- 1Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, Jiangxi Agricultural University, Nanchang, China
- 2Southern Regional Collaborative Innovation Center for Grain and Oil Crops in China, Nanchang, China
- 3College of Agronomy, Jiangxi Agricultural University, Nanchang, China
- 4Huazhong Agricultural University, Wuhan, China
Cystathionine γ-synthase (CGS), methionine γ-lyase (MGL), cystathionine β-lyase (CBL) and cystathionine γ-lyase (CGL) share the Cys_Met_Meta_PP domain and play important roles in plant stress response and development. In this study, we defined the genes containing the Cys_Met_Meta_PP domain (PF01053.20) as CBL-like genes (CBLL). Twenty-nine CBLL genes were identified in the peanut genome, including 12 from cultivated peanut and 17 from wild species. These genes were distributed unevenly at the ends of different chromosomes. Evolution, gene structure, and motif analysis revealed that CBLL proteins were composed of five different evolutionary branches. Chromosome distribution pattern and synteny analysis strongly indicated that whole-genome duplication (allopolyploidization) contributed to the expansion of CBLL genes. Comparative genomics analysis showed that there were three common collinear CBLL gene pairs among peanut, Arabidopsis, grape, and soybean, but no collinear CBLL gene pairs between peanut and rice. The prediction results of cis-acting elements showed that AhCBLLs, AdCBLLs, and AiCBLLs contained different proportions of plant growth, abiotic stress, plant hormones, and light response elements. Spatial expression profiles revealed that almost all AhCBLLs had significantly higher expression in pods and seeds. All AhCBLLs could respond to heat stress, and some of them could be rapidly induced by cold, salt, submergence, heat and drought stress. Furthermore, one polymorphic site in AiCBLL7 was identified by association analysis which was closely associated with pod length (PL), pod width (PW), hundred pod weight (HPW) and hundred seed weight (HSW). The results of this study provide a foundation for further research on the function of the CBLL gene family in peanut.
Introduction
Sulfur-containing amino acids play an important role in the growth and development of plants and animals (Lorraine et al., 1986). When plants lack sulfur-containing amino acid, their metabolic processes will be abnormal and growth will be affected (Kery et al., 1994). Methionine (Met) is a sulfur-containing amino acid that is essential to all organisms and indirectly regulates a variety of cellular processes through S-adenosine methionine (SAM) (Amir, 2010). SAM is also a methyl donor for the methylation of proteins, lipids, DNA, and RNA, and is a precursor to biosynthesis of the plant hormones ethylene, polyamines, and biotin (Benkova et al., 2003). A recent study showed that Met activates GLR (glutamate receptor), thereby activating Ca2+ channels that regulate stomatal movement and plant growth (Galili et al., 2016). Met also promotes root morphogenesis and enhances chlorophyll content to enhance photosynthesis (Sarropoulou et al., 2013).
Met biosynthesis requires cystathionine γ-synthase (CGS), cystathionine β-lyase (CBL), and Met synthase enzymes (Messerschmidt et al., 2003). In plants, CGS first converts cystine (Cys) and activates the conversion of homoserine to cystathionine. Cystathionine is then cleaved by CBL to yield homoCys, pyruvate, and ammonia. Met synthase finally methylates homo-Cys to produce Met (Ravanel et al., 1998). Animals and some microorganisms can metabolize Met back to Cys by a reverse trans-sulfuration pathway that involves cystathionine β-synthase (CBS) and cystathionine γ-lyase (CGL). Plants and certain bacteria are proposed to have an alternative route (or routes) to convert methionine to cysteine, of which the first step is mediated by methionine γ-lyase (MGL). The subsequent steps in this pathway(s) have not been definitively established (Goyer et al., 2007). Arabidopsis mutants perturbed in Met metabolism have been described,mto alleles carried single base-pair mutation in a conserved domain led to over-accumulation of Met (Goto et al., 2002). Over-expression of AtCGS caused an increased level of Met, which induced the up-regulation of genes involved in ethylene and abscisic acid homeostasis and light, sucrose, salt and osmotic stresses regulation (Hacham et al., 2013; Cohen et al., 2014, 2017; Whitcomb et al., 2018). Pharmacological treatment of plants with inhibitors against CGS or CBL induced a deficiency in Met biosynthesis and caused growth inhibition (Ravanel et al., 1998). Knockdown of CGS and CBL in Arabidopsis led to abnormal leaf development stunted (Kim and Leustek., 2000; Levin et al., 2000). CBL is crucial for embryo patterning and the maintenance of the root stem cell niche in Arabidopsis (Liu et al., 2019). The Met homeostasis gene METHIONINE GAMMA LYASE (AtMGL) is up-regulated by dual stress in leaves, conferring resistance to nematodes when overexpressed, AtMGL regulates Met metabolism under conditions of multiple stressors, Met degradation, and plays a subordinate role to threonine deaminase (Goyer et al., 2007; Joshi and , 2009; Atkinson et al., 2013). A similar strategy has been applied to produce transgenic plants with reduced CBL levels in potato. These CBL antisense plants exhibited a short bushy stature, altered leaf morphology, and small tuber size (Maimann et al., 2000). In soybean, the overexpress of AtD-CGS notably increased the level of soluble Met in developing green seeds (3.8–7-fold), and these soybean seeds also showed high levels of other amino acids; furthermore, the total Met content, which included Met incorporated into proteins, notably increased in the mature dry seeds of these two transgenic lines by 1.8- and 2.3-fold, respectively (Song et al., 2013).
Peanut is an annual leguminous herb, is widely cultivated around the world as an important source of oil and protein for humans (Yang et al., 2014; Liao et al., 2018; Bertioli et al., 2019). Peanut kernels are rich in natural nutrients such as proteins, fatty acids (FAs), vitamins, minerals, and fiber (Chen et al., 2010). Studies have demostrated that the protein content of peanut seeds is around 23–33%; however, the content of sulfur-containing Met and Cy is low, especially for the former (Yang et al., 2001). Met and the enzyme coding genes involved in the Met/Cys interconversion pathway play inhibitory roles in both amino acid composition and content of storage protein, and also in tissue development and stress response regulation. Therefore, it is of great significance to further explore the CBLL gene family in peanut and identifythe function of family members in improving peanut yield, quality, and stress tolerance. In this study, we defined the genes containing the Cys_Met_Meta_PP domain (PF01053.20) as CBL-like genes (CBLL). Twenty-nine CBLL genes were identified from the peanut genome, whose conserved domains, phylogenetic tree, chromosome distribution, gene structure and expression pattern were analyzed. The results provided a basis for the role of peanut CBLL in the development and formation of peanut pods, and stress response regulation. This research laid a foundation for the identification and utilization of peanut CBLL genes, which is of great significance for the molecular based breeding of cultivars with multi-resistance, high yield, and good quality.
