- 1School of Clinical Medicine, Shanghai University of Medicine & Health Sciences, Shanghai, China
- 2Department of Respiratory and Critical Care Medicine, Shanghai General Hospital, Shanghai Jiaotong University, Shanghai, China
Background: The overexpression of human antigen R (HuR) has been proven in various types of cancer and is associated with the poor survival lung cancer patients. HuR overexpression stabilizes the mRNA of tumor-promoting genes by binding with 3′-UTR AU-rich elements. However, the role of HuR in the proliferation of lung cancer is unclear.
Methods: HuR expression was assessed using immunohistochemistry of tumor tissue samples from ten patients with lung cancer and ten patients with benign lung disease. Gene, protein, mRNA, and lncRNA changes in A549 HuR knockdown (KD) cells were assessed by single-cell RNA sequencing analysis. Furthermore, cell proliferation, migration, and invasion were determined by Cell Counting Kit-8 (CCK-8) assays and Transwell assays with or without Matrigel. The cell cycle was assessed by propidium iodide staining. The protein level, mRNA level and half-life of PLK1 were detected by western blotting and RT-qPCR.
Results: In clinical patients, the expression of HuR was significantly higher in lung cancer patients than in patients with benign lung disease. RNA sequencing analysis of A549 HuR knockdown cells revealed that the main function of HuR was related to ribonucleoprotein complex biogenesis. HuR was found to regulate signaling pathways mainly related to the spliceosome, RNA transport and the cell cycle. HuR KD suppressed the proliferation, migration and invasion of A549 cells, indicating its promotive role in these processes.
Conclusion: These results demonstrate that HuR plays an important role in the progression of lung cancer.
Background
Lung cancer is a systemic disease that causes more than one-fourth of all cancer-related deaths worldwide (American Cancer Society Cancer Statistics, 2021). Thus, the identification of novel therapeutic molecular targets to inhibit lung cancer proliferation is urgently needed to improve patient survival.
The human antigen R (HuR), an ELAV protein contains 3′ RNA-binding domains and RNA stabilization by the AU-rich elements (AREs) (Papatheofani et al., 2021). This makes HuR an important RNA-binding protein that exerts pleiotropic effects on tumorigenesis and tumor development (Brody and Dixon, 2018; Dhir et al., 2019). HuR can alter the cellular function to proliferation (Yu et al., 2020), heat or hypoxia stress (Reglero et al., 2020; Deka and Saha, 2021), apoptotic pathway (Chen et al., 2020), immune cell differentiation (Yu et al., 2021), senescence (Liebig et al., 2020), pro-inflammatory gene (Ke et al., 2021), and the immune regulation (Liu et al., 2021a; Goutas et al., 2021) by its posttranscriptional influence on specific target mRNAs. HuR overexpression in lung cancer may regulate progression by controlling mRNA stability (Dixon et al., 2001; Denkert et al., 2006; Lim et al., 2009). However, the role of HuR in the proliferation of lung cancer is still unclear. Polo-like kinase 1 (PLK1) plays an important role in the initiation, maintenance, and completion of mitosis. Dysfunction of PLK1 may promote cancerous transformation and drive cancer progression (Liu et al., 2017; Montaudon et al., 2020; Shin et al., 2020).
Therefore, in this study, we assessed the expression of HuR in primary lung cancer patients and benign lung disease patients. In addition, we used a human lung adenocarcinoma cancer cell line (A549) to knockdown (KD) and overexpress HuR to determine whether HuR can influence the function of those cells by single-cell RNA sequencing analysis and in vitro experiments.
Materials and Methods
Patients and Clinical Samples
The present study was approved by the Ethics Review Board of Shanghai General Hospital affiliated with Shanghai Jiaotong University (Shanghai, China). Written informed consent was obtained from all patients before enrollment in the study. All records were anonymized to protect individual confidentiality. The tumor tissues were retrieved from paraffin-embedded blocks. The clinical-stage was determined according to the recommendations of the eighth International Association for the Study of Lung Cancer (Ettinger et al., 2021).
Immunohistochemical Staining and Immunoreactivity Scoring
Immunohistochemical staining and immunoreactivity scoring procedures were performed as described previously (Xiao et al., 2022). In brief, Resected specimens (lung cancer tissue and benign lung disease tissue) were fixed in formalin and embedded in paraffin. The primary antibody, HuR, was incubated overnight, and the secondary antibody was for 30 min. The expression of HuR in cancer cells was evaluated by immunoreactivity scoring based on the intensity and extent of staining.
Cell Lines and Culture Conditions
The human lung adenocarcinoma cell line (A549) was purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. It was cultured in Ham’s F-12K medium (Gibco) with 10% fetal bovine serum (Gibco) and maintained at 37°C in a humidified incubator with 5% CO2.
Cell Transfection
Cells (3×105 cells/well) were plated in a 12-well plate with a growth medium and without antibiotics 1 day before transfection. Twenty pmol siRNA was diluted in 50 µL Opti-MEM™ Reduced Serum Medium (Gibco). Next, we diluted 1.5 µL Lipofectamine 2000 in 50 µL Opti-MEM Reduced Serum Medium. After 5 min of incubation at room temperature, the diluted siRNA was combined with the diluted Lipofectamine 2000 and incubated for 15 min at room temperature. The cultured cells were added to the siRNA-liposome complexes. The cells were collected 72 h later, and the gene expression of HuR was detected by real-time PCR to verify the interference effect. We chose three different siRNA sequences to find the most effective one; the primer sequences used were as follows: HuR siRNA 610 (forward 5′- CGUUUAUCCGGUUUGACAATT -3′, reverse 5′- UUGUCAAACCGGAUAAACGTT -3′), siRNA 1024 (forward 5′- GCUUUGUGACCAUGACAAATT -3′, reverse 5′- UUUGUCAUGGUCACAAAGCTT -3′), siRNA 1239 (forward 5′- CAAGUGUUUGUCUUUGUCUTT -3′, reverse 5′- AGACAAAGACAAACACUUGTT -3′).
