1,4
Nisha Bhatia1,4 Dalamu Dalamu1
Dalamu Dalamu1 Sharmistha Naik1,5
Sharmistha Naik1,5 Anuj K. Poonia4
Anuj K. Poonia4 Rajesh K. Singh1
Rajesh K. Singh1 Satish K. Luthra6
Satish K. Luthra6 Vinod Kumar1
Vinod Kumar1 Manoj Kumar6
Manoj Kumar6Potato (Solanum tuberosum L.) (2n = 4x = 48) is the third most important food crop after rice and wheat in terms of human consumption. Potato is considered as the staple food in Europe and parts of Americas. In 2018, the world total potato production was 368.17 million tonnes led by China (90.26 mt) followed by India (48.53 mt) (FAOSTAT, 2018). The increasing world population from the now 7.7 to the expected 9.7 billion by 2050 has posed a great challenge of food availability (United Nations, 2019). Potato suffers from various pathogens, insect pests, and environmental abiotic stresses. The condition is worsening under the climate change scenario. In India, the mean potato productivity in major potato-growing states, which together account for about 90% of the national potato production, is likely to decline by 2.0% in 2050s and 6.4% in 2080s (Rana et al., 2020). To address these issues, conventional breeding has shown key roles in varietal development programs combined with the deployment of marker-assisted selection mainly for late blight, viruses, and potato cyst nematode–resistant varieties the world over such as Kufri Karan in India (ICAR-CPRI Annual Report, 2018-19). Later, potato transgenics have also been developed for resistance to diseases (e.g., late blight and viruses), abiotic stresses (e.g., heat and drought), insect pest (e.g., potato cyst nematode and potato tuber moth), processing quality (e.g., reduced cold-induced sweetening), but none of them are being applied at the field level. Hence, with the advancement of sequencing technologies and availability of the potato genome sequence (Potato Genome Sequencing Consortium, 2011), it is possible to modulate the target genes applying genomics tools like genome editing.
Genome editing is an advanced genomics tool which can be deployed for crop improvement by gene knock-out and insertion/deletion mutagenesis (Hameed et al., 2018). It allows double-stranded breaks (DSBs) at specific sites in the genome and repairs via naturally occurring DNA repair mechanisms, namely, nonhomologous end joining (NHEJ) or homologous recombination (HR). In the past, this system was earlier facilitated by protein-guided nucleases such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). But now, attention has been driven on the new RNA-guided nuclease called clustered regularly interspaced short palindromic repeats (CRISPR)—CRISPR associated (Cas) (Nadakuduti et al., 2018). The TALENs and ZFNs require particular expertise, longer timelines, and higher costs than those needed for assembling CRISPR/Cas. Indeed, a tremendous progress has been reported on the utility of CRISPR/Cas in crops. In potato, CRISPR/Cas has been demonstrated for tuber quality, disease resistance (late blight and potato virus Y), phenotype, and other traits (Dangol et al., 2019; Hameed et al., 2020; Hofvander et al., 2021). This article provides the current status of CRISPR/Cas, future perspectives, and challenges in potato.
CRISPR/Cas is the most powerful biological tool to create targeted modification in the genome, which allows easy designing and construction of gene-specific single guide RNA (sgRNA). The sgRNA vectors are easily reprogrammable to direct Streptococcus pyogenes Cas9 (SpCas9) to generate DSBs and are then repaired endogenously by the error-prone NHEJ or HR pathways. CRISPR/Cas is divided into two distinct classes based on the sequence, structure, and functions of the Cas proteins. Class 1 consists of types I, III, and IV andutilizes a multi-protein effector complex, whereas class 2 includes types II, V, and VI and uses a single effector protein; of which, type II and V target DNA, whereas type VI targets RNA. CRISPR/Cas9 (class 2, type-II) is the most commonly exploited machinery for DNA target. Remarkable innovations in CRISPR/Cas9 variant FnCas9 (Francisella novicida) (Price et al., 2015) and CRISPR/Cas13a (type VI, LshCas13a from Leptotrichia shahii) (Aman et al., 2018) have opened new avenues for RNA targets also. The SpCas9 and RNase III ribonucleases generate the Cas9/guide RNA complex that recognizes and cleaves DNA sequences adjacent to the 5′-NGG protospacer adjacent motif (PAM) and induces site-specific DSBs (Khatodia et al., 2016; Cao et al., 2020). Currently, CRISPAR/Cas9 has revolutionized plant research due to its simplicity, multiplexing, cost-effectiveness, high efficiency, and minimum off targets. Unlike genetically modified organisms, CRISPR/Cas creates alterations in the existing genome without the introduction of foreign genes, particularly site-directed nucleases (SDN1 and SDN2). Hence, CRISPR/Cas is expected to be transgene free, and biosafety regulations are under consideration in various countries (Schmidt et al., 2020).