Materials and Methods
Identification of the CBLL Genes in Peanut
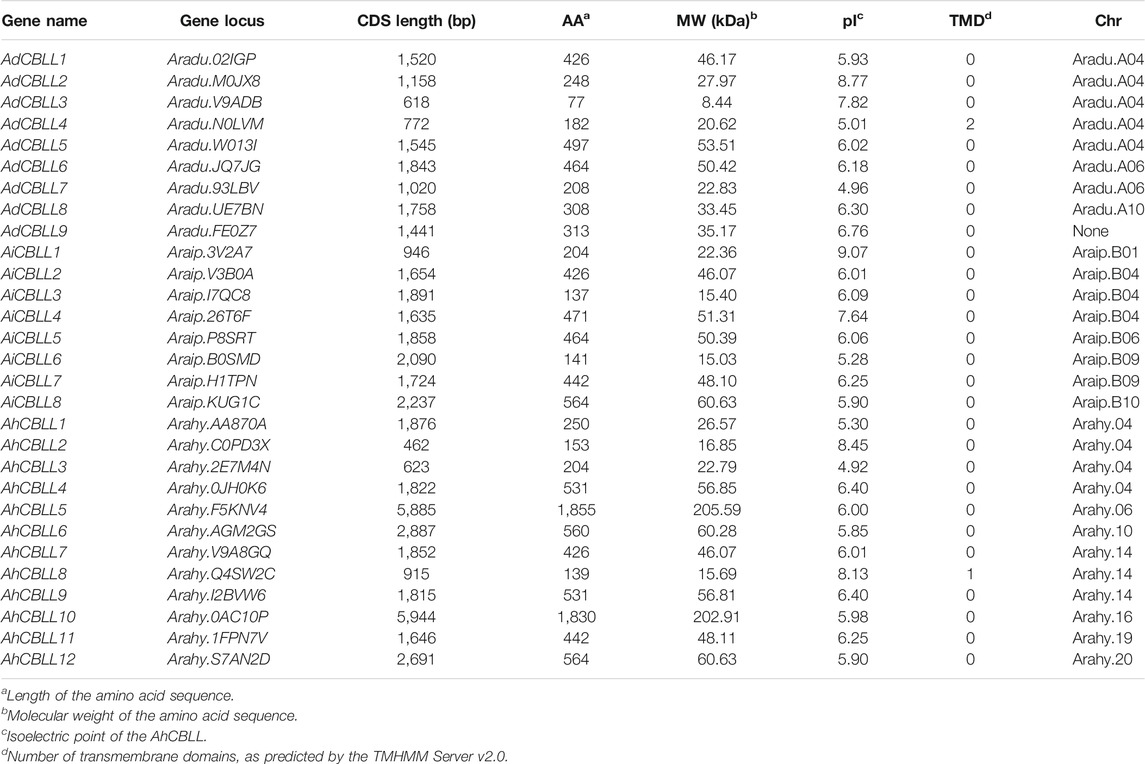
To identify CBLL genes in peanut, the predicted protein sequences were downloaded from PeanutBase (https://peanutbase.org) (Arachis Duranensis V14167: A-genome; Arachis Ipaensis K30076: B-genome). The Cys_Met_Meta_PP domain (PF01053.20) was identified from the protein sequences by using the HMMER 3.0 program at a standard E-value < 1 × 10−5 (Deng et al., 2019; Misra et al., 2019). Conserved domain searches were performed against the conserved domain database in NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Members with incomplete conserved functional domains were removed. The CBLL genes were named as AdCBLL1 to AdCBLL9, AiCBLL1 to AiCBLL8, and AhCBLL1 to AhCBLL12 according to their positions on peanut chromosomes. Physicochemical parameters of peanut CBLL proteins were then generated by ProtParam Tools, including theoretical isoelectric points (pI) and molecular weights (MW) (Gasteiger et al., 2005).
Analyses of Phylogeny, Gene Structure, and Conserved Motifs
Phylogenetic analysis was carried out based on the protein sequences of the CBLL genes in peanut (A.hypogaea L., A.duranensis, A.ipaensis) and Arabidopsis. The protein sequences were aligned by ClustalW, and the unrooted Neighbor-Joining (NJ) phylogenetic tree was constructed by MEGA 5.2 software with 1,000 bootstrap replicates. Gene annotation information was downloaded from PeanutBase (http://www.peanutbase.org/) and GSDS 2.0 (http://gsds.gao-lab.org/) was used to visualize the gene structure. The composition of conserved motifs was searched by the Multiple EM for Motif Elicitation (MEME) online tool by setting a maximum number as 20 (http://meme-suite.org/tools/meme).
Gene Duplication and Synteny Analysis of the Peanut CBLL Genes
MCScan (http://chibba.agtec.uga.edu/duplication/mcscan) was used to identify AhCBLs duplications and the synteny block of CBLL genes of peanut and the other four species (Arabidopsis, rice, grape and soybean). BLASTP was applied to find homologous sequences of CBLL genes between peanut and Arabidopsis, after that literatures were reviewed to explore the function of published CBLL genes in Arabidopsis. Tandem duplications were defined as adjacent homologous genes on the same chromosome with a distance of <50 kb (Cannon et al., 2004). If they were paralogs located on duplicated chromosomal blocks, they were defined as a segmental duplication event (Guo et al., 2016). Non-synonymous (Ka) and synonymous (Ks) substitution of each duplicated genes were calculated using the PAL2NAL program (Suyama et al., 2006), which was based on the codon model program in PAML (Yang, 2007).
Cis-Acting Element Analysis in the Promoters of Peanut CBLLs
Regulatory elements of promoter sequences can control gene expression. The 2-kb promoter sequences of 29 CBLL genes were downloaded from PeanutBase (http://www.peanutbase.org/) and used to predict the cis-regulatory element through the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot, 2002). The radar figures were manually generated by R3.5.1 scripts.
Expression Profiles of AhCBLL Genes in Different Tissues
RNA-seq data sets of 22 peanut tissues were downloaded from PeanutBase and the NCBI SRA database to explore the expression profiles of AhCBLL genes in different tissues, which were submitted by Clevenger et al. (Clevenger et al., 2016).
Plant Materials, Growth Conditions and Treatments
Different experimental treatments were carried out for 10-day-old seedlings of a Chinese elite peanut cultivar Changhua18 which were planted in vermiculite and irrigated with sterilized water (26°C, 16-h light/8-h dark). For hormone treatments, seedlings were sprayed with solutions containing 6-benzyl amino purine (6-BA) (25 μM), indole-3-acetic acid (IAA) (50 μM), gibberellic acid (GA) (100 μM), salicylic acid (SA) (100 μM), abscisic acid (ABA) (100 μM), ethylene (ACC) (500 μM) and methyl jasmonate (MJ) (100 µM)(Jain et al., 2006; Wang et al., 2016). For heat and cold stresses, the 10-day-old seedlings were transferred to two environmental temperatures of 40°C (H40) and 4°C (Cold 4) respectively (Jain et al., 2006; Wang et al., 2016). Seedling samples were collected at 0, 1, 3, 6, 9 and 12 h after the above treatment. For submergence (Sub), the seedlings were soak in water to a depth of 5 cm from tip to surface, and samples were collected at 0, 6, 12, 24, 48, and 72 h after treatment (Wang et al., 2016). For the NaCl and polyethylene glycol (PEG) treatments, the 10-day-old seedlings were immersed in NaCl solution (200 mM) and PEG6000 (20%, w/v) (Jain et al., 2006; Song et al., 2009), and samples were collected after 0, 0.5, 1, 3, 6, 12, and 24 h after seedling treatment. Three biological replicates were performed; each sample included around eight seedlings. All samples were placed in liquid nitrogen during sampling and stored at −80°C to preserve RNA integrity.
The genotype data of the CBLL genes used here were obtained from transcriptome sequencing data of a peanut germplasm population with 146 accessions (unpublished data). Each line of the peanut populations was planted in five different environments (Wuhan 2016, Wuhan 2017, Yangluo 2016, Yangluo 2017, and Zhanjiang 2016). All seedlings were planted within the experimental plot with 12 plants in a line in each environment.
RNA Isolation and qRT-PCR Analysis
Total RNA of samples were isolated using TRIzol reagent (Invitrogen) according to the manufacture’s requirements. M-MLV reverse transcriptase (Promega) was used to synthesize the first chain of cDNA from 5 µg total RNA. Quantitative Real-Time PCR (qRT-PCR) was performed using 2×SYBR Green Master Mix (Bio-Rad) on a 96-well plate with a gene-specific primer (Supplementary Table S1). The thermal cycle was as follows: 95°C for 5 min; 40 cycles of 95°C for 10 s, primer-specific annealing temperature of, 72°C for 10 s, for 15 cycles; then the melt curve was from 65 to 95°C.