RNA Extraction and RT-PCR
Total RNA was extracted by using the TRIzol method. First-strand cDNA was synthesized using the PrimeScriptTM RT Reagent Kit (TAKARA). The primer sequences used were as follows: HuR (forward 5′-TAATCGCCATAGCCTTCCTAA-3′, reverse 5′-GGCGTCTGCAAATGGTTGTA-3′), PLK1 (forward 5′-TGTGCTGTCGGTATGGAGAG-3′, reverse 5′-TCTGGTACACAGGACTTGCG-3′) and GAPDH (forward 5′-CAA TGA CCC CTT CAT TGA CC-3′, reverse 5′-GAC AAG CTT CCC GTT CTC AG-3′). RT-PCRs were performed using SYBR® Premix Ex Taq™ II (TAKARA). Three independent experiments were performed, and the data are shown as the mean ± SEM.
Western Blotting
SDS-PAGE was performed, and antibodies specific for HuR/ELAVL1 (1:1,000, Abcam), PLK1 (1:1,000, Abcam), and β-actin (1:10,000, Proteintech) were applied for western blotting. Goat anti-mouse IgG (H + L) and goat anti-rabbit IgG (H + L) with HRP were used as the secondary antibodies (1:1,000 Beyotime). The results were visualized using an enhanced chemiluminescence detection system (Biosharp). β-Actin functioned as the reference protein.
Cell Proliferation Assay
Then, 100 μL of cell suspension of the A549 normal group and siRNA interfering group (5,000 cells/well) was seeded in a 96-well plate. The cells were preincubated for 24 h in a humidified incubator (37°C, 5% CO2) and incubated for an appropriate length of time (0, 24, 48, 72 h) in the incubator. Then, 10 μL of CCK-8 solution was added to each well (Beyotime, Shanghai, China). The plate was incubated for 4 h in an incubator, and the absorbance was measured at 450 nm with a microplate reader.
Cell Migration and Invasion Detection
The Transwell assay was performed with the Transwell system of Corning Co., Ltd. Inserts containing filters were uncoated for the migration assays or coated with Matrigel for the invasion assays. Then, 500 µL complete medium with 10% serum was added to the lower chamber, and a total of 1 × 105 cells were added to 200 μL serum-free medium in the upper chamber. After 24 h of incubation at 37°C, the cells that had migrated through the filters were fixed with 800 µL of 4% methanol for 10 min, stained with 800 µL of crystal violet for 15–30 min, and counted under a microscope.
Cell Cycle
A549 cells and A549 HuR KD cells incubated with F-12K medium without FBS were seeded in 6-well plates overnight. The F-12K medium was changed to 10% FBS, and the cells were incubated for 24 h. The cells were harvested, and cellular DNA was stained with propidium iodide (PI) solution. Fluorescence was measured with a flow cytometer (FACS Aria II, BD Bioscience, United States). Different cell cycle phases were distinguished based on the DNA content. DNA histograms were analyzed by ModFit 5.0 software.
RNA Sequencing Analysis
RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using a NanoPhotometer® spectrophotometer.
(IMPLEN, CA, United States). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2,100 system (Agilent Technologies, CA, United States). A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample.
Statistical Analysis
All values are expressed as the mean ± SEM for each experiment or a representative cell culture-based experiment. All statistical analyses were performed using GraphPad Prism 6.0. The student’s t-test was used to analyze the difference between the two groups. The Pearson’s test was used to analyze the correlation between the two samples. For all tests, the significance threshold was set at p ≤ 0.05.