Several complex traits of agronomic importance are considered in potato while breeding a new variety. The multigenic-controlled biotic/abiotic stresses are difficult to improve through conventional breeding in less time, which could be possible by using CRISPR/Cas9. The gene knockout mechanism has been applied in potato for late blight resistance using susceptibility (S) genes (StDND1, StCHL1, and StDMR6-1) (Kieu et al., 2021). A few successful examples are discussed later for biotic/abiotic stress resistance/tolerance, tuber quality, and phenotype traits improvement in potato (Table 1, and Supplementary Figures 1 and 2).
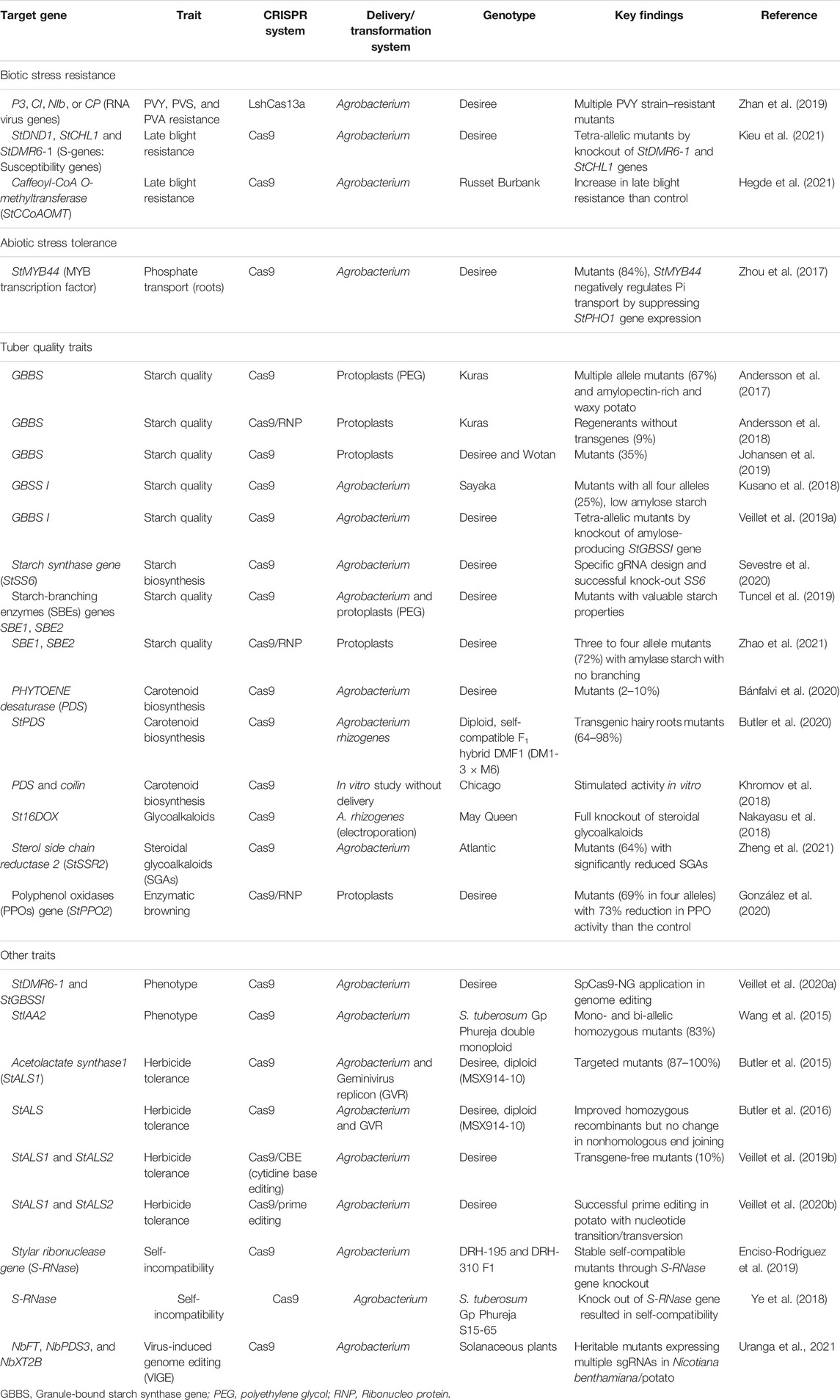
TABLE 1. Successful examples of application of CRISPR/Cas genome editing technology for biotic and abiotic stress resistance/tolerance, tuber quality, and phenotype and other traits in potato.