Results
Identification of the CBLL Gene Family in Peanut
In order to identify CBLL gene families in peanut, we downloaded the published peanut genome sequence from PeanutBase (https://peanutbase.org/). The Cys_Met_Meta_PP domain (PF01053.20) containing proteins were identified by HMMER 3.0 with a standard E-value < 1 × 10−5; further, we removed the incomplete sequences, and identified 29 AhCBLL members (Table 1) from the cultivated peanut (A. hypogaea L.) and its diploid progenitors (A. duranensis, A. ipaensis). Table 1 summarizes genes with complete sequences, among which 17 CBLLs were from A-genome, and 12 CBLLs were from B-genome; they were distributed unevenly across chromosomes. The 12 members from A. hypogaea L. were named as AhCBLL1∼AhCBLL12, the nine AhCBLLs from A.duranensis were named as AdCBLL1∼AdCBLL9, and the eight AhCBLLs from A.ipaensis were named as AiCBLL2∼AiCBLL8, respectively, according to their chromosomal order. We then determined the chromosome location, amino acid number (AA), mRNA length, theoretical isoelectric points (pI) and other information of peanut CBLLs (Table 1). The open reading frame (ORF) lengths of the CBLL genes ranged from 462 bps to 5,944 bps (Table 1). The protein sequences of the peanut CBLL genes were significantly different; sequence lengths ranged from 77 to 1855 aa. The molecular weights (MWs) of AdCBLLs varied from 8.44 kDa (AdCBLL3) to 53.51 kDa (AdCBLL5); AiCBLLs ranged from 15.03 kDa (AiCBLL6) to 60.63 kDa (AiCBLL8); and AhCBLLs varied from 15.69 kDa (AhCBLL8) to 205.59 kDa (AhCBLL5). The pI was small for the overwhelming majority of CBLLs, ranging from 4.92 (AhCBLL3) to 9.07 (AiCBLL1). AhCBLL8 carried one conservative transmembrane domain (TMDs), AdCBLL4 carried two TMDs, and other peanut CBLLs did not contain TMDs.
Phylogeny, Gene Structure, and Conserved Motifs of the CBLL Gene Family in Peanut
Arabidopsis thaliana emerged is the model organism of choice for in plant biology research, we screened CBLL members through the A. thaliana genome, and four members were detected. To investigate the evolutionary relationships of the CBLL family genes in peanut, we conducted an NJ-phylogenetic tree, and analyzed the gene exon/intron structural and conserved motifs. The results demonstrated that the 29 peanut and four Arabidopsis CBLL proteins could be integrated into five clades (Figure 1A). Eight, six, one, four, and 10 CBLLs pertained to clade I (CGS clade), clade II (MGL clade), clade III (new clade), clade Ⅳ (CBL clade) and clade Ⅴ (new clade), respectively (Figure 1A). All the four large clades included CBLLs from the cultivated peanut and its two diploid progenitors. Interestingly, these five distinct groups have different gene structure and motif arrangement. The CGS clade had approximately 11 exons, and only AiCBLL3 had 10 exons. In sharp contrast to those in clade I, the MGL clade possessed two-three exons with one exception (AiCBLL1, 10 exon); the CBL clade had the greatest exon number, ranging from 12 to 29. For the two new clades, the only gene in clade III had 13 exons and clade Ⅴ had 2–13 exons (Figure 1B). Further, we identified 20 different conserved motifs (Figure 1C). In general, clade I, clade II, and clade Ⅳ had more motifs than clade III and clade Ⅴ. Motif 1 was the most common, present in all CBLL genes except AdCBLL8, AiCBLL5, and AhdCBLL3. Otherwise, the vast majority of CBLLs included motif 2, 3, 4, 6, 8, and 11. Motif 18, and 19 were clade-specific elements in clade I, motif 13 and 14 only existed in clade II, and motif 15 only existed in clade Ⅳ.
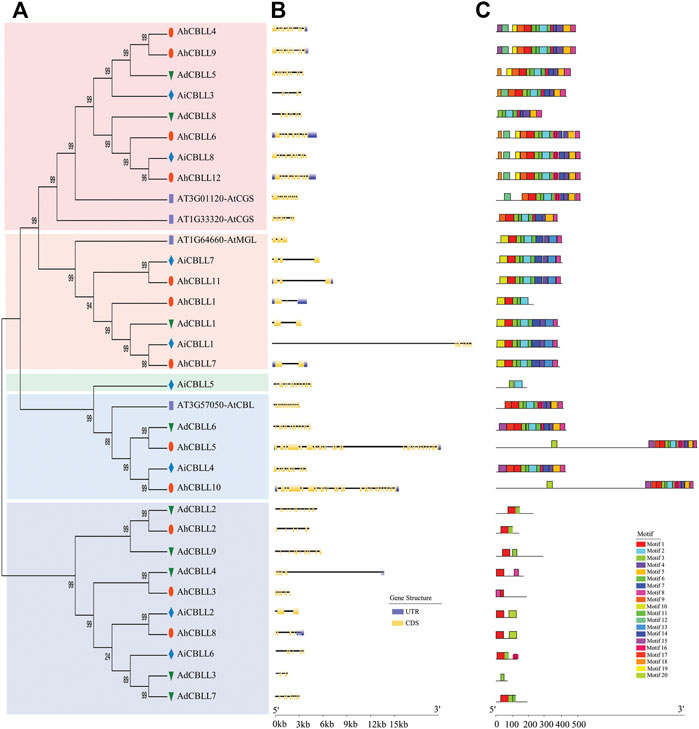
FIGURE 1. Comparison of the gene structure and motif of 29 CBLL genes in peanut. (A) Unrooted phylogenetic tree with over 50% bootstrap value above the branch. The clade I, II, III, Ⅳ and Ⅴ were displayed in pink, orange, green, blue and dark blue colors, separately. The names of species were abbreviated to two letters, named as Arachis duranensis (Ad), Arachis ipaensis (Ai), Arachis hypogaea L. Tifrunner (Ah). (B) Exon/introns and untranslated regions (UTRs) of CBLLs. Green boxes denote UTR (untranslated region); yellow boxes denote CDS (coding sequence); black lines denote introns. The length of protein can be estimated using the scale at the bottom. (C) Motif architectures of all CBLL genes. Each motif is illustrated with a specific color, and the distribution of identified motifs corresponds to their positions.
Gene Duplication and Synteny Analyses of Peanut CBLLs
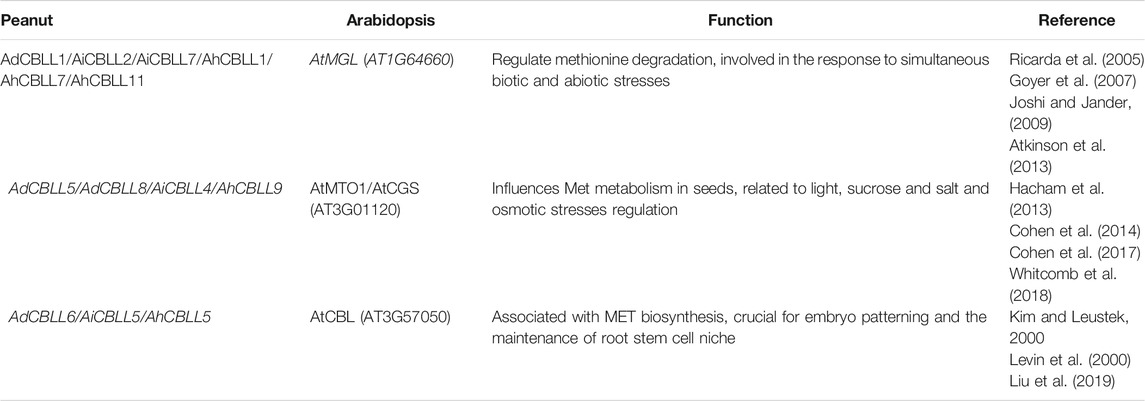
Chromosomal location analyses revealed that the 12 AhCBLLs were distributed unevenly on seven chromosomes (chromosomes 04, 06, 10, 14, 16, 19 and 20); nine AdCBLLs were present on chromosomes A04, A06 and A10; and eight AiCBLLs were distributed on chromosomes B01, B04, B06, B09, and B10 (Figure 2). A total of 17 chromosomal fragment repeat gene pairs were identified without tandem repeats (Figure 3 and Supplementary Tables S2–S5). Genomic synteny analyses between cultivated and wild peanut species uncovered five (AiCBLL1, AiCBLL6, AdCBLL3, AdCBLL7 and AdCBLL9) wild species-specific CBLL members (Figures 2, 3 and Supplementary Tables S2–S5). Further, we calculated the Ks (synonymous) and Ka (non-synonymous) values of the duplicated gene pairs and found that the Ka/Ks ratio for duplicated AhCBLL gene pairs ranged from 0.03 to 0.95 with an average of 0.25 (Supplementary Table S6). The ω values of all duplicated gene pairs were less than one, domostrated that purifying selection occurred on these duplicated gene pairs. The whole genome-wide collinear analysis identified that 41.38% (six AhCBLL genes, six pairs, Supplementary Table S7; three AdCBLL genes, three pairs; three AiCBLL genes, three pairs, Supplementary Table S8), 41.38% (six AhCBLL genes, eight pairs, Supplementary Table S9; three AdCBLL genes, three pairs; three AiCBLL genes, three pairs, Supplementary Table S10), and 48.28% (six AhCBLL genes, six pairs, Supplementary Table S11; four AdCBLL genes, seven pairs; four AiCBLL genes, seven pairs, Supplementary Table S12) of the AhCBLLs were orthologous with Arabidopsis, grape, and soybean CBLLs, but none with rice, respectively (Figure 4). Synteny analysis with soybean, Arabidopsis, and grape revealed three conserved CBLL genes (AhCBLL4, AhCBLL6, and AhCBLL9) in these species. Both collinear and BLAST methods were used to identify AhCBLL gene orthologs between peanut and Arabidopsis; 13 orthologous pairs were found (Table 2). The orthologs in Arabidopsis included AtMGL participating in Met degradation and plant defense (Ricarda et al., 2005; Goyer et al., 2007; Joshi and Jander, 2009; Atkinson et al., 2013), AtMOT1/CGS relating to the regulation of seed growth and metabolism in Arabidopsis (Goto et al., 2002; Hacham et al., 2013; Cohen et al., 2014, 2017; Whitcomb et al., 2018), and AtCBL relating to the synthesis of plant hormones (Kim and Leustek., 2000; Levin et al., 2000; Liu et al., 2019). Therefore, we speculated that these AhCBLL homologous genes might play multiple roles in peanut growth, development, and stress resistance.