Results
HuR is Overexpressed in Primary Lung Cancer Patients and Associated With a Poor Prognosis
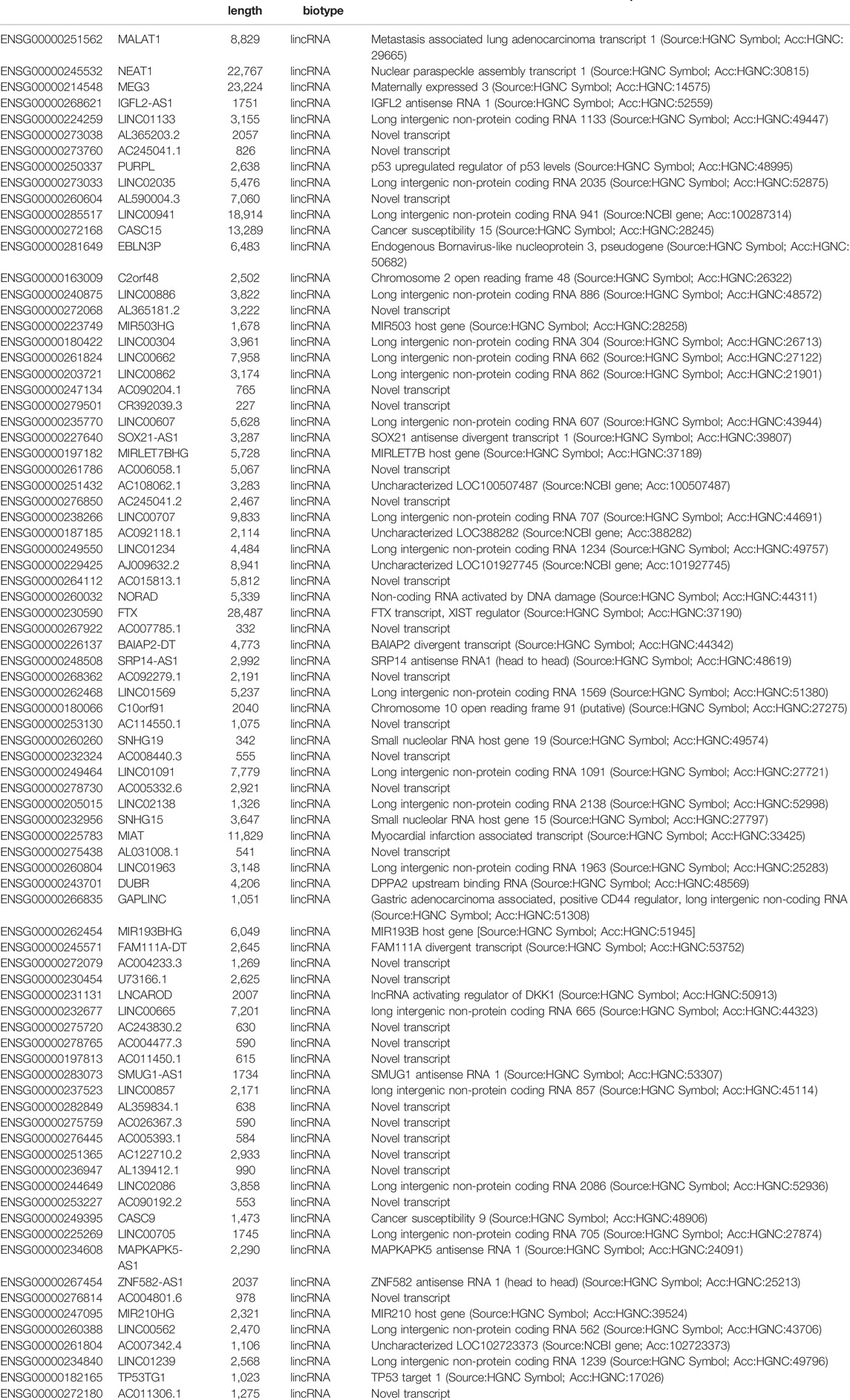
The immunohistochemical expression of HuR and IRS is shown in Figure 1, comparing representative samples of benign lung disease (Figure 1A) and primary lung cancer (Figure 1B). In the lung cancer slide, several cells that were positive for HuR were observed. Furthermore, Figure 1C shows the IRS of HuR expression in patients with lung cancer using IHC compared with benign lung disease (p < 0.01). Therefore, the overexpression of HuR is correlated with cancer. Analysis of The Cancer Genome Atlas (TCGA) database and Kaplan-Meier analysis of overall survival of 962 lung cancer patients with high HuR expression compared with 964 lung cancer patients with low HuR expression revealed that high expression of HuR indicated a poor prognosis (p = 1.8e-05) (Figure 1D).
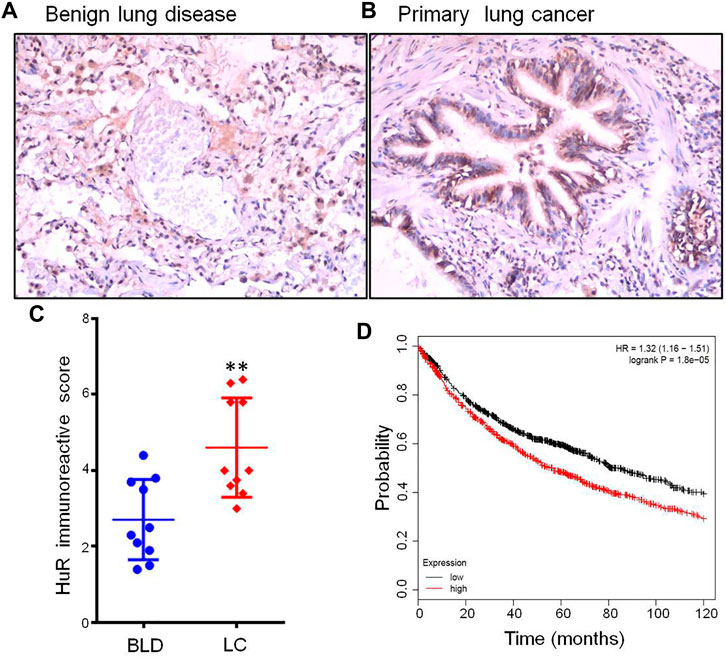
FIGURE 1. HuR expression in lung cancer patients and benign lung disease patients was detected using immunohistochemistry. HuR was stained in samples from patients with (A) benign lung disease (200 × magnification) and (B) lung cancer (200 × magnification) by IHC. (C) The IRS of HuR in benign lung disease and lung cancer samples. (D) Kaplan-Meier curve of overall survival according to HuR expression in the TCGA. There were 962 lung cancer patients with high expression among a total of 1926 patients, and lung cancer patients with high expression of HuR had a poor prognosis (p = 1.8e-05). **p < 0.01. BLD, benign lung disease; LC, lung cancer.