CRISPR/Cas has emerged as an alternative and efficient technology to accelerate potato breeding (Table 1). It has been demonstrated for potato virus Y (PVY) and late blight (Phytophthora infestans) resistance in potato. Cas13a protein was deployed to confer resistance to three PVY strains (RNA virus) by targeting P3, CI, Nib, and CP viral genes (Zhan et al., 2019). Host genes like the eukaryotic translation initiation factor eIF4E and coilin have also been found very effective for PVY resistance (Makhotenko et al., 2019). Recently, late blight resistance was demonstrated in potato by the knockout of susceptibility genes StDMR6-1 and StCHL1 (Kieu et al., 2021) and Caffeoyl-CoA O-methyltransferase (StCCoAOMT) (Hegde et al., 2021).
Abiotic stresses such as heat, drought, salinity, and cold are very important in potato, but with meagre work that is available in potato so far. Zhou et al. (2017) developed mutants (84%) by manipulating potato MYB transcription factor gene StMYB44, which negatively regulates phosphate transport in potato by suppressing StPHO1 gene expression (Table 1). Considerable research work on abiotic stress has been reported in cereals and other crops, but not in potato. Recently, we have proposed the use of CRISPR/Cas to manipulate N metabolism genes for improving nitrogen use efficiency in potato (Tiwari et al., 2020).
CRISPR/Cas studies have been reported in potato for traits like improved tuber starch quality (Andersson et al., 2017, 2018; Kusano et al., 2018; Johansen et al., 2019; Tuncel et al., 2019; Veillet et al., 2019a; Sevestre et al., 2020; Zhao et al., 2021), carotenoid biosynthesis (Khromov et al., 2018; Bánfalvi et al., 2020; Butler et al., 2020), glycoalkaloids (Nakayasu et al., 2018; Zheng et al., 2021), and enzymatic browning (González et al., 2020) (Table 1). Functional mutants were developed for variations in phenotype (Wang et al., 2015; Veillet et al., 2020a) and herbicide tolerance (Butler et al., 2015, 2016). Self-compatible regenerants were also produced using Cas9 via Agrobacterium (Ye et al., 2018; Enciso-Rodriguez et al., 2019) or virus-induced genome editing (VIGE) (Uranga et al., 2021a; 2021b). Researchers have demonstrated the utility of Cas9 base editing and prime editing tools for herbicide tolerance in potato (Veillet et al., 2019b; 2020a; 2020b; 2020c).