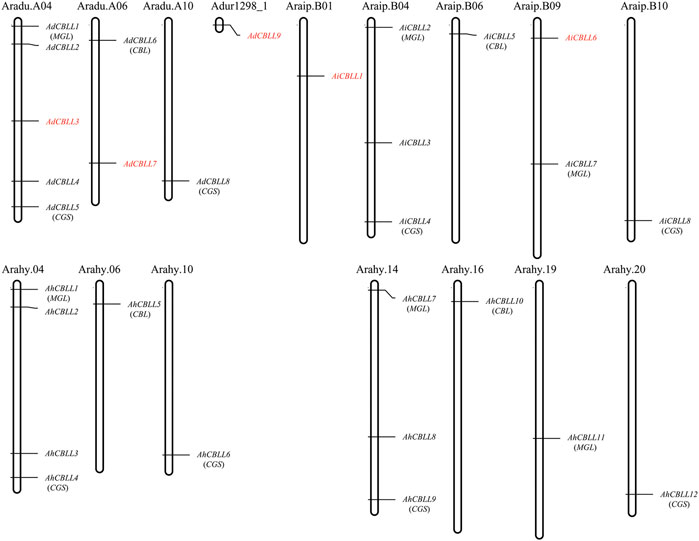
FIGURE 2. Chromosomal locations of peanut CBLL genes. Chromosomal positions of the peanut CBLL genes were mapped based on data from PeanutBase. The chromosome number was indicated above each chromosome. Genes in red indicated wild species specific.
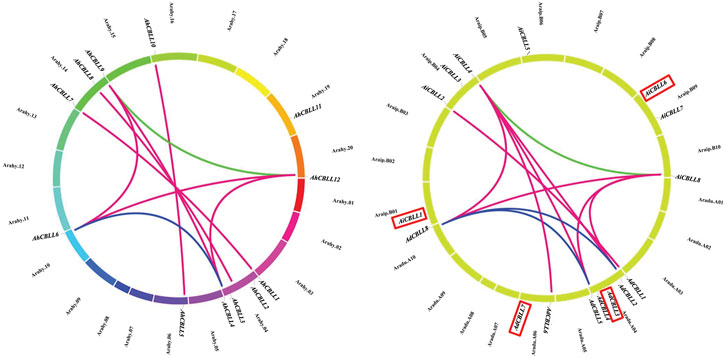
FIGURE 3. Chromosomal distribution and gene duplications of the AdCBLL, AiCBLL, and AhCBLL genes. The scales on the circle were in Megabases. Each colored bar represented a chromosome as indicated. Gene IDs were labeled on the basis of their positions on the chromosomes. Red frames indicated wild species specific CBLL genes.
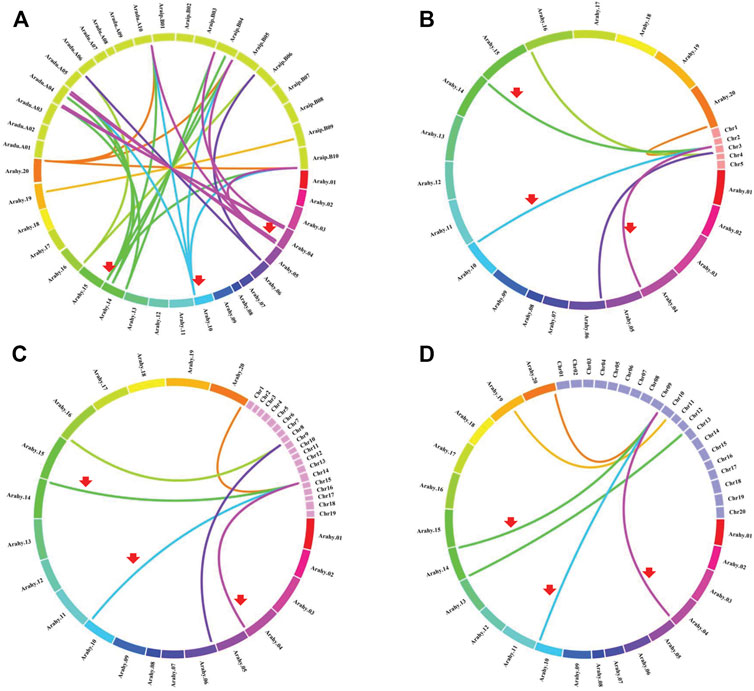
FIGURE 4. Comparative physical mapping showed the degree of orthologous relationships of AhCBLL genes with (A) peanut, (B) Arabidopsis, (C) grape and (D) soybean. Red arrows indicated common collinear CBLL gene pairs.
Cis-Acting Element Prediction of Peanut CBLLs
We analyzed and predicted cis-acting elements in the 2-kb upstream sequences of CBLL genes via the PlantCARE database. In total, 51 cis-regulatory elements were detected (Figures 5A–C), 11 subclasses and four main categories were defined as plant growth, abiotic stress, phytohormone responsiveness, and light responsiveness element groups (Figures 5D–F). In the promoter region of the AdCBLLs, the largest subdivision was the light responsiveness group, containing 44.1% predicted cis-elements, phytohormone responsiveness elements ranked second (34.1%); abiotic stress response elements were 14.7%, and elements involved in plant growth accounted for 7.1%. AdCBLL5 had the greatest number of elements with 33 in total (Figure 5D). For AiCBLLs, the percentage of light, phytohormone, abiotic stress, and plant growth responsiveness cis-elements was 43.6, 25.8, 22.1, and 8.6% (Figure 5E). AiCBLL7 had the greatest number of elements at 35 in total. In AhCBLLs, the similar proportions were 43.7, 28.7, 21.6, and 6.0% (Figure 5F). In the light response category, Box 4tbox4 (light-responsive element) and GT1-motif (part of a module for light response) were the most dominant. Meanwhile, cis-acting elements responding to auxin, abscisic acid, gibberellin, flavonoids, methyl jasmonate and salicylic acid were involved in the phytohormone responsiveness group. The CGTCA-motif and TGACG-motif (methyl jasmonate response elements) were followed by ABRE (related to the abscisic acid response). In the abiotic stress response category, ARE (elements regarding oxygen-deficient induction) covered the largest portion, and both WUN-motif (wound-responsive element) and LTR (relating to low-temperature responsiveness) exited in the promoters of AdCBLLs, AiCBLLs, and AhCBLLs. GCN4 motif (elements related to endosperm expression) and CAT-box (referred to meristem expression) were the largest motifs in plant growth regulation. Intriguingly, all types of cis-regulatory elements were distributed widely throughout the promoter regions of CBLL genes.