The Main Function of HuR in A549 Cells According to Single-Cell RNA Sequencing Analysis
To examine the function of HuR in lung adenocarcinoma cells, the lung adenocarcinoma cell line A549 was used, and we performed HuR KD for single-cell RNA sequencing analysis. First, the correlation between the two samples was checked by Pearson’s test. We found that 2,466 genes were upregulated and 3,114 genes were downregulated in HuR KD A549 cells versus control cells, as shown in the heatmap (Figure 2). Figure 3 indicates that the main functions of HuR in A549 cells were related to ribonucleoprotein complex biogenesis. The involved signaling pathways were included those related to spliceosomes, RNA transport, and the cell cycle (Figure 4). Table 1 and Table 2 show the changes in lncRNAs and microRNAs in A549 HuR KD cells.
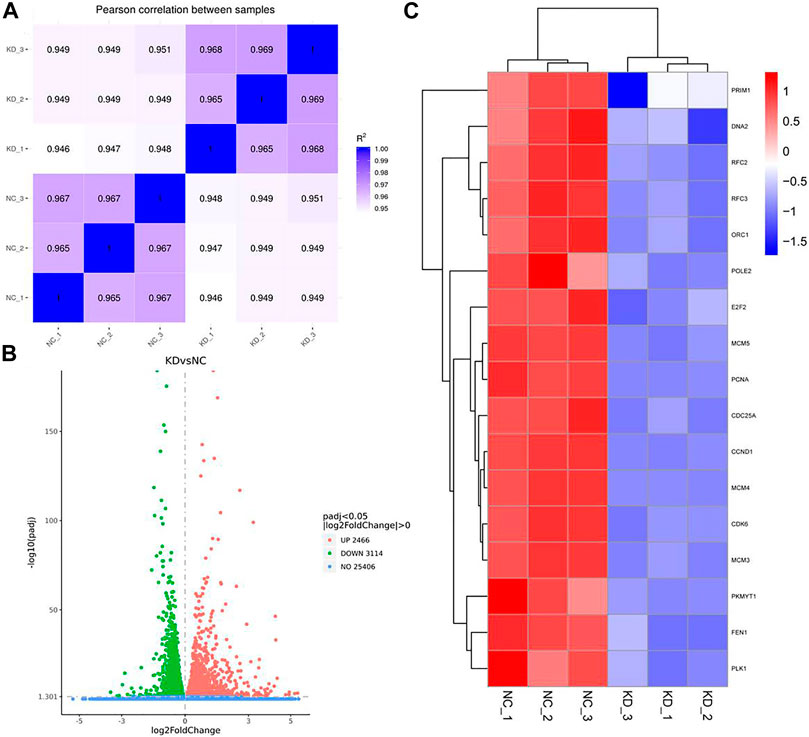
FIGURE 2. Heatmap showing the expression of genes in the lung cancer cell line A549 according to single-cell RNA sequencing. Cells are grouped by HuR KD status. (A). Pearson correlation between HuR KD A549 cells and the control group. (B). HuR KD A549 cells were analyzed by RNA sequencing, which revealed 2,466 upregulated genes and 3,114 downregulated genes. (C) The gene heatmap shows the expression of major genes involved in the cell cycle between the two groups.
FIGURE 3. The cell functions enriched for significant differentially expressed genes between the HuR KD A549 cells and control cells. The histogram of GO analysis (A) combined with the bubble chart (B) shows that HuR mainly regulates the synthesis of ribonucleoprotein protein complexes.
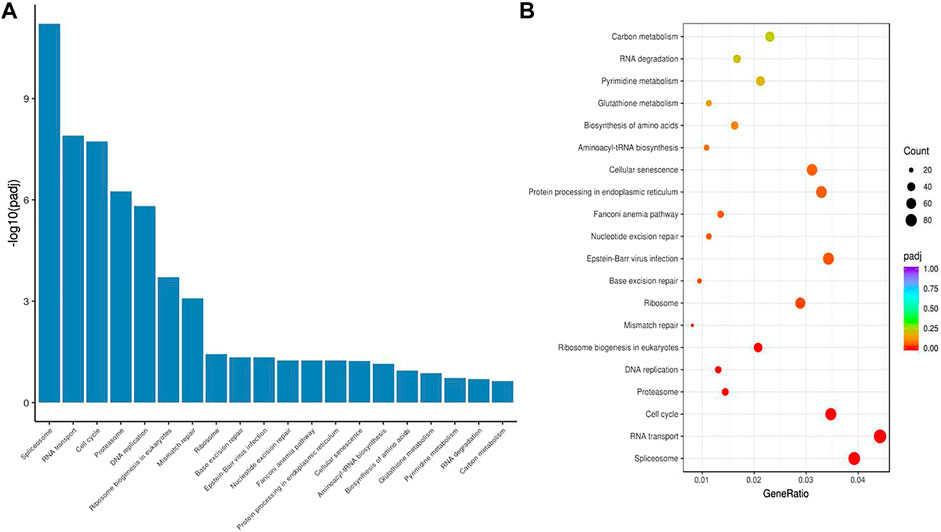
FIGURE 4. The signaling pathways enriched for significant differentially expressed genes in HuR KD A549 cells versus control cells. The histogram (A) and bubble chart (B) of the KEGG results show that HuR mainly regulates pathways related to the spliceosome, RNA transport, and the cell cycle.