Because potato is a highly amenable crop to tissue culture, transformation methods such as Agrobacterium, particle bombardment or biolistic, floral-dip, and protoplasts have been applied to it (Sandhya et al., 2020). The most common Agrobacterium-mediated transformation and protoplasts that have been successfully deployed in CRISPR/Cas in potato are sgRNA dicot-origin promoters like Arabidopsis (AtUp)/potato (StU6p)/U3p and plant promoters like CaMV 35S (Belhaj et al., 2013). However, the Agrobacterium-mediated method cannot be used to deliver ribonucleoprotein (RNP) complexes, and elimination of the Cas9 assembly from the plant genome via selfing or backcrossing is more complicated in genetically complex and vegetatively propagated tetraploid potato (Koltun et al., 2018). In potato, each botanical seed called True Potato Seed (TPS), which is a product of the meiosis process, is genetically different from another seed, hence the maintenance of the clonal identity is very crucial.
To address the above issues, the DNA-free delivery system is an ideal approach using somatic cells, i.e. protoplast. Polyethylene glycol (PEG)–mediated protoplast transformation has been found to be an excellent alternative for the efficient delivery of Cas9/gRNA-RNPs in potato (Andersson et al., 2017). DNA-free preassembled Cas9/gRNA-RNPs were directly delivered into the plant cells to induce mutations (Park and Choe, 2019) and were also demonstrated in lipofection-mediated DNA-free delivery (Liu et al., 2020). But with the establishment of suspension culture, protoplast isolation and regeneration into whole plants are the associated problems of the protoplast system (Sandhya et al., 2020).
VIGE is an emerging approach for CRISPR/Cas9 delivery. VIGE involving plant virus–derived vector such as geminivirus replicon has been demonstrated for fast and efficient delivery of sgRNAs in potato (Butler et al., 2015, 2016). This VIGE system bypasses the requirement of transformation and regeneration of plants which is a time-consuming and tedious process. But the large size of a Cas9 assembly challenges the use of the virus vector, as the length of a foreign insert negatively correlates with the stability of the vector.
Recently, base editing and prime editing are the upgraded and more efficient approaches of Cas9. The programmable base editing technology, like the adenine base editor that coverts A.T to G.C without DNA cleavage, has emerged as a boon for crop improvement (Gaudelli et al., 2017). Catalytically inactive Cas9 variant dCas9 or Cas9-nickase is fused with cytosine or adenosine deaminase domain to introduce the desired point mutations (C to T or A to G) in the target region (Mishra et al., 2020). Veillet et al. (2020c) deployed Staphylococcus aureus–cytosine base editor (CRISPR-SaCas9 CBE) to edit StDMR6-1 in potato. Similarly, herbicide tolerance genes Acetolactate synthase1 and Acetolactate synthase2 (StALS2) were targeted through Cas9 cytidine base editing and Cas9 prime editing technologies, respectively (Veillet et al., 2019b; 2020b). Ariga et al. (2020) used the potato virus X vector to express a base editor consisting of modified Cas9 fused with cytidine deaminase to introduce the targeted nucleotide substitution in Nicotiana benthamiana. However, the size of the base editor is larger than Cas9 and this hindered the delivery into cells by the viral vectors.
Overall, high heterozygosity, tetrasomic inheritance, severe inbreeding depression, and vegetative propagation caused difficulties in the successful application of CRISPR/Cas in tetraploid potato. Furthermore, the selection of suitable sgRNA, robust CRISPR/Cas, and efficient transformation protocols and phenotypes without off targets are the main decisive factors in potato. Currently, gene knockout is a preferred mechanism in plants and even all four alleles were mutated through Cas9 in potato StGBSS gene (Andersson et al., 2017). PAM limitation (NGG) is one of the drawbacks of SpCas9, and therefore more diversity in CRISPR/Cas toolbox is necessary (Veillet et al., 2020a).
Desirable plant phenotypes, biotic/abiotic stress resistance/tolerance, and improved tuber quality traits play key roles in potato. The availability of robust CRISPR/Cas arrays, target genes selection, efficient plant transformation protocols, and minimum off-target mutants are the major issues in tetraploid potato. It is a fact that improvement of multigenic traits is difficult than that of the monogenic traits, particularly in potato, due to polyploidy and clonal propagation. Despite this, considerable success has been achieved in potato for some traits and mostly through the gene knockout or insertion/deletion process. Studies have suggested that the use of multiplexing SpCas9 that can handle single or multiple sgRNA/RNPs via targeting conserved sequences combined with protoplast-mediated transformation is an ideal option in potato. Apart from this, awareness among people and policy makers/regulators would be necessary for the success of genome editing research. Collectively, CRISPR-Cas provides an effective next-generation toolbox for fast potato breeding to achieve sustainable crop yield.