FIGURE 5. Identification of cis-acting elements in all the peanut CBLL genes. (A) Distribution of cis-acting elements in 2 kb upstream of each AdCBLLs. (B) Distribution of cis-acting elements in 2 kb upstream of each AiCBLLs. (C) Distribution of cis-acting elements in 2 kb upstream of each AhCBLLs. The different colored boxes indicate distinct promoter elements. (D) Assessment AdCBLL different subclass and category proportions in a radar chart. (E) Assessment AiCBLL different subclass and category proportions in a radar chart. (F) Assessment AhCBLL different subclass and category proportions in a radar chart. The lengths of the petals are proportional to the number of elements in each subclass qualitatively. Purple, blue, green and red petals represent plant growth regulation, abiotic stress responses, phytohormone and light responsiveness, respectively.
Expression Profile of AhCBLLs in Different Tissues of Peanut
Previous studies showed the involvement of CBLL family genes during different development stages (Kim and Leustek., 2000; Levin et al., 2000; Maimann et al., 2000; Goto et al., 2002; Joshi and Jander, 2009; Atkinson et al., 2013; Hacham et al., 2013; Song et al., 2013; Cohen et al., 2014, 2017; Whitcomb et al., 2018; Liu et al., 2019). Thus, the holistic expression patterns of peanut CBLLs in different tissues are needed to provide more insight into their roles during plant growth and development. Tissue analyses of 12 CBLL genes showed distinct tissue-specific expression patterns across the 22 tissues (leaf, stem, root, flower, pod and seed) (Figure 6). AhCBLL2, 4, and 9 showed higher expression level in almost all the sink tissues; AhCBLL5 and 10 were expressed mostly in the early seed developmental stages (seed pattee 5 and 6); AhCBLL1 and 7 had strong expression during the relatively later seed developmental stages (seed pattee 7, 8, and 10), especially in seed pattee 8. AhCBLL6 and 12 were highly expressed in root, nodule, and peg tip; Additionally, AhCBLL6 was enriched in peg tip pat 1 and AhCBLL11 was expressed highly in the peg and fruit tissues.
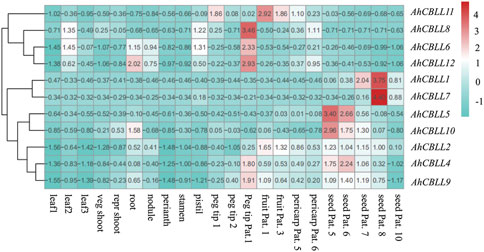
FIGURE 6. The expression profiles of AhCBLL genes. The heat map of the AhCBLL gene expression levels was hierarchically clustered using the R package “pheatmap” with the data normalization method of Z-score standardization. The color scale bar ranging from blue to red represents low and high expression, respectively. Abbreviation of the 22 tissues used in the expression profiles of AhCBLL genes were as following: seedling leaf 10 days post emergence (leaf 1), main stem leaf (leaf 2), lateral stem leaf (leaf 3), vegetative shoot tip from main stem (veg shoot), reproductive shoot tip from first lateral (repr shoot), 10-day roots (root), 25-day nodules (nodule), perianth, stamen, pistil, aerial gynophore tip (peg tip 1), subterranean peg tip (peg tip 2), Pattee 1 stalk (peg tip Pat. 1), Patte 1 pod (fruit Pat. 1), Pattee 3 pod (fruit Pat.3), Pattee 5 pericarp (pericarp Pat.5), Pattee 6 pericarp (pericarp Pat.6), Pattee 5 seed (seed Pat. 5), Pattee 6 seed (seed Pat. 6), Pattee 7 seed (seed Pat.7), Pattee 8 seed (seed Pat. 8), Pattee 10 seed (seed Pat.10).
Expression Pattern of the AhCBLLs in Plant Hormone Response
To uncover the possible functions of AhCBLLs in response to hormone stress, we conducted qRT-PCR to analyze their relative expressions under 6-BA, NAA, ACC, GA, MeJA, and ABA treatments (Figure 7). In this study, a two-fold change (|log2| > 1) was considered as significantly different for gene expression under each treatment. Of all the 12 AhCBLLs, nine genes showed increased expression under all treatments, while the remaining three (AhCBLL2, 4 and 6) had reduced expression at least one treatment (ABA, ACC, or 6-BA). For instance, AhCBLL6, AhCBLL4, and AhCBLL2 were down-regulated under ABA, ACC, and 6-BA treatments, respectively. Interestingly, the responses of the AhCBLLs to these plant hormones were different; AhCBLLs responded to NAA, ACC, and MeJA in the early time series, later to 6-BA, and were to slow responded to GA and ABA (mostly at 24 h after treatment). The results indicated that these AhCBLL genes might regulate relevant hormone signaling pathways.
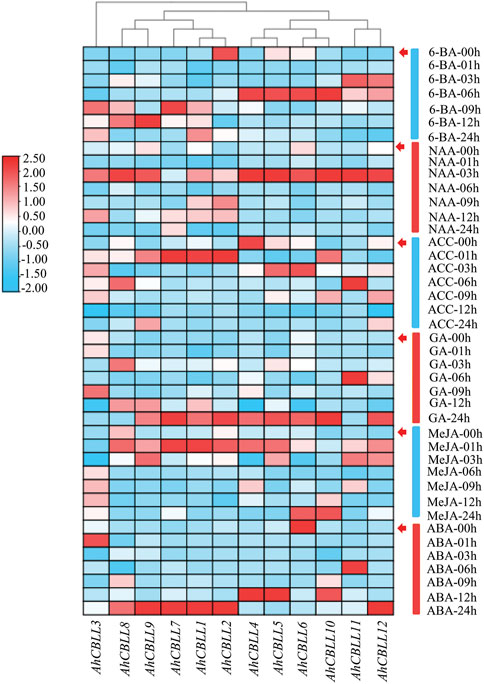
FIGURE 7. Relative expression of AhCBLLs under 6-BA, NAA, ACC, GA, MeJA and ABA treatments. Expression characteristics of AhCBLLs in response to different phytohormone at six-time points (00, 01, 03, 06, 09 and 12 h) were normalized to 00 h treatment. The fold changes values were calculated by the 2−△△Ct method and log2 and represented in color scale legend at the left of the heatmap: red indicated up-regulation and blue showed down-regulated expression.
Expression Pattern of the AhCBLLs Under Abiotic Stresses
Plants suffer from a wide variety of environmental stressors under natural conditions. We investigated the expression of AhCBLL genes, responding to five abiotic stressors (Figure 8). Results showed that the accumulation of AhCBLL1, AhCBLL2, AhCBLL3, AhCBLL8, AhCBLL9, and AhCBLL11 transcripts occurred at different time points after the submergence treatment, while the remaining six showed obviously down or up regulated curves. Expression levels of AhCBLL3, AhCBLL8, and AhCBLL9 were elevated starting at 6 h under cold stress and the other nine members reached their highest expression at 24 h. Interestingly, all analyzed AhCBLL genes showed a positive response to heat stress. AhCBLL 1, 5, 7, and 8 were up-regulated rapidly after 1 h of treatment, while AhCBLL 3, 4, 6, 10, 11, and 12 were strongly upregulated after 3 h of treatment. Moreover, three AhCBLL genes, AhCBLL1, -2, and 3 were inhibited under NaCl stress, and the remaining nine were up-regulated after NaCl treatment at different time points. Surprisingly, all AhCBLLs were induced after PEG stress. In summary, AhCBLL genes might play important roles in various types of environmental stress regulation.