HuR Promotes Cell Proliferation, Migration, and Invasion
To find the most effective siRNA for knocking down HuR expression, we chose three different siRNA sequences and detected their efficiencies by real-time PCR (Figure 5A) and western blotting (Figure 5B). We then explored the role of HuR and found that it can promote the migration and invasion of A549 cells compared with that of A549 HuR KD cells (Figures 5C,D). Statistical results are shown in Figures 5E,F. Next, we evaluated the effect of HuR on the proliferation of A549 cells (Figure 6C).
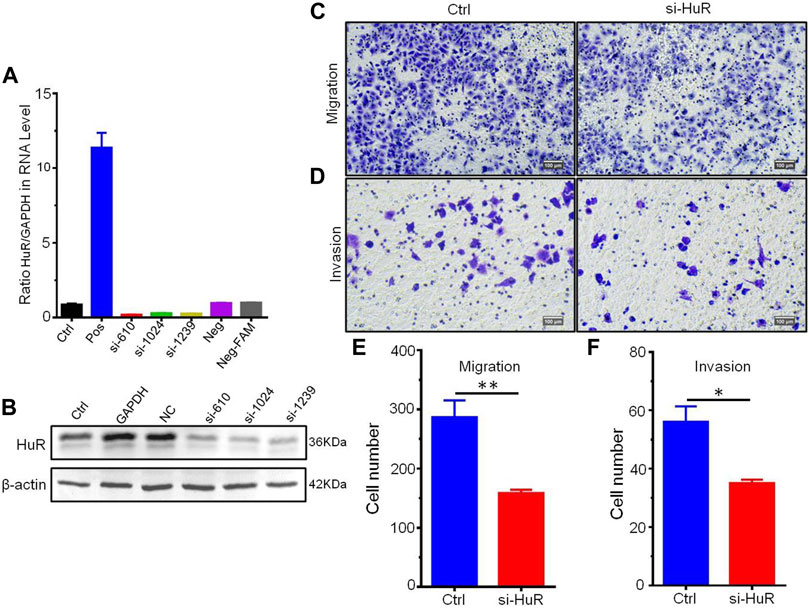
FIGURE 5. HuR promotes the migration and invasion of A549 cells. Migration assay of A549 HuR KD cells (1*105/well) compared with A549 cells as a control. The expression of HuR genes was detected by western blotting and real-time PCR to verify the interference effect of siHuRs in A549 cells (A,B). Cells that migrated into the lower chamber were photographed (C) and quantified (E). Matrigel cell invasion assay of A549 HuR KD cells (1*105/well) compared with A549 cells as a control for 24 h. Cells that invaded through the Matrigel were photographed (D) and quantified (F). Scale bar 100 μm *p < 0.05, **p < 0.01, ***p < 0.001. Data are represented as the mean ± SEM. All experiments were repeated at least three times.
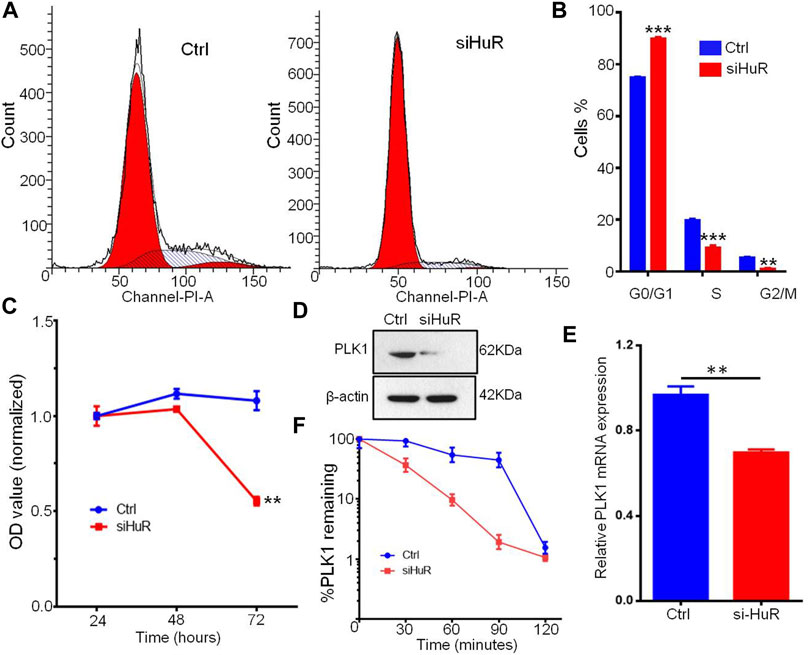
FIGURE 6. HuR promotes the cell cycle progression and proliferation of A549 cells. (A,B) Cell cycle histograms of A549 HuR KD cells compared with A549 cells. (C). CCK-8 assay of the proliferation of A549 cells and HuR KD A549 cells. (D). PLK1 was measured by western blotting, and to normalize protein loading, the blots were also probed for β-actin. (E). Relative PLK1 mRNA expression in A549 cells with HuR KD for 24 h (F). The decay rates of the PLK1 and GAPDH mRNA were assessed in A549 cells by RT-qPCR at different time points (30, 60, 90, 120 min) after transcription inhibition with actinomycin D. Data are represented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. n = 3.