JT conceived the idea and wrote the manuscript. All authors performed research, literature reviewing, and editing, and approved the manuscript for publication.
This work is funded by the Biotechnology programme by ICAR-CPRI, Shimla, CABin Scheme, and ICAR-LBS Outstanding Young Scientist Award project to JT.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank ICAR-CPRI, CABin Scheme, and ICAR-LBS Outstanding Young Scientist Award project for support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.827808/full#supplementary-material
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018). RNA Virus Interference via CRISPR/Cas13a System in Plants. Genome Biol. 19, 1. doi:10.1186/s13059-017-1381-1
Andersson, M., Turesson, H., Nicolia, A., Fält, A. S., Samuelsson, M., and Hofvander, P. (2017). Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum tuberosum) by Transient CRISPR-Cas9 Expression in Protoplasts. Plant Cel Rep 36, 117–128. doi:10.1007/s00299-016-2062-3
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome Editing in Potato via CRISPR-Cas9 Ribonucleoprotein Delivery. Physiol. Plant 164 (4), 378–384. doi:10.1111/ppl.12731
Ariga, H., Toki, S., and Ishibashi, K. (2020). Potato Virus X Vector-Mediated DNA-free Genome Editing in Plants. Plant Cel Physiol 61 (11), 1946–1953. doi:10.1093/pcp/pcaa123
Bánfalvi, Z., Csákvári, E., Villányi, V., and Kondrák, M. (2020). Generation of Transgene-free PDS Mutants in Potato by Agrobacterium-Mediated Transformation. BMC Biotechnol. 20 (1), 25. doi:10.1186/s12896-020-00621-2
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and Nekrasov, V. (2013). Plant Genome Editing Made Easy: Targeted Mutagenesis in Model and Crop Plants Using the CRISPR/Cas System. Plant Methods 9, 39–47. doi:10.1186/1746-4811-9-39
Butler, N. M., Atkins, P. A., Voytas, D. F., and Douches, D. S. (2015). Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS One 10, e0144591. doi:10.1371/journal.pone.0144591
Butler, N. M., Baltes, N. J., Voytas, D. F., and Douches, D. S. (2016). Geminivirus-mediated Genome Editing in Potato (Solanum tuberosum L) Using Sequence-specific Nucleases. Front. Plant Sci. 7, 1045. doi:10.3389/fpls.2016.01045
Butler, N. M., Jansky, S. H., and Jiang, J. (2020). First-generation Genome Editing in Potato Using Hairy Root Transformation. Plant Biotechnol. J. 18 (11), 2201–2209. doi:10.1111/pbi.13376
Cao, Y., Zhou, H., Zhou, X., and Li, F. (2020). Control of Plant Viruses by CRISPR/Cas System-Mediated Adaptive Immunity. Front. Microbiol. 11, 593700. doi:10.3389/fmicb.2020.593700
Dangol, S. D., Barakate, A., Stephens, J., Çalıskan, M. E., and Bakhsh, A. (2019). Genome Editing of Potato Using CRISPR Technologies: Current Development and Future Prospective. Plant Cel Tissue Organ. Cult 139, 403–416. doi:10.1007/s11240-019-01662-y
Enciso-Rodriguez, F., Manrique-Carpintero, N. C., Nadakuduti, S. S., Buell, C. R., Zarka, D., and Douches, D. (2019). Overcoming Self-Incompatibility in Diploid Potato Using CRISPR-Cas9. Front. Plant Sci. 10, 376. doi:10.3389/fpls.2019.00376
FAOSTAT (2018). Available at: https://www.fao.org/faostat/en/ Accessed: November 20, 2021.