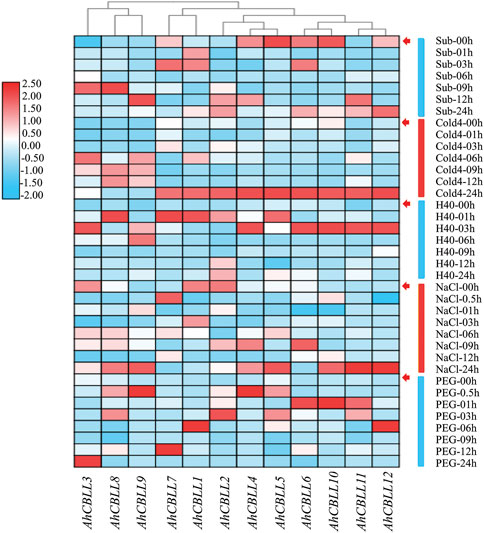
FIGURE 8. Expression changes of AhCBLLs under different abiotic stresses. The abbreviation represented as following, Sub: submergence, Cold4: cold of 4°C, H40: the heat of 40°C, NaCl: 200 mM NaCl, and PEG, polyethylene glycol.
Association Analysis of Peanut CBLLs with 104 Traits of Peanut
To uncover the roles of CBLL genes in peanut development and stress response, we performed candidate gene association analysis using five SNPs (single nucleotide polymorphisms) in AhCBLLs from transcriptome data of 146 peanut lines, and 104 phenotypes related to peanut development and stress response were collected from the five environments. The results indicated that one polymorphic site [B09_90283818(C/M/A)] was significantly associated with PL, PW, HPW, and HSW traits (Figure 9A and Supplementary Tables S13–S15). The site B09_90283818 mainly formed three haplotypes [B09_90283818(C/M/A)) (Figure 9B) in the population and were located in the predicted exon region of AiCBLL7 (Figure 9C). Results showed that PL, PW, HPW, and HSW in haplotype A were significantly higher than those in haplotype C (Figure 9D).
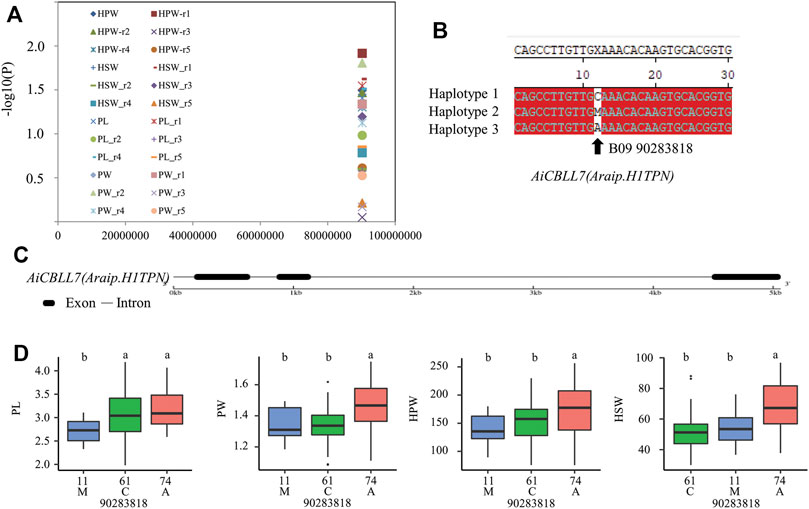
FIGURE 9. Association mapping results and the phenotypes of the polymorphic sites of the peanut CBLLs associated with HPW/HSW/PL/PW variation. (A) Association results between HPW/HSW/PL/PW and the polymorphisms in peanut CBLLs. (B) Sequences of site significantly associated with HPW/HSW/PL/PW variation. (C) The gene structures of AiCBLL7. (D) Phenotypic comparison of haplotypes of the associated site with HPW/HSW/PL/PW in five environments of the population.
Discussion
Duplication Contributed to AhCBLLs Expansion
Gene duplication contributes significantly to the proliferation of genes in plant species (Davidson et al., 2013; Hou et al., 2014), and leads to gene diversification or drives the evolution of genes. Based on synteny analyses, nine of the 12 AhCBLLs had syntenic relationships, all of them were inherited from A. duranensis and A. ipaensis genomes (Figures 2, 3 and Supplementary Table S2); the main expansion mechanism of AhCBLLs was whole-genome duplication (allopolyploidization), while none‐segmental and tandem duplication events occurred in the tetraploid stage.
Duplicated genes often evolve to lose their original functions and/or obtain new functions to enhance the adaptability of plants (Dias et al., 2003). Previous research demonstrates that a diversified expression pattern might be a significant reason for retaining duplicated genes in the genome (Gu et al., 2002). The duplicated genes lead to gene-dose effects or result in functional diversity. We found that all the AhCBLL gene pairs from allopolyploid species displayed a similar tissues expression pattern. For example, gene pair AhCBLL1 (Arahy.04)-AhCBLL7 (Arahy.14) was mainly expressed in the later seed development stage. Pat.8; AhCBLL4 (Arahy.04)-AhCBLL9 (Arahy.14) showed a higher expression level in peg. tip. Pat.1, AhCBLL5 (Arahy.06)–AhCBLL10 (Arahy.16) were enriched in the earlier seed development stage. Pat.5 and 6, and Ah CBLL6 (Arahy.10)-AhCBLL12 (Arahy.20) were highly expressed in root, nodule, and peg tip. Pat.1 (Figure 6). Most of the four allopolyploidy duplicated gene pairs demonstrated similar expression patterns to hormone treatments and abiotic stresses that might result in the way of gene-dose-effect; however, the expression of AhCBLL4 was inhibited under submergence and ACC treatment while AhCBLL9 was active ated; AhCBLL6 was down-regulated under ABA treatment while AhCBLL12 was up-regulated (Figures 7, 8); this opposite expression profile of the allopolyploidy duplicated gene pairs implied functional diversity. These results suggested that gene duplication of the CBLL family in peanut provided intricate regulation of signal transduction.
Subfunctionalization of the AhCBLL Genes in Peanut
The phylogenetic tree divided all the 29 peanut CBLL proteins into four clades (Figure 1A). Eight, six, one, four, and 10 CBLLs pertained to clade I (CGS clade), clade II (MGL clade), clade III (new clade), clade IV (CBL clade), clade V (new clade), respectively. The members in the CGS clade, MGL clade and CBL clade might have similar functions to the corresponding Arabidopsis genes; however, the function of the members in clade III and clade V was uncertain. The annotation of the one gene of clade III and most genes of clade V were CBLs; however, the gene structure and motif arrangement were quite different from those in the CBL clade, indicating divergence in gene function.
The prediction of cis-acting elements can provide important clues for the study of gene expression regulation (Zhu et al., 2017). In this study, analysis of the 2-kb upstream sequences of the initial codon of AhCBLL genes showed that the AhCBLLs contained multiple stress and hormone response elements, but the types and numbers were different. The percentage of cis-acting elements to phytohormone in A. duranensis was approximately 12%, which was more than that in A. ipaensis, indicating that the transcriptomic regulation of AdCBLLs might be more complex than in AiCBLLs. Most AhCBLL genes not only had cis-acting elements respond to abiotic adversity, but also had elements that responded to hormonal signals such as gibberellin, abscisic acid, salicylic acid and methyl jasmonate (Figure 5). For example, TCA-element, LTR, circadian and TC-rich repeats were exited in the promotor region of AhCBLL1, implying that AhCBLL1 might respond to salicylic acid signals and participate in the regulation of cold stress, circadian rhythm and plant defense response. The positive response of most AhCBLLs genes to hormones was the mainstream regardless of the AhCBL2 and AhCBL6 were down-regulated in 6-BA and ABA, respectively (Figure 7), implying their relevant roles in these hormone signal pathways. Surprising finding was that all analyzed AhCBLL genes showed positive response to heat and PEG stresses (Figure 8), suggesting that the AhCBLL may be important for peanut resistance to heat stress (Figure 8). Similarly, AhCBLL1, AhCBLL2, AhCBLL3, AhCBLL8, AhCBLL9, and AhCBLL11 were found to up-regulated by submergence (Figure 8), indicating the physiological function of these genes in peanut water logging stress tolerance mechanisms. Additinally, the expression of the nine AhCBLL genes except AhCBLL1, 2, and 3 were obviously enhanced after NaCl treatment at different time points (Figure 8). Considering the common positive response of AhCBLL8, AhCBLL9, and AhCBLL11 under heat, drought, submergence, and salt stresses, overexpression of these three genes in peanut may be an effective method to improve the peanut comprehensive abiotic stress resistance. Many important genes were selectively expressed in specific tissues during various physiological and developmental processes (Wan et al., 2014). Tissue expression of the AhCBLLs showed multiple tissue expression patterns suggesting subfunctionalization of this family.