HuR Regulates PLK1 mRNA Levels and Protein Expression
PLK1, an essential cell cycle regulator, plays a key role in regulating the cell cycle by compromising cell cycle checkpoints and increasing the expression of chromosomal instability (CIN)-related genes. We assumed that the changes in PLK1 expression might be a possible mechanism by which G1/S and G2/M transition occurs during cell growth induced by HuR. Flow cytometry analysis showed that HuR increased the number of cells in the S phase of the cell cycle compared to that in the untreated group (Figures 6A,B). Next, we examined whether the decrease in PLK1 protein expression mediated by HuR was correlated with a decrease in the PLK1 transcript level. To this end, quantitative RT-PCR of PLK1 was performed on control or HuR KD A549 cells 2 days after transfection. HuR KD decreased the expression of PLK1, as indicated by western blot analysis (Figure 6D). In treated cells, we showed that HuR treatment led to a decrease in PLK1 mRNA levels by 2-fold (Figure 6E).
Next, to examine the PLK1 mRNA half-life (t1/2) in A549 cells treated with HuR KD, actinomycin D inhibitor was applied, and the level of PLK1 mRNA was measured by RT-qPCR at 0, 30, 60, 90 and 120 min. In HuR KD A549 cells, the PLK1 mRNA t1/2 was 33 min, whereas, in control A549 cells, PLK1 mRNA t1/2 was 97 min. The mRNA t1/2 of GAPDH (the control) was not changed.
Discussion
Lung cancer is a leading cause of cancer-associated mortality worldwide, with a 5-year survival rate of <15% (Siegel et al., 2021). Proliferation is the leading factor influencing cancer development (Liu et al., 2021b; Hou et al., 2021; Wang et al., 2021). HuR is the key molecule in lung cancer progression (Liao et al., 2011; Li et al., 2013; Zhang et al., 2020). However, little is known about how HuR might act to specifically promote lung cancer proliferation. In this study, the expression of HuR was significantly associated with lung cancer aggressiveness; for example, lung patients with high expression of HuR showed a poor prognosis in the TCGA database. Here, we also found that the main function of HuR in the lung adenocarcinoma cell line A549 was related to ribonucleoprotein complex biogenesis. The signaling pathways that regulated HuR in A549 cells were mainly related to the spliceosome, RNA transport, and the cell cycle. Furthermore, our results suggest that HuR can suppress the protein and mRNA expression of PLK1 in A549 cells, which may promote cell cycle progression and cell proliferation, migration, and invasion. Overall, these data suggest that HuR is overexpressed in lung cancer patients and related to poor prognosis. HuR plays an important role in lung cancer progression as well.
In this study, we confirmed that HuR was overexpressed in patients with lung cancer versus those with benign lung diseases, such as benign pulmonary nodules and tuberculosis. In addition, the IRS of HuR in lung cancer tissue was found to differ markedly from that in tissue from patients with benign lung diseases. These findings emphasize that lung cancer is a complex disease; in particular, lung proliferation is a very important step in the progression of cancer. Kaplan-Meier analysis of overall survival according to HuR expression was performed by employing the TCGA. Lung cancer patients with high expression of HuR had a poor prognosis. This suggests that the overexpression of HuR in lung cancer increases the risk of metastasis. Wang et al proposed that UDP-glucose regulates lung cancer metastasis and uncovered a mechanism by which UDP-glucose 6-dehydrogenase promotes tumor metastasis by increasing the association of HuR with SNAI1 mRNA and therefore stabilizing SNAI1 mRNA (Wang et al., 2019).
Our results show for the first time the function of HuR in lung adenocarcinoma cells, and HuR KD A549 cells were analyzed by single-cell RNA sequencing analysis. HuR KD in A549 cells yielded 2,466 upregulated genes and 3,114 downregulated genes. The main functions of HuR in A549 cells were related to ribonucleoprotein complex biogenesis. The signaling pathways were mainly related to the spliceosome, RNA transport, and the cell cycle.
Little is known about the functionality of HuR in lung cancer. In the current study, we assessed the proliferation, migration, and invasion of A549 cells after HuR KD. We confirmed that HuR can enhance the proliferation of A549 cells and promote their migration and invasion. The KD of HuR did not dramatically enhance proliferation, migration, or invasion. These results indicate the enhancing effect of HuR on lung cancer cell proliferative abilities. This finding is consistent with that of Muralidharan R et al, who found that targeted inhibition of HuR in cancer cells suppresses several HuR-regulated oncoproteins, increasing anticancer treatment efficacy (Muralidharan et al., 2016).
We detected the cell cycle distribution in A549 cells and explored potential related mechanisms. A549 control cells showed accelerated S phase transition from the G1 phase compared with HuR KD cells, which were arrested in the G1 phase. PLK1 is a well-known regulator of the cell cycle and can arrest cells in the G1/S phase by blocking the G2/M transition. In this study, we showed that KD of HuR led to the downregulation of PLK1. The HuR can stable PLK1 mRNA was shown by western blotting, RT-PCR, and mRNA decay assays. However, a study by de Carcer G et al. showed that PLK1 overexpression can prevent the development of Kras-induced and Her2-induced mammary gland tumors, which exhibit high levels of CIN (de Cárcer et al., 2018). Overall, our data imply that HuR can influence the cell cycle signaling pathway in lung cancer cells. This further corroborated the findings of our study. However, this hypothesis regarding the downstream function of PLK1 is beyond the scope of this study.
This study does have some shortcomings. In the clinical specimen analysis, we did not conduct further analysis of clinical data for patients with high HuR expression. In vitro experiments, the detailed mechanism of the downregulation of PLK1 and inhibition of cell cycle progression after HuR KD was not further explored.