Gaudelli, N. M., Komor, A. C., Rees, H., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable Base Editing of A.T to G.C in Genomic DNA without DNA Cleavage. Nature 551, 464–471. doi:10.1038/nature24644
González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A. S., et al. (2020). Reduced Enzymatic browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 10, 1649. doi:10.3389/fpls.2019.01649
Hameed, A., Mehmood, M. A., Shahid, M., Fatma, S., Khan, A., and Ali, S. (2020). Prospects for Potato Genome Editing to Engineer Resistance against Viruses and Cold-Induced Sweetening. GM Crops Food 11 (4), 185–205. doi:10.1080/21645698.2019.1631115
Hameed, A., Zaidi, S. S., Shakir, S., and Mansoor, S. (2018). Applications of New Breeding Technologies for Potato Improvement. Front. Plant Sci. 9, 925. doi:10.3389/fpls.2018.00925
Hegde, N., Joshi, S., Soni, N., and Kushalappa, A. C. (2021). The Caffeoyl-CoA O-Methyltransferase Gene SNP Replacement in Russet Burbank Potato Variety Enhances Late Blight Resistance through Cell wall Reinforcement. Plant Cel Rep 40 (1), 237–254. doi:10.1007/s00299-020-02629-6
Hofvander, P., Andreasson, E., and Andersson, M. (2021). Potato Trait Development Going Fast-Forward with Genome Editing. Trends Genet. S0168-9525 (21), 00288–292. doi:10.1016/j.tig.2021.10.004
ICAR-CPRI Annual Report (2018). Annual Report. Shimla, Himachal Pradesh, India: ICAR-Central Potato Research Institute.
Johansen, I. E., Liu, Y., Jørgensen, B., Bennett, E. P., Andreasson, E., Nielsen, K. L., et al. (2019). High Efficacy Full Allelic CRISPR/Cas9 Gene Editing in Tetraploid Potato. Sci. Rep. 9, 17715. doi:10.1038/s41598-019-54126-w
Khatodia, S., Bhatotia, K., Passricha, N., Khurana, S. M., and Tuteja, N. (2016). The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 7, 506. doi:10.3389/fpls.2016.00506
Khromov, A. V., Gushchin, V. A., Timberbaev, V. I., Kalinina, N. O., Taliansky, M. E., and Makarov, V. V. (2018). Guide RNA Design for CRISPR/Cas9-mediated Potato Genome Editing. Dokl Biochem. Biophys. 479 (1), 90–94. doi:10.1134/s1607672918020084
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, B. L., and Andreasson, E. (2021). Mutations Introduced in Susceptibility Genes through CRISPR/Cas9 Genome Editing Confer Increased Late Blight Resistance in Potatoes. Sci. Rep. 11 (1), 4487. doi:10.1038/s41598-021-83972-w
Koltun, A., Corte, L. E. –D., Mertz-Henning, L. M., and Gonçalves, L. S. A. (2018). Genetic Improvement of Horticultural Crops Mediated by CRISPR/Cas: a New Horizon of Possibilities. Hortic. Brasil. 36, 290–298. doi:10.1590/s0102-053620180302
Kusano, H., Ohnuma, M., Mutsuro-Aoki, H., Asahi, T., Ichinosawa, D., Onodera, H., et al. (2018). Establishment of a Modified CRISPR/Cas9 System with Increased Mutagenesis Frequency Using the Translational Enhancer dMac3 and Multiple Guide RNAs in Potato. Sci. Rep. 8, 13753. doi:10.1038/s41598-018-32049-2
Liu, W., Rudis, M. R., Cheplick, M. H., Millwood, R. J., Yang, J.-P., Ondzighi-Assoume, C. A., et al. (2020). Lipofection-mediated Genome Editing Using DNA-free Delivery of the Cas9/gRNA Ribonucleoprotein into Plant Cells. Plant Cel Rep 39, 245–257. doi:10.1007/s00299-019-02488-w