Among the 13 CBLL orthologous pairs between peanut and Arabidopsis (Table 2), the functions of the corresponding ortholog genes in Arabidopsis have been determined; they functioned in influencing flowering, cell elongation, pollen tube growth, and played an important role in seed germination (Table 2). AtMOT1/AtCGS have been found to affect the physiological and behavioral processes of seeds, related to the biosynthesis of ethylene and polyamines (Goto et al., 2002; Hacham et al., 2013; Cohen et al., 2014, 2017; Whitcomb et al., 2018). Furthermore, AtMGL, regulates Met degradation, involved in the response to simultaneous biotic and abiotic stresses (Ricarda et al., 2005; Goyer et al., 2007; Joshi and Jander, 2009; Atkinson et al., 2013). Therefore, these AhCBLL orthologous genes may also play multiple roles in peanut development, and plant hormone synthesis or response.
AhCBLL Gene Plays an Important Role in Peanut Pod and Seed Development
The single-nucleotide polymorphic sites in AiCBLL7 (corresponding to AhCBLL11), were significantly associated with PL, PW, HPW and HSW variation. The polymorphic site in AiCBLL7, [B09_90283818(C/M/A)], located in the predicted exon region of the gene, B09_90283818(C/M/A) led a 268E to 268 K amino acid transition in the peanut population. These results indicated that B09_90283818(C/M/A) sequence polymorphisms might be the actual functional sites. Further, AhCBLL11 was mainly expressed in the peg and fruit, especially in early fruit development stages, which provided additional evidence for its function in peanut pod development. Most AhCBLLs exhibited tissue-specific expression patterns, almost all the AhCBLLs were expressed in higher levels in the peg, fruit, or seed (Figure 6). Further investigation was needed to confirm the roles of AiCBLL7 (AhCBLL11) in the pod and seed development of peanut.
Conclusion
In summary, this genome-wide identification, characterization and expression analysis of peanut CBLL genes provides valuable information for understanding the evolution and molecular functions of the peanut CBLL gene family, and highlights potential CBLL genes involved in peanut pod (seed) development and abiotic stress responses. The results of this study provide a foundation for further research regarding the function of the peanut CBLL gene family.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WR and LW carried out all the experiments and data analyses. WR and ZZ prepared the figures and tables. SW performed the qRT-PCR experiments. LW, JF, and JZ made modifications to the article. All authors read and approved the final manuscript.
Funding
This research was funded by grants from the National Key Research and Development Program of China (No. 2018YFD1000900) and the Jiangxi Agriculture Research System (No. JCARS-18). Funders did not participate in the design of the study, analysis of the results and writing of the manuscript, but provided financial support for the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.821163/full#supplementary-material
References
Amir, R. (2010). Current Understanding of the Factors Regulating Methionine Content in Vegetative Tissues of Higher Plants. Amino Acids 39, 917–931. doi:10.1007/s00726-010-0482-x
Atkinson, N. J., Lilley, C. J., and Urwin, P. E. (2013). Identification of Genes Involved in the Response of Arabidopsis to Simultaneous Biotic and Abiotic Stresses. Plant Physiol. 162 (4), 2028–2041. doi:10.1104/pp.113.222372
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, Efflux-dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 115, 591–602. doi:10.1016/s0092-8674(03)00924-3
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The Genome Sequence of Segmental Allotetraploid Peanut Arachis hypogaea. Nat. Genet. 51 (5), 877–884. doi:10.1038/s41588-019-0405-z
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 4, 10–21. doi:10.1186/1471-2229-4-10
Chen, Z., Zou, X., Song, L., Zou, X., Zhang, J., Chen, L., et al. (2010). Present Situation and Development Countermeasures of Peanut Production in Jiangxi Province. Bull. Agric. Sci. Technol. 06, 18–21.
Clevenger, J., Chu, Y., Scheffler, B., and Ozias-Akins, P. (2016). A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 7, 1446. doi:10.3389/fpls.2016.01446
Cohen, H., Hacham, Y., Panizel, I., Rogachev, I., Aharoni, A., and Amir, R. (2017). Repression of CYSTATHIONINE γ-SYNTHASE in Seeds Recruits the S-Methylmethionine Cycle. Plant Physiol. 174 (3), 1322–1333. doi:10.1104/pp.17.00579
Cohen, H., Israeli, H., Matityahu, I., and Amir, R. (2014). Seed-Specific Expression of a Feedback-Insensitive Form of CYSTATHIONINE-γ-SYNTHASE in Arabidopsis Stimulates Metabolic and Transcriptomic Responses Associated with Desiccation Stress. Plant Physiol. 166 (3), 1575–1592. doi:10.1104/pp.114.246058
Davidson, C. J., Guthrie, E. E., and Lipsick, J. S. (2013). Duplication and Maintenance of the Myb Genes of Vertebrate Animals. Biol. Open 2 (2), 101–110. doi:10.1242/bio.20123152
Deng, X., An, B., Zhong, H., Yang, J., Kong, W., and Li, Y. (2019). A Novel Insight into Functional Divergence of the MST Gene Family in rice Based on Comprehensive Expression Patterns. Genes 10 (3), 239. doi:10.3390/genes10030239
Dias, A. P., Braun, E. L., McMullen, M. D., and Grotewold, E. (2003). Recently Duplicated maize R2R3 Myb Genes Provide Evidence for Distinct Mechanisms of Evolutionary Divergence after Duplication. Plant Physiol. 131 (2), 610–620. doi:10.1104/pp.012047
Galili, G., Amir, R., and Fernie, A. R. (2016). The Regulation of Essential Amino Acid Synthesis and Accumulation in Plants. Annu. Rev. Plant Biol. 67, 153–178. doi:10.1146/annurev-arplant-043015-112213
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. e., Wilkins, M. R., Appel, R. D., et al. (2005). Protein Identification and Analysis Tools on the ExPASy Server. 2-D Proteome Anal. Protoc.pp, 571–607. doi:10.1385/1-59259-890-0:571
Goto, D. B., Ogi, M., Kijima, F., Kumagai, T., Werven, F. v., Onouchi, H., et al. (2002). A Single-Nucleotide Mutation in a Gene Encoding S-Adenosylmethionine Synthetase Is Associated with Methionine Over-accumulation Phenotype in Arabidopsis thaliana. Genes Genet. Syst. 77 (2), 89–95. doi:10.1266/ggs.77.89
Goyer, A., Collakova, E., Shachar-Hill, Y., and Hanson, A. D. (2007). Functional Characterization of a Methionine -Lyase in Arabidopsis and its Implication in an Alternative to the Reverse Trans-sulfuration Pathway. Plant Cel Physiol. 48 (2), 232–242. doi:10.1093/pcp/pcl055
Gu, Z., Cavalcanti, A., Chen, F.-C., Bouman, P., and Li, W.-H. (2002). Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 19, 256–262. doi:10.1093/oxfordjournals.molbev.a004079
Guo, M., Liu, J.-H., Ma, X., Zhai, Y.-F., Gong, Z.-H., and Lu, M.-H. (2016). Genome-wide Analysis of the Hsp70 Family Genes in Pepper (Capsicum Annuum L.) and Functional Identification of CaHsp70-2 Involvement in Heat Stress. Plant Sci. 252, 246–256. doi:10.1016/j.plantsci.2016.07.001
Hacham, Y., Matityahu, I., and Amir, R. (2013). Light and Sucrose Up-Regulate the Expression Level of Arabidopsis Cystathionine γ-synthase, the Key Enzyme of Methionine Biosynthesis Pathway. Amino Acids 45 (5), 1179–1190. doi:10.1007/s00726-013-1576-z
Holowach, L. P., Madison, J. T., and Thompson, J. F. (1986). Studies on the Mechanism of Regulation of the mRNA Level for a Soybean Storage Protein Subunit by Exogenous L-Methionine. Plant Physiol. 80, 561–567. doi:10.1104/pp.80.2.561
Hou, X.-J., Li, S.-B., Liu, S.-R., Hu, C.-G., and Zhang, J.-Z. (2014). Genome-wide Classification and Evolutionary and Expression Analyses of Citrus MYB Transcription Factor Families in Sweet orange. PLoS One 9 (911), e112375. doi:10.1371/journal.pone.0112375
Jain, M., Nijhawan, A., Tyagi, A. K., and Khurana, J. P. (2006). Validation of Housekeeping Genes as Internal Control for Studying Gene Expression in rice by Quantitative Real-Time PCR. Biochem. Biophysical Res. Commun. 345, 646–651. doi:10.1016/j.bbrc.2006.04.140
Joshi, V., and Jander, G. (2009). Arabidopsis Methionine γ-Lyase Is Regulated According to Isoleucine Biosynthesis Needs but Plays a Subordinate Role to Threonine Deaminase. Plant Physiol. 151 (1), 367–378. doi:10.1104/pp.109.138651
Jost, R., Altschmied, L., Bloem, E., Bogs, J., Gershenzon, J., Hähnel, U., et al. (2005). Expression Profiling of Metabolic Genes in Response to Methyl Jasmonate Reveals Regulation of Genes of Primary and Secondary Sulfur-Related Pathways in Arabidopsis thaliana. Photosynth Res. 86, 491–508. doi:10.1007/s11120-005-7386-8
Kery, V., Bukovska, G., and Kraus, J. P. (1994). Transsulfuration Depends on Heme in Addition to Pyridoxal 5'-phosphate. Cystathionine Beta-Synthase Is a Heme Protein. J. Biol. Chem. 269 (41), 25283–25288. doi:10.1016/s0021-9258(18)47244-4
Kim, J., and Leustek, T. (2000). Repression of Cystathionine γ-synthase in Arabidopsis thaliana Produces Partial Methionine Auxotrophy and Developmental Abnormalities. Plant Sci. 151, 9–18. doi:10.1016/s0168-9452(99)00188-0
Lescot, M. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Levin, J. Z., Framond, A. J. D., Tuttle, A., Bauer, M. W., and Heifetz, P. B. (2000). Methods of Double-Stranded RNA-Mediated Gene Inactivation in Arabidopsis and Their Use to Define an Essential Gene in Methionine Biosynthesis. Plant Mol. Biol. 44, 759–775. doi:10.1023/a:1026584607941
Liao, B., Yin, Y., and Ma, N. (2018). Review and prospect of Development of Oil Crop Industry in China. J. Agric. 8 (1), 115–120.