In summary, our data showed that HuR overexpression in lung cancer was related to a poor prognosis. Furthermore, our study showed that HuR can enhance cell proliferation, migration, and invasion, which could enhance the expression of PLK1 in A549 cells. Based on our results, further studies are necessary to explore how HuR effect on the downstream molecular in lung cancer.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov; PRJNA767760.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical approval was granted by Shanghai General Hospital (research protocol number 2022KY011). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XY, QF, and HX contributed to the design of the study, analysis of the data, and drafting of the manuscript. All authors critically revised the paper and agreed to be accountable for all aspects of the work.
Funding
This work was sponsored by the National Natural Science Foundation of China (No. 82172692), Shanghai Pujiang Program (No. 2020PJD049) and a local high-level university construction project (No. E1-2602-21-201006-9).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank our patients, who willingly gave their time to participate in this research.
Abbreviations
FBS, fetal bovine serum; HuR, human antigen R; IRS, immunoreactivity score; 3′-UTR 3′, untranslated region; KD, knockdown.
References
American Cancer Society Cancer Statistics, (2021). American Cancer Society Cancer Statistics 2021 Report. J. Nucl. Med. 62 (3), 12N.
Brody, J. R., and Dixon, D. A. (2018). Complex HuR Function in Pancreatic Cancer Cells. Wiley Interdiscip. Rev. RNA 9 (3), e1469. doi:10.1002/wrna.1469
Chen, T., Dai, X., Dai, J., Ding, C., Zhang, Z., Lin, Z., et al. (2020). AFP Promotes HCC Progression by Suppressing the HuR-Mediated Fas/FADD Apoptotic Pathway. Cell Death Dis 11 (10), 822. doi:10.1038/s41419-020-03030-7
de Cárcer, G., Venkateswaran, S. V., Salgueiro, L., El Bakkali, A., Somogyi, K., Rowald, K., et al. (2018). Plk1 Overexpression Induces Chromosomal Instability and Suppresses Tumor Development. Nat. Commun. 9 (1), 3012. doi:10.1038/s41467-018-05429-5
Deka, K., and Saha, S. (2021). Heat Stress Induced Arginylation of HuR Promotes Alternative Polyadenylation of Hsp70.3 by Regulating HuR Stability and RNA Binding. Cell Death Differ 28 (2), 730–747. doi:10.1038/s41418-020-00619-5
Denkert, C., Koch, I., von Keyserlingk, N., Noske, A., Niesporek, S., Dietel, M., et al. (2006). Expression of the ELAV-like Protein HuR in Human colon Cancer: Association with Tumor Stage and Cyclooxygenase-2. Mod. Pathol. 19 (9), 1261–1269. doi:10.1038/modpathol.3800645
Dhir, T., Schultz, C. W., Jain, A., Brown, S. Z., Haber, A., Goetz, A., et al. (2019). Abemaciclib Is Effective against Pancreatic Cancer Cells and Synergizes with HuR and YAP1 Inhibition. Mol. Cancer Res. 17 (10), 2029–2041. doi:10.1158/1541-7786.mcr-19-0589
Dixon, D. A., Tolley, N. D., King, P. H., Nabors, L. B., McIntyre, T. M., Zimmerman, G. A., et al. (2001). Altered Expression of the mRNA Stability Factor HuR Promotes Cyclooxygenase-2 Expression in colon Cancer Cells. J. Clin. Invest. 108 (11), 1657–1665. doi:10.1172/jci12973
Ettinger, D. S., Wood, D. E., Aisner, D. L., Akerley, W., Bauman, J. R., Bharat, A., et al. (2021). NCCN Guidelines Insights: Non-small Cell Lung Cancer, Version 2.2021. J. Natl. Compr. Canc Netw. 19 (3), 254–266.
Goutas, D., Pergaris, A., Giaginis, C., and Theocharis, S. (2021). HuR as Therapeutic Target in Cancer: What the Future Holds. Curr. Med. Chem. 29 (1), 56–65. doi:10.2174/0929867328666210628143430
Hou, G., Yang, J., Tang, J., and He, Y. (2021). LncRNA GAS6-AS2 Promotes Non-small-cell Lung Cancer Cell Proliferation via Regulating miR-144-3p/MAPK6 axis. Cell Cycle 20 (2), 179–193. doi:10.1080/15384101.2020.1867782
Ke, Y., Lv, X., Fu, X., Zhang, J., Bohio, A. A., Zeng, X., et al. (2021). Poly(ADP-ribosyl)ation Enhances HuR Oligomerization and Contributes to Pro-inflammatory Gene mRNA Stabilization. Cell. Mol. Life Sci. 78 (4), 1817–1835. doi:10.1007/s00018-020-03618-4
Li, Y.-J., Wang, C.-H., Zhou, Y., Liao, Z.-Y., Zhu, S.-F., Hu, Y., et al. (2013). TLR9 Signaling Repressed Tumor Suppressor miR-7 Expression through Up-Regulation of HuR in Human Lung Cancer Cells. Cancer Cel Int 13 (1), 90. doi:10.1186/1475-2867-13-90
Liao, W.-L., Wang, W.-C., Chang, W.-C., and Tseng, J. T. (2011). The RNA-Binding Protein HuR Stabilizes Cytosolic Phospholipase A2α mRNA under Interleukin-1β Treatment in Non-small Cell Lung Cancer A549 Cells. J. Biol. Chem. 286 (41), 35499–35508. doi:10.1074/jbc.m111.263582