Makhotenko, A. V., Khromov, A. V., Snigir, E. A., Makarova, S. S., Makarova, V. V., Suprunova, T. P., et al. (2019). Functional Analysis of Coilin in Virus Resistance and Stress Tolerance of Potato Solanum tuberosum Using CRISPR-Cas9 Editing. Dokl Biochem. Biophys. 484, 88–91. doi:10.1134/s1607672919010241
Mishra, R., Joshi, R. K., and Zhao, K. (2020). Base Editing in Crops: Current Advances, Limitations and Future Implications. Plant Biotechnol. J. 18, 20–31. doi:10.1111/pbi.13225
Nadakuduti, S. S., Buell, C. R., Voytas, D. F., Starker, C. G., and Douches, D. S. (2018). Genome Editing for Crop Improvement-Applications in Clonally Propagated Polyploids with a Focus on Potato (Solanum tuberosum L.). Front. Plant Sci. 9, 1607. doi:10.3389/fpls.2018.01607
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free Hairy Roots of Potato by CRISPR/Cas9 Mediated Genome Editing of the St16DOX Gene. Plant Physiol. Biochem. 131, 70–77. doi:10.1016/j.plaphy.2018.04.026
Park, J., and Choe, S. (2019). DNA-free Genome Editing with Preassembled CRISPR/Cas9 Ribonucleoproteins in Plants. Transgenic Res. 28 (Suppl. 2), 61–64. doi:10.1007/s11248-019-00136-3
Potato Genome Sequencing Consortium (2011). Genome Sequence and Analysis of the Tuber Crop Potato. Nature 475, 189–195. doi:10.1038/nature10158
Price, A. A., Sampson, T. R., Ratner, H. K., Grakoui, A., and Weiss, D. S. (2015). Cas9-mediated Targeting of Viral RNA in Eukaryotic Cells. Procd. Natl. Acad. Sci. U. S. A. 112, 6164–6169. doi:10.1073/pnas.1422340112
Rana, A., Dua, V. K., Chauhan, S., and Sharma, J. (2020). Climate Change and Potato Productivity in Punjab—Impacts and Adaptation. Potato Res. 63, 597–613. doi:10.1007/s11540-020-09460-2
Sandhya, D., Jogam, P., Allini, V. R., Abbagani, S., and Alok, A. (2020). The Present and Potential Future Methods for Delivering CRISPR/Cas9 Components in Plants. J. Genet. Eng Biotechnol 18, 25. doi:10.1186/s43141-020-00036-8
Schmidt, S. M., Belisle, M., and Frommer, W. B. (2020). The Evolving Landscape Around Genome Editing in Agriculture. EMBO Rep. 21, e50680. doi:10.15252/embr.202050680
Sevestre, F., Facon, M., Wattebled, F., and Szydlowski, N. (2020). Facilitating Gene Editing in Potato: a Single-Nucleotide Polymorphism (SNP) Map of the Solanum tuberosum L. Cv. Desiree Genome. Sci. Rep. 10 (1), 2045. doi:10.1038/s41598-020-58985-6
Tiwari, J. K., Buckseth, T., Singh, R. K., Kumar, M., and Kant, S. (2020). Prospects of Improving Nitrogen Use Efficiency in Potato: Lessons from Transgenics to Genome Editing Strategies in Plants. Front. Plant Sci. 11, 597481. doi:10.3389/fpls.2020.597481
Tuncel, A., Corbin, K. R., Ahn-Jarvis, J., Harris, S., Hawkins, E., Smedley, M. A., et al. (2019). Cas9-mediated Mutagenesis of Potato Starch-Branching Enzymes Generates a Range of Tuber Starch Phenotypes. Plant Biotechnol. J. 17 (12), 2259–2271. doi:10.1111/pbi.13137
United Nations, Department of Economic and Social Affairs, Population Division (2019). World Population Prospects 2019: Highlights (ST/ESA/SER.A/423).