Liu, G., Yang, W., Zhang, X., Peng, T., Zou, Y., Zhang, T., et al. (2019). Cystathionine Beta‐lyase Is Crucial for Embryo Patterning and the Maintenance of Root Stem Cell Niche in Arabidopsis. Plant J. 99 (3), 536–555. doi:10.1111/tpj.14343
Maimann, S., Wagner, C., Kreft, O., Zeh, M., Willmitzer, L., Hofgen, R., et al. (2000). Transgenic Potato Plants Reveal the Indispensable Role of Cystathionine Beta-Lyase in Plant Growth and Development. Plant J. 23, 747–758. doi:10.1046/j.1365-313x.2000.00842.x
Messerschmidt, A., Worbs, M., Steegborn, C., Wahl, M. C., Huber, R., Laber, B., et al. (2003). Determinants of Enzymatic Specificity in the Cys-Met-Metabolism PLP-dependent Enzyme Family: Crystal Structure of Cystathionine γ-Lyase from Yeast and Intrafamiliar Structure Comparison. Biol. Chem. 384 (3), 373–386. doi:10.1515/BC.2003.043
Misra, V. A., Wafula, E. K., Wang, Y., dePamphilis, C. W., and Timko, M. P. (2019). Genome-wide Identification of MST, SUT and SWEET Family Sugar Transporters in Root Parasitic Angiosperms and Analysis of Their Expression during Host Parasitism. BMC Plant Biol. 19 (1), 196. doi:10.1186/s12870-019-1786-y
Ravanel, S., Gakiere, B., Job, D., and Douce, R. (1998). The Specific Features of Methionine Biosynthesis and Metabolism in Plants. Proc. Natl. Acad. Sci. 95, 7805–7812. doi:10.1073/pnas.95.13.7805
Song, S., Hou, W., Godo, I., Wu, C., Yu, Y., Matityahu, I., et al. (2013). Soybean Seeds Expressing Feedback-Insensitive Cystathionine γ-synthase Exhibit a Higher Content of Methionine. J. Exp. Bot. 64 (7), 1917–1926. doi:10.1093/jxb/ert053
Song, Y., Wang, L., and Xiong, L. (2009). Comprehensive Expression Profiling Analysis of OsIAA Gene Family in Developmental Processes and in Response to Phytohormone and Stress Treatments. Planta 229, 577–591. doi:10.1007/s00425-008-0853-7
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: Robust Conversion of Protein Sequence Alignments into the Corresponding Codon Alignments. Nucleic Acids Res. 34 (Web Server), W609–W612. doi:10.1093/nar/gkl315
Wan, L., Wu, Y., Huang, J., Dai, X., Lei, Y., Yan, L., et al. (2014). Identification of ERF Genes in Peanuts and Functional Analysis of AhERF008 and AhERF019 in Abiotic Stress Response. Funct. Integr. Genomics 14 (3), 467–477. doi:10.1007/s10142-014-0381-4
Wang, Z., Wang, Y., Yang, J., Hu, K., An, B., Deng, X., et al. (2016). Reliable Selection and Holistic Stability Evaluation of Reference Genes for Rice under 22 Different Experimental Conditions. Appl. Biochem. Biotechnol. 179, 753–775. doi:10.1007/s12010-016-2029-4
Whitcomb, S. J., Nguyen, H. C., Brückner, F., Hesse, H., and Hoefgen, R. (2018). CYSTATHIONINE GAMMA-SYNTHASE Activity in rice Is Developmentally Regulated and Strongly Correlated with Sulfate. Plant Sci. 270, 234–244. doi:10.1016/j.plantsci.2018.02.016
Yang, J., Zhang, Y., Yang, P., and Ji, M. (2014). Analysis of Peanut Industry Development in Shandong Province. Jiangsu Agric. Sci. 42 (9), 433–436.
Yang, X., Chen, Z., and Zhao, M. (2001). Isolation and Some Properties of Peanut Protein. J. Chin. Cereals Oils Assoc. 16 (05), 25–28. doi:10.3901/jme.2001.03.025
Yang, Z. (2007). PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 24 (8), 1586–1591. doi:10.1093/molbev/msm088
Zhu, Q., Zhao, Y., Lv, H., Luan, F., and Gao, P. (2017). Identification and Characterization of CBL Family Genes in Watermelon. North. Hortic. (15), 18–24.
Sarropoulou, V., Dimassi-Theriou, K., and Therios, L. (2013). L-Methionine Influences in vitro Root Regeneration, Total Chlorophyll, Total Carbohydrate and Endogenous Proline Content in the Sweet Cherry Rootstock MxM 14 (Prunus avium × Prunus mahaleb). Sci. Horti. 161, 88–94. doi:10.1016/j.scienta.2013.06.012
Keywords: peanut, cis-acting element, genome-wide association analysis, CBLL gene family, expression pattern
Citation: Ren W, Zeng Z, Wang S, Zhang J, Fang J and Wan L (2022) Global Survey, Expressions and Association Analysis of CBLL Genes in Peanut. Front. Genet. 13:821163. doi: 10.3389/fgene.2022.821163
Received: 23 November 2021; Accepted: 28 January 2022;
Published: 09 March 2022.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Swarup Roy Choudhury, Indian Institute of Science Education and Research, IndiaSantosh Kumar Upadhyay, Panjab University, India
Zulfiqar Ali, Muhammad Nawaz Shareef University of Agriculture, Pakistan
Copyright © 2022 Ren, Zeng, Wang, Zhang, Fang and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiahai Fang, ZmFuZ2ppYWhhaTIwMTlAMTI2LmNvbQ==; Liyun Wan, d2FubGl5dW4yMDE5QDE2My5jb20=
 Weifang Ren1,2,3
Weifang Ren1,2,3 Juncheng Zhang
Juncheng Zhang Liyun Wan
Liyun Wan