Liebig, J. K., Kuphal, S., and Bosserhoff, A. K. (2020). HuRdling Senescence: HuR Breaks BRAF-Induced Senescence in Melanocytes and Supports Melanoma Growth. Cancers (Basel) 12 (5). doi:10.3390/cancers12051299
Lim, S.-J., Lee, S.-H., Joo, S. H., Song, J. Y., and Choi, S. I. (2009). Cytoplasmic Expression of HuR Is Related to Cyclooxygenase-2 Expression in colon Cancer. Cancer Res. Treat. 41 (2), 87–92. doi:10.4143/crt.2009.41.2.87
Liu, Y., Hu, F., and Zhao, L. (2021). Effect of Nano-Platinum on Proliferation and Apoptosis of Non-small Cell Lung Cancer Cells via P53 Pathway. J. Nanosci Nanotechnol 21 (2), 903–908. doi:10.1166/jnn.2021.18629
Liu, Y., Li, X., Zhang, H., Zhang, M., and Wei, Y. (2021). HuR Up-Regulates Cell Surface PD-L1 via Stabilizing CMTM6 Transcript in Cancer. Oncogene 40 (12), 2230–2242. doi:10.1038/s41388-021-01689-6
Liu, Z., Sun, Q., and Wang, X. (2017). PLK1, A Potential Target for Cancer Therapy. Translational Oncol. 10 (1), 22–32. doi:10.1016/j.tranon.2016.10.003
Montaudon, E., Nikitorowicz-Buniak, J., Sourd, L., Morisset, L., El Botty, R., Huguet, L., et al. (2020). PLK1 Inhibition Exhibits strong Anti-tumoral Activity in CCND1-Driven Breast Cancer Metastases with Acquired Palbociclib Resistance. Nat. Commun. 11 (1), 4053. doi:10.1038/s41467-020-17697-1
Muralidharan, R., Babu, A., Amreddy, N., Basalingappa, K., Mehta, M., Chen, A., et al. (2016). Folate Receptor-Targeted Nanoparticle Delivery of HuR-RNAi Suppresses Lung Cancer Cell Proliferation and Migration. J. Nanobiotechnol 14 (1), 47. doi:10.1186/s12951-016-0201-1
Papatheofani, V., Levidou, G., Sarantis, P., Koustas, E., Karamouzis, M. V., Pergaris, A., et al. (2021). HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment. Biomedicines 9 (2). doi:10.3390/biomedicines9020119
Reglero, C., Lafarga, V., Rivas, V., Albitre, Á., Ramos, P., Berciano, S. R., et al. (2020). GRK2-Dependent HuR Phosphorylation Regulates HIF1α Activation under Hypoxia or Adrenergic Stress. Cancers (Basel) 12 (5). doi:10.3390/cancers12051216
Shin, S.-B., Jang, H.-R., Xu, R., Won, J.-Y., and Yim, H. (2020). Active PLK1-Driven Metastasis Is Amplified by TGF-β Signaling that Forms a Positive Feedback Loop in Non-small Cell Lung Cancer. Oncogene 39 (4), 767–785. doi:10.1038/s41388-019-1023-z
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer Statistics, 2021. CA A. Cancer J. Clin. 71 (1), 7–33. doi:10.3322/caac.21654
Wang, J., Sun, N., Han, W., Tong, L., Xu, T., and Li, G. (2021). Long Non‐coding RNA CCAT1 Sponges miR‐490 to Enhance Cell Proliferation and Migration of Non‐small Cell Lung Cancer. Thorac. Cancer 12 (3), 364–371. doi:10.1111/1759-7714.13758
Wang, X., Liu, R., Zhu, W., Chu, H., Yu, H., Wei, P., et al. (2019). UDP-glucose Accelerates SNAI1 mRNA Decay and Impairs Lung Cancer Metastasis. Nature 571 (7763), 127–131. doi:10.1038/s41586-019-1340-y
Xiao, H., Ye, X., Vishwakarma, V., Preet, R., and Dixon, D. A. (2022). CRC-derived Exosomes Containing the RNA Binding Protein HuR Promote Lung Cell Proliferation by Stabilizing C-Myc mRNA. Cancer Biol. Ther. 23 (1), 139–149. doi:10.1080/15384047.2022.2034455
Yu, S., Tripod, M., Atasoy, U., and Chen, J. (2021). HuR Plays a Positive Role to Strengthen the Signaling Pathways of CD4+ T Cell Activation and Th17 Cell Differentiation. J. Immunol. Res. 2021, 9937243. doi:10.1155/2021/9937243
Yu, X., Li, Y., Ding, Y., Zhang, H., Ding, N., and Lu, M. (2020). HuR Promotes Ovarian Cancer Cell Proliferation by Regulating TIMM44 mRNA Stability. Cell Biochem Biophys 78 (4), 447–453. doi:10.1007/s12013-020-00939-w
Keywords: lung cancer, human antigen R, proliferation, RNA sequencing analysis, prognosis
Citation: Ye X, Fu Q and Xiao H (2022) The Role of RNA-Binding Protein HuR in Lung Cancer by RNA Sequencing Analysis. Front. Genet. 13:813268. doi: 10.3389/fgene.2022.813268
Received: 11 November 2021; Accepted: 09 March 2022;
Published: 05 April 2022.
Edited by:
Rifat Hamoudi, University of Sharjah, United Arab EmiratesReviewed by:
Khalid S. A. Khabar, King Faisal Specialist Hospital & Research Centre, Saudi ArabiaJack D. Keene, Duke University, United States
Copyright © 2022 Ye, Fu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Xiao, eGlhb2h1aTc3MTIxMEAxNjMuY29t
†These authors have contributed equally to this work
 Xiong Ye
Xiong Ye Qiang Fu
Qiang Fu Hui Xiao
Hui Xiao