Uranga, M., Aragonés, V., Selma, S., Vázquez-Vilar, M., Orzaez, D., and Daros, J.-A. (2021a). Efficient Cas9 Multiplex Editing Using Unspaced sgRNA Arrays Engineering in a Potato Virus X Vector. Plant J. 106, 555–565. doi:10.1111/tpj.15164
Uranga, M., Vazquez-Vilar, M., Orzáez, D., and Daròs, J. A. (2021b). CRISPR-Cas12a Genome Editing at the Whole-Plant Level Using Two Compatible RNA Virus Vectors. CRISPR J. 4 (5), 761–769. doi:10.1089/crispr.2021.0049
Veillet, F., Chauvin, L., Kermarrec, M. P., Sevestre, F., Merrer, M., Terret, Z., et al. (2019a). The Solanum tuberosum GBSSI Gene: a Target for Assessing Gene and Base Editing in Tetraploid Potato. Plant Cel Rep 38 (9), 1065–1080. doi:10.1007/s00299-019-02426-w
Veillet, F., Kermarrec, M. -P., Chauvin, L., Guyon-Debast, A., Chauvin, J. E., Gallois, J. –L., et al. (2020b). Prime Editing Is Achievable in the Tetraploid Potato, but Needs Improvement. bioRxiv. doi:10.1101/2020.06.18.159111
Veillet, F., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., and Nogué, F. (2020a). CRISPR-induced Indels and Base Editing Using the Staphylococcus aureus Cas9 in Potato. PLoS One 15, e0235942. doi:10.1371/journal.pone.0235942
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019b). Transgene-free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Inter. J. Mol. Sci. 20 (2), 402. doi:10.3390/ijms20020402
Veillet, F., Perrot, L., Guyon-Debast, A., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., et al. (2020c). Expanding the CRISPR Toolbox in P. patens Using SpCas9-NG Variant and Application for Gene and Base Editing in Solanaceae Crops. Inter. J. Mol. Sci. 21 (3), 1024. doi:10.3390/ijms21031024
Wang, S., Zhang, S., Wang, W., Xiong, X., Meng, F., and Cui, X. (2015). Efficient Targeted Mutagenesis in Potato by the CRISPR/Cas9 System. Plant Cel Rep 34, 1473–1476. doi:10.1007/s00299-015-1816-7
Ye, M., Peng, Z., Tang, D., Yang, Z., Li, D., Xu, Y., et al. (2018). Generation of Self-Compatible Diploid Potato by Knockout of. S-RNaseNature Plant 4 (9), 651–654. doi:10.1038/s41477-018-0218-6
Zhan, X., Zhang, F., Zhong, Z., Chen, R., Wang, Y., Chang, L., et al. (2019). Generation of Virus-Resistant Potato Plants by RNA Genome Targeting. Plant Biotechnol. J. 17, 1814–1822. doi:10.1111/pbi.13102
Zhao, X., Jayarathna, S., Turesson, H., Fält, A. S., Nestor, G., González, M. N., et al. (2021). Amylose Starch with No Detectable Branching Developed through DNA-free CRISPR-Cas9 Mediated Mutagenesis of Two Starch Branching Enzymes in Potato. Sci. Rep. 11 (1), 4311. doi:10.1038/s41598-021-83462-z
Zheng, Z., Ye, G., Zhou, Y., Pu, X., Su, W., and Wang, J. (2021). Editing Sterol Side Chain Reductase 2 Gene (StSSR2) via CRISPR/Cas9 Reduces the Total Steroidal Glycoalkaloids in Potato. All Life 14, 401–413. doi:10.1080/26895293.2021.1925358
Keywords: abiotic, biotic, CRiSPR/Cas, genome editing, Potato, quality
Citation: Tiwari JK, Buckseth T, Challam C, Zinta R, Bhatia N, Dalamu D, Naik S, Poonia AK, Singh RK, Luthra SK, Kumar V and Kumar M (2022) CRISPR/Cas Genome Editing in Potato: Current Status and Future Perspectives. Front. Genet. 13:827808. doi: 10.3389/fgene.2022.827808
Received: 02 December 2021; Accepted: 11 January 2022;
Published: 02 February 2022.
Edited by:
Deepmala Sehgal, International Maize and Wheat Improvement Center, MexicoReviewed by:
Muntazir Mushtaq, National Bureau of Plant Genetic Resources (ICAR), IndiaCopyright © 2022 Tiwari, Buckseth, Challam, Zinta, Bhatia, Dalamu, Naik, Poonia, Singh, Luthra, Kumar and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jagesh Kumar Tiwari, amFnZXNodGl3YXJpQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.